Crohn’s disease (CD) is a chronic inflammatory disease predominantly affecting the terminal ileum. The ~200 CD-risk loci identified by genome-wide association studies are enriched for genes involved in T-cell signaling, highlighting the importance of T cells in CD pathology.1,2 It is crucial to study T cells in their disease-relevant context, namely the intestinal mucosa. Although human single-cell atlases are in development,3 location-and disease-specific single-cell RNA sequencing (scRNAseq) datasets are still scarce. In this study, we used scRNAseq of disease-relevant cells to examine pathomechanisms and identify potential drug targets on a cellular level in CD.
Methods
We performed flow cytometry and scRNAseq of 5292 CD3-positive T lymphocytes isolated from peripheral blood (PBLs) and ileal biopsy specimens from 3 patients with CD with mild to moderate disease activity (Figure 1, step 1). Biopsy specimens were dissociated and separated into intraepithelial T lymphocytes (IELs) and lamina propria T lymphocytes (LPLs).4 Then, we integrated T-cell transcriptomes with genome-wide association study CD-risk loci and drug-target identification resources. Through a literature search and online database analysis, we identified 179 CD-risk genes and 2712 drug-target genes that we aligned to differentially expressed genes. Then, we selected genes encoding proteins targeted by drugs currently available for (clinical trials in) humans (Supplementary Methods).
Figure 1.
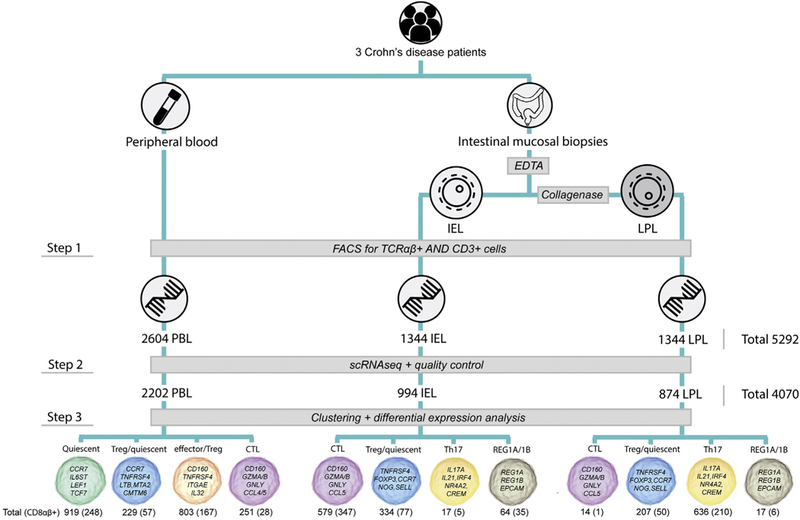
Experimental flowchart. EDTA and collagenase are treatments used to isolate IELs and LPLs, respectively. T-cell subtype characterization was performed as described in the Supplementary Methods. The number of epitopic CD8αβ-positive T cells measured by flow cytometry is presented within brackets. FACS, fluorescence-activated cell sorting; TCR, T-cell receptor.
Results
After quality control, 4070 T cells remained for analysis. These cells expressed 966 genes per cell, on average, and 41,134 distinct genes in total (Figure 1, step 2). IELs, LPLs, and PBLs showed markedly different expression profiles (Supplementary Figure 1 and Supplementary Table 1).
Unsupervised clustering of scRNAseq data identified 6 distinct T-cell types that have different distributions in IELs, LPLs, and PBLs: cytotoxic T lymphocytes (CTLs) dominate IELs, quiescent T cells dominate the PBL reservoir, and T-helper 17 (Th17) cells dominate LPLs (Figure 1, step 3). In peripheral blood, T-regulatory (Treg) cells intermixed with effector T cells (effector/Treg cells) and quiescent T cells (Treg/quiescent cells). Treg/quiescent cells also were present among IELs and LPLs. Among IELs and LPLs, we identified a cluster of cells of a previously undefined cell type characterized by expression of CD3 and REG1A/B and confirmed this with immunofluorescence staining. All T-cell subtypes were present in all patients.
Strikingly, all cell-subtype clusters consisted of epitopic CD8αβ-positive and -negative cells. Moreover, CD8A and CD8B transcripts were expressed in epitopic CD8αβ-positive and -negative cells, which was confirmed in a publicly available naïve CD4+ T-cell scRNAseq dataset.5
Using permutation analysis, we found that IELs and LPLs expressed significantly more CD-risk genes than expected by chance (P = .00389 and P < .00001, respectively), suggesting that cells within these compartments play a role in CD inflammation.2 PBLs were not enriched for CD-risk gene expression (P = .45954). Th17 cells showed the largest number of over-expressed CD-risk genes and were most specifically enriched for CD-risk gene expression (P < .00001). Mucosal CTL and Treg/quiescent and peripheral blood effector/Treg cells also were significantly enriched for CD-risk gene expression (P = .0278, P = .02477, and P = .00081, respectively).
We investigated which drug targets are expressed by Th17 cells and CTLs, cell types with well-characterized expression signatures that play a central role in CD pathogenesis (Table 1). Th17 cells showed up-regulation of IL17A, whose gene product is targeted by secukinumab. ITGAE, whose gene product is targeted by etrolizumab, was upregulated in mucosal CTLs. S1PR5, up-regulated in peripheral blood CTLs, is a known drug target for ozanimod. Potential targets for drug repositioning include PDE4D in mucosal Th17 cells, a target for apremilast, under investigation for treatment of ulcerative colitis; ITGB2 in peripheral blood CTLs, a target for lifitegrast, approved for keratoconjunctivitis sicca; and ALOX5AP in mucosal CTLs, a target for fiboflapon, under investigation for asthma.
Table 1.
Overexpression of CD-Risk Genes and Genes Encoding Potential Drug Targets in Ileal Mucosal Th17 Cells and CTLs
| Mucosal Th17 cells |
Peripheral blood CTL |
Mucosal CTL |
||||
|---|---|---|---|---|---|---|
| Gene | Drug or compound | Gene | Drug or compound | Gene | Drug or compound | |
| CD-risk gene | CCL20 | Chemoattractant for various immune cells | CTSW | Regulation of T-cell cytolytic activity | PLCG2 | Transmembrane signaling of immune system receptors |
| DNAJB4 | Heat shock protein, involved in protein folding | LSP1 | Adhesion and transendothelial migration | PTPN22 | Negative regulator of TCR signaling, positive regulator of TLR signaling | |
| IFNG | Cytokine, involved in adaptive–innate immunity | PRKCB | Apoptosis regulation | SOCS1 | Cytokine-inducible negative regulation of cytokine signaling | |
| IRF4 | Regulation of mucosal Th17 cell differentiation | PTPRC | T-cell antigen receptor signaling regulation | |||
| MAP3K8 | T-helper cell differentiation and IFN gamma expression | |||||
| Known CD-drug target | IL17A | Secukinumab | S1PR5 | Ozanimod | CD3D/E/G | NI-0401 |
| FCGR3A | IgG class mAbsa | ITGAE CCR9 TGFBR1 | Etrolizumab Vercirnon Mongersenc | |||
| Candidate for drug repositioning | DNAJB1 | Apatorsen | CCL5 | Heparin compounds | CCL5 | Heparin compounds |
| SIK1 | Dabrafanib | FGR | Dasatinib | ADRB1 | STD-101-D1 | |
| HSP90AA1 | Nedocromil | FCGR3A | IgG class mAbsb | ALOX5AP | Fiboflapon | |
| PTGER4 | Rivenprost; limaprost; dinoproston | ITGB2 | Lifitegrast | SLAMF7 | Elotuzumab | |
| PDE4D | Apremilast | ADRB2 | β2-adrenergic receptor blockers | MDM4 | ALRN-6924 | |
NOTE. Ileal mucosal Th17 cells, peripheral blood CTLs, and ileal mucosal CTLs show the largest number of significantly overexpressed CD-risk genes. The top 5 most significantly overexpressed CD-risk genes are listed in the first section of this table. In the second section, the top 5 most significantly overexpressed known CD drug targets and candidates for drug repositioning, per cell type, are listed. Selection of CD-risk genes is explained in the CD-Risk Genes section in the supplement. “Known CD-drug target” refers to genes encoding targets for drugs currently approved or under investigation for treatment of CD in humans. “Candidate for drug repositioning” refers to genes encoding targets for drugs currently approved for or under investigation in humans for other diseases than CD (Drug-Target Genes section in supplement).
IFN, interferon; IgG, immunoglobulin G; mAbs, monoclonal antibodies; TCR, T-cell receptor; TLR, toll-like receptor.
For example, adalimumab, etanercept, and natalizumab.
For example, alefacept and alemtuzumab.
Indirect target.
Discussion
We have demonstrated that multiple ileal mucosal T-cell subtypes and 1 peripheral blood T-cell subtype from patients with CD are enriched for CD-risk gene expression. T-cell subtypes known to be involved in CD pathogenesis provide promising targets for future cell type-specific therapies in patients with CD. A limitation of our study is the small sample, which might decrease the amount of variation covered. However, most cell type-specific gene expression signatures remained after correcting for inter-individual differences. Because location-and disease-specific scRNAseq data are still limited, detailed datasets like ours are an important reference for furthering our understanding of the molecular processes leading to health and disease and identifying potential targets for drug development.
Supplementary Material
Acknowledgments
The authors thank the patients-participants of the 1000 inflammatory bowel disease cohort for contributing blood and intestinal biopsy samples; Tjasso Blokzijl, Desiree Brandenbrug-Weening, Jelkje de Boer, Kim de Lange, Tim Raine, and Pieter van der Vlies for laboratory support; Patricia Rogers, Wayel Abdulahad, and Geert Mesander for technical assistance with fluorescence-activated cell sorting; Rudi Alberts for data management support; the UMCG Genomics Coordination center, the UG Center for Information Technology and their sponsors BBMRI-NL and TarGet for storage and computer infrastructure; and Timothy Tickle and Mark Daly for contributing to the scientific discussion.
Funding
Cisca Wijmenga is supported by a European Research Council (ERC) advanced grant (FP/2007–2013/ERC grant 2012–322698), a Netherlands Organisation for Scientific Research (NWO) Spinoza prize (NwO SPI 92–266), the NWO Gravitation Netherlands Organ-on-Chip Initiative (024.003.001), the Stiftelsen Kristian Gerhard Jebsen foundation (Norway), and a RuG Investment Agenda Grant for Personalized Health. Rinse K. Weersma is supported by an NWO VIDI grant (016.136.308). Eleonora A. Festen is supported by a Dutch Digestive Foundation Career Development grant (CDG 14–04). Werna T. Uniken Venema is supported by the Foundation “De Drie Lichten,” the Netherlands, and the Boehringer Ingelheim Fund. The Department of Genetics, Section Research and Development, University of Groningen, University Medical Center Groningen contributed to laboratory expenses for this study.
Abbreviations used in this paper:
- CD
Crohn’s disease
- CTL
cytotoxic T lymphocyte
- IEL
intraepithelial T lymphocyte
- LPL
lamina propria T lymphocyte
- PBL
peripheral blood T lymphocyte
- scRNAseq
single-cell RNA sequencing
- Th17
T-helper 17 cell
- Treg
T-regulatory cell
Footnotes
Conflicts of interest
There are no (potential) conflicts of interest to declare.
Writing assistance
This articlewas edited for language and formatting by Kate Mc Intyre, Scientific Editor in the Department of Genetics, University Medical Center Groningen.
Supplementary Material
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org and at https://doi.org/10.1053/j.gastro.2018.10.046.
References
- 1.Liu JZ, van Sommeren S, Huang H, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 2015;47:979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farh KK-H, Marson A, Zhu J, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 2015;518:337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Regev A, Teichmann SA, Lander ES, et al. The human cell atlas. Elife 2017;6:e27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raine T, Liu JZ, Anderson CA, et al. Generation of primary human intestinal T cell transcriptomes reveals differential expression at genetic risk loci for immune-mediated disease. Gut 2015;64:250–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng GXY, Terry JM, Belgrader P, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun 2017;8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


