Abstract
Transporters are important therapeutic but yet understudied targets due to lack of available assays. Here we describe a novel label-free, whole-cell method for the functional assessment of Solute Carrier (SLC) inhibitors. As many SLC substrates are also ligands for G protein-coupled receptors (GPCRs), transporter inhibition may affect GPCR signalling due to a change in extracellular concentration of the substrate/ligand, which can be monitored by an impedance-based label-free assay. For this study, a prototypical SLC/GPCR pair was selected, i.e. the equilibrative nucleoside transporter-1 (SLC29A1/ENT1) and an adenosine receptor (AR), for which adenosine is the substrate/ligand. ENT1 inhibition with three reference compounds was monitored sensitively via AR activation on human osteosarcoma cells. Firstly, the inhibitor addition resulted in an increased apparent potency of adenosine. Secondly, all inhibitors concentration-dependently increased the extracellular adenosine concentration, resulting in an indirect quantitative assessment of their potencies. Additionally, AR activation was abolished by AR antagonists, confirming that the monitored impedance was AR-mediated. In summary, we developed a novel assay as an in vitro model system that reliably assessed the potency of SLC29A1 inhibitors via AR signalling. As such, the method may be applied broadly as it has the potential to study a multitude of SLCs via concomitant GPCR signalling.
Subject terms: Membrane proteins, Extracellular signalling molecules, Phenotypic screening, Receptor pharmacology
Introduction
Solute carriers (SLCs) are transmembrane transport proteins that control a cell’s intra- and extracellular communication with its environment by regulating the translocation of small molecules, inorganic ions or other proteins across biological membranes. Their function as gatekeepers, their ubiquitous presence in the human body, and the large number of SLCs (more than 400) render them potential therapeutic targets1. However, their in-depth investigation has been limited as not many cellular assays are available to study transporter activity, making drug discovery difficult. The main and most well-known assay format currently used is the uptake assay, which measures the accumulation of a radiolabeled substrate in cells expressing the transporter under investigation2. Such assay however, presents limitations as only the end-point of the assay can be measured and/or washing steps are needed, which make this type of assay laborious and prone to artefacts. Moreover, the need for radioactive substrates has disadvantages, such as high costs, handling of radioactivity and storage of radioactive waste3. In addition, label-free electrophysiological methods have been applied and many electrogenic SLCs, i.e. transporters the function of which results in a positive or negative intracellular net charge, have been studied as such (Grewer et al. 2013)4. The best-known electrophysiological technique is patch-clamp, a well-studied concept that measures the direct current generated by ions that flow through the transporters, with a plethora of instruments being commercially available (Priest et al. 2004)5. In addition to patch-clamp, solid supported membrane (SSM)-based electrophysiology has been applied to transporters as a more sensitive approach, where SURFE²R N1 (Nanion Technologies) is the main instrumentation used6. However, no label-free assays applicable to non-electrogenic membrane transporters, which represent the majority of SLCs are available. Therefore, the development of a homogenous, kinetic and label-free assay in the field of non-electrogenic membrane transporter research is of utmost interest.
Label-free whole cell assays typically use a biosensor that detects physical properties of cells, such as size, adhesion and morphology, in order to measure cellular responses upon ligand stimulation7. The main advantage of using biosensors and cell morphology to functionally assess a ligand-target interaction is that cells are monitored in real time and in high sensitivity, making it possible to study cell systems with endogenous target expression8,9. As a result, no modification of the cells, for example transfection of an (engineered) target into an artificial cell line, or the compounds, i.e. with a fluorescent or radioactive tag, is necessary, which avoids potential artefacts10. Over the last decade, the number of available label-free cellular assays has significantly increased, and many of these have been used to study G protein-coupled receptor (GPCR) signalling11,12. Examples are the EPIC (Corning Inc.) and xCELLigence (ACEA Biosciences), which measure cell morphology optically and by impedance, respectively. However, the use of label-free assays are still rather unexploited, if at all, for membrane transporters.
ENT1 (also known as SLC29A1) is the most abundant nucleoside transport (NT) protein in the human body13. This SLC plays a crucial role in the provision of nucleosides, as it participates in the salvage pathways of nucleotide synthesis in cells lacking de novo biosynthetic pathways14. Therapeutically, ENT1 can be targeted by drugs that are both substrates and inhibitors. In the cases of viral infections and in some types of cancer, ENT1 transports well-known drugs inside the cell in order to exert their action, e.g. gemcitabine and ribavirin, respectively15,16. As far as ENT1 inhibition is concerned, molecules that diminish ENT1 activity are proposed as an add-on treatment of cancer, whenever ENT1 is overexpressed17. Moreover, ENT1 inhibitors can potentially be used in the treatment of ischemic heart disease18, stroke19 and inflammatory diseases20. Of note, in many cases the therapeutic effect of ENT1 inhibitors is induced by adenosine21, as its increased extracellular concentration can potentiate neuroprotective and cardioprotective actions resulting from the activation of neighbouring adenosine receptors (ARs).
In the current study, we describe the development of a novel cellular assay for the functional assessment of SLC activity by using the label-free impedance-based xCELLigence instrument. Many endogenous substrates of membrane transporters are also ligands for GPCRs, e.g glutamate, dopamine and adenosine22,23. Thus, we hypothesized that by inhibiting a transporter, the substrate concentration would increase outside of the cell, resulting in increased GPCR signalling that is subsequently monitored with the xCELLigence (Fig. 1). For proof-of-principle, we investigated the inhibition of ENT1 transporters by well-known ENT1 inhibitors, and monitored concomitant adenosine receptor signalling. For validation purposes we performed radioligand binding studies on SLC29A1 as well.
Figure 1.
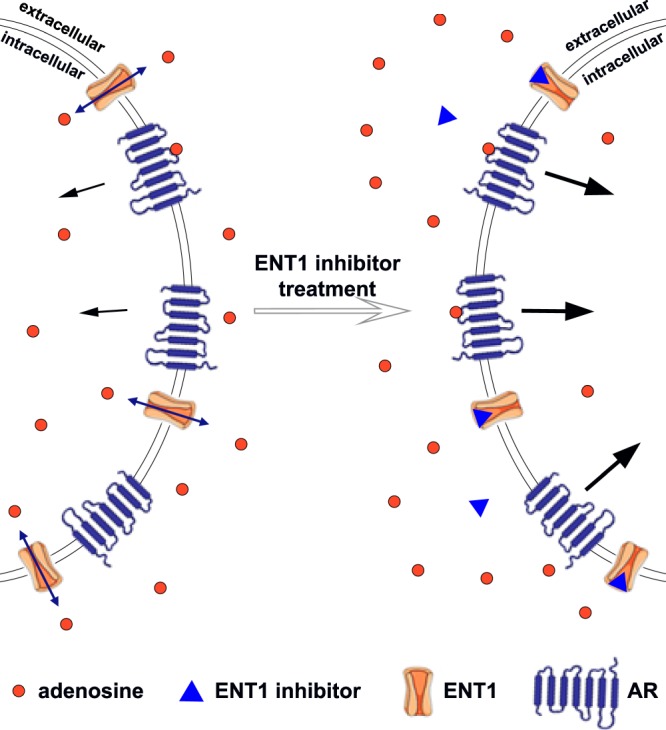
Assay principle. ENT1 equilibrates adenosine concentrations inside and outside of the cell membrane. Extracellular adenosine binds to ARs and causes their activation and signalling (black arrows). After treatment with an ENT1 inhibitor, adenosine cannot be translocated intracellularly with the same efficiency, depending on inhibitor’s inhibitory potency and concentration. The resulting higher extracellular adenosine concentration will cause increased AR activation (thicker arrows).
Results
Assay development and optimization
To confirm the suitability of U-2 OS cells for studying ENT1 function via AR signalling, we performed a radioligand binding assay on U-2 OS membranes. U-2 OS membranes were incubated with [3H]NBTI and increasing concentrations of reference inhibitors, i.e. NBTI, dilazep and dipyridamole. All inhibitors fully displaced the radioligand from the ENT1. NBTI had the highest affinity (pKi = 8.7 ± 0.02), followed by dilazep (pKi = 8.5 ± 0.1) and dipyridamole (pKi = 7.2 ± 0.1) (Supplementary Material; Fig. S1 and Table S1).
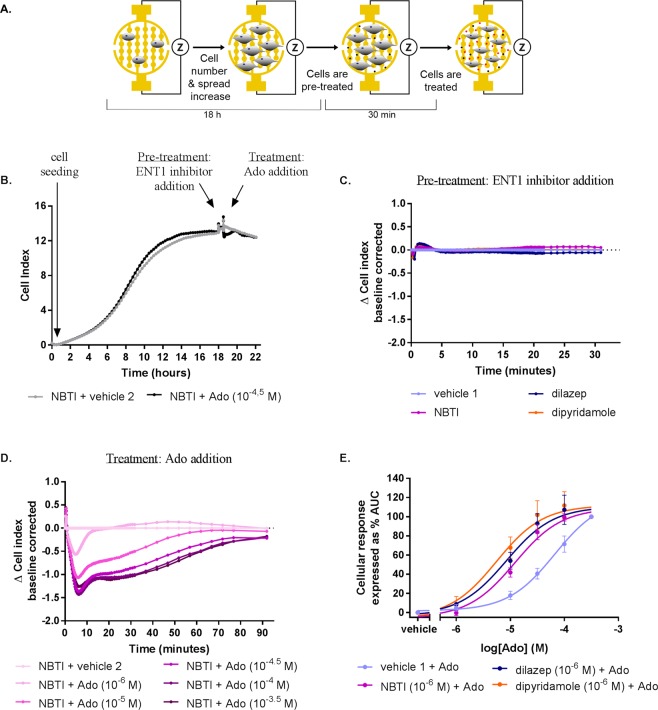
Subsequently, U-2 OS whole cells were used to monitor the inhibitors’ activity in the impedance-based label-free technology. U-2 OS cells adhered strongly to the bottom of the wells and thus the gold-coated electrodes of the E-plates, and therefore no additional coating was necessary to obtain a signal. Various concentrations of cells per well were tested in order to achieve a uniform cell monolayer (Supplementary Material; Fig. S2), which was the case for a concentration of 20,000 cells/well. After cell seeding, attachment, spreading and overnight proliferation, this concentration resulted in a cell index (CI) ranging from 10.0 to 12.0 (Fig. 2A,B). Thus, 20,000 cells/well was selected for all further experiments, as it allowed reliable and reproducible measurements of ENT1 inhibition and subsequent AR activation.
Figure 2.
Concentration-dependent effects of adenosine (Ado) after cell pre-treatment with a single concentration of ENT1 inhibitors (“Format 1”). (A) Graphic representation of cell seeding, spreading, pre-treatment and treatment protocol. (B) Representative xCELLigence traces of a full experiment when cells were pre-treated with NBTI (10−6 M) and subsequently stimulated with adenosine. A representative response (C) after NBTI, dilazep, dipyridamole pre-treatment and (D) after adenosine treatment of cells pre-treated with NBTI. (E) Concentration-response curves for adenosine with or without ENT1 inhibitors pre-treatment (adenosine 10−3.5 M response as 100%). Data shown are mean ± SEM from at least three separate experiments performed in duplicate.
Validation of the Assay Principle
For the development of the label-free assay, we hypothesized that ENT1 inhibition can be detected via adenosine receptor signalling, due to changes in the extracellular concentration of adenosine.
Firstly, it was examined if ENT1 inhibition would lead to an increased extracellular adenosine concentration. Hence, cells were pre-treated with a single concentration (10−6 M) of an ENT1 inhibitor and subsequently treated with several concentrations of adenosine (i.e. “Format 1”). Upon pre-treatment of cells for 30 min with a single concentration of an inhibitor (10−6 M) no change of impedance (expressed as ΔCl) was detected (Fig. 2C). On the contrary, subsequent treatment of cells with various concentrations of adenosine led to a decrease of ΔCl (Fig. 2D). Specifically, the signal decreased in a concentration-dependent manner reaching a minimum ranging from −0.5 to −1.5 ΔCl after approximately 8 to 10 min. The initial decrease in ΔCI was followed by a slow increase back to baseline within 90 min. From these impedance changes, the AUC was determined and a concentration-response curve was obtained providing apparent potency values of adenosine for AR signalling after pre-treatment with vehicle and NBTI, dilazep and dipyridamole, i.e. pEC50 values of 4.2 ± 0.1, 4.9 ± 0.1, 5.1 ± 0.1, 5.2 ± 0.1, respectively (Fig. 2E and Table 1).
Table 1.
Potency of adenosine obtained with whole cell impedance-based experiments performed with U-2 OS cells after pre-treatment with different ENT1 inhibitors at 1 μM (“Format 1”), and potency (pIC50) and inhibitory efficacy of ENT1 inhibitors obtained with whole cell impedance-based experiments performed with U-2 OS cells followed by adenosine treatment at 10−4.5 M (“Format 2”).
| Format 1 | Format 2 | ||
|---|---|---|---|
| Adenosine pEC50 ± SEMa | pIC50 ± SEM (IC50 (nM)) | Inhibitory efficacyb (%) ± SEM | |
| vehicle 1 | 4.2 ± 0.1 | n.a. | 100 ± 5.6 |
| +NBTI | 4.9 ± 0.1 **** | 8.3 ± 0.3 (2.6) | 219 ± 7.3**** |
| +Dilazep | 5.1 ± 0.1 **** | 10.1 ± 0.1 (0.1) | 286 ± 19**** |
| +Dipyridamole | 5.2 ± 0.1 **** | 8.6 ± 0.5 (8.1) | 525 ± 7.3**** |
Values are mean ± SEM of at least three separate experiments performed in duplicate.
aSignificance compared to vehicle’s 1 pEC50 was tested using one-way ANOVA with Dunnett’s post-hoc test. ****p < 0.0001.
bData are normalized to maximal response of adenosine (10−4.5 M) of vehicle 1 (100%). Significance compared to vehicle 1 was tested using one-way ANOVA with Dunnett’s post-hoc test. ****p < 0.0001.
n.a.: not applicable.
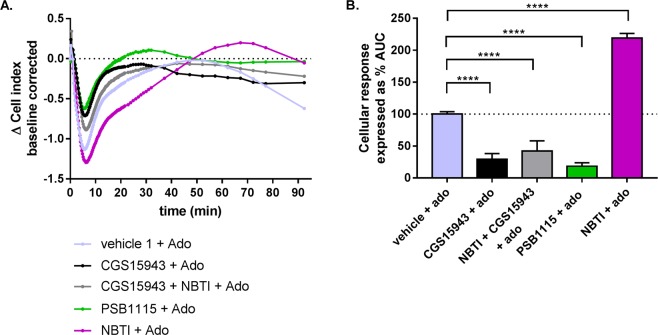
Next, to confirm that the observed changes in impedance and the pEC50 values of adenosine are AR-specific, we used CGS 15943 as a non-selective AR antagonist and PSB 1115 as a selective A2B AR antagonist. Cells were pre-treated with either CGS 15943, NBTI and PSB 1115 or a combination of the first two. Upon pre-treatment for 30 min with each of the compounds no change of impedance was detected (data not shown). As observed in Fig. 2D, pre-treatment with NBTI and the consecutive treatment with adenosine led to an increased AUC (219 ± 7.3%) (Fig. 3; Table 1). In contrast, pre-treatment with CGS 15943 followed by adenosine stimulation resulted in a cellular response with a strongly diminished AUC (29.4 ± 9%) (Fig. 3). Interestingly, when cells were simultaneously pre-treated with CGS 15943 and NBTI, an AUC similar to pre-treatment with CGS 15943 alone was observed (42.2 ± 16%) (Fig. 3). This result indicates that the increased extracellular concentration of adenosine, caused by NBTI inhibition of ENT1, cannot generate an increased cellular response when ARs are blocked by CGS 15943. Similarly, pre-treatment of cells with PSB 1115 resulted in a decreased AUC (18.5 ± 5.4%) (Fig. 3), indicating the A2B AR to be the most prevalent AR. Taken together, this shows that inhibition of ENT1 results in an increased apparent potency for adenosine-mediated AR signalling.
Figure 3.
The response measured is adenosine receptor AR-mediated. (A) A representative response of adenosine (10−4.5 M) after pre-treatment with 10−6 M NBTI, CGS15943, PSB1115 or NBTI and CGS15943 simultaneously. (B) Bar graphs represent the AUC after adenosine addition (normalized to vehicle 1 as 100%). Data shown are mean ± SEM from at least three separate experiments performed in duplicate. Significance compared to vehicle was tested using one-way ANOVA with Tukey’s multiple comparison post-test. ****p < 0.0001.
Pharmacological characterization of ENT1 inhibitors in U-2 OS cells using the xCELLigence
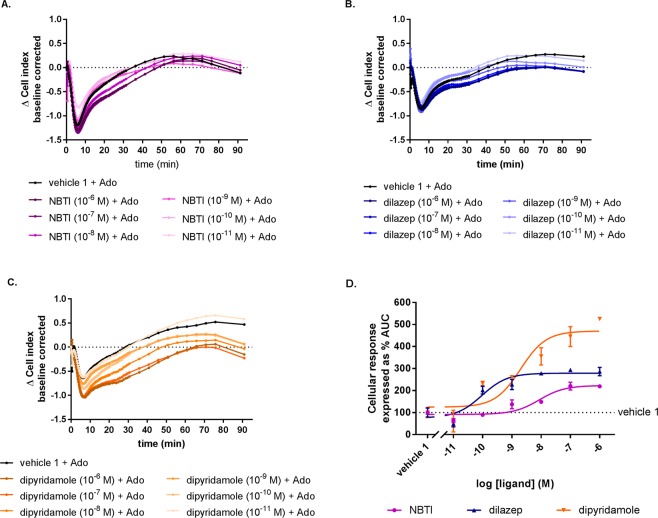
Lastly, the inhibitory effect of different known ENT1 inhibitors was evaluated in a different assay set up (i.e. “Format 2”). Cells were pre-treated with increasing concentrations of NBTI, dilazep and dipyridamole ranging from 10−11 to 10−6 M. Successive addition of a fixed concentration of adenosine (30 μM) led to a negative response with a peak around 8 to 10 minutes depending on the concentration of the inhibitor. This phase was followed by an increase of the response that in all cases reached plateau within 90 minutes after the adenosine addition (Fig. 4A–C). Of note, the shape of these traces were similar for all experimental set-ups, indicative for a typical signature of adenosine-mediated AR signalling in U-2 OS cells. Moreover, all compounds appeared to enhance the adenosine-mediated AR signalling in a concentration-dependent manner as higher concentrations of the inhibitors seem to decrease further the ΔCI especially after the initial negative peak is reached. Interestingly, pre-treatment with various concentrations of dipyridamole generated a negative response with a significantly higher inhibitory efficacy (% AUC at 1 µM) compared to dilazep and NBTI. In conclusion, using this assay set up an ENT1 inhibitor’s potency and inhibitory efficacy could be obtained. Furthermore, different ENT1 inhibitors have different potencies for indirectly inhibiting adenosine-mediated AR signalling.
Figure 4.
Concentration-dependent effects of ENT1 inhibitors after subsequent AR stimulation with a single adenosine concentration (“Format 2”). A representative adenosine response (10−4.5 M) after pre-treatment with ENT1 inhibitors, i.e. (A) NBTI, (B) dilazep and (C) dipyridamole. (D) Concentration-response curves of ENT1 inhibitors (normalized to the maximal response of adenosine alone (10−4.5 M, 100%)). Data shown are mean ± SEM from at least three separate experiments performed in duplicate.
Discussion
The significance of membrane transporters in drug discovery is widely recognized24 and their in-depth investigation is essential as they play a vital role both in physiological conditions and in many diseases. Unfortunately, membrane transporters are vastly understudied compared to other groups of target proteins, and the attention they are getting from the scientific community is limited and not indicative of their biological relevance25. One of the main reasons is that the available assays to study transporter activity have limitations, as already introduced above. For another large membrane-bound protein family, GPCRs, label-free technologies are nowadays frequently used to assess receptor pharmacology26–28. Specifically, it has been shown that agonist-induced adenosine receptor signalling causes cell morphology changes that can be monitored via both optical- and impedance-based label-free assays29–32. In addition, many efforts to study ion channels with label-free technologies are reported. Typical examples are the monitor of TRP ion channel activity with an impedance based assay33, as well as the study of GIRK channel and GABAA receptor with an optical biosensor34,35. Of note, as far as membrane transporters are concerned, there is currently a single report in which an optical-based label-free assay (i.e. Epic by Corning) is used to study the electrogenic transporter SLC34A22. However, this method has not been applied to other electrogenic transporters so far, and still leaves a need for the many non-electrogenic transporters. We therefore established an impedance-based assay suitable for investigating the inhibition of non-electrogenic membrane transporters via the indirect measurement of GPCR-mediated cell signalling on cells that, in this case, endogenously express both targets.
Initial experiments with U-2 OS cells demonstrated that U-2 OS cell line can be used for xCELLigence experiments as they strongly adhere to the bottom of the wells, which is a requirement for impedance measurements36. This resulted in a CI, and thus provided a good window of detection for the assay (Fig. 2B). Additionally, radioligand binding assays validated the presence of ENT1 on U-2 OS cell membranes, since, among others, the determined affinity for all ENT1 inhibitors was found to be in good agreement with available literature data37,38. The presence of ARs on U-2 OS cells has already been reported previously, hence it was not re-examined39,40.
As adenosine is the endogenous substrate and agonist of ENT1 and ARs, respectively, it was chosen as the compound for assay development. Concentration-response curves of adenosine to cells pre-treated with or without a single concentration of ENT1 inhibitors yielded significant differences on the apparent pEC50 value of adenosine (Fig. 2, Table 1). The low pEC50 value of adenosine measured in the absence of ENT1 inhibitor (4.2 ± 0.1), might indicate that mainly A2BAR is expressed in this cell line, as potency values for the other ARs are reported to be at least 100-fold higher41. This hypothesis is compatible with the data publically available from “The Human Protein Atlas” showing that A2B is the most prevalently expressed AR subtype on U-2 OS cells42–45. Moreover, pre-treatment of cells with the non-selective ARs antagonist CGS 1594346, led to a large decrease of subsequent adenosine signalling, evidence that an AR-specific signal had been captured (Fig. 3), while pre-treatment with the selective A2B AR antagonist PSB 1115, resulted in a further decrease of adenosine signalling, confirming the hypothesis of A2B AR being the main player in the signalling pathway. It is important to note that intracellular adenosine concentrations are strictly regulated to be kept low47,48. Specifically, adenosine kinase (ADK), is the principle enzyme to regulate intracellular adenosine and metabolise it to adenosine monophosphate (AMP), which could be further phosphorylated to adenosine diphosphate (ADP) and triphosphate (ATP). Consequently, the intracellular and extracellular adenosine concentrations will not be equilibrated, leading to a fast and continuous adenosine transportation inside the cell by ENT1. As a result, the extracellular adenosine concentration are significantly higher when an ENT1 inhibitor is present.
Subsequent experiments established that the current assay allowed the inhibitory effect of ENT1 inhibitors to be measured in two different assay formats: (1) pre-treatment with a fixed concentration of ENT1 inhibitors and subsequent addition of different concentrations of adenosine (Fig. 2) and (2) pre-treatment with different concentrations of ENT1 inhibitors and subsequent addition of a single concentration of adenosine (Fig. 4). In case of the first format, apparent pEC50 values were obtained for adenosine in the absence and presence of different inhibitors, while in the second format the changes of the apparent pEC50 values are caused by a concentration-dependent effect of ENT1 inhibitors, which thus are best quantified as pIC50 values for the inhibitors. For NBTI, a selective inhibitor of ENT1, a potency value (8.3 ± 0.3) comparable to the radioligand binding data (8.7 ± 0.02) (Supplementary Material; Table S1) as well as previously reported data was found49. Dilazep appeared to be approximately 10-fold more potent compared to the literature49 and the radioligand binding data, which may be explained by the fact that dilazep is a not selective ENT1 and ENT2 inhibitor50. The second format also provides information on the inhibitory efficacy of ENT1 inhibitors (i.e. the efficacy of adenosine signalling via AR that results from ENT1 inhibition) on adenosine-mediated receptor signalling (Fig. 4D). Here it was found that dipyridamole had the highest efficacy, which might be explained by the fact that dipyridamole is a non-selective ENT inhibitor, i.e. it has been shown to have high affinity to ENT1, ENT2, ENT3 transporters51 and lower affinity to ENT452 as well. Together, this could lead to significantly higher extracellular concentrations of adenosine that explain dipyridamole’s increased inhibitory efficacy compared to an ENT1 selective inhibitor, such as NBTI. In addition, dilazep seems to follow the same trend of increased inhibitory efficacy as it is also a non-selective inhibitors for ENT1. Such hypothesis is reinforced by the fact that it has been shown that all ENTs are endogenously expressed on U-2 OS cells53–56. The inhibition of ENT2, ENT3 and ENT4 on U-2 OS cells using this novel method merits further investigation.
Conclusions
In conclusion, we developed and validated an assay to detect SLC activity via GPCR signalling in living cells, using a label-free whole cell impedance-based system (xCELLigence), which is to our knowledge the first case where a biosensor technology is used to study the activity of inhibitors for non-electrogenic membrane transporters. As a proof-of-concept the inhibition of ENT1 via subsequent AR signalling in human osteosarcoma cells (U-2 OS) that endogenously express these targets, was studied. We were able to show that its inhibition could be monitored sensitively and quantified accurately via the indirect measurement of adenosine-mediated ARs signalling. Thus, this approach offers the possibility to study membrane transporters that are or can be linked to a GPCR via their common substrate/ligand. Ultimately, this label-free whole-cell assay technology opens novel opportunities for membrane transporter drug discovery.
Methods
Materials and reagents
Dipyridamole was kindly provided by Janssen Pharmaceutics and dilazep was obtained from Asta-Werke (Degussa Pharma Gruppe, Bielefeld, Germany). NBTI and adenosine were purchased from Sigma-Aldrich (St. Louis, MO, USA), while CGS 15943 was purchased from Tocris Bioscience (Bristol, UK). PET E-plates 16 and 96 for the xCELLigence DP and SP system (ACEA Biosciences, San Diego, CA, USA) were obtained from Bioké (Leiden, the Netherlands). Homo sapiens bone osteosarcoma cells (U-2 OS) were a kind gift from Mr. Hans den Dulk (Leiden Institute of Chemistry, department of Molecular Physiology, Leiden University, the Netherlands). All other compounds and materials were obtained from standard commercial sources.
Cell culture
U-2 OS cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with stable glutamine 10% (v/v) New Born Calf Serum (NBCS), 100 IU/ml penicillin and 100 mg/ml streptomycin at 37 °C and 7% CO2. Cells were cultured as a monolayer on 10 cm ø plates and used for whole cell experiments when a confluency of ~90% was reached.
Membrane preparation
U-2 OS cells were grown as a monolayer in 15 cm ø plates to 80–90% confluency. Then they were detached by scraping into 5 ml of phosphate-buffered saline (PBS) and subsequently centrifuged for 5 min at 1500 rpm to remove PBS. The pellets were resuspended in ice-cold 50 mM Tris-HCl buffer, pH 7.4 and homogenized with an Ultra Turrax homogenizer (IKA-Werke GmbH & Co.KG, Staufen, Germany). Membranes and the cytosolic fraction were separated by centrifugation at 31,000 rpm in an Optima LE-80 K ultracentrifuge (Beckman Coulter, Fullerton, CA) at 4 °C for 20 min. The pellet was resuspended in 10 mL of Tris-HCl buffer and the homogenization and centrifugation step were repeated. Finally the membrane pellet was resuspended in 50 mM Tris-HCl buffer, pH 7.4, and aliquots were stored at −80 °C. Membrane protein concentrations were measured using a BCA protein determination57.
Radioligand binding assay
U-2 OS membranes were thawed, homogenized using an Ultra Turrax homogenizer at 24,000 rpm (IKA-Werke GmbH & Co.KG, Staufen, Germany) and diluted to the desired concentration (6 μg/well) using assay buffer (50 mM Tris-HCl pH 7.4). Displacement experiments were performed using 10 concentrations of competing ligand in the presence of a final concentration of 4 nM [3H]NBTI. At this concentration, total binding (TB) did not exceeded 10% of the radioligand present in the assay in order to prevent ligand depletion. Nonspecific (NSB) binding was determined in the presence of 10−5 M NBTI. Total reaction volume was 100 μL and final concentrations of DMSO were ≤0.25%. The experiment was initiated by addition of membranes. Samples were incubated at 25 °C for 60 min to reach equilibrium. The incubation was terminated by rapid vacuum filtration over GF/C filter using Brandel Harvester 24w (Brandel, Gaithersburg, MD, USA). Filters were subsequently washed three times using ice-cold wash buffer (50 mM Tris-HCl, pH 7.4). After drying the filters at 55 °C for 30 min, filter-bound radioactivity was determined by liquid scintillation spectrometry using a Tri-Carb liquid scintillation counter (PerkinElmer, Groningen, The Netherlands).
Label-free whole-cell assay
Detection principle
Label-free whole-cell assays were performed using the xCELLigence real-time cell analyser (RTCA) system8,9, as described previously26. In short, the RTCA system measures the electrical impedance generated by cells adhering to gold-coated electrodes embedded on the bottom of microelectronic E-plates. Variations in number, degree of adhesion, cellular viability and morphology of cells result in relative changes in impedance (Z), which are recorded continuously at 10 kHz and displayed in the unitless parameter coined Cell Index (CI)7,8. CI is a relative measure defined as (Zi-Z0) Ω/15 Ω, where Zi is the impedance at each individual time point and Z0 represents the baseline impedance in the absence of cells, which is measured prior to the start of the experiment.
General Protocol
U-2 OS cells were harvested by re-suspending in cell culture medium after brief trypsinization (treatment with trypsin/EDTA for about 5 min) and centrifuged once at 200 × g (1500 rpm) for 5 min. Background impedance (Z0) was measured after the addition of 40 μL culture media to 16 or 96 well E-plates. Cells were seeded by adding 50 μL of cell suspension containing 20,000 cells per well. After resting at room temperature for at least 30 min, the E-plate was placed into the recording station situated in a 37 °C and 5% CO2 incubator. Impedance was measured every 15 minutes overnight.
Cell pre-treatment: Cells were pre-treated by an ENT1 inhibitor (10−6 M or 10−6 to 10−11 M, depending on the assay), adenosine receptor antagonist or vehicle control (0.25% dimethylsulfoxide (DMSO) in Phosphate-buffered saline (PBS)) in 5 µl after 18 h. CI was recorded for at least 30 minutes with a recording schedule of 15 second intervals for 20 minutes, followed by intervals of 1 minute.
Cell treatment: Afterwards, cells were stimulated with adenosine (10−4.5 or 10−6 to 10−3.5 M depending on the assay) or vehicle control (0.125% DMSO in PBS) in 5 μL and CI was recorded for at least 90 minutes with a recording schedule of 15 second intervals for 20 minutes, followed by intervals of 1 minute for 10 minutes and finally 5 minutes. In all cases, final well volumes and DMSO concentrations upon cell and ligands addition were 100 μL and 0.375%, respectively, for all wells and assays.
Data analysis
Radioligand binding assay
Data analyses were performed using GraphPad Prism 7.0 software (GraphPad Software Inc., San Diego, CA, USA). pIC50 values in radioligand displacement assays were obtained by non-linear regression curve fitting into a sigmoidal concentration-response curve using the equation:
pKi values were acquired from pIC50 values using the Cheng-Prusoff equation58:
The KD value of 1.89 nM for [3H] NBTI, was obtained by fitting the data from homologous displacement experiments (Supplementary Material; Fig. S1) to the model “One site – Homologous”.
Label-free whole-cell assay
RTCA software 2.0 (ACEA Biosciences, Inc.) was used to obtain the experimental data. All data were analysed using GraphPad Prism 7.00 (GraphPad software, San Diego, CA, USA). After subtracting baseline (vehicle control), ligand responses were normalized at the time of ligand addition to obtain Δ Cell Index (Δ CI) values to correct for ligand-independent responses. Cells that were not pre-treated were used as control (vehicle 1) for the ENT1 inhibitor addition, while cells that were not treated with adenosine (vehicle 2) were used as control for the adenosine addition. The time of normalization was either 18 h or 18 h 30 min after cell seeding for analysis of ENT1 inhibitor/ARs antagonist or adenosine effects, respectively.
The absolute values of Total Area Under the Curve (AUC) up to 90 min after adenosine addition were exported to Graphpad Prism for further analysis yielding bar graphs or concentration–response curves. pEC50 values of adenosine after pre-treatment of cells with one concentration of ENT1 inhibitors (format 1; Table 1) and pIC50 values of ENT1 inhibitors (format 2; Table 1) were obtained using non-linear regression curve fitting of Total AUC data into a sigmoidal dose-response curve with variable slope. Data shown are the mean ± SEM of at least three individual experiments performed in duplicate.
Statistical significance was determined using one-way ANOVA, followed by a Tukey’s post test for comparison of all columns or a Dunnett’s post test when comparing to vehicle. If p-values were below 0.05, observed differences were considered statistically significant.
Supplementary information
Acknowledgements
We thank Dr. Sylvia Le Dévédec (LACDR, Leiden, The Netherlands) for her kind help in obtaining cell images (Fig. S2).
Author Contributions
Participated in research design; A.V., A.P.I.J., L.H.H., Conducted experiments; A.V., Performed data analysis; A.V., Wrote or contributed to the writing of the manuscript; A.V., A.P.I.J., L.H.H.
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-48829-3.
References
- 1.Hediger MA, Clémençon B, Burrier RE, Bruford EA. The ABCs of membrane transporters in health and disease (SLC series): Introduction. Molecular Aspects of Medicine. 2013;34:95–107. doi: 10.1016/j.mam.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wong Soo-Hang, Gao Alice, Ward Sabrina, Henley Charles, Lee Paul H. Development of a Label-free Assay for Sodium-Dependent Phosphate Transporter NaPi-IIb. Journal of Biomolecular Screening. 2012;17(6):829–834. doi: 10.1177/1087057112442961. [DOI] [PubMed] [Google Scholar]
- 3.Blass, B. Basic Principles of Drug Discovery and Development. (Elsevier Science, 2015).
- 4.Grewer, C., Gameiro, A., Mager, T. & Fendler, K. Electrophysiological characterization of membrane transport proteins. Annual review of biophysics 42, 95–120, 10.1146/annurevbiophys-083012-130312 (2013). [DOI] [PubMed]
- 5.Priest, B. T. et al. Automated Electrophysiology Assays. (Eli Lilly & Company and the National Center for Advancing Translational Sciences, Bethesda (MD), 2004). [PubMed]
- 6.Weinglass A, Kaczorowski G, Garcia M. Technologies for transporter drug discovery. Channels. 2008;2:312–321. doi: 10.4161/chan.2.5.6896. [DOI] [PubMed] [Google Scholar]
- 7.Fang Y. The development of label-free cellular assays for drug discovery. Expert Opinion on Drug Discovery. 2011;6:1285–1298. doi: 10.1517/17460441.2012.642360. [DOI] [PubMed] [Google Scholar]
- 8.Yu N, et al. Real-Time Monitoring of Morphological Changes in Living Cells by Electronic Cell Sensor Arrays: An Approach To Study G Protein-Coupled Receptors. Analytical Chemistry. 2006;78:35–43. doi: 10.1021/ac051695v. [DOI] [PubMed] [Google Scholar]
- 9.Xi B, Yu N, Wang X, Xu X, Abassi YA. The application of cell-based label-free technology in drug discovery. Biotechnology journal. 2008;3:484–495. doi: 10.1002/biot.200800020. [DOI] [PubMed] [Google Scholar]
- 10.Halai R, Cooper MA. Using label-free screening technology to improve efficiency in drug discovery. Expert Opin Drug Discov. 2012;7:123–131. doi: 10.1517/17460441.2012.651121. [DOI] [PubMed] [Google Scholar]
- 11.Scott CW, Peters MF. Label-free whole-cell assays: expanding the scope of GPCR screening. Drug discovery today. 2010;15:704–716. doi: 10.1016/j.drudis.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 12.Zhang R, Xie X. Tools for GPCR drug discovery. Acta Pharmacologica Sinica. 2012;33:372–384. doi: 10.1038/aps.2011.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baldwin SA, et al. The equilibrative nucleoside transporter family, SLC29. Pflugers Arch. 2004;447:735–743. doi: 10.1007/s00424-003-1103-2. [DOI] [PubMed] [Google Scholar]
- 14.Yao SYM, Ng AML, Cass CE, Baldwin SA, Young JD. Nucleobase Transport by Human Equilibrative Nucleoside Transporter 1 (hENT1) The Journal of Biological Chemistry. 2011;286:32552–32562. doi: 10.1074/jbc.M111.236117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagmann W, Jesnowski R, Löhr JM. Interdependence of Gemcitabine Treatment, Transporter Expression, and Resistance in Human Pancreatic Carcinoma Cells. Neoplasia (New York, N.Y.) 2010;12:740–747. doi: 10.1593/neo.10576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iikura M, et al. ENT1, a Ribavirin Transporter, Plays a Pivotal Role in Antiviral Efficacy of Ribavirin in a Hepatitis C Virus Replication Cell System. Antimicrobial Agents and Chemotherapy. 2012;56:1407–1413. doi: 10.1128/AAC.05762-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pennycooke M, Chaudary N, Shuralyova I, Zhang Y, Coe IR. Differential Expression of Human Nucleoside Transporters in Normal and Tumor Tissue. Biochemical and Biophysical Research Communications. 2001;280:951–959. doi: 10.1006/bbrc.2000.4205. [DOI] [PubMed] [Google Scholar]
- 18.Rose JB, et al. Equilibrative nucleoside transporter 1 plays an essential role in cardioprotection. American Journal of Physiology - Heart and Circulatory Physiology. 2010;298:H771–H777. doi: 10.1152/ajpheart.00711.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abd-Elfattah AS, Wechsler AS. Separation Between Ischemic and Reperfusion Injury by Site Specific Entrapment of Endogenous Adenosine and Inosine Using NBMPR and EHNA. Journal of Cardiac Surgery. 1994;9:387–396. doi: 10.1111/jocs.1994.9.3s.387. [DOI] [PubMed] [Google Scholar]
- 20.Lin W, Buolamwini JK. Synthesis, flow cytometric evaluation, and identification of highly potent dipyridamole analogues as equilibrative nucleoside transporter 1 inhibitors. J Med Chem. 2007;50:3906–3920. doi: 10.1021/jm070311l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bone DBJ, Robillard KR, Stolk M, Hammond JR. Differential regulation of mouse equilibrative nucleoside transporter 1 (mENT1) splice variants by protein kinase CK2. Molecular Membrane Biology. 2007;24:294–303. doi: 10.1080/09687860701210617. [DOI] [PubMed] [Google Scholar]
- 22.Boeck CR, et al. Effects of glutamate transporter and receptor ligands on neuronal glutamate uptake. Neuroscience Research. 2005;53:77–83. doi: 10.1016/j.neures.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 23.Gulley JM, Zahniser NR. Rapid regulation of dopamine transporter function by substrates, blockers and presynaptic receptor ligands. European Journal of Pharmacology. 2003;479:139–152. doi: 10.1016/j.ejphar.2003.08.064. [DOI] [PubMed] [Google Scholar]
- 24.The International Transporter Consortium. Membrane transporters in drug development. Nature Reviews. Drug Discovery9, 215–236, 10.1038/nrd3028 (2010). [DOI] [PMC free article] [PubMed]
- 25.César-Razquin A, et al. A Call for Systematic Research on Solute Carriers. Cell. 2015;162:478–487. doi: 10.1016/j.cell.2015.07.022. [DOI] [PubMed] [Google Scholar]
- 26.Hillger JM, et al. Whole-cell biosensor for label-free detection of GPCR-mediated drug responses in personal cell lines. Biosensors and Bioelectronics. 2015;74:233–242. doi: 10.1016/j.bios.2015.06.031. [DOI] [PubMed] [Google Scholar]
- 27.Rocheville, M., Martin, J., Jerman, J. & Kostenis, E. In Progress in Molecular Biology and Translational Science Vol. 115 (ed. Terry Kenakin) 123–142 (Academic Press, 2013). [DOI] [PubMed]
- 28.Nederpelt I, Vergroesen RD, IJzerman AP, Heitman LH. Persistent GnRH receptor activation in pituitary αT3-1 cells analyzed with a label-free technology. Biosensors and Bioelectronics. 2016;79:721–727. doi: 10.1016/j.bios.2015.12.066. [DOI] [PubMed] [Google Scholar]
- 29.Guo D, Mulder-Krieger T, IJzerman AP, Heitman LH. Functional efficacy of adenosine A2A receptor agonists is positively correlated to their receptor residence time. British journal of pharmacology. 2012;166:1846–1859. doi: 10.1111/j.1476-5381.2012.01897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hillger JM, et al. Getting personal: Endogenous adenosine receptor signaling in lymphoblastoid cell lines. Biochemical Pharmacology. 2016;115:114–122. doi: 10.1016/j.bcp.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 31.McGuinness R. Label-free cell-based assays bring greater biorelevance to early drug discovery. Assay Drug Dev Technol. 2009;7:191–193. doi: 10.1089/adt.2009.9993. [DOI] [PubMed] [Google Scholar]
- 32.Hinz S, et al. Adenosine A2A receptor ligand recognition and signaling is blocked by A2B receptors. Oncotarget. 2018;9:13593–13611. doi: 10.18632/oncotarget.24423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pänke O, Weigel W, Schmidt S, Steude A, Robitzki AA. A cell-based impedance assay for monitoring transient receptor potential (TRP) ion channel activity. Biosensors and Bioelectronics. 2011;26:2376–2382. doi: 10.1016/j.bios.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 34.Klein AB, Nittegaard-Nielsen M, Christensen JT, Al-Khawaja A, Wellendorph P. Demonstration of the dynamic mass redistribution label-free technology as a useful cell-based pharmacological assay for endogenously expressed GABAA receptors. MedChemComm. 2016;7:426–432. doi: 10.1039/C5MD00442J. [DOI] [Google Scholar]
- 35.Krebs KM, et al. Label-Free Whole Cell Biosensing for High-Throughput Discovery of Activators and Inhibitors Targeting G Protein-Activated Inwardly Rectifying Potassium Channels. ACS omega. 2018;3:14814–14823. doi: 10.1021/acsomega.8b02254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lieb S, et al. Label-free analysis of GPCR-stimulation: The critical impact of cell adhesion. Pharmacological Research. 2016;108:65–74. doi: 10.1016/j.phrs.2016.04.026. [DOI] [PubMed] [Google Scholar]
- 37.Tromp RA, Spanjersberg RF, von Frijtag Drabbe Künzel JK, IJzerman AP. Inhibition of Nucleoside Transport Proteins by C8-Alkylamine-Substituted Purines. Journal of Medicinal Chemistry. 2005;48:321–329. doi: 10.1021/jm049303k. [DOI] [PubMed] [Google Scholar]
- 38.Rehan S, Ashok Y, Nanekar R, Jaakola V-P. Thermodynamics and kinetics of inhibitor binding to human equilibrative nucleoside transporter subtype-1. Biochemical Pharmacology. 2015;98:681–689. doi: 10.1016/j.bcp.2015.09.019. [DOI] [PubMed] [Google Scholar]
- 39.Langemeijer EV, Verzijl D, Dekker SJ, IJzerman AP. Functional selectivity of adenosine A(1) receptor ligands? Purinergic Signalling. 2013;9:91–100. doi: 10.1007/s11302-012-9334-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, Eishingdrelo A, Kongsamut S, Eishingdrelo H. G-protein-coupled receptors mediate 14-3-3 signal transduction. Signal Transduction And Targeted Therapy. 2016;1:16018. doi: 10.1038/sigtrans.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan L, Burbiel JC, Maaß A, Müller CE. Adenosine receptor agonists: from basic medicinal chemistry to clinical development. Expert Opinion on Emerging Drugs. 2003;8:537–576. doi: 10.1517/14728214.8.2.537. [DOI] [PubMed] [Google Scholar]
- 42.The Human Protein Atlas, https://www.proteinatlas.org/ENSG00000170425-ADORA2B/cell#rna – ADORA2B, (Accessed on 25/01/2019).
- 43.The Human Protein Atlas, https://www.proteinatlas.org/ENSG00000128271-ADORA2A/cell#rna – ADORA2A, (Accessed on 25/01/2019).
- 44.The Human Protein Atlas, https://www.proteinatlas.org/ENSG00000282608-ADORA3/cell#rna – ADORA3, (Accessed on 25/01/2019).
- 45.The Human Protein Atlas, https://www.proteinatlas.org/ENSG00000163485-ADORA1/cell#rna - ADORA1 (Accessed on 25/01/2019).
- 46.IUPHAR/BPS Guide to PHARMACOLOGY, http://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=3 - Adenosine receptors, http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?tab=biology&ligandId=384 (Accessed on 25/01/2019).
- 47.Xu Y, et al. Regulation of endothelial intracellular adenosine via adenosine kinase epigenetically modulates vascular inflammation. Nature Communications. 2017;8:943. doi: 10.1038/s41467-017-00986-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boison D. Adenosine kinase: exploitation for therapeutic gain. Pharmacol Rev. 2013;65:906–943. doi: 10.1124/pr.112.006361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pirovano IM, Van Belle H, IJzerman AP. Inhibition of nucleoside uptake in human erythrocytes by a new series of compounds related to lidoflazine and mioflazine. Eur J Pharmacol. 1990;189:419–422. doi: 10.1016/0922-4106(90)90040-5. [DOI] [PubMed] [Google Scholar]
- 50.Playa H, et al. Dilazep analogues for the study of equilibrative nucleoside transporters 1 and 2 (ENT1 and ENT2) Bioorg Med Chem Lett. 2014;24:5801–5804. doi: 10.1016/j.bmcl.2014.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Chunmei, Lin Wenwei, Playa Hilaire, Sun Shan, Cameron Keyuna, Buolamwini John K. Dipyridamole analogs as pharmacological inhibitors of equilibrative nucleoside transporters. Identification of novel potent and selective inhibitors of the adenosine transporter function of human equilibrative nucleoside transporter 4 (hENT4) Biochemical Pharmacology. 2013;86(11):1531–1540. doi: 10.1016/j.bcp.2013.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou M, Duan H, Engel K, Xia L, Wang J. Adenosine Transport by Plasma Membrane Monoamine Transporter: Reinvestigation and Comparison with Organic Cations. Drug Metabolism and Disposition. 2010;38:1798–1805. doi: 10.1124/dmd.110.032987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.The Human Protein Atlas, https://www.proteinatlas.org/ENSG00000112759-SLC29A1/cell#rna - SLC29A1 (Accessed on 25/01/2019).
- 54.The Human Protein Atlas, https://www.proteinatlas.org/ENSG00000174669-SLC29A2/cell#rna - SLC29A2 (Accessed on 25/01/2019).
- 55.The Human Protein Atlas, https://www.proteinatlas.org/ENSG00000198246-SLC29A3/cell#rna - SLC29A3 (Accessed on 25/01/2019).
- 56.The Human Protein Atlas, https://www.proteinatlas.org/ENSG00000164638-SLC29A4/cell#rna - SLC29A4 (Accessed on 25/01/2019).
- 57.Smith PK, et al. Measurement of protein using bicinchoninic acid. Analytical biochemistry. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 58.Cheng Y, Prusoff WH. Relationship between the inhibition constant (KI) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochemical Pharmacology. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.