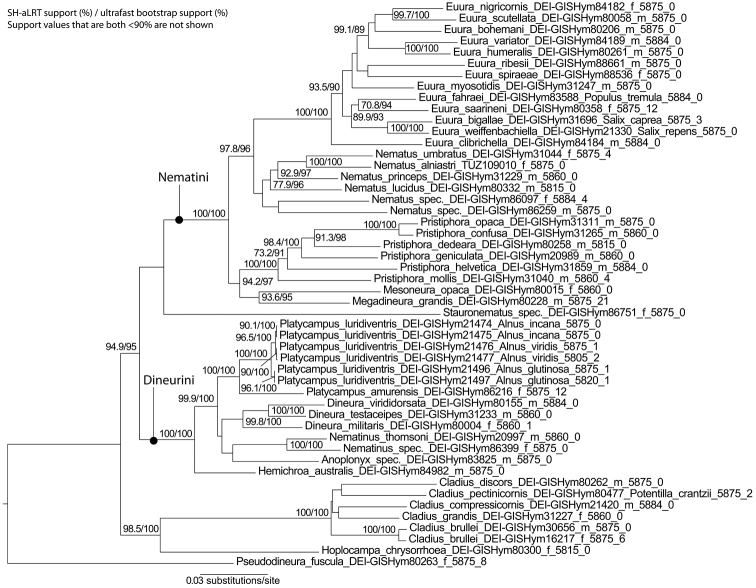
Figure 1.
Maximum likelihood tree of Nematinae based on four genes (COI, NaK, POL2, TPI). Only specimens sequenced for all four genes were included. Short introns from POL2 and TPI were excluded. The best-fit model chosen according to Bayesian information criterion was GTR+R4. Numbers at branches show SH-aLRT support (%) / ultrafast bootstrap support (%) values. Support values for weakly supported branches (<90) are not shown. Letters “f” and “m” stand for “female” and “male”, and are not given for larvae. Numbers at the end of the tip labels refer to the length of the sequence and the number of ambiguous positions (e.g., heterozygosities). The number of ambiguous positions given for two males are due to variation in mitochondrial COI because of possible heteroplasmy. The tree was rooted as in Prous et al. (2014). The scale bar shows the number of estimated substitutions per nucleotide position.