Abstract
The Sec61 protein-conducting channel mediates transport of many proteins, such as secretory proteins, across the endoplasmic reticulum (ER) membrane during or after translation. Posttranslational transport is enabled by two additional membrane proteins associated with the channel, Sec63 and Sec62, but its mechanism was poorly understood. Here we determined a structure of the Sec complex (Sec61-Sec63-Sec71-Sec72) from Saccharomyces cerevisiae by cryo-electron microscopy (cryo-EM). The structure shows that Sec63 tightly associates with Sec61 though interactions in cytosolic, transmembrane, and ER-luminal domains, prying open Sec61’s lateral gate and translocation pore and thus activating the channel for substrate engagement. Furthermore, Sec63 optimally positions binding sites for cytosolic and luminal chaperones in the complex to enable efficient polypeptide translocation. Our study provides mechanistic insights into eukaryotic posttranslational protein translocation.
The eukaryotic Sec61 or prokaryotic SecY complex forms a universally conserved protein-conducting channel that is essential for biogenesis of many proteins (1-3). The channel mediates transport of soluble (e.g., secretory) proteins across the eukaryotic ER membrane or the prokaryotic plasma membrane through its water-filled pore and integration of membrane proteins into the lipid phase through its lateral gate. The Sec61/SecY channel consists of an hourglass-shaped α-subunit, which contains 10 transmembrane segments (TMs 1–10), and two small β- and γ-subunits, which are single-pass membrane proteins in eukaryotes (4). Often, translocation is coupled with translation (i.e., cotranslational translocation) by direct docking of a translating ribosome onto the channel. The channel also translocates many proteins in a posttranslational manner, the mechanisms of which differ between eukaryotes and prokaryotes. In eukaryotes, posttranslational translocation requires two essential membrane proteins Sec63 and Sec62, which associate with the channel (5-8), and the ER-resident Hsp70 chaperone BiP, which grasps the substrate polypeptide in the ER lumen and prevents it from backsliding to the cytosol (9-12). In fungal species, the complex (hereafter referred to as the Sec complex) is further associated with the nonessential Sec71 and Sec72 subunits (10, 11, 13). The molecular architecture of the Sec complex and the functions of its subunits are poorly defined.
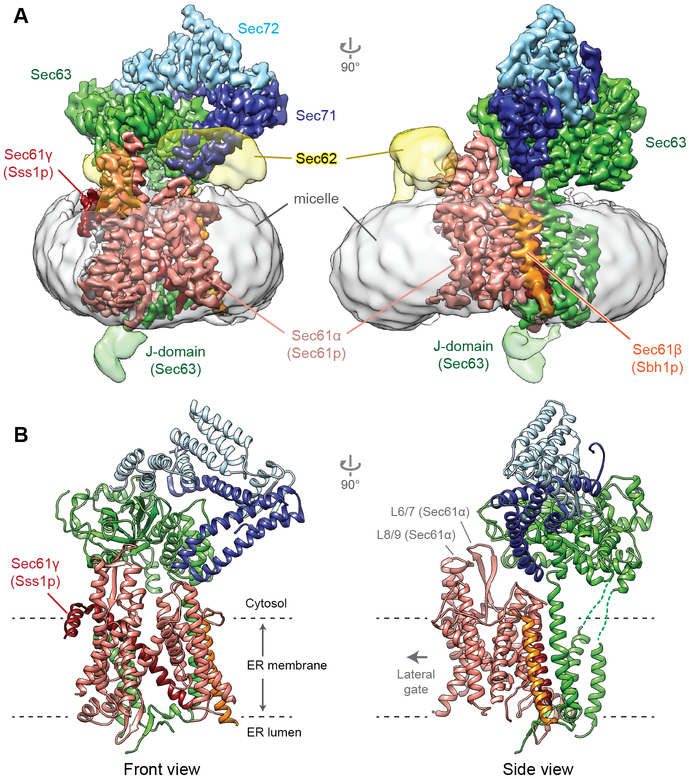
To gain insight into Sec-mediated protein translocation, we determined a structure of the Saccharomyces cerevisiae Sec complex at 3.7-Å resolution by cryo-EM (Fig. 1, and figs S1 and S2). Many side chains are clearly visible in the density map, enabling modeling of an accurate atomic structure (Fig. 1B, and fig S2C). The map also allowed us to improve the model for the eukaryotic Sec61 channel, which was previously built into maps at ~4–5-Å local resolutions (14, 15). However, Sec62 and the ER-luminal J-domain of Sec63, which transiently interacts with BiP (9-11, 16), were not sufficiently resolved for model building, likely due to their flexible motions (Fig. 1A). The structure reveals that Sec63 together with Sec71-72 forms a large soluble domain, which sits on the cytosolic side of the Sec61 channel (Fig. 1). Sec63 consists of an N-terminal domain containing 3 TMs and a J-domain between the second and third TMs, and a C-terminal cytosolic domain (Fig. 2, A and B). The cytosolic domain contains two α-helical domains (HD1 and HD2) and an immunoglobulin-like (fibronectin type-III; shortly FN3) domain, which are arranged similarly to the homologous region of the Brr2 RNA helicase (17) (fig. S3). Sec71-Sec72, the structure of which is similar to a recent crystal structure of Chaetomium thermophilum Sec71-72 (18), clamps Sec63’s cytosolic domain like ‘tongs’ (fig. S4).
Figure 1. Structure of the yeast Sec complex.
Cryo-EM density map (A) and atomic model (B) of the yeast posttranslational protein translocation complex. Front view, view into the lateral gate.
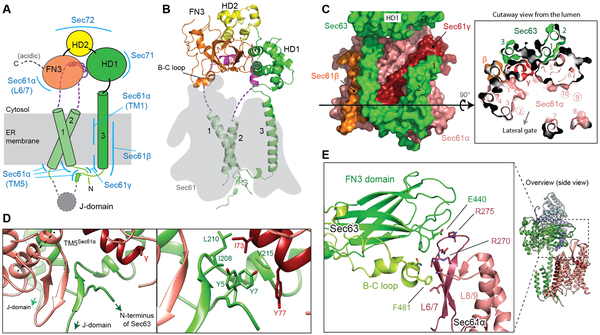
Figure 2. Structure of Sec63 and interactions with the channel.
(A) A schematic of Sec63 domains. Regions interacting with other parts of the complex are indicated by blue lines. Unmodeled regions are shown in dashed lines. (B) Structure of Sec63 (front view). The position of Sec61 is shown by a gray shade. (C) Interactions between TMs of Sec63 and Sec61. Left, a view from the back; right, a cutaway view from the ER lumen. Black arrowed line, the cross-sectional plane. Note that TMs 2, 9, and 10 of Sec61α are located above the cross-sectional plane. (D) Interactions between Sec63 and Sec61 in the luminal side. Left, a β-sheet formed between Sec61α (TM5 indicated by a dashed line) and the segment between Sec63 TM3 and the J-domain. Right, a magnified view with side chains in sticks. (E) Interactions between the FN3 domain and the cytosolic loop L6/7 of Sec61α (also see Fig. 1B).
Sec63 makes extensive contacts with the channel through its transmembrane, cytosolic, and luminal domains, indicative of a major role in regulating the channel’s function (Fig. 2 C-E). In the membrane region, the TMs of Sec63 are located at the back (opposite from the lateral gate) of the Sec61 channel, interacting with the TMs of Sec61β and Sec61γ as well as TM1 and TM5 of Sec61α (Fig. 2C). Considering the extensive interactions between these elements, the TMs of Sec63 likely makes a main contribution to the association between Sec61 and the rest of the Sec complex. In the cytosolic region, the FN3 domain of Sec63 interacts with the loop between TM6 and TM7 (L6/7) of Sec61α through antigen-antibody-like binding. Like other FN3 domains, FN3 of Sec63 has a canonical β-sandwich fold comprised of 7 β-stands (referred to as A to G) but contains unusually long A-B, B-C, and D-E inter-strand loops (fig. S3 B and C). With both A-B and B-C loops, FN3 creates a binding surface for L6/7 using a combination of surface complementarity and electrostatic and hydrophobic interactions (Fig. 2E, and fig. S3D). Although sequence conservation is not obvious, metazoan Sec63s have similar extensions in the A-B and B-C loops. We expect analogous interactions between Sec63 and Sec61 in other eukaryotes. The interaction between FN3 and L6/7 is noteworthy because L6/7, together with L8/9, forms a docking site for the ribosome (14, 19, 20) (fig. S5A). Accordingly, superimposition of the Sec complex with a ribosome-bound Sec61 structure shows massive steric clashes between the ribosome and the cytosolic domains of Sec63 and Sec62 (fig. S5B), explaining why Sec61 in the Sec complex cannot bind to the ribosome (7, 11). In the ER luminal side, a segment preceding TM3 of Sec63 is directed into the luminal funnel of the Sec61 channel through the crevice present between TM5 of Sec61α and the TM of Sec61γ (Fig. 2D). Notably, this segment makes an antiparallel β-sheet together with a β-hairpin looping out in the middle of Sec61α’s TM5. This β-augmentation is further buttressed by hydrophobic interactions with the N-terminal segment of Sec63. These features are highly conserved throughout eukaryotes and thus likely play an important role in optimal positioning of the J-domain.
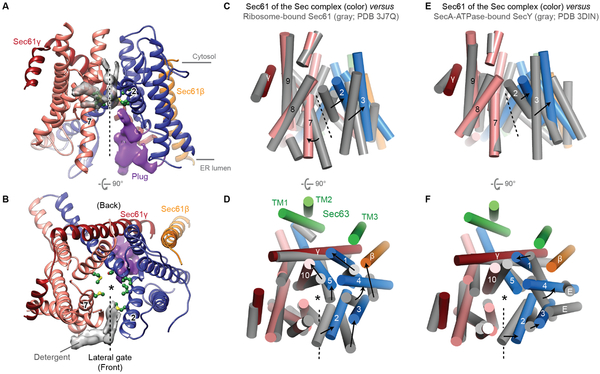
One striking feature of the Sec complex structure is a fully open channel (Fig. 3, A and B). The Sec61/SecY channel has a characteristic ‘clamshell’-like topology, in which its central pore can open towards the lipid phase through the lateral gate formed between TM2 and TM7. Compared to previous Sec61/SecY structures (4, 14, 21-24), the channel in the Sec complex displays a substantially wider opening at its lateral gate, through which a signal sequence can readily pass as an α-helix. (Fig. 3, and fig. S6). This contrasts with structures of channels associated with the ribosome or the bacterial posttranslational translocation motor SecA (14, 21-24), where the channel shows an only partially open lateral gate (Fig. 3, C–F), which was proposed to be further opened by interaction with the hydrophobic signal sequence during the initial substrate insertion. The opening is achieved by a largely rigid-body movement between the two halves (TMs 1–5 and 6–10) of Sec61α and additional motions of the lateral gate helices. The fully open conformation appears to be a result of the extensive interactions with Sec63. For example, binding between FN3 and L6/7 perhaps pulls the C-terminal half of Sec61α to open the lateral gate. However, further investigation will be necessary to understand the precise mechanism and the dynamics of channel gating in the native membrane environment. At the open lateral gate slit, there is a weak density feature, which likely represents bound detergent molecules (Fig. 3, A and B). In the native membrane, lipid molecules may occupy this site and facilitate initial binding of signal sequences.
Figure 3. A fully opened Sec61 channel in the Sec complex.
(A and B) Structure of the Sec61 channel. The N- and C- terminal halves of Sec61α are in blue and salmon, respectively. Gray density feature is presumed detergent molecules. Pore-lining residues are shown as green balls and sticks. Density feature for the plug is in purple. ‘2’ and ‘7’ indicate TM2 and TM7 respectively. (C–F) Comparison of Sec61 of the Sec complex (colored) with Sec61 of the cotranslational ribosome-Sec61 complex (gray; C and D) or SecY of a bacterial posttranslational SecA-SecY channel complex (gray; E and F). The structures are aligned with respect to the C-terminal half of Sec61α (C–F). Shown are the front (A, C, and E) and cytosolic (B, D, and F) views. Numbers indicate corresponding TMs. Dashed line, lateral gate. Asterisk, translocation pore. For simplicity, L6/7 and L8/9 of Sec61α were not shown. In D and F, TMs of Sec63 are also shown (green). Also see fig. S6 for comparisons to archaeal SecY and substrate-engaged channels.
Our channel structure likely also represents a fully open state of the translocation pore (Fig. 3B, and fig. S7). The radius of the pore constriction is ~3 Å, large enough to readily pass an extended polypeptide chain. The opening would also permit passage of small hydrated ions and polar molecules in the absence of a translocating polypeptide (25, 26), although the relatively positive electrostatic potential around the pore may disfavor permeation of positively charged species (fig. S7C). Yeast Sec61 has a relatively less hydrophobic pore constriction compared to non-fungal Sec61 and prokaryotic SecY (fig. S7D). In prokaryotes, reduction of hydrophobicity in the pore constriction has been shown to lead to membrane potential dissipation (26), and similarly, in higher eukaryotes it might cause calcium leakage from ER. However, yeast may tolerate ion leakage because calcium is stored primarily in the vacuole. In resting or primed channels, the pore is closed or narrow (<2 Å in radius), and further blocked by a small α-helical plug in the luminal funnel (4, 14, 21). By contrast, in our structure the plug seems flexible and displaced from the pore (Fig 3, A and B).
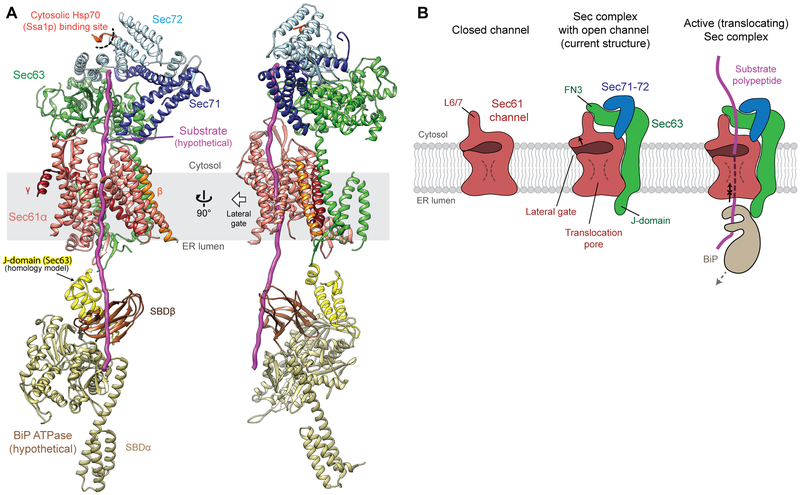
The spatial arrangement of Sec63 and Sec71-72 with respect to the Sec61 channel suggests how these components play roles in accepting a polypeptide substrate from a cytosolic chaperone and handing it over to the channel and subsequently to BiP. Studies of C. thermophilum Sec72 have suggested that Sec72 provides a docking site for the cytosolic Hsp70 chaperone Ssa1p (18), which prevents substrates from premature folding or aggregation before translocation (6). Superimposition of the co-crystal structure of Sec72 and an Ssa1p C-terminal tail shows that the Ssa1p-binding site is ~60 Å above the channel’s pore (Fig. 4A). While the cytosolic domain of Sec63-71-72 sits on top of Sec61, its position is tilted such that the polypeptide can insert straight down to the pore. Similarly, Sec62 is also positioned off the translocation path (Fig. 1A). Therefore, upon release from Ssa1p, a substrate would efficiently engage with the pore without obstruction. The structure also allows us to propose how BiP Hsp70 may catch the substrate in the ER lumen. Despite the low resolution of the J-domain (Fig. 1A), we could dock a homology model into the EM density map based on the shape of the feature and the orientations of the flanking segments (Fig. 4A). We then superimposed a recent crystal structure of a bacterial J-domain–Hsp70 complex (27) to our EM structure (Fig. 4A). Strikingly, this modeling exercise showed a peptide binding cleft of the Hsp70 (called substrate-binding domain β or SBDβ) is placed directly below the translocation pore. Thus, the J-domain seems optimally positioned to allow BiP to grasp the substrate polypeptide as it emerges from the channel.
Figure 4. Model of an active translocation complex.
(A) The Sec complex structure superimposed with a Ssa1p C-terminal peptide (red orange; PDB ID: 5L0Y) and DnaK Hsp70 as a model for BiP (yellow and brown; PDB ID: 5RNO). (B) Schematics for a closed Sec61 channel in isolation (left), an open channel in association with Sec63 (middle), and an active Sec complex engaged with a substrate (right; corresponding to the model in (A)). For the full translocation cycle, see fig S8.
Our structure offers a model for how Sec63 enables posttranslational translocation (Fig. 4B, and fig. S8) and provides a more complete picture of how the Sec61/SecY channel works together with different binding partners (i.e., ribosomes, Sec63, or SecA) to enable transport of a range of substrates. Association of Sec63 seems to induce full opening of the channel, a conformation in which the channel can readily accept a substrate polypeptide. Such a conformation, compared to a partially open channel seen with the other modes, is likely advantageous for many posttranslational-specific substrates, which tend to have a less hydrophobic signal sequence (28-30).
Supplementary Material
Acknowledgments:
We thank D. Toso for help with electron microscope operation, and J. Hurley, S. Brohawn, and K. Tucker for critical reading of manuscript.
Funding: This work was funded by UC Berkeley (E.P.) and an NIH training grant (T32GM008295; S.I.).
Footnotes
Competing interests: None declared.
Data and materials availability: The cryo-EM density maps and atomic model have been deposited in EM Data Bank (accession code: EMD-0336) and Protein Data Bank (accession code: 6N3Q), respectively.
References and Notes
- 1.Park E, Rapoport TA, Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu Rev Biophys 41, 21–40 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Voorhees RM, Hegde RS, Toward a structural understanding of co-translational protein translocation. Curr Opin Cell Biol 41, 91–99 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Mandon EC, Trueman SF, Gilmore R, Protein translocation across the rough endoplasmic reticulum. Cold Spring Harb Perspect Biol 5, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van den Berg B et al. , X-ray structure of a protein-conducting channel. Nature 427, 36–44 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Rothblatt JA, Deshaies RJ, Sanders SL, Daum G, Schekman R, Multiple genes are required for proper insertion of secretory proteins into the endoplasmic reticulum in yeast. J Cell Biol 109, 2641–2652 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deshaies RJ, Sanders SL, Feldheim DA, Schekman R, Assembly of yeast Sec proteins involved in translocation into the endoplasmic reticulum into a membrane-bound multisubunit complex. Nature 349, 806–808 (1991). [DOI] [PubMed] [Google Scholar]
- 7.Meyer HA et al. , Mammalian Sec61 is associated with Sec62 and Sec63. J Biol Chem 275, 14550–14557 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Tyedmers J et al. , Homologs of the yeast Sec complex subunits Sec62p and Sec63p are abundant proteins in dog pancreas microsomes. Proc Natl Acad Sci U S A 97, 7214–7219 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feldheim D, Rothblatt J, Schekman R, Topology and functional domains of Sec63p, an endoplasmic reticulum membrane protein required for secretory protein translocation. Mol Cell Biol 12, 3288–3296 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brodsky JL, Schekman R, A Sec63p-BiP complex from yeast is required for protein translocation in a reconstituted proteoliposome. J Cell Biol 123, 1355–1363 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Panzner S, Dreier L, Hartmann E, Kostka S, Rapoport TA, Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell 81, 561–570 (1995). [DOI] [PubMed] [Google Scholar]
- 12.Matlack KE, Misselwitz B, Plath K, Rapoport TA, BiP acts as a molecular ratchet during posttranslational transport of prepro-alpha factor across the ER membrane. Cell 97, 553–564 (1999). [DOI] [PubMed] [Google Scholar]
- 13.Green N, Fang H, Walter P, Mutants in three novel complementation groups inhibit membrane protein insertion into and soluble protein translocation across the endoplasmic reticulum membrane of Saccharomyces cerevisiae. J Cell Biol 116, 597–604 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voorhees RM, Fernandez IS, Scheres SH, Hegde RS, Structure of the mammalian ribosome-Sec61 complex to 3.4 A resolution. Cell 157, 1632–1643 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Braunger K et al. , Structural basis for coupling protein transport and N-glycosylation at the mammalian endoplasmic reticulum. Science 360, 215–219 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matlack KE, Plath K, Misselwitz B, Rapoport TA, Protein transport by purified yeast Sec complex and Kar2p without membranes. Science 277, 938–941 (1997). [DOI] [PubMed] [Google Scholar]
- 17.Nguyen TH et al. , Structural basis of Brr2-Prp8 interactions and implications for U5 snRNP biogenesis and the spliceosome active site. Structure 21, 910–919 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tripathi A, Mandon EC, Gilmore R, Rapoport TA, Two alternative binding mechanisms connect the protein translocation Sec71-Sec72 complex with heat shock proteins. J Biol Chem 292, 8007–8018 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng Z, Jiang Y, Mandon EC, Gilmore R, Identification of cytoplasmic residues of Sec61p involved in ribosome binding and cotranslational translocation. J Cell Biol 168, 67–77 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Becker T et al. , Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science 326, 1369–1373 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zimmer J, Nam Y, Rapoport TA, Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature 455, 936–943 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Egea PF, Stroud RM, Lateral opening of a translocon upon entry of protein suggests the mechanism of insertion into membranes. Proc Natl Acad Sci U S A 107, 17182–17187 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voorhees RM, Hegde RS, Structure of the Sec61 channel opened by a signal sequence. Science 351, 88–91 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L et al. , Crystal structure of a substrate-engaged SecY protein-translocation channel. Nature 531, 395–399 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heritage D, Wonderlin WF, Translocon pores in the endoplasmic reticulum are permeable to a neutral, polar molecule. J Biol Chem 276, 22655–22662 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Park E, Rapoport TA, Preserving the membrane barrier for small molecules during bacterial protein translocation. Nature 473, 239–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kityk R, Kopp J, Mayer MP, Molecular Mechanism of J-Domain-Triggered ATP Hydrolysis by Hsp70 Chaperones. Mol Cell 69, 227–237 e224 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Ng DT, Brown JD, Walter P, Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J Cell Biol 134, 269–278 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith MA, Clemons WM Jr., DeMars CJ, Flower AM, Modeling the effects of prl mutations on the Escherichia coli SecY complex. J Bacteriol 187, 6454–6465 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trueman SF, Mandon EC, Gilmore R, A gating motif in the translocation channel sets the hydrophobicity threshold for signal sequence function. J Cell Biol 199, 907–918 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mastronarde DN, Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Zheng SQ et al. , MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA, cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Rohou A, Grigorieff N, CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Afonine PV et al. , Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen VB et al. , MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barad BA et al. , EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat Methods 12, 943–946 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA, Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A 98, 10037–10041 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pettersen EF et al. , UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.