Abstract
Heat shock factors regulate responses to high temperature, salinity, water deprivation, or heavy metals. Their function in combinations of stresses is, however, not known. Arabidopsis HEAT SHOCK FACTOR A4A (HSFA4A) was previously reported to regulate responses to salt and oxidative stresses. Here we show, that the HSFA4A gene is induced by salt, elevated temperature, and a combination of these conditions. Fast translocation of HSFA4A tagged with yellow fluorescent protein from cytosol to nuclei takes place in salt-treated cells. HSFA4A can be phosphorylated not only by mitogen-activated protein (MAP) kinases MPK3 and MPK6 but also by MPK4, and Ser309 is the dominant MAP kinase phosphorylation site. In vivo data suggest that HSFA4A can be the substrate of other kinases as well. Changing Ser309 to Asp or Ala alters intramolecular multimerization. Chromatin immunoprecipitation assays confirmed binding of HSFA4A to promoters of target genes encoding the small heat shock protein HSP17.6A and transcription factors WRKY30 and ZAT12. HSFA4A overexpression enhanced tolerance to individually and simultaneously applied heat and salt stresses through reduction of oxidative damage. Our results suggest that this heat shock factor is a component of a complex stress regulatory pathway, connecting upstream signals mediated by MAP kinases MPK3/6 and MPK4 with transcription regulation of a set of stress-induced target genes.
Keywords: Arabidopsis, combined stress, heat, heat shock factor A4A, MAP kinases, phosphorylation, promoter binding, salinity, transcription regulation
Heat shock factor A4A is phosphorylated by MPK4; it directly binds to promoters of various chaperon and transcription factor genes and controls responses to combined salt and heat stresses.
Introduction
In nature, a number of simultaneously acting environmental effects determine plant growth and development. Extreme conditions require the capability of particular adaptations to face challenges of several, often simultaneous abiotic stresses. Combinations of adverse conditions have a more serious impact on plants than separately imposed stresses and physiological consequences are more dramatic than effects of individual stresses (Rizhsky et al., 2004b; Suzuki et al., 2014). System biology analysis has revealed that combinations of stresses lead to novel transcript and metabolome profiles, which are not just the sum of responses present in plants under individual stresses (Rasmussen et al., 2013; Sewelam et al., 2014; Georgii et al., 2017). A combination of salinity and heat resulted in specific metabolite profiles that included modulation of ion balance, water status, and photosynthetic activity, along with production of protective compounds, which are not typical for salinity or heat alone (Rivero et al., 2014). Experiments with combined salt, mannitol, and heat stresses revealed that heat- and salt-induced genes have usually higher, while osmotically induced genes have lower, expression levels (Sewelam et al., 2014). More than half of the transcriptome changes in combined treatments could not be predicted from single stresses (Rasmussen et al., 2013).
Production of reactive oxygen species (ROS) is a consequence of aerobic metabolism that is enhanced by most adverse conditions, generating oxidative damage as a secondary stress. Maintaining ROS homeostasis in drought and salt stress is essential for plant survival and acclimation, especially in combination with heat or high light (Miller et al., 2010). Besides being damaging, ROS and in particular hydrogen peroxide (H2O2) are important components of stress signaling that are essential to coordinate responses to a number of single or combined biotic and abiotic stress conditions (Volkov et al., 2006; Baxter et al., 2014; Choudhury et al., 2017).
Heat shock factors (HSFs) are essential regulators of responses to heat and a number of other adverse conditions. In the absence of stress, HSFs form complexes with heat shock proteins (mainly HSP90) in cytosol, which are dissolved during stress when HSFs relocalize to nuclei. HSFs activate target genes in trimeric form, recognizing heat shock elements (HSEs) comprising palindromic transcription factor (TF) binding domains (5′-AGAAnnTTCT-3′) (Akerfelt et al., 2010; Anckar and Sistonen, 2011). While in yeast and animals one or a few genes encode HSFs, plants have large HSF gene families, which are composed of 21 genes in Arabidopsis (Nover et al., 2001), 25 genes in rice (Chauhan et al., 2011), and 64 genes in rapeseed (Zhu et al., 2017), representing broad functional diversification. Plant HSFs regulate responses not only to high temperatures but also to diverse environmental stresses (Scharf et al., 2012; Albihlal et al., 2018). Genome-wide transcript profiling experiments have revealed that plant HSFs not only regulate the expression of heat shock protein (HSP) and chaperon genes but control genes that are implicated in transcriptional regulation, protein biosynthesis, metabolism, development, transport, and signal transduction (Busch et al., 2005). A recent study identified almost a thousand target genes of HSFA1b factor in Arabidopsis and established a hierarchical network of 27 HSFA1b-controlled TFs, which regulate the activity of 1780 genes (Albihlal et al., 2018). Earlier we showed that ROS-controlled HSFA4A (AT4G18880) of Arabidopsis promotes the expression of a wide set of defense genes including ZnF, MYB and WRKY-type transcription factors (TFs), and regulates tolerance to salinity (Pérez-Salamó et al., 2014). HSFA4A-type factors were shown to confer cadmium tolerance to rice (Shim et al., 2009) and desiccation tolerance to sunflower (Carranco et al., 2017) and rapeseed (Lang et al., 2017). Rapeseed HSFA4A is upregulated by both drought and heat, confirming that it is implicated in multiple stress responses (Zhu et al., 2017).
HSFs can undergo multiple post-translational modifications, such as phosphorylation, acetylation, or sumoylation, which can modulate their activity and stability (Akerfelt et al., 2010). Phosphorylation of human HSF1 by the mitogen-activated protein (MAP) kinase extracellular signal-regulated kinase 1 (ERK1) and glycogen synthase kinase 3 (GSK3) was shown to inhibit HSF1’s function and repress the heat shock response in non-stress conditions (Chu et al., 1996). Such inhibitory phosphorylation can promote interaction with HSP90 and is required for sumoylation, which represses HSF1-dependent activation of target genes (Hietakangas et al., 2003; Wang et al., 2006). In contrast to ERK1 or GSK3, hyperphosphorylation of the human HSF1 on particular Ser residues by calcium/calmodulin (CaM)-dependent protein kinase II (CaMKII) or polo-like kinase 1 (PLK1) has a stimulatory effect as it enhances nuclear translocation and promotes transcription (Holmberg et al., 2001; Kim et al., 2005). Yeast HSF1 is hyperphosphorylated upon heat shock, which is required for recognition of HSEs of target promoters and is a prerequisite for the activation (Hashikawa and Sakurai, 2004). In plants, HSF phosphorylation is more complex due to the large gene families of heat shock factors and protein kinases. In Arabidopsis the cyclin-dependent CDC2s kinase was shown to phosphorylate HSF1, which prevented binding to target DNA (Reindl et al., 1997). An example of positive regulation is the calcium-dependent heat activated MAP kinase of tomato that phosphorylates and activates HSFA3 (Link et al., 2002). CaM-binding protein kinase 3 (AtCBK3) can interact with and phosphorylate AtHSFA1 of Arabidopsis, promoting binding to HSEs and transcription of HSP genes (Liu et al., 2008). Heat stress can activate MAP kinase 6 (MPK6) in Arabidopsis, which forms a complex with and phosphorylates HSFA2 (Evrard et al., 2013). MPK6 and the closely related MPK3 are key components of cellular defenses regulating resistance against various pathogens, oxidative stress responses and ethylene or ABA signaling (Rasmussen et al., 2012; Su et al., 2017; Sun et al., 2018; Bigeard and Hirt, 2018). MPK3 and MPK6 can phosphorylate and control the activity of various TFs including ZAT10, which controls oxidative stress responses (Nguyen et al., 2012b), MYB44 involved in ABA signaling (Nguyen et al., 2012a), and pathogen-related WRKY33 or ERF6 (Mao et al., 2011; Meng et al., 2013) and ICE1 implicated in cold tolerance (Li et al., 2017). Earlier we reported that the Arabidopsis HSFA4A is a substrate of MAP kinases MPK3 and MPK6 and determined phosphorylation sites by mass spectrometry (Pérez-Salamó et al., 2014). The biological or molecular function of such MAP kinase-mediated phosphorylation was, however, not studied. Here we describe that HSFA4A is phosphorylated by MPK4, another key stress-related MAP kinase in Arabidopsis, and show that HSFA4A is implicated in responses not only to salinity but also to combined salt and heat stresses.
Materials and methods
Plant material and growth conditions
Arabidopsis Col-0 ecotype was used in all experiment. HSFA4A cDNA was overexpressed under the control of estradiol-inducible promoter of pER8GW vector (Papdi et al., 2008). In pHSFA4A::HSFA4A-YFP lines, expression of yellow fluorescent protein (YFP)-tagged HSFA4A cDNA was controlled by the 2 kb HSFA4A promoter.
Arabidopsis plants were grown in sterile conditions on half-strength Murashige and Skoog (½MS) medium in growth chambers with an 8 h light–16 h dark light cycle at 22 °C and 100 µE m−2 s−1 light intensity (control condition). To induce the transcription, 5 µM estradiol was added to the culture medium 24 h before and during stress treatments (Pérez-Salamó et al., 2014).
Stress tolerance was evaluated in vitro. In one test system, seeds were germinated and plantlets were grown on agar-solidified ½MS medium supplemented by 50–150 mM NaCl and 5 µM estradiol. Heat stress was imposed by incubating 10-day-old plants in high temperatures: 37 °C in light and 30 °C in dark for 2–8 d. In the second test system seeds were germinated and plants were grown on nylon mesh (SEFAR 07-20/13) on agar-solidified standard ½MS medium for 10 d and then transferred to the surface of liquid culture medium (10 ml medium in a 13 cm diameter Petri dish). Salt stress was imposed by supplementing liquid medium with 100 or 150 mM NaCl. Heat stress was 37 °C in light and 30 °C in dark for 2–4 d. Stress combinations were implemented by simultaneous application of salt and heat treatments. Plants were subsequently removed and transferred to standard ½MS culture medium for recovery. Plant survival and growth was evaluated 10 d later. Transgenic Arabidopsis lines were generated by the floral dip method as described (Clough and Bent, 1998). Agrobacterium-mediated transformation of cell suspension culture was made as described (Pérez-Salamó et al., 2014).
Stress treatments for transcript and western analysis, phosphorylation studies, and chromatin immunoprecipitation (ChIP) assays were performed on 10-day-old in vitro-grown seedlings using the sterile hydroponic system described above. Unless otherwise stated, salt stress was imposed with 150 mM NaCl and heat stress was generated with 37 °C in light and 30 °C in dark (8/16 h of light/dark cycle). Stress treatments were initiated 2 h after the start of light period (time 0). Transcript profiling and western analysis were performed with samples collected after 2, 6 and 24 h of stress, while in vivo phosphorylation and chromatin binding (ChIP assay) were tested on plants treated for 6 h.
Gene cloning
To generate pHSFA4A promoter and HSFA4A–YFP gene fusion, 2kb-long promoter fragment of the HSFA4A gene (AT4G18880) was cloned and fused to the YFP-tagged HSFA4A of the pPCV-HSFA4A-YFP construct (Pérez-Salamó et al., 2014). After sequencing, the pHSFA4A::HSFA4A-YFP fragment was moved in the pMDC99 binary Gateway vector by a Gateway LR clonase reaction (Curtis and Grossniklaus, 2003).
To generate a phosphorylation-mimicking version of HSFA4A, the Ser309 residue was replaced by Asp309 as described (Pérez-Salamó et al., 2014). The sequenced S309D-HSFA4A cDNA was cloned into the pENTR2b Gateway vector using BamHI and XhoI restriction sites, and moved into the binary destination vector pER8GW (Papdi et al., 2008) with Gateway LR clonase reaction, producing the estradiol-inducible pER8-HSFA4A-S309D.
To generate mutant gene constructs for a bimolecular fluorescence complementation (BiFC) assay, the HSFA4A-S309A (Pérez-Salamó et al., 2014) and the newly generated HSFA4A-S309D fragments were PCR amplified from pENTR2b-HSFA4A-S309A and pENTR2b-HSFA4A-S309D plasmids using the HSFA4A-HindIII and HSFA4A-SmaI-NoStop primers. The PCR fragments were subsequently cloned into the HindIII and SmaI sites of pSAT1A-nEYFP-N1a and pSAT1A-cEYFP-N1 BiFC vectors (http://www.bio.purdue.edu/people/faculty/gelvin/nsf/index.htm). Oligonucleotides used in this study are listed in Table 1.
Table 1.
List of oligonucleotides used in this study
| Code | 5′–3′ sequence | Use | Reference |
|---|---|---|---|
| HsfA4A-qF | CTTTGAACCTATCGCCGTGT | HSFA4A (AT4G18880) qRT-PCR | Pérez-Salamó et al., 2014 |
| HsfA4A-qR | TGTGTGTGAAGAAGGGCTTG | HSFA4A (AT4G18880) qRT-PCR | Pérez-Salamó et al., 2014 |
| ZAT12-qF | GACGCTTTGTCGTCTGGATT | Zat12 (AT5G59820) qRT-PCR | Pérez-Salamó et al., 2014 |
| ZAT12-qR | GTGTCCTCCCAAAGCTTGTC | Zat12 (AT5G59820) qRT-PCR | Pérez-Salamó et al., 2014 |
| WRKY30-qF | AGAGCGATGATTCCGATCAAG | WRKY30 (AT5G24110) qRT-PCR | Besseau et al., 2012 |
| WRKY30-qR | CATCGTCCAGCGTTCTATCAA | WRKY30 (AT5G24110) qRT-PCR | Besseau et al., 2012 |
| Hsp17.6A-qF | CCAAAGAAAAAGCCAAGAAGC | Hsp17.6A (AT5G12030) qRT-PCR | Pérez-Salamó et al., 2014 |
| Hsp17.6A-qR | TGGAAACCTTCCAAACTCCA | Hsp17.6A (AT5G12030) qRT-PCR | Pérez-Salamó et al., 2014 |
| ER8A | GCTTGGGCTGCAGGTCGAGGCTAA | Amplification of inserts in pER8 | Papdi et al., 2008 |
| ER8B | CTGGTGTGTGGGCAATGAAACTGATGC | Amplification of inserts in pER8 | Papdi et al., 2008 |
| GAPDH2_qF | AATGGAAAATTGACCGGAATGT | GAPDH2 (AT1G13440) qRT-PCR | Papdi et al., 2008 |
| GAPDH2_qR | CGGTGAGATCAACAACTGAGACA | GAPDH2 (AT1G13440) qRT-PCR | Papdi et al., 2008 |
| ZAT12-chipF | GTTGTCATGCCTTTGCTGG | Zat12 (AT5G59820) ChIP qPCR | This study |
| ZAT12-chipR | GGTCAGGTGTATTCTAGAAAGTGG | Zat12 (AT5G59820) ChIP qPCR | This study |
| WRKY30-chipF | AAAGGATCGAGAAGCAGAGAAC | WRKY30 (AT5G24110) ChIP qPCR | This study |
| WRKY30-chipR | CGAAGAAAGCTGATGTGGTTTG | WRKY30 (AT5G24110) ChIP qPCR | This study |
| Hsp17.6A-chipF | ACTTCTCCAACGATCAAGACG | Hsp17.6A (AT5G12030) ChIP qPCR | This study |
| Hsp17.6A-chipR | AGCAATGTAGAGAGAGCGATTG | Hsp17.6A (AT5G12030) ChIP qPCR | This study |
| TUA3-chipF | TTTCCTAAGCGGTTTCAGATTAAAG | TUA3 (AT5G19770) ChIP qPCR | This study |
| TUA3-chipR | GAATTGCTACTTAAGCTATCTTCCTAAC | TUA3 (AT5G19770) ChIP qPCR | This study |
| T7 promoter | TAATACGACTCACTATAGGG | HSFA4A (AT4G18880) cloning | This study |
| T7 terminator | TATGCTAGTTATTGCTCAG | HSFA4A (AT4G18880) cloning | This study |
| HSFA4A-S309D | CAAATCTCCTCCTGATCCAAGGATCATC | HSFA4A (AT4G18880) mutagenesis | This study |
| M13 F | GTAAAACGACGGCCAGTG | pBluescript II SK+, sequencing | |
| M13 R | GGAAACAGCTATGACCATG | pBluescript II SK+, sequencing | |
| HSFA4A-HindIII | TGAGCAAGCTTATGGATGAGAATAATCATGGAGTTTC | BiFC vector cloning | Pérez-Salamó et al., 2014 |
| HSFA4A-SmaI-NoStop | TGATTCCCGGGACTTCTCTCTGAAGAAGTCAGAT | BiFC vector cloning | Pérez-Salamó et al., 2014 |
Physiological parameters
Lipid peroxidation was determined in 10-day-old plants exposed to single or combined stresses for 48 h, measuring the malondialdehyde (MDA) content, using a thiobarbituric acid (TBA) test (Hodges et al., 1999). The amount of MDA–TBA complex was calculated according to the following equation: X (%)=100×(OD532−OD600) (Zsigmond et al., 2012).
Microscopic techniques
To study intracellular localization of HSFA4A, 5-day-old seedlings expressing the pHSFA4A::HSFA4A-YFP construct were imaged using an Olympus FV1000 confocal laser scanning microscope. To follow changes in intracellular localization, the YFP signal was recorded in roots by taking pictures of the same cells in every 5 min, up to 30 min. Plantlets were placed on slides and immersed either in standard ½MS culture medium or medium supplemented by 100 mM NaCl. BiFC assays were performed as described (Pérez-Salamó et al., 2014). Fluorescence intensities were analysed in nuclear regions and cytoplasm using ImageJ2 (https://imagej.net/ImageJ2).
Chromatin immunoprecipitation assay
A ChIP assay was used to verify the interaction of HSFA4A protein with three selected gene promoters: ZAT12 (AT5G59820), HSP17.6A (AT5G12030), and WRKY30 (AT5G24110). The chromatin was isolated from stress-treated and control pHSFA4A::HSFA4A-YFP expressing plants, following the Abcam ChIP protocol (Haring et al., 2007). The immunoprecipitation was carried out with anti-green fluorescent protein (GFP) antibody (Roche) and Dynabeads Protein G (Invitrogen/Thermo Fisher Scientific). Control ChIP experiment was carried out with Dynabeads Protein G without anti-GFP antibody. Reverse cross-linking and DNA purification were carried out by the Abcam ChIP protocol. Fragments of immunoprecipitated DNA were amplified by quantitative PCR using specific primers for the HSE-containing promoter regions of ZAT12, HSP17.6A, and WRKY30 genes. For reference, the tubulin α-3 (TUA3, AT5G19770) promoter fragment was used, which has no HSE. qPCR data were normalized with values obtained on TUA3. TF binding was calculated as fold enrichment over the control (Aleksza et al., 2017).
SDS-PAGE, non-denaturing PAGE and western blot
Total protein was isolated from 100 mg seedlings with 50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1% Triton X-100 and 1× Protease Inhibitor Cocktail (Sigma-Aldrich/Merck). For the multimerization studies, DTT was added to the 100 µg total protein extract in different final concentrations (0, 25, 50, 75 mM) and incubated for 10 min in room temperature. The samples were size separated on 7% non-denaturating SDS-PAGE and transferred to Immobilon polyvinylidene difluoride (PVDF) membrane (Merck Millipore) for western detection. For denaturing electrophoresis, 25 µg total protein was size separated on 8% SDS-PAGE, transferred onto Immobilon PVDF membrane. For western hybridization membranes were incubated in 1×TBST blocking buffer (50 mM Tris–HCl pH 8.0, 150 mM NaCl, 0.05% Tween-20, 5% dry skimmed milk) for 1 h and with anti-GFP antibody (Roche, 1:2000 dilution) in blocking buffer for 1.5 h. After washing with 1× TBST three times for 10 min, the membranes were incubated for 1.5 h with an anti-mouse-POD secondary antibody (Pierce, dilution 1:5000), washed with 1× TBST as before. The membrane was subsequently overlaid with Immobilon Western Chemiluminescent HRP Substrate (Merck Millipore) and chemiluminescence was detected with a Fusion FX5 camera system (Vilber Lourmat).
RNA isolation and RT-PCR (qPCR)
Total RNA was isolated from 100 mg Arabidopsis seedlings, using a Nucleospin Plant RNA Kit (Macherey-Nagel). Isolated RNA was DNase treated with TURBO DNA-free™ Kit (Invitrogen/Thermo Fisher Scientific), and first-strand cDNA synthesis of 1 µg of total RNA was carried out with a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems/Thermo Fisher Scientific), using random hexamers. Real-time PCR was carried out with the ABI 7900 Fast Real Time System (Applied Biosystems) with the following protocol: 40 cycles at 95 °C for 15 s, 60 °C for 60 s, using Maxima SYBR Green qPCR Master Mix (Thermo Fisher Scientific). GAPDH2 (AT1G13440) was used as reference gene. The normalized relative transcript levels were calculated with the 2−ΔΔCt method (Czechowski et al., 2005).
Protein phosphorylation
All gene constructs used in phosphorylation experiments (His6-MPK3, maltose-binding protein (MBP)–HSFA4A, MBP, glutathione S-transferase (GST)–MPK4) were expressed in BL21DE39 Rosetta cells (Novagen). The GST–MPK4 construct was obtained from Robert Dóczi (Martonvásár, Hungary). Proteins were purified by affinity chromatography, following the manufacturer’s instructions (Dóczi et al., 2007). Proteins were dialysed overnight in 50 mM NaCl, 10% glycerin, 2 mM β-mercaptoethanol, 10 mM Tris–HCl pH 7.5. Protein integrity was checked by SDS-PAGE. In vitro phosphorylation was performed with purified proteins as described (Pérez-Salamó et al., 2014); 1–2 µg of proteins were used in each phosphorylation reaction, which was performed in 20 mM Tris–HCl (pH 8.0), 5 mM MgCl2, 1 mM DTT, containing 185 kBq [γ-32P]ATP) at room temperature (23–25 °C) for 1 h. Proteins were separated on 12% SDS-PAGE and the polyacrylamide gel was stained with Coomassie Blue (Thermo Fisher Scientific). Autoradiography was performed with AGFA Medical X-ray Blue Film (AGFA, Germany).
Identification of MPK4 phosphorylation sites in HSFA4A
The phosphorylation reaction for mass spectrometry was identical to that described above, except that non-radiolabeled 1 µM ATP (Thermo Fisher Scientific) was used. After the reaction, samples were separated by SDS-PAGE, and protein bands were visualized by Coomassie Blue staining. The protein band corresponding to MBP–HSFA4A was excised and analysed by mass spectrometry in the Laboratory of Proteomics Research of the Biological Research Centre (www.szbk.u-szeged.hu/services_proteomics_research.php). Proteins were in-gel digested by trypsin at 37 °C for 6 h as described (https://msf.ucsf.edu/protocols.html). The digest was split and half of the sample subjected to Fe(III)-immobilized metal affinity chromatography phosphopeptide enrichment (Ficarro et al., 2009). The digest with and without enrichment was analysed on an Orbitrap Elite (Thermo Scientific) mass spectrometer, online coupled to a Waters nanoAcquity HPLC. Mass spectrometry (MS) data acquisition was performed in a data-dependent fashion; in each survey scan the five most abundant multiply charged precursor ions were selected for high-energy collision dissociation fragmentation at 30% normalized collision energy. Both, MS and tandem mass spectrometry (MS/MS) spectra were acquired in the orbitrap at a resolution of 60 000 and 15 000, respectively. Dynamic exclusion was 30 s.
Data were processed by Protein Discoverer (v1.3), and the generated peak lists were subjected to a database search using Protein Prospector (v5.22.0) against the Uniprot 1.11.2017 database, considering also the protein sequence of the expressed MBP–HSFA4A and the MPK4 kinase. Monoisotopic masses were used with a precursor mass tolerance of ±5 ppm and a fragment mass tolerance of ±20 ppm. Carbamidomethylation of Cys residues was set as fixed modification; acetylation of protein N-termini, cyclization of peptide N-terminal Gln residues, Met oxidation and phosphorylation of Ser, Thr, and Tyr residues were selected as variable modifications and a maximum two variable modifications were permitted per peptide. Search results were accepted at a false discovery rate of 5% at the protein level and 1% at the peptide level. Modification site assignments with a minimum site localization in peptide (SLIP) score of six were accepted (Baker et al., 2011). MS/MS spectra were manually inspected.
Relative quantification of the phosphorylation at a given site was performed at the MS1 level from the liquid chromatography–MS analysis of the non-enriched tryptic digest. The peak areas of the non-phosphorylated and phosphorylated peptides were calculated by Pinnacle (v1.0.83.0) using the isotopic cluster of the corresponding precursor ions:
where PP is phosphopeptide.
Identification of phosphorylation sites in vivo
To test HSFA4A phosphorylation in vivo, protein extracts were isolated from 10-day-old salt-treated (150 mM NaCl, 6 h) and control pHSFA4A::HSFA4A-YFP expressing plants. Seedlings (500 mg) were harvested, frozen in liquid nitrogen and ground with a TissueLyser at 30 Hz. Total proteins were extracted using the manufacturer’s Lysis buffer supplemented with 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 1× Sigma protease inhibitor cocktail, 3 mM p-nitrophenyl phosphate, 1 µM MG132. Protein extracts (3 mg/immunoprecipitate) were immunopurified using anti-GFP antibody-coupled very small (50 nm) magnetic beads (MACS® Technology, Miltenyi), digested in-column with trypsin, and analysed by MS as described (Horvath et al., 2017).
Data analysis, bioinformatics
Protein sequences were obtained from Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html). Multiple sequence alignment was performed with Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). Transcription factor binding sites were identified with AthaMap (http://www.athamap.de). DAP-seq data were compiled manually from online data of the Neomorph database: http://neomorph.salk.edu/PlantCistromeDB.
Results
Regulation of HSFA4A
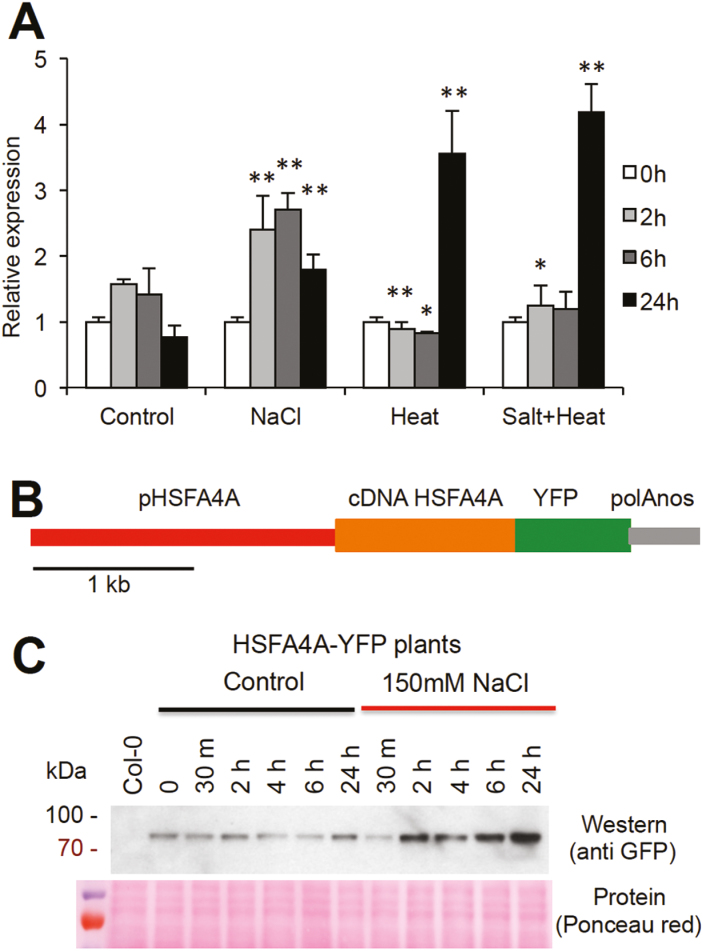
The HSFA4A gene was shown to be induced by a number of abiotic and biotic stresses (Pérez-Salamó et al., 2014; see eFP Browser: http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi?primaryGene=AT4G18880&modeInput=Absolute). Expression of this factor with a stress combination was, however, not studied. When wild-type Arabidopsis plants were treated by high salinity, high temperature, and a combination of these conditions, HSFA4A expression changed dramatically, but induction followed different kinetics. Transcription in control plants was slightly and temporally enhanced, probably as consequence of lid opening and touching these plants during transfer to fresh medium. Salt (150 mM NaCl) induced transcription in 2 h and remained 2- to 3-fold elevated for up to 24 h. Heat (37 °C) and the combination of heat and salt stress had no effect on transcript levels in the first hours but enhanced HSFA4A transcription after 24 h (Fig. 1A). These results suggest that heat and salinity regulate HSFA4A through different signaling pathways. To study the HSFA4A protein in vivo, a transgenic Arabidopsis line was generated that expresses the YFP-tagged HSFA4A under the control of its own 2 kb-long promoter (pHSFA4A::HSFA4A-YFP, Fig. 1B). Western hybridization confirmed the presence of the HSFA4A–YFP chimeric protein in transgenic plants, which was more abundant in salt, heat, and combined salt and heat stressed plants correlating with stress-dependent induction of the endogenous HSFA4A gene (Fig. 1; Supplementary Fig. S1 at JXB online).
Fig. 1.
Regulation of HSFA4A. (A) Transcriptional regulation of HSFA4A gene in wild-type Arabidopsis plants treated with salt (150 mM NaCl), heat stress (37 °C in light and 30 °C in dark), and their combination for 2, 6, and 24 h. Relative expression is shown where 1 corresponds to transcript level at 0 h. Error bars indicate standard error; asterisks indicate significant differences from control: *P<0.05 and **P<0.01 (Student’s t-test). (B) Schematic map of the pHSFA4A::HSFA4A-YFP gene construct. (C) Detection of HSFA4A–YFP fusion protein in 10-day-old control and salt-stressed plants (150 mM NaCl, 0–24 h) transformed with the pHSFA4A::HSFA4A-YFP gene construct. Salt treatment led to enhanced HSFA4A–YFP specific western signal. (This figure is available in color at JXB online.)
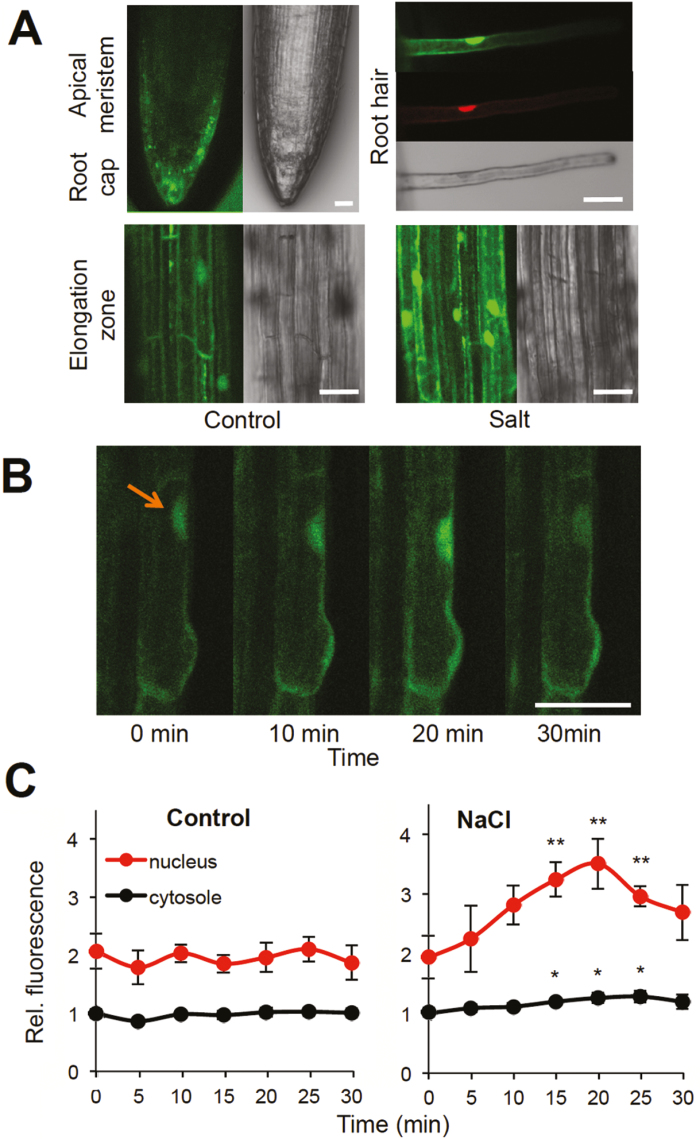
Confocal microscopic observations revealed weak HSFA4A–YFP-derived fluorescence in green parts of the plants (not shown) while it was well detectable in roots: the signal was strong in cells of root caps, elongation and differentiation zones, and root hairs. In root cells and root hairs HSFA4A–YFP was present in both cytoplasm and nuclei. Parallel with enhanced western signal, several hours of salt treatment led to stronger fluorescence in root cells, which became particularly strong in nuclei (Fig. 2A). Several hours of stress enhanced content of HSFA4–YFP protein, which could be detected either by western assay or visualization with confocal microscopy (Figs 1B, 2A; Supplementary Fig. S1).
Fig. 2.
Intracellular localization and transfer of HSFA4A. (A) Confocal microscopic detection of the HSFA4A–YFP fusion protein in different segments of roots. Root hair is stained with propidium iodide to demonstrate nuclear localization of the YFP signal. Segments of elongation zone are shown with and without salt treatment (100 mM NaCl, 2 h). (B) HSFA4A is transported into nuclei during salt stress. Roots were treated with 100 mM NaCl, and HSFA4A–YFP-derived fluorescence was monitored in individual cells at regular intervals. Arrow indicate position of a nucleus. (C) Quantitative evaluation of YFP fluorescence in cytosol and nuclei. Relative fluorescence is shown, where 1 corresponds to intensity measured in cytosol at time 0. YFP-derived fluorescence was rapidly enhanced in nuclei of salt-treated cells, while it did not change in control cells. Scale bar on images indicates 20 µm. Error bars indicate standard error; asterisks indicate significant differences from time 0: *P<0.05 and **P<0.01 (Student’s t-test).
Heat shock factors are shuttling proteins with predominant cytoplasmic localization in non-stressed conditions and nuclear accumulation upon heat and other stress (Scharf et al., 1998; Heerklotz et al., 2001; Akerfelt et al., 2010). To study intracellular shifts of HSFA4A during stress, pHSFA4A::HSFA4A-YFP expressing roots were treated with salt (100 mM NaCl) and change of YFP-derived fluorescence was monitored in the same root cells. Salt stress led to a fast and temporal accumulation of HSFA4A–YFP in nuclei while the fluorescence pattern did not change significantly in non-treated control cells (Fig. 2B, C). In nuclei, YFP-derived fluorescence reached its maximum 20 min after the initiation of salt treatment, which was followed by a gradual decrease (Fig. 2C). In cytosol only minimal change in YFP-derived fluorescence could be observed. These results suggest rapid transfer of HSFA4A to nuclei that starts within minutes upon onset of salt stress and most probably takes place before gene activation and de novo protein biosynthesis.
Binding of HSFA4A to promoter elements of target genes
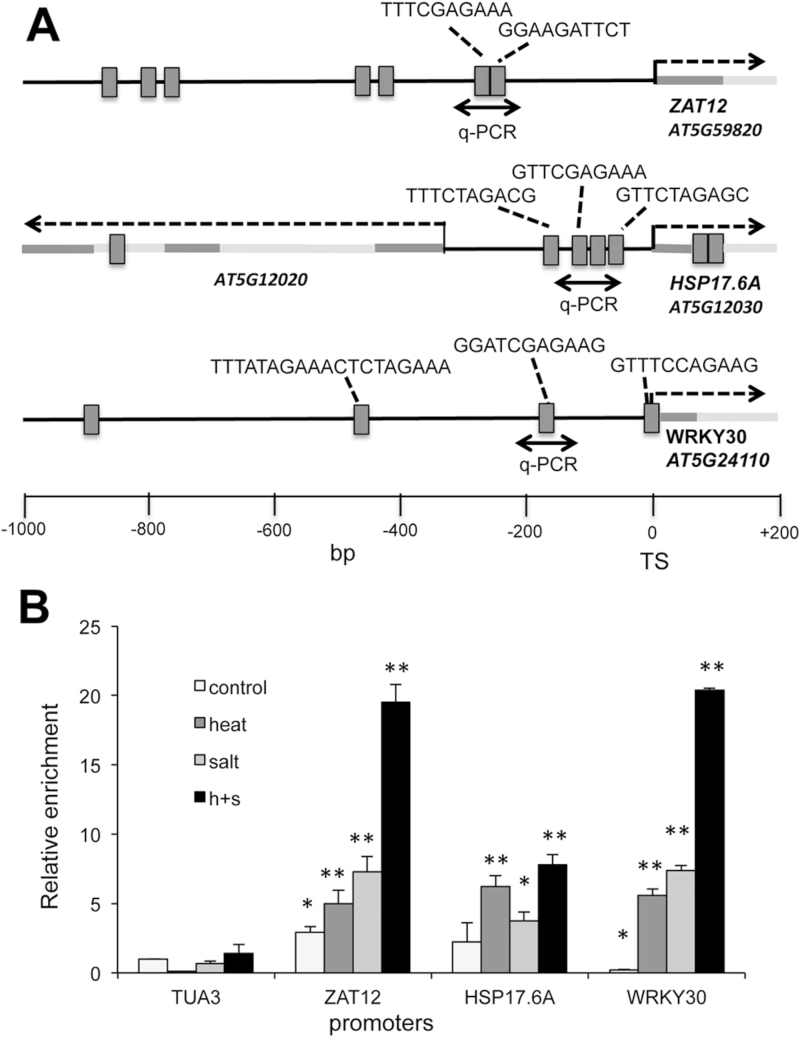
Whole-genome transcript profiling has identified genes that were upregulated by HSFA4A overexpression (Pérez-Salamó et al., 2014). These genes can be direct targets of this heat shock factor, binding to their cis-acting HSEs, or can indirectly be induced by TFs, which are themselves regulated by HSFA4A. Three HSFA4A-induced genes were selected to test promoter binding: HSP17.6A, ZAT12, and WRKY30, encoding a small heat shock protein, a zinc finger and a WRKY-type TF, respectively (Pérez-Salamó et al., 2014). Promoter regions of these genes contain several HSE motifs, suggesting that they can be direct targets of HSFs (Fig. 3A). Promoter binding was tested in vivo by ChIP assays, using transgenic plants expressing the pHSFA4A::HSFA4A-YFP gene construct, treated with salt (150 mM NaCl), high temperature (37 °C), or both stresses for 6 h before chromatin extraction. The ChIP assay revealed specific enrichment of HSE-containing promoter regions of all three tested genes (Fig. 3B). Heat treatment significantly enhanced enrichment on all three promoters, while salt promoted HSFA4A binding to ZAT12 and WRKY30 promoters but only slightly influenced binding to HSP17.6A. The combination of both stresses was additive on promoter binding on ZAT12 and WRKY30 genes, while on the HSP17.6A promoter it was similar to heat treatment (Fig. 3B). Binding to the TUA3 promoter, which lacked HSE motifs, was not altered and was used as a non-specific control. These results demonstrate that the HSFA4A factor can directly bind to the promoters of the three target genes, which is enhanced depending on the type of stress.
Fig. 3.
Binding of HSFA4A on target gene promoters. (A) Schematic map of ZAT12, HSP17.6A, and WRKY30 promoters according to AthaMap. Promoter regions between −1000 and +200 bp are shown. Black line indicates promoter, dark grey corresponds to 5′-UTR and exon while light grey is intron sequence. HSE motifs are indicated by grey boxes and sequences connected to the amplified regions are shown above the target region. Dashed arrows indicates transcription initiation. Amplified target sequences by qPCR are indicated by black double arrows. (B) ChIP assay with YFP-tagged HSFA4A using transgenic plant expressing the pHSFA4A::HSFA4A-YFP gene construct (see Fig. 1B, C). Plants were treated by salt (150 mM NaCl, 6 h), heat stress (37 °C, 6 h), and their combination before ChIP assay. ChIP results are shown as relative enrichment by qPCR, where reference (value 1) is the qPCR value of the TUA3 promoter, which lacks any HSE motif, at control conditions. Note enrichments on different promoter regions, which can be enhanced by salt or heat treatments. Error bars indicate standard error; asterisks indicate significant differences from ChIP values of TUA3: *P<0.05 and **P<0.01 (Student’s t-test).
HSFA4A is phosphorylated by MAP kinase 4
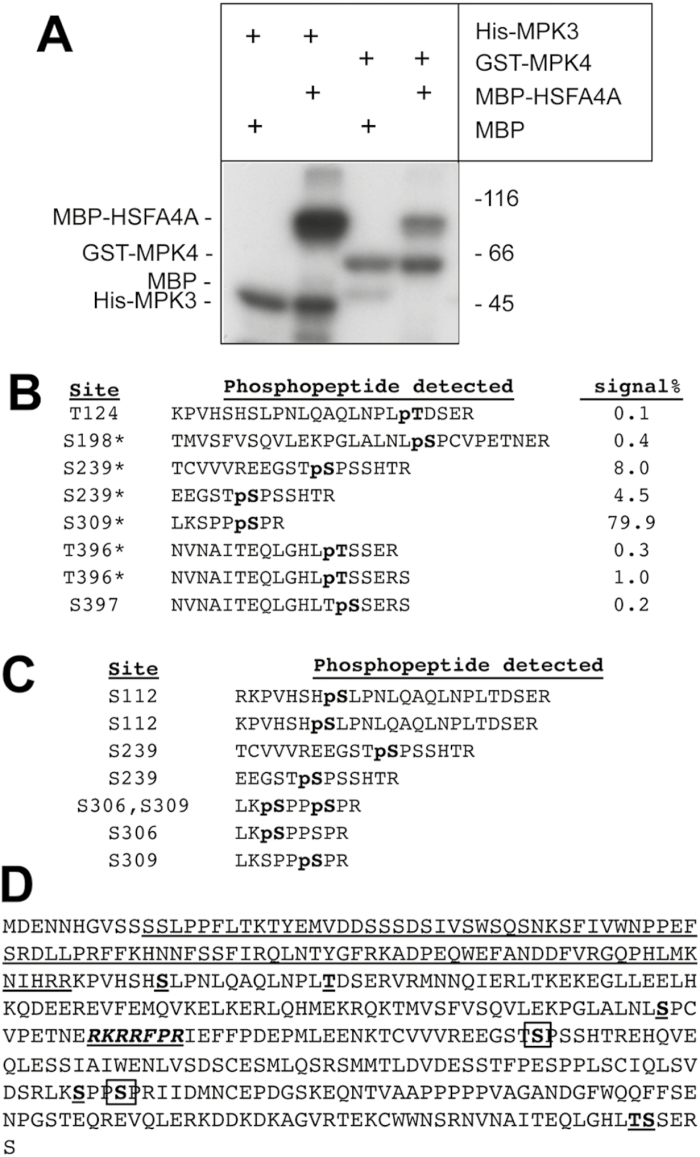
Earlier we showed that MAP kinases MPK3 and MPK6 can interact with and phosphorylate HSFA4A. Five phosphorylated amino acid residues were identified by MS, and Ser309 was found to be the dominant MPK3 and MPK6 phosphorylation site (Pérez-Salamó et al., 2014). An alternative stress-related MAP kinase signaling pathway is controlled by MPK4, which is implicated in pathogen responses and ROS homeostasis (Pitzschke et al., 2009; Dóczi and Bögre, 2018). To reveal possible involvement of HSFA4A in MPK4-controlled stress signaling, phosphorylation of HSFA4A by MPK4 was tested in vitro. We found that HSFA4A can be phosphorylated not only by MPK3 but also by MPK4 (Fig. 4A). Subsequent analysis by mass spectrometry identified six amino acid residues of HSFA4A which were phosphorylated by MPK4: Thr124, Ser198, Ser239, Ser309, Thr396, and Ser397 (Fig. 4B; Supplementary Dataset S1). Four of the identified sites coincided with amino acid residues phosphorylated also by MPK3 (Ser198, Ser239, Ser309, Thr396) (Pérez-Salamó et al., 2014). The two phospho-isoforms modified at Thr396 and Ser397 could be distinguished based on differences in retention time and fragmentation pattern. Calculation of phosphopeptide signal frequencies showed that 80% of the Ser309 residues can be phosphorylated by MPK4, in contrast to the other Ser or Thr residues, which were phosphorylated with much lower frequencies (0.1–8%, Fig. 4B), suggesting that Ser309 is the primary phosphorylation site for MPK4. Ser309 is the dominant phosphorylation site also for MPK3 and MPK6 (Pérez-Salamó et al., 2014), suggesting that it is the primary target of MAP kinases.
Fig. 4.
Phosphorylation of HSFA4A. (A) In vitro phosphorylation of HSFA4A by MAP kinases MPK3 and MPK4. MBP-tagged HSFA4A was phosphorylated in vitro by His-MPK3 or GST–MPK4. (B) List of phosphopeptides identified by MS. Phosphorylated amino acids are indicated with bold letters (pT, pS). MBP-tagged HSFA4A was phosphorylated in vitro by MPK4, in-gel digested by trypsin, and analysed by mass spectrometry. The modified sites within the detected tryptic peptides were determined from MS/MS spectra acquired following ferric nitrilotriacetate chelate (Fe(III)-NTA) phosphopeptide enrichment (Supplemental Dataset S1). Phosphopeptide signal% was calculated from MS signal areas of the unmodified and phosphorylated peptides detected in the tryptic digest without phosphopeptide enrichment. Note that these values are not absolute phosphorylation ratios. (C) Detection of phosphopeptides in vivo. HSFA4A–YFP fusion protein was immunoprecipitated from transgenic plants, and phosphopeptides were detected by mass spectrometry. Bold letters indicate phosphorylated amino acids. (D) Amino acid sequence of HSFA4A. Amino acids phosphorylated by MPK3 (Pérez-Salamó et al., 2014) and MPK4 (this study) or detected in immunoprecipitated samples are shown with bold and underlined letters. Boxed letters indicate amino acids that were detected in both in vitro and in vivo phosphorylation assays. Underlined letters in italics indicate predicted MAPK docking motif (RKRRFPR). Conserved DNA binding domain is underlined. GST, glutathione S-transferase; His, polyhistidine tag; MBP, maltose binding protein.
To test phosphorylation of HSFA4A in vivo, YFP-tagged HSFA4A was purified from control and salt-treated transgenic plants expressing the pHSFA4A::HSFA4A-YFP gene construct (Fig. 1B, C), and phosphopeptides were identified by mass spectrometry (Fig. 4C). Phosphorylation of Ser239 and Ser309 was observed in both in vivo phosphorylation detection assays and in vitro MPK3 and MPK4 phosphorylation experiments (Fig. 4D; Supplementary Table S1) confirming that these amino acids are indeed in vivo targets of MAP kinases. Phosphorylation of Ser112 and Ser306 was also revealed in vivo; they were not phosphorylated by MAP kinases (Fig. 4C). Computer prediction suggested that these amino acid residues can be phosphorylated by protein kinases such as protein kinase A (PKA), cyclin dependent kinase (CDK), casein kinase 1 (CK1), casein kinase 2 (CK2), and GSK3 (Supplementary Table S1). These results suggest that HSFA4A is under complex post-translational control as it can be phosphorylated not only by MAP kinases but also by other classes of protein kinases.
Multiple alignment of amino acid sequences of 33 HSFA4-type TFs from 27 plant species revealed that the identified phosphorylation sites can be assigned to three groups (Supplementary Dataset S2). Ser112 is present in all plant HSFA4 proteins, while Thr124, Ser306, and Thr396 are conserved in most of the proteins. Ser198, Ser239, and Ser309 are present only in HSFA4A-type proteins of closely related plant species that belong to the Brassicaceae family (Arabidopsis, Arabis halleri, Capsella rubella, Brassica rapa, and Eutrema salsugineum). On the other hand, it is intriguing that Thr396 was missing in the Arabidopsis HSFA4C and five proteins most related to this HSF in Brassicaceae species (Supplementary Dataset S2; Supplementary Fig. S2). A conserved MAP kinase docking domain is present in all HSFA4-type kinases (Fig. 4D; Supplementary Dataset S2). These results suggest that the most conserved phosphorylation sites are present in all plant HSFA4-type TFs, but MPK3-, MPK4-, and MPK6-mediated phosphorylation is characteristic only of HSFA4 factors of the Brassicaceae species.
Phosphorylation affects intramolecular interactions of HSFA4A
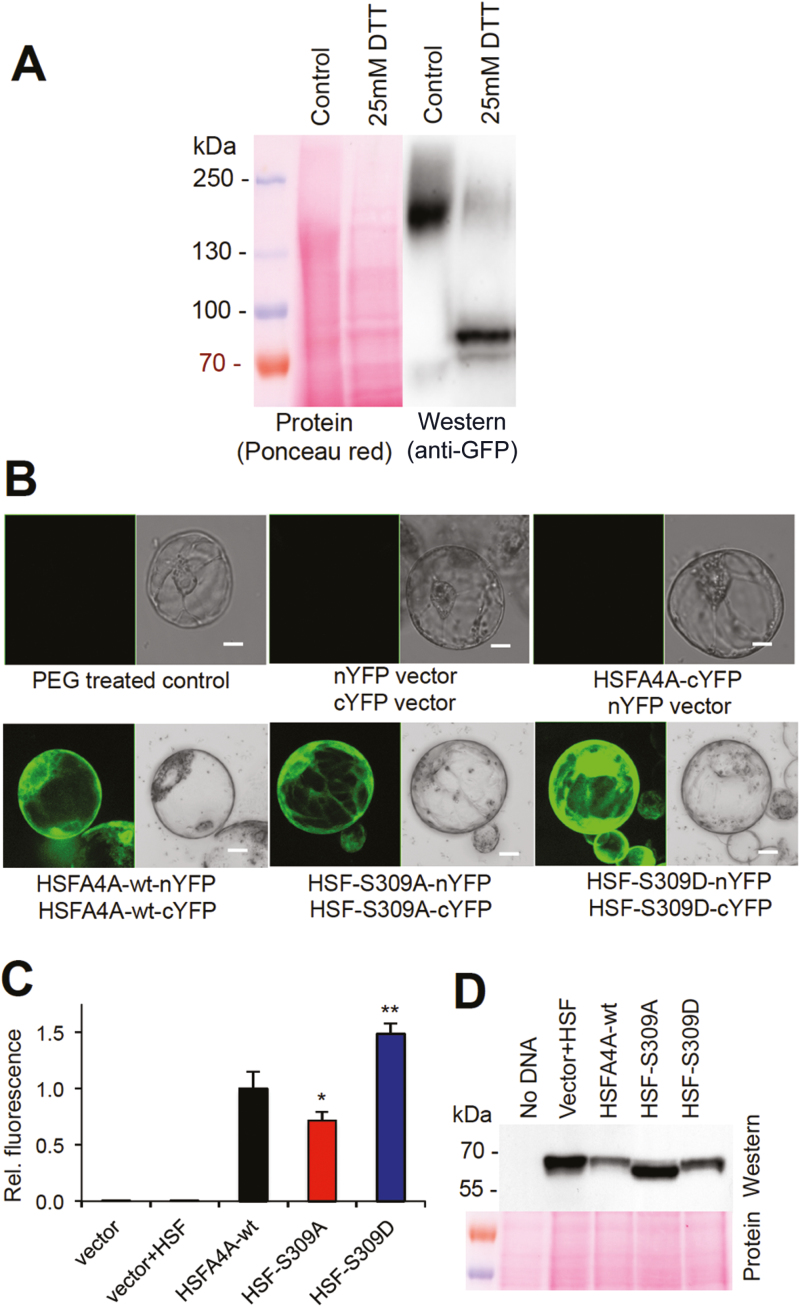
Recognition of heat shock elements and transcriptional activation of target genes requires trimerization of heat shock factors (Miller and Mittler, 2006; Akerfelt et al., 2010). Intramolecular dimerization of HSFA4A has previously been demonstrated, and conserved Cys residues were shown to be essential to stabilize such interactions (Pérez-Salamó et al., 2014). To study multimer formation of HSFA4A in vivo, protein extracts from HSFA4A–YFP-expressing plants were separated on non-denaturing gels and the YFP-tagged TF was detected by western hybridization using anti-GFP antibody. In crude extracts a high molecular mass complex was detected with molecular mass of approximately 200 kDa. DTT treatment reduced the abundance of the high molecular mass complex while a 70–80 kDa band appeared, suggesting that in reducing conditions the HSFA4A-containing complex is disassembled and the monomer HSFA4A is released (Fig. 5A). Similar results were obtained with protein extracts isolated from HSFA4A–YFP-expressing cell suspension cultures (Supplementary Fig. S3A). In plant cells most HSFA4A protein seem to exist in high molecular mass complexes, which can be monomerized in a reducing environment. In vitro assay showed that oxidative conditions favor HSFA4A multimerization while in a reducing environment monomers are predominantly formed (Supplementary Fig. S3B).
Fig. 5.
Multimerization of HSFA4A. (A) Detection of HSFA4A–YFP multimers in Arabidopsis plants transformed with the pHSFA4A::HSFA4A-YFP gene construct. Protein extracts were treated with or without DTT and separated on non-denaturing polyacrylamide gels. HSFA4A–YFP was detected by western hybridization with anti-GFP antibody. Separated and membrane-blotted proteins were stained with Ponceau Red. (B) BiFC assay of wild-type HSFA4A (HSFA4A-wt), and mutants in which Ser309 was changed to Ala (HSF-S309A) or Asp (HSF-S309D). nYFP and cYFP indicates N- and C-terminal half of YFP protein. Controls include polyethylene glycol-treated protoplasts without plasmids, protoplasts transformed with plasmids having nYFP and cYFP fragments, or protoplasts expressing HSFA4A-cYFP in combination with the empty nYFP plasmid (upper row). Typical BiFC images are shown. (C) Quantitative evaluation of fluorescence signals in YFP-expressing transformed protoplasts in BiFC experiments. Relative fluorescence intensities are shown, where 1 equals signals of protoplasts expressing the wild-type HSFA4A constructs (HSFA4A-wt) while HSF-S309A and HSF-S309D indicate S309A and S309D mutants, respectively. (D) Western detection of HSFA4A–YFP fusions in BiFC experiment. Anti-GFP antibody was used to detect the proteins in transformed protoplasts. Note that comparable amount of HSFA4A–YFP was produced in each BiFC samples. In fact, slightly lower amount of wild-type and higher amount of S309A version of HSFA4A was produced. Error bars indicate standard error; asterisks indicate significant differences from HSFA4A-wt: *P<0.05 and **P<0.01 (Student’s t-test). Scale bar on images indicates 10 μm.
To test the effect of MAP kinase-mediated phosphorylation on HSFA4A multimerization in vivo, protein–protein interactions of wild-type and point mutants of HSFA4A proteins were compared in BiFC assays using a protoplast-based transient expression system. The dominant phosphorylation site Ser309 was replaced by either Ala (non-phosphorylating, S309A) or Asp (phosphorylation mimicking, S309D) amino acids. Dimerization of wild-type and mutant HSFA4A could be confirmed by detecting YFP-derived fluorescence in all three versions of HSFA4A, while the BiFC signal was missing in the control samples transformed with empty vectors alone or in combination with one of the HSFA4A–YFP partners (Fig. 5B). When BiFC fluorescence intensities with wild-type and mutant HSFA4A were compared, S309A displayed 30% weaker, while the S309D mutant had 50% stronger, fluorescence than the wild-type TF (Fig. 5C). Western detection of YFP with anti-GFP antibody confirmed the efficient production of all HSFA4A–YFP protein forms in our transient expression system (Fig. 5D). These data suggest that MAP kinase-mediated phosphorylation is not essential for HSFA4A dimerization but has a positive influence on it.
HSFA4A can enhance tolerance to combined salt and heat stresses
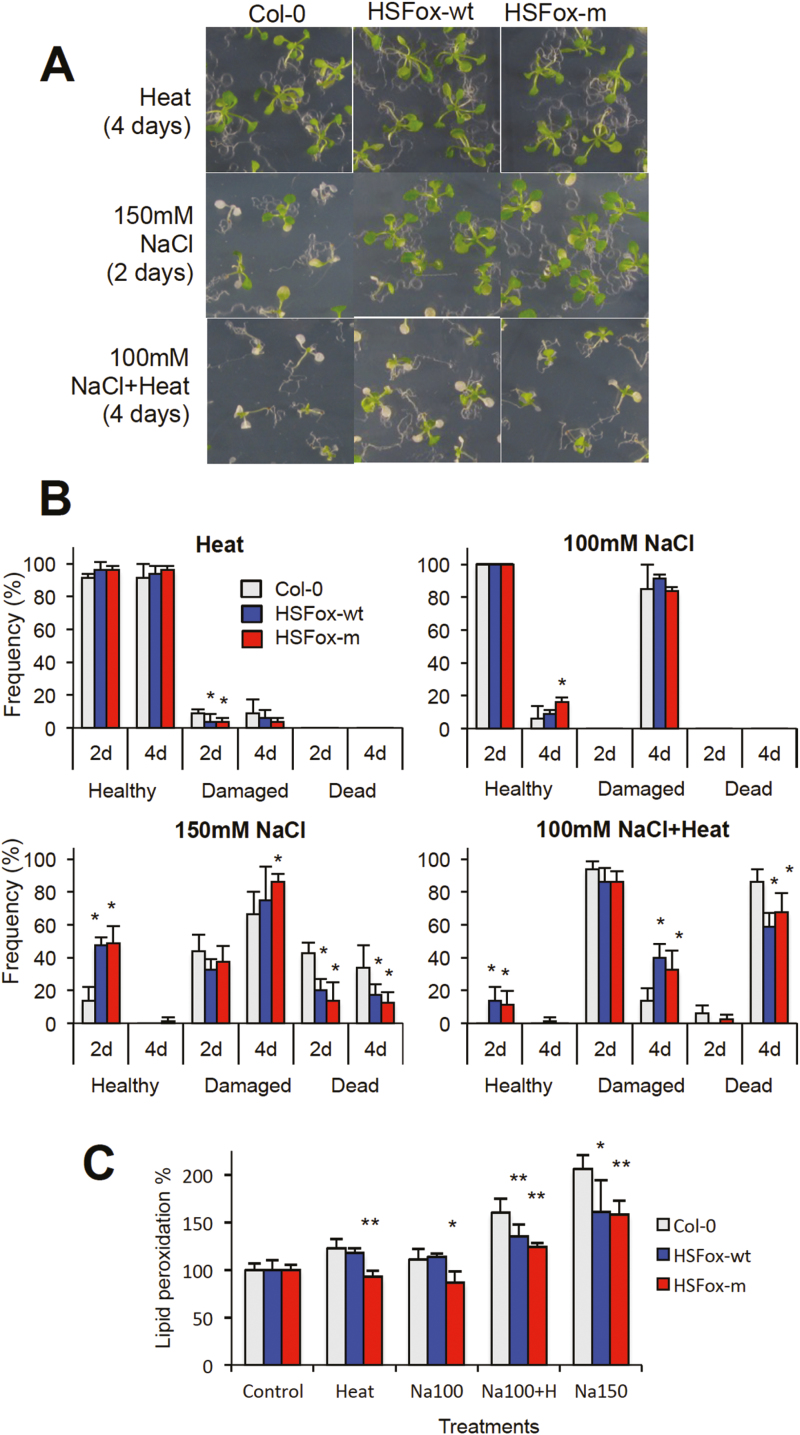
Overexpression of HSFA4A in Arabidopsis could enhance tolerance to salt, heavy metal, and oxidative agents while the knockout mutant showed hypersensitivity to salt (Pérez-Salamó et al., 2014; Faragó et al., 2018). Whether this TF could modulate responses to combined stresses is, however, not known. To evaluate responses to combined salt and heat stresses, tolerance of wild-type and HSFA4A-overexpressing Arabidopsis lines were tested in two experimental systems. In the first trials, seedlings were germinated and grown on culture media containing 0, 50, 75, 100, 125, and 150 mM NaCl, and 10-day-old plantlets were treated by 4 or 8 d of heat stress (37 °C in light and 30 °C in dark). Survival rates of HSFA4A-overexpressing plants were higher than wild-type plants on salt-containing media with or without heat stress (Supplementary Fig. S4). In the second set of experiments, wild-type seeds (Col-0) and transgenic lines overexpressing wild-type and S309D mutant versions of HSFA4A were germinated on standard culture medium and 10-day-old plantlets were exposed to different doses of salt, heat, and combined stresses followed by transfer to standard culture medium to allow recovery. Heat stress had only a minor effect on plant viability in these conditions, while salinity affected plants in a concentration-dependent manner. Damage was clearly alleviated by overexpression of both forms of HSFA4A, especially when higher salt doses (150 mM NaCl) were used. Around 50% of HSFA4A-overexpressing and 15% of wild-type plants recovered completely, while 15–20% of HSFA4A-overexpressing and 40% of wild-type plants died after exposure to 150 mM NaCl for 2 d (Fig. 6A, B). When 100 mM NaCl was combined with high temperature for 2 d, 15% of the transgenic but none of the wild-type plants recovered completely. After 4 d of combined stress, 10% of the wild-type and 30–40% of transgenic plants survived (Fig. 6A, B). In these conditions both HSFA4A forms alleviated damage to a similar extent. Combination of higher doses of salt (150 mM NaCl) with heat led to complete lethality (not shown). These results indicate that overexpression of HSFA4A not only improved tolerance to salt but could increase viability under simultaneous heat and salt stresses.
Fig. 6.
HSFA4A overexpression enhances tolerance to heat and salt stresses. Ten-day-old in vitro-grown plantlets were treated by salt (100 mM, 150 mM NaCl), heat (37 °C in light, 30 °C in dark) or their combination for 2 or 4 d. Rates of surviving healthy (vigorous growth with several new green leaves), damaged (small plants with retarded growth and/or chlorotic leaves), and dead plants (completely chlorotic with no green leaves) were scored 10 d after recovery. Similar results were obtained with independent transgenic lines of both constructs and one representative transgenic line was used for each construct in this experiments. (A) Growth of wild-type (Col-0) and transgenic plants overexpressing the wild-type (HSFox-wt) and S309D mutant (HSFox-m) forms of HSFA4A after heat, 150 mM NaCl, and combined 100 mM NaCl and heat treatments. (B) Frequencies of healthy, damaged, and dead plants after heat, salt, and combined heat and salt stresses applied for 2 or 4 d. Survival frequencies of control, non-stressed plants (all survived and healthy) and plants treated by 150 mM NaCl and heat (all dead) are not shown. (C) Lipid peroxidation rates of wild-type and HSFA4A-overexpressing lines. Values are normalized to control, non-treated plants. Error bars indicate standard deviation; asterisks indicate significant differences to Col-0 wild-type plants: *P<0.05 and **P<0.01 (Student’s t-test).
Reactive oxygen species (ROS) are generated by many adverse environmental conditions imposing oxidative damage to stressed plants. One of the deleterious effect of ROS is lipid peroxidation, which damages membranes and is a good indicator of oxidative damage. To assess the effect of HSFA4A on ROS-triggered damage, lipid peroxidation rates were compared in wild-type and HSFA4A-overexpressing plants. All individual or combined stress treatments enhanced lipid peroxidation in wild-type plants and the damage was proportional to the severity of the stress (Fig. 6C). Overexpression of both forms of HSFA4A reduced lipid peroxidation when 150 mM NaCl or combined 100 mM NaCl and heat stress was imposed. In mild conditions (heat or 100 mM NaCl alone) the S309D mutant was slightly more efficient in reducing lipid peroxidation (Fig. 6C). These results indicate, that HSFA4A can reduce oxidative damage imposed not only by individual salt or heat stresses, but also by stress combinations.
Discussion
Heat shock factors in plants are components of complex regulatory networks with various levels of control including transcription regulation, posttranslational modifications, intracellular transport, intra- and intermolecular interactions, and homo- and heteromeric trimer formation (Akerfelt et al., 2010; Scharf et al., 2012). HSFA4A responds to various stress conditions including salinity, heavy metals, oxidative agents, high temperatures, and treatments that generate protein misfolding (Pérez-Salamó et al., 2014; Lin et al., 2018). Our results show that salt and heat induction of HSFA4A follow a different pattern: expression is upregulated by salt in 2 h while heat alone or in combination with salt stress promotes expression only after 24 h (Fig. 1). Change in protein abundance roughly correlated with alterations in transcript levels (Fig. 1; Supplementary Fig. S1). Information on expression control of plant HSFs is scarce and no TF has been described that regulates the HSFA4A gene. Binding of different classes of TFs to the promoter and 5′-untranslated region (UTR) of HSFA4A could be revealed by online data mining of genome-wide DNA affinity purification sequencing (DAP-seq) (O’Malley et al., 2016) or ChIP-seq (Albihlal et al., 2018) studies. ChIP-seq data suggest HSFA1B binding, while DAP-seq revealed binding of bZIP, WRKY, C2H2, MYB, and HSF-type TFs to HSFA4A promoter, which has conserved recognition motifs for such factors (Supplementary Figs S5, S6). These TFs can regulate HSFA4A expression in different conditions.
Regulation at the post-translational level includes various types of protein modifications such as phosphorylation, sumoylation, or modulation of intracellular localization, which can profoundly affect the activity of a regulatory protein. These features were studied with YFP-tagged HSFA4A, expressed in transgenic Arabidopsis plants under the control of its own promoter. HSFA4A–YFP accumulated in a salt- and heat-dependent manner (Fig. 1; Supplementary Fig. S1), confirming that transcriptional activation by several stresses in fact leads to enhanced HSFA4A protein levels. Subcellular distribution of HSFs is regulated by the balance between nuclear import/export processes (Scharf et al., 2012). In non-stressed roots cells, HSFA4A–YFP could be detected in cytosol and nuclei while salt treatment led to fast nuclear accumulation (Fig. 2). These observations correlate with earlier reports describing the cytoplasmic localization of inactive HSFs, which move to nuclei in stress conditions allowing HSE recognition and activation of target genes (Scharf et al., 1998, 2012; Heerklotz et al., 2001). Nuclear localization signals are among the most conserved sequence elements of plant HSFs, composed by basic amino acid residues (RKRRF/LPR, Supplementary Dataset S2). While precise transport of plant HSFs is not known, animal models suggest, that nuclear import is mediated by special transport proteins (Wang and Lindquist, 1998). Interaction of tomato A2- and A1-type HSFs was shown to be important for efficient nuclear import (Scharf et al., 1998). In plant cells HSFA4A–YFP protein was predominantly detected in high molecular mass complexes. Such complexes could be disrupted with reducing treatments generating monomer proteins (Fig. 5A; Supplementary Fig. S3). Sensitivity of HSFs to redox changes has been reported, suggesting that ROS accumulation during stress conditions, particularly H2O2, can stabilize the transcriptionally active trimers (Miller and Mittler, 2006; von Koskull-Döring et al., 2007). Conserved Cys residues were suggested to stabilize intramolecular interactions of HSFA4A through redox-sensitive disulfide bonds (Pérez-Salamó et al., 2014). Our data suggest that the high molecular mass complex formation of HSFA4A depends on the redox environment, and reducing conditions favor monomerization (Fig. 5; Supplementary Fig. S3).
ROS accumulation is an important physiological consequence of environmental stress, which generates additional oxidative damage (Apel and Hirt, 2004; Choudhury et al., 2017). Maintenance of ROS homeostasis is essential in adverse conditions and plants have a sophisticated ROS sensing, signaling, and detoxification system to avoid or reduce ROS-generated damage (Miller et al., 2010). H2O2 is a common secondary signaling messenger, which can activate MAP kinase phosphorylation cascades, namely the MKK4/5–MPK3/6 and the MKK1/2–MPK4 modules, which are important stress signaling pathways in Arabidopsis (Nakagami et al., 2006; Colcombet and Hirt, 2008; Smékalová et al., 2014). Our results suggest, that ROS signals converge on HSFA4A through MPK4 and MPK3/6-mediated phosphorylation (Fig. 4; Pérez-Salamó et al., 2014). It is intriguing that the Ser309 residue is the dominant phosphorylation site for both MPK3/6 and MPK4 (Fig. 4; Pérez-Salamó et al., 2014). A recent phosphoproteomic analysis identified sets of common and specific targets for MPK3, MPK4, and MPK6 kinases and suggested that substrate specificities depend on particular Ser/Thr phosphorylation sites on the target proteins (Rayapuram et al., 2018). In HSFA4A and in the related heat shock factors of the Brassicaceae family, the conserved Ser198, Ser239, and Ser309 residues are followed by Pro, forming an authentic (S/T)P phosphorylation site for all three MAP kinases. It is interesting that the other phosphorylated amino acids identified by MS/MS analysis (Thr124, Thr238, Thr396, Ser397) are not followed by Pro in HSFA4A, nor in closely related HSFs (Fig. 4; Supplementary Dataset S2). Sequence analysis identified a single MAP kinase docking domain that does not overlap with the phosphorylation sites of HSFA4A (Fig. 4; Pitzschke, 2015; Dóczi and Bögre, 2018). The DNA binding domain (DBD), and MAP kinase docking domains are the most conserved motifs in the HSFA4-type factors (Supplementary Dataset S2) suggesting that MAP kinase-mediated phosphorylation of HSFA4-type TFs is evolutionarily conserved in both monocots and dicots. MAP kinase substrates are phosphorylated either by MPK3/6 or MPK4 kinases but not both (Dóczi and Bögre, 2018). Rare exceptions include WRKY33 and HSFA4A, which are substrates of both MPK4 and MPK3/6 (Fig. 4; Pérez-Salamó et al., 2014; Leissing et al., 2016). These TFs seem to be controlled by both the MKK4/5–MPK3/6 and the MKK1/2–MPK4 regulatory modules (Colcombet and Hirt, 2008; Smékalová et al., 2014; Xu and Zhang, 2015; Dóczi and Bögre, 2018). While MPK3 and MPK6 are predominantly positive regulators of plant defenses, MPK4 has multiple functions and can play a negative role in stress signaling (Pitzschke et al., 2009). Both MAP kinase modules coordinate defenses against different plant pathogens and abiotic stresses such as high salinity, osmotic and oxidative stresses, and extreme temperatures (Colcombet and Hirt, 2008; Rasmussen et al., 2012; Smékalová et al., 2014; Dóczi and Bögre, 2018). HSFs can be phosphorylated by different kinases, exerting either negative or positive effects on their activity (Chu et al., 1996; Holmberg et al., 2001; Hietakangas et al., 2003; Evrard et al., 2013). Mass spectrometry identified novel phosphorylation sites of HSFA4A, which are not substrates of the studied MAP kinases (Fig. 4; Supplementary Table S1). HSFA4A can therefore be regulated by several classes of kinases through multiple post-translational modifications. Identity and function of these kinases remains to be determined by further studies.
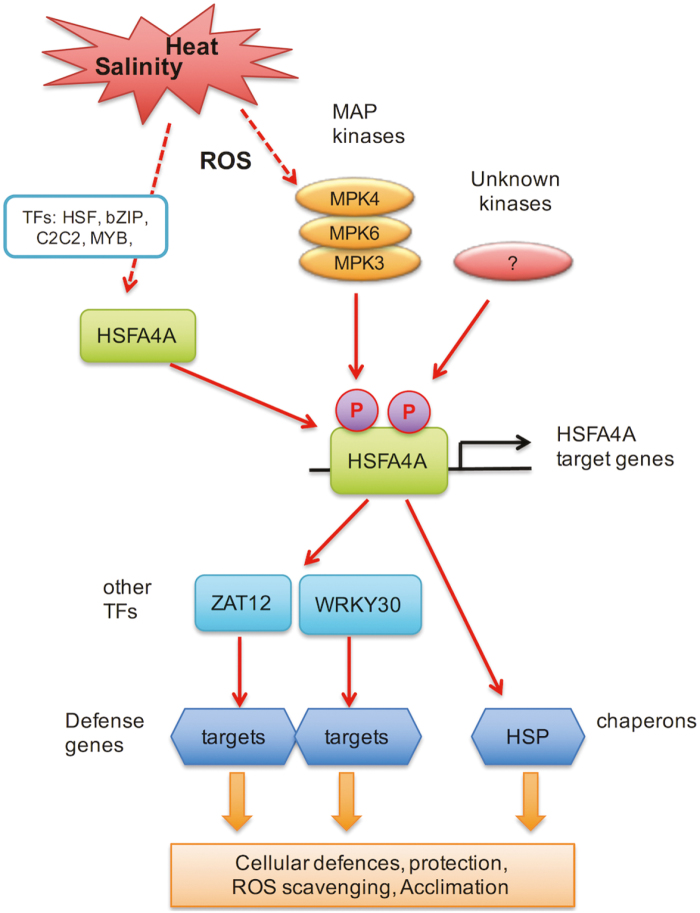
HSFA4-type TFs are implicated in responses to salinity, heavy metals, or desiccation, which generate ROS and cause oxidative damage in plant cells (Shim et al., 2009; Pérez-Salamó et al., 2014; Lang et al., 2017; Li et al., 2018). Oxidative stress can be more damaging when plants are exposed to a combination of adverse conditions, and therefore ROS control is particularly important in extreme environments. Overexpression of HSFA4A improved plant survival and reduced lipid peroxidation in individual and combined heat and salt stresses suggesting that this TF is important to reduce oxidative stress (Fig. 6). HSFA4A can alleviate cellular damage either by directly activating effector genes with protective functions such as chaperones and ROS scavengers or enhance expression of other TFs that regulate the expression of different target genes (Fig. 7). HSFA4A directly binds to the promoters of a set of stress-induced target genes including HSP17.6A, ZAT12, and WRKY30 (Fig. 3). HSP17.6A belongs to the molecular chaperones, which are critical to maintain protein homeostasis during stress conditions (Wang et al., 2004). Overproduction of HSP17.6A in Arabidopsis increased salt and drought tolerance (Sun et al., 2001) and reduced ABA sensitivity in germinating seeds (Papdi et al., 2008). ZAT12 and WRKY30 are important TFs in stress signaling. ZAT12 controls ROS signaling and promotes transcription of genes involved in redox control such as ROS scavenging ascorbate peroxidase 1 (APX1) (Rizhsky et al., 2004a; Davletova et al., 2005). ZAT12 reduces iron uptake by responding to peroxide signals thereby alleviating Fe-promoted oxidative stress (Le et al., 2016). In tomato ZAT12 can reduce heat-derived oxidative stress by activating various defense genes encoding small HSPs and antioxidant enzymes (Shah et al., 2013). WRKY-type TFs are principal regulators of defenses against pathogen attack (Eulgem et al., 2000). WRKY30 is induced by salt and oxidative conditions and upon overexpression enhances tolerance to salt and oxidative stress (Scarpeci et al., 2013) and controls leaf senescence through salicylic acid-dependent signals (Besseau et al., 2012). Rice WRKY30 responds to SA and jasmonic acid and promotes defenses against fungal pathogens (Peng et al., 2012). ZAT12 and WRKY30 can induce the expression of different sets of target genes, amplifying the defense response mediated by HSFA4A. HSFA4A, therefore, can function as a regulatory hub that connects upstream signaling mediated by ROS, MAP kinase, and other kinases with a downstream transcription regulatory network, comprising different classes of TFs and effector genes (Fig. 7).
Fig. 7.
Model of stress signal transduction and transcription regulation mediated by HSFA4A. Environmental stresses such as salt and heat generate reactive oxygen species, which in turn can activate MAP kinases MPK3/6 and MPK4. Expression of HSFA4A is activated by stress conditions, in which different classes of TFs (e.g. HSF, bZIP, C2H2, and MYB) are implicated. Phosphorylation of HSFA4A by MPK3/6, MPK4, and other unknown kinases modulates its activity and induction of target genes. HSFA4A binds to promoters of effector genes such as chaperones (e.g. HSP17.6A) or other TFs (e.g. WRKY30 and ZAT12) in a stress-dependent manner and activate their transcription. Induction of these target genes contributes to stress tolerance, either directly producing protective proteins or indirectly through activation of other defense-related genes. (This figure is available in color at JXB online.)
Supplementary data
Supplementary data are available at JXB online.
Dataset S1. Mass spectrometry data of HSFA4A phosphorylation by MPK4.
Dataset S2. Multiple sequence alignment of HSFA4-type TFs.
Fig. S1. Western detection of HSFA4A–YFP.
Fig. S2. Phylogenetic tree of HSFA4-type proteins.
Fig. S3. Multimerization of HSFA4A in cell cultures and in vitro.
Fig. S4. Plant survival in combined stress.
Fig. S5. Promoter structure of HSFA4A.
Fig. S6. TF binding on HSFA4A locus.
Table S1. HSFA4A phosphorylation summary.
Acknowledgements
Authors are indebted to Robert Dóczi for providing MPK4 construct, and to Annamária Király and Mihály Dobó for excellent technical assistance. Research was supported by OTKA NN-110962, NKFI NN-118089 and GINOP 2.3.2-15-2016-00001 grants. NA was supported by Ph.D. fellowship of the University of Szeged, GR was supported by OTKA project PD115502, and APSZ was supported by GINOP 2.3.2-15-2016-00032.
References
- Akerfelt M, Morimoto RI, Sistonen L. 2010. Heat shock factors: integrators of cell stress, development and lifespan. Nature Reviews. Molecular Cell Biology 11, 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albihlal WS, Obomighie I, Blein T, Persad R, Chernukhin I, Crespi M, Bechtold U, Mullineaux PM. 2018. Arabidopsis HEAT SHOCK TRANSCRIPTION FACTORA1b regulates multiple developmental genes under benign and stress conditions. Journal of Experimental Botany 69, 2847–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleksza D, Horváth GV, Sándor G, Szabados L. 2017. Proline accumulation is regulated by transcription factors associated with phosphate starvation. Plant Physiology 175, 555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anckar J, Sistonen L. 2011. Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annual Review of Biochemistry 80, 1089–1115. [DOI] [PubMed] [Google Scholar]
- Apel K, Hirt H. 2004. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annual Review of Plant Biology 55, 373–399. [DOI] [PubMed] [Google Scholar]
- Baker PR, Trinidad JC, Chalkley RJ. 2011. Modification site localization scoring integrated into a search engine. Molecular & Cellular Proteomics 10, M111.008078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter A, Mittler R, Suzuki N. 2014. ROS as key players in plant stress signalling. Journal of Experimental Botany 65, 1229–1240. [DOI] [PubMed] [Google Scholar]
- Besseau S, Li J, Palva ET. 2012. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. Journal of Experimental Botany 63, 2667–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigeard J, Hirt H. 2018. Nuclear Signaling of Plant MAPKs. Frontiers in Plant Science 9, 469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch W, Wunderlich M, Schöffl F. 2005. Identification of novel heat shock factor-dependent genes and biochemical pathways in Arabidopsis thaliana. The Plant Journal 41, 1–14. [DOI] [PubMed] [Google Scholar]
- Carranco R, Prieto-Dapena P, Almoguera C, Jordano J. 2017. SUMO-dependent synergism involving heat shock transcription factors with functions linked to seed longevity and desiccation tolerance. Frontiers in Plant Science 8, 974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan H, Khurana N, Agarwal P, Khurana P. 2011. Heat shock factors in rice (Oryza sativa L.): genome-wide expression analysis during reproductive development and abiotic stress. Molecular Genetics and Genomics 286, 171–187. [DOI] [PubMed] [Google Scholar]
- Choudhury FK, Rivero RM, Blumwald E, Mittler R. 2017. Reactive oxygen species, abiotic stress and stress combination. The Plant Journal 90, 856–867. [DOI] [PubMed] [Google Scholar]
- Chu B, Soncin F, Price BD, Stevenson MA, Calderwood SK. 1996. Sequential phosphorylation by mitogen-activated protein kinase and glycogen synthase kinase 3 represses transcriptional activation by heat shock factor-1. The Journal of Biological Chemistry 271, 30847–30857. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Colcombet J, Hirt H. 2008. Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. The Biochemical Journal 413, 217–226. [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. 2003. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiology 133, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. 2005. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology 139, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S, Schlauch K, Coutu J, Mittler R. 2005. The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiology 139, 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dóczi R, Bögre L. 2018. The quest for MAP kinase substrates: gaining momentum. Trends in Plant Science 23, 918–932. [DOI] [PubMed] [Google Scholar]
- Dóczi R, Brader G, Pettkó-Szandtner A, Rajh I, Djamei A, Pitzschke A, Teige M, Hirt H. 2007. The Arabidopsis mitogen-activated protein kinase kinase MKK3 is upstream of group C mitogen-activated protein kinases and participates in pathogen signaling. The Plant Cell 19, 3266–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. 2000. The WRKY superfamily of plant transcription factors. Trends in Plant Science 5, 199–206. [DOI] [PubMed] [Google Scholar]
- Evrard A, Kumar M, Lecourieux D, Lucks J, von Koskull-Döring P, Hirt H. 2013. Regulation of the heat stress response in Arabidopsis by MPK6-targeted phosphorylation of the heat stress factor HsfA2. PeerJ 1, e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faragó D, Sass L, Valkai I, Andrási N, Szabados L. 2018. PlantSize offers an affordable, non-destructive method to measure plant size and color in vitro. Frontiers in Plant Science 9, 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficarro SB, Adelmant G, Tomar MN, Zhang Y, Cheng VJ, Marto JA. 2009. Magnetic bead processor for rapid evaluation and optimization of parameters for phosphopeptide enrichment. Analytical Chemistry 81, 4566–4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgii E, Jin M, Zhao J, Kanawati B, Schmitt-Kopplin P, Albert A, Winkler JB, Schäffner AR. 2017. Relationships between drought, heat and air humidity responses revealed by transcriptome-metabolome co-analysis. BMC Plant Biology 17, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring M, Offermann S, Danker T, Horst I, Peterhansel C, Stam M. 2007. Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods 3, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashikawa N, Sakurai H. 2004. Phosphorylation of the yeast heat shock transcription factor is implicated in gene-specific activation dependent on the architecture of the heat shock element. Molecular and Cellular Biology 24, 3648–3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heerklotz D, Döring P, Bonzelius F, Winkelhaus S, Nover L. 2001. The balance of nuclear import and export determines the intracellular distribution and function of tomato heat stress transcription factor HsfA2. Molecular and Cellular Biology 21, 1759–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietakangas V, Ahlskog JK, Jakobsson AM, Hellesuo M, Sahlberg NM, Holmberg CI, Mikhailov A, Palvimo JJ, Pirkkala L, Sistonen L. 2003. Phosphorylation of serine 303 is a prerequisite for the stress-inducible SUMO modification of heat shock factor 1. Molecular and Cellular Biology 23, 2953–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges DM, DeLong JM, Forney CF, Prange RK. 1999. Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 207, 604–611. [DOI] [PubMed] [Google Scholar]
- Holmberg CI, Hietakangas V, Mikhailov A, et al. 2001. Phosphorylation of serine 230 promotes inducible transcriptional activity of heat shock factor 1. The EMBO Journal 20, 3800–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath BM, Kourova H, Nagy S, et al. 2017. Arabidopsis RETINOBLASTOMA RELATED directly regulates DNA damage responses through functions beyond cell cycle control. The EMBO Journal 36, 1261–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SA, Yoon JH, Lee SH, Ahn SG. 2005. Polo-like kinase 1 phosphorylates heat shock transcription factor 1 and mediates its nuclear translocation during heat stress. The Journal of Biological Chemistry 280, 12653–12657. [DOI] [PubMed] [Google Scholar]
- Lang S, Liu X, Xue H, Li X, Wang X. 2017. Functional characterization of BnHSFA4a as a heat shock transcription factor in controlling the re-establishment of desiccation tolerance in seeds. Journal of Experimental Botany 68, 2361–2375. [DOI] [PubMed] [Google Scholar]
- Le CT, Brumbarova T, Ivanov R, Stoof C, Weber E, Mohrbacher J, Fink-Straube C, Bauer P. 2016. ZINC FINGER OF ARABIDOPSIS THALIANA12 (ZAT12) interacts with FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (FIT) linking iron deficiency and oxidative stress responses. Plant Physiology 170, 540–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leissing F, Nomoto M, Bocola M, Schwaneberg U, Tada Y, Conrath U, Beckers GJ. 2016. Substrate thiophosphorylation by Arabidopsis mitogen-activated protein kinases. BMC Plant Biology 16, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Zhang H, Zhao H, Gao T, Song A, Jiang J, Chen F, Chen S. 2018. Chrysanthemum CmHSFA4 gene positively regulates salt stress tolerance in transgenic chrysanthemum. Plant Biotechnology Journal 16, 1311–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ding Y, Shi Y, Zhang X, Zhang S, Gong Z, Yang S. 2017. MPK3- and MPK6-mediated ICE1 phosphorylation negatively regulates ICE1 stability and freezing tolerance in Arabidopsis. Developmental Cell 43, 630–642.e4. [DOI] [PubMed] [Google Scholar]
- Lin KF, Tsai MY, Lu CA, Wu SJ, Yeh CH. 2018. The roles of Arabidopsis HSFA2, HSFA4a, and HSFA7a in the heat shock response and cytosolic protein response. Botanical Studies 59, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link V, Sinha AK, Vashista P, Hofmann MG, Proels RK, Ehness R, Roitsch T. 2002. A heat-activated MAP kinase in tomato: a possible regulator of the heat stress response. FEBS Letters 531, 179–183. [DOI] [PubMed] [Google Scholar]
- Liu HT, Gao F, Li GL, Han JL, Liu DL, Sun DY, Zhou RG. 2008. The calmodulin-binding protein kinase 3 is part of heat-shock signal transduction in Arabidopsis thaliana. The Plant Journal 55, 760–773. [DOI] [PubMed] [Google Scholar]
- Mao G, Meng X, Liu Y, Zheng Z, Chen Z, Zhang S. 2011. Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. The Plant Cell 23, 1639–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Xu J, He Y, Yang KY, Mordorski B, Liu Y, Zhang S. 2013. Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. The Plant Cell 25, 1126–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Mittler R. 2006. Could heat shock transcription factors function as hydrogen peroxide sensors in plants? Annals of Botany 98, 279–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R. 2010. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant, Cell & Environment 33, 453–467. [DOI] [PubMed] [Google Scholar]
- Nakagami H, Soukupová H, Schikora A, Zárský V, Hirt H. 2006. A Mitogen-activated protein kinase kinase kinase mediates reactive oxygen species homeostasis in Arabidopsis. The Journal of Biological Chemistry 281, 38697–38704. [DOI] [PubMed] [Google Scholar]
- Nguyen XC, Hoang MH, Kim HS, Lee K, Liu XM, Kim SH, Bahk S, Park HC, Chung WS. 2012a Phosphorylation of the transcriptional regulator MYB44 by mitogen activated protein kinase regulates Arabidopsis seed germination. Biochemical and Biophysical Research Communications 423, 703–708. [DOI] [PubMed] [Google Scholar]
- Nguyen XC, Kim SH, Lee K, et al. 2012. b Identification of a C2H2-type zinc finger transcription factor (ZAT10) from Arabidopsis as a substrate of MAP kinase. Plant Cell Reports 31, 737–745. [DOI] [PubMed] [Google Scholar]
- Nover L, Bharti K, Döring P, Mishra SK, Ganguli A, Scharf KD. 2001. Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress & Chaperones 6, 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley RC, Huang SC, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. 2016. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165, 1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papdi C, Abrahám E, Joseph MP, Popescu C, Koncz C, Szabados L. 2008. Functional identification of Arabidopsis stress regulatory genes using the controlled cDNA overexpression system. Plant Physiology 147, 528–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Hu Y, Tang X, Zhou P, Deng X, Wang H, Guo Z. 2012. Constitutive expression of rice WRKY30 gene increases the endogenous jasmonic acid accumulation, PR gene expression and resistance to fungal pathogens in rice. Planta 236, 1485–1498. [DOI] [PubMed] [Google Scholar]
- Pérez-Salamó I, Papdi C, Rigó G, et al. 2014. The heat shock factor A4A confers salt tolerance and is regulated by oxidative stress and the mitogen-activated protein kinases MPK3 and MPK6. Plant Physiology 165, 319–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke A. 2015. Modes of MAPK substrate recognition and control. Trends in Plant Science 20, 49–55. [DOI] [PubMed] [Google Scholar]
- Pitzschke A, Djamei A, Bitton F, Hirt H. 2009. A major role of the MEKK1-MKK1/2-MPK4 pathway in ROS signalling. Molecular Plant 2, 120–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen MW, Roux M, Petersen M, Mundy J. 2012. MAP kinase cascades in Arabidopsis innate immunity. Frontiers in Plant Science 3, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S, Barah P, Suarez-Rodriguez MC, Bressendorff S, Friis P, Costantino P, Bones AM, Nielsen HB, Mundy J. 2013. Transcriptome responses to combinations of stresses in Arabidopsis. Plant Physiology 161, 1783–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayapuram N, Bigeard J, Alhoraibi H, Bonhomme L, Hesse AM, Vinh J, Hirt H, Pflieger D. 2018. Quantitative phosphoproteomic analysis reveals shared and specific targets of arabidopsis mitogen-activated protein kinases (MAPKs) MPK3, MPK4, and MPK6. Molecular & Cellular Proteomics 17, 61–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reindl A, Schöffl F, Schell J, Koncz C, Bakó L. 1997. Phosphorylation by a cyclin-dependent kinase modulates DNA binding of the Arabidopsis heat-shock transcription factor HSF1 in vitro. Plant Physiology 115, 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero RM, Mestre TC, Mittler R, Rubio F, Garcia-Sanchez F, Martinez V. 2014. The combined effect of salinity and heat reveals a specific physiological, biochemical and molecular response in tomato plants. Plant, Cell & Environment 37, 1059–1073. [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Davletova S, Liang H, Mittler R. 2004a The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. The Journal of Biological Chemistry 279, 11736–11743. [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Shuman J, Shulaev V, Davletova S, Mittler R. 2004b When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiology 134, 1683–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpeci TE, Zanor MI, Mueller-Roeber B, Valle EM. 2013. Overexpression of AtWRKY30 enhances abiotic stress tolerance during early growth stages in Arabidopsis thaliana. Plant Molecular Biology 83, 265–277. [DOI] [PubMed] [Google Scholar]
- Scharf KD, Berberich T, Ebersberger I, Nover L. 2012. The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochimica et Biophysica Acta 1819, 104–119. [DOI] [PubMed] [Google Scholar]
- Scharf KD, Heider H, Höhfeld I, Lyck R, Schmidt E, Nover L. 1998. The tomato Hsf system: HsfA2 needs interaction with HsfA1 for efficient nuclear import and may be localized in cytoplasmic heat stress granules. Molecular and Cellular Biology 18, 2240–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewelam N, Oshima Y, Mitsuda N, Ohme-Takagi M. 2014. A step towards understanding plant responses to multiple environmental stresses: a genome-wide study. Plant, Cell & Environment 37, 2024–2035. [DOI] [PubMed] [Google Scholar]
- Shah K, Singh M, Rai AC. 2013. Effect of heat-shock induced oxidative stress is suppressed in BcZAT12 expressing drought tolerant tomato. Phytochemistry 95, 109–117. [DOI] [PubMed] [Google Scholar]
- Shim D, Hwang JU, Lee J, Lee S, Choi Y, An G, Martinoia E, Lee Y. 2009. Orthologs of the class A4 heat shock transcription factor HsfA4a confer cadmium tolerance in wheat and rice. The Plant Cell 21, 4031–4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smékalová V, Doskočilová A, Komis G, Samaj J. 2014. Crosstalk between secondary messengers, hormones and MAPK modules during abiotic stress signalling in plants. Biotechnology Advances 32, 2–11. [DOI] [PubMed] [Google Scholar]
- Su J, Zhang M, Zhang L, Sun T, Liu Y, Lukowitz W, Xu J, Zhang S. 2017. Regulation of stomatal immunity by interdependent functions of a pathogen-responsive MPK3/MPK6 cascade and abscisic acid. The Plant Cell 29, 526–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Nitta Y, Zhang Q, Wu D, Tian H, Lee JS, Zhang Y. 2018. Antagonistic interactions between two MAP kinase cascades in plant development and immune signaling. EMBO Reports 19, e45324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Bernard C, van de Cotte B, Van Montagu M, Verbruggen N. 2001. At-HSP17.6A, encoding a small heat-shock protein in Arabidopsis, can enhance osmotolerance upon overexpression. The Plant Journal 27, 407–415. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Rivero RM, Shulaev V, Blumwald E, Mittler R. 2014. Abiotic and biotic stress combinations. New Phytologist 203, 32–43. [DOI] [PubMed] [Google Scholar]
- Volkov RA, Panchuk II, Mullineaux PM, Schöffl F. 2006. Heat stress-induced H2O2 is required for effective expression of heat shock genes in Arabidopsis. Plant Molecular Biology 61, 733–746. [DOI] [PubMed] [Google Scholar]
- von Koskull-Döring P, Scharf KD, Nover L. 2007. The diversity of plant heat stress transcription factors. Trends in Plant Science 12, 452–457. [DOI] [PubMed] [Google Scholar]
- Wang Z, Lindquist S. 1998. Developmentally regulated nuclear transport of transcription factors in Drosophila embryos enable the heat shock response. Development 125, 4841–4850. [DOI] [PubMed] [Google Scholar]
- Wang W, Vinocur B, Shoseyov O, Altman A. 2004. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends in Plant Science 9, 244–252. [DOI] [PubMed] [Google Scholar]
- Wang X, Khaleque MA, Zhao MJ, Zhong R, Gaestel M, Calderwood SK. 2006. Phosphorylation of HSF1 by MAPK-activated protein kinase 2 on serine 121, inhibits transcriptional activity and promotes HSP90 binding. The Journal of Biological Chemistry 281, 782–791. [DOI] [PubMed] [Google Scholar]
- Xu J, Zhang S. 2015. Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends in Plant Science 20, 56–64. [DOI] [PubMed] [Google Scholar]
- Zhu X, Huang C, Zhang L, Liu H, Yu J, Hu Z, Hua W. 2017. Systematic analysis of Hsf family genes in the Brassica napus genome reveals novel responses to heat, drought and high CO2 stresses. Frontiers in Plant Science 8, 1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zsigmond L, Szepesi A, Tari I, Rigó G, Király A, Szabados L. 2012. Overexpression of the mitochondrial PPR40 gene improves salt tolerance in Arabidopsis. Plant Science 182, 87–93. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.