miRNA-mediated silencing of R2R3 MYB transcription factor promotes the production of specific secondary metabolites in grapes.
Keywords: Anthocyanins, flavonols, grapes, miRNA, R2R3 MYB, secondary silencing
Abstract
MicroRNAs are a class of non-coding small RNAs involved in the negative regulation of gene expression, which play critical roles in developmental and metabolic pathways. Studies in several plants have identified a few microRNAs and other small RNAs that target regulators of the phenylpropanoid metabolic pathway called the MYB transcription factors. However, it is not well understood how sRNA-mediated regulation of MYBs influences the accumulation of specific secondary metabolites. Using sRNA sequencing, degradome analysis, mRNA sequencing, and proteomic analysis, we establish that grape lines with high anthocyanin content express two MYB-targeting microRNAs abundantly, resulting in the differential expression of specific MYB proteins. miR828 and miR858 target coding sequences of specific helix motifs in the mRNA sequences of MYB proteins. Targeting by miR828 caused MYB RNA decay and the production of a cascade of secondary siRNAs that depend on RNA-dependent RNA polymerase 6. MYB suppression and cascade silencing was more robust in grape lines with high anthocyanin content than in a flavonol-rich grape line. We establish that microRNA-mediated silencing targeted the repressor class of MYBs to promote anthocyanin biosynthesis in grape lines with high anthocyanins. We propose that this process regulates the expression of appropriate MYBs in grape lines to produce specific secondary metabolites.
Introduction
MicroRNAs (miRNAs) are key regulators of gene expression in both plants and animals. Plant miRNAs are processed by a specific Dicer-like nuclease from long RNA precursors with base-paired foldback structures to an active form consisting of 21- to 24-nucleotide (nt) RNAs (Baulcombe, 2004). These miRNAs form a ribonucleoprotein complex with the Argonaute (AGO) family member AGO1, which can subsequently bind by base pairing to a target RNA with high complementarity (Baumberger and Baulcombe, 2005; Bartel, 2009). The consequence of targeting by the ribonucleoprotein complex depends on the nature of the target RNA as well as the extent of its complementarity with the miRNA. Usually, the target RNA is cleaved to result in reduced protein accumulation if there is near-complete complementarity between the miRNA and its target. Such an outcome may also require complete complementarity at positions 10 and 11 of the miRNA. A lower complementation percentage between the miRNA and its target also leads to reduced protein production through translational suppression, usually without turnover of the target RNA (Brodersen et al. 2008). A third outcome can be miRNA-mediated targeting of chromatin-associated RNAs that leads to specific epigenetic modifications (Khraiwesh et al., 2010; Wu et al., 2010; Creasey et al., 2014).
However, most striking among the functions of plant miRNAs is in their ability to initiate regulatory cascades with multiple mRNA targets (MacLean et al., 2010). The first step in these cascades requires the conversion of an otherwise non-functional miRNA-targeted RNA fragment into a double-stranded RNA molecule by an RNA-dependent RNA polymerase (RDR; usually RDR6 in Arabidopsis thaliana and other eudicots) (Howell et al., 2007; Qu et al., 2008). Not all miRNA-mediated targeting results in this cascade silencing. There are at least three known reasons for the initiation of cascade silencing: (i) when the initiator miRNA duplex structure is asymmetrical (Manavella et al., 2012), (ii) if the initiator miRNA is an atypical 22 nt in length rather than 21 nt (Chen et al., 2010; Cuperus et al., 2010), or (iii) if there are two distinct target sites for miRNA within the target RNA (Axtell et al., 2006). Once initiated, the cascade silencing generates abundant secondary small interfering RNAs (siRNAs), usually of 21 or rarely 24 nt length. These siRNAs associate with specific AGO proteins based on their 5′ nucleotide and/or the length bias of specific AGOs to target RNAs with high complementarity (Chen et al., 2007).
The most conserved secondary siRNA locus that is seen across distantly related plants is a locus named TAS3 (Cuperus et al., 2011). TheTAS3-derived secondary siRNAs target auxin response factor (ARF) mRNAs in a process that likely evolved in a common ancestor of seed plants (Axtell and Bartel, 2005). TAS3-derived siRNAs play a crucial role in plant development by regulating auxin signaling (Axtell et al., 2006; Felippes and Weigel, 2009). Other secondary siRNA loci in rice (Song et al., 2012), tomato (Shivaprasad et al., 2012a), soybean (Arikit et al., 2014; Fei et al., 2015), maize (Dotto et al., 2014; Zhai et al., 2015), strawberry (Xia et al., 2015), wheat (Yao et al., 2007), and Brachypodium are species-specific and are likely to have arisen more recently (Johnson et al., 2009; Vogel et al., 2010).
In dicots, secondary siRNAs known as transacting siRNAs (tasi-RNAs) or phased siRNAs (phasi-RNAs) target pentatricopeptide repeat (PPR) and ARF mRNAs (Axtell et al., 2006). Surprisingly, both PPR and ARF mRNAs are targeted separately by secondary siRNAs and also by miRNAs such as miR161, miR160, or miR167 (Rhoades et al., 2002; Allen et al., 2005; Yoshikawa et al., 2005). Although it is not entirely clear why cascade silencing targets are targeted by multiple small RNA species, it is possible to speculate that the secondary siRNAs play a role in either reinforcing or coordinating the action of primary sRNAs or miRNAs.
In addition to their role in development and disease resistance (Nonogaki, 2010), cascade loci have been implicated in specific metabolic processes by regulating MYB transcription factors. The MYB superfamily is a diverse group of transcription factors represented in eukaryotes that are known to control processes such as epidermal cell differentiation, flavonoid biosynthesis, stress tolerance, and pathogen resistance. MYB proteins are classified into four classes, 1R, 2R (R2R3), 3R (R1R2R3), and 4R (Dubos et al., 2010), depending on the number of adjacent repeats homologous to animal c-MYB (Klempnauer et al., 1982). In plants, the MYB family has selectively expanded through the R2R3-MYBs. R2R3-MYB transcription factors regulate secondary metabolism, while other MYBs regulate stress responses and development. The R2R3-MYBs, along with two other proteins, bHLH and WD40, can act as both activators and repressors of the phenylpropanoid pathway (Ramsay and Glover, 2005). The phenylpropanoid pathway is an essential pathway in higher plants required for the production of metabolites such as lignin, flavonols, proanthocyanins, and anthocyanins (Fraser and Chapple, 2011).
A well-conserved low-abundant secondary siRNA locus named TAS4, initiated by miR828, regulates the expression of transcription factors of the MYB family in Arabidopsis and apple (Rajagopalan et al., 2006; Espley et al., 2007; Xia et al., 2012). Studies have suggested that a few Arabidopsis MYBs, such as MYB11, MYB12, and MYB113, are targeted by miR156 (Gou et al., 2011), and MYB111 is targeted by miR858 (Sharma et al., 2016), thereby regulating anthocyanin accumulation. In Arabidopsis (Rajagopalan et al., 2006; Hsieh et al., 2009), lotus (Zheng et al., 2014), and apple (Xia et al., 2012), siR81(-) (Luo et al., 2012), one of the TAS4-derived siRNAs, has been shown to target RNAs coding for MYBs such as MYB75 and MYB113 (as in Arabidopsis), or MYB90 and MYB113 (as in potato), that are associated with anthocyanin accumulation (Luo et al., 2012; Rock, 2013; Li et al., 2016; Liu et al., 2016). Due to the absence of a clear phylogenetic relationship among MYB proteins in different plant species, it is not possible to predict which subgroup of MYBs is being targeted in these plants. It has been speculated that these miRNAs and secondary siRNAs regulate the phenylpropanoid pathway during normal development as well as being part of responses to stresses (Shin et al., 2013). There is a strong correlation between the number of MYBs in a given plant and the number of small RNAs that are capable of targeting some of the MYBs. However, it is not known how miRNA/siRNA-mediated regulation of MYBs and other regulators of the phenylpropanoid pathway results in the production of specific secondary metabolites. This is largely due to a lack of studies involving contrasting genotypes producing diverse secondary metabolites, as well as a lack of genetic approaches.
The objective of this study was to understand the functional significance of miRNA-mediated regulation of MYB RNAs in grape. We performed differential sRNA, RNA-seq, and proteome analysis of three closely related grape varieties with varied anthocyanin and flavonol contents. Our results indicated that grape varieties with high anthocyanin levels have two previously identified miRNAs abundantly expressed in both young leaf and fruit peel tissues. During the analysis we also identified a novel MYB repressor, named VvMYB114, which is a target of miRNAs in anthocyanin-rich grape varieties. When expressed in tobacco, VvMYB114 repressed the expression of enzymes such as DFR and UFGT, which promote anthocyanin accumulation. Since MYB114 homologs with EAR (ERF-associated amphiphilic repression motif) repressor motifs are well known among dicots, often as targets of miRNAs, we predict that targeting of MYB repressors might be a general strategy to allow activator MYBs to shift the pathway towards anthocyanin biosynthesis.
Materials and methods
Plant materials
Three grape lines with contrasting phenotypes, Dilkhush (DK), Red Globe (RG), and Bangalore Blue (BB), were used in this study. The plant materials were collected from the Indian Institute of Horticultural Research (IIHR) Campus, Bangalore, and grown in a greenhouse under natural light conditions and 70% relative humidity at 23 °C. Nicotiana benthamiana wild-type (WT) and RDR6i plants and Nicotiana tabacum (Wisconsin 35) were grown in a growth chamber (Panasonic) at 23 °C under a light/dark cycle of 16/8 h and 70% relative humidity.
Plasmid clones and constructs
For generating overexpression constructs, cDNA was prepared from grape young leaf samples using Superscript III (Invitrogen), according to the manufacturer’s instructions. Amplified products were cloned into modified pBIN19 vectors (Bevan, 1984) under the CaMV 35S promoter and NOS terminator. Primer pairs used for cloning are listed in Supplementary Table S4 at JXB online. For the generation of mutations in the EAR motif, to substitute the 87th L residue of MYB114 with A or I residues, a PCR-based ligation method was performed using primer pairs incorporating the respective mutations. The binary vectors were then transformed into Agrobacterium tumefaciens strain LBA4404 with pSB1 (pSB1 harbors extra copies of vir genes for better efficiency; Stachel and Nester, 1986), and used for further transient and stable transformation experiments. Details of the restriction sites incorporated and used for cloning and mutations are given in Supplementary Table S4.
Multiple sequence alignments and phylogenetic tree
The protein sequences of MYBs belonging to the R2R3 family were retrieved from NCBI and a multiple sequence alignment was performed using Geneious 8. The aligned peptides from the R2R3 regions were extracted and a phylogenetic tree was constructed using MEGA 6 with the maximum likelihood algorithm and bootstrap 100.
Total RNA extraction and library preparation for RNA-seq and sRNA-seq
Isolation of total RNA from young leaves and fruit peels of grape was performed using the Qiagen RNeasy Plant Mini Kit according to the manufacturer’s instructions with slight modifications as described by Čepin et al. (2010). The concentration of RNA and purity of the samples were estimated using a Nanodrop spectrophotometer (Thermo Fisher Scientific) and Qubit fluorometer (Thermo Fisher Scientific). An aliquot of the samples from young leaves and fruit peels was separated on an Agilent RNA Bioanalyzer chip to check for integrity. RNA sequencing (RNA-seq) libraries with two biological replicates were constructed according to the NEXT flex Rapid directional mRNA-seq bundle library protocol outlined by Trapnell et al. (2012), at Genotypic Technology Pvt Ltd, Bangalore, India, and sequenced on the Illumina NextSeq500 platform. sRNA sequencing (sRNA-seq) libraries with two biological replicates were prepared according to the TruSeq Small RNA Sample Preparation Guide (Illumina, San Diego, California, USA) at Genotypic Technology Pvt Ltd, Bangalore, India, and sequenced on the Illumina NextSeq500 platform.
RNA blot analysis
RNA blots were performed as described by Akbergenov et al. (2006) and Shivaprasad et al. (2012b). Briefly, approximately 10 or 15 µg of total RNA extracted as described above was air dried and resuspended in 8 µl loading buffer (0.10% bromophenol blue, 0.10% xylene cyanol in 100% deionized formamide), heated at 95 °C for 1 min, and loaded on to a 15% denaturing polyacrylamide gel (19:1 acrylamide:bisacrylamide and 8 M urea). The gel was run at 100 V for 3 h and then transferred to a Hybond N+ membrane (GE Healthcare) by electroblotting (Bio Rad) at 10 V overnight at 4 °C. RNA hybridization was performed at 35 °C for 12 h using UltraHyb-oligo buffer (Ambion) containing appropriate radiolabeled short DNA oligo probes (Supplementary Table S3), end-labeled with 32P-ATP (BRIT, India) by polynucleotide kinase (NEB), and purified through MicroSpin G-25 columns (GE Healthcare). The blot was washed twice with 2× SSC buffer plus 0.5% SDS for 30 min at 35 °C. The signal was detected after exposure on a phosphorimager screen using a Molecular Imager (GE Healthcare). For repeated hybridization, the membrane was stripped and re-probed.
Quantitative RT–PCR analysis of phenylpropanoid genes
Isolation of total RNA from tobacco leaves was performed using TRIzol Reagent (Ambion) as per the manufacturer’s instructions. cDNA was prepared using random hexamers and Superscript III (Invitrogen), according to the manufacturer’s protocol. Primer pairs used for the amplification are listed in Supplementary Table S4. GAPDH was used as the control for each gene.
Degradome library preparation
Approximately 35 μg of total RNA was used to prepare a degradome library as described by Zhai et al. (2014) with the following modification: approximately 120 µg of total RNA was taken for the analysis. For the size selection of 128 bp PARE-ligated products, Agencourt® AMPure® XP, Beckman-Coulter beads were used instead of PAGE purification.
Processing of Illumina sRNA-seq reads and expression analysis
The sRNA-seq reads were processed for adaptor removal and filtered for a length range of 16–35 nt using the UEA small RNA Workbench version 3 (Moxon et al., 2008; Stocks et al., 2012). miRNA expression analysis was performed using the miRProf tool in the UEA small RNA Workbench. A cut-off of log2(fold change) >2 was used for differential expression analysis of miRNAs. Novel miRNAs were identified using the miRcat tool from the UEA small RNA Workbench.
Processing and mapping of Illumina mRNA-seq reads and gene expression analysis
The RNA-seq reads produced by the Illumina sequencer were processed to remove adaptors and low-quality bases. The reads obtained after pre-processing were aligned to the grape transcriptome (downloaded from https://urgi.versailles.inra.fr/Species/Vitis) and analyzed further as described by Trapnell et al. (2012).
miRNA target prediction, sequence alignments, and phased sRNA analysis
The target of the miRNAs was predicted using the psRNA tool (Dai and Zhao, 2011) and TAPIR tool (Bonnet et al., 2010). The sequences in the target regions were extracted, and the sequence logos of the miRNA target region were generated with WebLogo (Schneider and Stephens, 1990). Sequence alignments and phylogenetic analysis were performed using the maximum likelihood method.
Loci producing phased siRNAs were identified using the UEA small RNA Workbench version 3. Using the TASI prediction tool, phasing analysis of sRNA sequencing datasets was performed with a stringent cutoff of P≤0.01.
Transient and stable expression
Leaves of 3- to 4-week-old N. tabacum Wisconsin 35 plants were infiltrated with Agrobacterium strain LBA4404 (pSB1) harboring constructs to express genes of interest. The cultures of Agrobacterium were adjusted to an OD600 of 1.0 using 10 mM MgCl2 (pH 5.6). Before infiltration of the leaves, 60 µM acetosyringone was added to the bacterial culture and incubated for 30 min. Infiltrated tissues were collected after 3 days for RNA analysis. Transformation of tobacco leaf discs was performed using A. tumefaciens (LBA4404 with pSB1 and appropriate gene of interest) as described previously (Sunilkumar et al., 1999). Transgenic plants were regenerated from kanamycin-resistant calli and maintained in a growth chamber (70% relative humidity at 28 °C).
Anthocyanin estimation
Anthocyanin estimation was performed as described by Nakata and Ohme-Takagi (2014). Approximately 5 volumes of extraction buffer (45% methanol and 5% acetic acid) were added to the plant tissues and mixed thoroughly. Centrifugation was performed twice at 12 000 g for 5 minutes at room temperature. The absorbance (Abs) of the supernatant was measured at 530 nm and 657 nm, and the anthocyanin content, expressed as Abs530 g FW–1, was calculated by using the formula:
Total flavonol estimation
Total flavonol estimation was performed as described by Koyama et al. (2012) with modifications as described below. The samples (infiltrated tobacco leaves and grape fruit peels; 0.5 g) were ground into powder in liquid nitrogen and extracted with 10 ml of 75% ethanol for 3 h. The filtrate was dried and dissolved in 5 ml of 75% ethanol. To 0.1 ml of this extract, 0.5 ml of NaNO2 (50 mg ml–1) was added and mixed vigorously, followed by incubation for 6 min. Next, 0.5 ml of aluminum nitrate was added to the reaction mixture and further incubated for 6 min. The reaction was stopped by the addition of 2.5 ml 1 M NaOH and the absorbance was measured at 510 nm after 15 min of incubation. A standard curve with varying amounts of quercetin from 0 to 100 µg was used to quantify the amount of quercetin produced in the samples by measuring the absorbance.
4-Dimethylaminocinnamaldehyde and toluidine blue O staining
Grape berries at different developmental stages were collected (Supplementary Fig. S1) and hand sectioned for staining for the detection of proanthocyanidins and their precursor molecules (Jun et al., 2015). Sections were stained using 0.01% (w/v) 4-dimethylaminocinnamaldehyde (DMACA) in absolute ethanol containing 0.8% w/v hydrochloric acid for 20 min. DMACA stains proanthocyanidins and flavan-3-ols green or blue (Abeynayake et al., 2011). The sections were also stained for 5 sec using 0.5% (w/v) toluidine blue O (TBO) in 0.1 M phosphate buffer, pH 7.2. TBO stains polyphenolic compounds such as proanthocyanidins a greenish-blue color (Debeaujon et al., 2003).
Protein extraction and total proteome analysis
Extraction of protein was performed according to Wang et al. (2006), with some modifications specifically optimized for grape tissues. Briefly, leaf and fruit peel samples were ground in a mortar in liquid nitrogen. Ground samples were washed with 10% trichloroacetic acid/acetone mixture, vortexed, and centrifuged at 16 000 g for 10 min. This washing step was repeated until the maximum color from the samples disappeared. Samples were further washed with 80% methanol containing 0.1 M ammonium acetate, followed by centrifugation at 16 000 g for 10 min. The final wash was performed with 80% acetone followed by 10 min centrifugation at 16 000 g. The resulting pellet was air dried at room temperature for 20 min to remove residual acetone. For protein precipitation, a 1:1 ratio of phenol (pH 8.0) and SDS buffer was added to the air-dried pellet, mixed thoroughly, and incubated at room temperature for 5 min. The mixture was then centrifuged at 16 000 g for 7 min, after which the upper phenol phase was transferred to a fresh tube. To the collected supernatant, five times the volume of methanol containing 0.1 M ammonium acetate was added. Total proteins were precipitated overnight at –20 °C and pelleted the next morning by centrifugation at 16 000g for 10 min. The resulting pellet was carefully washed with 100% methanol followed by 80% acetone, air dried, and then dissolved in 0.1 M ammonium bicarbonate buffer for further analysis. In-solution digestion of the total protein was performed using 13 ng µl–1 trypsin in 10 mM ammonium bicarbonate containing 10% (v/v) acetonitrile.
Approximately 500 ng of the prepared samples was subjected to liquid chromatography/tandem mass spectrometry (LC-MS/MS), using a LTQ Orbitrap XL (Thermo Scientific), C-18 column, 15 cm length, for 180 minutes. The raw LC-MS/MS data for the three replicates were imported into Progenesis QI P version 2.0 (Nonlinear Dynamics Limited). All six tissues, with three biological replicates, were included in the analysis. The young leaf and fruit peel sample of DK was chosen as the reference, and all other ion intensity maps from the other grape lines were automatically aligned to the reference. Considering the good initial quality of the alignment, the dataset was not subjected to any further manual correction such as vector editing. Relative quantitation using Hi-3 was selected for automatic processing in the software. After successful alignment, no further filtering was applied for subsequent quantification steps in the software. Default settings, such as no protein grouping and quantitation from non-conflicting features, were used for protein building. For peptide identification, the Mascot protein search engine was used against the combined target-decoy database with the following parameters: enzyme: trypsin; maximum missed cleavage: three; variable modifications: deamidation, oxidation. Mascot search tolerance parameters were as follows: peptide tolerance, auto; fragment tolerance, auto; false discovery rate, <1%. Ion matching requirements were two fragments per peptide, five fragments per protein, and one peptide per protein. The results obtained are presented in Supplementary Tables S5 and S6.
Results
Profiling of sRNAs from three grape lines
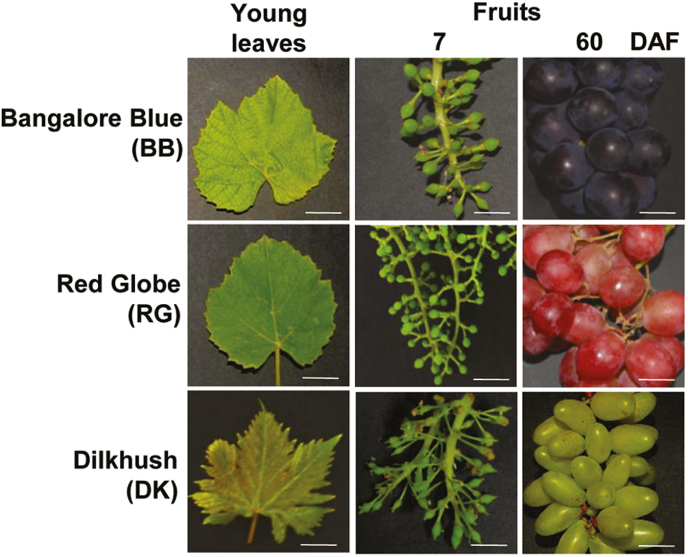
We performed sRNA profiling using three grape varieties commonly grown in the Indian subcontinent: Dilkhush (DK), a selection from Anab-e-shahi that produces berries with negligible anthocyanins; Bangalore Blue (BB), a variety well known for high anthocyanin levels; and Red Globe (RG), a variety that has medium levels of anthocyanins in between those of DK and BB (Fig. 1, Supplementary Fig. S1). Anthocyanin accumulation began 6 weeks before fruit ripening in these grape lines. Until ripening, berries of these lines were morphologically indistinguishable, other than slight variations in shape and size (Supplementary Fig. S1C). DK accumulated very high levels of flavonols, while BB accumulated the lowest amount of flavonols (Supplementary Fig. S1B). In order to understand the regulation of anthocyanin development in grape berries (Supplementary Fig. S1A), we carried out detailed staining of anthocyanin intermediates during various stages of berry development. Accumulation of proanthocyanin intermediates (Supplementary Fig. S2) and precursors of proanthocyanins was higher in various stages of berry development in BB than in the grape lines with reduced anthocyanin levels (Supplementary Fig. S3).
Fig. 1.
Phenotypes of the grape lines used in this study. DAF, Days after fertilization.
We profiled sRNAs from young leaves and fruit peels from all three grape lines. More than 60 million reads were obtained from deep sequencing of young leaf and fruit peel-derived sRNA libraries (Supplementary Table S1). For young leaves, the BB sample had abundant 21-nt sRNAs and a minor peak at 24-nt sRNAs, a profile similar to most dicots (Supplementary Fig. S4A) including grapes (Pantaleo et al., 2010; Picardi et al., 2010; Sun et al., 2015). However, in the DK and RG samples, there was a major peak at 21 nt and a prominent peak at 23 nt, with comparatively less accumulation of 24-nt sRNAs. Previous reports have recorded few grape varieties with a major peak at 23 nt instead of a prominent 24-nt peak (Belli Kullan et al., 2015). In our analysis, a major 23-nt peak was not observed in fruit peel tissues, indicating that their accumulation is spatiotemporally regulated (Supplementary Fig. S4B). In order to identify the source of the 23-nt peak in the RG and DK samples, we mapped all 23-nt sRNA reads to the grape genome. We identified a single 23-nt read that corresponded to the 23-nt peak at the genomic locus Chr17:65148–65171.
Similar to other plants, the 5′ terminals of the 21-nt sRNA reads a had prominent bias for the nucleotide U, and those of the 24-nt sRNAs had a bias for A (Supplementary Fig. S4C, D). In all three grape varieties, most sRNAs originated from intergenic regions, introns, and transposons (Supplementary Fig. S4E, F). Unlike most plant genomes, the grape genome has a significantly higher percentage covering introns and transposons (Supplementary Fig. S4G), as observed previously (Pantaleo et al., 2010). The distribution of sRNAs was also identical across the grape varieties and matched published reports for grapes (Pantaleo et al., 2010).
MYB-targeting miRNAs accumulate at higher levels in grape varieties with high anthocyanin levels
In order to understand the role of miRNAs in the development of grapes, we identified conserved and less-conserved miRNAs using bioinformatics tools. Grape lines exhibited high accumulation of well-conserved miRNAs such as miR159, miR166, miR168, and miR319 (Supplementary Table S2). Expression levels, measured as reads per million genome matching reads, varied significantly between the young leaves and fruit peels, as expected. However, there was minor variation between grape varieties in the abundance of most conserved miRNAs in a given tissue.
Analysis of miRNA abundance using miRProf (Moxon et al., 2008; Stocks et al., 2012) indicated that there were many miRNAs that were comparable among grape lines, such as miR168, miR166, and miR319. However, several miRNAs were differentially expressed between the grape lines. Among the miRNAs that accumulated substantially differently between lines, few were associated with secondary metabolic pathways. miR828, miR3632, miR403, and miR858 predominantly accumulated at higher levels in anthocyanin-rich BB compared with RG- and DK-derived tissues (Supplementary Fig. S5). These differentially expressed miRNAs have diverse functions, ranging from developmental transition and organogenesis to metabolism and nutrient deprivation (Chen, 2005; ().
Among the miRNAs that were differentially expressed between grape lines for a given tissue, miR828, miR858, miR156, miR159, and miR827 targeted MYB transcription factors, with TAPIR (Bonnet et al., 2010) scores ranging from 3 to 10 (Supplementary Fig. S6). The expression of miR827, miR156, and miR159 was not significantly different between grape peels, and no reads matching to the degradome were observed; this led us to narrow down to the other two MYB targeting miRNAs for further analysis.
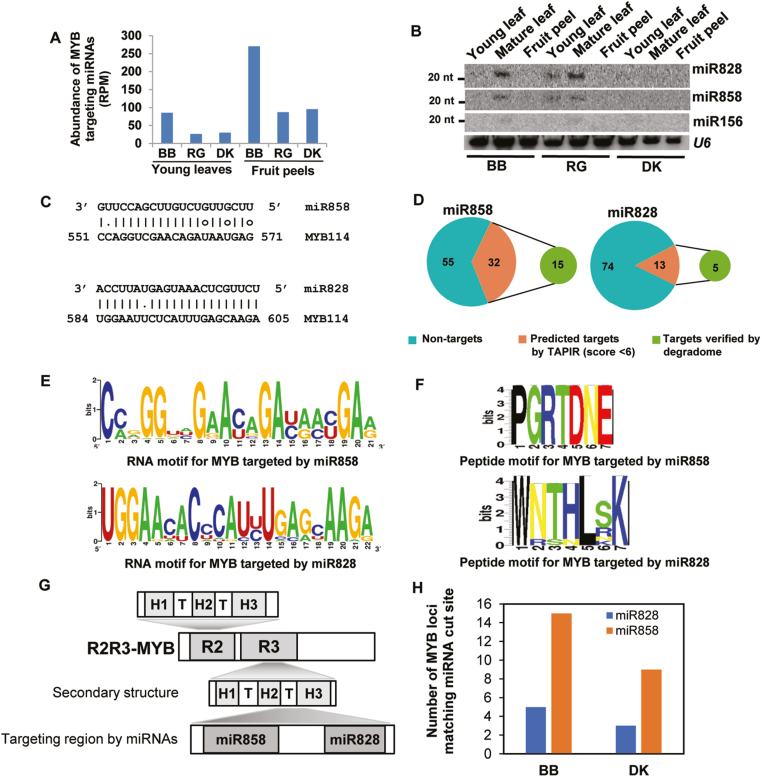
Among the selected miRNAs that are capable of targeting MYBs, miR858 has the most conserved sequence (Supplementary Fig. S7A). The 22-nt miRNA initiator of the TAS4 cascade silencing pathway, miR828, is well expressed, but is not a well-conserved miRNA (Supplementary Fig. S7B). miR828-mediated regulation of MYBs works at two different levels. miR828 itself targets multiple MYBs; in addition, a tasi-RNA derived from miR828 targeting of TAS4 non-coding RNA, named TAS4 D4-siRNA, also targets MYB RNAs (Rock, 2013). The absence of miR828 in related clade members indicates either a case of independent evolution in specific plants to target specific, well-conserved target regions, and/or the loss of this miRNA is related clades of plants. The combined accumulation of miR858 and miR828 was plotted for each sample in order to understand the extent of MYB targeting (Fig. 2A). We also performed northern blot analysis for the abundance of MYB-targeting miRNAs in the three grape varieties under study (Fig. 2B).
Fig. 2.
MYB targeting miRNAs accumulate in grape varieties with high anthocyanin levels. (A) Bar graph showing the combined expression of miR828 and miR858 among grape lines. RPM values are averages of replicates. (B) Northern blot analysis confirming the accumulation of MYB targeting miRNAs in grapes tissues. U6 was used as a loading control. The 20 nt position of the marker is indicated. (C) Alignment of MYB-targeting miRNAs, miR828 and miR858, with a representative MYB mRNA. Analysis was performed using TAPIR. (D) MYB-targeting abilities of miR828 and miR858 represented as pie charts. Numbers indicate the R2R3 MYB sequences considered for the analysis. (E) Conserved RNA and (F) peptide motifs of MYBs targeted by miR858 and miR828, respectively. (G) Schematic diagram of the MYB domains targeted by miRNAs. H1–3 are helices. (H) Bar graph showing the number of MYB loci whose cleavage was confirmed by degradome analysis.
miRNAs target unique motifs of MYB transcription factors
The grape genome encodes nearly 108 R2R3 MYBs (Matus et al., 2008; Hichri et al., 2011). Among the annotated 87 grape MYBs, nearly half are potential targets of miRNAs. R2R3 MYB members are predominantly represented among the MYBs that are targets of miRNAs. Among the miRNAs, miR858 can target 36% of MYBs and miR828 can target 15%, with a TAPIR target prediction score of 6 (Fig. 2C, D). Degradome analysis was performed using selected grape lines, which confirmed the MYB targeting abilities (Fig. 2D, H) of miR828 and miR858. Abundant degradome reads matched to the AGO1-mediated cleavage sites of many R2R3 MYBs, such as VvMYB114, WER, and MYB3, confirming that they are targeted by multiple miRNAs (Supplementary Fig. S8).
These miRNAs target MYB mRNAs at unique RNA sequence motifs coding for specific protein domains (Fig. 2E, F). An RNA motif coding for the R3 repeat of MYBs was a major region where miRNAs targeted these RNAs. Within R3, conserved motifs coding for helix 3 are targeted by miRNAs (Fig. 2G). The RNA motifs targeted by miRNAs do not overlap with each other. The protein domains coding for these motifs are conserved only among MYB family members. Our results are similar to predictions in apple and peach (Xia et al., 2012; Zhu et al., 2012) and reinforce the observation that MYBs are routinely targeted by multiple miRNAs.
Differential accumulation of MYB RNAs and proteins between grape cultivars
We performed whole-genome strand-specific RNA-seq to understand the dynamics of miRNA-mediated regulation of MYBs. We used two different tissues (young leaves and fruit peels) of the three grape lines for this analysis. As expected, most of the tissue-specific RNAs were expressed in all three grape varieties (Supplementary Fig. S9A, B), confirming the similarity in their gene expression patterns. However, in leaves and fruit peel samples, approximately 300–500 mRNAs were expressed differentially with a cut-off of log2(fold change) >2. A full list of the differentially expressed mRNAs deduced from this analysis is provided in Supplementary Datasets S1 and S2).
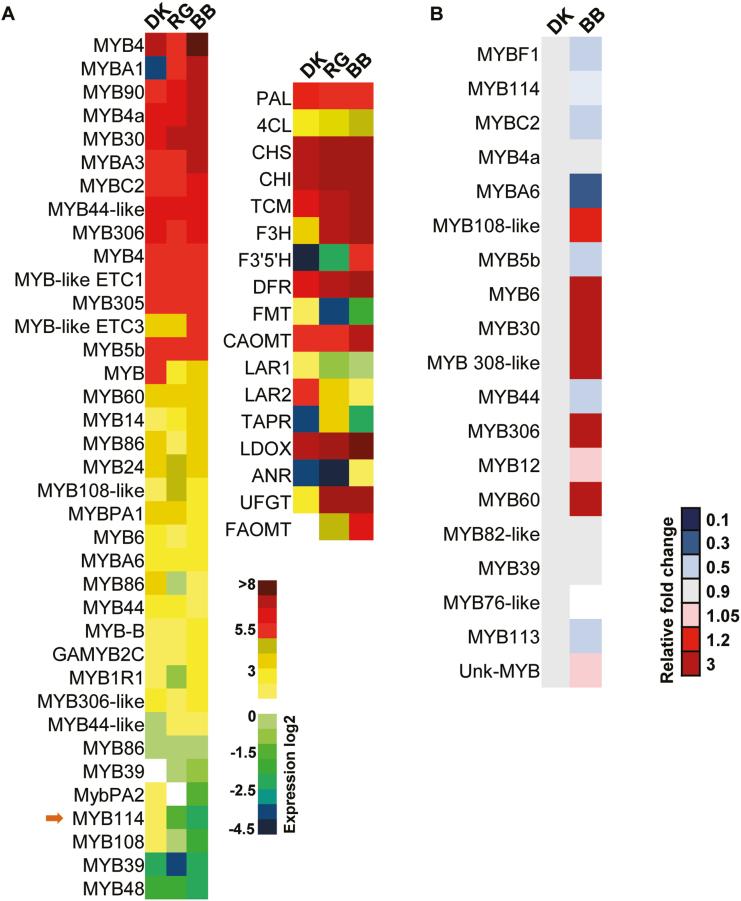
Among the differentially expressed genes, several MYBs, including targets of miRNAs, accumulated at significantly different levels (P=0.05) between the grape lines (Supplementary Datasets S1 and S2). The colored grape varieties BB and RG had low expression of specific MYBs, such as VvMYB114, VvMYB48, VvMYB39, and VvMYB108. It is important to note that among the differentially expressed RNAs, only VvMYB114 is targeted by both miR828 and miR858. In addition to MYBs, genes involved in the phenylpropanoid pathway were significantly differentially expressed between the grape lines. Several key genes, such as UFGT (UDP glucose flavonoid-3-O-glucosyltransferase) and FAOMT (flavonoid 3′,5′-methyltransferase), were expressed at higher levels in BB and RG, which also have a higher accumulation of anthocyanins (Fig. 3A). RNA-seq analysis was verified by RT–PCR analysis for selected members of the phenylpropanoid pathway (Supplementary Fig. S9 and Table S4). Taken together, these results indicate surprisingly diverse expression patterns for several key genes previously reported to be essential for anthocyanin accumulation. Most MYB genes that are targeted by miRNAs exhibited negative correlation in expression in the anthocyanin-rich BB line, as expected.
Fig. 3.
MYB RNAs accumulate differentially between grape lines. (A) Differential expression analysis (log2) of MYB gene family members and phenylpropanoid pathway genes. (B) Comparative expression levels of the MYB proteins across fruit peel tissues of grapes.
In order to verify whether RNA-seq analysis can indeed be correlated with the corresponding accumulation of proteins in these lines, we performed a high-throughput proteome analysis using LC-MS/MS, specifically optimized for the grape tissues used in this study. Similar to the results of RNA-seq, in this analysis most proteins displayed comparable levels of abundance, while few candidates showed differential accumulation (Supplementary Fig. S10A). Several MYB proteins and phenylpropanoid pathway enzymes were differentially expressed between grape lines (Supplementary Table S5). Many proteins involved in the phenylpropanoid pathway also accumulated at different levels in the different grape lines, and these are likely downstream of key MYB regulators (Supplementary Fig. S10B). This result was anticipated, since MYBs regulate the phenylpropanoid pathway at multiple levels, and any deviation in the accumulation of a single MYB could potentially alter the expression patterns of multiple phenylpropanoid pathway genes. Most strikingly, VvMYB114 and other MYBs had lower accumulation in the colored grape lines BB compared with DK (Fig. 3B), consistent with the high sRNA abundance and reduced RNA expression. Thus, the expression pattern of MYBs in terms of both RNA and protein levels was negatively correlated with the expression of miRNAs in grape lines with high anthocyanin accumulation.
miR828 initiates RDR6-dependent secondary silencing in MYB114 loci
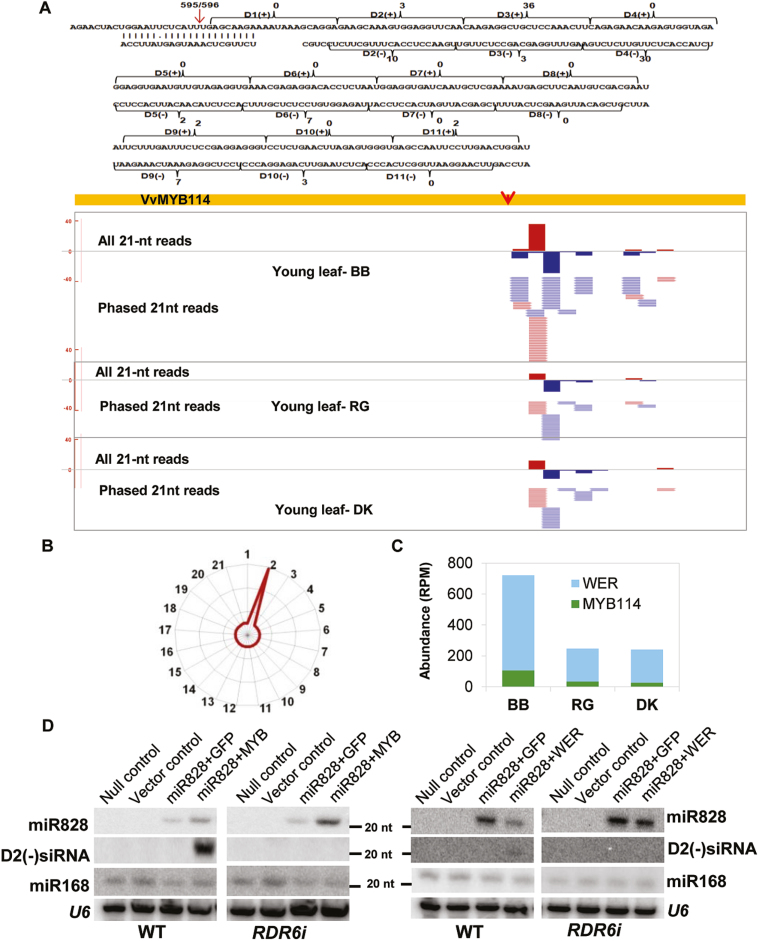
It has been well documented that MYBs regulate the expression of other MYBs at the transcriptional and post-transcriptional levels (Ramsay and Glover, 2005). Among the miRNAs targeting MYBs, miR828 is a 22-nt miRNA that is capable of targeting many MYBs, including MYB114 and WER. It has been shown independently that miRNAs 22 nt in length are capable of initiating secondary siRNAs in their target loci that are capable of targeting other members of the target gene family (Chen et al., 2010; Cuperus et al., 2010; Shivaprasad et al., 2012a). miR828 is highly conserved across dicots and initiates the TAS4 secondary silencing pathway. We hypothesized that miR828-targeted MYBs might be sources of secondary siRNAs that might modulate the expression of other regulators of anthocyanin biosynthesis, including MYBs.
We predicted all secondary silencing loci among grape datasets (Moxon et al. 2008). Many conserved TAS loci, such as TAS3 and TAS4, were abundantly represented in all grape varieties, as expected (Table 1). There were unique secondary loci among the grape lines; for example, VvMYB114 was a strong secondary silencing locus in BB when compared with DK (Fig. 4A). BB had a greater than 4-fold higher abundance of MYB114-derived secondary siRNAs than DK. Although the TAS4 pathway is well conserved across dicots, VvMYB114 was not identified previously as a phased secondary siRNA locus. The MYB114 locus produced siRNAs that are characteristic of TAS loci in Arabidopsis and other dicots. For example, the phasing register (Fig. 4B) and size of secondary siRNAs fulfilled criteria set for such loci in model and non-model plants. Other secondary loci derived from MYBs, such as WER, also accumulated varied levels of secondary siRNAs across the grape lines and tissues studied (Fig. 4C).
Table 1.
List of secondary silencing loci across the three grape lines
| Line | Locus ID | Chromosome | Start position | End position | Number of phased sequences | P |
|---|---|---|---|---|---|---|
| BB | TAS3 | 5 | 359 230 | 359 481 | 9 | 0.004993 |
| TAS4 | 14 | 21 607 855 | 21 608 106 | 17 | 1.46E-13 | |
| MYB114 | 9 | 1 161 821 | 1 162 720 | 11 | 2.00E-08 | |
| WER | 17 | 9 703 392 | 9 704 368 | 15 | 1.14E-13 | |
| RG | TAS3 | 5 | 359 237 | 359 488 | 10 | 4.68E-05 |
| TAS4 | 14 | 21 607 855 | 21 608 106 | 16 | 6.50E-11 | |
| MYB114 | 9 | 1 161 821 | 1 162 720 | 6 | 7.87E-07 | |
| WER | 17 | 9 703 392 | 9 704 368 | 10 | 4.65E-09 | |
| DK | TAS3 | 5 | 359 237 | 359 488 | 7 | 0.0014 |
| TAS4 | 14 | 21 607 876 | 21 608 127 | 16 | 1.63E-11 | |
| MYB114 | 9 | 1 161 821 | 1 162 720 | 6 | 3.24E-07 | |
| WER | 17 | 9 703 392 | 9 704 368 | 11 | 2.65E-10 |
Fig. 4.
MicroRNA 828 initiates RDR6 dependent secondary silencing in MYB114 locus. (A) Mapped phased reads matching the MYB114 locus. Abundance of individual phased reads for each position (D1–D11) has been mentioned. The miR828 target site is indicated with a red arrow. A genome browser screenshot of this locus with phased sRNAs matching from three grape lines is shown in the bottom panel. (B) Phasing register of the secondary siRNA reads produced from the VvMYB114 locus. (C) Bar graph displaying the abundance of secondary siRNAs derived from MYB114 and WER loci among grape lines. (D) Northern analysis to confirm the involvement of RDR6 in the generation of phased secondary siRNAs. MYB114 (right) or WER (left) was infiltrated into N. benthamiana WT and RDR6i lines, either with empty binary vector or with miR828. U6 and miR168 serve as controls.
Since MYB114 was a novel secondary siRNA locus, we explored whether the biogenesis of secondary siRNAs derived from this RNA required silencing-associated genes such as RDR6 (Yoshikawa et al., 2005). Grape is an unusual model system for which tools such as mutant lines and T-DNA insertion lines are not readily available. In addition, the transformation of the grape lines used in this study has not been achieved in spite of efforts from the host laboratory. Hence, we used tobacco as a model system to study whether RDR6 is indeed required for the production of secondary siRNAs from VvMYB114 loci targeted by miR828. We cloned miRNAs and several of their target MYBs, such as MYB114 and WER, in binary vectors (Supplementary Fig. S11). We introduced miR828 and MYB114 or WER genes to WT N. benthamiana and an RDR6-silenced line (Schwach et al., 2005). Similar experiments have been carried out previously to uncover the role of RDR6 in miRNA-mediated silencing (Shivaprasad et al., 2012a; Chenna et al., 2018).
miR828 was extremely abundant in transiently infiltrated tobacco leaves, while levels of endogenous tobacco miR828 accumulated at negligible, often below detection levels, as seen in control uninfiltrated leaves. WT plants, where RDR6 is active, produced secondary siRNAs from MYB114 loci abundantly, while RDR6i plants were unable to generate secondary siRNAs. Multiple probes looking at several secondary siRNAs did not produce any positive signal in RDR6i plants (Fig. 4D). Similar results were obtained for another miR828 target MYB, WER, which exhibited all the characteristic features of secondary loci (Supplementary Fig. S12). These results indicate that cascade silencing in grape is an RDR6-dependent cascade silencing mechanism, similar to that identified in other, previously studied plants.
MYBs targeted by miRNAs are repressors of anthocyanin accumulation
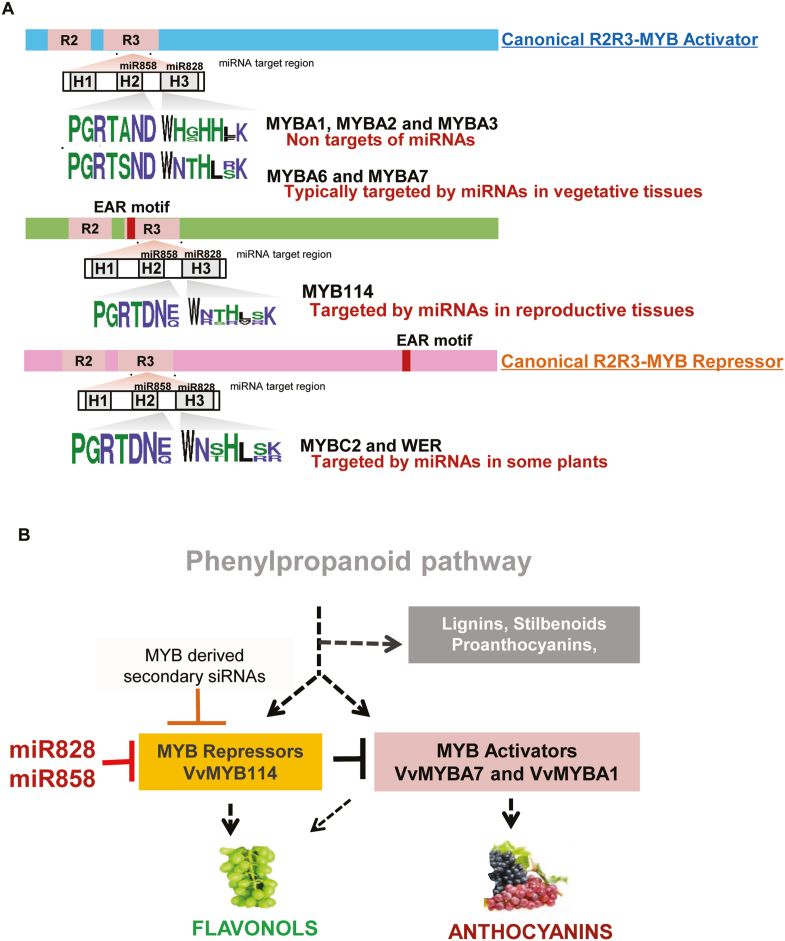
The means by which miRNAs efficiently target RNAs coding for MYBs has been studied extensively in several plant species. Many such interactions resulted in secondary silencing (Hsieh et al., 2009), such as those observed in this study. However, it is not known why MYB transcription factors that are generally known to promote anthocyanin accumulation are also targets of miRNAs. Several landmark studies have proposed that some MYBs act as repressors of anthocyanin accumulation (Jin et al., 2000; Aharoni et al., 2001; Deluc, 2006; Matsui et al., 2008; Albert et al., 2011, 2014b, 2015). Since VvMYB114 is a novel candidate that accumulates at varied levels in different grape lines, we hypothesized that MYB114 might be a repressor of anthocyanin accumulation. This prediction was mainly based on the observation that MYB114 is targeted at higher levels in anthocyanin-rich grape lines and its RNA and protein accumulated at lower levels. Using phylogenetic analysis, we found that VvMYB114 falls into a clade with other known MYB repressors. A characteristic feature of MYB repressors is the presence of a unique EAR motif at the C-terminal end (Albert, 2015). Unlike canonical repressor MYBs, VvMYB114 had a consensus LxLxL-type active repressor EAR motif in its N-terminal end. Among repressor transcription factors, the presence of an EAR motif is more common at the N-terminal end than at the C-terminal end (Kagale and Rozwadowski, 2011). It is important to note that peach MYB114, which was studied previously as a promoter of anthocyanin (Yao et al., 2017), is in a different clade along with MYB activators such as MYBA1, and not with VvMYB114 (Supplementary Fig. S13).
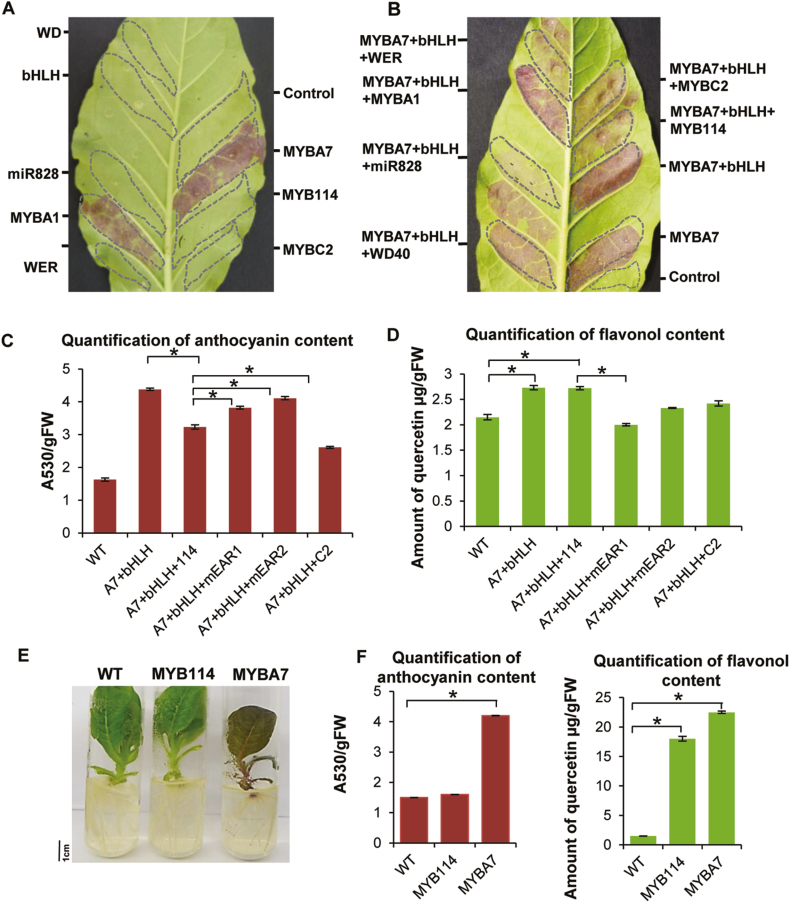
We introduced multiple MYBs that might act as activators or repressors of anthocyanins, based on previous reports, to tobacco. Well-known MYB activators such as VvMYBA1 and VvMYBA7 (Matus et al., 2008, 2017) produced abundant anthocyanins when introduced into tobacco, while MYBs such as VvMYB114 and VvMYBC2 did not result in the accumulation of anthocyanins (Fig. 5A, Supplementary Fig. S14A, B). This result indicates that VvMYB114 is not a promoter of anthocyanin accumulation, and is likely a repressor class of MYB. We further explored whether VvMYB114 is indeed a repressor that can potentially alter and compete with the existing activator MYB:bHLH:WD40 complex. We co-infiltrated VvMYB114 along with activator MYBs and other known repressors (Fig. 5B) into tobacco leaves. MYBC2, a repressor of anthocyanin biosynthesis, reduced anthocyanin levels when co-infiltrated with the anthocyanin activator VvMYBA7. Interestingly, VvMYB114 also reduced anthocyanin accumulation when co-infiltrated with the MYB activator MYBA7 (Fig. 5B). This reduction was observed along with the co-expression of bHLH, a cofactor in the MYB complex. These results establish that VvMYB114 is a novel repressor of anthocyanin accumulation in grapes. It is not clear exactly how VvMYB114 acts as a repressor, although it has the canonical EAR motif. Albert et al. (2015a) and others have proposed that MYB repressors might work in several ways. These repressor MYBs have the potential to compete with MYB activators for c-factors such as bHLH and WD40 (Hartmann et al., 2005; Gonzalez et al., 2008; An et al., 2012; Albert et al., 2015). Additionally, repressor MYBs have been implicated in destabilizing activator MYB complexes (Albert, 2015).
Fig. 5.
VvMYB114 is a repressor of anthocyanins to promote flavonol accumulation. (A) Transient expression of MYB-expressing constructs in tobacco introduced through agroinfiltration. MYBA7 and MYBA1 infiltration led to high accumulation of anthocyanin, while MYB114 did not alter anthocyanin content. Dotted lines indicate agroinfiltrated regions. (B) Activator MYBA7 with bHLH partners was infiltrated with and without MYB114. MYBC2, which reduces anthocyanin resulting from MYBA7, served as a control for a repressor. (C) Estimation of anthocyanin in leaves transiently expressing MYBs and WT plants. Data are the mean ±SE of three biological replicates. *P<0.005 (Student’s t-test). Two mutants of the EAR motif in MYB114 were used in this experiment. (D) Estimation of flavonols in leaves transiently expressing MYBs in WT plants. Data are the mean ±SE of three biological replicates. *P<0.005 (Student’s t-test). (E) Phenotypes of tobacco plants stably expressing grape MYBA7 and MYB114. (F) Estimation of anthocyanin and flavonol contents from transgenic leaf tissues overexpressing MYB114 and MYBA7. WT tobacco was used as a control. Data are the mean ±SE of three biological replicates (three independent transgenic lines). *P<0.005 (Student’s t-test).
In order to understand the specific role of repressor MYBs with an N-terminal EAR motif in regulating secondary metabolites, we constructed two EAR mutants in which the residues LxLxL in the EAR motif were mutated to LxAxL (mEAR1) and LxIxL (mEAR2), respectively. The EAR motif in VvMYB114 is at the N-terminal, a region that is involved in interaction with bHLH. We constructed the mEAR2 mutant to understand the significance of lysine in the middle position (LxLxL), specifically to see whether bHLH interaction and repression activity are coupled. We co-infiltrated MYB114 and the MYB114 EAR mutants along with MYBA7 and bHLH complex into WT tobacco leaves (Fig. 5C, D). The mEAR1 mutant showed increased anthocyanin content and reduced flavonols compared with WT MYB114, suggesting that VvMYB114 is likely to be a repressor of anthocyanin due to the presence of the N-terminal EAR domain. VvMYB114 repressed anthocyanin accumulation, indicating that it is a crucial factor in the phenylpropanoid pathway that is capable of shifting the final product even in tissues that are producing abundant anthocyanins. To further understand the role of the N-terminal EAR motif in changing the flux for the flavonol pathway, we estimated the flavonol content in the same tissues (Fig. 5D). The mEAR2 mutant showed increased anthocyanin content compared with WT MYB114 but did not show significantly reduced flavonol levels, suggesting the importance of lysine for its anthocyanin repression activity. Based on these findings, we hypothesized that VvMYB114 might be directly or indirectly activating the promoters of genes involved in flavonol biosynthesis.
Overexpression of VvMYB114 shifts metabolism to flavonol biosynthesis
To understand the role of VvMYB114 in the flavonol pathway, we generated transgenic plants expressing MYB114 and measured anthocyanins and flavonols in these plants (Fig. 5E, F; Supplementary Fig. S15). The activator VvMYBA7 served as a control. Surprisingly, there was high accumulation of flavonols in MYB114-expressing transgenic plants, while MYBA7-expressing plants were enriched in both anthocyanins and flavonols (Fig. 5E, F). This indicated that VvMYB114 induces flavonoid biosynthesis in transgenic plants and might be a promoter of flavonol biosynthesis from dihydroxyflavonol intermediates. Several activator MYBs act as upstream factors to promote the production of both anthocyanins and flavonols. VvMYBA7 promoted the phenylpropanoid pathway to activate production of both anthocyanins and flavonols, clearly indicating that it is an upstream regulator of the phenylpropanoid pathway. On the other hand, VvMYB114 promoted the production of only flavonols such as quercetin, indicating that it is a regulator specific to the branch point of the phenylpropanoid pathway that regulates flavonols (Supplementary Fig. S15C). The qRT–PCR analysis of phenylpropanoid pathway genes from leaf tissues showed that MYB114 induced the up-regulation of genes involved specifically in flavonol production from dihydroxyflavonol intermediates, such as flavonoid 3′-monooxygenase (F3MO), to quercetin and myceretin derivatives (Supplementary Fig. S15C). It is intriguing and worth speculating how exactly a MYB that has the EAR motif for transcriptional repression represses some promoters while promoting expression from a few others. It is possible that the up-regulation of flavonol-promoting genes is not via the direct action of MYB114, but through feedback regulation or other mechanisms that are still unknown. However, these results collectively indicate that the expression of VvMYB114 in the absence of miRNA targeting in specific grape lines results in abundant accumulation of flavonols.
Discussion
In this study, we show that targeting of MYBs by miRNAs and siRNAs has the potential to specifically alter the end-product of the phenylpropanoid pathway in grapes. Although it was well known that several MYBs from various model and non-model plants were targets of miRNAs and tasi-RNAs, it was not known how exactly such targeting might influence changes in the anthocyanin and flavonol levels in reproductive tissues such as berries. Our results indicate that miRNA-mediated MYB regulation shifts the pathway towards either anthocyanins or flavonols, and match predictions made previously (Rock, 2013). We anticipate that these results will help us understand how the phenylpropanoid pathway is regulated at the molecular level to fine-tune the production of specific metabolites in specific plant tissues.
miRNAs and evolution of their target MYBs
In many plants rich in secondary metabolites there is a high representation of R2R3-type MYBs as well as abundant expression of miRNAs that have the potential to target them. It has previously been proposed that this correlation might have been a result of co-evolution. Indeed, many miRNAs that target MYBs reside in close proximity to their targets (Allen et al., 2004; Du et al., 2012a).
Several plants that have colored fruits were specifically favored during domestication (Aharoni et al., 2004; Feng et al., 2010). It is interesting to note that plants with anthocyanin-rich fruits have higher numbers of MYB genes; this has been reported for apple (Xia et al., 2012), grape (Azuma et al. 2008; this report), peach (Rahim et al., 2014), soybean (Du et al., 2012b), maize (Du et al., 2012a), and strawberry (Lin-Wang et al., 2010). In many genera such as Medicago, Citrus, and Aquilegia, by contrast, the number of MYB genes is low (Martin and Paz-Ares, 1997; Du et al., 2012a). It has been previously proposed that the enrichment of MYB genes in certain species is likely due to selection for colored fruits during domestication (Dias et al., 2003; Feller et al., 2011). In agreement with this hypothesis, anthocyanin-rich fruit plants such as grape and apple encode some of the highest numbers of MYBs (Espley et al., 2007; Allan et al., 2008). This type of selection might result in targeted evolution of MYBs that regulate anthocyanin production along with specific miRNAs and siRNAs to regulate their expression. Most fruit crops such as grapes depend on coloration for fruit dispersal. It is possible that the domestication of grapes might also have led to artificial evolution of MYBs and miRNAs to induce the production of specific secondary metabolites. Artificial selection might have also worked due to other animals, in addition to humans, involved in fruit dispersal. For example, birds are well known to identify black and red-colored fruits more readily than those that are not very colorful (Duan et al., 2014; Renoult et al., 2014).
Identification of a novel repressor MYB in grape
A major target of miRNAs in our analysis is a relatively unknown MYB gene named VvMYB114. MYB114 expression was significantly lower in anthocyanin-rich grape lines due to targeting by miRNAs and potentially by secondary siRNAs, and such an outcome was likely reinforced due to RDR6-dependent cascade silencing. VvMYB114 has a classical LxLxL-type EAR motif that has been previously implicated in transcriptional repression (Albert et al., 2014a). The EAR motif was one of the first repressor motifs to be discovered in plants a decade ago and is present in more than 20% of transcriptional repressors in plants. The EAR motif has been implicated in repressor activity through its interaction with TOPLESS, a cofactor of histone deacetylase (HDAC) that is responsible for chromatin remodeling (Kazan, 2006; Kagale and Rozwadowski, 2011). However, this EAR motif is in the N-terminal end of VvMYB114, unlike other canonical repressor MYBs such as MYBC2 and WER, in which EAR motifs are present in the C-terminal end (Fig. 6A). However, in many well-studied transcriptional repressors, such as an AUX/IAA protein, NINJA, and AFP, EAR motifs are observed in the N-terminal end (Kagale and Rozwadowski, 2011). It appears that activator MYBs are not active targets of miRNAs, unlike repressor MYBs in reproductive tissues. The miRNA target regions in helix 3 of MYB repressors such as MYB114 and WER are slightly different in both nucleotide and amino acid sequence from those of MYB activators, indicating that the presence of an EAR motif and a specific RNA motif coding for helix 3 might have co-evolved in MYB repressors to ensure their targeting by sRNAs to promoter anthocyanin accumulation. An alternative possibility is that VvMYB114-like repressors alone co-evolved with miRNAs to specifically activate flavonols. Our results do not exclude the presence of other layers of MYB regulation. Further studies are required to understand why this targeting prominently selects MYB repressors in grapes and other plants.
Fig. 6.
A proposed model for the regulation of secondary metabolites in grapes. (A) Schematic representation of activator and repressor MYB classes in grapes. VvMYB114 is targeted by miRNAs due to unique amino acids in miRNA target regions. It has a typical repressor EAR motif (red box) at the N-terminal end, unlike most repressors. (B) A proposed model for the regulation of secondary metabolites in grapes and likely in other plants.
In our study, MYB activators such as MYBA7 and MYBA1 acted as inducers of both anthocyanins and flavonols. This observation indicates that MYB activators are upstream of dihydroxyflavonol conversion and have the potential to enhance accumulation of both flavonols and anthocyanins. While the BB line had negligible levels of flavonols, the RG line had appreciable levels of both anthocyanins and flavonols. In the DK line, there was very low accumulation of activator MYBs, while the MYB114 repressor was the most abundant. In transgenic tobacco expressing MYB114 there was a higher accumulation of flavonols. Taken together, these results suggest that MYB114 is a key regulator mediating the production of flavonols by converting dihydroxyflavonols to flavonols, thereby shifting the phenylpropanoid pathway to flavonol biosynthesis. In line with this suggestion, regulators of flavonol biosynthesis from dihydroxyflavonols such as F3MO were highly up-regulated in MYB114-expressing transgenic plants. VvMYBF1 has been reported to induce flavonols in grape tissues by inducing the transcription of the flavonol synthase (FLS) gene (Czemmel et al., 2009). However, VvMYB114 induces flavonols by inducing the expression of FLS and F3MO, a regulator downstream of FLS. Transgenic plants expressing MYB114 showed significant induction of FLS and F3MO expression. It will be of great interest to understand exactly how MYB114 promotes flavonol pathway genes and directly or indirectly negatively regulates anthocyanin pathway genes.
Fine-tuning of MYB expression in grape tissues
It is also intriguing that many MYBs are targeted by two classes of sRNAs, namely, miRNAs and siRNAs, in plants. This raises the question of why this regulation requires a network of multiple miRNAs and multiple siRNAs to target MYBs redundantly. The answer might not be simple. It appears that such a system arose due to feedback systems that commonly operate in MYB-mediated regulation. Having multiple means to regulate a single MYB RNA might ensure robust targeting to permit the right combination of MYBs to be expressed in a given tissue. When we analyzed MYBs that are targeted by miR828 and miR858, we found at least five MYBs that are common targets of both miRNAs: MYB114, MYB82, MYBCS1, MYBPA1, and MYBPA2. We studied MYB114 in some detail; however, other candidates are equally interesting, since all five of these MYBs are expressed at higher levels in flavonol-rich DK fruit peels (Fig. 3A, Supplementary Dataset S1), strongly correlating with reduced miRNA levels in this tissue. MYBPA1 and MYBPA2 promote ANR and LAR1, candidates that are involved in the accumulation of precursors of flavonoids (which include flavonols, proanthocyanins, and anthocyanins). The functions of MYBCS1 and MYB82 are relatively unknown; however, MYBCS1 has an N-terminal EAR motif, similar to MYB114. Co-targeting by multiple miRNAs, in addition to strengthening silencing at two different levels, provides robustness and probably spatiotemporal regulation if the miRNAs are tissue-specific. In this case, all five common targets achieve a common goal of producing higher levels of flavonols collectively, as in the DK line, thus strengthening the idea that co-targeting by miRNAs achieves robustness.
Targeting of single mRNAs by both miRNAs and multiple phased siRNAs is not uncommon in plants. In fact, most phased pathway genes, both coding and non-coding, are targeted by multiple miRNAs and multiple siRNAs. A common feature of phased siRNAs is that the initiator miRNA is capable of targeting multiple targets of a single gene family to generate a pool of target-derived siRNAs that ultimately target members of the same family or, rarely, other targets. In tomato, secondary siRNAs derived from nucleotide binding site-leucine rich repeat (NBS-LRR) RNAs targeted other regulators of the resistance pathway, so that a single miRNA ensures the silencing of multiple targets that can be reversed by a single switch when needed (Shivaprasad et al., 2012a). Detailed functional analysis of secondary siRNAs, including their targeting abilities in multiple cascade silencing pathways in plants, is likely to answer these questions.
In our analysis, we also observed up-regulation of miR828 by certain MYBs, such as MYB114 and MYBA1 (Supplementary Fig. S14C), indicating that MYB expression is constantly regulated through feedback mechanisms. Such a feedback loop involving miR828 has also been identified in Arabidopsis (Hsieh et al., 2009). In grape, induced expression of miR828 by MYBs such as MYB114 might provide robust secondary targeting. In such a scenario, targeting of MYBs by miRNAs ultimately results in a reduction in the expression of inducer miRNA.
Surprisingly, we observed that the abundance of miRNAs that target R2R3 MYBs responsible for anthocyanin and flavonol accumulation in reproductive tissues was very high in some plants, while related species within the clades and families expressed these miRNAs at lower levels. Most of these species produce high levels of anthocyanins or flavonols, either naturally or as a result of domestication-associated selection. Based on the observations made here for grape lines, it is tempting to speculate that in other species of plants too, miRNAs might be targeting MYB114-like MYB genes or related species-specific members of MYBs that regulate branch points of the phenylpropanoid pathway (Fig. 6B). It is also possible, as described above, that a strong selection for colorful fruits might have led to targeting of specific MYBs to promote the production of specific metabolites in some plants, while retaining the ability to produce flavonols in others.
In multiple plant species, spectacular increases in anthocyanin expression have been achieved. It is also well known that in some plants it is possible to enhance the accumulation of flavonols. Our results open up a new avenue where it is possible to regulate secondary metabolites by specifically expressing activator or repressor MYBs or through the engineering of specific miRNAs. This understanding might pave the way for further fine-tuning of flavonol and anthocyanin synthesis in crops and other plants, to promote nutraceutical fortification.
While this manuscript was under review, Bonar et al. (2018) reported a strong correlation between high anthocyanin content in potato tuber and the abundance of miR828, in agreement with the findings of the present study. They propose that the target of miR828 is a MYB repressor.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Estimation of total anthocyanins and flavonols in fruit peels of grape lines used in this study.
Fig. S2. DMACA staining of berries at various developmental stages.
Fig. S3.TBO staining of berries at various developmental stages.
Fig. S4. Small RNA profiling across selected grape lines.
Fig. S5. Differential expression of known microRNAs across grape lines.
Fig. S6. Phylogenetic tree of MYBs from grapes and targeting abilities of miR828 and miR858.
Fig. S7. Sequence variations of miR828 and miR858 among plants.
Fig. S8. Schematic diagram of miRNA-mediated cleavage in MYB mRNAs through degradome analysis.
Fig. S9. MYB RNAs accumulate differentially between grape lines.
Fig. S10. MYB proteins accumulate differentially between grape tissues and lines.
Fig. S11. Linear diagrams of the constructs used in this study.
Fig. S12. Analysis of the secondary silencing cascade in the WER locus.
Fig. S13. Phylogenetic tree of MYB activators and repressors from various plant species.
Fig. S14. Quantification of anthocyanin content and RT–PCR from samples transiently expressing MYB genes and miRNAs.
Fig. S15. Phenotypes of MYB-expressing transgenic plants.
Table S1. Summary of small RNA statistics.
Table S2. Abundance and targets of conserved miRNAs from grape lines.
Table S3. List of oligonucleotides used as probes.
Table S4. Details of primer pairs used for cloning and qRT–PCR.
Table S5. Mass spectrometric analysis of total proteins from reproductive tissues of the DK and BB grape lines.
Table S6. Mass spectrometric analysis of total proteins from reproductive tissues of the DK and RG grape lines.
Dataset S1. Differentially expressed genes among the fruit peel samples.
Dataset S2. Differentially expressed genes among grape young leaf samples.
Acknowledgements
The authors acknowledge the access to proteomics, radiation laboratory, central imaging, greenhouse, and sequencing facilities provided by the host institute. sRNA and RNA-seq was performed at Genotypic Technologies, Bangalore. Thanks go to Prof. David Baulcombe for RDR6i seeds, Prof. K. Veluthambi for discussions and for the pBIN19 vector and Agrobacterium strain LBA4404 (pSB1), Dr Vikas Kumar for guidance with LC/MS-MS, and N. D. Sunitha for comments. Thanks are due to Vikram Jha, Umashree, and Anupama Nair for help with the subcloning. We thank the scientists at IIHR, Bangalore and the farmers for the grape lines. PVS acknowledges support from the Ramanujan Fellowship (SR/S2/RJN-109/2012; Department of Science and Technology, Government of India). PVS’s laboratory is supported by National Center for Biological Sciences, Tata Institute for Fundamental Research core funding and a grant (BT/PR12394/AGIII/103/891/2014) from the Department of Biotechnology, Government of India. CS acknowledges a research fellowship from the Department of Biotechnology.
Data deposition
Small RNA sequencing data for grape lines are available in Gene Expression Omnibus (GEO) series GSE107907, from GSM2883166 to GSM2883177 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE107907). RNA sequencing data for the grape lines are available in GEO series GSE107905, from GSM2883154 to GSM2883165 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE107905). Degradome data are available in GSE118701 series (from GSM3336816 to GSM333619) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE118701). Accession numbers of the MYB genes used for cloning can be found in Supplementary Table S4 and those of other MYB sequences can be found in Supplementary Tables S5 and S6. The raw spectrum files obtained from the LC-MS/MS are available in the GNPS MASSIVE database (accession number: MSV000083722, ftp://MSV000083722@massive.ucsd.edu).
Author contributions
PVS designed all experiments, discussed results, and wrote the manuscript. VT performed most of the experiments and co-wrote the manuscript. CS performed bioinformatic analysis. AN helped with proteome analysis. AP performed degradome sequencing.
References
- Abeynayake SW, Panter S, Mouradov A, Spangenberg G. 2017. A high-resolution method for the localization of proanthocyanidins in plant tissues. Plant Methods 17, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharoni A, De Vos CH, Wein M, Sun Z, Greco R, Kroon A, Mol JN, O’Connell AP. 2001. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. The Plant Journal 28, 319–332. [DOI] [PubMed] [Google Scholar]
- Aharoni A, Giri AP, Verstappen FW, Bertea CM, Sevenier R, Sun Z, Jongsma MA, Schwab W, Bouwmeester HJ. 2004. Gain and loss of fruit flavor compounds produced by wild and cultivated strawberry species. The Plant Cell 16, 3110–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbergenov R, Si-Ammour A, Blevins T, et al. . 2006. Molecular characterization of geminivirus-derived small RNAs in different plant species. Nucleic Acids Research 34, 462–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert NW. 2015. Subspecialization of R2R3-MYB repressors for anthocyanin and proanthocyanidin regulation in forage legumes. Frontiers in Plant Science 6, 1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert NW, Davies KM, Lewis DH, Zhang H, Montefiori M, Brendolise C, Boase MR, Ngo H, Jameson PE, Schwinn KE. 2014a. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. The Plant Cell 26, 962–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert NW, Davies KM, Schwinn KE. 2014b. Repression – the dark side of anthocyanin regulation? Acta Horticulturae 1048, 129–136. [Google Scholar]
- Albert NW, Griffiths AG, Cousins GR, Verry IM, Williams WM. 2015. Anthocyanin leaf markings are regulated by a family of R2R3-MYB genes in the genus Trifolium. New Phytologist 205, 882–893. [DOI] [PubMed] [Google Scholar]
- Albert NW, Lewis DH, Zhang H, Schwinn KE, Jameson PE, Davies KM. 2011. Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. The Plant Journal 65, 771–784. [DOI] [PubMed] [Google Scholar]
- Allan AC, Hellens RP, Laing WA. 2008. MYB transcription factors that colour our fruit. Trends in Plant Science 13, 99–102. [DOI] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC. 2005. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221. [DOI] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Sung GH, Spatafora JW, Carrington JC. 2004. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nature Genetics 36, 1282–1290. [DOI] [PubMed] [Google Scholar]
- An XH, Tian Y, Chen KQ, Wang XF, Hao YJ. 2012. The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation. Journal of Plant Physiology 169, 710–717. [DOI] [PubMed] [Google Scholar]
- Arikit S, Xia R, Kakrana A, et al. . 2014. An atlas of soybean small RNAs identifies phased siRNAs from hundreds of coding genes. The Plant Cell 26, 4584–4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Bartel DP. 2005. Antiquity of microRNAs and their targets in land plants. The Plant Cell 17, 1658–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Jan C, Rajagopalan R, Bartel DP. 2006. A two-hit trigger for siRNA biogenesis in plants. Cell 127, 565–577. [DOI] [PubMed] [Google Scholar]
- Azuma A, Kobayashi S, Mitani N, Shiraishi M, Yamada M, Ueno T, Kono A, Yakushiji H, Koshita Y. 2008. Genomic and genetic analysis of Myb-related genes that regulate anthocyanin biosynthesis in grape berry skin. Theoretical and Applied Genetics 117, 1009–1019. [DOI] [PubMed] [Google Scholar]
- Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D. 2004. RNA silencing in plants. Nature 431, 356–363. [DOI] [PubMed] [Google Scholar]
- Baumberger N, Baulcombe DC. 2005. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proceedings of the National Academy of Sciences, USA 102, 11928–11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belli Kullan J, Lopes Paim Pinto D, Bertolini E, et al. . 2015. miRVine: a microRNA expression atlas of grapevine based on small RNA sequencing. BMC Genomics 16, 393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan M. 1984. Binary Agrobacterium vectors for plant transformation. Nucleic Acids Research 12, 8711–8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonar N, Liney M, Zhang R, et al. . 2018. Potato miR828 is associated with purple tuber skin and flesh color. Frontiers in Plant Science 9, 1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet E, He Y, Billiau K, Van de Peer Y. 2010. TAPIR, a web server for the prediction of plant microRNA targets, including target mimics. Bioinformatics 26, 1566–1568. [DOI] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. 2008. Widespread translational inhibition by plant miRNAs and siRNAs. Science 320, 1185–1190. [DOI] [PubMed] [Google Scholar]
- Cepin U, Gutiérrez-Aguirre I, Balažic L, Pompe-Novak M, Gruden K, Ravnikar M. 2010. A one-step reverse transcription real-time PCR assay for the detection and quantitation of Grapevine fanleaf virus. Journal of Virological Methods 170, 47–56. [DOI] [PubMed] [Google Scholar]
- Chen H-M, Chen L-T, Patel K, Li Y-H, Baulcombe DC, Wu S-H. 2010. 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proceedings of the National Academy of Sciences, USA 107, 15269–15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H-M, Li Y-H, Wu S-H. 2007. Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proceedings of the National Academy of Sciences, USA 104, 3318–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. 2005. MicroRNA biogenesis and function in plants. FEBS Letters 579, 5923–5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna S, Debjani B, Kannan P, Varsha T, Ashwin N, Melvin P, Shivaprasad PV. 2018. Major domestication-related phenotypes in Indica rice are due to loss of miRNA-mediated laccase silencing. The Plant Cell 30, 2649–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creasey KM, Zhai J, Borges F, Van Ex F, Regulski M, Meyers BC, Martienssen RA. 2014. miRNAs trigger widespread epigenetically activated siRNAs from transposons in Arabidopsis. Nature 508, 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus JT, Carbonell A, Fahlgren N, Garcia-Ruiz H, Burke RT, Takeda A, Sullivan CM, Gilbert SD, Montgomery TA, Carrington JC. 2010. Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nature Structural & Molecular Biology 17, 997–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus JT, Fahlgren N, Carrington JC. 2011. Evolution and functional diversification of MIRNA genes. The Plant Cell 23, 431–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czemmel S, Stracke R, Weisshaar B, Cordon N, Harris NN, Walker AR, Robinson SP, Bogs J. 2009. The grapevine R2R3-MYB transcription factor VvMYBF1 regulates flavonol synthesis in developing grape berries. Plant Physiology 151, 1513–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Zhao PX. 2011. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Research 39, W155–W159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Nesi N, Perez P, Devic M, Grandjean O, Caboche M, Lepiniec L. 2003. Proanthocyanidin-accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development. The Plant Cell 11, 2514–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluc L, Barrieu F, Marchive C, Lauvergeat V, Decendit A, Richard T, Carde JP, Mérillon JM, Hamdi S. 2006. Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiology 140, 499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias AP, Braun EL, McMullen MD, Grotewold E. 2003. Recently duplicated maize R2R3 Myb genes provide evidence for distinct mechanisms of evolutionary divergence after duplication. Plant Physiology 131, 610–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto MC, Petsch KA, Aukerman MJ, Beatty M, Hammell M, Timmermans MC. 2014. Genome-wide analysis of leafbladeless1-regulated and phased small RNAs underscores the importance of the TAS3 ta-siRNA pathway to maize development. PLoS Genetics 10, e1004826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Feng BR, Yang SS, Huang YB, Tang YX. 2012a. The R2R3-MYB transcription factor gene family in maize. PLoS ONE 7, e37463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Yang SS, Liang Z, Feng BR, Liu L, Huang YB, Tang YX. 2012b. Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biology 12, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Q, Goodale E, Quan RC. 2014. Bird fruit preferences match the frequency of fruit colours in tropical Asia. Scientific Reports 4, 5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. 2010. MYB transcription factors in Arabidopsis. Trends in Plant Science 15, 573–581. [DOI] [PubMed] [Google Scholar]
- Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC. 2007. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. The Plant Journal 49, 414–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Q, Li P, Teng C, Meyers BC. 2015. Secondary siRNAs from Medicago NB-LRRs modulated via miRNA-target interactions and their abundances. The Plant Journal 83, 451–465. [DOI] [PubMed] [Google Scholar]
- Felippes FF, Weigel D. 2009. Triggering the formation of tasiRNAs in Arabidopsis thaliana: the role of microRNA miR173. EMBO Reports 10, 264–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller A, Machemer K, Braun EL, Grotewold E. 2011. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. The Plant Journal 66, 94–116. [DOI] [PubMed] [Google Scholar]
- Feng S, Wang Y, Yang S, Xu Y, Chen X. 2010. Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta 232, 245–255. [DOI] [PubMed] [Google Scholar]
- Fraser CM, Chapple C. 2011. The phenylpropanoid pathway in Arabidopsis. The Arabidopsis Book 9, e0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. 2008. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. The Plant Journal 53, 814–827. [DOI] [PubMed] [Google Scholar]
- Gou JY, Felippes FF, Liu CJ, Weigel D, Wang JW. 2011. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. The Plant Cell 23, 1512–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann U, Sagasser M, Mehrtens F, Stracke R, Weisshaar B. 2005. Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Molecular Biology 57, 155–171. [DOI] [PubMed] [Google Scholar]
- Hichri I, Barrieu F, Bogs J, Kappel C, Delrot S, Lauvergeat V. 2011. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. Journal of Experimental Botany 62, 2465–2483. [DOI] [PubMed] [Google Scholar]
- Howell MD, Fahlgren N, Chapman EJ, Cumbie JS, Sullivan CM, Givan SA, Kasschau KD, Carrington JC. 2007. Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. The Plant Cell 19, 926–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh LC, Lin SI, Shih AC, Chen JW, Lin WY, Tseng CY, Li WH, Chiou TJ. 2009. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiology 151, 2120–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C. 2000. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. The EMBO Journal 19, 6150–6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Kasprzewska A, Tennessen K, Fernandes J, Nan GL, Walbot V, Sundaresan V, Vance V, Bowman LH. 2009. Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Research 19, 1429–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun JH, Liu C, Xiao X, Dixon RA. 2015. The transcriptional repressor MYB2 regulates both spatial and temporal patterns of proanthocyandin and anthocyanin pigmentation in Medicago truncatula. The Plant Cell 27, 2860–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S, Rozwadowski K. 2011. EAR motif-mediated transcriptional repression in plants: an underlying mechanism for epigenetic regulation of gene expression. Epigenetics 6, 141–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K. 2006. Negative regulation of defence and stress genes by EAR-motif-containing repressors. Trends in Plant Science 11, 109–112. [DOI] [PubMed] [Google Scholar]
- Khraiwesh B, Arif MA, Seumel GI, Ossowski S, Weigel D, Reski R, Frank W. 2010. Transcriptional control of gene expression by microRNAs. Cell 140, 111–122. [DOI] [PubMed] [Google Scholar]
- Klempnauer KH, Gonda TJ, Bishop JM. 1982. Nucleotide sequence of the retroviral leukemia gene v-myb and its cellular progenitor c-myb: the architecture of a transduced oncogene. Cell 31, 453–463. [DOI] [PubMed] [Google Scholar]
- Koyama K, Ikeda H, Poudel PR, Goto-Yamamoto N. 2012. Light quality affects flavonoid biosynthesis in young berries of Cabernet Sauvignon grape. Phytochemistry 78, 54–64. [DOI] [PubMed] [Google Scholar]
- Li S, Wang W, Gao J, Yin K, Wang R, Wang C, Petersen M, Mundy J, Qiu JL. 2016. MYB75 phosphorylation by MPK4 is required for light-induced anthocyanin accumulation in Arabidopsis. The Plant Cell 28, 2866–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Wang K, Bolitho K, Grafton K, Kortstee A, Karunairetnam S, McGhie TK, Espley RV, Hellens RP, Allan AC. 2010. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biology 10, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Lin-Wang K, Espley RV, et al. . 2016. Functional diversification of the potato R2R3 MYB anthocyanin activators AN1, MYBA1, and MYB113 and their interaction with basic helix-loop-helix cofactors. Journal of Experimental Botany 67, 2159–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo QJ, Mittal A, Jia F, Rock CD. 2012. An autoregulatory feedback loop involving PAP1 and TAS4 in response to sugars in Arabidopsis. Plant Molecular Biology 80, 117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean D, Elina N, Havecker ER, Heimstaedt SB, Studholme DJ, Baulcombe DC. 2010. Evidence for large complex networks of plant short silencing RNAs. PLoS ONE 5, e9901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavella PA, Koenig D, Weigel D. 2012. Plant secondary siRNA production determined by microRNA-duplex structure. Proceedings of the National Academy of Sciences, USA 109, 2461–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Paz-Ares J. 1997. MYB transcription factors in plants. Trends in Genetics 13, 67–73. [DOI] [PubMed] [Google Scholar]
- Matsui K, Umemura Y, Ohme-Takagi M. 2008. AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. The Plant Journal 55, 954–967. [DOI] [PubMed] [Google Scholar]
- Matus JT, Aquea F, Arce-Johnson P. 2008. Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biology 8, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus JT, Cavallini E, Loyola R, et al. . 2017. A group of grapevine MYBA transcription factors located in chromosome 14 control anthocyanin synthesis in vegetative organs with different specificities compared with the berry color locus. The Plant Journal 91, 220–236. [DOI] [PubMed] [Google Scholar]
- Moxon S, Schwach F, Dalmay T, Maclean D, Studholme DJ, Moulton V. 2008. A toolkit for analysing large-scale plant small RNA datasets. Bioinformatics 24, 2252–2253. [DOI] [PubMed] [Google Scholar]
- Nakata M, Ohme-Takagi M. 2014. Quantification of anthocyanin content. Bio-Protocol 4, 1428–1440. [Google Scholar]
- Nonogaki H. 2010. MicroRNA gene regulation cascades during early stages of plant development. Plant & Cell Physiology 51, 1840–1846. [DOI] [PubMed] [Google Scholar]
- Pantaleo V, Szittya G, Moxon S, Miozzi L, Moulton V, Dalmay T, Burgyan J. 2010. Identification of grapevine microRNAs and their targets using high-throughput sequencing and degradome analysis. The Plant Journal 62, 960–976. [DOI] [PubMed] [Google Scholar]
- Picardi E, Horner DS, Chiara M, Schiavon R, Valle G, Pesole G. 2010. Large-scale detection and analysis of RNA editing in grape mtDNA by RNA deep-sequencing. Nucleic Acids Research 38, 4755–4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F, Ye X, Morris TJ. 2008. Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proceedings of the National Academy of Sciences, USA 105, 14732–14737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahim MA, Busatto N, Trainotti L. 2014. Regulation of anthocyanin biosynthesis in peach fruits. Planta 240, 913–929. [DOI] [PubMed] [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. 2006. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes & Development 20, 3407–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay NA, Glover BJ. 2005. MYB–bHLH–WD40 protein complex and the evolution of cellular diversity. Trends in Plant Science 10, 63–70. [DOI] [PubMed] [Google Scholar]
- Renoult JP, Valido A, Jordano P, Schaefer HM. 2014. Adaptation of flower and fruit colours to multiple, distinct mutualists. New Phytologist 201, 678–686. [DOI] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. 2002. Prediction of plant microRNA targets. Cell 110, 513–520. [DOI] [PubMed] [Google Scholar]
- Rock CD. 2013. Trans-acting small interfering RNA4: key to nutraceutical synthesis in grape development? Trends in Plant Science 18, 601–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Research 18, 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwach F, Vaistij FE, Jones L, Baulcombe DC. 2005. An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiology 138, 1842–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J-K, Wu J, Lii Y, Li Y, Jin H. 2013. Contribution of small RNA pathway components in plant immunity. Molecular Plant-Microbe Interactions 6, 617–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D, Tiwari M, Pandey A, Bhatia C, Sharma A, Trivedi PK. 2016. MicroRNA858 is a potential regulator of phenylpropanoid pathway and plant development. Plant Physiology 171, 944–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin DH, Choi M, Kim K, Bang G, Cho M, Choi SB, Choi G, Park YI. 2013. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Letters 587, 1543–1547. [DOI] [PubMed] [Google Scholar]
- Shivaprasad PV, Chen HM, Patel K, Bond DM, Santos BA, Baulcombe DC. 2012a. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. The Plant Cell 24, 859–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad PV, Dunn RM, Santos BA, Bassett A, Baulcombe DC. 2012b. Extraordinary transgressive phenotypes of hybrid tomato are influenced by epigenetics and small silencing RNAs. The EMBO Journal 31, 257–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad PV, Thillaichidambaram P, Balaji V, Veluthambi K. 2006. Expression of full-length and truncated Rep genes from Mungbean yellow mosaic virus-Vigna inhibits viral replication in transgenic tobacco. Virus Genes 33, 365–374. [DOI] [PubMed] [Google Scholar]
- Song X, Li P, Zhai J, et al. . 2012. Roles of DCL4 and DCL3b in rice phased small RNA biogenesis. The Plant Journal 69, 462–474. [DOI] [PubMed] [Google Scholar]
- Stachel SE, Nester EW. 1986. The genetic and transcriptional organization of the vir region of the A6 Ti plasmid of Agrobacterium tumefaciens. The EMBO Journal 5, 1445–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocks MB, Moxon S, Mapleson D, Woolfenden HC, Mohorianu I, Folkes L, Schwach F, Dalmay T, Moulton V. 2012. The UEA sRNA workbench: a suite of tools for analysing and visualizing next generation sequencing microRNA and small RNA datasets. Bioinformatics 28, 2059–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Fan G, Su L, Wang W, Liang Z, Li S, Xin H. 2015. Identification of cold-inducible microRNAs in grapevine. Frontiers in Plant Science 6, 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunilkumar G, Vijayachandra K, Veluthambi K. 1999. Preincubation of cut tobacco leaf explants promotes Agrobacterium-mediated transformation by increasing vir gene induction. Plant Science 141, 51–58. [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Garvin DF, Mockler TC, et al. . 2010. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463, 763–768. [DOI] [PubMed] [Google Scholar]
- Wang W, Vignani R, Scali M, Cresti M. 2006. A universal and rapid protocol for protein extraction from recalcitrant plant tissues for proteomic analysis. Electrophoresis 27, 2782–2786. [DOI] [PubMed] [Google Scholar]
- Wu L, Zhou H, Zhang Q, Zhang J, Ni F, Liu C, Qi Y. 2010. DNA methylation mediated by a microRNA pathway. Molecular Cell 38, 465–475. [DOI] [PubMed] [Google Scholar]
- Xia R, Ye S, Liu Z, Meyers BC, Liu Z. 2015. Novel and recently evolved microRNA clusters regulate expansive F-BOX gene networks through phased small interfering RNAs in wild diploid strawberry. Plant Physiology 169, 594–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia R, Zhu H, An YQ, Beers EP, Liu Z. 2012. Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biology 13, R47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao G, Ming M, Allan AC, et al. . 2017. Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. The Plant Journal 92, 437–451. [DOI] [PubMed] [Google Scholar]
- Yao Y, Guo G, Ni Z, Sunkar R, Du J, Zhu JK, Sun Q. 2007. Cloning and characterization of microRNAs from wheat (Triticum aestivum L.). Genome Biology 8, R96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, Poethig RS. 2005. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes & Development 19, 2164–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Arikit S, Simon SA, Kingham BF, Meyers BC. 2014. Rapid construction of parallel analysis of RNA end (PARE) libraries for Illumina sequencing. Methods 67, 84–90. [DOI] [PubMed] [Google Scholar]
- Zhai J, Zhang H, Arikit S, Huang K, Nan G-L, Walbot V, Meyers BC. 2015. Spatiotemporally dynamic, cell-type–dependent premeiotic and meiotic phasiRNAs in maize anthers. Proceedings of the National Academy of Sciences, USA 112, 3146–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Wang S, Sunkar R. 2014. Genome-wide discovery and analysis of phased small interfering RNAs in Chinese sacred lotus. PLoS ONE 9, e113790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Xia R, Zhao B, An YQ, Dardick CD, Callahan AM, Liu Z. 2012. Unique expression, processing regulation, and regulatory network of peach (Prunus persica) miRNAs. BMC Plant Biology 12, 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.