Abstract
Laboratorial and epidemiological research has established a relationship between paraquat (PQ) exposure and a risk for Parkinson’s disease. Previously, we have investigated the effects of nuclear factor erythroid 2 related factor 2 (Nrf2) and microRNAs in PQ-induced neurotoxicity, addressing the function of miR-380-3p, a microRNA dysregulated by PQ, as well as Nrf2 deficiency. Nrf2 is known to mediate the expression of a variety of genes, including noncoding genes. By chromatin immunoprecipitation, we identified the relationship between Nrf2 and miR-380-3p in transcriptional regulation. qRT-PCR, Western blots, and dual-luciferase reporter gene assay showed that miR-380-3p blocked the translation of the transcription factor specificity protein-3 (Sp3) in the absence of degradation of Sp3 mRNA. Results based on cell counting analysis, annexin v-fluorescein isothiocyanate/propidium iodide double-staining assay, and propidium iodide staining showed that overexpression of miR-380-3p inhibited cell proliferation, increased the apoptotic rate, induced cell cycle arrest, and intensified the toxicity of PQ in mouse neuroblastoma (N2a [Neuro2a]) cells. Knockdown of Sp3 inhibited cell proliferation and eclipsed the alterations induced by miR-380-3p in cell proliferation. Two mediators of apoptosis and cell cycle identified in previous studies as Sp3-regulated, namely cyclin-dependent kinase inhibitor 1 (p21) and calmodulin (CaM), were dysregulated by PQ, but not Sp3 deficiency. In conclusion, Nrf2-regulated miR-380-3p inhibited cell proliferation and enhanced the PQ-induced toxicity in N2a cells potentially by blocking the translation Sp3 mRNA. We conclude that CaM and p21 were involved in PQ-induced toxicity.
Keywords: paraquat, neurotoxicity, Nrf2, miR-380-3p, Sp3, p21, CaM
Paraquat (PQ) has been used worldwide for more than half a century as an efficient agrichemical, yet its application remains highly controversial given its documented toxicity. PQ causes a widespread oxidative/nitrosative stress in cells, after 1-electron reduction chemistry. The dopaminergic neurons readily take up PQ via the dopamine transporter and the organic cation transporter-3 (Barlow et al., 2003; Rappold et al., 2011), and are highly sensitive to PQ-induced oxidative stress (Dröge and Schipper, 2007; Fitsanakis et al., 2002), resulting in loss of dopaminergic neurons in the substantia nigra, one of the cardinal clinical features of Parkinson’s disease (PD). Neurodegeneration models established for PQ toxicity both in vivo and in vitro have thus been widely employed to address mechanisms associated with the etiology of PD (Berry et al., 2010).
PD is a common complex syndrome ascribed to complicated pathogenesis, encompassing motor deficits, static tremors, bradykinesia, rigidity, and postural/gait disturbance, as well as other nonmotor neurological deficits to various degrees, which are generally considered to be manifestation of loss in dopaminergic neurons. The etiology of PD involves both genetic and environmental factors. Genes, such as alpha-synuclein and parkin (Kuwahara et al., 2006; Shimura et al., 2000) have been shown to be associated with the pathogenesis of familial PD. In addition, environmental factors, including chemicals may also trigger PD pathogenesis. Epidemiological studies have established PQ exposure is a risk factor for the development of PD (Dinis-Oliveira et al., 2006; Kieburtz and Wunderle, 2013; Tanner et al., 2013).
As mentioned above, the neurotoxicity of PQ has been mainly attributed to redox cycling in dopaminergic neurons or/and glial cells (Bonneh-Barkay et al., 2005a,b; McCormack et al., 2006), with the most effective redox cyclers inducing the highest degree of dopaminergic cell injury (Bonneh-Barkay et al., 2005a). The generation of 4-hydroxynonenals identified in vivo also suggests that oxidative stress is the predominant mechanism by which PQ induces toxicity (McCormack et al., 2005). Nuclear factor erythroid 2 related factor 2 (Nrf2) regulates the expression of numerous antioxidant proteins, such as superoxide dismutase 1and 2 (SOD1, SOD2), glutathione peroxidase 1 and 5 (GPX-1, GPX-5), and NAD(P)H quinone oxidoreductase (NQO1) (De Long et al., 1986; Johnson and Giulivi, 2005; Liddell et al., 2006; Siegel and Ross, 2000; Song et al., 2014). In our previous study, Nrf2 was found to be protective in PQ-induced neurotoxicity (Li et al., 2012), establishing a critical role for oxidative stress in mediating PQ neurotoxicity.
MicroRNAs (miRNAs) are small noncoding regulatory RNAs of 18–25 nucleotides in length that regulate gene expression at posttranscriptional level by sequence-specifically binding to the 3′-untranslated (3′UTR) region of the target mRNAs to inhibit the translation or induce the degradation of these mRNAs. miRNAs are involved in survival and differentiation of dopaminergic neurons and are relevant to the pathogenesis and progress of PD (Anderegg et al., 2013; de Mena et al., 2010; Hébert and De Strooper, 2007; Goodall et al., 2013). Apart from the direct association with PD etiology, miRNAs are also closely related to Nrf2: various interactions between the transcription factor Nrf2 and miRNAs have been revealed. For example, miR-200a and miR-141 target the mRNA of Kelch-like ECH-associated protein 1 (Keap1), a master regulator of Nrf2 by deleteriously binding to it, reducing the cytoplasmic abundance of Keap1 protein, thus releasing Nrf2 proteins from sequestration and further degradation (van Jaarsveld et al., 2013; Yang et al., 2014). Furthermore, some miRNAs, such as miR-146a, miR-132, and miR-28, can directly bind to Nrf2 mRNA and repress its translation (Smith et al., 2015; Stachurska et al., 2013; Yang et al., 2011).
Previous studies have revealed 3 mechanisms for the regulation of the transcription of miRs. (1) Methylation level of the upstream CpG rich region of miRs was found to be inversely related to miR expression (Fiaschetti et al., 2014). (2) Nucleotide polymorphism occurs in the promotor region of miRs and induces a variety in their expression level (Zhang et al., 2014). (3) The activity of the transcription factor bound to the promotor region of miRs likely plays another determinant role in miR expression (Liang et al., 2009). The transcription factor Nrf2 has been shown to regulate the expression of many miRNAs, such as miR-29 and miR-1 (Kurinna et al., 2014; Shah et al., 2014; Singh et al., 2013). Accordingly, it is reasonable to suggest that Nrf2 might regulate the expression of certain miRNAs, concomitant with its well-documented antioxidant effect. In the present study, Nrf2 was shown to promote the expression of miR-380-3p whereas PQ inhibiting the same miR. miR-380-3p (5′-uauguaguaugguccacaucuu-3′) is encoded within chromosome 12 (109711803–109711863, precursor) in the mouse, and is relatively conserved among various species. The expression of miR-380-3p was found restricted to embryos and brain in adults (Seitz et al., 2004), implying a special role that miR-380-3p may have in neuronal function, differentiation and survival.
Previously, a set of miRNAs in mouse substantia nigra was found dysregulated by PQ (Wang et al., 2017). Because the potency of miRNAs has been widely recognized, altered miRNA expression profile is likely to exert significant health effects. In the present study, we addressed one of these dysregulated miRNAs, miR-380-3p, and its function in PQ-induced nerve cell injury. To decipher the mechanism underlying the function miR-380-3p, apart from its direct target, namely specificity protein-3 (Sp3), some genes potentially regulated by Sp3 (Gartel et al., 2000; Pan et al., 2000; Sowa et al., 1999) were also studied. Specifically, we examined the expression of 2 ubiquitously active molecules, namely cyclin-dependent kinase inhibitor 1 (p21) and calmodulin (CaM). Furthermore, we explored its interaction with Nrf2 and the regulatory role Nrf2 exerts over miR-380-3p. This research was predominantly carried out in Neuro2a (N2a) cells, a mouse neuroblastoma cell line that has a long history of serving in studies on dopamine-associated neurotoxicity (LePage et al., 2005).
MATERIALS AND METHODS
Quantitative RT-PCR for validation of miRNA and mRNA expression
Total RNA samples were extracted from N2a cells and reversely transcribed to cDNA using Prime Script RT reagent kit RR037a for the miRNAs, and kit RR047a for the long RNAs (TAKARA, Japan), following the manufacturer’s instructions. The reverse transcription of miRNAs entailed specific stem-loop primers obtained from Ribobio (Guangzhou, China). The subsequent real-time PCR mixture was established with SYBRPremix Ex Taq RR820a (TAKARA), following the manufacturer’s instructions, and performed in 7500 real-time PCR system (ABI) or Light cycler 480 (Roche, Switzerland). The reaction tubes were incubated at 95°C for 30 s, followed by 40 cycles at an interval of 5 s at 96°C and an interval of 34 s at 60°C. For melting curve analysis, the amplifications were incubated at 95°C for 15 s, 60°C for 1 min and 95°C for 15 s. Data were analyzed by 2−ΔΔCT. The expression level of miRNA was normalized to U6, and long RNAs were normalized to GAPDH.
Dual-luciferase assay
Luciferase assays were carried out to confirm the interaction between miR-380-3p and the 3′UTR of Sp3 mRNA. Two hundred and ninety-three T cells, which are frequently used in dual-luciferase reporter assay and which do not contain endogenous mmu-miR-380-3p, were seeded into 24-well plates, cotransfected with 40 nM miR-380-3p mimic or negative control sequences and 100 nM dual-luciferase vector (psiCHECK2, Promega) carrying the wild-type or mutant Sp3 3′UTR sequences. After 24 h, the cells were lysed and measured for luciferase activities using GloMax 96 (Promega, Madison, Wisconsin), according to the manufacturer’s protocol. The ratio of Renilla to firefly luciferase signal was used to normalize Renilla luciferase activity. The sequences of wild-type or mutant Sp3 3′UTR were listed in Table 1.
Table 1.
The Sequences Containing Wild-type or Mutant Binding Site (Red) of miR-380-3p, Obtained From the 3′-Untranslated (3′UTR) of Specificity Protein-3 (Sp3) mRNA
| ID | Sequences (5′-3′) |
|---|---|
| mmu-Sp3 mRNA 3′UTR (WT) | ATTATGCA TAAC TGACAAA TCAAG T T TCCAAGCAAA TG T TACA TAG TG TAGGC T T TAC T TAGC T TA TCAA T T TG TCA T T T TGAAGC TAA T TA T T T TAA T TAGG T TAAC TG TACAA TA T T T TAAGCA T TAC TC T TG TAAGA T T T TGAAAAC TACA T T T TAACA TGGACC T T TAGGGA TAG TCACC T T T TAAA TCC TG T TGAAAAGCCA TG T T TAAGA T T TAA T T TGCCAAAA TAA TG TC T TG T TAA TA T TC T T TCAA TAA TGAAG T TGGGCAA T |
| mmu-Sp3 mRNA 3′U TR (mutant) | A T TA TGCA TAAC TGACAAA TCAAG T T TCCAAGCAAA TG T TACA TAG TG TAGGC T T TAC T TAGC T TA TCAA T T TG TCA T T T TGAAGC TAA T TA T T T TAA T TAGG T TAAC TG TACAA TA T T T TAAGCA T TAC TC T TG TAAGA T T T TGAAAAC TGAGC T T TAACA TGGACC T T TAGGGA TAG TCACC T T T TAAA TCC TG T TGAAAAGCCA TG T T TAAGA T T TAA T T TGCCAAAA TAA TG TC T TG T TAA TA T TC T T TCAA TAA TGAAG T TGGGCAA T |
Chromatin immunoprecipitation assay
N2a cells were cultured as described above. Prior to cross-linking, a group of cells were treated with 100 μM PQ for 48 h. Cross-linking was performed with formaldehyde at a final concentration of 1% (wt/wt) and terminated after 5 min by addition of glycine at a final concentration of 0.125 M. Cells were harvested with lysis buffers in chromatin immunoprecipitation (ChIP) kit (BersinBio, Guangzhou, China) according to the manufacturer’s instructions. Chromatin was sonicated with UCD300 (Diagenode, Belgium) to generate DNA fragments with an average size of 500 bp. Then 4 h incubation with monoclonal Nrf2 (CST) or IgG (Millipore) antibody-beads was performed in vertical mixer at 4°C. Next, washing and elution were performed (washing and elution buffer, respectively) with a ChIP kit, according to the manufacturer’s instructions. After reversal cross-linking, the DNA was collected by centrifugations and dissolved in ddH2O. Immunoprecipitated DNA was analyzed by qPCR and the enrichment was expressed as percentage of the input for each condition. The sequences of primers used in the following PCR were listed in Table 2.
Table 2.
The Sequences of Primers Used in the qPCR Following Chromatin Immunoprecipitation
| Gene | No. | F/R | Primer Sequence | Site | Product (bp) |
|---|---|---|---|---|---|
| mir379 | 1 | F | CAACCCTGTCCTCTCTGCTC | 323 | 238 |
| R | GAAGCAGGTGGAACCAGAAG | ||||
| mir379 | 2 | F | GGCAGCCAGTTGTTTGATTT | 1271 | 227 |
| R | GAGGTTCCTGGGTATGAGCA | ||||
| Beta-actin | 3 | F | AACAGCCGGAGCTACACACT | 213 | |
| R | GGGCTCAGCCTATGAGTCAG |
Western blotting
Protein level of Sp3 in N2a cells was assessed by Western blot and normalized to GAPDH. Total protein samples were extracted from N2a cells with lysis buffer. Protein concentrations were determined by BCA protein assay kit, standardized by bovine serum albumin. Equal amounts of protein were loaded in each lane for sodium dodecyl sulfate polyacrylamide gel electrophoresis (5% polyacrylamide for compression and 10% for separation, wt/wt). Next, the separated proteins were transferred to nitrocellulose membranes. The membranes were blocked with 5% (wt/wt) skimmed milk for 1 h before the overnight incubation with primary antibodies in 5% (wt/wt) skimmed milk (anti-SP3, 1:200, Santa Cruz; anti-BCL2, 1:1000, Abcam; anti-BAX, 1:2000, Abcam; anti-GAPDH, 1:10000, Abcam) at 4°C. Membranes were rinsed with TBST for 6 times with 5 min for each time before incubating with horseradish peroxidase-conjugated goat antirabbit IgG (1:10000 in TBST, Millipore) for 1 h, followed by 5 min * 5 times rinse with TBST and 5 min with TBS. The protein bands were detected by chemiluminescence and analyzed with ImageJ 2.1.
Bisulfite sequencing PCR
The primer sets of bisulfite sequencing PCR (BSP) were used to target the CpG: 29 located between Dlk1 and Gtl2 as shown in Table 3. Extracted DNA samples from N2a cells were converted using a Bisulfite DNA Modification Kit (BersinBio), following the manufacturer’s instructions. PCR amplification with BSP primers was then predenatured at 95°C for 5 min, followed by 30 cycles at an interval of 10 s at 95°C, an interval of 15 s at 58°C and an interval of 15 s at 72°C, and 72°C for 10 min after the cycles. To ensure amplification efficiency, BSP products were analyzed by 3% (wt/wt) agarose gel electrophoresis. The amplifications were inserted into pMD 18-T Vectors (TAKARA) and then transferred into Trans1-T1 (TAKARA), following the manufacturer’s instructions. The successfully transformed bacterial colonies were lysed after 20 h incubation at 37°C and the bacteria solution was verified by PCR with primers targeting CpG: 29 and 3% (wt/wt) agarose gel electrophoresis. Ten colonies were collected for each group and then the sequencing of bacteria solutions was entrusted to Sangon Biotech (Shanghai).
Table 3.
The Sequences of Primers Targeting CpG: 29
| Gene | Primer Name | Primer Sequence (5′-3′) | Products | CpGs |
|---|---|---|---|---|
| CpG: 29 | Forward | TAGAGATTGAGGATTATTGGAGTAAG | 384 bp | 31 |
| Reverse | CCAAACCTAACTAAAAACACATTAC |
Cell culture
The murine neuroblastoma cell line N2a was purchased from Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China), and maintained in Dulbecco’s Modified Eagle’s Medium (DMEM, HyClone, Utah) containing 10% (wt/wt) fetal bovine serum (HyClone) and 1% (wt/wt) penicillin and streptomycin (Gibco, Massachusetts) at 37°C in a humidified atmosphere containing 5% (vol/vol) CO2. For resuspension, the cells were trypsinized by Trypsin-EDTA (Gibco), and the cells were passaged when attaining 80% confluence.
Cell treatments
For PQ treatment, the cells were maintained in complete medium containing 50, 100, and 200 µM PQ, alone or following a 6 h pretreatment with 40 µM tBHQ. Cells without any treatment were used as control. The N2a cells were transfected with miRIDIAN miR-380-3p mimics, hairpin inhibitors (Dharmacon, Colorado) or siRNAs (Genepharma, Shanghai, China) using DharmaFECT reagent (Dharmacon), and the corresponding scrambled sequences were also transfected in cells as negative controls. The transfections were carried out in Reduced Serum Media (Opti-MEM, Gibco) and limited to 6 h to reduce cytotoxicity. The transfection of siRNA and corresponding scrambled sequences was after the same protocol. The mimics and siRNAs were applied at 40 nM, and inhibitors at 100 nM, under conditions analogous to the corresponding negative controls, respectively.
Cell proliferation assay
Cell proliferation was measured using Cell Counting Kit-8 (CCK-8) reagent (Solarbio, Beijing, China) according to the manufacturer’s protocol. The N2a cells were seeded into 96-well plates (5 × 103 cells per well) and incubated. Upon reaching 40% confluency, the cells were transfected with miRIDIAN miR-380-3p mimic or hairpin inhibitor. The corresponding scrambled sequences were adopted in negative control groups. The transfected cells were then exposed to 0, 50, 100, or 200 µM of PQ for 48 h, followed by cell proliferation assessment. A mixture of 90 μl medium and 10 μl CCK-8 solution was used to replace the PQ-stained medium in each well, incubated for 2 h and the absorbance values were measured at 450 nm using a microplate reader.
Cell cycle analysis
Cells were trypsinized, harvested, washed twice with cold PBS, fixed with 75% (wt/wt) ice-cold ethanol at −20°C overnight, washed twice with cold PBS, and then incubated with PI/RNase Staining Buffer (BD Biosciences, New Jersey) for 15 min at room temperature. The tubes were stored at 4°C protected from light prior to analyzing and the PI signal was examined by a flow cytometry (FACSVerse, BD Biosciences) within 1 h.
Apoptosis assay
Apoptosis was quantified using FITC Annexin V Apoptosis Detection Kit I (BD Biosciences) following the manufacturer’s protocol, and with flow cytometry (FACSVerse, BD Biosciences). Cells were trypsinized and collected 48 h after transfection, washed twice with cold PBS, and resuspended at 1 × 106 cells/ml. Hundred microliters of the cell solution was transferred to a new tube and mixed with 5 µl of FITC Annexin V and 5 µl of PI, gently vortexed. After incubation for15 min in the dark, 400 μl of 1× Binding Buffer was added to each tube, and the cells were analyzed by flow cytometry within 1 h.
Statistical analysis
The results were analyzed by SPSS 22 and expressed as mean ± SD. One-way analysis of variance (ANOVA) and Student-Newman-Keuls test for pairwise comparisons were used to detect significant differences among the groups under varying experimental treatments. The differences between 2 groups were determined by t test or Welch’s t test when the variances were unequal. The interactions between factors were determined by two-way ANOVA. p < .05 was the criterion for statistical significance.
RESULTS
Nrf2 Transcriptionally Regulates miR-380-3p Expression in N2a Murine Neuroblastoma Cell Line
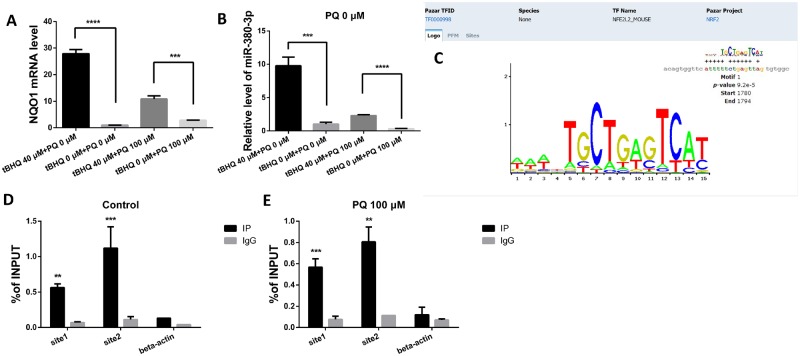
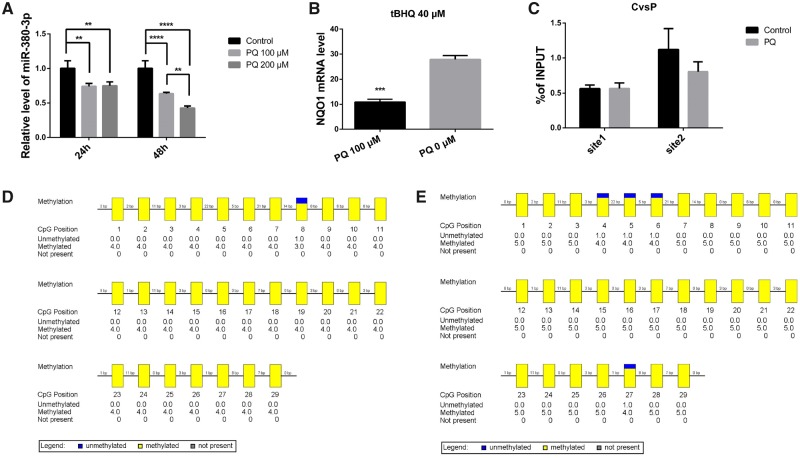
Nrf2 is sequestered by Keap1, anchored to the cytoplasm and degraded by Cullin 3, and seldom been activated in the absence of particular triggers, such as oxidative stress (Itoh et al., 1999). Herein, tert-Butylhydroquinone (tBHQ), a specific activator of Nrf2 (Li et al., 2005, 2012, 2014), was applied to N2a cells at 40 µM alone or followed by 100 µM PQ. The former releases Nrf2 from the sequestration of Keap1, and thus enhances the transcriptional function of Nrf2. The NAD(P)H dehydrogenase [quinone] 1 (NQO1) mRNA level, which was already verified as transcriptionally regulated by Nrf2 (Dinkova-Kostova and Talalay, 2010), was increased by treatment with tBHQ (Figure 1A, p < .0001 or p < .001, respectively). The miR-380-3p level increased along with the tBHQ-induced activation of Nrf2 (Figure 1B, p < .001 or p < .0001, respectively).
Figure 1.
Nuclear factor erythroid 2 related factor 2 (Nrf2) upregulated miR-380-3p expression. A, The expression of NQO1 in Neuro2a (N2a) cells increased 48 h after a 6 h pretreatment with 40 µM tBHQ (tert-Butylhydroquinone), n = 3. ****p < .0001 versus the corresponding control group. Six hours pretreatment with 40 µM tBHQ upregulated the expression of NQO1 in N2a cells exposed to 100 µM PQ for 48 h, n = 3. ***p < .001 versus the corresponding control group. B, The 6 h pretreatment with 40 µM tBHQ upregulated the expression of miR-380-3p in N2a cells, n = 3. ***p < .001 versus the corresponding control group. Pretreatment with 40 µM tBHQ for 6 h upregulated the expression of miR-380-3p in N2a cells exposed to 100 µM PQ for the following 48 h, n = 3. ****p < .0001 versus the corresponding control group. C, The binding site motif was obtained from the transcriptional factor encyclopedia established by center for molecular medicine and therapeutics (CMMT). The motif-based sequence analysis was conducted on the MEME suite, a motif-based sequence analysis tool funded by National Institute of Health, United States. D and E, Nrf2-specific antibodies captured over 6 times of fragments of miR-379 promoter region as the negative control IgG did, n = 3. ***p < .001 or **p < .01 versus the corresponding IgG group respectively. The chromatin fragments in (D) was extracted from untreated N2a cells and that of (E) was from cells exposed to 100 µM PQ (paraquat) for 48 h.
To validate the hypothesis that Nrf2 transcriptionally regulates miR-380-3p, we obtained the binding site motif of mmu-Nrf2 (Figure 1C) from the transcriptional factor encyclopedia, a database established by the center for molecular medicine and therapeutics (CMMT). Many eukaryotic miRs, just like miR-380, are encoded in clusters resembling the operons common in prokaryotes and all miRs within 1 cluster share the upstream promoter region. The 2000 bp upstream of miR-379, the first miR in the cluster according to genome mapping, should contain the potential promoters that regulate the expression of miR-380-3p and thus was searched for possible binding sites. By motif-based analysis, an -ATTTTTCTGAGTTAG- (1780–1794) was predicted as a possible Nrf2 binding site within the 2000 bp sequence.
Next, we adopted ChIP to confirm the prediction. Using ChIP followed by qPCR covering 2 properly selected sites in the potential promoter region, the combination of Nrf2 and the promoter region of miR-380-3p (Figs. 1D and 1E, p < .01 or p < .001, respectively) was identified.
miR-380-3p Suppresses the Translation of Sp3 mRNA
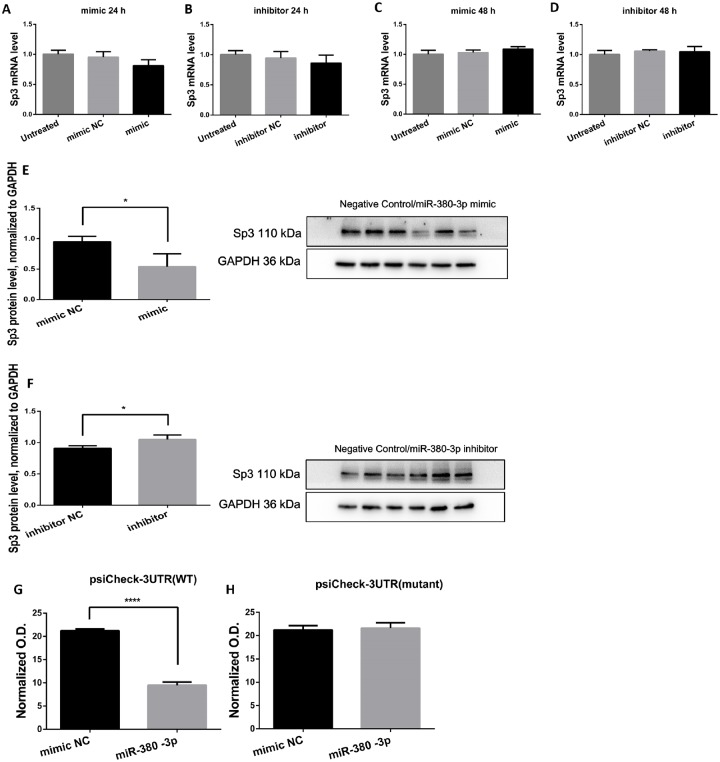
A location pairing the seed sequence of miR-380-3p was found in the 3′UTR region of Sp3 mRNA (-ttgaaaactacattttaaca-, 3117–3123), indicating a canonical match likely to regulate the translation of Sp3 mRNA (Wang, 2014). Accordingly, Sp3 mRNA and protein levels after alterations in miR-380-3p expression were determined as to test our prediction.
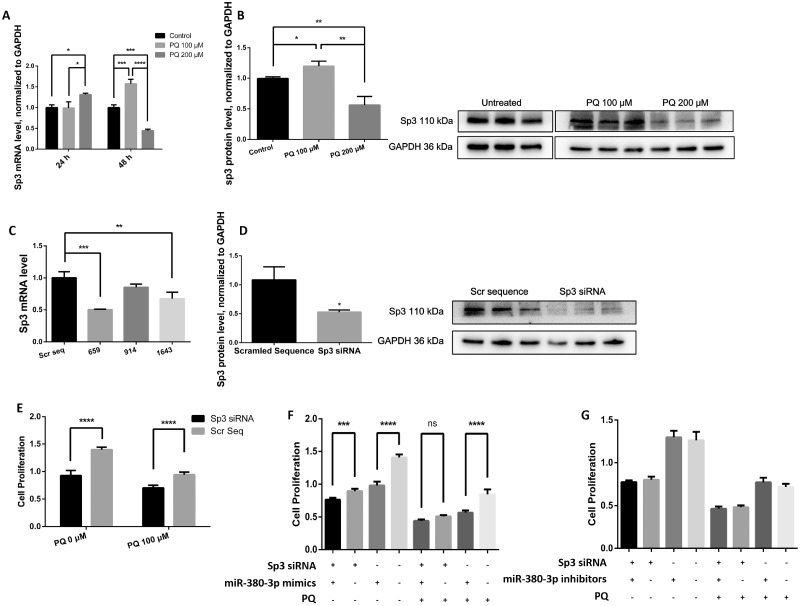
Though miR-induced mRNA degradation mostly occurs in plants, it has also been reported in vertebrates. qRT-PCR was performed to measure the Sp3 mRNA level in N2a cells after transfection of miR-380-3p mimics/inhibitors. The Sp3 mRNA abundance was indistinguishable from control after the transient transfections (Figs. 2A–D, p > .05), whereas its protein level was changed in response to alterations in miR-380-3p expression (Figs. 2E and 2F, p < .05).
Figure 2.
miR-380-3p targets specificity protein-3 (Sp3) mRNA and suppresses its translation, n = 3. A and B, The Sp3 mRNA level in Neuro2a (N2a) cells was not changed by miR-380-3p mimics or inhibitors 24 h after transient transfection. p > .05 versus the corresponding negative control groups, respectively. C and D, Sp3 mRNA level was still not changed by miR-380-3p mimics or inhibitors 48 h after the transient transfection. p > .05 versus the corresponding negative control groups, respectively. E, Forty-eight hours after transfection, the Sp3 protein level in N2a cells was decreased by miR-380-3p mimics. *p < .05 versus the negative control group. F, Forty-eight hours after transfection, the Sp3 protein level was elevated by miR-380-3p inhibitors. *p < .05 versus the negative control group. G, Two hundred and ninety-three T cells were lysed and examined for luciferase activity 24 h after the cotransfection of the reporter carrying wild-type binding site and miR-380-3p mimic. The mimics repressed the relative activity of Renilla luciferase (normalized to firefly luciferase). ****p < .0001 versus the negative control group. H, The mutation in predicted binding site abolished the difference between mimics group and negative control group. p > .05 versus the negative control group.
However, the changes in Sp3 protein level were insufficient to properly validate the regulation or to locate the exact binding site in the 3′UTR of Sp3 mRNA. Accordingly, we cotransfected the dual-luciferase reporter (psiCHECKTM-2 Vector) carrying the predicted binding site (Table 1), along with miR-380-3p mimics or the corresponding negative control sequence, into 293 T cells. We found that miR-380-3p mimics suppressed the translation of Renilla luciferase (Figure 2G, p < .0001). Furthermore, mutation (Table 1) in the predicted binding site abolished the suppression by the mimics (Figure 2H, p > .05).
Overexpression of miR-380-3p Enhances the Toxicity of PQ in N2a Cells
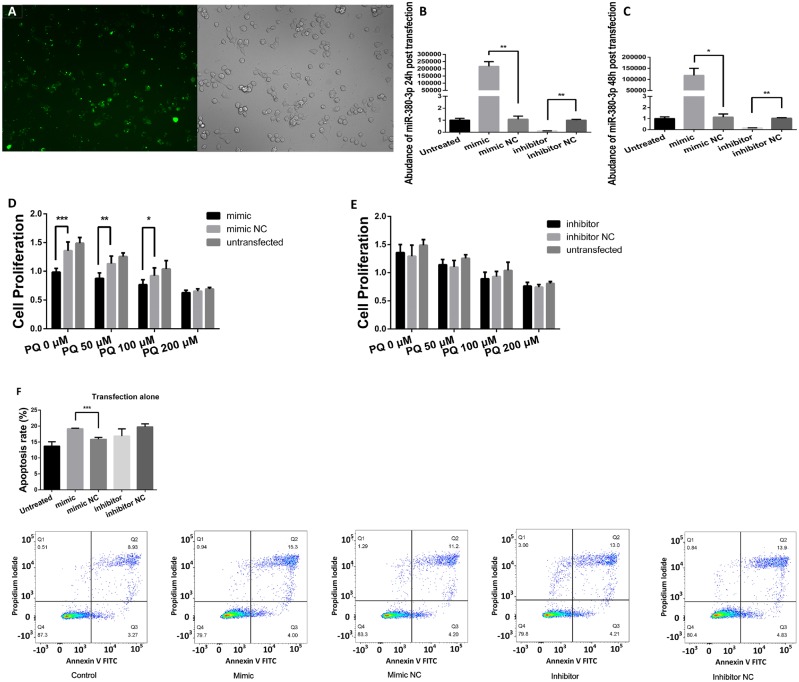
By transient transfection of miR-380-3p mimics/inhibitors, synthetics resembling or counteracting mature miRNAs, we achieved immediate functional gain/loss of miR-380-3p (Figs. 3A–C, p < .05 or p < .01, respectively), and observed the ensuing effects. Cell proliferation, apoptosis rate, and cell cycle distribution assays were carried out, to establish the role of miR-380-3p in nerve cell survival and proliferation.
Figure 3.
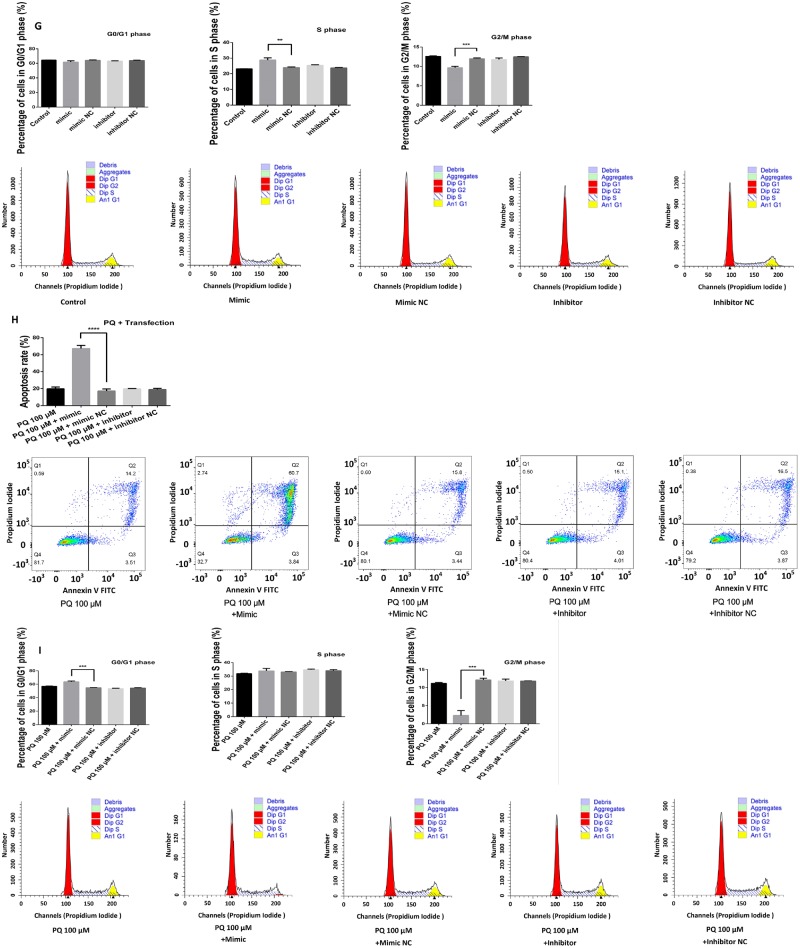
Aberrant increase in miR-380-3p enhanced the toxicity of paraquat (PQ). A, Oligos modified by 5-FAM were transferred into Neuro2a (N2a) cells by Dharmacon, according to manufacturer’s instruction. The view was excited at 492 nm and observed at 518 nm. B and C, RNA samples were extracted 24 h (B) or 48 h (C) after the transient transfection and examined for miR-380-3p abundance by qRT-PCR. The mimics increased miR-380-3p abundance in N2a cells, whereas the inhibitors brought it down, n = 3. *p < .05 or **p < .01 versus the corresponding negative control groups as labeled by the zig-zag lines. D, N2a cells were transfected with miR-380-3p mimics or corresponding negative control sequences before exposure to PQ at 50, 100, or 200 μM for the upcoming 48 h and then examined for proliferation. The cells transfected with mimics exhibited lower proliferation than corresponding negative controls when the concentration of PQ in the medium was relatively low (0, 50, or 100 μM), n = 6. *p < .05, **p < .01, or ***p < .001 versus the corresponding negative control groups as labeled by the zig-zag lines. E, N2a cells were transfected with miR-380-3p inhibitors or corresponding negative control sequences before treatments similar to (D), and then examined cell proliferation. The cell proliferation of inhibited groups was not significantly different from the negative control groups, n = 6. F and G, N2a cells had been transfected with miR-380-3p mimics, inhibitors, or corresponding negative control sequences and, 48 h later, were tested for apoptosis rate (F) and cell cycle proportion (G). The miR-380-3p mimics increased the apoptosis rate and altered the cell cycle proportion of N2a cells, whereas inhibitors did not cause obvious alteration, n = 3. **p < .01 or ***p < .001 versus the corresponding negative control groups as labeled by the zig-zag lines. H and I, N2a were exposed to 100 μM PQ for 48 h after treatment of miR-380-3p mimics, inhibitors, or corresponding negative control sequences and then tested for apoptosis rate (H) and cell cycle proportion (I). Similar to (G), the apoptosis rate of miR-380-3p-overexpresssed group was higher than the corresponding control group and the proportion of G2/M phase was smaller, whereas inhibitors did not cause any obvious alterations, n = 3. ***p < .001 or ****p < .0001 versus the corresponding negative control groups as labeled by the zig-zag lines.
Figure 3.
Continued
The miR-380-3p mimics inhibited cell proliferation (Figure 3D, p < .001, p < .01, or p < .05, respectively) and increased apoptosis rate of N2a cells (Figure 3F, p < .001), altering the cell phase proportion, reducing the mitotic cells and keeping more cells in S phase (Figure 3G, p < .01 or p < .001, respectively). However, the inhibition of miR-380-3p did not cause detectable alterations in any of the above endpoints (Figs. 3E–G, p > .05). Upon exposure to PQ, miR-380-3p mimics caused lower cell proliferation, higher apoptosis, and cycle arrest in N2a cells (Figs. 3D, 3H, and 3I, p < .0001, p < .001, p < .01, or p < .05, respectively), whereas miR-380-3p inhibition failed to induce any detectable alterations (Figs. 3E, 3H, and 3I, p > .05).
PQ Reduces the Expression of miR-380-3p in N2a Cells
PQ reduced the expression of miR-380-3p in N2a cells (Figure 4A, p < .0001 or p < .01, respectively), and longer exposure or higher concentration intensified this effect. Furthermore, the expression of NQO1 was also reduced by PQ when Nrf2 was already activated by tBHQ (Figure 4B, p < .001) and the binding between Nrf2 and the miR-380-3p promoter region was also slightly weakened by PQ, though the difference did not attain statistical significance (Figure 4C, p > 0.05).
Figure 4.
Paraquat (PQ) reduced the expression of miR-380-3p in Neuro2a (N2a) cells. A, N2a cells were treated with PQ at 100 or 200 μM for 24 or 48 h, and the treatments basically reduced miR-380-3p level in N2a cells, n = 3. **p < .01 or****p < .0001 for the comparisons labeled by the zig-zag lines. B, Both groups were pretreated with 40 μM tBHQ (tert-Butylhydroquinone) for 6 h, and the group exposed to 100 μM PQ for 48 h showed lower NQO1 mRNA abundance, n = 3. ***p < .001 versus the control group. C, Comparison of nuclear factor erythroid 2 related factor 2 (Nrf2)-specific coprecipitation efficiency between the 2 groups in (Figs. 1F and 1G), for 2 sites respectively, n = 3. p > .05 for the group exposed to 100 µM PQ versus the control group at both sites. D and E, Methylation condition of CpG: 29 in untreated N2a cells (D) and those cultured in 100 μM PQ for 48 h (E). The data were analyzed by BIQ analyzer and the probably erroneous or clone sequences were automatically removed before calculation.
The methylation level of an intergenic germ-line derived differentially methylated region (IG-DMR) between Dlk1 and Gtl2 was previously found to be relevant to the regulation of the expression of miR-380-3p and other miRNAs within the cluster (Lin et al., 2003; Seitz et al., 2004). To further determine the mechanism by which PQ influences the miR-380-3p expression, we examined whether PQ changes the methylation level of the IG-DMR. But exposure to PQ did not induce substantial alterations in the methylation of IG-DMR (Figs. 4D and 4E).
PQ Dysregulates Sp3 Expression in N2a Cells
Among all the potential targets of miR-380-3p, Sp3 had been recognized as functional in many systems (Gandhy et al., 2015; Göllner et al., 2001; Krüger et al., 2007; van Loo et al., 2007). Thus, we investigated the involvement of Sp3 in PQ-induced neurotoxicity. First, PQ was found to cause disordered Sp3 expression. At the early stage of exposure or at lower concentration, the Sp3 mRNA and protein level in N2a cells were elevated in response to PQ exposure, but subsequently reduced to levels lower than the untreated group as the treatment proceeded both in time or concentration (Figs. 5A and 5B, p < .05, p < .001, or p < .0001, respectively).
Figure 5.
Specificity protein-3 (Sp3) is involved in the paraquat (PQ)-induced neurotoxicity cascade in Neuro2a (N2a) cells. A, Total RNA samples were extracted from N2a cells after exposure to 100 or 200 μM PQ for 24 or 48 h. After 24 h, 200 μM PQ increased the Sp3 mRNA level in N2a cells, n = 3. *p < .05 for the comparisons labeled by the zig-zag lines. At 48 h post treatment, the Sp3 mRNA level was reduced by 200 μM PQ, but elevated by 100 μM PQ, n = 3. *p < .05, ***p < .001, or ****p < .0001 for the contrasts labeled by zig-zag lines. B, Total protein samples were extracted from N2a cells after exposure to 100 or 200 μM PQ for 48 h and semiquantitatively analyzed by Western blots. The Sp3 protein level was reduced by 200 μM PQ but elevated by 100 μM PQ, n = 3. *p < .05 or **p < .01 for the contrasts labeled by zig-zag lines. C, Total RNA samples were extracted from N2a cells 24 h after transfection of siRNAs targeting Sp3 mRNA. The numbers 659, 914, and 1643 represent the location of siRNA binding sites on the Sp3 mRNA consensus sequence. siRNA 659 was the most efficient among the 3, n = 3. **p < .01 or ***p < .001 for the contrasts against the scrambled sequence group. D, Total protein samples were extracted from N2a cells 48 h after transient transfection of siRNA 659 and semiquantitatively analyzed by Western blots, n = 3. *p < .05 versus the scrambled sequence group. E, Cell proliferation was measured 48 h after transfection and found decreased by siRNA 659, n = 3. ****p < .0001 versus the corresponding negative control group. F, Cell proliferation was measured 48 h after cotransfection of miR-380-3p mimics/NC sequence and siRNA 659/scrambled control, n = 6. nsp > .05, ***p < .001, or ****p < .0001 for the contrasts labeled by zig-zag lines. G, Cell proliferation was measured 48 h after cotransfection of miR-380-3p inhibitors/NC sequence and siRNA 659/scrambled control, n = 6. p > .05 for the contrasts between miR-380-3p inhibition groups and the corresponding NC groups.
Next, we determined whether Sp3 had an effect on PQ-induced neurotoxicity. Several siRNAs targeting Sp3 mRNA were designed (Table 4) and the knockdown efficiency was examined (Figs. 5C and 5D, p < .05, p < .01, or p < .001, respectively). After specific knockdown of Sp3, the proliferation of N2a cells, compared to the groups transfected with negative control sequence, was found generally lower (Figure 5E, p < .0001), analogous to the effect of miR-380-3p overexpression.
Table 4.
The Sequences of Designed siRNAs Targeting Specificity Protein-3 (Sp3) mRNA
| ID | Sense (5′-3′) | Antisense (5′-3′) |
|---|---|---|
| mmu-Sp3-659 | GCACCAGGAUCAGAUUCAUTT | AUGAAUCUGAUCCUGGUGCTT |
| mmu-Sp3-914 | GCAAUUGGUGGCUCAUCAUTT | AUGAUGAGCCACCAAUUGCTT |
| mmu-Sp3-1643 | GCUGCCCAACAAAUUACUUTT | AAGUAAUUUGUUGGGCAGCTT |
| Negative control | UUCUCCGAACGUGUCACGUTT | ACGUGACACGUUCGGAGAATT |
Because miR-380-3p targets Sp3 mRNA and exerts an effect similar to Sp3 siRNA, it is probable that the effect of miR-380-3p occurs predominantly by regulating Sp3.To assess if Sp3 deficiency would abolish the effects of miR-380-3p and thus to determine whether miR-380-3p was mediated via Sp3, we knocked down the Sp3 mRNA prior to the application of miR-380-3p mimics or inhibitors and examined cell proliferation. Knockdown of Sp3 seemed to narrow the gap between cells transfected with mimics and the corresponding negative control groups (Figure 5F, p < .001, p < .0001, or p > .05, respectively). However, cell proliferation was not altered by miR-380-3p inhibition (Figure 5G, p > .05).
CaM and p21 Are Dysregulated by PQ
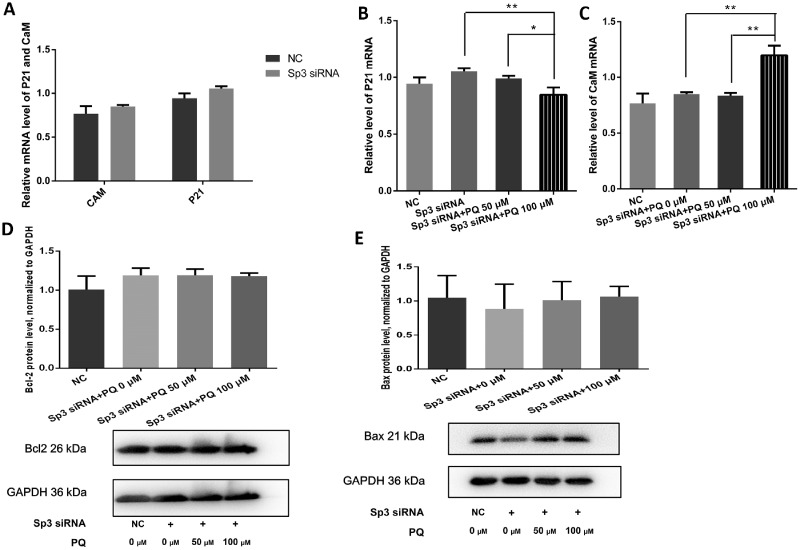
As well as being regulated by Sp3 (Pan et al., 2000; Sowa et al., 1999), p21 is a potent cyclin-dependent kinase inhibitor and a key regulator in cell cycle progression (Gartel and Radhakrishnan, 2005) and apoptosis (Almond and Cohen, 2002), whereas CaM plays a role in many crucial cellular processes, such as metabolism, inflammation, apoptosis, short- and long-term memory, and immune response (Chin and Means, 2000; Lledo et al., 1995; Nishizawa et al., 1988). In this study decreased expression of Sp3, as well as lower concentrations of PQ (50 μM), failed to significantly alter the expression of CaM or p21 (Figure 6A, p > .05). However, the p21 mRNA abundance was Sp3-independently reduced by 100 μM PQ (Figure 6B, p < .05 or p < .01, respectively), whereas the CaM mRNA level increased following exposure to 100 μM PQ (Figure 6C, p < .01). Sp3 knockdown did not alter CaM and p21 expression levels.
Figure 6.
Calmodulin (CaM) and cyclin-dependent kinase inhibitor 1 (p21) were dysregulated by paraquat (PQ). A, Total RNA samples were extracted from Neuro2a (N2a) cells 48 h after transient transfection. Knockdown of specificity protein-3 (Sp3) did not alter the p21 and CaM mRNA level in N2a cells, n = 3, p > .05. B and C, Total RNA samples were collected 48 h after transient transfection and exposure to PQ. p21 and CaM mRNA level were altered by Sp3 siRNA but PQ, n = 3. *p < .05 or **p < .01 for the contrasts labeled by zig-zag lines. D, Western blots of Bcl2 protein from N2a cells 48 h posttransient transfection and treatment with PQ, n = 3, p > .05. E, Western blots of Bax protein from N2a cells 48 h posttransient transfection and treatment with PQ, n = 3, p > .05.
To explore other possible pathways through which Sp3 may cast its effects, protein levels of 2 critical apoptosis mediators, Bcl2 and Bax (Cimmino et al., 2005; Czabotar et al., 2009), were examined. The expression of Bax or Bcl2 was not significantly altered by either PQ treatment or Sp3 interference (Figs. 6D and 6E, p > .05).
DISCUSSION
Various hypotheses have been advanced to the mechanism/s of PQ-induced dopaminergic neuron injury. Apart from causing long-lasting dopamine overflow that keeps dopaminergic neuron in constant excitement and leads to excitotoxic injury (Shimizu et al., 2003), PQ also inhibits mitochondrial respiratory complex I and causes mitochondrial dysfunction, thus inducing neuronal apoptosis or necrosis (Hauser and Hastings, 2013). Furthermore, the synthesis and fibrosis of alpha-synuclein in murine dopaminergic neuron has been shown to be accelerated by PQ, contributing to the formation of Lewy bodies, a cardinal feature of PD (Manning-Bog et al., 2002). In addition, widespread oxidative/nitrosative stress underlies its neurotoxicity, attesting to the critical role of Nrf2 as a key mediator of PQ-induced cell injury.
As critical mediators, potentially regulating the translation of at least 30% of vertebrate proteins (Ellwanger et al., 2011; Krek et al., 2005), miRNAs are highly relevant to neurodegeneration (Bicchi et al., 2013; Nelson et al., 2008; Roth et al., 2016). In our previous study, the expression of a set of miRNAs in mouse substantia nigra was dysregulated by PQ, and miR-380-3p, which was restricted to brains of adult mice (Seitz et al., 2004), was downregulated as evidenced by ChIP analysis (Wang et al., 2017).Yet, the biological function of miR-380-3p has yet to be determined. Accordingly, herein, we focused on potential target genes of miR-380-3p, such as Sp3.Though the negative effect of miR-380-3p on cell survival and proliferation has been verified in the present study, it has yet to be established that miR-380-3p increases the risk of PD or other neurodegenerative disorders.
Sp3, better known as transcriptional factor Sp3, is a main member of Sp-like transcriptional factor family, and it is ubiquitously maintained in a wide spectrum of species, ranging from nematodes to human beings (Philipsen and Suske, 1999). Sp3 deficiency causes ossification impairment (Göllner et al., 2001), cardiac malfunction (van Loo et al., 2007), erythropoiesis impairment, and placental defects (Krüger et al., 2007). Specific knockdown of Sp3 also reduces cell proliferation in HepG2 cells (Gandhy et al., 2015). Furthermore, Sp3 is an established competitor of Sp1, and moderately reduces the transcriptional function of Sp1 (Yu et al., 2003), the latter promoting the synthesis of microtubule-associated protein tau (Maloney and Lahiri, 2012) and amyloid precursor protein (Docagne et al., 2004), key molecules associated with the progression of PD and Alzheimer’s disease (AD). In the present study, Sp3 deficiency induced by transfected miR-380-3p mimics and small interference RNAs also played a negative role in nerve cell survival and proliferation.
Though all of the above suggest a potential role of Sp3 in neurodegeneration, few studies have addressed the mechanism underlying the role of Sp3 in neurological disorders. In this study, p21 and CaM expression were altered by exposure to PQ, corroborating their role in PQ-induced neurotoxicity. Knockdown of Sp3 failed to alter the regulation of p21 and CaM, implying that Sp3, though verified in previous studies as being transcriptionally regulating p21 and CaM, probably failed to exert a major influence on p21 and CaM expression. As it was previously shown, in addition to its abundance, phosphorylation of Sp3 is positively correlated with its binding to the promoter (Chu et al., 2003). It has also been reported that SUMOylation (SUMO, small ubiquitin-related modifier) of Sp3 is one of the main regulatory events controlling the activity of this transcription factor, and potentially the molecular switch that determines whether Sp3 functions as an transcriptional activator or a repressor (Stielow et al., 2010). Such regulatory events may also be involved in the toxic cascade induced by PQ, altering the function of Sp3. As a main member of the Sp family, Sp3 has its binding box in the promotor region in a wide range of genes, which in fact provides it access to the control room of all kinds of downstream molecules via which Sp3 conducts its function. But in the present study, p21, CaM, and 2 well-known apoptosis mediators, Bcl2 and Bax, were unlikely to be among these downstream molecules. However, the N2a cells adopted in the present study was found to give responses different from other neuron or neuron-like cells (LePage et al., 2005), so it is too early to exclude these molecules from all situations (Figure 7).
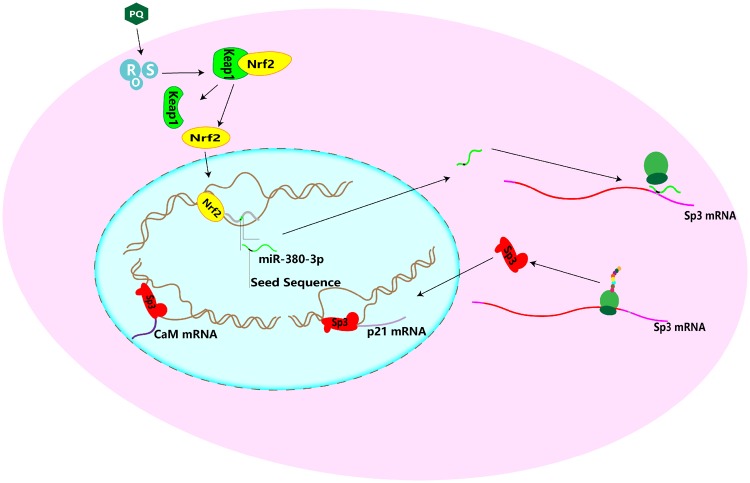
Figure 7.
Proposed scheme of Nrf2/miR380-3p/Sp3 pathway. Paraquat translocates nuclear factor erythroid 2 related factor 2 (Nrf2) to the nucleus (the dashed oval) upon dislocation from Kelch-like ECH-associated protein 1 (Keap1). Next, Nrf2 actives the expression of miR-380-3p. It further attenuates the translation of specificity protein-3 (Sp3) mRNA. Sp3 might regulate the expression of downstream molecules such as Calmodulin (CaM) and cyclin-dependent kinase inhibitor 1 (p21).
Our novel studies established miR-380-3p bound to the 3′UTR of Sp3 and restrained the translation of Sp3 protein, suppressing N2a cell proliferation and enhancing the toxicity of PQ. In addition, knockdown of Sp3 altered N2a cell proliferation in a fashion analogous to miR-380-3p, and narrowed, or even eliminated, the gap between cells overexpressing miR-380-3p and the negative control groups. Surprisingly, inhibition of miR-380-3p did not induce detectable alterations in cell proliferation, apoptosis rate or cell cycle proportion, which is probably due to the low level of endogenous miR-380-3p in N2a cells; miR-380-3p was only detected in embryos and the adult brain at a fairly low level (Seitz et al., 2004). The low abundance of miR-380-3p likely makes it more difficult for inhibitors to exert their effects, mandating higher concentrations of inhibitors to achieve measurable inhibition. Thus, the Sp3 protein level was not elevated to a degree that was sufficiently high to impart obvious alterations in cell proliferation, apoptosis rate, or cell cycle proportion. In addition, N2a cells have been found by previous researchers to be much less sensitive to certain neurotoxins than some other neuron or neuron-like cells, mandating caution to extrapolate in capacity of miR-380-3p deficiency in vivo or in other cell lines from negative results in N2a cells (LePage et al., 2005).
Nrf2 is a transcriptional factor of many antioxidant enzymes. It is recognized as a crucial protective mediator in oxidant-induced neuronopathies. Earlier studies have established that several miRs are transcriptionally regulated by Nrf2 (Kurinna et al., 2014; Shah et al., 2014; Singh et al., 2013). Here, we report, for the first time, that miR-380-3p transcriptionally regulated by Nrf2, and inhibited the translation of Sp3 mRNA, suggesting that Nrf2 regulates Sp3 posttranscriptionally via miR-380-3p. This seems to counter the guardian-like portrait of Nrf2. In fact, in addition to Sp3 deficiency, Sp3 overexpression was also found to induce neuron necrosis (Citron et al., 2008). Notably, Sp3 overexpression has already been identified in the brains of patients suffering from neurodegenerations (Boutillier et al., 2007). All the documented results above suggest the importance to keep Sp3 at an appropriate level that is neither too high nor low. Thus, we posit that Nrf2 likely serves to facilitate neuron survival by restraining detrimental overexpression of Sp3 through miR-380-3p. Tempered enhancement of miR-380-3p, such as one induced by Nrf2, may function to restore Sp3 homeostasis, and thus offer protection against neurodegeneration. Additional studies are clearly mandated to better characterize the function of Sp3 in more specific situations, given the intricate regulation that Sp3 is under.
Noteworthy, the fact that miR-380-3p is encoded within a miRNA cluster, suggests that other miRs, such as miR-379, miR-411, miR-329, to name a few, within the cluster are likely to be transcriptionally regulated by Nrf2; all these miRs are probably functional. For example, miR-380-5p, a miR sharing precursor with miR-380-3p, was found to target p53, a potent molecule known as the guardian of the genome, and function in cellular survival of neuroblastoma (Swarbrick et al., 2010), implying that Nrf2 may repress the activity of p53 via miR-380-5p. Additionally, a particular miRNA in vertebrates can regulate the translation of as many as hundreds of different mRNAs (Krek et al., 2005). Thus, it is feasible that some other mRNAs, such as Cu5, Clcn5, Mdga2, Tceb3, Wapal, and Zbtb20, predicted by sequence-based analysis, might be targeted by miR-380-3p and contribute to its effects. However, the Nrf2/miR-380-3p/Sp3 pathway has yet to be determined as specific for PQ or widely involved oxidative stress-induced neurotoxicity.
PQ was demonstrated to attenuate the expression of miR-380-3p. In earlier studies, oxidative stress was shown to increase the expression of miR-153 and miR-146a, and these miRs subsequently inhibited the translation of Nrf2 (Jiao et al., 2015; Narasimhan et al., 2014; Smith et al., 2015). These effects may represent possible mechanisms by which impairs the expression of miR-380-3p. We also explored other possible pathways by which PQ might decrease miR-380-3p levels, such as changes in methylation in regions regulating the expression of miR-380-3p, namely the intergenic differentially methylated regions (IG-DMRs). The IG-DMRs were invariably highly methylated in response to treatment with various concentrations of PQ, suggesting that changes in methylation are unlikely to account for mechanisms by which PQ reduces the expression of miR-380-3p in N2a cells. However, the high methylation level may explain the restriction of miR-380-3p abundance, because high methylation level in the IG-DMR silenced the miRNA gene cluster encoding miR-380-3p (Seitz et al., 2004). The underlying mechanisms of PQ-induced neurotoxicity remain to be fully understood. Our future work should include further exploration on how PQ attenuates miR-380-3p expression, and a better characterization of the function of miR-380-3p.
AUTHOR CONTRIBUTIONS
H.L. and S.W. for conceptualized and supervised the study; Z.C. and F.Z. performed most of the experiments and analyzed the data; Y.D. contributed to experiments; Z.C. wrote the original draft; Y.Z., R.G., and J.L. contributed to discussion; F.Z., M.A., and Q.Z. reviewed and edited the manuscript. All authors have read and approved the final manuscript.
DECLARATION OF CONFLICTING INTERESTS
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING
This work was supported by the National Natural Science Foundation in China (81573195, 81172715, 30800936); the Provincial Natural Science Foundation of Fujian Province (2017J01523); the Project of Health and Family Planning Commission of Fujian Province (2017-1-63); and the Joint Funds for the Innovation of Science and Technology, Fujian Province (2017Y9105). M.A. was supported in part by grants from the National Institute of Environmental Health Sciences (R01ES07331, R01ES10563, R01ES020852).
REFERENCES
- Almond J. B., Cohen G. M. (2002). The proteasome: A novel target for cancer chemotherapy. Leukemia 16, 433–443. [DOI] [PubMed] [Google Scholar]
- Anderegg A., Lin H. P., Chen J. A., Caronia-Brown G., Cherepanova N., Yun B., Joksimovic M., Rock J., Harfe B. D., Johnson R., et al. (2013). An Lmx1b-miR135a2 regulatory circuit modulates Wnt1/Wnt signaling and determines the size of the midbrain dopaminergic progenitor pool. PLoS Genet. 9, e1003973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow B. K., Thiruchelvam M. J., Bennice L., Cory-Slechta D. A., Ballatori N., Richfield E. K. (2003). Increased synaptosomal dopamine content and brain concentration of paraquat produced by selective dithiocarbamates. J. Neurochem. 85, 1075–1086. [DOI] [PubMed] [Google Scholar]
- Berry C., La Vecchia C., Nicotera P. (2010). Paraquat and Parkinson’s disease. Cell Death Differ. 17, 1115–1125. [DOI] [PubMed] [Google Scholar]
- Bicchi I., Morena F., Montesano S., Polidoro M., Martino S. (2013). MicroRNAs and molecular mechanisms of neurodegeneration. Genes (Basel) 4, 244–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneh-Barkay D., Langston W. J., Di Monte D. A. (2005a). Toxicity of redox cycling pesticides in primary mesencephalic cultures. Antioxid. Redox Signal. 7, 649–653. [DOI] [PubMed] [Google Scholar]
- Bonneh-Barkay D., Reaney S. H., Langston W. J., Di Monte D. A. (2005b). Redox cycling of the herbicide paraquat in microglial cultures. Mol. Brain Res. 134, 52–56. [DOI] [PubMed] [Google Scholar]
- Boutillier S., Lannes B., Buée L., Delacourte A., Rouaux C., Mohr M., Bellocq J. P., Sellal F., Larmet Y., Boutillier A. L., et al. (2007). Sp3 and Sp4 transcription factor levels are increased in brains of patients with Alzheimer’s disease. Neurodegener. Dis. 4, 413–423. [DOI] [PubMed] [Google Scholar]
- Chin D., Means A. R. (2000). Calmodulin: A prototypical calcium sensor. Trends Cell Biol. 10, 322–328. [DOI] [PubMed] [Google Scholar]
- Chu S., Cockrell C. A., Ferro T. J. (2003). Expression of α-ENaC2 is dependent on an upstream Sp1 binding motif and is modulated by protein phosphatase 1 in lung epithelial cells. Biochem. Biophys. Res. Commun. 303, 1159–1168. [DOI] [PubMed] [Google Scholar]
- Cimmino A., Calin G. A., Fabbri M., Iorio M. V., Ferracin M., Shimizu M., Wojcik S. E., Aqeilan R. I., Zupo S., Dono M., et al. (2005). miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. U.S.A. 102, 13944–13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citron B. A., Dennis J. S., Zeitlin R. S., Echeverria V. (2008). Transcription factor Sp1 dysregulation in Alzheimer’s disease. J. Neurosci. Res. 86, 2499–2504. [DOI] [PubMed] [Google Scholar]
- Czabotar P. E., Colman P. M., Huang D. C. S. (2009). Bax activation by Bim? Cell Death Differ 16, 1187–1191. [DOI] [PubMed] [Google Scholar]
- De Long M. J., Prochaska H. J., Talalay P. (1986). Induction of NAD(P)H: Quinone reductase in murine hepatoma cells by phenolic antioxidants, azo dyes, and other chemoprotectors: A model system for the study of anticarcinogens. Proc. Natl. Acad. Sci. U.S.A. 83, 787–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mena L., Cardo L. F., Coto E., Miar A., Diaz M., Corao A. I., Alonso B., Ribacoba R., Salvador C., Menendez M., et al. (2010). FGF20 rs12720208 SNP and microRNA-433 variation: No association with Parkinson’s disease in Spanish patients. Neurosci. Lett. 479, 22–25. [DOI] [PubMed] [Google Scholar]
- Dinis-Oliveira R. J., Remião F., Carmo H., Duarte J. A., Navarro A. S., Bastos M. L., Carvalho F. (2006). Paraquat exposure as an etiological factor of Parkinson’s disease. Neurotoxicology 27, 1110–1122. [DOI] [PubMed] [Google Scholar]
- Dinkova-Kostova A. T., Talalay P. (2010). NAD(P)H: Quinone acceptor oxidoreductase 1 (NQO1), a multifunctional antioxidant enzyme and exceptionally versatile cytoprotector. Arch. Biochem. Biophys. 501, 116–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docagne F., Gabriel C., Lebeurrier N., Lesné S., Hommet Y., Plawinski L., Mackenzie E. T., Vivien D. (2004). Sp1 and Smad transcription factors co-operate to mediate TGF-beta-dependent activation of amyloid-beta precursor protein gene transcription. Biochem. J. 383, 393–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dröge W., Schipper H. M. (2007). Oxidative stress and aberrant signaling in aging and cognitive decline. Aging Cell 6, 361–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellwanger D. C., Büttner F. A., Mewes H. W., Stümpflen V. (2011). The sufficient minimal set of miRNA seed types. Bioinformatics 27, 1346–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiaschetti G., Abela L., Nonoguchi N., Dubuc A. M., Remke M., Boro A., Grunder E., Siler U., Ohgaki H., Taylor M. D., et al. (2014). Epigenetic silencing of miRNA-9 is associated with HES1 oncogenic activity and poor prognosis of medulloblastoma. Br. J. Cancer 110, 636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitsanakis V. A., Amarnath V., Moore J. T., Montine K. S., Zhang J., Montine T. J. (2002). Catalysis of catechol oxidation by metal-dithiocarbamate complexes in pesticides. Free Radic. Biol. Med. 33, 1714–1723. [DOI] [PubMed] [Google Scholar]
- Gandhy S. U., Imanirad P., Jin U.-H., Nair V., Hedrick E., Cheng Y., Corton C. J., Kim K., Safe S. (2015). Specificity protein (Sp) transcription factors and metformin regulate expression of the long non-coding RNA HULC. Oncotarget 6, 26359–26372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartel A. L., Goufman E., Najmabadi F., Tyner A. L. (2000). Sp1 and Sp3 activate p21 (WAF1/CIP1) gene transcription in the Caco-2 colon adenocarcinoma cell line. Oncogene 19, 5182–5188. [DOI] [PubMed] [Google Scholar]
- Gartel A. L., Radhakrishnan S. K. (2005). Lost in transcription: P21 repression, mechanisms, and consequences. Cancer Res. 65, 3980–3985. [DOI] [PubMed] [Google Scholar]
- Göllner H., Dani C., Phillips B., Philipsen S., Suske G. (2001). Impaired ossification in mice lacking the transcription factor Sp3. Mech. Dev. 106, 77–83. [DOI] [PubMed] [Google Scholar]
- Goodall E. F., Heath P. R., Bandmann O., Kirby J., Shaw P. J. (2013). Neuronal dark matter: The emerging role of microRNAs in neurodegeneration. Front. Cell. Neurosci. 7, 178.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser D. N., Hastings T. G. (2013). Mitochondrial dysfunction and oxidative stress in Parkinson’s disease and monogenic parkinsonism. Neurobiol. Dis. 51, 35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hébert S. S., De Strooper B. (2007). Molecular biology. miRNAs in neurodegeneration. Science 317, 1179–1180. [DOI] [PubMed] [Google Scholar]
- Itoh K., Wakabayashi N., Katoh Y., Ishii T., Igarashi K., Engel J. D., Yamamoto M. (1999). Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 13, 76–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao G., Pan B., Zhou Z., Zhou L., Li Z., Zhang Z. (2015). MicroRNA-21 regulates cell proliferation and apoptosis in H2O2-stimulated rat spinal cord neurons. Mol. Med. Rep. 12, 7011–7016. [DOI] [PubMed] [Google Scholar]
- Johnson F., Giulivi C. (2005). Superoxide dismutases and their impact upon human health. Mol. Aspects Med. 26, 340–352. [DOI] [PubMed] [Google Scholar]
- Kieburtz K., Wunderle K. B. (2013). Parkinson’s disease: Evidence for environmental risk factors. Mov. Disord. 28, 8–13. [DOI] [PubMed] [Google Scholar]
- Krek A., Grün D., Poy M. N., Wolf R., Rosenberg L., Epstein E. J., MacMenamin P., da Piedade I., Gunsalus K. C., Stoffel M., et al. (2005). Combinatorial microRNA target predictions. Nat. Genet. 37, 495–500. [DOI] [PubMed] [Google Scholar]
- Krüger I., Vollmer M., Simmons D., Elsässer H. P., Philipsen S., Suske G. (2007). Sp1/Sp3 compound heterozygous mice are not viable: Impaired erythropoiesis and severe placental defects. Dev. Dyn. 236, 2235–2244. [DOI] [PubMed] [Google Scholar]
- Kurinna S., Schäfer M., Ostano P., Karouzakis E., Chiorino G., Bloch W., Bachmann A., Gay S., Garrod D., Lefort K., et al. (2014). A novel Nrf2-miR-29-desmocollin-2 axis regulates desmosome function in keratinocytes. Nat. Commun. 5, 5099. [DOI] [PubMed] [Google Scholar]
- Kuwahara T., Koyama A., Gengyo-Ando K., Masuda M., Kowa H., Tsunoda M., Mitani S., Iwatsubo T. (2006). Familial Parkinson mutant α-synuclein causes dopamine neuron dysfunction in transgenic Caenorhabditis elegans. J. Biol. Chem. 281, 334–340. [DOI] [PubMed] [Google Scholar]
- LePage K. T., Dickey R. W., Gerwick W. H., Jester E. L., Murray T. F. (2005). On the use of neuro-2a neuroblastoma cells versus intact neurons in primary culture for neurotoxicity studies. Crit. Rev. Neurobiol. 17, 27–50. [DOI] [PubMed] [Google Scholar]
- Li H., Wu S., Wang Z., Lin W., Zhang C., Huang B. (2012). Neuroprotective effects of tert-butylhydroquinone on paraquat-induced dopaminergic cell degeneration in C57BL/6 mice and in PC12 cells. Arch. Toxicol. 86, 1729–1740. [DOI] [PubMed] [Google Scholar]
- Li J., Johnson D., Calkins M., Wright L., Svendsen C., Johnson J. (2005). Stabilization of Nrf2 by tBHQ confers protection against oxidative stress-induced cell death in human neural stem cells. Toxicol. Sci. 83, 313–328. [DOI] [PubMed] [Google Scholar]
- Li S., Li J., Shen C., Zhang X., Sun S., Cho M., Sun C., Song Z. (2014). tert-Butylhydroquinone (tBHQ) protects hepatocytes against lipotoxicity via inducing autophagy independently of Nrf2 activation. Biochim. Biophys. Acta 1841, 22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang R., Bates D., Wang E. (2009). Epigenetic control of microRNA expression and aging. Curr. Genomics 10, 184–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddell J. R., Dringen R., Crack P. J., Robinson S. R. (2006). Glutathione peroxidase 1 and a high cellular glutathione concentration are essential for effective organic hydroperoxide detoxification in astrocytes. Glia 54, 873–879. [DOI] [PubMed] [Google Scholar]
- Lin S. P., Youngson N., Takada S., Seitz H., Reik W., Paulsen M., Cavaille J., Ferguson-Smith A. C. (2003). Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat. Genet. 35, 97–102. [DOI] [PubMed] [Google Scholar]
- Lledo P. M., Hjelmstad G. O., Mukherji S., Soderling T. R., Malenka R. C., Nicoll R. A. (1995). Calcium/calmodulin-dependent kinase II and long-term potentiation enhance synaptic transmission by the same mechanism. Proc. Natl. Acad. Sci. U.S.A. 92, 11175–11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney B., Lahiri D. K. (2012). Structural and functional characterization of H2 haplotype MAPT promoter: Unique neurospecific domains and a hypoxia-inducible element would enhance rationally targeted tauopathy research for Alzheimer’s disease. Gene 501, 63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning-Bog A. B., McCormack A. L., Li J., Uversky V. N., Fink A. L., Di Monte D. A. (2002). The herbicide paraquat causes up-regulation and aggregation of alpha-synuclein in mice: Paraquat and alpha-synuclein. J. Biol. Chem. 277, 1641–1644. [DOI] [PubMed] [Google Scholar]
- McCormack A. L., Atienza J. G., Johnston L. C., Andersen J. K., Vu S., Di Monte D. A. (2005). Role of oxidative stress in paraquat-induced dopaminergic cell degeneration. J. Neurochem. 93, 1030–1037. [DOI] [PubMed] [Google Scholar]
- McCormack A. L., Atienza J. G., Langston J. W., Di Monte D. A. (2006). Decreased susceptibility to oxidative stress underlies the resistance of specific dopaminergic cell populations to paraquat-induced degeneration. Neuroscience 141, 929–937. [DOI] [PubMed] [Google Scholar]
- Narasimhan M., Riar A. K., Rathinam M. L., Vedpathak D., Henderson G., Mahimainathan L. (2014). Hydrogen peroxide responsive miR153 targets Nrf2/ARE cytoprotection in paraquat induced dopaminergic neurotoxicity. Toxicol. Lett. 228, 179–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson P. T., Wang W. X., Rajeev B. W. (2008). MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol. 18, 130–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa Y., Okui Y., Inaba M., Okuno S., Yukioka K., Miki T., Watanabe Y., Morii H. (1988). Calcium/calmodulin-mediated action of calcitonin on lipid metabolism in rats. J. Clin. Invest. 82, 1165–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Solomon S. S., Shah R. J., Palazzolo M. R., Raghow R. S. (2000). Members of the Sp transcription factor family regulate rat calmodulin gene expression. J. Lab. Clin. Med. 136, 157–163. [DOI] [PubMed] [Google Scholar]
- Philipsen S., Suske G. (1999). A tale of three fingers: The family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 27, 2991–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappold P. M., Cui M., Chesser A. S., Tibbett J., Grima J. C., Duan L., Sen N., Javitch J. A., Tieu K. (2011). Paraquat neurotoxicity is mediated by the dopamine transporter and organic cation transporter-3. Proc. Natl. Acad. Sci. U.S.A. 108, 20766–20771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth W., Hecker D., Fava E. (2016). Systems biology approaches to the study of biological networks underlying Alzheimer’s disease: Role of miRNAs. Methods Mol. Biol. 1303, 349–377. [DOI] [PubMed] [Google Scholar]
- Seitz H., Royo H., Bortolin M. L., Lin S. P., Ferguson-Smith A. C., Cavaillé J. (2004). A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res. 14, 1741–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah N. M., Zaitseva L., Bowles K. M., MacEwan D. J., Rushworth S. A. (2014). NRF2-driven miR-125B1 and miR-29B1 transcriptional regulation controls a novel anti-apoptotic miRNA regulatory network for AML survival. Cell Death Differ. 44, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K., Matsubara K., Ohtaki K., Fujimaru S., Saito O., Shiono H. (2003). Paraquat induces long-lasting dopamine overflow through the excitotoxic pathway in the striatum of freely moving rats. Brain Res. 976, 243–252. [DOI] [PubMed] [Google Scholar]
- Shimura H., Hattori N., Kubo S. I., Mizuno Y., Asakawa S., Minoshima S., Shimizu N., Iwai K., Chiba T., Tanaka K., et al. (2000). Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat. Genet. 25, 302–305. [DOI] [PubMed] [Google Scholar]
- Siegel D., Ross D. (2000). Immunodetection of NAD(P)H: Quinone oxidoreductase 1 (NQO1) in human tissues. Free Radic. Biol. Med. 29, 246–253. [DOI] [PubMed] [Google Scholar]
- Singh A., Happel C., Manna S. K., Acquaah-Mensah G., Carrerero J., Kumar S., Nasipuri P., Krausz K. W., Wakabayashi N., Dewi R., et al. (2013). Transcription factor NRF2 regulates miR-1 and miR-206 to drive tumorigenesis. J. Clin. Invest. 123, 2921–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E. J., Shay K. P., Thomas N. O., Butler J. A., Finlay L. F., Hagen T. M. (2015). Age-related loss of hepatic Nrf2 protein homeostasis: Potential role for heightened expression of miR-146a. Free Radic. Biol. Med. 89, 1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Kang S. M., Lee W. T., Park K. A., Lee K. M., Lee J. E. (2014). Glutathione protects brain endothelial cells from hydrogen peroxide-induced oxidative stress by increasing Nrf2 expression. Exp. Neurobiol. 23, 93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa Y., Orita T., Minamikawa-Hiranabe S., Mizuno T., Nomura H., Sakai T. (1999). Sp3, but not Sp1, mediates the transcriptional activation of the p21/WAF1/Cip1 gene promoter by histone deacetylase inhibitor. Cancer Res. 59, 4266–4270. [PubMed] [Google Scholar]
- Stachurska A., Ciesla M., Kozakowska M., Wolffram S., Boesch-Saadatmandi C., Rimbach G., Jozkowicz A., Dulak J., Loboda A. (2013). Cross-talk between microRNAs, nuclear factor E2-related factor 2, and heme oxygenase-1 in ochratoxin A-induced toxic effects in renal proximal tubular epithelial cells. Mol. Nutr. Food Res. 57, 504–515. [DOI] [PubMed] [Google Scholar]
- Stielow B., Krüger I., Diezko R., Finkernagel F., Gillemans N., Kong-a-San J., Philipsen S., Suske G. (2010). Epigenetic silencing of spermatocyte-specific and neuronal genes by SUMO modification of the transcription factor Sp3. PLoS Genet. 6, e1001203.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbrick A., Woods S. L., Shaw A., Balakrishnan A., Phua Y., Nguyen A., Chanthery Y., Lim L., Ashton L. J., Judson R. L., et al. (2010). miR-380-5p represses p53 to control cellular survival and is associated with poor outcome in MYCN-amplified neuroblastoma. Nat. Med. 16, 1134–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner C. M., Kame F., Ross G. W., Hoppin J. A., Goldman S. M., Korell M., Marras C., Bhudhikanok G. S., Kasten M., Chade A. R., et al. (2013). Rotenone, paraquat, and Parkinson’s disease. Mov. Disord. 23(Suppl. 2), 8–13. [Google Scholar]
- van Jaarsveld M. T. M., Helleman J., Boersma A. W. M., van Kuijk P. F., van Ijcken W. F., Despierre E., Vergote I., Mathijssen R. H. J., Berns E. M. J. J., Verweij J., et al. (2013). miR-141 regulates KEAP1 and modulates cisplatin sensitivity in ovarian cancer cells. Oncogene 32, 4284–4293. [DOI] [PubMed] [Google Scholar]
- van Loo P. F., Mahtab E. A. F., Wisse L. J., Hou J., Grosveld F., Suske G., Philipsen S., Gittenberger-de Groot A. C. (2007). Transcription factor Sp3 knockout mice display serious cardiac malformations. Mol. Cell. Biol. 27, 8571–8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Ren N., Cai Z., Lin Q., Wang Z., Zhang Q., Wu S., Li H. (2017). Paraquat and MPTP induce neurodegeneration and alteration in the expression profile of microRNAs: The role of transcription factor Nrf2. NPJ Parkinsons Dis. 3, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. (2014). Composition of seed sequence is a major determinant of microRNA targeting patterns. Bioinformatics 30, 1377–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. J., Tao H., Hu W., Liu L. P., Shi K. H., Deng Z. Y., Li J. (2014). MicroRNA-200a controls Nrf2 activation by target Keap1 in hepatic stellate cell proliferation and fibrosis. Cell. Signal. 26, 2381–2389. [DOI] [PubMed] [Google Scholar]
- Yang M., Yao Y., Eades G., Zhang Y., Zhou Q. (2011). MiR-28 regulates Nrf2 expression through a Keap1-independent mechanism. Breast Cancer Res. Treat. 129, 983–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Datta P. K., Bagchi S. (2003). Stability of the Sp3-DNA complex is promoter-specific: Sp3 efficiently competes with Sp1 for binding to promoters containing multiple Sp-sites. Nucleic Acids Res. 31, 5368–5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Qian J., Cao Q., Li P., Wang M., Wang J., Ju X., Meng X., Lu Q., Shao P., et al. (2014). A potentially functional polymorphism in the promoter region of miR-34b/c is associated with renal cell cancer risk in a Chinese population. Mutagenesis 29, 149–154. [DOI] [PubMed] [Google Scholar]