Glasshouse-based metabolic markers predict yield gap-based drought tolerance determined from multiseason and site field trials which could serve as a potential breeding tool for selecting drought-resilient crops.
Keywords: Amino acids, drought stress, grain yield, metabolomics, RWC, stomatal conductance, wheat
Abstract
Water limits crop productivity, so selecting for a minimal yield gap in drier environments is critical to mitigate against climate change and land-use pressure. We investigated the responses of relative water content (RWC), stomatal conductance, chlorophyll content, and metabolites in flag leaves of commercial wheat (Triticum aestivum L.) cultivars to three drought treatments in the glasshouse and in field environments. We observed strong genetic associations between glasshouse-based RWC, metabolites, and yield gap-based drought tolerance (YDT; the ratio of yield in water-limited versus well-watered conditions) across 18 field environments spanning sites and seasons. Critically, RWC response to glasshouse drought was strongly associated with both YDT (r2=0.85, P<8E-6) and RWC under field drought (r2=0.77, P<0.05). Moreover, multiple regression analyses revealed that 98% of genetic YDT variance was explained by drought responses of four metabolites: serine, asparagine, methionine, and lysine (R2=0.98; P<0.01). Fitted coefficients suggested that, for given levels of serine and asparagine, stronger methionine and lysine accumulation was associated with higher YDT. Collectively, our results demonstrate that high-throughput, targeted metabolic phenotyping of glasshouse-grown plants may be an effective tool for selection of wheat cultivars with high field-derived YDT.
Introduction
Wheat (Triticum aestivum L.) has been a staple food for at least 8000 years (Colledge et al., 2004). It remains one of the most important food crops in terms of harvested area and trade value, and is a major source of energy and nutrition for ~4.5 billion people (Curtis, 2002; United Nations, 2013). With the world population predicted to increase to 9.6 billion by 2050, the demand for wheat is only going to increase (United Nations, 2013). Water availability is critical for wheat production, and drought is the major cause of yield losses. For example, during the ‘Millennium Drought’ (2002–2009), wheat yields in Australia were up to 25% lower than average (van Dijk et al., 2013). With rapidly growing demands on available land and water resources, and the possibility that climate change will increase the frequency and severity of drought events in wheat-growing areas (Trenberth et al., 2014), the generation of wheat cultivars with greater water-use efficiency is of the utmost urgency.
Breeding crops for improved yield gap-based drought tolerance (YDT; the ratio of yield in rainfed versus irrigated field conditions) requires the selection of genotypes with improved yield performance under field drought conditions (Rebetzke et al., 2013). However, field-based drought experiments spanning entire crop cycles are resource intensive, subject to seasonal variability, and thus largely reserved for testing of late-generation breeding lines. Effective high-throughput methods are therefore required to screen large numbers of early-generation lines for their potential drought tolerance. For this purpose, a variety of screening methods have been employed, such as predicting YDT on the basis of growth and physiology of seedlings under osmotic stress in laboratory culture (Munns et al., 2010). Critically, many findings from controlled-environment experiments show little relevance when translated to the field (Passioura, 2006; Rebetzke et al., 2014). To be of real-world significance, it is critical that any drought tolerance marker developed in glasshouse conditions be predictive of YDT in field environments.
Thermographic and hyperspectral leaf reflectance imaging techniques have been employed in high-throughput screening of drought-tolerant crops by estimating traits such as canopy temperature depression (thermographic imaging) and photosynthetic capacity (chlorophyll fluorescence), aerial biomass, and leaf water contents (hyperspectral reflectance) (Blum et al., 1982; Babar et al., 2006). Canopy temperature depression is positively correlated with yield under water stress (Rashid et al., 1999; Rebetzke et al., 2012). However, canopy temperature alone typically explains less than half of variation in YDT (or similar metrics, such as drought susceptibility index) (Blum et al., 1989), leaving the opportunity for improvement. Indeed, early assessments of hyperspectral reflectance indices suggested that some of them are better predictors of grain yield under drought (R2≤0.8) than thermographic imaging (r2<0.5) (Babar et al., 2006). However, hyperspectral systems are usually very expensive, and the data analysis and modelling are not trivial.
An emerging approach for selection of crops with improved performance under water-limiting conditions is to use metabolite-based markers (Degenkolbe et al., 2013). Metabolites are renowned for their information contents (Fiehn, 2002), and some properties associated with metabolite-based markers for drought tolerance make them complementary to or even more powerful than other marker types. High-throughput and cost-effective metabolome-assisted selection methods could enhance the efficiency of breeding not only by providing an indication of drought tolerance but also by pointing to mechanisms of differential tolerance and giving information about nutritional and milling qualities (Shewry et al., 2002). Several studies have investigated associations between metabolite markers and drought tolerance-related traits. For example, Degenkolbe et al. (2013) identified associations between metabolic and physiological traits, both measured in the glasshouse, highlighting a negative association between asparagine levels and water-use efficiency (r2=0.41) and yield (r2=0.50) under drought stress. A recent study in a maize (Zea mays L.) hybrid identified a number of associations between metabolites and yield when both traits were measured in the field (r2≤0.29 for individual metabolites and ≤99% with seven metabolite multiple regression) (Obata et al., 2015). To our knowledge, there has been one attempt (in maize) to identify associations between metabolic traits measured in the glasshouse and drought tolerance determined in the field (Witt et al., 2012), but this did not uncover any significant associations.
In wheat, several metabolomics studies have been performed, such as a targeted metabolite profiling of four wheat cultivars under controlled osmotic stress to show a positive relationship between soluble carbohydrate accumulation and drought tolerance (Kerepesi and Galiba, 2000), glasshouse-based drought experiments comparing the temporal drought responses of three wheat cultivars (Bowne et al., 2012), and quantitative trait locus (QTL) analysis using a set of 179 cv Excalibur/Kukri doubled-haploid wheat lines grown in the field, which revealed significant associations between metabolite levels and grain yield under water-limited conditions (Hill et al., 2013). However, to our knowledge, there have been no broad-spectrum metabolite profiling studies directly analysing associations between the responses of wheat cultivars to drought treatments in the glasshouse- and field-derived YDT.
To provide new insights into potential links between field-based measures and glasshouse experiments, we determined the mean proportional yield deficits (i.e. YDT) of eight commercial wheat cultivars in multi-location/season field experiments. In parallel, we measured relative water content (RWC), stomatal conductance (gs), and untargeted GC/MS metabolomics under glasshouse- and field-based drought stress treatments. From these data, we identified highly significant associations for metabolite profiles and significant associations for the drought-responsive physiological traits in glasshouse-grown wheat and field-based YDT.
Materials and methods
Plant materials, experimental design, and drought treatment under a controlled environment
Seeds from eight wheat cultivars (Kukri, Excalibur, Gladius, Wyalkatchem, Yitpi, RAC875, Drysdale, and Weebill; Supplementary Table S1 at JXB online) were obtained from CSIRO, Black Mountain, Australian Capital Territory, Australia, and grown in the CSIRO controlled-temperature greenhouse, set at 24/16 °C day/night temperature and relative humidity of 60% between June and November 2012. Four seeds from each cultivar of 45–50 mg were sown in large round plastic pots of a dimension 25 cm (bottom diameter)×30 cm (height). A total of 54 pots were filled with fertile, compost-based potting mix, and plants were thinned to two seedlings per pot. There were six pots (12 plants) of each cultivar including a drought-susceptible control line ‘Yenda’. Pots were arranged on five benches in a randomized complete block design in the glasshouse. Day-length extension to 16 h was provided to hasten development in all cultivars so that the anthesis date was approximately the same. Plants were sprayed with pesticides as necessary to avoid damage from fungal or insect pests. All pots were watered daily to pot capacity until 50 d after sowing. From this stage, water was withheld in drought-treated plants until day 15 of the first drought regime when Kukri and Yenda (a drought-susceptible cultivars) started showing severe wilting signs based on visual assessment and then plants were rewatered to 100% soil water capacity (~1.4 litres) to match the sporadic rainfall pattern (Bowne et al., 2012). Drought-treated plants were then subjected to a second drought by withholding water for a further 13 d, making the total drought period 28 d. The control plants were maintained at 100% soil water capacity. An illustration of the glasshouse cyclic drought is presented in Supplementary Fig. S3A. Separate pots within glasshouse experiments were considered independent replicates. Effects were considered significant at P<0.05.
Field-based drought treatment in a managed environment facility
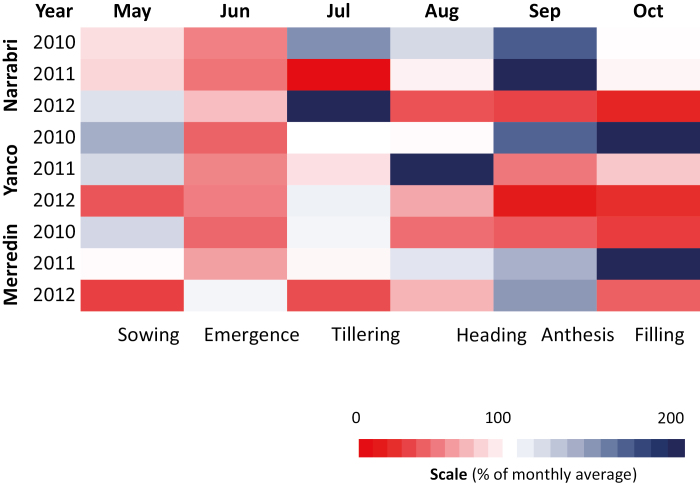
The assessment of variety performance and determination of YDT under terminal drought conditions was undertaken across three seasons at three national managed environment facilities (MEFs) located across Australia. Details of the facilities are summarized in Rebetzke et al. (2013). Briefly, a set of five known drought-susceptible and drought-tolerant wheat cultivars (Gladius, Wyalkatchem, Yitpi, RAC875, and Weebill) were assembled and sown in 2010–2012 at the MEFs in Merredin (Western Australia), Narrabri, and Yanco (New South Wales, Australia) (Rebetzke et al., 2013). At each MEF, the response of the individual cultivars was ascertained in both a well-managed rainfed and an irrigated experiment. The irrigated field trial was designed to supply 25% more water than an average season. The drought in the rainfed plots was terminal in all field environments tested. Precisely, plants experienced severe early-stage drought in all rainfed environments and seasons, and moderate to severe drought during reproductive and grain-filling stages in four of the nine seasons (Fig. 1; Supplementary Table S2). Soil types were a red-brown earth of slightly acid to neutral soil pH, except at Narrabri where the soil is a black vertisol. Crops were commonly sown after a canola (Brassica napus L.) or pea (Pisum sativa L.) break crop to minimize the incidence of root disease, and managed with adequate nutrition and pesticides to control weeds and leaf diseases. Experiments were partially replicated (‘p-rep’) designs averaging 1.4 replicates where each line is replicated an average 1.4 times across the irrigated and rainfed treatments in each MEF (Smith et al., 2006). In all experiments, all the plots were treated with 30 ml of pre-sowing irrigation and entries were sown at an optimal 3–5 cm sowing depth into 6 m long, 0.17 m spaced, eight row plots at seeding rates consistent with local practice (i.e. 120 seeds m–2 at Merredin and Narrabri, and 180 seeds m–2 at Yanco). Nutrients were supplied at sowing as Starter 15® (14% N:12.7% P:11% S) applied at 103 kg ha–1. Additional nitrogen was applied as needed to meet crop demand and ensure grain protein was achieved at industry standards of ≥11.5% (data not shown). Crops were largely reliant on stored and in-crop rainfall, with only enough irrigation supplied to produce ~20% yield benefit in the irrigated compared with the rainfed treatments (Rebetzke et al., 2013). Plots were end-trimmed at maturity to ~5.4 m length and the outside border rows were removed before machine harvesting to obtain estimates of plot yield for each cultivar. Samples were harvested for metabolite profiling and RWC measurement from five cultivars (Gladius, Wyalkatchem, Yitpi, RAC875, and Weebill) grown in the MEFs under the aforementioned growth conditions.
Fig. 1.
Rainfall patterns of the three field sites and three years from which yield gap-based drought tolerance (YDT) of the wheat cultivars was calculated. The monthly rainfall for each site and year was expressed as a percentage of the 100 year monthly average and then plotted as a heat map on the colour scale of red (0% rainfall) to blue (200% rainfall). The approximate developmental stages corresponding to the months are indicated as sowing, emergence of leaf, tillering, formation of lateral shoot, heading, anthesis, and grain filling.
Genotyping by sequencing (GBS)
Genomic DNA was extracted from leaf tissue of five biological replicates from each cultivar using methods described in CIMMYT-ICAR laboratory protocols with small modifications. After incubation with CTAB extraction buffer [100 mM Tris, 700 mM NaCl, 50 mM EDTA, 1% CTAB (mixed alkyltrimethyl-ammonium bromide), 140 mM β-mercaptoethanol], DNA was extracted using an equal volume of chloroform. The supernatant was treated with 10 µg µl–1 RNase. DNA was precipitated with an equal volume of isopropanol, followed by two washes of the pellet with ice-cold 80% ethanol, resuspended in Tris-EDTA (10 mM Tris–HCl pH 8.0, and 0.1 mM EDTA). DNA quality was assessed on a 1% agarose gel and quantified using NanoDrop ND-2000 (Thermo Scientific NanoDrop Products). Each DNA sample was normalized to 50 ng µl–1 and GBS was carried out by Diversity Array Technology Pty Ltd (DArT PL), Canberra, Australia. In brief, DArT PL uses the combinations of complexity reduction methods followed by sequencing on a HiSeq Illumina platform (Illumina Inc., San Diego, CA, USA). Single nucleotide polymorphism (SNP) calling was done as described previously (Li et al., 2015)
DartSeq identified 19 101 SNPs across 40 samples (eight wheat cultivars and five biological replicates). The genotype matrix has 12% missing data and 16% ‘het’ calls which may include homoeologues and paralogues, and are unexpected in inbred lines; ‘het’ calls were further treated as missing data. We excluded 5386 SNPs which had call rates of <30 of 40 samples. We further excluded 1442 singleton and 1688 doubleton variants, leaving 10 585 valid SNPs (Supplementary Table S3). The genotypes for each replicate were averaged, and pairwise genetic correlation among cultivars was calculated and used for further analysis. A Euclidian distance matrix among 40 samples was generated and plotted using hierarchical clustering (Supplementary Fig. S1) using R-script and RStudio software (www.rstudio.com).
Measurement of soil water content and relative water content
Soil water content (SWC) was measured gravimetrically on five cultivars by weighing three replicates/cultivar/treatment on every alternate day throughout the stress period in the glasshouse under irrigated and water-stressed conditions. Flag leaf RWC was measured with three biological replicates per cultivar per treatment under both environmental (glasshouse and field) and treatment (irrigated and water stressed) groups. In brief, the youngest, fully expanded leaf was excised at the base of the lamina and its fresh weight was determined. TWs were obtained after soaking the leaf segment in deionized water for 24 h at room temperature under low-light conditions. After soaking, leaves were quickly and carefully blotted dry with tissue paper, and turgid was measured. Dry weights were determined after oven drying the leaf samples at 70 °C for 72 h to a constant weight. The RWC was calculated using following equations:
where FW is the fresh weight of the leaf sample, TW is the turgid weight after rehydrating the leaf sample for 24 h, and DW is the leaf weight after oven drying for 72 h at 70 °C.
Chlorophyll measurement
Chlorophyll content was measured with an auto-calibrating chlorophyll meter (SPAD 502, Spectrum Technologies, Plainfield, IL, USA) from the middle portions of fully expanded flag leaves with six biological replicates/cultivar/treatment in the glasshouse condition. The measurement of each plant was calculated from the average of five readings from different parts of the flag leaf.
Stomatal conductance measurement
Stomatal conductance was measured in situ on the abaxial surface of fully expanded young leaves at midday using a dynamic diffusion porometer (AP4, Delta-T Devices, Cambridge, UK). Measurements were taken from six biological replicates/cultivar/treatment. The porometer was calibrated to the glasshouse conditions at the beginning of each measurement session.
Metabolite extraction and derivatization
Metabolites of high and intermediate polarity were extracted from leaf tissue of eight cultivars of glasshouse- and five cultivars of field-grown plants, using a hot methanol protocol. In brief, the youngest fully expanded leaf was harvested and immediately frozen in liquid N2. A 100 mg aliquot of frozen tissue was ground in a TissueLyzer (Qiagen) for 2 min at 20 Hz. A 500 µl aliquot of metabolite extraction buffer (100% methanol, 86 µg ml–1 norleucine, and 8.6 µg ml–1 ribitol) was added and vortexed. Sample tubes were incubated with shaking on an Eppendorf ThermoMixer set at 65 °C/14 000 g for 15 min. Samples were centrifuged for 2×10 min at 20 000 g and the supernatant was transferred into a new tube after each centrifugation. A ‘pooled reference’ control extract was then prepared by combining equal volume aliquots of each extract so that it could be run with each GC/MS batch to provide a universal control for interbatch variations in instrument response for each metabolite, enabling us to make meaningful comparisons between metabolite signals recorded in different analytical batches. Dried metabolite extracts were chemically derivatized by methoximation and trimethylsilylation on a Gerstel MPS2XL Multipurpose Sampler (Gerstel) operating in the PrepAhead mode for automated online derivatization and sample injection. The derivatization procedure consisted of the following steps: (i) addition of 10 µl of 20 mg ml−1 methoxyamine hydrochloride (Supelco, Cat. # 33045-U) in anhydrous derivatization grade pyridine (Sigma-Aldrich, Cat. # 270970) and incubation at 37 °C for 90 min with agitation at 750 rpm; (ii) addition of 15 µl of derivatization grade N-methyl-N-(trimethylsilyl) trifluoroacetamide (MSTFA; Sigma-Aldrich; Cat. No. 394866) and incubation at 37 °C for 30 min with agitation at 750 rpm; and (iii) addition of 5 µl of alkane mix [0.2% (w/v) each of n-dodecane, n-pentadecane, n-nonadecane, n-docosane, n-octacosane, n-dotriacontane, and n-hexatriacontane dissolved in anhydrous pyridine] and incubation for 1 min at 37 °C with agitation at 750 rpm. Samples were injected into the GC/MS instrument immediately after derivatization.
GS/MS metabolomic analysis
Derivatized metabolite samples were analysed on an Agilent 5975C GC/MSD system comprised of an Agilent GC 7890N gas chromatograph and 5975C Inert MSD quadrupole MS detector (Agilent Technologies, Palo Alto, CA, USA). The GC was fitted with a 0.25 mm ID, 0.25 μm film thickness, 30 m Varian FactorFour VF-5ms capillary column with a 10 m integrated guard column (Varian Inc., Palo Alto, CA, USA; Product No. CP9013). Samples were injected into the split/splitless injector operating in splitless mode with an injection volume of 1 μl, an initial septum purge flow of 3 ml min−1 increasing to 20 ml min−1 after 1 min, and a constant inlet temperature of 230 °C. Helium carrier gas flow rate was held constant at 1 ml min−1. The GC column oven was held at the initial temperature of 70 °C for 1 min before being increased to 325 °C at 15 °C min−1 and then being held at 325 °C for 3 min. Total run time was 21 min, with transfer line temperature and MS source temperature at 250 °C and quadrupole temperature at 150 °C. Electron impact ionization energy was 70 eV and the MS detector was operated in full-scan mode in the range 40–600 m/z with a scan rate of 3.6 spectra s−1. The MSD was pre-tuned against perfluorotributylamine (PFTBA) mass calibrant using the ‘atune.u’ autotune method provided with Agilent GC/MSD Productivity ChemStation Software (Revision E.02.01.1177; Agilent Technologies; Product No. G1701EA).
Quantitative and statistical analyses of MS data
All GC/MS data were processed using the online MetabolomeExpress data processing pipeline (www.metabolome-express.org) (Carroll et al., 2010). Raw GC/MS files were exported to NetCDF format using Agilent MSD ChemStation software (Revision E.02.01.1177; Agilent Technologies; Product No. G1701EA) and NetCDF files were uploaded to the ANU_Pogson MetabolomeExpress data repository. Peak detection settings were: Slope threshold=200; Min. Peak Area=1000; Min. Peak Height=500; Min. Peak Purity Factor=2; Min. Peak Width (Scans)=5; Extract Peaks=on. Peaks were identified by mass spectral and retention index (MSRI) library matching which used retention index and mass spectral similarity as identification criteria. MSRI library matching parameters were as follows: RI Window= ±2 RI Units; MST Centroid Distance= ±1 RI Unit; Min. Peak Area (for peak import): 5000; MS Qualifier Ion Ratio Error Tolerance=30%; Min. Number of Correct Ratio Qualifier Ions=2; Max. Average MS Ratio Error=70%; Remove qualifier ion not time-correlated with quantifier ion=OFF; Primary MSRI Library=‘Yadav_2016_WheatDroughtToleranceExperiment. MSRI’; Add Unidentified Peaks to Custom MSRI Library=ON; Use RI calibration file specified in metadata file=ON; Carry out per-sample fine RI calibration using internal RI standards=OFF. Library matching results were then used to construct a metabolite×sample data matrix, with peak areas being normalized to an internal standard (i.e. ribitol).
To remove non-biological batch to batch variation in signals and allow data from different analytical batches to be compared, each normalized metabolite signal was normalized again to the mean signal intensity in five technical replicate runs of a pooled reference extract (made by mixing together equal volume aliquots of all the extracts in the study) analysed in the same analytical batch. As a quality control filter, samples were checked for the presence of a strong ribitol peak with a peak area of at least 1×105 and a deviation from the median internal standard peak area (for that GC/MS batch sequence) of <70% of the median value. Statistical normalization to tissue mass was not required because chemical normalization to tissue mass had already been carried out by adjusting the extraction solvent volume proportionally to tissue mass. For determination of metabolic phenotypes, the treatment/control signal intensity ratio of each metabolite was calculated by dividing the mean (normalized) signal intensity of each metabolite in each set of treatment plants by its mean (normalized) signal intensity in its associated set of control plants.
Statistical significances were calculated by two-tailed Welch’s t-tests (n=5) in the MetabolomeExpress Comparative Statistics tool. The full data-set has been uploaded to the MetabolomeExpress MetaPhen Database (MetabolomeExpress Dataset IDs 112 and 127) and will be made publicly accessible upon publication of this article.
Calculation of adjusted conserved response (ACR) score
This ACR score was calculated as follows:
where nincreases, ndecreases, nsignificant_increases, and nsignificant_decreases are the numbers of cultivars in which: fold change (FC) >1, FC<1, FC>1 and P<0.05, and FC<1 and P<0.05, respectively. To test whether the consistency of the direction of response of each metabolite across the genotypes was statistically significant, a sign test was performed by input of nincreases and ntotal (where ntotal was the total number of cultivars, i.e. eight) as parameters into the binom.test function of R. The resulting P-value is referred to as pbinom.
The response value (RV) of a metabolite was calculated from its treatment/control FC as follows:
This transformation places metabolite responses on a linear scale, with increases and decreases represented as positive and negative values, respectively.
Regression analyses of metabolites, YDT, and RWC
Metabolite–YDT and metabolite–RWC associations were analysed using MetabolomeExpress MetaAnalyser, a web-based software tool for aligning, comparing, and identifying metabolites with patterns of interest across the results of multiple experiments (Carroll et al., 2010). It was used to generate the metabolite response heat map (Fig. 4), and to perform single metabolite–YDT and metabolite–physiological parameter correlation and multiple linear regression analysis. This was done by selecting the relevant responses in the control panel and performing an analysis with the following settings: Include metabolites that are missing data in some class comparisons=ON, Filter metabolites of unknown structure=ON, Transformation of signal intensity ratio=‘Natural Logarithm’, Min. Pearson’s r=0.3, Max. Exact P-value of r=0.05, Max q=0.4, Display charts=ON.
Fig. 4.
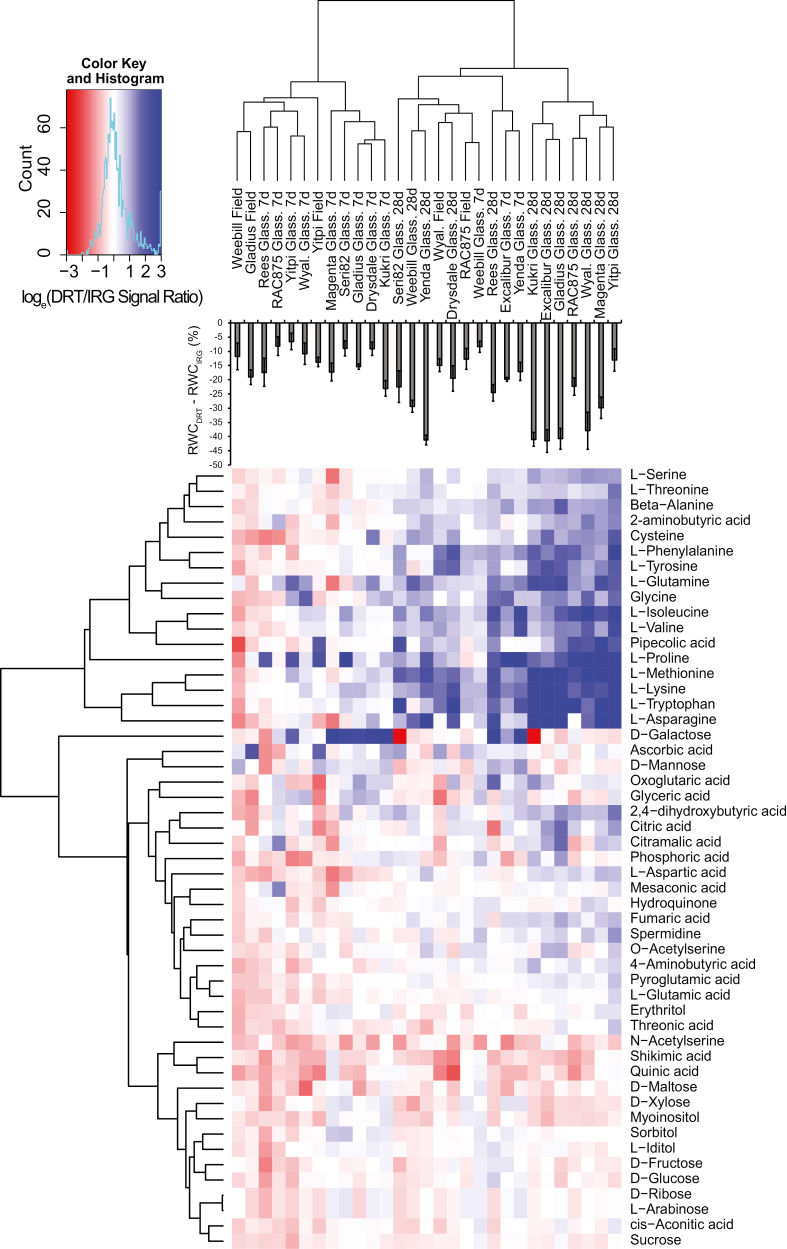
Heat map showing hierarchically clustered responses of metabolites to 7 d and 28 d glasshouse and a field drought condition. The response values, represented as natural log-transformed GC/MS signal intensity ratios, were clustered. Increases and decreases in the response values are displayed as blue and red coloured, respectively. RWC responses are overlaid on the heat map for visual comparisons. DRT and IRG represent drought and irrigation treatment groups, respectively.
This led to the calculation of the following. (i) ACR scores indicating the tendency of each metabolite to respond consistently and significantly in the same direction across the selected experiments (see above for definition of ACR). (ii) The two-tailed P-value of a binomial sign test (pbinom) treating increases and decreases of any P-value as successes or failures, respectively, to test the null hypothesis that increases and decreases occurred with equal probability. (iii) A table of statistical information for significant linear associations between natural log-transformed treatment/control metabolite signal ratios and the meta-variables (stomatal conductance, RWC, chlorophyll, YDT) including Pearson’s correlation coefficient (r), the exact P-value of r, the slope of the line of best fit, and the q-value returned by Benjamini–Hochberg false discovery rate (FDR) correction with the p.adjust function in R (number of tests=number of metabolites). (iv) Combinatorial multiple regression results fitting multi-metabolite linear regression models against the meta-variables. The combinatorial multiple regression testing process performed by the MetaAnalyser tool executed the following steps.
Every combination of two, three, or four metabolites drawn from the set of metabolites that were individually associated with YDT (with r>0.5 and P<0.12) in MetaAnalyser analysis were automatically computed in PHP, and a code-generation approach was used to write an R script testing each metabolite combination in a multiple regression model against the meta-variables (YDT and RWC) using the ‘lm ()’ function of R. The values for metabolite levels were given as natural log-transformed treatment/control GC/MS signal intensity ratios. Ordinary bootstrapping with 250-fold replicated using the ‘boot()’ and ‘boot.ci()’ functions of the ‘boot’ package (version 1.3-18) was performed on the R2 value returned for each multiple regression model fitting to estimate 95% confidence intervals. Metabolome data are deposited at MetabolomeExpress MetaPhenDB (MetabolomeExpress Dataset IDs 112 and 127).
Orthogonal partial least squares (OPLS) analysis
OPLS analysis was performed in R using the package, ‘ropls’ (Thevenot et al., 2015). The input data matrix was a [metabolite×sample] matrix of values representing the relative signal intensities of metabolites in each sample, normalized to their mean signal intensity in the pooled reference samples analysed in the same analytical GC/MS batch. Default settings were used unless stated otherwise below. Cross-validation was performed with the default of seven cross-validation segments, while significance testing of the models was performed by permutation testing with 1000 permutations. See the online documentation of the ‘ropls’ package for further algorithmic details.
Routine statistical analyses
Detailed statistical methods for association and phenotypic similarity analyses are provided above. Binomial sign tests were performed in R version 3.2.3 using the binom.test() function. Figures were prepared in Adobe Illustrator Creative Cloud (v. 18.1.0) and OriginPro 2017 software. General statistical analyses such as ANOVA were performed using GenStat 17th Edition.
Statistical comparisons of metabolic traits between field and glasshouse experiments
Metabolic responses to field drought treatments were statistically compared with responses to glasshouse drought treatments using the PhenoMeter web application (Carroll et al., 2015) associated with MetabolomeExpress (Carroll et al., 2010). The field drought response of each cultivar was searched separately against the responses of all 14 cultivars to both 7 d and 28 d glasshouse drought treatments. Settings were as follows: Minimum Absolute Metabolite Fold Change=1.5, Minimum r2=0, Maximum Fisher’s Exact P-value=0.1, Maximum pnon-bio=1, Include metabolites of unknown structure=OFF. Note that, by default, hits require a pnon-bio<0.05 to be considered statistically significant. ‘Directional Overlap p-value’ is the P-value of a binomial sign test in which co-directional metabolite responses (a metabolite responding in the same direction in field and drought response) were considered successes and incongruent responses were considered failures.
The correlation (r) between field and glasshouse response represents the Pearson correlation of the two sets of response values. The PhenoMeter (PM) score was calculated as –sgn(R)×r2×log10DOP where DOP is the directional overlap P-value described above. pnon-bio is an estimate of the likelihood of obtaining a PM score greater than or equal to the observed score when the metabolite labels of the query response are randomly shuffled, as determined by a permutation test.
Results
Determining field-based YDT and analysing genetic relatedness of wheat cultivars
Yields from nine rainfed and nine irrigated field environments were assessed in eight commercial wheat cultivars (Kukri, Excalibur, Gladius, Wyalkatchem, Yitpi, RAC875, Drysdale, and Weebill; Supplementary Table S1) at multiple locations across Australia (3 field sites×3 seasons). YDT, a measure of a cultivar’s capacity to moderate the yield gap between actual and potential yield, was calculated as the ratio of mean grain yield between rainfed and irrigated environments. (Table 1). Cultivars with the highest YDT scores (i.e. lowest yield gaps between two environments) were Weebill (0.94), Drysdale (0.93), and RAC875 (0.88), whereas the lowest YDT score of Kukri (0.71) reflects its reduced ability to mitigate yield gap (Table 1).
Table 1.
Grain yield and yield gap-based drought tolerance (YDT)
| Cultivars | Mean grain yield (t ha–1) | YDT | |
|---|---|---|---|
| Rainfed | Irrigated | ||
| Kukri | 2.76 | 3.89** | 0.71 |
| Excalibur | 2.77 | 3.52** | 0.79 |
| Gladius | 3.04 | 3.65** | 0.83 |
| Wyalkatchem | 3.02 | 3.61** | 0.84 |
| Yitpi | 3.03 | 3.54* | 0.86 |
| RAC875 | 3.24 | 3.70* | 0.88 |
| Drysdale | 3.17 | 3.40 | 0.93 |
| Weebill | 3.24 | 3.46 | 0.94 |
| LSD (0.05) | 0.42 | 0.49 | – |
YDTs were calculated as the ratio of mean grain yields under rainfed and irrigated (rainfed+supplementary irrigation) conditions from 18 field environments across Australia (3 sites×3 seasons×2 irrigation treatments). LSD, least significant difference of the means.
**, *Significant difference between rainfed and irrigated for each cultivars at 0.01 and 0.05, respectively.
Rainfall patterns compared with 100 year averages during the growing season provide an indicative measure of potential drought, but in and of themselves are not a measure of SWC. The rainfall patterns indicate that the crops were potentially exposed to droughts ranging from moderate to severe during early growth stages (leaf emergence and tillering) and to severe drought in four out of nine environments during anthesis until grain-filling stages, a critical period during which grain yield is determined (Fig. 1; Supplementary Table S2).
GBS using DArTseq was employed to evaluate these eight wheat cultivars for evidence of population structure and genetic purity. We identified ~19 101 SNPs across 40 samples (eight cultivars and five biological replicates; Supplementary Table S3). SNP calling and filtering of false calls were performed as described in the methods. A Euclidian distance matrix among 40 samples was generated and plotted using hierarchical clustering (Supplementary Fig. S1). No strong genetic relatedness structure was present within the population, ruling out major selection bias from the current study. Selection of wheat cultivars and assessment of various metabolic and physiological traits have been summarized in a flow chart (Supplementary Fig. S2).
Effect of glasshouse drought stress on SWC and leaf wilting
Cyclic drought treatment in the glasshouse began 50 d after sowing (at the booting stage) and was achieved by withholding watering for 15 d followed by rewatering once to saturation and then withholding water for a further 13 d, bringing the total period of drought up to 28 d (Supplementary Fig. S3). Measurements of leaf RWC and metabolite levels (eight cultivars: Kukri, Excalibur, Gladius, Wyalkatchem, Yitpi, RAC875, Drysdale, and Weebill) of the flag leaves were made at 7 d and 28 d after initiation of the drought treatment for both control (irrigated, IRG) and drought- (DRT) treated plants. Measurements of SWC, gs, and chlorophyll were undertaken on a subset of cultivars; Kukri, Yitpi, RAC875, Drysdale, and Yenda (refer to Supplementary Table S4, for raw data). Yenda was included in glasshouse trials as a ‘control’ to validate the drought regime as it is reported to be a drought-susceptible cultivar. Indeed, it was among the most susceptible lines under the imposed drought treatments (Figs 2A, 3).
Fig. 2.
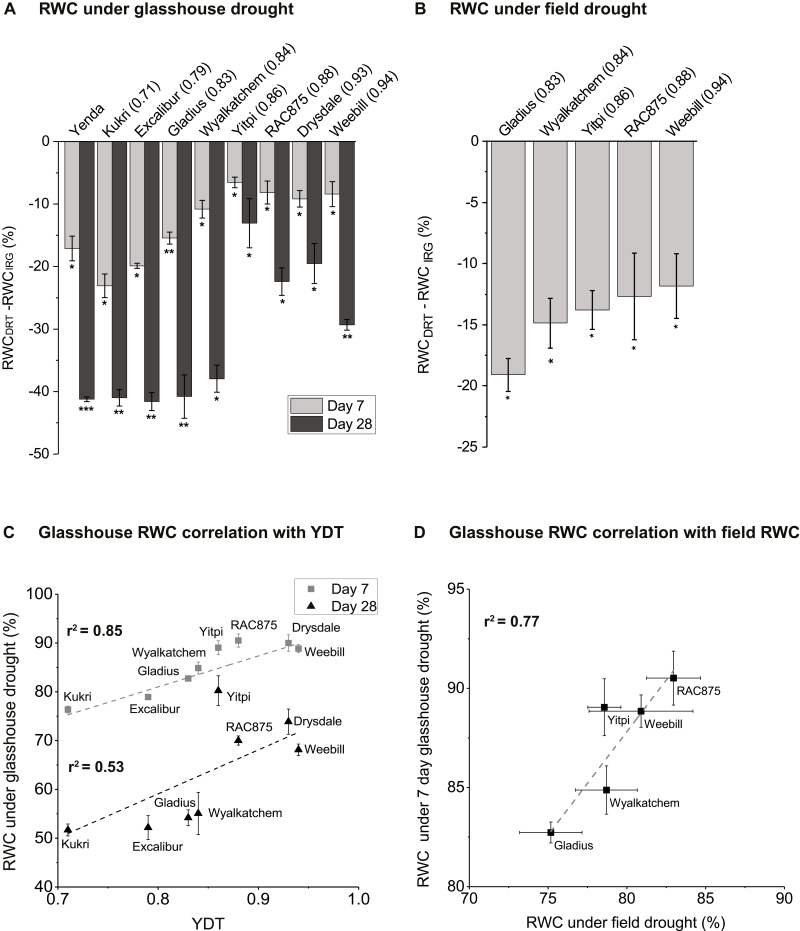
Flag leaf relative water content (RWC), its correlation with YDT and between glasshouse and field drought conditions. (A) RWC in all the wheat cultivars under early (day 7) and late (day 28) glasshouse drought relative to the irrigated conditions [RWC (DRT– IRG)]. (B) RWC under rainfed (field drought) relative to the irrigated field conditions. (C) Correlation of RWC under glasshouse drought with field-based YDT. (D) Correlation of RWC between 7 d glasshouse drought and field drought conditions. Error bars represent ±SE (n=3). Asterisks represent statistically significant differences from the respective irrigated control at *P<0.05, **P<0.01, and ***P<0.001. YDT values are shown in parentheses next to the cultivar, except for Yenda which was used as a drought-susceptible control line. The line of the best fit (not taking errors into account) is displayed as a dashed line. DRT and IRG represent drought and irrigation treatment groups, respectively. YDT, yield gap-based drought tolerance.
Fig. 3.
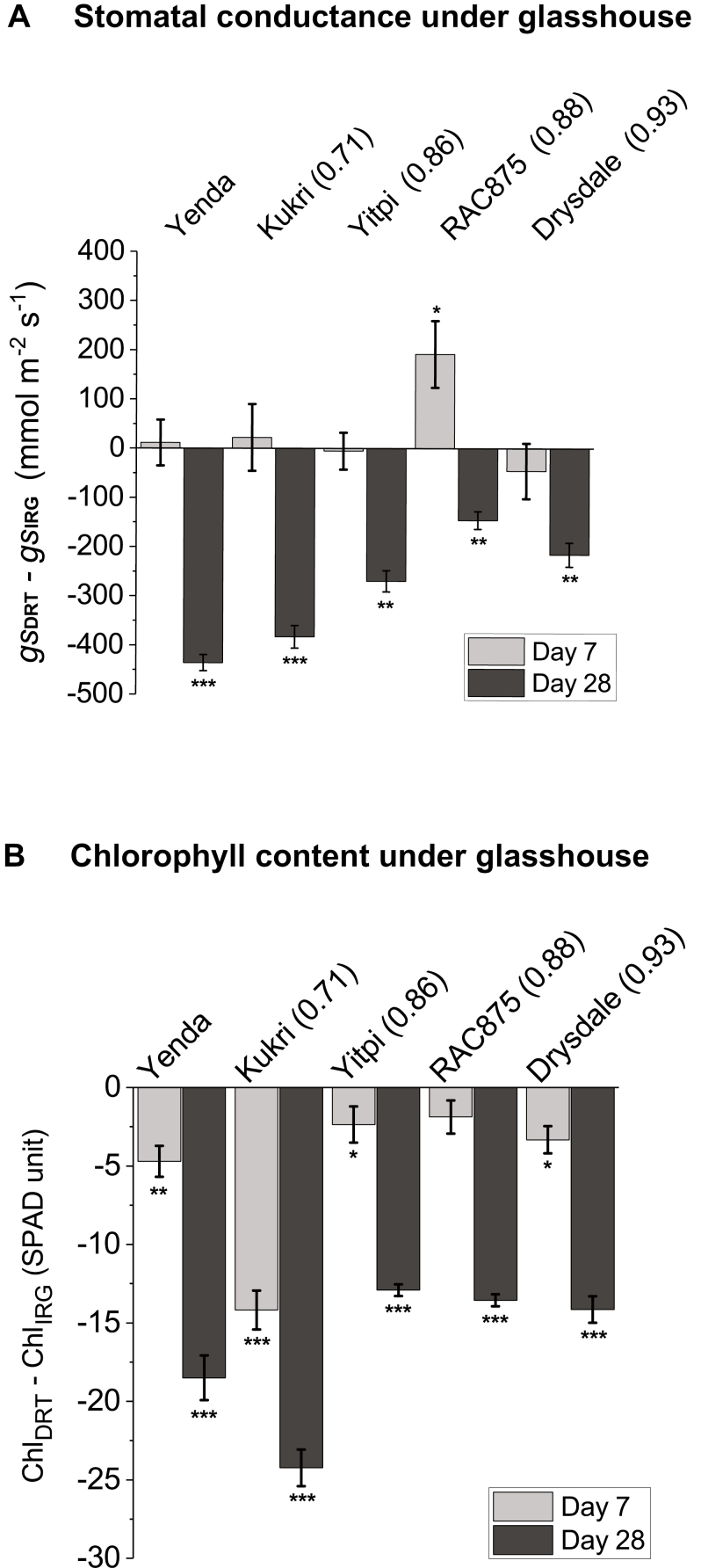
Stomatal conductance (Pornsiriwong et al., 2017) and chlorophyll content (Munns et al., 2006) in response to glasshouse drought treatments. (A) Flag leaf gs, under drought relative to the irrigated conditions [gs (DRT–IRG)] at 7 d and 28 d after the initiation of water stress. (B) Flag leaf chlorophyll content (SPAD units) in drought-stressed relative to the irrigated conditions at 7 d and 28 d [Chl (DRT–IRG)] after the initiation of water stress. YDT values are shown in parentheses next to the cultivar. Error bars represent ±SE (n=6). Asterisks represent statistically significant differences from the respective irrigated control at *P<0.05, **P<0.01, and ***P<0.001 by Welch’s t-test. DRT and IRG represent drought and irrigation treatment groups, respectively. YDT, yield gap-based drought tolerance.
Periodic measurements of SWC were performed to assess the severity and homogeneity of soil water loss on a subset representing drought-tolerant (Drysdale, RAC875, and Yitpi) and drought-susceptible cultivars (Kukri and Yenda). At day 7 of drought, SWC ranged from 50% to 70% in all cultivars, and at day 15 it dropped down further, ranging from 20% to 30% among cultivars. Rewatering brought SWC up to an average level of 80% in all cultivars and, upon imposition of recurring drought, it further decreased to a level between 25% and 35% at day 28 (Supplementary Fig. S3B). Most cultivars showed no significant differences in SWC across the drought regimes, except that Drysdale and Yenda maintained slightly lower SWC than the others. However, at the end time point of cyclic drought, all cultivars reached a similar SWC level. Irrigated plants across the cultivars and time points remained turgid.
The visual impact of drought varied across cultivars and, as expected, drought- tolerant cultivars maintained lower leaf wilting than drought-susceptible cultivars throughout the cyclic drought regime (Supplementary Fig. S4). At the conclusion of the drought, all plants were rewatered and then received the same watering regime as the irrigated pots. All drought-treated plants recovered and set viable seed (data not shown).
Physiological responses to glasshouse drought were associated with YDT
All water-stressed cultivars had a significant reduction in flag leaf RWC at days 7 and 28, expressed as a net decrease in RWC (RWCDRT–RWCIRG) (Fig. 2A). However, drought-tolerant cultivars (Drysdale, RWC875, and Yitpi) maintained a higher RWC compared with drought-susceptible cultivars (Kukri and Yenda) (Fig. 2A). Five cultivars characterized in the field environment had significantly reduced flag leaf RWC under rainfed conditions as compared with irrigated conditions (Fig. 2B).
Flag leaf RWC of the plants was positively associated with YDT under both the 7 d (r2=0.85, P=7.4E-6) and the 28 d (r2=0.53, P=7E-4) glasshouse drought treatments (Fig. 2C). The r2 between RWC and YDT under rainfed field conditions was similar to that after 28 d of glasshouse drought, but the association was not statistically significant, possibly due to smaller sample size (r2=0.52, P=0.16). Notably, RWC values under the 7 d glasshouse and field drought treatments were significantly positively associated with one another (r2=0.77, P=0.011; Fig. 2D).
To probe the physiological impact of the drought treatments, we undertook leaf stomatal conductance measurements in five cultivars (Drysdale, RAC875, Yitpi, Kukri and Yenda) (Fig. 3A). There were no significant reductions in gs at 7 d of drought. In fact, RAC875, the cultivar with the highest leaf RWC under the 7 d drought condition, even displayed a marginally significant increase in gs. In contrast, gs decreased significantly in all five cultivars at day 28 of the drought treatment. However, the extent of decline in gs of drought-tolerant cultivars (Drysdale, RWC875, and Yitpi) was lower than that of drought-susceptible cultivars (Kukri and Yenda). As expected (Pradhan et al., 2012), significant declines in chlorophyll content (measured as SPAD index) were observed in four of five cultivars at day 7 and in all five at day 28 of the drought treatment (Fig. 3B).
Associations of individual metabolites with YDT and RWC
Having confirmed that the drought treatments had elicited the expected physiological responses, we performed untargeted GC/MS metabolomics analyses with the aim of identifying metabolites that were correlated with YDT. The study detected a variety of amino acids, organic acids, sugars, polyols, and polyamines including many unidentified components (Supplementary Table S5). Overlay of the net decrease in RWC in drought versus irrigated treatments onto a hierarchical clustering of the responses of identified metabolites to glasshouse and field drought treatments showed that the metabolite responses of severely water-stressed plants generally clustered together (Fig. 4).
To test for evidence of metabolic pre-acclimation against drought stress, we first tested whether the levels of any individual metabolites in the leaves of irrigated, unstressed plants were significantly associated with YDT. In leaves harvested from irrigated plants at day 7 of the treatment period (Supplementary Table S6A), fructose-6-phosphate (r= –0.83, P=0.01) and isoleucine (r= –0.72, P=0.04) levels were negatively associated with YDT. At day 28 (Supplementary Table S6B), the levels of shikimate and myo-inositol were marginally significantly negatively associated with YDT (r= –0.61 and –0.69, respectively; P<0.073), while 2,4-dihydroxybutanoate was marginally positively associated (r=0.69, P<0.06) in irrigated plants. Under irrigated field conditions, organic acids such as itaconate (r=0.89, P<0.05) were significantly positively associated with YDT whereas citramalate was marginally positively associated (r=0.82, P<0.1). On the other hand, glycerate, 2-oxoglutarate (r= –0.94, P<0.05), and xylose (r= –0.91, P<0.05) were negatively associated with YDT (Supplementary Table S6C).
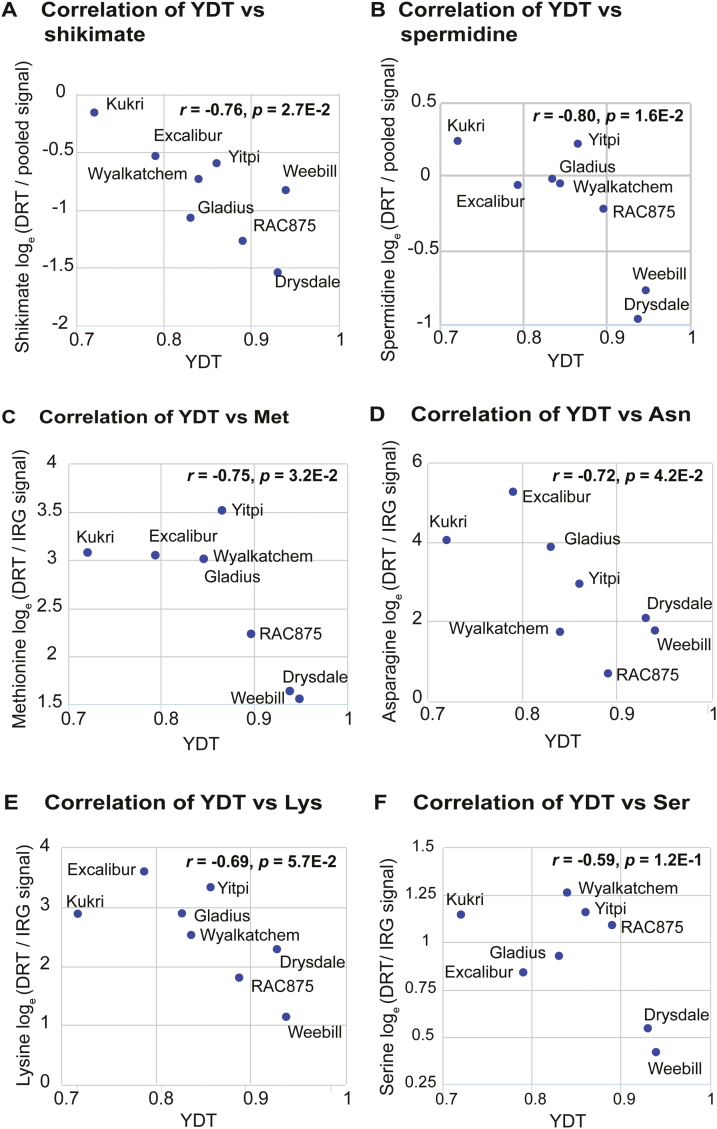
We next examined associations between YDT and individual metabolite levels under drought stress. After the short-term (7 d) glasshouse drought treatment, marginally significant negative associations between YDT and levels of ascorbate (r= –0.68) and 2,4-dihydroxybutanoate (r= –0.66) were observed (P<0.08; Supplementary Table S6D). After 28 d of drought, the levels of the organic acid, shikimate, and the polyamines, putrescine and spermidine (Supplementary Table S6E; Fig. 5A, B), were significantly negatively correlated with YDT (r= –0.82 to –0.76, P<0.05). Marginally significant negative associations were observed for 2-hydroxycinnamate, tyrosine, serine, and xylose (P<0.1; Supplementary Table S6E). Under field drought, 4-aminobutyrate was significantly negatively associated with YDT (r= –0.89, P<0.05), while marginally significant associations were observed between YDT and glycine (r=0.87), glutamate (r= –0.85), and serine (r= –0.78) levels (P=0.055–0.12; Supplementary Table S6F). Methionine and asparagine responses to glasshouse drought showed significant negative associations with YDT (Supplementary Table S6G).
Fig. 5.
Scatter plots for selected metabolite and YDT correlations under the 28 d glasshouse drought treatment. (A and B) Levels of shikimate and spermidine, represented as the natural log-transformed signal intensity ratios between drought-treated (DRT) samples and a universal pooled control extract are plotted against YDT. (C–F) Responses of methionine, asparagine, lysine, and serine represented as natural log-transformed signal intensity ratios of the drought-treated (DRT) to irrigated (IRG) samples are plotted against YDT (yield gap-based drought tolerance) scores. (This figure is available in colour at JXB online.)
While simple metabolite measurements under a single set of environmental conditions may prove to be useful predictors of YDT, we were interested to explore whether the responses of metabolites to drought stress (expressed as natural logarithms of metabolite mean signal intensity ratios between drought-treated and control samples) would be more predictive of YDT. Under 28 d drought, the responses of methionine (r= –0.75; Fig. 5C) and asparagine (r= –0.73; Fig. 5D) were significantly negatively associated with YDT, while marginally significant negative associations were observed for lysine (Fig. 5E), serine (Fig. 5F), glutamine, 2-hydroxycinnamic acid, and glucuronate (r= –0.60 to –0.69 with SE of 0.03–0.07, P<0.12), and a marginal positive association was observed for galactose (r=0.69±0.07; P<0.06; Supplementary Table S6G).
We next analysed associations between metabolite levels and flag leaf RWC. Metabolite–RWC associations were anticipated given that RWC–YDT (Fig. 2C) and metabolite–YDT associations had already been observed (Supplementary Table S6; Fig. 5). Indeed, significant metabolite–RWC associations were observed under all environmental treatments except for irrigated field conditions (Supplementary Table S7). Interestingly, a particularly high number of metabolite–RWC associations were observed under the 7 d glasshouse drought (Supplementary Table S7)—the treatment associated with the most significant RWC–YDT correlation (Fig. 2C). RWC was strongly negatively associated with the responses of the following metabolites under 28 d drought stress: serine (r= –0.84, P=3.1E-3), N-acetylserine (r= –0.89, P=3.1E-3), and asparagine (r= –0.81, P=1.5E-2) (Supplementary Fig. S5). Notably, asparagine levels at 28 d drought were also negatively associated with RWC under 7 d drought (r= –0.81, P=1.4E-2; Supplementary Fig. S5D). In contrast, myoinositol was the only metabolite that was positively associated with RWC under 28 d drought treatment (r=0.73, P=4E-2; Supplementary Fig. S5E). This was due to a pattern of stronger depletion of myoinositol levels in the drought-susceptible cultivars. Conversely, a negative association between phosphate response and RWC was linked to greater increases in the drought-susceptible cultivars (r= –0.72, P=4.5E-2; Supplementary Fig. S5F). Comprehensive tables of all metabolite–YDT and metabolite–RWC association results are provided in Supplementary Table S7.
Metabolome–YDT orthogonal partial least squares analyses
We next used the supervised multivariate technique, OPLS (Thevenot et al., 2015), to identify multivariate models explaining as much YDT variation as possible. In four separate analyses, we applied OPLS to the metabolite levels of the eight wheat cultivars to investigate whether global, untargeted metabolic profiles could reliably predict YDT under any of the four glasshouse environmental conditions (7 d irrigated, 7 d drought, 28 d irrigated, or 28 d drought). With 7-fold cross-validation and 1000 permutation significance testing, significant models were generated from metabolite levels under all four conditions (pR2Y<0.05 and pQ2<0.05). However, cross-validated predictive ability of the model (Q2) only exceeded the generally accepted threshold of 0.5 under 7 d irrigated (Q2= 0.78) and 28 d drought conditions (Q2=0.63) (Supplementary Table S8;Supplementary Fig. S6). Metabolite variable loadings and variable importance for the projection (VIP) values are presented in Supplementary Table S9. In summary, the YDT-predictive performance of global GC/MS metabolite profiles was close to that of RWC.
Metabolite–YDT multiple regression analyses
Aiming to identify metabolite–YDT statistical models explaining more of the YDT variation, we explored a third approach: multiple regression of selected sets of metabolites. This approach has an advantage over the previous two in that it exploits the complementarity of predictive information provided by different metabolites while excluding variables unrelated to YDT, thereby reducing noise. We performed multiple regression analyses between YDT and every combination of two, three, and four metabolites that were individually associated with YDT (with r>0.5 and P<0.12), focusing on the 28 d time point of the glasshouse experiment. To filter out low confidence associations, we used bootstrapping to estimate 95% confidence intervals on R2 and removed models with lower limits of <0.9.
The 28 d irrigated glasshouse data set (metabolite levels represented as the natural logarithms of the signal intensity ratios between cultivar samples and the pooled reference extract) did not identify any multi-metabolite models with satisfactorily high R2 against YDT. A substantive improvement was achieved by analysing the glasshouse data set as a natural log-transformed FC ratio between 28 d drought and irrigated samples (lnFC). This identified a variety of multiple metabolite regression models with R2 values having lower 95% confidence limits, ≥0.9 (Table 2). All of these were based on various three or four metabolite combinations drawn from the set: serine, asparagine, methionine, lysine, glutamine, and galactose. The best (i.e. highest lower 95% confidence limit for R2) four metabolite model was that based on serine, asparagine, methionine, and galactose (YDT~lnFCSerine+lnFCAsparagine+ lnFCMethionine+lnFCGalactose; R2=0.99±0.01, P=3.5E-3) followed closely by YDT~lnFCSerine+lnFCAsparagine+lnFCMethionine+ lnFCLysine (R2=0.98, P=9E-3), and YDT~lnFCSerine+ lnFCAsparagine+lnFCGlutamine+lnFCLysine (R2=0.98, P=9E-3). The best three metabolite model was that based on serine, asparagine, and methionine (YDT~lnFCSerine+lnFCAsparagine+ lnFCMethionine; R2=0.94±0.001, P=6.2E-3). In general, replacing methionine with lysine gave models with only slightly lower performance (Table 2; Supplementary Table S7).
Table 2.
Multiple regression analysis showing that drought responses of four amino acids fully explain the genotypic variances in YDT
| Models | R 2 | P(R2) | Metabolites | r | P(r) |
|---|---|---|---|---|---|
| YDT~lnFCSerine+lnFCAsparagine+lnFCMethionine+lnFCGalactose | 0.99 | 3.5E-03 | l-Serine | –0.27 | 1.2E-02 |
| l-Asparagine | –0.05 | 7.4E-03 | |||
| l-Methionine | 0.09 | 4.2E-02 | |||
| d-Galactose | 0.02 | 4.2E-02 | |||
| YDT~lnFCSerine+lnFCAsparagine+lnFCMethionine | 0.94 | 6.9E-03 | l-Serine | –0.34 | 1.5E-02 |
| l-Asparagine | –0.06 | 7.5E-03 | |||
| l-Methionine | 0.12 | 5.8E-02 | |||
| YDT~lnFCSerine+lnFCAsparagine+lnFCGlutamine+lnFCLysine | 0.98 | 9.0E-03 | l-Serine | –0.28 | 6.6E-03 |
| l-Asparagine | –0.09 | 1.1E-02 | |||
| l-Glutamine | 0.05 | 9.5E-02 | |||
| l-Lysine | 0.11 | 2.4E-02 | |||
| YDT~lnFCSerine+lnFCAsparagine+lnFCLysine+lnFCMethionine | 0.98 | 9.0E-03 | l-Serine | –0.34 | 1.1E-02 |
| l-Asparagine | –0.07 | 5.7E-03 | |||
| l-Lysine | 0.05 | 1.2E-01 | |||
| l-Methionine | 0.08 | 9.5E-02 |
Multiple regression analysis was performed using the lm function in R with the models indicated. The linear and multiple correlation coefficients are indicated as r and R2, whereas their significance values at are indicated as P(r) and P(R2), respectively. lnFC represents the natural log-transformed fold changes of the respective amino acids (as subscripts) in drought-treated over control plants. r=the fitted linear effect coefficient in multiple regression; P(r)=the P-value associated with the fitted coefficient; n=8 for all the analysis.
Comparisons between glasshouse and field drought metabolite responses, and cross-study comparisons
To test whether metabolite responses observed in our glasshouse experiments were predictive of those in the field, we used the PhenoMeter phenotype comparison tool (Carroll et al., 2015) to statistically compare the field metabolic responses of five cultivars with their respective 7 d and 28 d glasshouse drought responses (Table 3). For Weebill and RAC875, there was no significant similarity between the field and either of the glasshouse time points, whereas the other three matched significantly for one or more time points. Yitpi matched significantly to both 7 d and 28 d of glasshouse drought response, whereas Wyalkatchem and Gladius closely resembled their 28 d glasshouse responses (Table 3).
Table 3.
Statistical comparisons of metabolic traits between field and glasshouse drought treatments
| Comparison with glasshouse DRT_7d | Comparison with glasshouse DRT_28d | |||||||
|---|---|---|---|---|---|---|---|---|
| Cultivars | Directional overlap P-value | R | PM score | p non-bio | Directional overlap P-value | R | PM score | p non-bio |
| Weebill | – | – | – | – | – | – | – | – |
| RAC875 | – | – | – | – | – | – | – | – |
| Yitpi | 7.4E-3 | 0.55 | 0.65 | 1.4E-239 | 0.1 | 0.5 | 0.25 | 1.6E-36 |
| Wyalkatchem | – | – | – | 1.2E-2 | 0.68 | 0.89 | 7.1E-28 | |
| Gladius | – | – | – | – | 1.3E-2 | –0.2 | –0.08 | 0.49 |
The cultivar names are hyperlinked to direct the reader to a PhenoMeter (see the Materials and Methods) search of that cultivar’s field drought response against the responses of all cultivars to DRT_7d and DRT_28d. Note that this search applies maximum binomial sign test and pnon-bio thresholds of 0.05, and therefore not all hits in this table will appear in the results. DRT=drought-treated; R=correlation coefficient, PM=phenometer phenotypic score (see the Materials and methods).
To gain a further insight into links between the observed metabolic traits and the drought treatment, we performed a comparison with a previous study reporting metabolite responses to short-term increases in [O2]/[CO2] in sunflower (Helianthus annuus L.) leaves, a condition increasing photorespiration (Abadie et al., 2016). The working hypothesis was that metabolic changes in our data might be associated with increased photorespiration arising from decreased stomatal conductance in response to dehydration-induced signals such as abscisic acid (ABA) and 3'-phosphoadenosine 5'- phosphate (PAP) (Pornsiriwong et al., 2017). Indeed, we found significant response overlap between high photorespiration and drought stress by binomial sign test. For example, of the 18 metabolites that responded >1.5-fold in both H. annuus under high [O2]/[CO2] and T. aestivum ‘Yenda’ under 7 d drought, 15 of them (2-oxoglutarate, fructose-6-phosphate, glucose-6-phosphate, fumarate, glycerate, glycine, homoserine, isoleucine, lysine, maleic acid, phenylalanine, threonine, tryptophan, valine, and xylose) responded in the same direction in both cases, while only three (alanine, β-alanine, and quinate) responded in opposite directions (P=7.5E-3).
Discussion
Phenotypes in the glasshouse predict drought tolerance in the field
While offering the potential for finer environmental control than field-based experiments, pot-based glasshouse experiments are reported not to accurately replicate the field environment, with observations made in glasshouse experiments often conflicting with measurements made in the field (Passioura, 2006; Rebetzke et al., 2014). However, glasshouse screening methods offer some major practical advantages over field-based methods, including fine biotic and abiotic environmental control—enabling better replication and lower costs. Here, we provide clear evidence that flag leaf RWC and easily measured metabolites of drought-stressed, glasshouse-grown wheat plants strongly associate with field-calculated YDT values (Figs 2C, 5; Supplementary Table S6). YDT was calculated from the plot yield data from extensive and expensive field trials of three consecutive seasons from 2010 to 2012 at three locations across Australia under rainfed and irrigated conditions. Herein, we have demonstrated that a set of three or four metabolites could be a powerful predictor of a dependent variable (i.e. YDT), which is a measure of the yield gap between rainfed (fluctuating seasonal rainfall) and irrigated environments (Table 2).
Drought limits grain yield and its impact varies with intensity and occurrence of drought during the various developmental stages of crop growth (Boonjung and Fukai, 1996). Therefore, it is critical to define the nature of drought specifically with reference to the performance in rainfed relative to irrigated wheat plots and the extent to which an average YDT would be reflective of the expected performance. Based on the seasonal rainfall and its distribution in the sites and seasons analysed, drought ranged from moderate to severe during early growth stages in all locations, whereas in four of nine environments the crops were exposed to severe drought during critical development stages (anthesis and grain filling) which influences grain yield, providing a range of yield gaps to produce the average YDT presented herein (Fig. 1; Table 1). A second consideration is the suitability of the glasshouse drought; in this respect, we observed significant genetic associations between our glasshouse- and field-based flag leaf RWC under moderate drought (Fig. 2D), and metabolite response profiles were often statistically significantly similar between glasshouse and field (Table 3).
Critically, the r2 values of glasshouse–field trait associations were highly dependent upon environmental conditions. For example, glasshouse RWC–YDT associations were much stronger under moderate 7 d drought, when genetic variation in RWC was more apparent, than under severe 28 d drought, when RWC values across the cultivars had partly flattened out due to ‘saturation’ of dehydration. Conversely, metabolite–YDT associations were stronger under 28 d drought when metabolic stress (e.g. high photorespiration) due to stomatal closure had a chance to influence metabolite levels in a cultivar-dependent manner.
Metabolites predict yield drought tolerance better than RWC
That RWC-explained 85% of YDT variance (Fig. 2C) is perhaps higher than one might expect given that the measure does not capture variation in the spatial and temporal impacts of drought across the plant. To understand where the residual variation in YDT that is unexplained by RWC might originate from, and identify markers to capture this variation, it is helpful to consider the structure of the causality network linking drought to grain yield. The core of this network may be simplified as: drought>dehydration>stomatal closure>decreased Ci>increased vo/vc>decreased A and increased flux into photorespiratory pathway>altered metabolite levels>decreased yield (where: Ci is the intercellular CO2 concentration; vo and vc are the rates of oxygenation and carboxylation by Rubisco, respectively; and A is the net rate of CO2 assimilation). Of course, it is important to recognize that signalling processes permeate across all levels of this network. That leaf water can explain ~85% of YDT variation highlights the considerable genetic variation existing in the complex first step of the network. However, it is obvious from the network structure that much opportunity remains for genetic variation to affect steps downstream of the initial responses; that is, those that translate dehydration into yield loss.
In the above causality network, metabolites lay penultimate to yield and are therefore affected by all upstream factors affecting the translation of drought into yield loss. For this reason, we expected the levels of certain metabolites (or changes thereof) to be more predictive of YDT than point-in-time measures of leaf water. Indeed, through a systematic combinatorial multiple regression approach, we have confirmed here that the responses to glasshouse drought of just three amino acids, namely methionine, serine, and asparagine, could predict YDT with R2 of 0.94±0.001, while the addition of lysine increased R2 to 0.98±0.01 (Table 2). These findings are in agreement with recent reports that multiple metabolites were able to segregate Brachypodium distachyon ecotypes according to drought tolerance under moderate water stress where growth and colour phenotypes failed to do so (Fisher et al., 2016).
It is noteworthy that five (83%) of the six metabolites identified among the top multi-metabolite–YDT associations (serine, asparagine, lysine, methionine, glutamine, and galactose) were amino acids, while amino acids comprised only 33% of the total set of identified metabolites; this is a significant over-representation (P<0.017; two-sided binomial sign test). Accumulation of amino acids has long been recognized as a hallmark response to abiotic stress (Stewart and Larher, 1980). Changes in amino acid profiles during drought stress have been attributed to various mechanisms, including decreased protein synthesis (Good and Zaplachinski, 1994), increased protein degradation (Huang and Jander, 2017), and enhanced biosynthesis driven by changes in substrate availability and enzyme regulation (Kishor et al., 2005). Amino acids play significant roles in stress tolerance (Rai, 2002), and accumulation of many amino acids has been observed in the leaves of different plant species under drought stress (Obata and Fernie, 2012; Hill et al., 2013) and osmotic stress (Huang and Jander, 2017).
So why might the combination of methionine, serine, and asparagine be so predictive of YDT? Clearly, these metabolites provide complementary (orthogonal) information pertinent to genetic variation in YDT.
Strong accumulation of asparagine in plants (including wheat under drought stress) and its possible role as an osmolyte have been reported (Carillo et al., 2005; Lea et al., 2007). However, our results clearly show that asparagine levels are negatively associated with drought tolerance in wheat, consistent with recent observations in rice (Degenkolbe et al., 2013). These findings suggest that asparagine accumulation may be associated with a biochemical response to dehydration that has negative consequences for yield. It is likely that this process is senescence. Asparagine is well known to accumulate in senescent leaves, and a comprehensive metabolomic dissection recently revealed that the asparagine/aspartate ratio increased steadily along a developmental senescence gradient (Watanabe et al., 2013). Asparagine accumulation during the onset of senescence has been repeatedly linked to increased expression of asparagine synthetase (Avila-Ospina et al., 2015; Moschen et al., 2016) while, in rice, both asparagine and asparagine synthetase transcript levels show negative associations with drought tolerance (Degenkolbe et al., 2013). Notably, artificially delaying drought-induced senescence by genetic manipulation of cytokinin metabolism in transgenic tobacco plants resulted in strongly increased drought tolerance (Rivero et al., 2007). Taken together with these previous observations, our results suggest that asparagine increases under drought may provide an informative readout of genetic variation in the sensitivity of senescence induction to drought stress and, thereby, of genetic variation in YDT.
That serine would accumulate more strongly in the more drought-sensitive cultivars is readily explained by higher levels of photorespiration expected in those cultivars since serine, produced by the mitochondrial glycine decarboxylase complex (GDC), is a major product of photorespiration that accumulates under high photorespiration (high [O2]/[CO2]) conditions in the absence of water stress (Abadie et al., 2016). Serine accumulation may therefore be considered as a readout of genetic variation in drought-induced photorespiratory load—a complex trait that is likely to be affected by many genetic loci but is nonetheless likely to be an important driver of YDT variation.
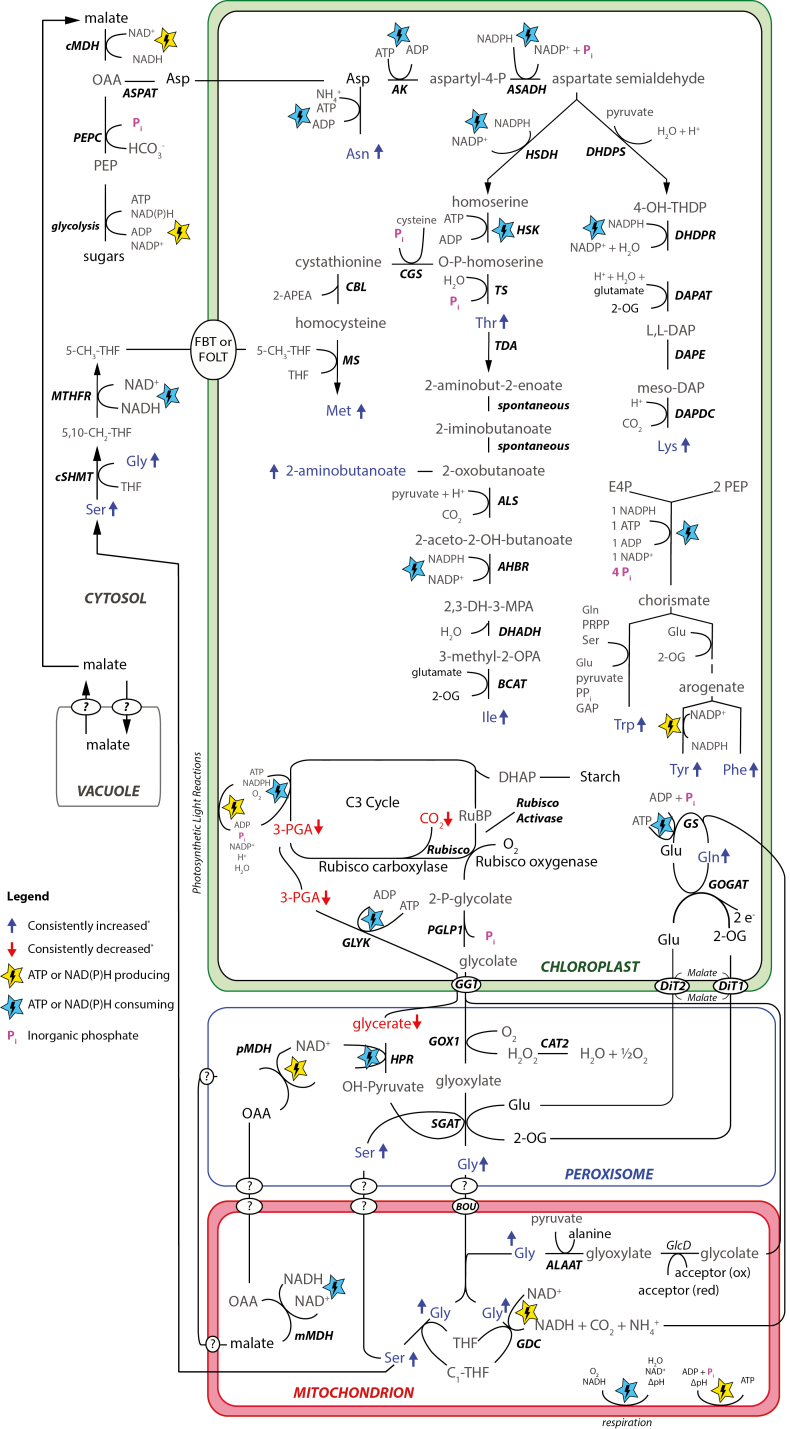
While methionine is not known as a core photorespiratory pathway metabolite, its biosynthetic pathway has obvious links to photorespiration through serine and 1-carbon metabolism (Fig. 6; see Supplementary Table S10 for abbreviations used in the pathway), and its biosynthesis is stimulated under high photorespiration conditions in the absence of water stress in H. annuus and Arabidopsis thaliana (Abadie et al., 2017).
Fig. 6.
Metabolic interactions between photorespiration and chloroplast amino acid metabolism. Metabolic pathways for chloroplast amino acid biosynthesis and photorespiration, and connecting pathways are illustrated schematically. Enzymes are labelled in black bold italics, and colour codes are explained in the key. Abbreviations used in the figure are defined in Supplementary Table S10. *Consistently increased or decreased metabolites were increased or decreased, respectively, in all lines under 28 d glasshouse drought except Trp and Tyr (increased in seven of eight lines) and glycerate (decreased in six of eight lines).
Interestingly, close examination of the fitted coefficients of the YDT~methionine+serine+asparagine multiple regression model revealed that while serine and asparagine had negative coefficients, methionine’s coefficient was positive (Table 2). A similar coefficient was observed for lysine. In other words, our results suggest that if two cultivars exhibit the same increases in serine and asparagine as one another, the cultivar with higher YDT would most probably be the one that accumulated methionine and/or lysine more strongly—pointing to possible links between stress-protective processes and methionine/lysine accumulation. Indeed, by consuming ATP and NAD(P)H, the biosynthesis of methionine and other aspartate-derived amino acids would, in principle, contribute to maintenance of energy homeostasis, protecting against over-reduction of PSII and consequent damage from oxidative stress (i.e. photo-inhibition). That aspartate family amino acid biosynthesis might function as a beneficial energy sink under chloroplast energy imbalance is supported by the observation that allosteric feedback inhibition by threonine of homoserine dehydrogenase (HSDH; Fig. 6), the enzyme thought to control flux of aspartate away from asparagine and into aspartate family amino acid biosynthesis, is strongly reduced under elevated chloroplastic NADPH/NADP+ ratios such as expected under drought stress (Bryan, 1990).
Another conceivable stress-protectant mechanism afforded by methionine biosynthesis is the dissipation of photorespiratory serine, since this would shift photorespiratory reaction equilibria to the right (away from 2-phosphoglycolate, glycolate, glyoxylate, glycine, and serine), thereby helping to keep the pool size of Calvin cycle-inhibiting photorespiratory intermediates (2-phosphoglycolate, glycolate, and glyoxylate) to a minimum (Fig. 6).
In summary, we have shown that complementary information from the responses of serine, asparagine, and methionine (or lysine) to glasshouse drought treatment explains up to 98% of genetic variance in multi-season, multi-site field YDT in a diverse set of wheat cultivars. We suggest that these metabolites may provide useful readouts of three important complementary dimensions of genetic variation in wheat drought tolerance: (i) the capacity to avoid severe dehydration and therefore stomatal closure and high rates of photorespiration (serine); (ii) the tendency to induce senescence upon the onset of severe stress (asparagine); and (iii) the capacity to divert aspartate away from asparagine and into biosynthesis of lysine and methionine. That three complementary metabolites can explain such a high proportion of YDT variance is not at all surprising given that leaf water alone can already explain up to 85% of YDT variance and that metabolites are positioned at the bottom of the causality network translating drought to yield loss, whereas leaf water status is positioned near the top. That is, dehydration represents just the beginning of drought’s impact on yield whereas metabolites are the ultimate building blocks of grain biomass, and altered metabolite profiles therefore represent the penultimate stage of impact of drought on yield. Metabolites truly are a ‘link between genotypes and phenotypes’ (Fiehn, 2002).
The network of processes leading to amino acid changes under drought are undoubtedly complex and multi-dimensional, and different amino acids will be influenced to different degrees by different processes, as described above. Therefore, while amino acids, as a group, may be particularly rich in drought tolerance-predictive metabolites, they provide complementary information about the different orthogonal drought stress responses. This probably explains why three or four amino acids measured in glasshouse experiments can, in fact, be strongly predictive of traits in the field provided the right environmental conditions are correctly matched to the right predictive trait. Although our results are encouraging, determining which combinations of screening conditions and marker traits provide the most efficient predictions will ultimately be a case of trial and error. To this end, further investigation into the applicability of metabolite marker screening to different germplasm, developmental stages, traits, and stress regimes is warranted. It is important to keep in mind that, while our sample of eight cultivars enabled metabolite–YDT associations to be detected with far greater confidence than possible with commonly used smaller sample sizes of 2–5 genotypes, far larger sample sizes will be necessary to overcome the problem of multiple hypothesis testing and confirm these associations beyond reasonable doubt in future association studies.
Conclusion
Our results demonstrate that a small set of metabolic traits measured in glasshouse drought experiments can, in fact, be strongly predictive of field-based measures of YDT and, to a lesser extent, RWC. To date, no markers measured in glasshouses have been reported to predict field-based drought tolerance. In the field, the best measure of drought tolerance is yield gap, but this requires multi-site trials that are an order of magnitude more resource intensive and can be impacted by environmental variation. Thus, high-throughput and standardized analysis of glasshouse-based markers in a less resource-intensive setting could potentially be an effective tool for selection of wheat cultivars with high field-based YDT for continued genetic gain in the face of increasing climate-and land-use pressures.
Supplementary data
Fig. S1. Dendogram showing hierarchical clustering of wheat cultivars.
Fig. S2. Flow chart summarizing selection of wheat cultivars and assessment of various traits in multiple environments.
Fig. S3. Experimental design and soil water content under cyclic drought treatment in a glasshouse.
Fig. S4. Effect of drought stress on leaf wilting in the glasshouse.
Fig. S5. Scatter plots of selected metabolites and RWC correlations under the 28 d glasshouse drought.
Fig. S6. Orthogonal partial least square (OPLS) multivariate model explaining YDT variation.
Table S1. Summary of quality, pedigree, breeding programme, area of adoption, morphology, and agronomic traits of wheat cultivars.
Table S2. Precipitation data of field trial sites across Australia.
Table S3. Raw data of genotyping by sequencing (GBS) of wheat cultivars.
Table S4. Raw data of the physiological traits.
Table S5. Metabolite responses of wheat cultivars to drought in the glasshouse and the field.
Table S6. Associations between metabolite responses to YDT.
Table S7. Metadata file containing metabolite–YDT and metabolite–RWC correlations under all time points and environments.
Table S8. OPLS analysis of metabolome and YDT associations.
Table S9. OPLS variable loadings for the metabolites levels.
Table S10. Definitions of the abbreviations used in Fig. 6.
Acknowledgements
We thank Professor Justin Borevitz for assistance with genotyping by sequencing, and Dr Teresa Neeman (Australian National University) for advice on experimental randomization and statistical modelling. We acknowledge the support of the Research School of Biology/Research School of Chemistry Joint Mass Spectrometry Facility and Diversity Arrays Technology, ACT, Australia and financial support by the Grains Research and Development Corporation Grant (ANU00020), Australian Research Council Centre of Excellence for Translational Photosynthesis (CE140100015), and Plant Energy Biology (CE140100008).
Author contributions
AKY and BJP conceived the original research plan and supervised the experiments; AKY, AJC, GJR, and GME designed and performed the research; AJC developed the software and statistical approaches, and performed metabolomics, physiological, and statistical association analyses. AKY, GJR, and GME analysed field-based yield and physiological data; AJC, AKY, and BJP wrote the paper with the contribution of GME and GJR. The authors declare no competing financial interests.
References
- Abadie C, Blanchet S, Carroll A, Tcherkez G. 2017. Metabolomics analysis of postphotosynthetic effects of gaseous O2 on primary metabolism in illuminated leaves. Functional Plant Biology 44, 929–940. [DOI] [PubMed] [Google Scholar]
- Abadie C, Boex-Fontvieille ER, Carroll AJ, Tcherkez G. 2016. In vivo stoichiometry of photorespiratory metabolism. Nature Plants 2, 15220. [DOI] [PubMed] [Google Scholar]
- Avila-Ospina L, Marmagne A, Talbotec J, Krupinska K, Masclaux-Daubresse C. 2015. The identification of new cytosolic glutamine synthetase and asparagine synthetase genes in barley (Hordeum vulgare L.), and their expression during leaf senescence. Journal of Experimental Botany 66, 2013–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babar MA, Reynolds MP, Van Ginkel M, Klatt AR, Raun WR, Stone ML. 2006. Spectral reflectance to estimate genetic variation for in-season biomass, leaf chlorophyll, and canopy temperature in wheat. Crop Science 46, 1046–1057. [Google Scholar]
- Blum A, Mayer J, Gozlan G. 1982. Infrared thermal sensing of plant canopies as a screening technique for dehydration avoidance in wheat. Field Crops Research 5, 137–146. [Google Scholar]
- Blum A, Shpiler L, Golan G, Mayer J. 1989. Yield stability and canopy temperature of wheat genotypes under drought-stress. Field Crops Research 22, 289–296. [Google Scholar]
- Boonjung H, Fukai S. 1996. Effects of soil water deficit at different growth stages on rice growth and yield under upland conditions. 2. Phenology, biomass production and yield. Field Crops Research 48, 47–55. [Google Scholar]
- Bowne JB, Erwin TA, Juttner J, Schnurbusch T, Langridge P, Bacic A, Roessner U. 2012. Drought responses of leaf tissues from wheat cultivars of differing drought tolerance at the metabolite level. Molecular Plant 5, 418–429. [DOI] [PubMed] [Google Scholar]
- Bryan JK. 1990. Differential regulation of maize homoserine dehydrogenase under physiological conditions. Plant Physiology 92, 785–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carillo P, Mastrolonardo G, Nacca F, Fuggi A. 2005. Nitrate reductase in durum wheat seedlings as affected by nitrate nutrition and salinity. Functional Plant Biology 32, 209–219. [DOI] [PubMed] [Google Scholar]
- Carroll AJ, Badger MR, Harvey Millar A. 2010. The MetabolomeExpress Project: enabling web-based processing, analysis and transparent dissemination of GC/MS metabolomics datasets. BMC Bioinformatics 11, 376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll AJ, Zhang P, Whitehead L, Kaines S, Tcherkez G, Badger MR. 2015. PhenoMeter: a metabolome database search tool using statistical similarity matching of metabolic phenotypes for high-confidence detection of functional links. Frontiers in Bioengineering and Biotechnology 3, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colledge S, Conolly J, Shennan S. 2004. Archaeobotanical evidence for the spread of farming in the Eastern Mediterranean. Current Anthropology 45, S35–S58. [Google Scholar]
- Curtis BC. 2002. Wheat in the world. Rome: Food and Agriculture Organization of the United Nations; http://www.fao.org/docrep/006/y4011e/y4011e04.htm. [Google Scholar]
- Degenkolbe T, Do PT, Kopka J, Zuther E, Hincha DK, Köhl KI. 2013. Identification of drought tolerance markers in a diverse population of rice cultivars by expression and metabolite profiling. PLoS One 8, e63637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O. 2002. Metabolomics—the link between genotypes and phenotypes. Plant Molecular Biology 48, 155–171. [PubMed] [Google Scholar]
- Fisher LH, Han J, Corke FM, Akinyemi A, Didion T, Nielsen KK, Doonan JH, Mur LA, Bosch M. 2016. Linking dynamic phenotyping with metabolite analysis to study natural variation in drought responses of Brachypodium distachyon. Frontiers in Plant Science 7, 1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good AG, Zaplachinski ST. 1994. The effects of drought stress on free amino acid accumulation and protein synthesis in Brassica napus. Physiologia Plantarum 90, 9–14. [Google Scholar]
- Hill CB, Taylor JD, Edwards J, Mather D, Bacic A, Langridge P, Roessner U. 2013. Whole-genome mapping of agronomic and metabolic traits to identify novel quantitative trait loci in bread wheat grown in a water-limited environment. Plant Physiology 162, 1266–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T, Jander G. 2017. Abscisic acid-regulated protein degradation causes osmotic stress-induced accumulation of branched-chain amino acids in Arabidopsis thaliana. Planta 246, 737–747. [DOI] [PubMed] [Google Scholar]
- Kerepesi I, Galiba G. 2000. Osmotic and salt stress-induced alteration in soluble carbohydrate content in wheat seedlings. Crop Science 40, 482–487. [Google Scholar]
- Kishor PK, Sangam S, Amrutha R, Laxmi PS, Naidu K, Rao K, Rao S, Reddy K, Theriappan P, Sreenivasulu N. 2005. Regulation of proline biosynthesis, degradation, uptake and transport in higher plants: its implications in plant growth and abiotic stress tolerance. Current Science 424–438. [Google Scholar]
- Lea PJ, Sodek L, Parry MAJ, Shewry PR, Halford NG. 2007. Asparagine in plants. Annals of Applied Biology 150, 1–26. [Google Scholar]
- Li H, Vikram P, Singh RP, et al. 2015. A high density GBS map of bread wheat and its application for dissecting complex disease resistance traits. BMC Genomics 16, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschen S, Bengoa Luoni S, Di Rienzo JA, et al. 2016. Integrating transcriptomic and metabolomic analysis to understand natural leaf senescence in sunflower. Plant Biotechnology Journal 14, 719–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R, James RA, Sirault XR, Furbank RT, Jones HG. 2010. New phenotyping methods for screening wheat and barley for beneficial responses to water deficit. Journal of Experimental Botany 61, 3499–3507. [DOI] [PubMed] [Google Scholar]
- Munns R, James RA, Läuchli A. 2006. Approaches to increasing the salt tolerance of wheat and other cereals. Journal of Experimental Botany 57, 1025–1043. [DOI] [PubMed] [Google Scholar]
- Obata T, Fernie AR. 2012. The use of metabolomics to dissect plant responses to abiotic stresses. Cellular and Molecular Life Sciences 69, 3225–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata T, Witt S, Lisec J, Palacios-Rojas N, Florez-Sarasa I, Yousfi S, Araus JL, Cairns JE, Fernie AR. 2015. Metabolite profiles of maize leaves in drought, heat, and combined stress field trials reveal the relationship between metabolism and grain yield. Plant Physiology 169, 2665–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passioura JB. 2006. The perils of pot experiments. Functional Plant Biology 33, 1075–1079. [DOI] [PubMed] [Google Scholar]
- Pornsiriwong W, Estavillo GM, Chan KX, et al. 2017. A chloroplast retrograde signal, 3'-phosphoadenosine 5'-phosphate, acts as a secondary messenger in abscisic acid signaling in stomatal closure and germination. eLife 6, e23361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan GP, Prasad PVV, Fritz AK, Kirkham MB, Gill BS. 2012. Effects of drought and high temperature stress on synthetic hexaploid wheat. Functional Plant Biology 39, 190. [DOI] [PubMed] [Google Scholar]
- Rai VK. 2002. Role of amino acids in plant responses to stresses. Biologia Plantarum 45, 481–487. [Google Scholar]
- Rashid A, Stark JC, Tanveer A, Mustafa T. 1999. Use of canopy temperature measurements as a screening tool for drought tolerance in spring wheat. Journal of Agronomy and Crop Science 182, 231–237. [Google Scholar]
- Rebetzke GJ, Chenu K, Biddulph B, Moeller C, Deery DM, Rattey AR, Bennett D, Barrett-Lennard EG, Mayer JE. 2013. A multisite managed environment facility for targeted trait and germplasm phenotyping. Functional Plant Biology 40, 1–13. [DOI] [PubMed] [Google Scholar]
- Rebetzke GJ, Fischer RA, van Herwaarden AF, Bonnett DG, Chenu K, Rattey AR, Fettell NA. 2014. Plot size matters: interference from intergenotypic competition in plant phenotyping studies. Functional Plant Biology 41, 107–118. [DOI] [PubMed] [Google Scholar]
- Rebetzke GJ, Rattey AR, Farquhar GD, Richards RA, Condon AG. 2012. Genomic regions for canopy temperature and their genetic association with stomatal conductance and grain yield in wheat. Functional Plant Biology 40, 14–33. [DOI] [PubMed] [Google Scholar]
- Rivero RM, Kojima M, Gepstein A, Sakakibara H, Mittler R, Gepstein S, Blumwald E. 2007. Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proceedings of the National Academy of Sciences, USA 104, 19631–19636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewry PR, Halford NG, Belton PS, Tatham AS. 2002. The structure and properties of gluten: an elastic protein from wheat grain. Philosophical Transactions of the Royal Society B: Biological Sciences 357, 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AB, Lim P, Cullis BR. 2006. The design and analysis of multi-phase plant breeding experiments. Journal of Agricultural Science 144, 393–409. [Google Scholar]
- Stewart GR, Larher F. 1980. Accumulation of amino acids and related compounds in relation to environmental stress. In: Miflin BJ, ed. Amino acids and derivatives. New York: Academic Press, 609–635. [Google Scholar]
- Thévenot EA, Roux A, Xu Y, Ezan E, Junot C. 2015. Analysis of the human adult urinary metabolome variations with age, body mass index, and gender by implementing a comprehensive workflow for univariate and OPLS statistical analyses. Journal of Proteome Research 14, 3322–3335. [DOI] [PubMed] [Google Scholar]
- Trenberth KE, Dai AG, van der Schrier G, Jones PD, Barichivich J, Briffa KR, Sheffield J. 2014. Global warming and changes in drought. Nature Climate Change 4, 17–22. [Google Scholar]
- United Nations 2013. World population prospects: the 2012 revision. New York: United Nations. [Google Scholar]
- van Dijk AIJM, Beck HE, Crosbie RS, de Jeu RAM, Liu YY, Podger GM, Timbal B, Viney NR. 2013. The Millennium drought in southeast Australia (2001–2009): natural and human causes and implications for water resources, ecosystems, economy, and society. Water Resources Research 49, 1040–1057. [Google Scholar]
- Watanabe M, Balazadeh S, Tohge T, Erban A, Giavalisco P, Kopka J, Mueller-Roeber B, Fernie AR, Hoefgen R. 2013. Comprehensive dissection of spatiotemporal metabolic shifts in primary, secondary, and lipid metabolism during developmental senescence in Arabidopsis. Plant Physiology 162, 1290–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt S, Galicia L, Lisec J, Cairns J, Tiessen A, Araus JL, Palacios-Rojas N, Fernie AR. 2012. Metabolic and phenotypic responses of greenhouse-grown maize hybrids to experimentally controlled drought stress. Molecular Plant 5, 401–417. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.