Abstract
Aim
To 3D print heart tissue, one must understand how the main two types of cardiac cells are affected by the printing process.
Materials & methods
Effects of gelatin methacryloyl (GelMA) concentration, extruder pressure and duration of UV exposure on survival of cardiac myocytes and fibroblasts were examined using lactate dehydrogenase and LIVE/DEAD assays, bioluminescence imaging and morphological assessment.
Results & conclusion
Cell survival within 3D printed cardiomyocyte-laden GelMA constructs was more sensitive to extruder pressure and GelMA concentrations than within 3D fibroblast-laden GelMA constructs. Cells within both types of constructs were adversely impacted by the UV curing step. Use of mixed cell populations and enrichment of bioink formulation with fibronectin led to an improvement of cardiomyocyte survival and spreading.
Keywords: bioluminescence imaging, cardiac fibroblasts, cardiomyocytes, cell viability, 3D bioprinting, GelMA
Cardiac myocytes (CMC) and cardiac fibroblasts (CFB) are the two main cell components of cardiac muscle. The CFBs provide essential structural support to CMCs and produce most of cardiac the extracellular matrix proteins [1]. Attempts to produce engineered strips of cardiac muscle using pure populations of CMCs failed, as the presence of CFBs is essential for cardiac muscle formation [2,3]. Indeed, multiple groups have shown that by combining these two cell populations, one can form small self-contracting cardiac muscle strips, also called engineered heart tissue (EHT) [4–7]. While the original, mold-based EHT technique works well, its ability to create complex free-standing shapes and/or distinct regions of EHT with specific cell content is limited. Recent advances in 3D bioprinting help to address this limitation as they offer an opportunity to create complex multicellular EHTs in a highly controlled manner [8,9]. To further advance this promising direction, one must understand how these two cardiac cell types are affected by the printing process. To the best of our knowledge, such a side-by-side comparison is yet to be reported.
The most common 3D bioprinting protocols involve extrusion-based printers that use cell-laden bioinks [10]. The main components of these bioinks have to fulfill three criteria: support cell adhesion, prompt cell-to-cell contact and allow for cell migration. Successful bioprinting also requires certain geometric accuracy and minimal effects on cell viability. Specifically, when it comes to cell-laden bioinks, thorough optimization of multiple printing parameters is needed in order to reduce shear stress on cells while producing high precision 3D constructs [11]. These parameters include the viscosity of bioink material, the gauge of the needle, extruder temperature and extrusion pressure. For photocurable bioinks, the duration of UV exposure also has to be considered.
In our study, we used gelatin methacryloyl (GelMA) as the cell carrier. The GelMA’s main component is gelatin which enhances tissue regeneration and printability, while the photopolymerizable methacrylamide group allows matrices to be covalently cross-linked by UV light after printing [12]. The GelMA is considered to be mechanically stable and systemically tunable based on its degree of methacrylation. Furthermore, the elastic modulus of GelMA can be adjusted by altering its concentration and printing parameters such as printing temperature, duration of UV exposure, type and amount of photoinitiator [10,13]. In addition, GelMA encapsulates cells, providing layer of protection during 3D printing process and associated shear-induced stress [14].
Different methods can be used to document viability of the cells within 3D printed constructs, each having its own advantages and shortcomings. To better understand the effects of 3D printing on cell viability, we used several well-known assays including the release of cytosolic lactate dehydrogenase (LDH assay), fluorescent dye based LIVE/DEAD assay and bioluminescence imaging (BLI). Although less common in bioprinting field, BLI provides a convenient way to monitor cell growth and proliferation longitudinally [15]. The use of BLI enabled us to examine long-term effect of printing steps and not just acute damage to cell membranes.
The use of two cardiac cell types allowed us to address the effects of extrusion-based bioprinting on cell survival and cell-to-cell interactions in GelMA. This information is a critical step for future efforts to print viable heart tissue.
Materials & methods
Cell preparation
Cells were isolated from the hearts of 2- to 3-day-old neonatal Sprague–Dawley rats of mixed sex according to a standard trypsin-collagenase digestion protocol [16]. All procedures were done in accordance to the guidelines of the Institutional Animal Care and Use Committee at the George Washington University and the NIH Guide for the Care and Use of Laboratory Animals. First-order separation of CMCs from CFBs was achieved by a preplating procedure, which relies on preferential attachment of CFBs to the bottom of T-flasks. After 1 h of preplating, the media containing floating CMCs was collected. The attached CFBs were then trypsinized and replated one- or more times to remove any remaining CMCs. Each cell population was then counted and combined with specified concentrations of GelMA and 0.5% photoinitiator (Biokey, LAP) at 1 × 106 cells/ml and triturated for 10 s to form a uniform viscous gel. Cell-laden bioink was then loaded into and placed in a 4°C ice bath for 10 min before printing.
Bioprinting protocol
Extrusion based printing was performed using a pneumatic 3D bioprinter (Biobot 1, Allevi). The CAD models were made in SolidWorks. The STL files were imported into Slic3r to obtain GCODE files with specified layer patterns, infills and print speeds. To obtain single, resolute printed struts, constructs were printed with 0% infill and a print speed of 2 mm/s. Before printing, the intended extruder was cooled to an initial temperature of 18–20°C. Constructs were printed in standard, coated well plates, cross-linked with UV light at 405 nm wavelength using the recommended manufacturer settings of 7 mW/cm2 for 2 min. Constructs were subsequently immersed in 1–2 ml media. After printing, well plates were cultured for 7 days using standard cell culture conditions.
In Vitro BLI
Bioluminescent images were obtained using an IVIS Lumina K Machine (PerkinElmer, MA, USA). After 24 h of printing (also called day 1), cells were infected with Firefly Luciferase (AdCMV-luc, 1.0 × 1011 PFU/ml, Iowa Gene Transfer Vector Core), a reporter virus that emits visible light when coupled with CycLuc-1. After 48 h of printing (i.e., day 2), constructs were imaged using bioluminescence at 550–650 nm wavelengths and analyzed based on average radiance using Xenogen Living Image v4.5.1 software. Blank wells were measured and subtracted from each construct’s region of interest. Measurements were acquired every 48 h yielding measurements on day 4 and 6. Excel slope function was used to calculate the daily change in the bioluminescence. The latter relies on linear regression between the numbers of days postprinting and the bioluminescence values. To ensure that luciferin-dependent signal was not cumulative, cells were washed with fresh media and reimaged between measurements to verify the lack of signal when luciferin substrate was not present [17].
Lactate dehydrogenase assay
Pierce LDH Cytotoxicity Assay Kit (Thermo Fisher Scientific) was used to quantify cytotoxicity in our 3D printed constructs. Cell media was collected every 48 h, and triplicate samples were prepared for analysis using a Thermo Fisher Scientific Varioskan Spectrophotometer. Absorbance readings were taken at 680 and 490 nm as per manufacturer’s protocol. Excel slope function was used to calculate the daily change in the LDH values. The latter relies on linear regression between the numbers of days postprinting and the LDH values.
LIVE/DEAD & organelle-specific staining
The 3D printed constructs were fixed using ice cold 1:1 acetone/methanol solution. Constructs were subsequently stained with a 1:1 mixture of DAPI (VECTASHIELD, #H-1200) and Phalloidin (VECTASHIELD, #H-1600) mounting medium. To confirm the presence of live and dead cells in 3D printed constructs, selected constructs were stained using a Thermo Fisher Scientific LIVE/DEAD cell viability kit (#L3224) containing calcein AM and Ethidium Homodimer-1. Depending on magnification and spectral properties of the dyes, an upright fluorescence microscope (Olympus DP80), Zeiss LSM510 confocal or PerkinElmer Nuance FX Hyperspectral system was used to take images of stained constructs.
Statistical analysis
Unless otherwise stated, all qualitative conclusions and quantitative values are based on at least three independent experiments. For LDH assays, each individual sample was run in triplicate. Differences between groups were determined by one-way ANOVA followed by a Student’s t-test with values of p < 0.05 considered significant.
Results
Study design
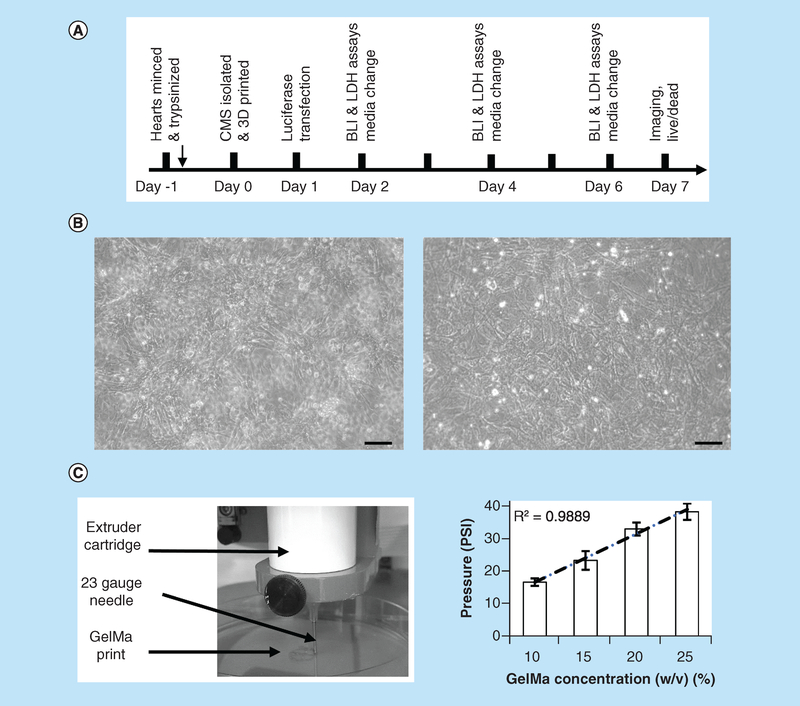
As illustrated in Figure 1A & B, on day 0, populations of freshly isolated CMCs and CFBs were used for 3D printing or plated as monolayer controls. Minimal pressure settings were used to extrude a specified percentage of GelMA bioink (Figure 1C). Transfection with luciferase was done after 24 h of 3D printing (day 1). Measurements were taken on days 2, 4 and 6; and images were acquired throughout 7 days of culture.
Figure 1. Outline of experimental design.
(A) CMCs and CFBs were isolated using standard procedure that involves overnight digestion with trypsin which followed by collagenase digestion next day. Cells were then mixed with GelMA, printed and placed in culture conditions for specified number of days. On days 2, 4, and 6, LDH and BLI measurements were taken, followed by imaging of live and fixed cells on day 7. (B) Sample phase-contrast images of CMCs (left) and CFBs (right) cultured in monolayers as parallel controls. Scale bar-100 micron. (C) Commercial 3D bioprinter (Allevi) was used to print cell-laden bioinks using specified values of extruder pressure and syringe temperatures. Graph on the right shows minimum pressures required to extrude different GelMA concentrations using 23-gauge needle at 20°C temperature. R stands for Pearson correlation coefficient.
BLI: Bioluminescence intensity; CFB: Cardiac fibroblast; CMC: Cardiac myocyte; GelMA: Gelatin methacryloyl; LDH: Lactate dehydrogenase.
Optimization of printing parameters
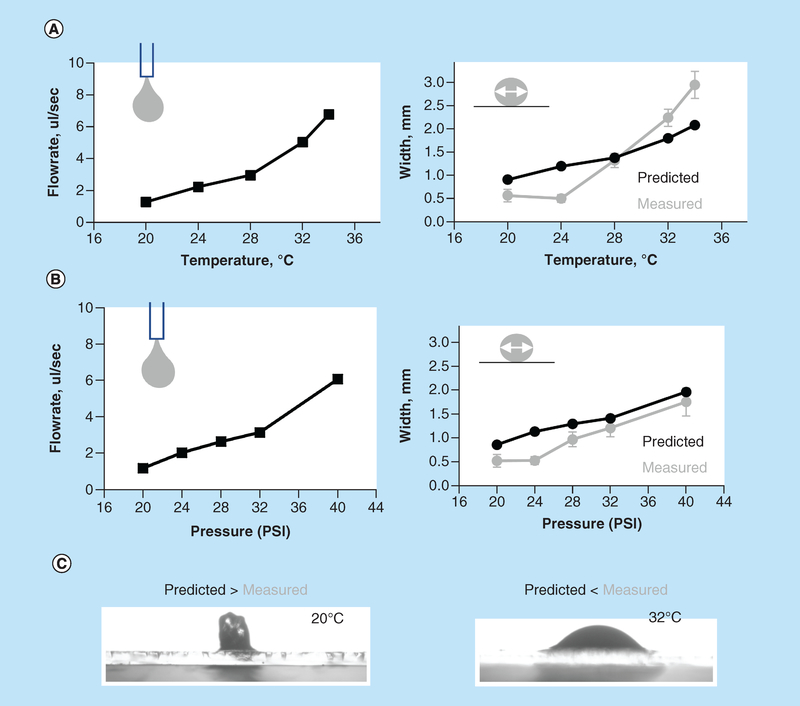
Pneumatic plunger-based 3D printers have simpler drive mechanisms than those with mechanical plungers and have an additional advantage of ease with which multiple cartridges can be used for printing. However, pneumatic-based printers have less control over the flow of extruded material [10]. Therefore, our first step was to evaluate how the rate of GelMA extrusion and the fidelity of printed struts are affected by temperature and pressure settings. As one would expect, and in agreement with previous reports, an increase in either bioink temperature, extruder pressure or diameter of the needle increases the volume of the extruded bioink (Figure 2) [13]. But when printing, it is the exact shape of the printed pattern that matters and not necessarily the extruded volume (an analogy being drawn a picture with a fine marker vs a thick one). Therefore, we looked into a relationship between the volume of the extruded material and the width of the GelMA struts. The predicted width value can be estimated assuming cylindrical shape of the strut using formula Q = 3.14 × r2 × v, where Q is bioink flowrate, r is the inner radius of the needle and v is linear velocity of the needle movement. At low extrusion pressures and temperatures, printed lines appeared narrower than instructed by their G-code, which corresponds to the cross-section shape of a printed strut being elongated. On the other hand, when temperature increased, the opposite was observed, with lines becoming wider than those defined by CAD settings. This corresponds to a less viscous gel that makes flatter structures. Illustrative images of both cases are shown in Figure 2C. Thus, if one wants to print GelMA based structures that conform to CAD designs with a high degree of precision, this nonlinear temperature effect on the shapbe of the printed lines in Z-dimension has also to be taken into account (in addition to all other well-known factors [18]).
Figure 2. Effects of extrusion pressure and temperature on width of printed gelatin methacryloyl struts.
(A) Predicted versus measured width of GelMA struts. Left panel: the flowrate of 15% GelMA printed using 23-gauge needle at specified temperatures was determined based on the weight of extruded hydrogel. Right panel: thickness of printed lines was then measured experimentally (grey symbols) and compared with the predicted thickness of cylindrically shaped struts (black symbols) based on flowrate values shown in A. Temperature above 28°C led to a wider than predicted struts, while temperature below led to a more narrow ones. (B) Similar measurements done at a constant temperature of 20°C while altering extruder pressure values. (C) Illustrative cross-sections of struts printed at 20° and 32°C are shown.
GelMA: Gelatin methacryloyl.
Effect of GelMA concentration of postprinting cell survival
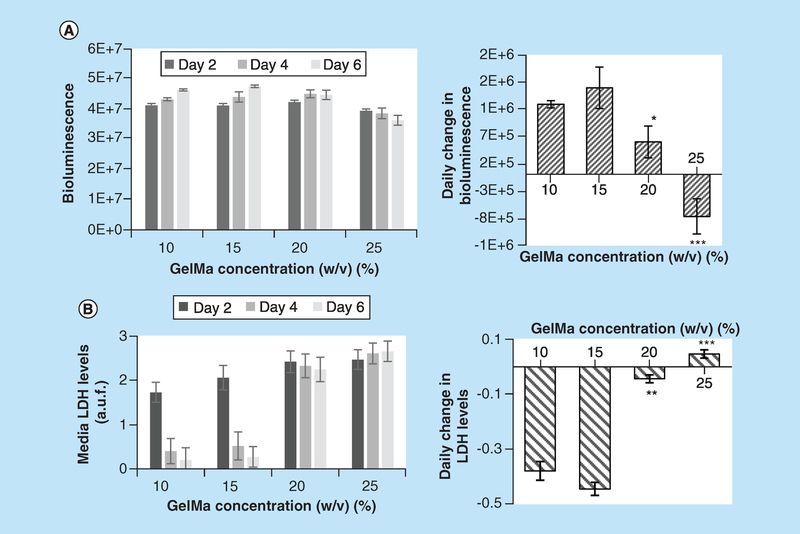
Upon UV polymerization, bioinks with high percentage of GelMA yield more structurally stable structures. However, high percentage of GelMA increases bioink viscosity, and thus the minimal pressure needed to extrude it (Figure 1C). High extrusion pressures or GelMA concentrations higher than 15% decreased long-term cell viability (Figure 3). The latter was determined based on BLI data with more ATP production linked to larger cell numbers (Figure 3A). The second index, a daily rate of LDH release was used as a first order estimate of cell damage (Figure 3B). Higher rate of LDH release in 20 and 25% GelMA constructs pointed to presence of damaged cells that continue to die off. Cell viability has previously been shown to decrease with GelMA concentrations [11–14] and may occur due to encapsulation stress, nutrient limitations or stress due to transient swelling after placement in media. Importantly, data shown in Figure 3 were collected from constructs that were kept in culture for 6 days and not immediately after printing. Thus, even if viable cell counts appear to be initially unaffected by the printing process, one must be concerned about its long-term effects. Based on cell viability data shown in Figure 3, we have resorted to use 10–15% GelMA for the rest of our studies.
Figure 3. Effects of different gelatin methacryloyl concentration on viability of cardiac fibroblast-laden constructs.
(A) Left panel: values of BLI from 3D printed constructs containing CFBs transfected with luciferin luciferase. The BLI values are expressed in ph/s/cm2/sr or photons per second per centimeter squared per steradian. Right panel: data shown in left panel recalculated as a daily change in BLI. Positive slopes for the 10 and 15% GelMA constructs point to progressive increase in BLI, while constructs printed with 25% GelMA show decline in BLI during 1 week in culture. Asterisks stand for difference with 10% GelMA constructs; *p < 0.05, **p < 0.01, ***p < 0.001. (B) Left panel: values of LDH release into the media from the same 3D printed constructs (arbitrary fluorescence units). Right panel: daily change in LDH release expressed as a slope between LDH and number of days in culture. Negative slopes for the 10 and 15% GelMA constructs point to increasing viability of the cells, while constructs printed using 20 and 25% GelMA show near zero or positive slopes. The latter points to a continuous loss of cells.
BLI: Bioluminescence intensity; CFB: Cardiac fibroblast; GelMA: Gelatin methacryloyl; LDH: Lactate dehydrogenase.
Effects of extruder pressure on postprinting cell survival
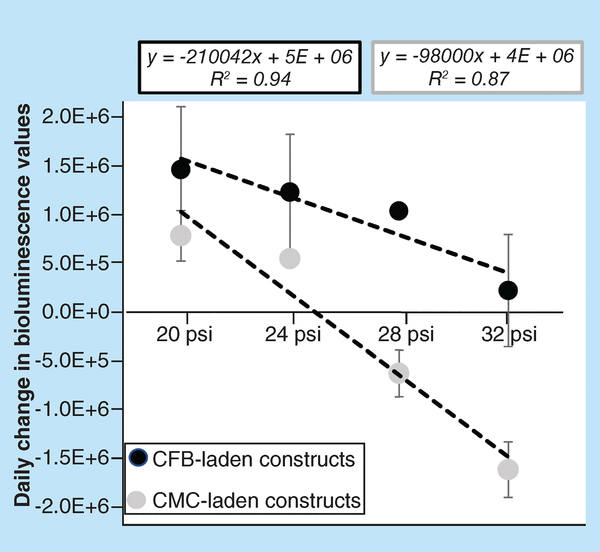
The BLI revealed that extruding pressure has more of an effect on the viability of CMC-laden constructs versus CFB-laden bioinks (Figure 4). Predictably, higher pressures yielded lower cell viability in constructs printed using either cell type. This was quantified using linear regression analysis of the relationship between the extrusion pressure (x) and the daily change in bioluminescence values (y). It revealed a significant correlation between the two variables for both types of constructs (R2 = 0.95 for CMC-laden, R2 = 0.88 for CFB-laden). However, the extent to which constructs printed with CMC-laden bioink deteriorated under high pressures was more pronounced. The comparison of the slopes of the regression lines (2.1 × 105 for CMC vs 0.98 × 105 for CFB) provided us with a quantitative measure of long-term susceptibility of cell-laden constructs to the effects of shear stress. The data also indicated that extruder pressures above 30 psi completely halt proliferation of cells in constructs printed using CFB-laden bioink (column at 32 psi shows nearly zero change in daily bioluminescence values). The same pressures caused a significant loss of viability in constructs printed using CMC-laden bioink, leading to negative values of daily BLI change.
Figure 4. Effects of extrusion pressure on viability of constructs printed using cardiac myocyte- and cardiac fibroblast-laden gelatin methacryloyl bioinks.
Daily change in values of BLI from 3D printed constructs containing cells transfected with luciferin luciferase. The BLI Units are ph/sec/cm2/sr. The slope of BLI/pressure relationship quantifies the difference between sensitivity of CMC- and CFB-laden constructs to sheer stress caused by high extrusion pressures. In CMC-laden constructs, high extrusion pressures led to a loss of viable cells which manifested as a negative value of daily BLI changes.
BLI: Bioluminescence intensity; CFB: Cardiac fibroblast; CMC: Cardiac myocyte; GelMA: Gelatin methacryloyl.
Effect of UV exposure time on postprinting cell survival
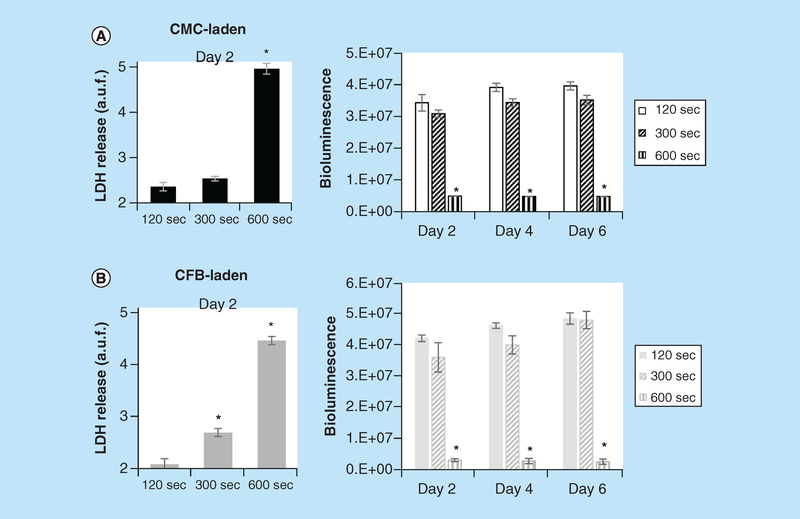
A critical step for many bioinks is photopolymerization after printing. The latter requires either UV or blue light exposure. We used both BLI and LDH assays to compare the negative effects of increased UV exposure time on cell viability of 3D printed constructs (Figure 5). Qualitative and quantitative analysis showed that extended exposure to 7 mW/cm2 UV light at 405 nm wavelength can have detrimental to cell viability and proliferation. For both types of constructs, 600 s exposure times resulted in halted cell proliferation and nearly complete cell death, while exposure times of less than 300 s had minimal effect on cell viability and proliferation. Thus, at tested exposure times (120, 300 and 600 s), UV affected viability of constructs printed with CFB- versus CMC-laden bioinks similarly. The UV exposures less than 2 min failed to fully polymerize GelMA.
Figure 5. Effects of UV exposure on viability of cardiac myocyte- and cardiac fibroblast-laden, 3D printed constructs.
Immediately after printing, GelMA constructs were exposed to 405 nm, 7 mW/cm2 Watt LED light for either 120, 300 or 600 s. Bioluminescence intensity values from cell-laden 3D printed CMC- and CFB-laden constructs taken at 2, 4 and 6 days of culturing. Initial UV exposures are indicated on x-axis. Shown are the BLI intensities and values of LDH release after 2 days of specified durations of UV exposure for CMC- (A) and CFB-laden constructs (B). Asterisks stand for difference with constructs exposed to UV for 120 s; *p < 0.001.
CFB: Cardiac fibroblast; CMC: Cardiac myocyte; LDH: Lactate dehydrogenase.
Verification of cell viability & proliferation using imaging
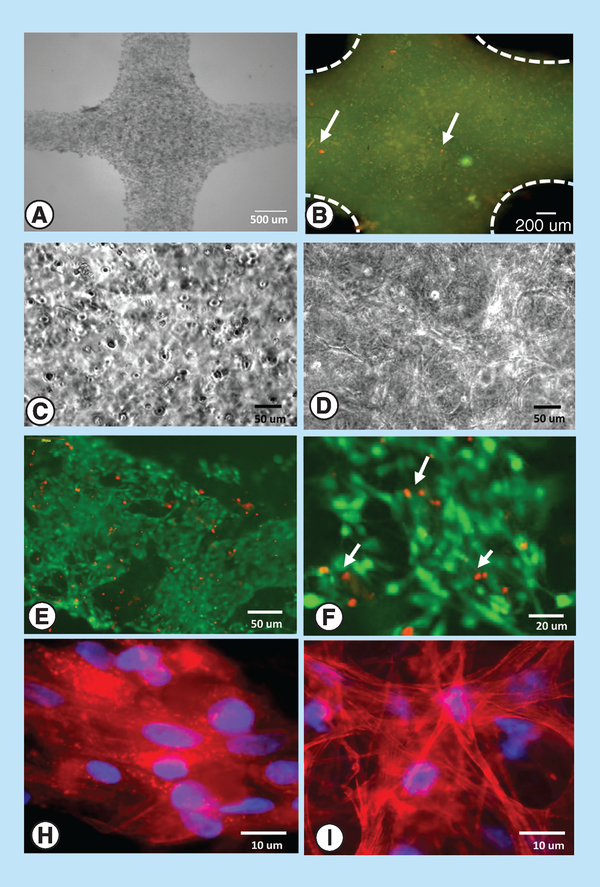
After 6–7 days in culture, we used phase contrast microscopy, LIVE/DEAD and phalloidin/DAPI staining to examine cell viability and shape within 3D printed 15% GelMA constructs (Figure 6A–I). In constructs printed using CFB-laden GelMA, cells were adhering, proliferating and making cell-to-cell connections (Figure 6D, E, F & I). This observation was consistent with BLI data shown in Figure 3, manual counts of cell nuclei and diminishing levels of LDH release from these constructs. In contrast, in 3D constructs printed using CMC-laden GelMA, cells were mostly constrained within clusters with minimal changes in total nuclei counts (Figure 6C). Addition of 10 μg/ml fibronectin and use of crude, nonpreplated preparations of CMC led to significant improvement of CMC spreading as shown in Figure 6H.
Figure 6. Cell appearance within 3D printed constructs.
Representative images of constructs printed using 15% GelMA bioink: (A) A phase contrast image of a CMC-laden construct, (B) LIVE/DEAD labeling of a freshly printed CMC construct showing diffuse GelMA staining, live cells in green and dead cells in red (arrows), (C) illustrative phase contrast image of a 3D construct with encapsulated CMC cells, (D) illustrative phase contrast image of a 3D construct with interconnected CFB cells, (E) LIVE/DEAD staining of a 7-day, CFB-laden construct showing abundance of interconnected live cells (green) and nuclei of dead cells (red), (F) a close-up of the same construct, (G) CMCs within 7-day old construct stained for actin (phalloidin, red) and nuclei (DAPI, blue), and (H) CFBs within 7-day old CFB-laden construct stained for actin (phalloidin, red) and nuclei (DAPI, blue).
CFB: Cardiac fibroblast; CMC: Cardiac myocyte; GelMA: Gelatin methacryloyl; LDH: Lactate dehydrogenase.
Dual extruder printing
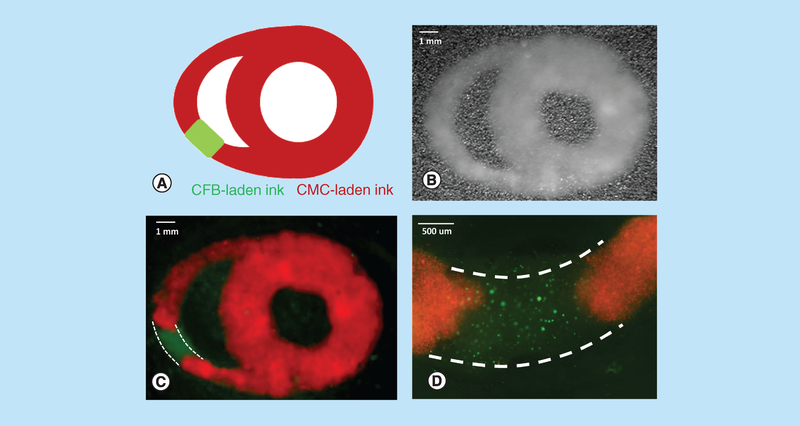
The ultimate goal of our studies was to understand the main differences 3D printing process has on survival of CMC versus CFB imbedded in GelMA bioink. Upon optimization of printer settings and use of minimal required UV and pressure values a wide variety of patterns could be printed using both cell types. An example of such pattern is shown in Figure 7. Two extruders loaded with CFB-(labeled in green) and CMC- (labeled in red) laden GelMa were used to print a pattern resembling cross-sectional view of the heart with a scar-like segment.
Figure 7. Dual extruder printing using cardiac myocyte- and cardiac fibroblast-laden gelatin methacryloyl.
(A) A cartoon of heart cross-section with scar-like segment which was then designed and assembled in SolidWorks. (B) Visual appearance of a freshly printed construct based on the above schematics using dual extruder printing with CMC- and CFB-laden 15% GelMA bioink formulations. (C) Multispectral imaging of the same construct using 360 nm LED source enabled clear demarcation of the two bioinks (CMCs were labeled with MitoTracker Red, while CFB with Calcein AM). Linear unmixing was applied as per manufacturer software (PerkinElmer Nuance FX 3.02). Dotted white lines outline the CFB-laden part of the construct. (D) A close-up image of CFB segment obtained using Olympus fluorescence microscope. Dotted white lines outline the CFB-laden part of the construct.
CFB: Cardiac fibroblast; CMC: Cardiac myocyte; GelMA: Gelatin methacryloyl.
GelMA shortcomings
Being made from gelatin, GelMA effectively absorbs most of the dyes, including Trypan Blue, primary or secondary antibodies. Therefore, immunostaining protocols that work well for monolayer cultures are not effective in staining cells within GelMA constructs even when one increases antibody concentration or incubation time. Therefore, after multiple attempts we had to resort to use low molecular weight stains such as phalloidin to identify shape of cells within printed GelMA structures (Figure 6). Another noticed shortcoming was the adverse effect of nonpolymerized GelMA + UV initiator mixture on cell viability, which occurs when cells are kept within extrusion syringe for extended time prior to cross-linking. The latter leads to poor quality of constructs, made toward the end of the printing process. Thus, it is recommended to use the smallest volume possible when printing and make a fresh mixture of cells and GelMA after minimal number of prints. At last, a general assumption that by lowering GelMA concentration cell spreading can be improved, it does not necessarily apply to all cell types. When we made constructs using 5% of GelMA bioink (using a support bath method [19]), CMC-laden constructs continued to show encapsulated and nonproliferating cells after 7 days of culture, much like in >10% GelMA formulations.
Discussion
Because of its hydrogel properties and mechanical stability, gelatin methacryloyl (GelMA) has been increasingly used for cell printing [13,20]. The GelMA forms covalent cross-links that are highly tunable making it particularly useful for tissue engineering applications [21]. When printing with GelMA, one essentially combines two techniques: cell encapsulation and bioprinting. Cell encapsulation is believed to provide a protective layer around engulfed cells, thus reducing the amount of shear stress cells undergo during printing and handling. On the other hand, encapsulation can adversely have ability of cells to spread and interconnect postprinting, something that we clearly observed in case of CMC-laden GelMA formulations. When cross-linked using photoinitiator such as Igacure of LAP (lithium phenyl-2,4,6-trimethylbenzoylphosphinate), GelMA was reported to have a negligible effect on the viability of the cells [22]. The latter is true barring several assumptions, in other words, that constructs are printed at short UV exposure times, low print speeds and low UV intensity. Once cross-linked, the 3D printing process became irreversible, allowing GelMA prints to sustain their shape in cell culture conditions for extended periods.
Despite general perception that different cells might be affected by printing process differently, only a few published studies provide quantitative assessment to support this statement. The Supplementary Table lists a handful of papers that directly compared different cell types as far as their postprinting survival with most of these studies reporting an on-time measurement using LIVE/DEAD assay. To the best of our knowledge, there are no published studies that compared, side by side, the effect of printing steps on survival of CMC and CFB.
We used neonatal rat CMC as they continue to be the most widely used cell sources in cardiac tissue engineering. Preplating procedure removes significant number of CFBs from crude whole heart digest, yielding a CMC-enriched fraction. The CFBs are still present in this fraction, but they constitute only 20–30% of total cell numbers [23]. Therefore, our study represents a first order approximation of the effects that UV exposure and extruder pressure might have on bioinks laden with these two main types of cardiac cells. One can envision a follow-up study in which a more defined population of CMCs, derived from either embryonic or induced pluripotent stem cells can be used to further define and investigate differential effects of 3D printing on cardiomyocyte survival. In this future study, an additional compounding factor to be considered will be a stage of CMC differentiation since both cell size and membrane composition are likely to be affected by the differentiation process.
It was important for us to compare the effects of bioprinting beyond the acute effects of shear stress on cell membranes. Instead we employed bi-daily bioluminescence measurements and recurrent assays of released LDH. The daily change in these variables then enabled us to quantitatively evaluate constructs’ postprinting proliferative status and viability. Because CMC cultures are known to deteriorate and redifferentiate during prolong periods of culture, while CFB continue proliferate, we did not extend these measurements beyond 1-week period since the above-noted differences in cell behavior make long-term comparison of printing effects difficult. Therefore, we focused only on a few day survival postprinting and normalized each cell type numbers to the appropriate controls.
Bioluminescence measurements are often used to monitor in vivo survival of implanted constructs [24]. They can also be useful for in vitro longitudinal monitoring, having the advantages of being inexpensive, easy-to-use, and effective method of tracking cell viability within 3D constructs. It is one limitation, however, might be that luciferin acts as a rate-limiting factor. In other words, for as long as there is luciferin present in cellular constructs, the constructs will continue to produce a signal. This runs the risk of overaccumulation of radiance and inaccurate measurements. This technical limitation can be easily overcome, particularly in vitro, by replacing media and reimaging constructs between measurements to confirm absence of background luciferin signal.
One of the visual observations from week-old printed constructs was a limited amount of CMC spreading and cell-to-cell connections, particularly when compared with constructs printed using CFB-laden bioink. This occurred even in constructs printed at lowest extrusion pressures using minimal duration of UV exposure. This lack of spreading most likely can be ascribed to the effect of cross-linked GelMA on the size of the pores within the biogel that can limit cell spread [25]. A possible solution to this might be incorporating a photodegradable functionality into GelMA and using UV light to manipulate CMC constructs’ mechanical properties and assist cellular alignment [26]. Another approach might be further lowering GelMA concentration while adding relevant adhesion proteins into bioink formulation. While using low GelMA concentrations have a benefit of yielding a more porous hydrogel, these prints are not capable of retaining their 3D shape without additional support. The latter can be provided by using an external support bath [19,27], by adding supplementary gelling steps [28,29] or by introducing alginate encasing [30].
Our studies have confirmed that increased UV exposure can adversely impact postprinting cell survival. The data also suggest that UV exposure affects viability of constructs printed using CFB- and CMC-laden bioinks to a similar extent. In radical chain polymerization, photoinitiatior molecules will absorb photons of light and dissociate into radicals that initiate polymerization. This reaction depends on the type of light used to dissociate the photoinitator molecules, and the duration for which the cells are exposed to that light (the UV exposure times noted in the results sections are of course not absolute values as they depend on both user equipment and UV-initiator type and concentration). While UV light has long been a preferred method of cross-linking, it runs a risk of damaging cells. Although a number of studies have shown the effectiveness of visible light to cross-link GelMA, this process requires longer exposures for gel to polymerize, so the overall outcome on cell survival is not immediately evident [31,32].
The end goal of our efforts is to print spatially defined 3D constructs that can include patterns of CMC and CFB mimicking different physiological and pathophysiological scenarios. As an example, we used two extruders printing to print a pattern resembling cross-sectional view of the heart with a scar-like segment (Figure 7). The 3D constructs like this can be used to study arrhythmia mechanisms [33,34], conduct personalized medicine testing using iPS-derived CMC or CFBs [35] or apply optogenetic tools to selectively excite specific segments of 3D printed cardiac constructs [36].
Conclusion
The long-term survival of cells within 3D constructs printed using CMC-laden GelMA bioink is significantly more sensitive to extruder pressure as compared with those printed using CFB. The CMC-laden constructs are also more affected by GelMA concentrations when it comes to their ability to form networks. The data also suggest that both cell types are adversely impacted by the UV curing step and the extent of such damage is similar. Additional strategies that can help to improve cardiomyocyte survival and network formation within 3D printed GelMA constructs include incorporation of adhesion molecules, use of CMC-CFB combination bioinks and lowering of GelMA concentration.
Future perspective
The prospect of having 3D printed on-demand biological tissues is both exciting and therapeutically promising. Yet despite the large number of research labs and a great amount of funding going into this field, printing viable structures using cell-laden bioinks remains a major challenge. The field of bioprinting is going through what is been deemed a ‘growing pains’ period – a familiar period of initial excitement based on a few, high profile proof-of-concept studies which followed by years of intensive effort to figure out the kinks of protocols and procedures. Therefore, both fundamental challenges but also many technical obstacles are expected to be solved within the next 5–10 years. The major challenge remains tissue vascularization – without it, 3D printed constructs will continue to be limited in both size and their long-term survivability. Another key challenge is creation of tissues with complex geometries using multiple cell types. A combined use of 3D printing and self-assembly principles from developmental biology is expected to help refine microscopic architecture of 3D printed constructs. On a more technical side is a matter of data reproducibility: the key outcomes of many 3D bioprinting studies are lab dependent and often hard to reproduce. In part, this can be explained by the large number of bioinks, cross-linkers and printer types on the market, as well as the different printer settings and cell types that each lab is using. This paper focused on one such detail, in other words, long-term effect of printer settings on survivability and connectivity of the two main types of cardiac cells.
Supplementary Material
Summary points.
Bioprinting of functional cardiac tissue requires multiple cell types, the main two being cardiac myocytes and cardiac fibroblasts.
Physical factors (such as extrusion pressure, shear stress, duration and strength of UV exposure) and chemical composition of bioinks affect different cell types differently.
The above factors can impact cell viability immediately after printing and have long-term effects on cell survival within 3D printed cell-laden constructs.
Bioluminescence can be used as a nondisruptive method to monitor cell presence and survival within printed constructs.
Extrusion pressure has a more pronounced effect of cardiomyocyte-laden 3D printed constructs when compared with the cardiofibroblast-laden ones.
Increased duration of UV exposure has negative effects on cell survival postprinting. The UV exposure settings used in our studies did not reveal significant differences between the survivability of cardiomyocyte-laden constructs versus their fibroblast-laden counterparts.
Understanding the similarities and differences between acute and long-term effects of extrusion printing on cardiomyocyte- versus fibroblast-laden gelatin methacryloyl constructs enables printing of multicellular designer 3D structures that can mimic different physiological and pathophysiological scenarios.
Acknowledgements
The authors thank Young and Simonyan for their help with bioluminescence measurements. Benjamin Holmes is gratefully acknowledged for his advice on 3D printing and useful discussions.
Financial & competing interests disclosure
This study was enabled by the financial support from the United States National Institutes of Health (grant number R21 HL122882 award) and GW OVPR Cross-Disciplinary Research Fund. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Footnotes
No writing assistance was utilized in the production of this manuscript.
Ethical disclosure
All procedures were done in accordance to the guidelines of the Institutional Animal Care and Use Committee at the George Washington University and the NIH Guide for the Care and Use of Laboratory Animals.
References
- 1.Camelliti P, Borg TK, Kohl P. Structural and functional characterisation of cardiac fibroblasts. Cardiovasc. Res 65, 40–51 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Christalla P, Didié M, Eschenhagen T, Tozakidou M, Ehmke M, Zimmermann WH. Fibroblasts are essential for the generation of embryonic stem cell derived bioengineered myocardium. Circulation 120, Abstract 2279, S615–a (2009). [Google Scholar]
- 3.Li Y, Asfour H, Bursac N. Age-dependent functional crosstalk between cardiac fibroblasts and cardiomyocytes in a 3D engineered cardiac tissue. Acta Biomater. 55, 120–130 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zimmermann WH K. Schneiderbanger K, Schubert P et al. Tissue engineering of a differentiated cardiac muscle construct. Circ. Res 90, 223–230 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Stoppel WL, Kaplan DL, Black LD. Electrical and mechanical stimulation of cardiac cells and tissue constructs. Adv. Drug Deliv. Rev 96, 135–155 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang D, Shadrin YD, Lam J, Xian HQ, Snodgrass HR, Bursac N. Tissue-engineered cardiac patch for advanced functional maturation of human ESC-derived cardiomyocytes. Biomaterials 34, 5813–5820 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tandon N, Marsano A, Maidhof R, Wan L, Park H, Vunjak-Novakovic G. Optimization of electrical stimulation parameters for cardiac tissue engineering. J. Tissue Eng. Regen. Med 5, e115–e225 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mandrycky C, Wang Z, Kim K, Kim DH. 3D bioprinting for engineering complex tissues. Biotechnol. Adv 34, 422–434 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vukicevic M, Mosadegh B, Min JK, Little SH. Cardiac 3D printing and its future directions. JACC Cardiovasc. Imaging 10, 171–184 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pati F, Jang J, Lee JW, Cho DW. Extrusion bioprinting In: Essentials of 3D Biofabrication and Translation. Atala A, Yoo JJ (Eds). Elsevier Inc., Amsterdam, The Netherlands, 123–152 (2015). [Google Scholar]
- 11.Webb B, Doyle BJ. Parameter optimization for 3D bioprinting of hydrogels. Bioprinting 8, 8–12 (2017). [Google Scholar]
- 12.Schuurman W, Levett PA, Pot MW et al. Gelatin-methacrylamide hydrogels as potential biomaterials for fabrication of tissue-engineered cartilage constructs. Macromol. Biosci 13, 551–561 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Billiet T, Gevaert E, De Schryver T, Cornelissen M, Dubruel P. The 3D printing of gelatin methacrylamide cell-laden tissue-engineered constructs with high cell viability. Biomaterials 35, 49–62 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Nichol JW, Koshy ST, Bae H, Hwang CM, Yamanlar S, Khademhosseini A. Cell-laden microengineered gelatin methacrylate hydrogels. Biomaterials 31, 5536–5544 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Contag CH, Bachmann MH. Advances in in vivo bioluminescence imaging of gene expression. Annu. Rev. Biomed. Eng 4, 235–260 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Arutunyan A, Swift LM, Sarvazyan NA. Initiation and propagation of ectopic waves: insights from an in vitro model of ischemia-reperfusion injury. Am. J. Physiol. Hear. Circ. Physiol 283, H741–H749 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simonyan H, Young C. A synthetic luciferin improves in vivo bioluminescence imaging of gene expression in cardiovascular brain regions. Physiol. Genomics 48, 762–770 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee BH, Lum N, Seow LY, Lim PQ, Tan LP. Synthesis and characterization of types A and B gelatin methacryloyl for bioink applications. Materials (Basel). 9(10), pii: E797 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinton TJ, Jallerat Q, Palchesko RN et al. Three-dimensional printing of complex biological structures by freeform reversible embedding of suspended hydrogels. Sci. Adv 1, e1500758–e1500758 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carrow JK, Kerativitayanan P, Jaiswal MK, Lokhande G, Gaharwar AK. Polymers for bioprinting In, Essentials of 3D Biofabrication and Translation.(Eds). Atala A, Yoo JJ. Elsevier Inc., Amsterdam, The Netherlands, 229–248, (2015). [Google Scholar]
- 21.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science 276, 1425–1428 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Williams CG, Malik AN, Kim TK, Manson PN, Elisseeff JH. Variable cytocompatibility of six cell lines with photoinitiators used for polymerizing hydrogels and cell encapsulation, Biomaterials 26, 1211–1218 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Radisic M, Park H, Martens TP et al. Pre-treatment of synthetic elastomeric scaffolds by cardiac fibroblasts improves engineered heart tissue. J. Biomed. Mater. Res. A 86, 713–724 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Close DM, Xu T, Sayler GS, Ripp S. In vivo bioluminescent imaging (BLI): noninvasive visualization and interrogation of biological processes in living animals, Sensors 11, 180–206 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan C, Wang DA. Macroporous hydrogel scaffolds for three-dimensional cell culture and tissue engineering. Tissue Eng. Part B. Rev 23, 451–461 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Tsang KMC, Annabi N, Ercole F et al. Facile one-step micropatterning using photodegradable gelatin hydrogels for improved cardiomyocyte organization and alignment. Adv. Funct. Mater 25, 977–986. (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhattacharjee T, Zehnder SM, Rowe KG et al. Writing in the granular gel medium. Sci. Adv 1(8), e1500655 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu W, Heinrich MA, Zhou Y et al. Extrusion bioprinting of shear-thinning gelatin methacryloyl bioinks. Adv. Healthc. Mater 6, 1601451 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yin J, Yan M, Wang Y, Fu J, Suo H. 3D Bioprinting of low-concentration cell-laden gelatin methacrylate (gelma) bioinks with a two-step cross-linking strategy. ACS Appl. Mater. Interfaces 10, 6849–6857 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Liu W, Zhong Z, Hu N et al. Coaxial extrusion bioprinting of 3D microfibrous constructs with cell-favorable gelatin methacryloyl microenvironments. Biofabrication 10, 24102 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bryant SJ, Nuttelman CR, Anseth KC. Cytocompatibility of UV and visible light photoinitiating systems on cultured NIH/3T3 fibroblasts in vitro. J. Biomater. Sci. Polym. Ed 11, 439–457 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Noshadi I, Hong S, Sullivan KE et al. In vitro and in vivo analysis of visible light crosslinkable gelatin methacryloyl (GelMA) hydrogels. Biomater. Sci 5, 2093–2105 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pumir A, Arutunyan A, Krinsky V, Sarvazyan NA. Genesis of ectopic waves: role of coupling, automaticity, and heterogeneity. Biophys. J 89, 2332–2349 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arutunyan A, Pumir A, Krinsky V, Swift LM, Sarvazyan NA. Behavior of ectopic surface: effects of ß-adrenergic stimulation and uncoupling. Am. J. Physiol 285, H2531–H2542 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tzatzalos E, Abilez OJ, Shukla P, Wu JC. Engineered heart tissues and induced pluripotent stem cells: macro- and microstructures for disease modeling, drug screening, and translational studies. Adv. Drug Deliv. Rev 96, 234–244 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu J, Entcheva E. Inscribing optical excitability to non-excitable cardiac cells: viral delivery of optogenetic tools in primary cardiac fibroblasts In: Kianianmomeni A (Ed.). Methods in Molecular Biology (Clifton, N.J.). Humana Press, NY, USA, 303–317 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.