Abstract
Flavonoids are secondary metabolites widely distributed among angiosperms, where they play diverse roles in plant growth, development, and evolution. The regulation of flavonoid biosynthesis in plants has been extensively studied at the transcriptional level, but post-transcriptional, translational, and post-translational control of flavonoid biosynthesis remain poorly understood. In this study, we analysed post-translational regulation of flavonoid biosynthesis in the ornamental plant Paeonia, using proteome and ubiquitylome profiling, in conjunction with transcriptome data. Three enzymes involved in flavonoid biosynthesis were identified as being putative targets of ubiquitin-mediated degradation. Among these, chalcone synthase (PhCHS) was shown to have the greatest number of ubiquitination sites. We examined PhCHS abundance in petals using PhCHS-specific antibody and found that its accumulation decreased at later developmental stages, resulting from 26S proteasome-mediated degradation. We further identified a ring domain-containing protein (PhRING-H2) that physically interacts with PhCHS and demonstrated that PhRING-H2 is required for PhCHS ubiquitination. Taken together, our results suggest that PhRING-H2-mediates PhCHS ubiquitination and degradation is an important mechanism of post-translational regulation of flavonoid biosynthesis in Paeonia, providing a theoretical basis for the manipulation of flavonoid biosynthesis in plants.
Keywords: Chalcone synthase, degradation, flavonoid biosynthesis, Paeonia, RING-H2, ubiquitination
RING-H2, a ring domain-containing protein, physically interacts with chalcone synthase in vivo and mediates its ubiquitination and degradation in peony.
Introduction
Flavonoids, a large family of phenolic secondary metabolites, are widely distributed among angiosperms, and include chalcones, flavones, flavanols, proanthocyanins (PAs), isoflavones, and anthocyanins (Winkel-Shirley, 2001; Grotewold, 2006). They have diverse biological functions in plant growth, development, and environmental adaptation, including UV protection, antioxidation, defense responses, and attraction of pollinators and animals for seed-dispersal (Winkel-Shirley, 2001; Muhlemann et al., 2018). They also represent major pigments in fruits, leaves, flowers, and seeds, and are beneficial components of the human diet, since they can contribute to the prevention of cardiovascular diseases, cancer (Hou, 2003; Amado et al., 2011), obesity, and diabetes, as well as improve visual function and promote antioxidant and anti-inflammatory activities (Tsuda, 2012; Wang et al., 2018b).
The biosynthesis of flavonoids in plants has been extensively characterized; they are derived from the general phenylpropanoid pathway, starting with the condensation of malonyl-CoA and 4-coumaroyl CoA to form chalcones, via the rate-limiting enzyme chalcone synthase (CHS; EC2.3.1.74) (Grotewold, 2006; Saito et al., 2007; Tanaka et al., 2008). CHS catalyses a reaction between three molecules of malonyl-CoA and one molecule of 4-coumaroyl CoA to form naringenin chalcone, which is rapidly converted into naringenin by chalcone isomerase (CHI). This provides the precursor for a variety of flavonoid derivatives through the activities of downstream enzymes in the pathway (Grotewold, 2006; Tanaka et al., 2008). CHS proteins have been isolated from a large taxonomic range of species, including dicots and monocots (Liou et al., 2018), and their functions in flavonoid biosynthesis have been confirmed through metabolic and molecular engineering (Forkmann and Martens 2001; Koes et al., 2005; Eloy et al., 2017; Nabavi et al., 2019). A mutation in the maize (Zea mays) COLORLESS2 (C2) gene, encoding a CHS, was reported to result in highly reduced levels of apigenin- and tricin-related flavonoids, which in turn caused a major reduction in the incorporation of tricin into the secondary cell wall polymer lignin (Eloy et al., 2017). Other studies of CHS genes have suggested that its expression and levels of protein accumulation are controlled at transcriptional, post-transcriptional, and post-translational levels (Koseki et al., 2005; Hosokawa et al., 2013; Zhang et al., 2017). At the transcriptional level, expression has been shown to be regulated by transcription factors belonging to the WD40, MYB, and basic helix–loop–helix (bHLH) families (Gonzalez et al., 2008; Shi and Xie, 2014; Xu et al., 2015). It has also been demonstrated that CHS is degraded by ubiquitination (Zhang et al., 2017); however, mechanisms that mediate its post-translational modification are not well understood.
Ubiquitination is a common post-translational modification in eukaryotic proteins (Hatakeyama and Nakayama, 2003; Ciechanover, 2005; Xie et al., 2015; Trujillo, 2018) and involves conjugating proteins with ubiquitin, a highly conserved 76-amino-acid polypeptide. In most cases, ubiquitination leads to protein degradation via the 26S proteasome system (Vierstra, 2012). In vivo, polyubiquitin chains are most frequently linked through Lys-48, and the canonical ubiquitin signal is recognized by the 26S proteasome, thereby targeting tagged proteins for degradation (Peng et al., 2003). Ubiquitination is a regulatory modification involved in diverse processes, including transcription, histone function, endocytosis, DNA repair, viral budding, and membrane trafficking (Schnell and Hicke, 2003; Passmore and Barford, 2004). The attachment of ubiquitin to proteins involve three classes of enzyme: ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin ligases (E3) (Hochstrasser, 1995). Ubiquitinated substrates may be degraded to peptides by the multiple subunits of the 26S proteasome system (Vierstra, 2012). Recently, protein degradation was shown to play important roles in anthocyanin biosynthesis (Zhang et al., 2013, 2015, 2017). Studies have shown that KFBCHS (F-box CHS) from Arabidopsis acts as a proteolytic negative regulator by mediating CHS degradation and coordinately controlling flavonoid biosynthesis in response to developmental cues and environmental stimuli (Zhang et al., 2017). It has also been reported that the apple (Malus domestica) BTB-TAZ protein, MdBT2, interacts with MdMYB1 and promotes its ubiquitination and degradation to regulate nitrate-mediated anthocyanin accumulation (Wang et al., 2018a). However, the significance and regulation of ubiquitination in controlling flavonoid biosynthesis have not been well characterized.
Plants in the genus of Paeonia are well-known ornamentals that are cultivated worldwide, including in China, which has a long history of their cultivation and breeding (Wang et al., 2001; Jia et al., 2008a; Li, 2010). In previous studies, we showed that flavonoids, especially anthocyanins, are the main color pigments in Paeonia plants, in particular, peonidin, pelargonidin, and cyanidin-derived anthocyanins (Zhang et al., 2007; Du et al., 2015; Gu et al., 2019). Flowers of Paeonia are also used in the diet due to their high levels of flavonoids, e.g. of quercetin, kaempferol, isorhamnetin, luteolin, and chalcone derivatives (Li et al., 2009). Much attention has been focused on the transcriptome analyses of differentially expressed genes or pigment chemical composition in flower petals in Paeonia (Wang et al., 2001; Zhang et al., 2007; Jia et al., 2008b; Zhang et al., 2015; Shi et al., 2017). Moreover, our recent study suggested a molecular mechanism for the transcriptional control of PsCHS by a MYB–bHLH–WD40 complex, during petal blotch color formation (Gu et al., 2019). However, little is known about the post-translational regulation of CHS, and the potential for its ubiquitin-mediated degradation in the peony model system.
In the current study, we used proteome and ubiquitylome profiling to investigate whether post-translational modification contributes to the regulation of flavonoid biosynthesis in Paeonia. We also report the identification of a PhRING-H2 protein, instead of F-box proteins, that interacts with peony CHS (PhCHS), thereby mediating its degradation. This revealed that for monitoring and manipulating flavonoid biosynthesis, diverse E3 ubiquitin–protein ligases need to be discovered in the future.
Materials and methods
Plant material
Paeonia ‘He Xie’ was used in this study. Plants were grown in Beijing Botanical Garden, Institute of Botany, and the Chinese Academy of Sciences. The collected petals or bud scales were immediately frozen in liquid nitrogen and stored at −80 °C for further analysis.
HPLC analysis
Flavonoid analysis was conducted as previously described (Li et al., 2009). Briefly, 0.1– 0.2 g of dried petals was placed in a 2% formic acid–methanol (v/v) solution for 24 h at 4 °C; it was centrifuged at 12 000 g for 5 min and the supernatant was filtered through a 0.22 µm membrane for further analysis. Flavonoids were detected using an Agilent 1100 HPLC with a Dionex diode array detector (Agilent Technologies Inc., Santa Clara, CA, USA) at 200–800 nm. Flavonoids in each sample were measured semi-quantitatively, using cyanidin-3-O-glucoside (Cy3G) and rutin as standards. Total content of anthocyanins and of flavones and flavanols was quantified by linear regression using a calibration curve and expressed as mg of Cy3G or rutin equivalents per gram dry weight (DW) (Wang et al., 2001).
Proteomic quantification and ubiquitous modification analysis
Petals at stages 1 and 3 of ‘He Xie’ were collected. Proteomic quantification and ubiquitous modification analysis were performed by Jingjie Biology Company (Hang Zhou, China).
For affinity enrichment, the tryptic peptides were dissolved in NETN buffer (100 mM NaCl, 1 mM EDTA, 50 mM Tris–HCl, 0.5% NP-40, pH 8.0) and incubated with pre-washed ubiquitin antibody beads (cat. no. PTM-1104, PTM-Biolabs) at 4°C overnight with gentle shaking. The beads were then washed four times with NETN buffer and twice with H2O. The bound peptides were eluted from the beads with 0.1% trifluoroacetic acid. Finally, the eluted fractions were combined and vacuum-dried. For tandem mass tag (TMT) and LC–tandem mass spectrometry (MS/MS) analysis, the resulting peptides were desalted with a ZipTipC18 (Millipore) according to the manufacturer’s instructions.
For the analysis of conserved ubiquitylated Lys residues and the degree of evolutionary conservation of ubiquitylation, we first used BLASTP to compare ubiquitylated protein sequences from Paeonia ‘He Xie’ against specified protein sequences in UniProtKB (http://www.uniprot.org/), which includes eight species: Oryza sativa subsp. japonica, Glycine max, Brachypodium distachyon, Vitis vinifera, Arabidopsis, Sorghum bicolor, Solanum lycopersicum, and Z. mays. By applying a reciprocal best BLAST hit approach, we determined the orthologous proteins among these genomes. For each orthologous group, we used MUSCLE v3.8.31 to create a multiple sequence alignment (Edgar, 2004). We then determined the lysine conservation for each species by counting the total number of conserved ubiquitylated lysine residues and the total number of conserved non-ubiquitylated lysine residues. If both the protein from Paeonia ‘He Xie’ and the query protein in the multiple sequence alignment were lysine residues at the aligned position, they were considered to be conserved. All lysine residues of proteins identified in this study were considered as a control. Mean conservation of ubiquitylated or control Lys site between Paeonia ‘He Xie’ sequences and sequences from other organisms in the specified protein sequences were plotted separately. P-values were calculated for each comparison using Fisher’s exact test.
For the protein–protein interaction network analyses, the interaction between ubiquitylation and the proteome network was analysed. We used STRING to define the possible interactions between proteins. We retained all interactions that had a confidence score ≥0.9 (higher confidence). Interaction networks predicted by STRING were visualized in Cytoscape. A graph of the theoretical clustering algorithm, molecular complex detection (MCODE), was utilized to analyse densely connected regions. MCODE is part of the plug-in tool kit of the network analysis and visualization software of Cytoscape.
Subcellular structure prediction and classification were performed using wolfsport (Colaert et al., 2009). To understand the properties of the identified Kub sites, we used the Motif-X program to compare the position-specific frequencies of the amino acid residues surrounding all ubiquitylated sites (Gnad et al., 2011). Analysis of the frequencies using neighboring amino acid residues was conducted for ubiquitylated Lys residues by iceLogo (Colaert et al., 2009). In addition, we performed protein secondary structure predictions using NetSurfP software to analyse the role of protein secondary structure in Kub site (Muller et al., 2010).
Identification of PsCHS-interacting proteins
The coding sequence (CDS) of PsCHS was cloned into the PEASY-E1 vector (TransGen Biotech). The method for purifying the recombinant protein was as previously described (Yuan et al., 2014). The purified His fusion protein (His–PsCHS) sample was incubated with Ni-NTA His Bind Resin (Merck). Crude proteins from ‘He Xie’ petals at stage 3 were extracted using a total extraction sample kit (cat. no. 786-259, Sango Biotech). Purified His–PsCHS was incubated with crude protein from petal for 2 h, and then the proteins that could not bind with His–PsCHS were washed off using wash buffer (300 mM NaCl, 50 mM NaH2PO4, 30 mM iminazole). The procedures were performed as previously described (Li et al., 2016). Finally, we identified PsCHS-interacting proteins by MS, which was completed by JingJie Biotechnology Co. Ltd (Hangzhou, China). The digested peptides were vacuum-dried, dissolved and desalted for nanoLC-MS/MS analysis. The primers are listed in Supplementary Table S1 at JXB online.
Expression profile analysis by qRT-PCR
Gene expression analysis was conducted by qRT-PCR as described by Gu et al. (2019). The relative quantification of mRNA transcripts was performed using the ΔΔCt method (Gu et al. 2019), with normalization to β-tubulin (Acc. No. EF608942). Primers used for qPCR analysis are listed in Supplementary Table S1.
Cell-free degradation assays
Cell-free protein degradation assays were conducted as previously described (Wang et al., 2009) with some modifications. Total petal proteins at stage 3 were extracted with degradation extraction buffer (25 mM Tris–MES, pH 7.5, 1 mM MgCl2, 4 mM PMSF, 5 mM DTT) and the cell debris was removed by centrifugation at 12 000 g at 4 °C. The supernatant was kept for the degradation assays. The purified PsCHS protein was divided into two equal parts and incubated with the total proteins at 25 °C. The specific 26S proteasome inhibitor MG132 (MedChemExpress) with a final concentration of 40 μM was added into one of the two parts and DMSO was added to the other as control. Samples were taken at different time intervals to measure PsCHS protein abundance by western blot analysis using an anti-His-tag monoclonal antibody from immunized mouse (1:5000, CWBIO, Beijing, China).
Bimolecular fluorescence complementation assays
The PhCHS coding sequence was fused with the C-terminus of pCambia 1300-YFPC to generate a PhCHS-YFPC plasmid, and the sequence of PhRING-H2 was fused with the N-terminus of pCambia 1300-YFPN to form PhRING-H2-YFPN. The resulting constructs were transiently expressed in tobacco (Nicotiana benthamiana) leaves by Agrobacterium infiltration as previously described (Schütze et al., 2009). Yellow fluorescent protein (YFP) fluorescence was imaged after transformation for 72 h using an Olympus BX61 confocal laser scanning microscope. The excitation wavelength for YFP fluorescence was 488 nm, and emission fluorescence was detected at 500–542 nm. Primers used for the bimolecular fluorescence complementation (BiFC) assays are listed in Supplementary Table S1.
Transfection of bud scale by agro-infiltration
For transient silencing of PhRING-H2 to characterize the ubiquitination of PhCHS, tobacco rattle virus (TRV) vector (TRV2:PhCHS) for virus-induced gene silencing was constructed and introduced into Agrobacterium as previously described (Tian et al., 2014) with TRV2 used as the control. Bud scales of 2–3 cm long from ‘He Xie’ were used for vacuum infiltration as described by Ma et al. (2008) and Du et al. (2015). The bud scales were submerged in infiltration mixture (1 M MES–KOH, 1 M MgCl2, 100 mM acetosyringone). The treated bud scales were placed in MS medium in the dark at 8 °C for 24 h, then, cultured at 23 °C for 48 h with 60% humidity. The bud scales were then used for ubiquitination analysis. Primers used for this experiment are listed in Supplementary Table S1.
For overexpression of PhRING-H2 to characterize the ubiquitination of PhCHS, OE-pCambia 1305-PhRING-H2 was constructed and introduced into Agrobacterium tumefaciens as previously described (Han et al., 2015). The A. tumefaciens cells containing PhRING-H2 were cultured and reached an OD of approximately 1.0 at 600 nm, and then were collected and suspended in infection buffer (10 mM MgCl2, 10 mM MES, pH 5.6, and 200 mM acetosyringone) as previously described (Han et al., 2015). Bud scales were submerged in infiltration mixture. The treated bud scales maintained in MS medium were cultured in the dark at 8 °C for 24 h, then kept at 23 °C for 48 h with 60% humidity. These bud scales were then used for ubiquitination analysis. Primers used for this experiment are listed in Supplementary Table S1.
Protein purification and antibody preparation
The CDSs of PhCHS and PhRING-H2 were cloned into the pEASY-E1 vector (Transgene Co., Beijing, China) with His tag and transformed into Escherichia coli BL 21 (DE3) for expression analysis. The BL 21 (DE3) cells containing His–PhCHS or His–PhRIN-H2 were cultured and reached an OD of approximately 0.6 at 600 nm, and were then induced by 1.0 mM isopropyl β-D-1-thiogalactopyranoside for 16 h at 16 °C. BL 21 (DE3) cells were collected and suspended in buffer (300 mM NaCl, 500 mM Na2HPO4). Ultrasound treatment was conducted for 15 min for cell lysis, followed by centrifugation for 1 h to collect the supernatant, followed by incubation with Ni-NTA His Bind Resin according to the manufacturer’s manual (Merck). The collected recombinant His–PhCHS protein was kept at −80 °C for further use.
For PhCHS-specific antibody preparation, the purified recombinant His–PhCHS protein was boiled for 5 min in a bath chamber, and then separated by 10% SDS-PAGE. Then, the PhCHS molecular band was cut and used to immunize rabbits at the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences (Beijing, China). The amino acid sequence of PhCHS used for antibody preparation is listed in Supplementary Table S2. PhCHS antibody specificity detection was performed as follows. Crude petal protein was incubated with PhCHS antibody overnight at 4 °C, then protein A+G agarose was added to the incubated proteins. After shaking at 4 °C for 3 h, the supernatant was removed by centrifugation at 1000 g for 5 min; then the cell debris was washed with PBS buffer for five times, and 20 µl of 1× SDS-PAGE sample loading buffer was added, followed by boiling at 100°C for 5 min, before it was used for SDS-PAGE. The protein band was cut and identified by MS, which was completed by Beijing Protein Innovation (Beijing, China). For β-actin antibody, we used anti-β-actin mouse monoclonal antibody, which was bought from CWBIO company (Beijing, China) and is widely suitable for plants. The target PhCHS protein could be identified and the coverage accounted for 59%, and therefore the PhCHS antibody was considered to be specific and could be used for further analysis (Supplementary Table S3).
Protein extraction and western blot
Total proteins were extracted from petals at different developmental stages according to the method described by Nagatomo et al. (2014), using extraction buffer (100 mM Tris–HCl, pH 8.0, 1 mM DTT containing 5% (w/v) polyvinylpolypyrrolidone and 0.1 mM phenylmethylsulfonyl fluoride). Then a BCA protein assay kit (CWBIO) was used for determining the concentration of total proteins. The denatured protein from petals was used for western blot according to the method described by Cai et al. (2018). PhCHS antibody (1:3000) was used for evaluating PhCHS expression abundance in petals of ‘He Xie’. Mouse monoclonal antibody of β-actin (anti-β-actin) (1:3000, CWBIO) was used for the reference protein.
Ubiquitination assay in vitro
For the in vitro ubiquitination assay, His–PhCHS and His–PhRING were purified using Ni-NTA His Bind Resin (Merck), as described (Cai et al., 2018). The ubiquitination assay was performed as described (Xie et al., 2002), namely, 300 ng of His–PhCHS fusion protein was mixed with an equal amount of PhRING-H2 fusion protein in the presence or absence of the following: 50 ng of E1 (Beyotime, Shanghai, China), 100 ng of E2 (Beyotime), and 5 µg of ubiquitin (Beyotime). The reaction system was used as described (Ding et al., 2015) and western blot were performed using ubiquitin polyclonal antibody from immunized rabbit (1:1000, Beyotime) to check ubiquitination levels of proteins.
Ubiquitination and degradation of PhCHS assay in vivo
To examine the effect of overexpression (PhRING-H2-OE) or down-regulation (PhRING-RNAi) on ubiquitination of PhCHS, bud scales with 2–3 cm in length were collected after transfection for 72 h. A ubiquitination assay was conducted by the above mentioned method, and anti-ubiquitin was used for western blotting and anti-β-actin was used as control. For the ubiquitination assay, PhCHS antibody (2 μg final concentration) was added to the extracted crude protein from bud scale incubated with PhCHS proteins for immunoprecipitation, after slowly shaking overnight at 4 °C. Then protein A+G agarose (Beyotime) was added to the above mixture at 4 °C for 3 h and centrifuged at 1000 g for 5 min. The supernatant was carefully removed and washed for five times with PBS buffer (8 mM Na2HPO4, 1.5 mM KH2PO4, 135 mM NaCl and 2.7 mM KCl); then 20 μl 1× SDS-PAGE loading buffer (CWBIO) was added to precipitates and boiled in a bath chamber for 5 min immediately. Western blotting was performed as described by Cai et al. (2018). Ubiquitin polyclonal antibody from immunized rabbit (1:1000, Beyotime) was used to evaluate the ubiquitination levels of PhCHS.
For the degradation of PhCHS assay, samples were collected immediately in liquid nitrogen at 0 or 3 h after treatment with MG132 (40 μM final concentration) or DMSO, respectively. Total proteins from bud scales were extracted according to the above described method (Nagatomo et al., 2014). The concentration of the total proteins was determined and adjusted at the same level for each treatment group.
Results
Flavonoid accumulation in petals of Paeonia ‘He Xie’
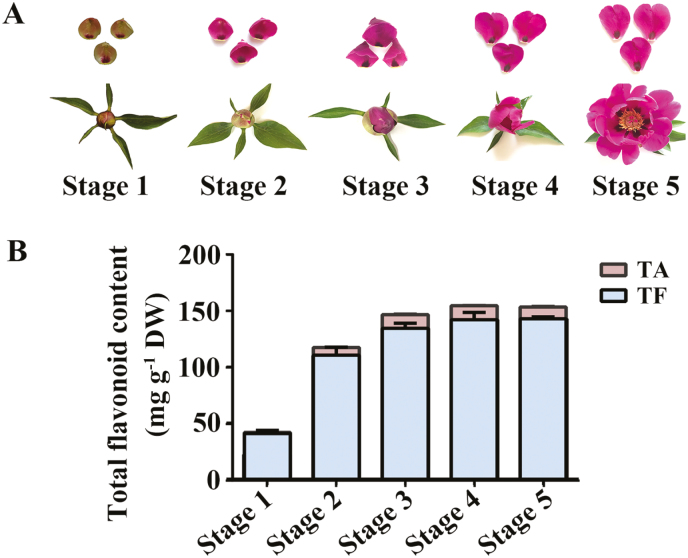
We first characterized the pattern of flavonoid accumulation in the petals from Paeonia ‘He Xie’ at five developmental stages (stages 1–5; Fig. 1A). Flavonoids accumulated increasingly from stage 1 to stage 4, peaked at stage 4, then decreased slowly at stage 5; the total content of flavone and flavanol was 40.3±2.9 mg g−1 at stage 1 and peaked with 142.9±1.7 mg g−1 at stage 5, while the total anthocyanin content in petals was 1.1±0.1 mg g−1 at stage 1 and reached the highest level of 10.4 ±0.5 mg g−1 at stage 4 (Fig. 1B).
Fig. 1.
Flavonoid accumulation during flower petal development. (A) Phenotypes of different developmental stages of petals of Paeonia ‘He Xie’. (B) Total flavonoid content from stage 1 to stage 5 in petals by HPLC-DAD analysis (mean ±SD; n=3). TA, total anthocyanin content, mg of Cy3G/100 mg dry weight (DW) of petals; TF, total flavone and flavanol content, mg of rutin/100 mg DW of petals. (This figure is available in color at JXB online.)
Proteome profiles in Paeonia ‘He Xie’
To further investigate the mechanism of flavonoid accumulation in Paeonia ‘He Xie’, the proteome profiles of petals at stage 1 and stage 3 were examined. We extracted proteins from the petals, and the trypsin enzyme-lysed peptides fractionated by high pH reverse-phase HPLC were subjected to electrospray ionization followed by MS/MS. Based on our formerly obtained transcriptome of petals, 5175 proteins in total were identified and the abundance of 3407 was quantified (Supplementary Table S4). Of these, the expression level of 513 proteins was up-regulated, and of 659 proteins was down-regulated (with a threshold of 1.5-fold) in stage 3 compared with stage 1 (Supplementary Table S5). These proteins/cognate genes were submitted to Gene Ontology (GO) enrichment analysis (Supplementary Fig. S1A, B), subcellular location prediction (Supplementary Fig. S1C, D), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (Supplementary Figs S2, S3). In the pathway of flavonoid biosynthesis, we identified that protein abundance of CHS, CHI, and flavanone 3-hydroxylase (F3H) were increased in stage 3 in comparison with stage 1 (Supplementary Table S6).
Ubiquitylome profiles in Paeonia ‘He Xie’ petals
The proteome analysis revealed substantial changes in the abundance of many proteins in petals during development. To investigate whether ubiquitination was associated with these changes, we performed a ubiquitin proteome analysis of two petal developmental stages (stages 1 and 3) using label-free quantitative ubiquitin enrichment techniques and high resolution LC-MS. In total, 5260 ubiquitin sites in 2267 proteins were identified, among which 3534 ubiquitin sites in 1692 proteins were accurately quantified. Following a normalization of the protein group filter, a total of 2605 ubiquitin sites in 1047 proteins were accurately quantified (Supplementary Table S7). Of these, 653 sites in 435 proteins were up-regulated targets and 767 sites in 471 proteins were down-regulated targets for putative ubiquitination in stage 3 as compared with stage 1, at a threshold of 1.5 (Supplementary Table S8). These results support the hypothesis that ubiquitination of proteins occurs in petals of peony ‘He Xie’ during the developmental process.
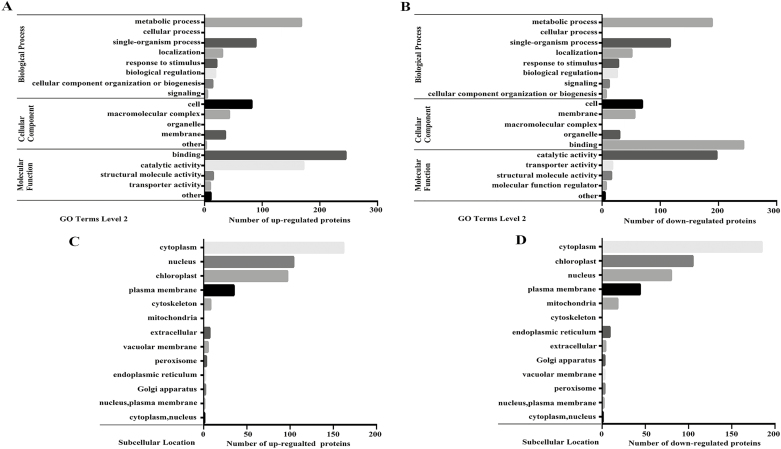
To elucidate the functional differences between the ubiquitinated proteins that we observed to be up-regulated or down-regulated, we performed GO enrichment analysis, which revealed that a total of 204 up-regulated or 193 down-regulated Kub (Lys (K) ubiquitination) proteins clustered in the ‘cellular component’ category. Furthermore, there were 480 up-regulated or 571 down-regulated Kub proteins in the ‘biological process’ category. In addition, 453 up-regulated or 486 down-regulated Kub proteins were in the ‘molecular function’ category. The subclasses of the enrichment proteins are shown in Fig. 2A, B and Supplementary Fig. S4.
Fig. 2.
Functional enrichment analysis of proteins with up-regulated or down-regulated Kub sites. (A, B) Gene Ontology (GO)-based enrichment analysis of proteins with up-regulated (A) or down-regulated (B) Kub sites during petal developmental stage 3 compared with stage 1. (C, D) Subcellular location analysis of up-regulated (C) or down-regulated (D) Kub sites during petal developmental stage 3 compared with stage 1.
In order to further analyse the functions of differentially expressed Kub proteins, subcellular structure prediction and classification were performed using wolfpsort. We observed that up-regulated proteins with ubiquitination sites were mainly enriched in cytoplasm, nucleus, chloroplast, and plasma membrane (162, 104, 97, and 35, respectively, which accounted for 37%, 24%, 22%, and 8% of the total identified proteins, respectively). The down-regulated proteins possessing ubiquitination sites were mainly enriched in the cytoplasm, chloroplast, nucleus, and plasma membrane (185, 105, 80, and 44, respectively, which accounted for 39%, 22%, 17%, and 9% of the total identified proteins, respectively) (Fig. 2C, D; Supplementary Fig. S4).
Sequence properties of ubiquitinated proteins
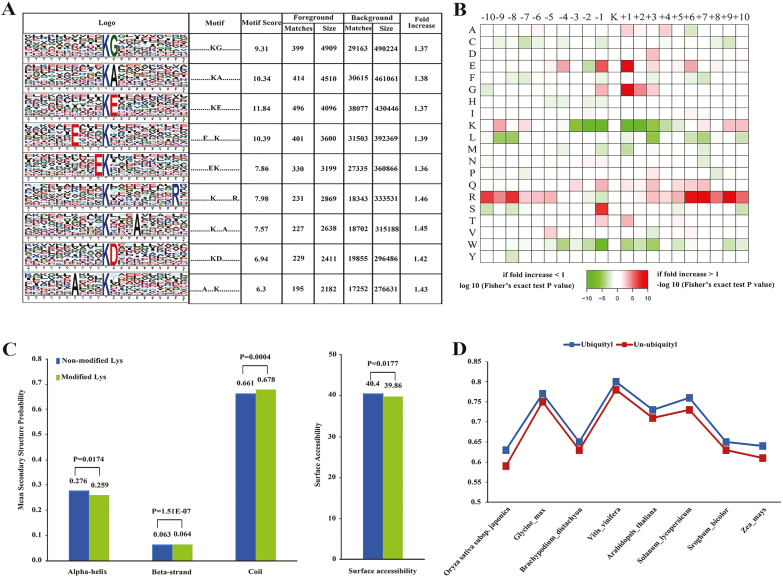
We analysed the position-specific frequencies of the amino acid residues surrounding all the ubiquitinated sites, and identified five unique sites that were designated as ..........KG........., ..........KA........., ..........KE........., ......E...K.........., and .........EK.......... (where a dot indicates a varying amino acid; Fig. 3A). Further analysis of the amino acids neighboring the ubiquitination sites indicated a rich frequency of hydrophilic residues (Fig. 3A), and a significant abundance in the frequency of hydrophilic residues, such as Cys and Trp, adjacent to ubiquitinated Lys residues (+1, +3, −1, and −3; Fig. 3B).
Fig. 3.
Motif analysis of identified Kub sites. (A) Ubiquitination motifs and the conservation of Kub sites. The height of each letter corresponds to the frequency of that amino acid residue at that position. The central K refers to the ubiquitinated Lys. (B) Amino acid sequence properties of ubiquitylation sites. The heat map shows significant position-specific under-representation or over-representation of amino acids flanking the modification sites. (C) Probabilities of different secondary structures (coil, α-helix, and β-strand) of modified and non-modified Lys residues were compared with the secondary structure probabilities of all Lys residues in this study. (D) Evolutionary conservation of ubiquitylated and non-ubiquitylated Lys residues on protein orthologs in selected eukaryotic species: Oryza sativa japonica, Glycine max, Brachypodium distachyon, Vitis vinifera, Arabidopsis thaliana, Solanum lycopersicum, Sorghum bicolor, and Zea mays. (This figure is available in color at JXB online.)
We investigated the likelihood of various secondary structures (α-helix, β-strand, and coil) near ubiquitinated Lys sites, compared with the secondary structure predictions of all Lys sites of proteins identified. Ubiquitinated Lys sites occurred significantly more frequently in unstructured regions of proteins (P=0.0004 for coil) and less frequently in structured regions (P=0.0174 for α-helix and P=1.51×10−7 for β-strand; Fig. 3C), while for surface accessibility P=0.0177.
To study the evolutionary conservation of ubiquitinated Lys or non-ubiquitinated Lys in plants, we aligned ‘He Xie’ proteins with their orthologs from eight other plant species. We observed that ubiquitinated Lys residues were significantly less conserved than non-ubiquitinated Lys residues, suggesting that ubiquitinated Lys residues do not maintain a stronger selective pressure compared with that of non-ubiquitinated Lys residues (Fig. 3D).
Correlation between protein abundance and ubiquitination
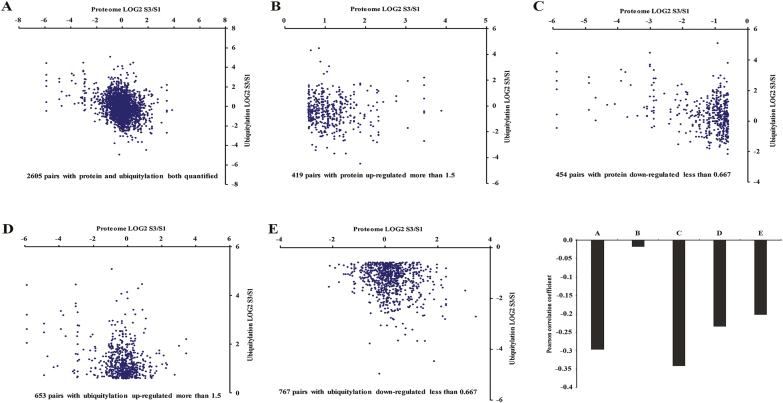
Ubiquitination plays an important role in proteasome-mediated protein degradation. We performed a correlation analysis comparing the whole proteome and ubiquitylome based on the quantitative results obtained in this study (Fig. 4). Pearson’s correlation coefficient was calculated as −0.296 when all significantly altered proteins were considered in terms of their ubiquitination, regardless of the direction of the change (Fig. 4A–F). We observed that the global abundance between proteome and ubiquitylome was negatively correlated in ‘He Xie’ petals at stage 3 versus stage 1. Restricting the analysis to pairs of up-regulated or down-regulated proteins resulted in an increased correlation (r=−0.017 and −0.341, respectively; Fig. 4B, C, F). For ubiquitination/protein pairs with significantly up- or down-regulated ubiquitination, two weak negative correlations were observed (r=−0.234 and −0.201, respectively; Fig. 4D–F). These results are consistent with protein expression levels being negatively regulated by ubiquitination. To further understand the function of the ubiquitinated proteins, we analysed protein–protein interactions between ubiquitylation and proteome networks, which indicated a complicated regulation occurring inside cells (see Supplementary Fig. S5).
Fig. 4.
Concordance between changes in proteins and their ubiquitination. (A–E) Correlation between protein and ubiquitination fold changes upon petal development for all ubiquitination-protein pairs: (A) 2605 pairs with both protein and ubiquitylation quantified; (B) 495 pairs were obtained with protein up-regulated more than 1.5; (C) 454 pairs were obtained with protein down-regulated less than 0.67; (D) 653 pairs with ubiquitylation were up-regulated more than 1.5; (E) 767 pairs with ubiquitylation were down-regulated less than 0.67. (F) Pearson correlations of the comparisons shown from (A–E). (This figure is available in color at JXB online.)
KEGG analysis of ubiquitinated proteins in the flavonoid biosynthesis pathway
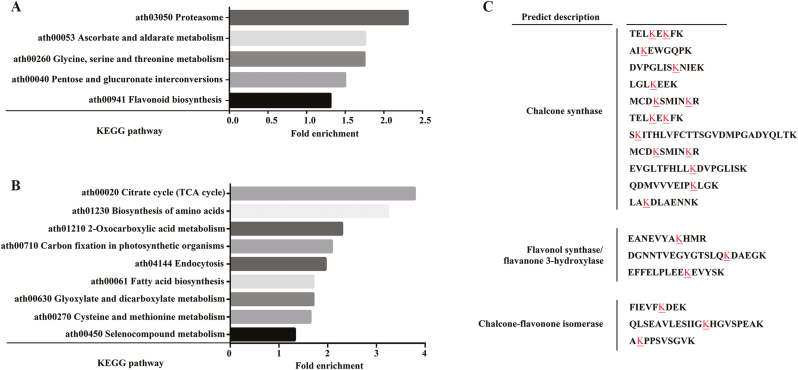
To further elucidate the function of proteins that underwent changes in ubiquitination, we performed KEGG pathway analysis. The categories ‘protein-processing pathways in the proteasome’, ‘ascorbate and aldarate metabolism’, ‘glycine, serine, and threonine metabolism’, ‘pentose and glucoronate interconversions’, and ‘flavonoid biosynthesis’ were all enriched by proteins with up-regulated Kub sites. Proteins with down-regulated Kub sites were enriched in pathways involving ‘citrate cycle (TCA cycle)’, ‘biosynthesis of amino acids’, ‘2-oxocarboxylic acid metabolism’, ‘carbon fixation in photosynthetic organisms’, ‘endocytosis’, ‘fatty acid biosynthesis’, ‘glyoxylate and dicarboxylate metabolism’, ‘cysteine and methionine metabolism’, and ‘selenocompound metabolism’ (Fig. 5A, B). We subsequently focused on the flavonoid metabolic pathway, which indicated a relationship with ubiquitination in Paeonia ‘He Xie’ petals (Fig. 5C). Specifically, we identified three enzymes (CHS, CHI, and F3H/flavonol synthase (FLS)) putatively with ubiquitination modifications, among which CHS had more modification sites than the other two (Fig. 5C).
Fig. 5.
KEGG analysis of ubiquitin-modified proteins involved in various pathways in petal development. (A) Pathways enriched in proteins with up-regulated Kub sites. (B) Pathways enriched in proteins with down-regulated Kub sites. (C) Ubiquitination modification sites of proteins involved in the flavonoid biosynthesis pathway, indicated by an underlined letter. (This figure is available in color at JXB online.)
PhCHS protein degradation is controlled by the 26S proteasome pathway
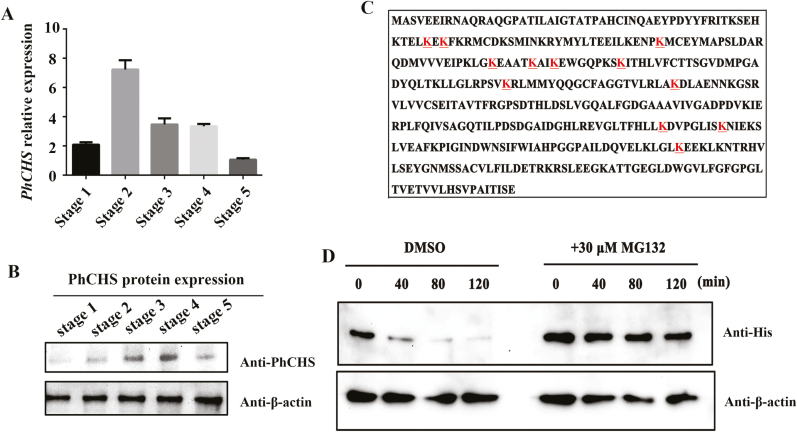
Western blot analysis was used to determine whether the abundance of PhCHS changed during different developmental stages of petals. We observed that PhCHS levels increased during petal development, peaking at stage 4, before decreasing at stage 5, indicating that PhCHS degradation might happen at a later developmental stage (Fig. 6A, B). To further confirm the ubiquitination of PhCHS, a recombinant histidine tagged form of the protein (His–PhCHS protein) was purified from E.coli and incubated with total protein extracted from petals at stage 3, and then MS analysis was performed. A total of 12 ubiquitination sites were identified in PhCHS, consistent with its degradation by the 26S proteasome system (Fig. 6C). In order to confirm that the abundance of PhCHS is controlled by the 26S proteasome system, proteins from ‘He Xie’ petals at stage 3 or bud scales were treated with the specific 26S proteasome inhibitor MG132 (Lyzenga et al., 2012) and then incubated with the purified PhCHS protein in a cell-free degradation assay or using PhCHS antibody for western blot analysis. We observed that the abundance of PhCHS was much higher in extracts from petals or bud scales treated with MG132 than in those from the control in vitro or in vivo (Fig. 6D; Supplementary Fig. S6). Taken together, the results supported the hypothesis that PhCHS was degraded via the 26S proteasome in ‘He Xie’ petals.
Fig. 6.
PhCHS accumulates in petals and its degradation is mediated by the 26S proteasome system. (A) PhCHS expression from stage 1 to stage 5 of petals (mean ±SD; n=3). (B) PhCHS protein accumulation from stage 1 to stage 5 of petals by western blot analysis; anti-β-actin was used as control. (C) The ubiquitination sites (underlined letters) in PhCHS identified by mass spectroscopy. (D) Cell-free degradation assay of recombinant His–PhCHS protein. Recombinant His–PhCHS was purified from Escherichia coli incubated with petal crude proteins at different stages and treated with specific 26S proteasome inhibitor MG132 at various time intervals. Western blot analysis was conducted using an anti-His antibody, and anti-β-actin protein concentration was used as a loading control. (This figure is available in color at JXB online.)
PhRING-H2 interacted with PhCHS and acted as an E3 ubiquitin ligase for ubiquitination of PhCHS in vitro
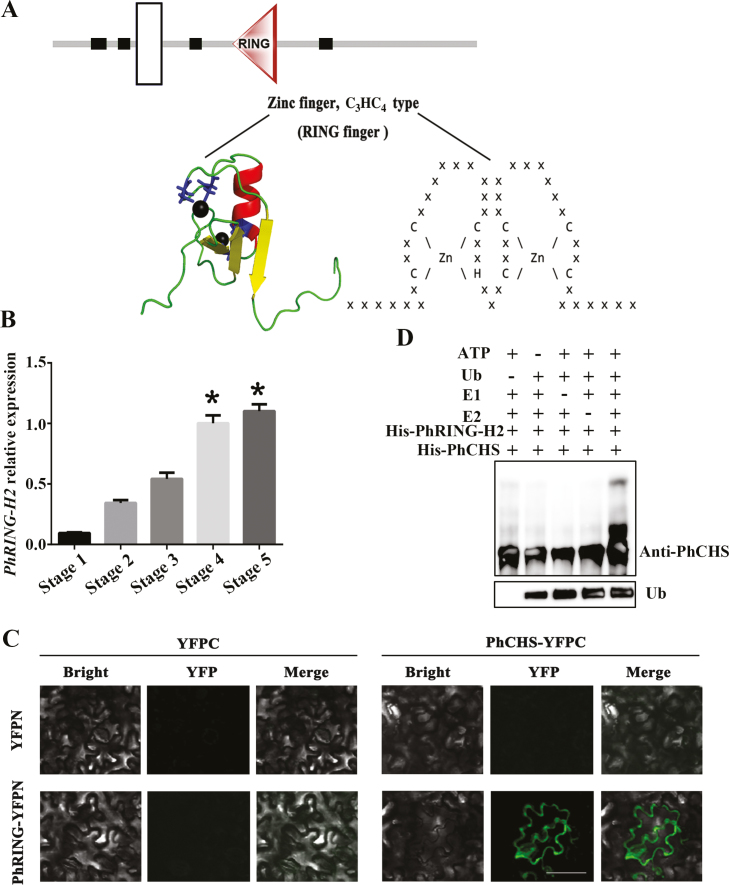
In order to better understand the PhCHS ubiquitination system, we performed a pull-down analysis of proteins from ‘He Xie’ petals at stage 5, using recombinant PhCHS as bait, and identified the interacting proteins by MS. A total of seven proteins were found in two independent experiments, including 50S ribosomal protein, elongation factor 2, CLP protease regulatory subunit CLPX1, RING-H2, histone H3.2-like, UDP-glucose 6-dehydrogenase 1-like, and histone-lysine N-methyltransferase (Supplementary Table S9). Of these, we focused on the RING-H2 protein (PhRING-H2) since many proteins with RING finger domains are known to simultaneously bind ubiquitination enzymes and their substrates, and function as important ligases (Lorick and Weissman, 1999; Joazeiro and Weissman, 2000). A RING finger domain is a structural domain of a zinc finger protein that contains a C3HC4 amino acid motif and binds two zinc cations. We first analysed the structure of PhRING-H2 (Fig. 7A) and identified a RING finger domain, indicating that PhRING-H2 acts as an E3 ubiquitin–protein ligase in ‘He Xie’. Using quantitative PCR, we showed that PhRING-H2 levels increased during petal development (Fig. 7B).
Fig. 7.
PhRING-H2 interacted with PhCHS and acted as an E3 ubiquitin ligase for ubiquitination of PhCHS in vitro. (A) Domain analysis of PhRING-H2. A RING finger domain is a structural domain of a zinc finger protein that contains a C3HC4 amino acid motif and binds two zinc cations. The structure prediction used the http://smart.embl.de/ website and tertiary structure prediction used the http://pfam.xfam.org website. White box indicates transmembrane region. Black boxes indicate low complexity region. Triangle indicates RING domain. (B) PhRING-H2 relative expression in petals during developmental stages (mean ±SD; n=3). (C) PhRING-H2 interacts with PhCHS in vivo. PhCHS fused with the C-terminus of YFPC (PhCHS–YFPC) was co-expressed with PhRING-H2 fused with the N-terminus of YFP (PhRING-H2–YFPN) in tobacco leaves. Images were collected after infiltration for 72 h. (D) PhRING-H2 acted as an E3 ubiquitin ligase of PhCHS in vitro. His–PhRING-H2 and His–PhCHS fusion protein were expressed in Escherichia coli and purified. His–PhRING-H2 fusion protein for E3 activity was measured in the presence or absence of ATP, ubiquitin, ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), His–PhCHS, or ubiquitin (Ub). Protein bands with ubiquitin attached were detected with PhCHS antibody from immunized rabbit. (This figure is available in color at JXB online.)
To confirm the interaction between PhCHS and PhRING-H2 in vivo, BiFC assays were performed in tobacco leaves. A strong fluorescence signal was detected following co-transformation with PhCHS fused at the C-terminus to yellow fluorescent protein (PhCHS–YFPC) and PhRING-H2 fused at the N-terminus to YFP (PhRING-H2–YFPN). However, no signals were detected with the combination of PhCHS–YFPC with YFPN, or YFPC with PhRING-H2–YFPN (Fig. 7C), indicating that PhCHS interacted with PhRING-H2 in vivo. Further, cell-free assays were conducted for examination of the ubiquitination of PhCHS mediated by PhRING-H2. The results indicated that PhCHS proteins could be ubiquitinated by addition of ATP, ubiquitin, ubiquitin-activating enzymes (E1), and ubiquitin-conjugating enzymes (E2) in the presence of PhRING-H2 (Fig. 7D). Taken together, our results suggested that PhCHS protein degradation was controlled by the 26S proteasome pathway via interaction with PhRING-H2.
PhRING-H2CHS acts as a proteolytic regulator of PhCHS in vivo
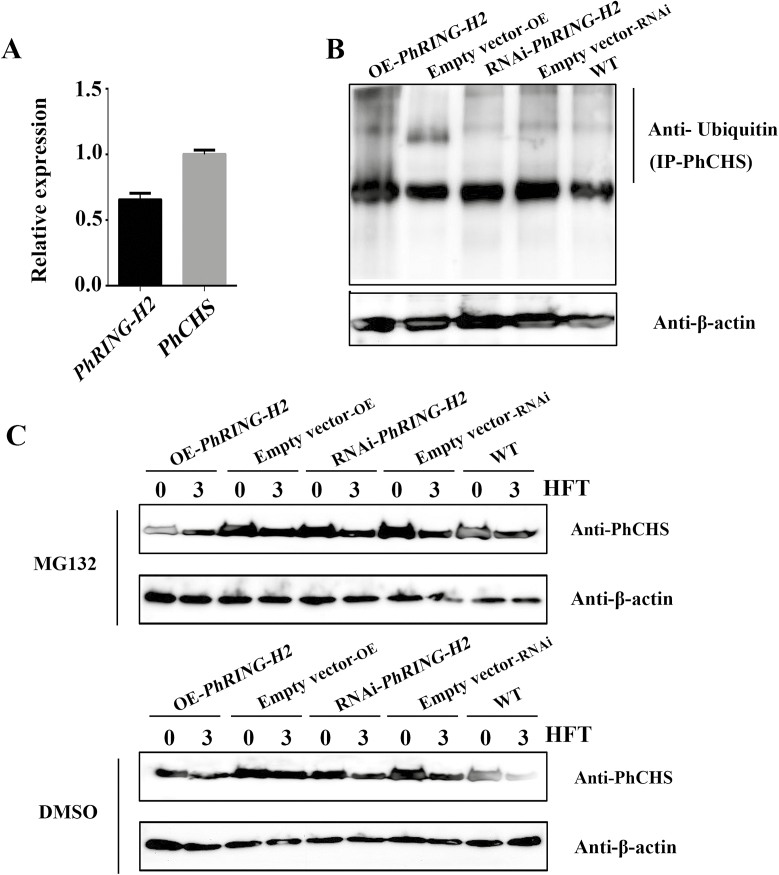
To confirm that PhCHS ubiquitin degradation is regulated by PhRING-H2 in vivo, we used ‘He Xie’ bud scales, which also have a red coloration, since petals are delicate, wilt easily, and are less amenable to such studies. We first confirmed by qRT-PCR that both PhCHS and PhRING-H2 are expressed in bud scales (Fig. 8A). To provide further evidence that PhRING-H2 can mediate the ubiquitination of PhCHS in vivo, we performed loss- or gain-of-function experiments using transient overexpression (OE) and RNA interference (RNAi) techniques. Relative expression levels of PhRING-H2 were measured using qRT-PCR in OE-PhRING-H2 and PhRING-H2-RNAi bud scales in comparison with control, and the results indicated that the expression levels were successfully up- or down-regulated and could be used for further analysis (see Supplementary Fig. S7). As shown in Fig. 8B, overexpression (OE-PhRING-H2) or down-regulation of the levels of PhRING-H2 (PhRING-H2-RNAi) could increase or decrease the level of ubiquitinated PhCHS, respectively (Fig. 8B). To confirm that the abundance of PhCHS is controlled by the 26S proteasome system, we used MG132 to treat the bud scales after transient transformation for 72 h to inhibit the degradation process. As shown in Fig. 8C, the degradation of PhCHS was reduced in bud scales transformed with OE-PhRING-H2 by addition of MG132 for 3 h as compared with that of control (DMSO), while PhCHS protein accumulated in bud scales transformed with PhRING-H2-RNAi in the presence of MG132 for 3 h (Fig. 8C). Taken together, we confirmed that PhCHS is degraded via the 26S proteasome and regulated by PhRING-H2 in Paeonia plants.
Fig. 8.
PhRING protein regulates PhCHS degradation in vivo. (A) PhCHS and PhRING-H2 relative expression in bud scale (mean ±SD; n=3). (B) Comparison of the abundance of ubiquitinated proteins from bud scale treated by transient transformation of PhRING-H2-OE or RNAi after injection for 72 h. PhRING-H2-OE and RNAi were performed as described in ‘Materials and methods’. Control samples were transfected with the empty vector. For the ubiquitination assay, PhCHS antibody (2 μg final concentration) was added to the extracted crude protein from bud scale and incubated with PhCHS proteins for immunoprecipitation, and the captured PhCHS proteins were assayed for ubiquitination analysis using ubiquitin antibody. Ubiquitin antibody (1:1000, from Beyotime, Shanghai, China) was used to evaluate ubiquitinated proteins. β-Actin antibody (1: 3000) was used as an internal control for evaluating the abundance of reference protein in total proteins. (C) Bud scales injected with PhRING-H2-OE or RNAi for 72 h following MG132 (40 μM final concentration) and DMSO treatment. PhCHS protein content was detected at 0 and 3 h after treatment with MG132 or DMSO. HFT, hours after treatment. PhCHS (1: 3000) antibody was used to evaluate PhCHS protein abundance in bud scales of ‘He Xie’. Control samples were transfected with the empty vector. β-Actin antibody (1: 3000) was used as an internal control for evaluating reference protein abundance in total protein content.
Discussion
Protein abundance is often tightly controlled, and protein ubiquitination and subsequent degradation is a well-established means to balance protein levels. This usually involves the linking of poly-ubiquitin chains to target proteins, and the subsequent recognition of the ubiquitin signal by the 26S proteasome, resulting in proteolysis (Wilkinson, 2000). We used ubiquitylome profiling by tandem mass tag (TMT) and LC-MS/MS to analyse the ubiquitin proteome of Paeonia ‘He Xie’ petal cells, and identified many proteins involved in petal development and secondary metabolism (Figs 1, 2). Notably we found three key enzymes involved in flavonoid biosynthesis to be ubiquitinated (Fig. 5). A number of iTRAQ (isobaric tag for relative and absolute quantification)/TMT-based quantitative analyses of plant proteomes and ubiquitylomes have been reported and have advantages over traditional 2D-gel based studies (Ma et al., 2014). In petunia (Petunia hybrida ‘Mitchell’), TMT-based quantitative proteome and ubiquitylome analysis, combined with transcriptome profiling, suggested that ubiquitination is associated with ethylene-induced petal senescence and the aging process (Guo et al., 2017). Hao et al. (2017) used quantitative proteomics to relate differentially expressed proteins to metabolic differences between different Japanese yew species (Taxus × media and Taxus mairei). In our study, we characterized the proteome and ubiquitylome of Paeonia petals to gain insights into the regulation of flavonoid biosynthesis at the protein level (Figs 3, 4).
Among the enzymes for flavonoid (anthocyanin) biosynthesis, CHS (EC2.3.1.74) is the rate-limiting enzyme and belongs to the type III polyketide synthase (PKS) superfamily, which also includes stilbene synthase and 2-pyrone synthase (Eckermann et al., 1998; Austin and Noel, 2003; Abe and Morita, 2010). Small modifications of only a few amino acids of CHS could significantly alter the binding pocket volume and modify enzyme function (Ferrer et al., 1999; Jez et al., 2000). Furthermore, it was reported that loss of CHS enzyme activity resulted in albino flowers in petunia (Petunia hybrida) and octoploid dahlia (Dahlia variabilis), or fruits of tomato (Solanum lycopersicum) and apple (Malus × domestica) (Napoli et al., 1990; Schijlen et al., 2007; Ohno et al., 2011; Waters et al., 2012; Dare et al., 2013). The transcriptional control of CHS has been extensively studied in many monocot and dicot species and its expression was shown to be tightly controlled in response to developmental and environmental signals (Feinbaum and Ausubel, 1988; Dao et al., 2011). In Arabidopsis, its transcription is regulated by AtMYB12/AtMYB11/AtMYB111, which belong to subgroup of the R2R3-MYB family, while, in maize (Zea mays) it is regulated by the MBW complex (Mehrtens et al., 2005; Petroni and Tonelli, 2011). However, post-translational regulation of CHS and associated protein modifications have not been extensively characterized. In Arabidopsis, it was demonstrated that CHS stability is controlled by proteolytic regulators through physical interaction with a kelch domain-containing F-box protein (KFB), which involves in ubiquitination and degradation (Zhang et al., 2017). However, the specific ubiquitinated sites remain unknown. In our study, the abundance of PhCHS was reduced at petal developmental stage 5 as compared with earlier stages (1–4), which demonstrated that degradation of PhCHS does occur. Further, we identified 12 putative ubiquitination sites in PhCHS, suggesting that it might be degraded by the 26S proteasome system (Fig. 6).
The ubiquitin–26S proteasome system (UPS) is emerging as a major post-translational control mechanism in plants (Zheng et al., 2016). It has been demonstrated that the key players for protein ubiquitination and degradation are the E3 ubiquitin ligases, which consist of a large and diverse family of proteins or protein complexes including single subunits (either a HECT domain or a RING-box domain) or multiple subunits (i.e. the Skp1–Cullin–F-box (SCF) complex, Anaphase Promoting Complex (APC) and Broad-complex, Tramtrack, and Bric-a-Brac (CUL3–BTB) (Moon et al., 2004). Up till now, several E3 ubiquitin ligases have been reported to be involved in regulation of flavonoid biosynthesis in Arabidopsis. The CONSTITUTIVELY PHOTOMORPHOGENIC1–SUPPRESSOR OF PHYA-105 (COP1–SPA) complex is an E3 ubiquitin ligase and the N-terminus of the COP1 protein has a RING finger domain that interacts with the respective coiled-coil domain between COP1 and SPA, which mediates the turnover of AtPAP1 and AtPAP2 and regulates their stability during anthocyanin biosynthesis (Deshaies and Joazeiro, 2009; Maier et al., 2013). The HECT domain-containing E3 ligase Ubiquitin Protein Ligase 3 (UPL3) interacts with key regulatory proteins GL3 and EGL3 in the Arabidopsis flavonoid pathway, and mediates their degradation through the UPS (Patra et al., 2013). In addition, proteolytic degradation of TRANSPARENT TESTA8 (TT8) and TRANSPARENT TESTA GLABRA1 (TTG1), as flavonoid regulators in Arabidopsis was also mediated by the UPS, although the E3 ligase protein that mediates the degradation of TT8 and TTG1 has not yet been identified (Patra et al., 2013). Therefore, the identification of E3 ubiquitin ligase components remains complicated since there are several hundred proteins in plants that help to specify appropriate targets for protein modification (Moon et al., 2004). In the current study, we identified a RING-box protein (PhRING-H2) that could interact with PhCHS and mediate its turnover (Figs 7, 8). A similar result was obtained in apple (Malus × domestica), where MdMYB1 was ubiquitinated and degraded through interaction with a RING E3 ligase, thereby reducing anthocyanin accumulation (An et al., 2017). The RING domain is a protein interaction domain that has been implicated in a range of diverse biological processes, while E3 ubiquitin–protein ligase activity is intrinsic to the RING domain and is likely to be a general function of this domain (Borden and Freemont, 1996; Freemont, 1993). Among E3 ubiquitin–protein ligases, the Skp1–Cullin–F-box (SCF) complex is the best characterized, and F-box protein regulates the specificity of the SCF complex and selectively interacts with target proteins that are subjected to protein modification (Zheng et al., 2002). KFBCHS was also confirmed to be a proteolytic regulator for AtCHS and controlled flavonoid biosynthesis in response to environmental or developmental signals in Arabidopsis (Zhang et al., 2017). In our study, we suggest that PhRING-H2 could mediate the ubiquitination of PhCHS, which contrasts with a previously reported mechanism involving an F-box protein. These results provide new insights into the post-translational regulation of CHS and provide a theoretical basis for the manipulation of flavonoid biosynthesis.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Gene Ontology (GO) enrichment analysis of up-regulated or down-regulated proteins and their subcellular location.
Fig. S2. KEGG analysis of up-regulated or down-regulated proteins.
Fig. S3. KEGG analysis of different expressed proteins involved in various pathways in petal development of Paeonia ‘He Xie’.
Fig. S4. Quantitative analysis of the distribution of differently expressed ubiquitinated proteins in GO terms level 2 and their subcellular location.
Fig. S5. Protein–protein interaction in the ubiquitination and proteome networks.
Fig. S6. PhCHS degradation is mediated by the 26S proteasome system in vivo.
Fig. S7. Effect of PhRING-H2-OE or PhRING-H2-RNAi on the expression levels of PhRING-H2 transcript.
Table S1. Primers list in this study.
Table S2. The amino acid sequence of PhCHS used for polyclonal antibody preparation.
Table S3. The results of mass spectrometry using PhCHS antibody.
Table S4. Summary of proteins identified by MS/MS spectrum.
Table S5. Summary of differentially expressed proteins (filtered with threshold value of expression fold change).
Table S6. Partial proteins involved in flavonoid biosynthesis pathway.
Table S7. Summary of MS/MS spectrum database search analysis.
Table S8. Differentially modified proteins after normalizing by proteome.
Table S9. Identification of PhRING-H2 as a candidate interactor with PhCHS.
Author contributions
LW, QS, and ZG designed the research; QS and ZG critically assessed the manuscript; SM, JZ, and NT performed the experiments; QS and ZG analysed the data; LW, QS, ZG, QH, Z-AL, and HZ wrote the paper with input from all the other authors. The authors declare no conflicts of interest.
Acknowledgements
This research work was supported by the National Natural Science Foundation of China (Grant No. 31471909 and 31772350). We also thank PlantScribe (www.plantscribe.com) for editing this manuscript.
References
- Abe I, Morita H. 2010. Structure and function of the chalcone synthase superfamily of plant type III polyketide synthases. Cheminform, 27, 809–838. [DOI] [PubMed] [Google Scholar]
- Amado NG, Fonseca BF, Cerqueira DM, Neto VM, Abreu JG. 2011. Flavonoids: potential Wnt/beta-catenin signaling modulators in cancer. Life Sciences 89, 545–554. [DOI] [PubMed] [Google Scholar]
- An JP, Liu X, Li HH, You CX, Wang XF, Hao YJ. 2017. Apple RING E3 ligase MdMIEL1 inhibits anthocyanin accumulation by ubiquitinating and degrading MdMYB1 protein. Plant & Cell Physiology 58, 1953–1962. [DOI] [PubMed] [Google Scholar]
- Austin MB, Noel JP. 2003. The chalcone synthase superfamily of type III polyketide synthases. Natural Product Reports 20, 79–110. [DOI] [PubMed] [Google Scholar]
- Borden KL, Freemont PS. 1996. The RING finger domain: a recent example of a sequence-structure family. Current Opinion in Structural Biology 6, 395–401. [DOI] [PubMed] [Google Scholar]
- Cai J, Qin G, Chen T, Tian S. 2018. The mode of action of remorin1 in regulating fruit ripening at transcriptional and post-transcriptional levels. New Phytologist 219, 1406–1420. [DOI] [PubMed] [Google Scholar]
- Ciechanover A. 2005. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nature Reviews Molecular Cell Biology 6, 79–87. [DOI] [PubMed] [Google Scholar]
- Colaert N, Helsens K, Martens L, Vandekerckhove J, Gevaert K. 2009. Improved visualization of protein consensus sequences by iceLogo. Nature Methods 6, 786–787. [DOI] [PubMed] [Google Scholar]
- Dao TT, Linthorst HJ, Verpoorte R. 2011. Chalcone synthase and its functions in plant resistance. Phytochemistry Reviews 10, 397–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dare AP, Tomes S, Jones M, McGhie TK, Stevenson DE, Johnson RA, Greenwood DR, Hellens RP. 2013. Phenotypic changes associated with RNA interference silencing of chalcone synthase in apple (Malus × domestica). The Plant Journal 74, 398–410. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. 2009. RING domain E3 ubiquitin ligases. Annual Review of Biochemistry 78, 399–434. [DOI] [PubMed] [Google Scholar]
- Ding S, Zhang B, Qin F. 2015. Arabidopsis RZFP34/CHYR1, a ubiquitin E3 ligase, regulates stomatal movement and drought tolerance via SnRK2.6-mediated phosphorylation. The Plant Cell 27, 3228–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Wu J, Ji KX, Zeng QY, Bhuiya MW, Su S, Shu QY, Ren HX, Liu ZA, Wang LS. 2015. Methylation mediated by an anthocyanin, O-methyltransferase, is involved in purple flower coloration in Paeonia. Journal of Experimental Botany 66, 6563–6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckermann S, Schröder G, Schmidt J, Strack D, Ru AE, Helariutta Y, Elomaa P, Kotilainen M, Kilpeläinen I, Proksch P. 1998. New pathway to polyketides in plants. Nature, 396, 387–390. [Google Scholar]
- Edgar R. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eloy NB, Voorend W, Lan W, et al. . 2017. Silencing CHALCONE SYNTHASE in maize impedes the incorporation of tricin into lignin and increases lignin content. Plant Physiology 173, 998–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinbaum RL, Ausubel FM. 1988. Transcriptional regulation of the Arabidopsis thaliana chalcone synthase gene. Molecular and Cellular Biology 8, 1985–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer JL, Jez JM, Bowman ME, Dixon RA, Noel JP. 1999. Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nature Structural Biology 6, 775–784. [DOI] [PubMed] [Google Scholar]
- Forkmann G, Martens S. 2001. Metabolic engineering and applications of flavonoids. Current Opinion in Biotechnology 12, 155–160. [DOI] [PubMed] [Google Scholar]
- Freemont PS. 1993. The RING finger. A novel protein sequence motif related to the zinc finger. Annals of the New York Academy of Sciences 684, 174–192. [DOI] [PubMed] [Google Scholar]
- Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. 2008. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. The Plant Journal 53, 814–827. [DOI] [PubMed] [Google Scholar]
- Gnad F, Gunawardena J, Mann M. 2011. PHOSIDA 2011: the posttranslational modification database. Nucleic Acids Research 39, D253–D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E. 2006. The genetics and biochemistry of floral pigments. Annual Review of Plant Biology 57, 761–780. [DOI] [PubMed] [Google Scholar]
- Gu ZY, Zhu J, Hao Q, Yuan YW, Duan YW, Men SQ, Wang QY, Hou QZ, Liu ZA, Shu QY, Wang LS. 2019. A novel R2R3-MYB transcription factor contributes to petal blotch formation by regulating organ-specific expression of PsCHS in tree peony (Paeonia suffruticosa). Plant Cell Physiology 60, 599–611. [DOI] [PubMed] [Google Scholar]
- Guo J, Liu J, Wei Q, Wang R, Yang W, Ma Y, Chen G, Yu Y. 2017. Proteomes and ubiquitylomes analysis reveals the involvement of ubiquitination in protein degradation in petunias. Plant Physiology 173, 668–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Dang R, Li J, Jiang J, Zhang N, Jia M, Wei L, Li Z, Li B, Jia W. 2015. SUCROSE NONFERMENTING1-RELATED PROTEIN KINASE2.6, an ortholog of OPEN STOMATA1, is a negative regulator of strawberry fruit development and ripening. Plant Physiology 167, 915–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama S, Nakayama KI. 2003. Ubiquitylation as a quality control system for intracellular proteins. Journal of Biochemistry 134, 1–8. [DOI] [PubMed] [Google Scholar]
- Hao J, Guo H, Shi X, Wang Y, Wan Q, Song YB, Zhang L, Dong M, Shen C. 2017. Comparative proteomic analyses of two Taxus species (Taxus × media and Taxus mairei) reveals variations in the metabolisms associated with paclitaxel and other metabolites. Plant and Cell Physiology 58, 1878–1890. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. 1995. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Current Opinion in Cell Biology 7, 215–223. [DOI] [PubMed] [Google Scholar]
- Hosokawa M, Yamauchi T, Takahama M, et al. . 2013. Phosphorus starvation induces post-transcriptional CHS gene silencing in Petunia corolla. Plant Cell Reports 32, 601–609. [DOI] [PubMed] [Google Scholar]
- Hou DX. 2003. Potential mechanisms of cancer chemoprevention by anthocyanins. Current Molecular Medicine 3, 149–159. [DOI] [PubMed] [Google Scholar]
- Jez JM, Austin MB, Ferrer J, Bowman ME, Schröder J, Noel JP. 2000. Structural control of polyketide formation in plant-specific polyketide synthases. Chemistry & Biology 7, 919–930. [DOI] [PubMed] [Google Scholar]
- Jia N, Shu QY, Wang LS, Du H, Xu YJ, Liu ZA. 2008a Analysis of petal anthocyanins to investigate coloration mechanism in herbaceous peony cultivars. Scientia Horticulturae 117, 167–173. [Google Scholar]
- Jia N, Shu QY, Wang LS, Liu ZA, Ren HX, Xu YJ, Tian DK, Tilt KM. 2008b Identification and characterization of anthocyanins by high-performance liquid chromatography-electrospray ionization-mass spectrometry in herbaceous peony species. Journal of the American Society for Horticulture Science, 133, 418–426. [Google Scholar]
- Joazeiro CA, Weissman AM. 2000. RING finger proteins: mediators of ubiquitin ligase activity. Cell 102, 549–552. [DOI] [PubMed] [Google Scholar]
- Koes R, Verweij W, Quattrocchio F. 2005. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends in Plant Science 10, 236–242. [DOI] [PubMed] [Google Scholar]
- Koseki M, Goto K, Masuta C, Kanazawa A. 2005. The star-type color pattern in Petunia hybrida ‘red Star’ flowers is induced by sequence-specific degradation of Chalcone Synthase RNA. Plant & Cell Physiology 46, 1879–1883. [DOI] [PubMed] [Google Scholar]
- Li C, Du H, Wang L, Shu Q, Zheng Y, Xu Y, Zhang J, Zhang J, Yang R, Ge Y. 2009. Flavonoid composition and antioxidant activity of tree peony (Paeonia section Moutan) yellow flowers. Journal of Agricultural and Food Chemistry 57, 8496–8503. [DOI] [PubMed] [Google Scholar]
- Li CH. 2010. The flavonoid composition in tree peony petals and their effects on the coloration. PhD thesis, University of Chinese Academy of Sciences, China. [Google Scholar]
- Li T, Jiang Z, Zhang L, Tan D, Wei Y, Yuan H, Li T, Wang A. 2016. Apple (Malus domestica) MdERF2 negatively affects ethylene biosynthesis during fruit ripening by suppressing MdACS1 transcription. The Plant Journal 88, 735–748. [DOI] [PubMed] [Google Scholar]
- Liou G, Chiang YC, Wang Y, Weng JK. 2018. Mechanistic basis for the evolution of chalcone synthase catalytic cysteine reactivity in land plants. The Journal of Biological Chemistry 293, 18601–18612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorick KL, Weissman AM. 1999. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proceedings of the National Academy of Sciences, USA 96, 11364–11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyzenga WJ, Stone SL. 2012. Abiotic stress tolerance mediated by protein ubiquitination. Journal Experimental Botany 63, 599–616. [DOI] [PubMed] [Google Scholar]
- Ma C, Zhou J, Chen G, Bian Y, Lv D, Li X, Wang Z, Yan Y. 2014. iTRAQ-based quantitative proteome and phosphoprotein characterization reveals the central metabolism changes involved in wheat grain development. BMC Genomics 15, 1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N, Xue J, Li Y, et al. . 2008. Rh-PIP2;1, a rose Aquaporin gene, is involved in ethylene-regulated petal expansion. Plant Physiology 148, 894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier A, Schrader A, Kokkelink L, et al. . 2013. Light and the E3 ubiquitin ligase COP 1/SPA control the protein stability of the MYB transcription factors PAP 1 and PAP 2 involved in anthocyanin accumulation in Arabidopsis. The Plant Journal 74, 638–651. [DOI] [PubMed] [Google Scholar]
- Mehrtens F, Kranz H, Bednarek P, Weisshaar B. 2005. The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiology 138, 1083–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Parry G, Estelle M. 2004. The ubiquitin-proteasome pathway and plant development. The Plant Cell 16, 3181–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlemann JK, Younts TLB, Muday GK. 2018. Flavonols control pollen tube growth and integrity by regulating ROS homeostasis during high-temperature stress. Proceedings of the National Academy of Sciences, USA 115, E11188–E11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Szklarczyk D, Julien P, et al. . 2010. eggNOG v2.0: extending the evolutionary genealogy of genes with enhanced nonsupervised orthologous groups, species and functional annotations. Nucleic Acids Research 38, D190–D195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabavi SM, Šamec D, Tomczyk M, et al. . 2019. Flavonoid biosynthetic pathways in plants: versatile targets for metabolic engineering. Biotechnology Advances, doi: 10.1016/j.biotechadv.2018.11.005. [DOI] [PubMed] [Google Scholar]
- Nagatomo Y, Usui S, Ito T, Kato A, Shimosaka M, Taguchi G. 2014. Purification, molecular cloning and functional characterization of flavonoid C-glucosyltransferases from Fagopyrum esculentum M. (buckwheat) cotyledon. The Plant Journal 80, 437–448. [DOI] [PubMed] [Google Scholar]
- Napoli C, Lemieux C, Jorgensen R. 1990. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. The Plant Cell 2, 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S, Hosokawa M, Kojima M, Kitamura Y, Hoshino A, Tatsuzawa F, Doi M, Yazawa S. 2011. Simultaneous post-transcriptional gene silencing of two different chalcone synthase genes resulting in pure white flowers in the octoploid dahlia. Planta 234, 945–958. [DOI] [PubMed] [Google Scholar]
- Passmore LA, Barford D. 2004. Getting into position: the catalytic mechanisms of protein ubiquitylation. The Biochemical Journal 379, 513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra B, Pattanaik S, Yuan L. 2013. Ubiquitin protein ligase 3 mediates the proteasomal degradation of GLABROUS 3 and ENHANCER OF GLABROUS 3, regulators of trichome development and flavonoid biosynthesis in Arabidopsis. The Plant Journal 74, 435–447. [DOI] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. 2003. A proteomics approach to understanding protein ubiquitination. Nature Biotechnology 21, 921–926. [DOI] [PubMed] [Google Scholar]
- Petroni K, Tonelli C. 2011. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Science 181, 219–229. [DOI] [PubMed] [Google Scholar]
- Saito R, Kuchitsu K, Ozeki Y, Nakayama M. 2007. Spatiotemporal metabolic regulation of anthocyanin and related compounds during the development of marginal picotee petals in Petunia hybrida (Solanaceae). Journal of Plant Research 120, 563–568. [DOI] [PubMed] [Google Scholar]
- Schijlen EG, de Vos CH, Martens S, Jonker HH, Rosin FM, Molthoff JW, Tikunov YM, Angenent GC, van Tunen AJ, Bovy AG. 2007. RNA interference silencing of chalcone synthase, the first step in the flavonoid biosynthesis pathway, leads to parthenocarpic tomato fruits. Plant Physiology 144, 1520–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell JD, Hicke L. 2003. Non-traditional functions of ubiquitin and ubiquitin-binding proteins. The Journal of Biological Chemistry 278, 35857–35860. [DOI] [PubMed] [Google Scholar]
- Schütze K, Harter K, Chaban C. 2009. Bimolecular fluorescence complementation (BiFC) to study protein–protein interactions in living plant cells. Methods in Molecular Biology 479, 189–202. [DOI] [PubMed] [Google Scholar]
- Shi MZ, Xie DY. 2014. Biosynthesis and metabolic engineering of anthocyanins in Arabidopsis thaliana. Recent Patents on Biotechnology 8, 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q, Li L, Zhang X, et al. . 2017. Biochemical and comparative transcriptomic analyses identify candidate genes related to variegation formation in Paeonia rockii. Molecules 22, 1364–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Nakamura N, Togami J. 2008. Altering flower color in transgenic plants by RNAi-mediated engineering of flavonoid biosynthetic pathway. Methods in Molecular Biology 442, 245–257. [DOI] [PubMed] [Google Scholar]
- Tian J, Pei H, Zhang S, et al. . 2014. TRV–GFP: a modified Tobacco rattle virus vector for efficient and visualizable analysis of gene function. Journal of Experimental Botany 65, 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo M. 2018. News from the PUB: plant U-box type E3 ubiquitin ligases. Journal of Experimental Botany 69, 371–384. [DOI] [PubMed] [Google Scholar]
- Vierstra RD. 2012. The expanding universe of ubiquitin and ubiquitin-like modifiers. Plant Physiology 160, 2–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Zhu D, Huang X, Li S, Gong Y, Yao Q, Fu X, Fan LM, Deng XW. 2009. Biochemical insights on degradation of Arabidopsis DELLA proteins gained from a cell-free assay system. The Plant Cell 21, 2378–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LS, Shiraishi A, Hashimoto F, Aoki N, Shimizu K, Sakata Y. 2001. Analysis of petal anthocyanins to investigate flower coloration of Zhongyuan (Chinese) and Daikon Island (Japanese) tree peony cultivars. Journal of Plant Research 114, 33–43. [Google Scholar]
- Wang XF, An JP, Liu X, Su L, You CX, Hao YJ. 2018a The nitrate-responsive protein MdBT2 regulates anthocyanin biosynthesis by interacting with the MdMYB1 transcription factor. Plant Physiology 178, 890–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Yu Q, Shen W, El Mohtar CA, Zhao X, Gmitter FG Jr. 2018b Functional study of CHS gene family members in citrus revealed a novel CHS gene affecting the production of flavonoids. BMC Plant Biology 18, 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters WF, Rubman S, Hurry MJ. 2012. Tandemly arranged chalcone synthase A genes contribute to the spatially regulated expression of siRNA and the natural bicolor floral phenotype in Petunia hybrida. Plant Journal for Cell & Molecular Biology 70, 739–749. [DOI] [PubMed] [Google Scholar]
- Wilkinson KD. 2000. Ubiquitination and deubiquitination: targeting of proteins for degradation by the proteasome. Seminars in Cell & Developmental Biology 11, 141–148. [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B. 2001. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiology 126, 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Guo HS, Dallman G, Fang S, Weissman AM, Chua NH. 2002. SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419, 167–170. [DOI] [PubMed] [Google Scholar]
- Xie X, Kang H, Liu W, Wang GL. 2015. Comprehensive profiling of the rice ubiquitome reveals the significance of lysine ubiquitination in young leaves. Journal of Proteome Research 14, 2017–2025. [DOI] [PubMed] [Google Scholar]
- Xu W, Dubos C, Lepiniec L. 2015. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends in Plant Science 20, 176–185. [DOI] [PubMed] [Google Scholar]
- Yuan H, Meng D, Gu Z, Li W, Wang A, Yang Q, Zhu Y, Li T. 2014. A novel gene, MdSSK1, as a component of the SCF complex rather than MdSBP1 can mediate the ubiquitination of S-RNase in apple. Journal of Experimental Botany 65, 3121–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JJ, Wang LS, Shu QY, Liu ZA, Li CH, Zhang J, Wei X, Tian D. 2007. Comparison of anthocyanins in non-blotches and blotches of the petals of Xibei tree peony. Scientia Horticulturae 114, 104–111. [Google Scholar]
- Zhang X, Abrahan C, Colquhoun TA, Liu CJ. 2017. A proteolytic regulator controlling chalcone synthase stability and flavonoid biosynthesis in Arabidopsis. The Plant Cell 29, 1157–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Gou M, Guo C, Yang H, Liu CJ. 2015. Down-regulation of Kelch domain-containing F-box protein in Arabidopsis enhances the production of (poly)phenols and tolerance to ultraviolet radiation. Plant Physiology 167, 337–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Gou M, Liu CJ. 2013. Arabidopsis Kelch repeat F-box proteins regulate phenylpropanoid biosynthesis via controlling the turnover of phenylalanine ammonia-lyase. The Plant Cell 25, 4994–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, et al. . 2002. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416, 703–709. [DOI] [PubMed] [Google Scholar]
- Zheng Q, Huang T, Zhang L, et al. . 2016. Dysregulation of ubiquitin-proteasome system in neurodegenerative diseases. Frontiers in Aging Neuroscience 8, 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.