AAI genes form an evolutionarily conserved large family showing the effects of genome duplication and lacking systematic study. GhAAI66 integrates multiple flower signaling pathways to induce early flowering.
Keywords: Early flowering, ectopic expression, floral integrators, gene duplication, Gossypium hirsutum, phylogenetic analysis
Abstract
Plants undergo a phase transition from vegetative to reproductive development that triggers floral induction. Genes containing an AAI (α-amylase inhibitor) domain form a large gene family, but there have been no comprehensive analyses of this gene family in any plant species. Here, we identified 336 AAI genes from nine plant species including122 AAI genes in cotton (Gossypium hirsutum). The AAI gene family has evolutionarily conserved amino acid residues throughout the plant kingdom. Phylogenetic analysis classified AAI genes into five major clades with significant polyploidization and showing effects of genome duplication. Our study identified 42 paralogous and 216 orthologous gene pairs resulting from segmental and whole-genome duplication, respectively, demonstrating significant contributions of gene duplication to expansion of the cotton AAI gene family. Further, GhAAI66 was preferentially expressed in flower tissue and as responses to phytohormone treatments. Ectopic expression of GhAAI66 in Arabidopsis and silencing in cotton revealed that GhAAI66 triggers a phase transition to induce early flowering. Further, GO (Gene Ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) analysis of RNA sequencing data and qRT–PCR (quantitative reverse transcription–PCR) analysis indicated that GhAAI66 integrates multiple flower signaling pathways including gibberellin, jasmonic acid, and floral integrators to trigger an early flowering cascade in Arabidopsis. Therefore, characterization of the AAI family provides invaluable insights for improving cotton breeding.
Introduction
Annual plants undergo a major phase transition triggering vegetative to reproductive development in their life cycle. The process of phase transition is rarely reversible; however, it ensures optimal timing of the transition for pollination as well as seed development (Boss et al., 2004). Of these, proper timing of flowering is most important for ensuring reproductive success (Lee and Lee, 2010). Genetic and physiological approaches to investigate flowering mechanisms have shown that various environmental as well as endogenous factors determine the phase transition and mediate the flowering signaling cascade (Boss et al., 2004). During the past decade, multiple studies have been carried out to provide a molecular understanding of control of flowering time by various comprehensive approaches (Mouradov et al., 2002; Ratcliffe and Riechmann, 2002; Simpson and Dean, 2002; Henderson et al., 2003; Yanovsky and Kay, 2003). The integration of multiple pathways collectively regulates a set of common targets that promotes the floral transition. These include light quality, photoperiod, ambient temperature, hormone signaling, and biosynthesis (Simpson and Dean, 2002; Boss et al., 2004) that induce or repress the expression of genes involved in the flowering signaling cascade. These floral pathway integrators include FLOWERING LOCUS T (FT), LEAFY (LFY), and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS (SOC1) (Nilsson et al., 1998; Kardailsky et al., 1999; Kobayashi et al., 1999; Lee et al., 2000; Samach et al., 2000). Further, gibberellin (GA) induces flowering signals in many plant species, such as GA-promoted flowering in Arabidopsis. Additionally, exogenous treatment with GA in Arabidopsis accelerates flowering under short-day conditions (Blazquez et al., 1998; Gocal et al., 2001). Previously it has been reported that GA and jasmonic acid (JA) antagonistically regulate flowering time in Arabidopsis as JA represses FT expression and mediates signaling cascades to delay flowering (Zhai et al., 2015).
Cotton is an important fiber crop and an ideal source of oilseed and feed. Although cotton is primarily cultivated as an annual crop, it is naturally a short-day photoperiodic perennial. Due to its perennial growth habit, crop management strategies are complicated. In this regard, more determinate cotton plant architecture is desired. Some genes, such as cotton GhTFL1-like genes, delay flowering due to ectopic expression in transgenic Arabidopsis (McGarry et al., 2016; Prewitt et al., 2018). Previously, expression of Arabidopsis FT in cotton led to altered plant architecture in terms of reducing indeterminate growth as well as perennial traits of cotton plant. Moreover, Arabidopsis FT expression uncoupled flowering from photoperiod in photoperiodic cotton and high florigen-synchronized flowering coupled with compressed growth habits in domesticated day-neutral cotton lines (McGarry and Ayre, 2012; McGarry et al., 2013). Cotton GhLFY, a homolog of floricaula/leafy (LFY), is expressed in the shoot apex and plays an important role in flower initiation (Li et al., 2013). Breeding cotton for plant architecture, growth habit, and tolerance to environmental and hormonal stresses is a high priority for plant breeders. The α-amylase inhibitor (AAI) domain is a plant lipid transfer protein (LTP), hydrophobic seed protein, and a trypsin α-amylase protein gene family with 68 members in Arabidopsis. In Arabidopsis, LTP3 served as a target of MYB96 and mediated freezing and drought tolerance (Guo et al., 2013). AtLTP2 played a structural role in maintaining the integrity of adhesion between the hydrophobic cuticle and the hydrophilic underlying cell wall below in Arabidopsis (Jacq et al., 2017). Another study reported that a gain-of-function mutation of AtLTP5 disturbed pollen tube tip growth and subsequent fertilization in Arabidopsis (Chae et al., 2009). Cotton GhLTPG1 was reported to regulate cotton fiber elongation by mediating transport of phosphatidylinositol monophosphates (Deng et al., 2016). However, the functions of plant AAI domain-containing genes remain largely unknown in cotton.
Here, we identified 336 AAI domain-containing genes in nine different plant species including monocots, dicots, moss, and ferns. We also exclusively identified 122 AAI genes in Gossypium hirsutum. We then conducted a phylogenetic analysis and identified conserved amino acid residues, protein motif distribution pattern, encoded proteins, gene structure, chromosomal location, gene duplication, synteny analysis, and Ka/Ks values. Moreover, spatial expression patterns of GhAAI genes and responses under various phytohormone treatments were monitored. We also performed ectopic expression of GhAAI66 in Arabidopsis and silencing by virus-induced gene silencing (VIGS), conducted Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) analysis of RNA sequencing (RNA-seq) data of OE-GhAAI66 lines, and confirmed our results using quantitative reverse transcription–PCR (qRT–PCR) analysis to produce a proposed working model of GhAAI66 in Arabidopsis. This work will provide a basic foundation to improve cotton breeding for hormonal responses and cotton plant growth habit.
Materials and methods
Gene identification and analysis of conserved residues
First we identified AAI domain-containing genes in Arabidopsis downloaded from TAIR 10 (http://www.arabidopsis.org). The retrieved Arabidopsis AAI genes were then used as a query to identify AAI genes in G. hirsutum (NAU, version 1.1), G. arboreum (ICR, version 1.0), G. raimondii (JGI, version 2.0), Theobroma cacao (version 10), Oryza sativa (version 10), Zea mays (version 10), Physcomitrella patens (version 3.3), Selaginella moellendorffii (version 1.0), and the alga Chlamydomonas reinhardtii (version 1.0). We downloaded the G. arboreum database from ftp://bioinfo.ayit.edu.cn/downloads/, and G. raimondii and G. hirsutum from COTTONGEN (https://www.cottongen.org/). The databases for other plant species were downloaded from Phytozome v11 (https://phytozome.jgi.doe.gov/pz/portal.html). Next, we confirmed all putative AAI gene sequences using Interproscan 63.0 (http://www.ebi.ac.uk/InterProScan/) (Jones et al., 2014) and SMART (http://smart.embl-heidelberg.de/) (Letunic et al., 2015). Basic properties of GhAAI genes were estimated using ExPASy ProtParam (http://us.expasy.org/tools/protparam.html) and we used softberry (www.softberry.com) to predict the subcellular localization. Further, conserved amino acid residues analysis was conducted using the online tool WEBLOG (Crooks et al., 2004) after multiple alignment of conserved domain regions by ClustalX with default parameters (Thompson et al., 1997).
Phylogenetic analysis and determination of protein motif distribution and gene structure
For phylogenetic analysis, AAI genes were aligned and a phylogenetic tree was constructed using MEGA 7.0 (Kumar et al., 2016) with Neighbor–Joining (NJ) and minimum evolution (ME) methods. Further, 1000 bootstrap replicates were used to determine support values for the inferred phylogenetic trees. The phylogenetic tree was then visualized using TreeView1.6 (http://etetoolkit.org/treeview/).
For determining the distribution pattern of protein motifs and gene structure analysis, a BED file of putative GhAAI sequences was used in Gene Structure Display Server 2.0 (http://gsds.cbi.pku.edu.cn/index.php) (Hu et al., 2015) to obtain gene structure. The online MEME program (http://meme-suite.org/tools/meme) (Bailey et al., 2006) was used for protein motifs as described previously (Li et al., 2019).
Chromosomal location, gene duplication, and synteny analysis
Chromosomal location information for all GhAAI genes was obtained from a gff3-file of cotton genome annotation data (ftp://ftp.bioinfo.wsu.edu/species/Gossypium_hirsutum/NAU-NBI_G) and genes were mapped on the chromosomes using MapInspect software (https://mapinspect.software.informer.com/) (Jia et al., 2018). Gene duplication and synteny analysis was performed based on a method described previously (Yang et al., 2017). MCScanX software was used to determine and analyze cotton AAI duplication and synteny, and a figure was generated using CIRCOS (Krzywinski et al., 2009). Ka/Ks values were estimated with PAL2NAL (http://www.bork.embl.de/pal2nal/) (Suyama et al., 2006) as well as the CODEML program of the PAML package (Yang, 2007).
Vector construction and generation of transgenic lines
For ectopic expression of GhAAI66, we used Arabidopsis thaliana Columbia-0 (Col-0) plants to generate transgenic OE-GhAAI66 lines. We amplified cDNA of full-length GhAAI66 genes by PCR with gene-specific primers. The amplified PCR product was then cloned into pCAMBIA-2301 to construct the vector driven by the constitutive Cauliflower mosaic virus (CaMV) 35S promoter and the construct was introduced into Agrobacterium tumefaciens strain GV3101. Transformation into Arabidopsis plants was employed using the floral dip method (Zhang et al., 2015). Arabidopsis seeds were germinated on 1/2 Murashige and Skoog (MS) medium and grown under long-day conditions (16 h light/8 h dark) at 23 °C. Transgenic plants were selected on solid 1/2 MS medium plates supplemented with 50 µg ml−1 kanamycin. For VIGS, a highly specific region of 300 bp from the coding sequence (CDS) was cloned into the CLCrV binary vector and transformed into GV3101. Inoculation in CRI24 cotton plants was performed as described previously (Gao et al., 2011).
RNA-seq, KEGG, and GO analysis
For RNA-seq analysis, whole plant seedlings of Arabidopsis Col-0/BRIS1 plants were collected 20 d after germination. Sequencing libraries of total extracted RNA were generated using a NEB-Next ® Ultra™ RNA Library Prep Kit for Illumina® (New England Biolabs, Hitchin, UK) and index codes were added to each sample. These libraries were then sequenced on the Illumina Hiseq 2000 platform, and 100 bp paired-end reads were generated. Each experiment was conducted using three biological repeats. We used TopHat (Trapnell et al., 2009) to map the reads to TAIR genome annotation (https://www.arabidopsis.org/). Read counts were generated using HTSeq with the union mode, and differentially expressed genes (DEGs) were identified by DEseq2 (Anders and Huber, 2010). Further KEGG and GO analyses were conducted in the KEGG database for enrichment (Kanehisa and Goto, 2000) and PlantGSEA software (Yi et al., 2013), respectively.
Plant material, hormone treatments, and qRT–PCR analysis
For determining spatial expression patterns and hormone treatments, we used G. hirsutum variety CRI24. Cotton plants were grown in field conditions with standard cultural practices to obtain different tissues [root, stem, leaf, flower, and ovules at 1, 3, 5, 7, 10, 15, and 20 days post-anthesis (DPA); and fiber at 7, 10, 15, and 20 DAP] for determining the spatial expression pattern. For hormone treatment, pre-germinated seeds were suspended in a container with liquid culture medium as described previously (Yang et al., 2014). Four-week-old seedlings of the 3–4 leaf stage were treated with brassinolide (BL; 0 µM), GA (100 µM), indole-3-acetic acid (IAA; 100 µM), salicylic acid (SA; 10 µM), and methyl jasmonate (MeJA; 10 µM for time points of 0.5, 1, 3, and 5 h. The collected tissues were frozen immediately in liquid nitrogen and stored at –80 °C for RNA extraction and qRT–PCR analysis.
Next, RNA was extracted using the RNAprep Pure Plant Kit (TIANGEN, Beijing, China). We synthesized cDNA from 1 μg of RNA using the Prime-Script® RT reagent kit (Takara, Dalian, China) with the manufacturer’s guidelines. As internal controls, cotton GhHis3 (GenBank accession no. AF024716) and Actin2 (AT3G18780.1) were used. qRT–PCR analysis was carried out using SYBR Green on a LightCycler 480 (Roche Diagnostics GmbH, Mannheim, Germany). Relative expression was calculated as described previously (Livak and Schmittgen, 2001). Primers used in this study for gene cloning and qPCR analysis are listed in Supplementary Table S6 at JXB online). For statistical analysis, the data were considered to have a normal distribution and we conducted two-tailed Student’s t-tests in Microsoft Excel 2011.
Results
Identification of AAI genes and analysis of conserved amino acid residues in AAI domains
In this study, we identified 122 AAI genes in G. hirsutum, 34 in G. arboreum, and 33 in G. raimondii. Additionally, we identified AAI genes in different dicotyledons (68 genes in Arabidopsis and 22 in T. cacao), monocotyledons (18 in O. sativa and 21 in Z. mays), moss (11 in P. patens), and fern (7 in S. moellendorffii). However, no AAI gene was identified in algae (C. reinhardtii) (Supplementary Table S1). In this study, our main focus was G. hirsutum, so we first compared AAI genes from BJI and NAU sequenced genomes and observed no difference. We thus proceeded with AAI genes retrieved from the NAU genome sequence database. Notably, G. hirsutum had more than double the number of GhAAI genes as compared with G. arboreum and G. raimondii, illustrating polyploidy and significant duplication events during hybridization.
Next, analysis of conserved amino acid residues within AAI domains of Arabidopsis, O. sativa, G. hirsutum, P. patens, and S. moellendorffii depicted the degree of conservation of each residue in the AAI domains of all studied species (Supplementary Fig. S1A–E). The distribution of AAI domain amino acid residues was highly conserved in all species. For instance, amino acid residues such as C, P, and L were highly conserved in the AAI domain across all species and cysteine (C) amino acid residue was equally distributed in the N- and C-terminal ends (four in each) of AAI genes; however, C exhibited enrichment in the middle as compared with the ends. Our results indicated that AAI domain sequences were highly conserved among dicots, monocots, moss, and fern.
Basic information including locus ID, chromosomal position, gene length, CDS, protein length, molecular weight (MW), isoelectric point (pl), GRAVY value, and subcellular localization of GhAAI proteins were predicted (Supplementary Table S2). GhAAI genes encoded proteins ranging from 86 (GhAAI9 and GhAAI67) to 567 (GhAAI5) amino acids, with MWs from 9088.41 Da (GhAAI67) to 60745.91 Da (GhAAI5) and pI values varying from 4.23 (GhAAI102) to 10.24 (GhAAI1). Moreover, extracellular (secreted) subcellular localization was predicted for all GhAAI members. Other estimated parameters are listed in Supplementary Table S2.
Phylogenetic analysis of AAI genes
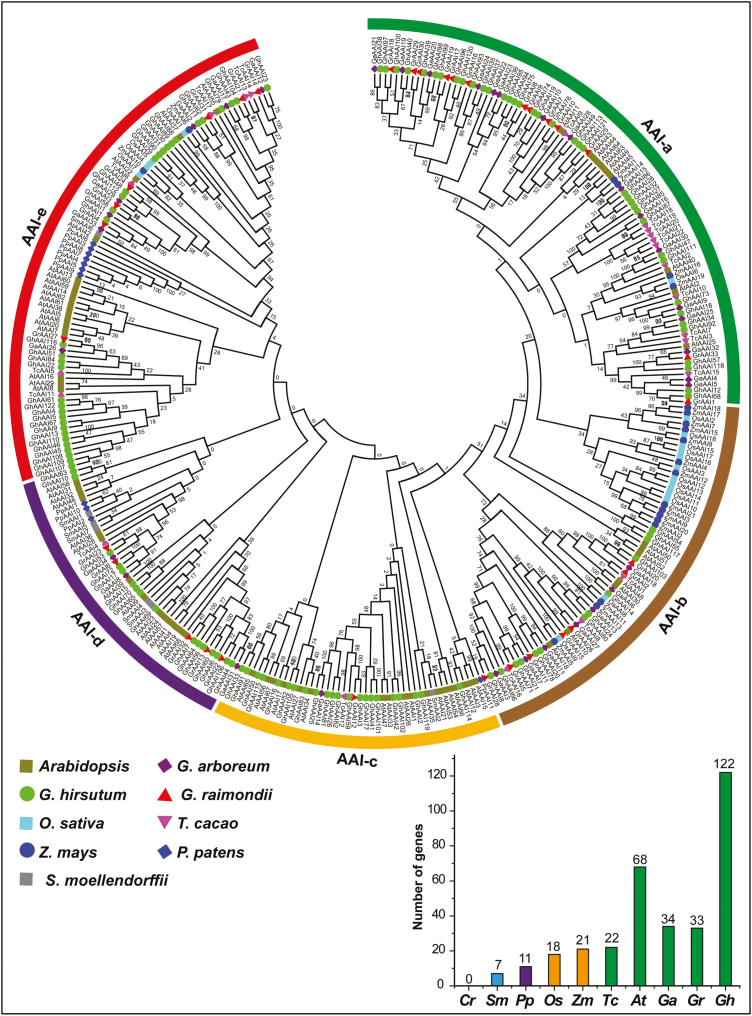
To investigate the evolutionary relationship among AAI genes of the nine aforementioned species, an unrooted phylogenetic tree was inferred using the NJ method. AAI genes were classified into five major groups (AAI-a to AAI-e) (Fig. 1). This phylogenetic analysis was then validated by constructing another tree using ME (see Supplementary Fig. S2); phylogenetic trees displayed consistent results including topologies of groups and numbers as well as positions of genes in corresponding groups. Group AAI-a was the largest with 93 members; AAI-b contained 65 members; AAI-c had 45 members; AAI-d had 48 members; and AAI-e contained 85 AAI genes. Most importantly, groups AAI-a and AAI-b were only present in dicotyledons and monocotyledons, with a lack of AAI genes from moss and fern. Similarly, groups AAI-c and AAI-d were present in dicotyledons, moss, and ferns, with a lack of AAI genes from monocotyledons. AAI-e was the only group containing AAI genes from moss, fern, dicotyledon, and monocotyledon plant species, illustrating that its evolution occurred before the separation of monocots, dicots, moss, and ferns. The other four AAI groups most probably emerged after the separation of moss, fern, monocot, and dicot plant species.
Fig. 1.
Phylogenetic analysis by the NJ method and number of AAI genes in nine plant species. The phylogenetic tree resolved all AAI genes from monocots, dicots, moss, and ferns into five major groups from AAI-a to AAI-e. The prefixes At, Ga, Gh, Gr, Tc, Os, Zm, Pp, and Sm were used before the names of A. thaliana, G. arboreum, G. hirsutum, G. raimondii, T. cacao, O. sativa, Z. mays, P. patens, and S. moellendorffii AAI genes, respectively. Bootstrap values are noted near nodes of each branch. (This figure is available in color at JXB online.)
According to the phylogenetic tree, most orthologous genes between allotetraploids and diploids were clustered close to each other in the same group, showing expansion of the AAI gene family. Further investigation indicated that there were ~71 orthologous gene pairs between allotetraploid and A-genome diploid cotton, while there were 74 orthologous pairs between allotetraploid and D-genome diploid cotton. Additionally, 113 orthologous pairs were identified within At and Dt subgenomes of allotetraploid cotton. Based on this analysis, we deduced that there were more than twice as many GhAAI genes than GaAAI and GrAAI genes, and that this might be the result of wide-ranging duplication events during polyploidization in G. hirsutum. Moreover, many closely clustered orthologous gene pairs were observed in all studied plant species including Arabidopsis, G. arboreum, G. raimondii, T. cacao, O. sativa, Z. mays, P. patens, and S. moellendorffii. These findings show that gene duplication was the main contributor to expansion of the AAI gene family in the plant kingdom.
It is an established fact that allotetraploid cotton (G. hirsutum) was derived as a result of polyploidization and hybridization between A (G. arboreum) and D (G. raimondii) diploid cotton genomes. To test this hypothesis, we constructed a phylogenetic NJ tree of AAI genes from three cotton species, namely G. hirsutum, G. arboreum, and G. raimondii (Supplementary Fig. S3). The phylogenetic tree divided cotton AAI genes into four main groups, with each group containing the AAI genes from all three cotton species closely clustered, forming orthologous and paralogous gene pairs among and within genomes, respectively. These results strengthen our hypothesis and validate our findings that G. hirsutum evolved from hybridization between G. arboreum and G. raimondii, and subsequent polyploidization.
Protein motifs and gene structure analysis
To better understand the evolutionary relationship among GhAAI gene family members, we generated a NJ tree of all 122 GhAAI genes along with protein motif distribution (Supplementary Fig. S4A) and gene structure (Supplementary Fig. S4B). The GhAAI genes with similar motif distribution patterns were clustered close to each other and made up one clade. Ten different motifs were distributed in all GhAAI proteins, ranging from three to six motifs in each protein. Further, all GhAAI genes had a conserved motif distribution pattern: for instance, motif 4 and motif 5 were found in all proteins. Gene structure analysis showed that GhAAI genes contained three different kinds of proteins, namely LTP2, hydrophobic seed protein, and trypsin α-amylase protein. The genes encoding similar kinds of proteins and intron–exon arrangements were closely clustered, occupying the position in one clade. Further, some genes encoding LTP2 proteins had one to multiple introns in their gene structure, except for a few genes. Introns were observed in the gene structures of 36 genes. Moreover, 51 GhAAI genes encoded hydrophobic seed protein while only one gene (GhAAI4) encoded trypsin α-amylase protein. However, GhAAI genes encoding hydrophobic seed protein and trypsin α-amylase protein lacked introns. Collectively, from 122 GhAAI genes, three kinds of proteins are encoded and only 36 genes had introns in their structures.
Chromosomal location, gene duplication, and synteny analysis of GhAAI genes
We investigated the chromosomal location of 122 identified GhAAI genes on their corresponding chromosomes. Mapping results indicated that 51 GhAAI genes were located on At subgenome chromosomes while 58 GhAAI genes were on Dt subgenome chromosomes (Supplementary Fig. S5). Moreover, 13 GhAAI genes were present in different scaffolds. The highest number of genes were located on A11 (11 GhAAI genes) and its orthologous chromosome D11 (14 GhAAI genes). However, no GhAAI genes were located on A06, D03, or D06 chromosomes. Absence of GhAAI genes on these chromosomes, uneven distributions, as well as scaffold locations of GhAAI genes illustrated translocation during evolution and incomplete genome sequencing.
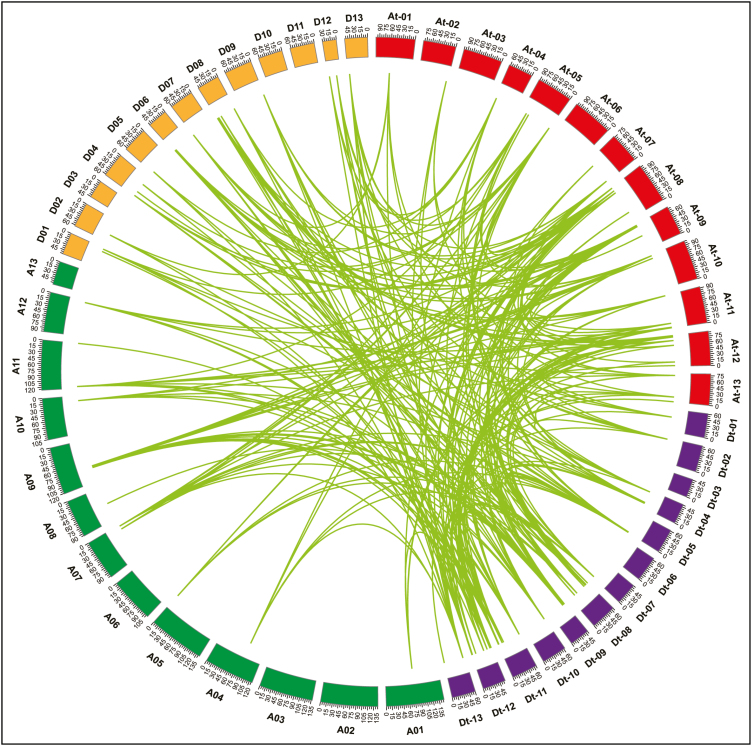
Phylogenetic analysis revealed the existence of numerous orthologous and paralogous gene pairs generated by gene duplication, so we further explored the locus relationships among and within At and Dt subgenomes as well as with A and D diploid cotton genomes. Based on syntenic relationships, there are 42 paralogous gene pairs identified as a result of segmental duplication (Fig. 2; Supplementary Table S3). Whole-genome duplication (WGD) resulted in 216 orthologous gene pairs and, of these, there were 32 pairs between the At subgenome and A genome, 36 pairs between the At subgenome and D genome, 39 pairs between the Dt subgenome and A genome, 38 pairs between the Dt subgenome and D genome, 23 pairs between orthologous chromosomes, and 48 orthologous gene pairs between non-orthologous chromosomes. However, no tandem duplication events were observed. Notably, most GhAAI genes formed orthologous or paralogous pairs with multiple genes. For instance, GhAAI66 and GhAAI18 accounted for 16 and 15 orthologous or paralogous pairs with other cotton AAI genes, respectively, demonstrating the significant contribution of gene duplication in cotton AAI gene family expansion. These results strengthen the idea that G. hirsutum was derived from hybridization of two diploids resembling G. arboreum and G. raimondii (Wendel and Cronn, 2003; Li et al., 2015). Here, we presumed that the high proportion of GhAAI genes from WGD as well as segmental duplication also contributed significantly in the expansion of the GhAAI gene family.
Fig. 2.
Gene duplication and collinearity analysis among cotton AAI genes including G. hirsutum (At and Dt subgenome), G. arboreum (A-genome), and G. raimondii (D-genome). Lines connecting genes depict ortholog pairs diverged from the same ancestor. At-01 to At-13indicate At subgenome chromosomes, and Dt-01 to Dt-13 display Dt subgenome chromosomes. Similarly, A01 to A13 and D01 to D13 depict G. arboreum and G. raimondii chromosomes, respectively. (This figure is available in color at JXB online.)
Next, we estimated the nature and extent of selection pressure on duplicated gene pairs as duplicated gene pairs underwent functional divergence and exhibited neofunctionalization, subfunctionalization, or non-functionalization during evolution (Prince and Pickett, 2002). Results of non-synonymous (Ka) and synonymous (Ks) values showed that 244 duplicated pairs had Ka/Ks <1.0 and, of these, 168 duplicated pairs exhibited a Ka/Ks value <0.5. However, 14 duplicated gene pairs showed Ka/Ks values >1.0 (Supplementary Table S3). Collectively, cotton AAI duplicated genes experienced strong purifying selection pressure as a high proportion of Ka/Ks values of duplicated gene pairs were <1.0, demonstrating positive selection pressure.
Tissue-specific expression pattern and responses to phytohormone treatment
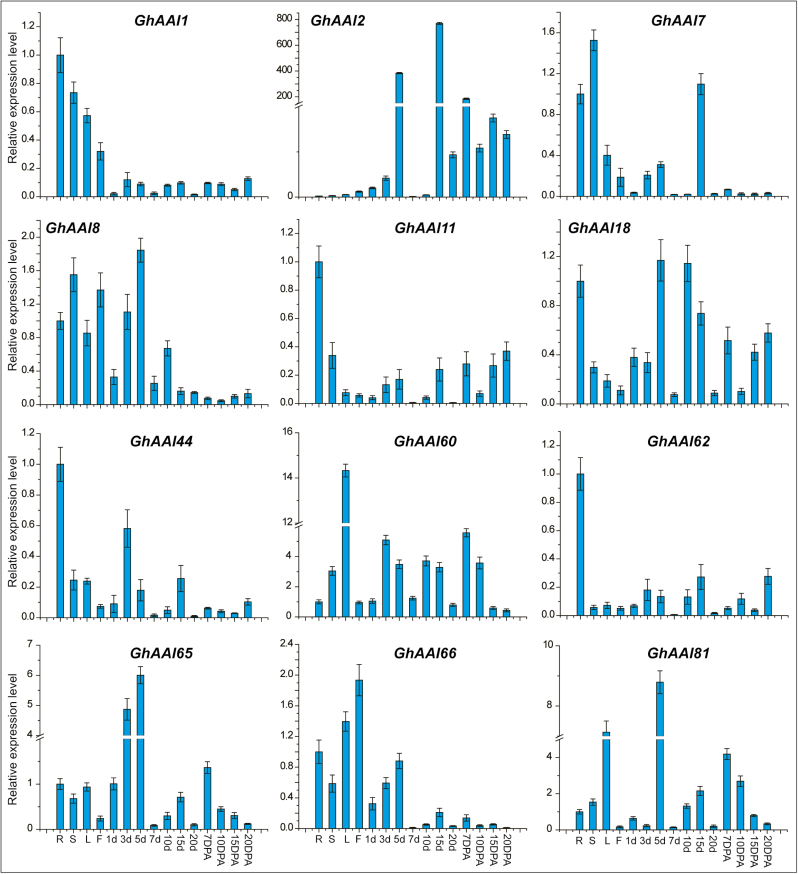
In order to determine the potential functions of GhAAI genes, we first identified the expression pattern of all 122 GhAAI genes in data of 22 different tissues obtained from published transcriptomes downloaded from NCBI, and generated a heat map (Supplementary Fig. S6). The results indicated that all genes exhibited ubiquitous expression with no specific pattern, and genes depicting similar expression patterns were closely clustered. However, only GhAAI6, GhAAI60, and GhAAI66 displayed significant expression values in all tissues. To validate these results, we selected 12 paralogous genes based on the fact that orthologous or paralogous genes usually exhibit similar functions (Altenhoff and Dessimoz, 2009). We analyzed the tissue-specific expression level of these 12 GhAAI genes in 15 different tissues including root, stem, leaf, flower, ovule (1, 3, 5, 7, 10, 15, and 20 DPA) and fiber (10, 7, 15, and 20 DPA) using qRT–PCR (Fig. 3). The results were in accordance with transcriptomic data, and the transcript levels of most genes were high in different vegetative tissues, except that expression of GhAAI2, GhAAI8, GhAAI18, GhAAI65, and GhAAI81 was high at different stages of ovule development. Importantly, for GhAAI66, which had maximum gene duplication, its expression level was high in flower, with a 2-fold increase, indicating its potential function during flower development.
Fig. 3.
Spatial expression pattern of GhAAI genes in different tissues using qRT–PCR analysis. Here, R, S, L, and F represent root, stem, leaf, and flower, respectively, while 1d, 3d, 5d, 7d, 10d, 15d, and 20d represent different days of ovule development. Further, 7, 10, 15, and 20 DPA indicate days of fiber development. Error bars indicate the SD of three independent biological repeats. (This figure is available in color at JXB online.)
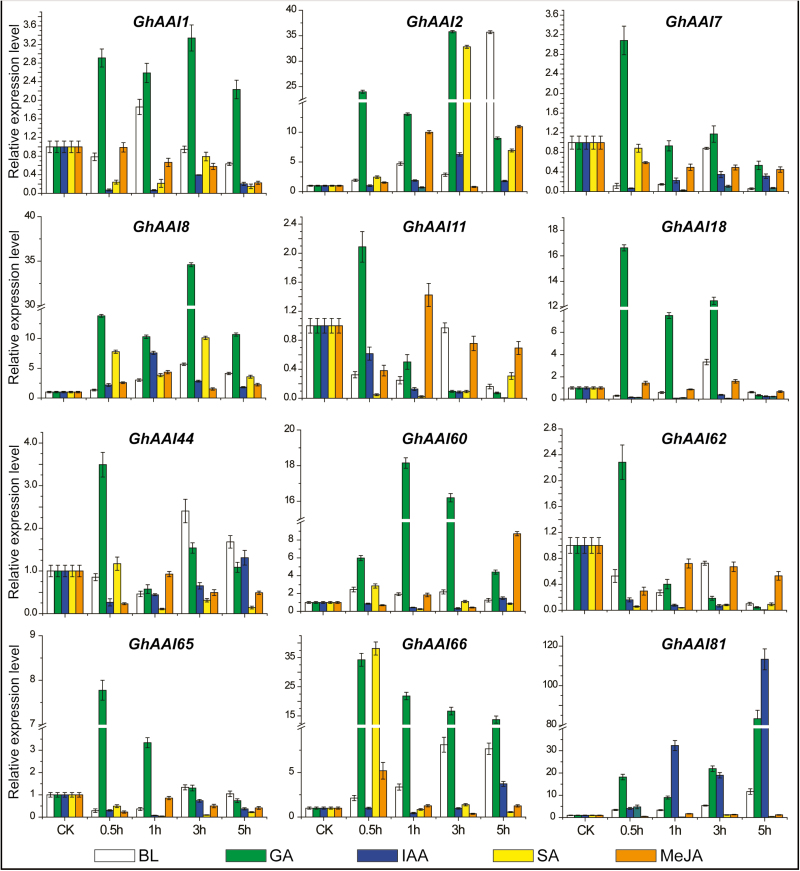
Next we investigated the responses of GhAAI genes under five different phytohormone treatments, namely BL, GA, IAA, SA, and MeJA, after 0.5, 1, 3, and 5 h using qRT–PCR (Fig. 4). GhAAI2, GhAAI8, and GhAAI66 were up-regulated across all hormonal treatments except at a few time points. Other observed genes showed ubiquitous responses without any specific pattern. Overall, all observed GhAAI genes were positively regulated at higher levels than the control following GA treatment, except at a few time points in some genes. Based on these findings, we assumed that GhAAI genes have potential functions in plant growth and development, and can be regulated by phytohormone treatments. Additionally, GhAAI66 exhibited higher transcript levels in flower tissues and under phytohormone responses, indicating that it might play an important role in plant growth, development, and phytohormonal response.
Fig. 4.
Responses of GhAAI genes after treating with BL, GA, IAA, SA, and MeJA at different time points performed by qRT–PCR analysis. The error bars show the SD of three independent biological repeats. (This figure is available in color at JXB online.)
Ectopic expression of GhAAI66 regulates phase transition to induce early flowering
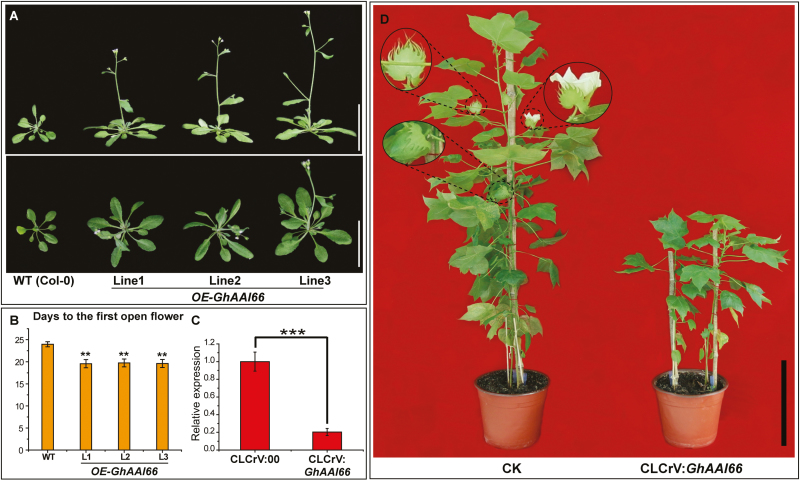
To determine the roles of GhAAI66 in flower development or regulation and transition from vegetative to reproductive growth, we overexpressed coding sequences under the control of the CaMV 35S promoter in Arabidopsis Col-0 plants. Compared with wild-type (WT) plants, transgenic plants overexpressing GhAAI66 showed early flowering (Fig. 5A). The statistical data indicated that OE-GhAAI66 lines opened their first flowers in 19.60±0.92 d as compared with WT plants where the first flower opened in 23.96±0.58 d (Fig. 5B), indicating the transition from vegetative to reproductive phase. Previously, the vegetative to reproductive phase transition was considered to have three phases: V-phase (vegetative rosette), I1-phase (inflorescence with cauline leaves subtending axillary branches), and I2-phase (an inflorescence bearing flowers) (Ratcliffe et al., 1998). As in OE-GhAAI66 lines, inflorescences bearing flowers were observed (Fig. 5A), so here we speculated that ectopic expression of GhAAI66 regulates the vegetative to reproductive phase transition through the I2-phase, which in turn induces early flowering.
Fig. 5.
Ectopic expression of GhAAI66 differentially impacts the phase transition in Arabidopsis. (A) Phenotypes of three independent OE-GhAAI66 transgenic lines revealed phase transition to induce early flowering. Scale bars are 5 cm. (B) Days from germination to first flower opening. Data are means (±SD) (n=20). (C) Relative expression level of GhAAI66 in control and CLCrV:GhAAI66 plants. Student’s t-test: *P<0.05, **P<0.01, ***P<0.001. (D) Phenotype of control and CLCrV:GhAAI66 plants. (This figure is available in color at JXB online.)
To identify the function of GhAAI66 for flowering in cotton, we conducted a VIGS experiment. First, we monitored the relative expression of GhAAI66 in CLCrV:00 and CLCrV:GhAAI66 plants to confirm the silenced expression of GhAAI66. The transcript level indicated that GhAAI66 expression was reduced in CLCrV:GhAAI66 plants (Fig. 5C). CLCrV:GhAAI66 plants had delayed flowering, while CLCrV:00 plants initiated flowering along with buds and developed cotton bolls; in addition, the plant height of CLCrV:GhAAI66 plants was suppressed significantly as compared with control plants (Fig 5D). These results strengthen our hypothesis that GhAAI66 regulates the vegetative to reproductive phase transition to induce early flowering.
GhAAI66 integrates multiple flower signaling pathways to induce early flowering
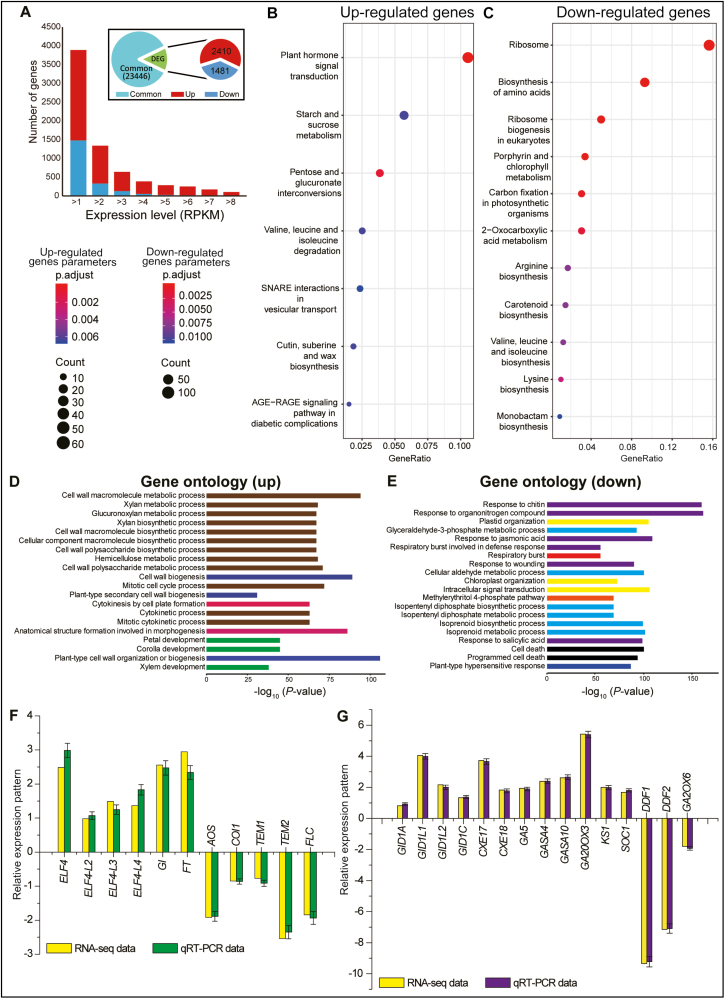
Next, to explore the mechanism by which ectopic expression of GhAAI66 regulates the vegetative to reproductive phase transition and induces early flowering, we conducted RNA-seq analysis of WT and OE-GhAAI66 lines. Among 27 337 expressed genes, ~3891 genes (14.23% of all expressed genes) were differentially expressed, comprising 2410 that were up-regulated (61.93% genes of all DEGs) and 1481 that were down-regulated (38.07% genes of all DEGs) in OE-GhAAI66 lines (Fig. 6A; Supplementary Table S4, S5). KEGG analysis of RNA-seq data indicated that genes for plant hormone signal transduction, starch and sucrose metabolism, and valine, leucine, and isoleucine (BCAA; branched chain amino acids) degradation were up-regulated (Fig. 6B). In contrast, genes for ribosomes, biosynthesis of amino acids, ribosome biogenesis, as well as valine, leucine, and isoleucine (BCAA) biosynthesis and many others were down-regulated (Fig. 6C). The up-regulation of genes for plant hormone signal transduction illustrated the involvement of hormone signaling pathways during the phase transition and early flowering induction. Findings during the past decade have elaborated that BCAA degradation provides energy for the early phase of germination and plays critical roles in maintaining amino acid homeostasis as well as normal seed development (Lu et al., 2011; Ding et al., 2012; Angelovici et al., 2013; Peng et al., 2015; Gipson et al., 2017). Here, genes for valine, leucine, and isoleucine degradation were up-regulated, while genes involved in their biosynthesis were down-regulated, demonstrating that BCAA homeostasis in OE-GhAAI66 lines might be an important contributor in the phase transition and early flowering. Further, GO enrichment showed that genes up-regulated in OE-GhAAI66 lines were mainly enriched in plant developmental processes as well as in cell wall metabolism, biosynthesis, and biogenesis processes (Fig. 6D). In contrast, down-regulated genes were involved in cell death and responses to chitin, JA, wounding, and SA (Fig. 6E).
Fig. 6.
Gene Ontology (GO) analysis of RNA-seq data of OE-GhAAI66 lines with respect to Col-0 Arabidopsis plants. (A) The expression map of the differentially expressed genes (DEGs). The bar graph shows the number of DEGs between Col-0 and OE-GhAAI66 plants. ‘Up’ and ‘down’ are the up-regulated and down-regulated genes in OE-GhAAI66 plants, respectively. (B) KEGG analysis of up-regulated genes from RNA-seq data. (C) KEGG analysis of down-regulated genes from RNA-seq data. Each experiment was conducted in three biological repeats. (D and E) Gene ontology enrichment of up- and down-regulated genes in OE-GhAAI66 plants. The lengths of the bars indicate the –log10-transformed P-values. (F, G) Relative expression pattern analysis of floral integrators, JA and GA receptors, biosynthesis, and catabolic genes by qRT–PCR analysis in order to validate RNA-seq data. (F) Up- and down-regulated floral integrators and JA receptor and biosynthesis genes. (G) Up- and down-regulated GA receptor and biosynthesis, responsive, and catabolic genes. Data are the log2fold change. Each experiment was conducted with three biological repeats. The error bars show the SD of three independent biological repeats. (This figure is available in color at JXB online.)
Deeper investigation indicated that ELF4 (EARLY FLOWERING 4), ELF4-L2 (ELF4-LIKE 2), ELF4-L3 (ELF4-LIKE 3), ELF4-L4 (ELF4-LIKE 4), GI (GIGANTEA), and FT (FLOWERING LOCUS T) were up-regulated in RNA-seq data (Fig. 6D). In contrast, the JA receptor COI1 (CORONATINE INSENSITIVE 1), biosynthesis gene AOS (ALLENE OXIDE SYNTHASE), and the three flowering repressors TEM1 (TEMPRANILLO 1), TEM2 (TEMPRANILLO 2), and FLC (FLOWERING LOCUS C) were down-regulated. These results were further validated by qRT–PCR analysis (Fig. 6F). As GA plays a positive role in flowering induction (Sun and Gubler, 2004) and because in our previous results most GhAAI genes including GhAAI66 had a positive response during GA treatment, we further investigated the mechanism to determine whether GA is also playing a part in the GhAAI66-induced phase transition and early flowering in OE-GhAAI66 lines as GA seemed to be acting upstream of AAI genes during GA treatment. We found that gibberellin receptor GID1A (GIBBERELLIN INSENSITIVE DWARF 1A), GID1C (GIBBERELLIN INSENSITIVE DWARF 1C), GID1L1 (GID1-LIKE1), GID1L2 (GID1-LIKE2), CXE17 (CARBOXYESTERASE 17), CXE18 (CARBOXYESTERASE 18), gibberellin biosynthetic GA5 (AtGA20ox1), GA2OX3 (GA 2-oxidase 3), KS1 (ent-Kaurene synthase 1), gibberellin responsive GASA4 (GIBBERELLIC ACID STIMULATED ARABIDOPSIS 4), GASA10 (GIBBERELLIC ACID STIMULATED ARABIDOPSIS 10), and SOC1 (SUPPRESSOR OF OVEREXPRESSION CONSTANS1) were up-regulated while DDF1 (DWARF AND DELAYED FLOWERING 1), DDF2 (DWARF AND DELAYED FLOWERING 2), and gibberellin catabolic GA2OX6 (GA 2-oxidase 6) were down-regulated in RNA-seq data. These results were also confirmed by qRT–PCR analysis (Fig. 6G).
Discussion
AAI genes form a large family in many plant species. For example, there are 68 AAIs in Arabidopsis and 122 in allotetraploid cotton G. hirsutum. The members of this gene family encode three domains, namely the LTP2 domain, the hydrophobic seed domain, and the trypsin α-amylase domain. Despite this, no systematic study on the AAI gene family in any species had been conducted until now. There are only a few studies about AAI genes containing the LTP2 domain (Hsu et al., 2005; Chae et al., 2009; Guo et al., 2013; Deng et al., 2016; Jacq et al., 2017); however, there are no studies about AAI genes carrying the hydrophobic seed domain or trypsin α-amylase domain. Cotton is an important fiber crop and is the main source of fiber for the textile industry (Bao et al., 2011). Advances in cotton genomics and genetics allowed us to perform a systematic study on cotton AAI genes and to investigate their potential functions. This study will provide basic information for further investigation of cotton AAI gene functions.
AAI genes were highly conserved during evolution
In the current study, we classified 336 AAI genes from different dicotyledons (Arabidopsis, G. hirsutum, G. arboreum, G. raimondii, and T. cacao), monocotyledons (O. sativa and Z. mays), moss (P. patens), and fern (S. moellendorffii) into five major groups. The results of phylogenetic analysis using NJ and ME methods were consistent and validated our findings. With the exception of group AAI-e, all AAI groups showed advanced evolution as groups AAI-a and AAI-b lack fern and moss AAI genes and groups AAI-c and AAI-d lack AAI genes from monocots, highlighting their evolution after the separation of fern and moss or monocots, respectively. Group AAI-e members evolved after the separation of moss, fern, monocot, and dicot species. WOX and YABBY gene families exhibited conserved amino acid residues and were found to be evolutionarily conserved (Yang et al., 2017, 2018). Similarly, in our study, AAI domain sequences were highly conserved among dicot (Arabidopsis and G. hirsutum), monocot (O. sativa), moss (P. patens), and fern (S. moellendorffii).
Ten different motifs were distributed in all GhAAI proteins, ranging from three to six motifs in each protein, demonstrating that GhAAI genes displayed a conserved motif distribution pattern. Tandem duplication has been reported always to result in more introns and new genes (Iwamoto et al., 1998). In our study, no tandem duplication events were observed, while 36 out of 122 GhAAI genes had one to multiple introns and, surprisingly, all 36 genes encoded LTP2 protein. Introns might play essential roles during the evolution of different species (Roy and Gilbert, 2006). It has been reported that genes contained more introns at early stages of expansion, with introns being lost with passage of time (Roy and Penny, 2007), suggesting that more advanced families had fewer introns in their genome (Roy and Gilbert, 2005). Despite the fact that conserved exon/intron motifs are functionally important even with low sequence conservation, exon/intron patterns of many gene families are conserved (Frugoli et al., 1998). We thus speculated that these introns were not lost during evolution, but diverged at early expansion stages of evolution, while other genes lost their introns over evolutionary time.
Previous findings documented many gene families lacking, or with fewer, introns in their genes (Serrano et al., 2006; Qanmber et al., 2018; Zhang et al., 2018). Insertion/deletion events contribute to exon/intron structural differences that might be helpful to estimate evolutionary mechanisms (Lecharny et al., 2003). Introns are under weak selection pressure, and genes with no introns might evolve at a rapid rate, while genes with larger or more introns contributed to gain of function in evolution. Further, gene loss/addition by segmental or WGD as well as incomplete sequencing of genomes are the main reasons for uneven distributions of GhAAI genes in At and Dt chromosomes of allotetraploid cotton.
Expansion and duplication of the GhAAI gene family during evolution
Allotetraploid cotton (G. hirsutum) evolved as a result of hybridization between A (G. arboreum) and D (G. raimondii) cotton genomes and subsequent polyploidization ~5–10 million years ago (mya) (Li et al., 2015) and provides the best model to study polyploidy (Wendel and Cronn, 2003). Generally, functional divergence is contributed by gene duplication that is important for environmental adaptability and evolutionary mechanisms (Conant and Wolfe, 2008). Segmental and salicoid duplications enlarged many gene families in ancestral plants ~65 mya (Barakat et al., 2009; Wang et al., 2013). Two large segmental and small-scale tandem duplications generated novel genes during evolution that accounted for genomic complexities in the plant kingdom (Cannon et al., 2004).
We identified 122 GhAAI genes, which is more than double the number of AAI genes in G. arboreum and G. raimondii, respectively, illustrating the effect of polyploidy which resulted in more gene duplication in the GhAAI gene family as compared with AAI genes in other cotton species (G. arboreum and G. raimondii). The dramatic increase in GhAAI family members can be evaluated from AAI genes in G. arboreum and G. raimondii. Although GhAAI genes were increased, gene loss after hybridization during genomic arrangements and chromosome doubling occurred (Paterson et al., 2012). Additionally, the cotton genome underwent fewer arrangements with respect to paleopolyploid maize and Brassica (Gaeta et al., 2007; Woodhouse et al., 2010). Our study identified 42 paralogous and 216 orthologous gene pairs as a result of segmental duplication and WGD, respectively. Interestingly, GhAAI66 and GhAAI18 accounted for 16 and 15 orthologous/paralogous pairs, demonstrating a significant contribution of gene duplication in cotton AAI gene family expansion.
Segmental duplication is important during evolution, as many plant species have multiple duplicated chromosomal blocks (Cannon et al., 2004) Further, it has been reported that Arabidopsis experienced WGD twice (Wang et al., 2011). Moreover, malvids (cotton and cacao) displayed a common ancestor and underwent ancient duplication ~18–58 mya (Li et al., 2014). Many Arabidopsis gene families also show gene family expansion (Baumberger et al., 2003; Wang et al., 2008). Additionally, cotton RH2FE3, YABBY, WOX, GRAS, and MIKC-Type MADS-Box, sesame heat shock proteins, and soybean WRKY show enlargement as the result of segmental duplication and WGD (Yin et al., 2013; Dossa et al., 2016; Ren et al., 2017; Yang et al., 2017, 2018; Qanmber et al., 2018; Zhang et al., 2018). Gene duplication analysis of AAI genes revealed a common ancestor between the A-genome and At subgenome similarly to D-genome and Dt subgenome. We concluded that segmental duplication and WGD are attributes of expansion in the GhAAI gene family. These findings will provide understanding of chromosomal interactions, genetic evolution, and intergenomic hereditary information transfer.
GhAAI genes have ubiquitous expression in tissues and regulated hormone treatments
Few investigations have been conducted to explore biological and physiological functions of AAI genes in plant biology. For instance, LTP2 is involved in cuticle–cell wall interface integrity and in permeability of the etiolated hypocotyl (Jacq et al., 2017). LTP3 was positively regulated by MYB96 as it directly binds to the LTP3 promoter and is involved in plant tolerance to freezing and drought stress in Arabidopsis (Guo et al., 2013). Further, GhMYB7/9 mediates transcriptional regulation of the LTP3 gene during fiber development in cotton (Hsu et al., 2005). GhLTPG1 was located on the cell membrane and was highly expressed in elongating fibers and the outer integument of cotton ovules (Deng et al., 2016). However, no studies have been conducted on AAI genes encoding the hydrophobic seed domain or trypsin α-amylase domain.
We determined that GhAAI gene expression was ubiquitous in different tissues. However, the transcript level of most genes was high in different vegetative tissues, except that in GhAAI2, GhAAI8, GhAAI18, GhAAI65, and GhAAI81, expression was high at different stages of ovule development. Importantly, the GhAAI66 expression level was high in flower, indicating its potential function during flower development. Our results from qRT–PCR analysis and previously published RNA-seq and transcriptomic data results were reasonable and validated our findings.
Additionally, all GhAAI genes exhibited up- or down-regulation when treated with various phytohormones. For instance, GhAAI2, GhAAI8, and GhAAI66 were up-regulated under all hormonal treatments with few exceptions. Overall, all estimated GhAAI genes were positively regulated following GA exposure except for a few time points with some genes. These findings strengthen our hypothesis that GhAAI genes might play important roles in signaling of different hormones in addition to the fact that GhAAI genes can be regulated by phytohormone treatments. Taken together, we speculated that GhAAI genes play diverse roles in plant growth and development under different hormonal treatments.
GhAAI66 regulates early flowering in Arabidopsis
In our study, ectopic expression of GhAAI66 regulated the phase transition from vegetative to reproductive growth to induce early flowering as OE-GhAAI66 lines opened their first flowers in fewer days as compared with WT plants that flowered 4 d later, indicating the transition from vegetative to reproductive phase. We deduced that ectopic expression of GhAAI66 regulates the vegetative to reproductive phase transition through the I2-phase, which in turn induces early flowering. Moreover, silenced CLCrV:GhAAI66 plants showed delayed flowering resulting from late phase transition from vegetative to reproductive growth, supporting our findings that GhAAI66 regulates the vegetative to reproductive phase transition to induce early flowering.
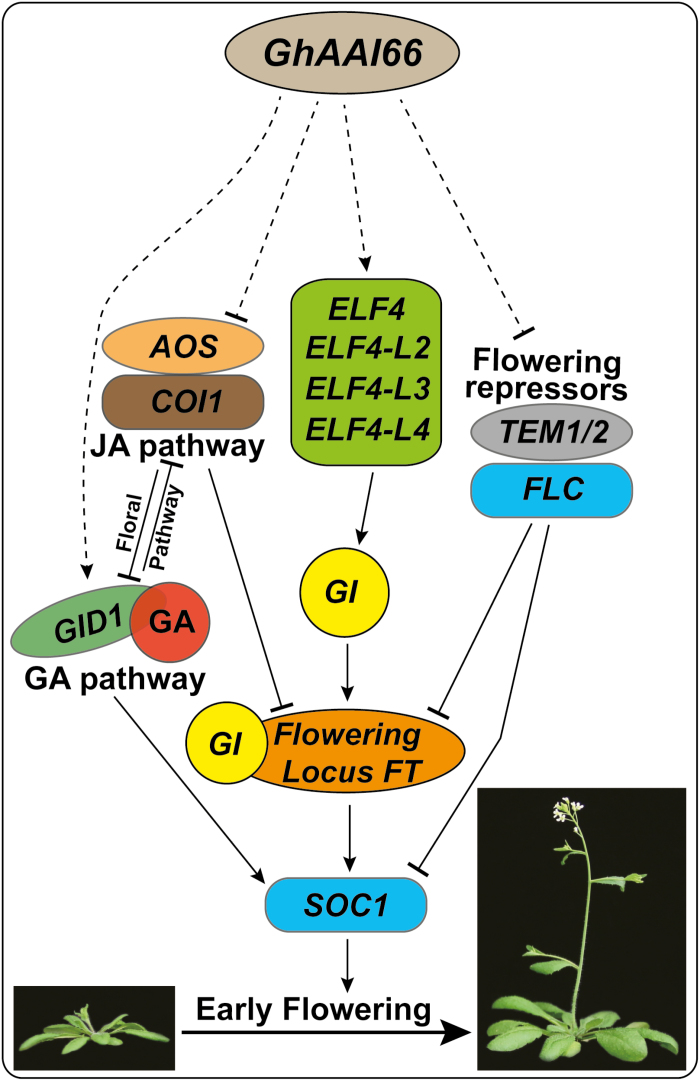
Decades of genetic research have revealed complex genetic networks for floral transition in Arabidopsis regulated by four genetic pathways: photoperiod, vernalization, autonomous, and GA-induced pathways (Simpson and Dean, 2002; Boss et al., 2004; Sung and Amasino, 2004; Baurle and Dean, 2006). The floral induction signals from these pathways are delivered to CONSTANS (CO) and FLC that antagonistically regulate flowering in Arabidopsis (Putterill et al., 1995; Samach et al., 2000). Here, GhAAI66 was shown to integrate multiple flower signaling pathways to induce early flowering in Arabidopsis, and we proposed a working model for GhAAI66 to induce flowering (Fig. 7). We found that expression of various GA receptor genes (GID1A, GID1C, GID1L1, GID1L2, CXE17, and CXE18), biosynthesis genes (GA5, GA2OX3, KS1, and SOC1), and enzymes was induced, while the expression of the GA catabolic gene GA2OX6 and JA receptor and biosynthesis genes (COI1 and AOS, respectively) was repressed. Previously, GA and JA were reported to antagonistically regulate flowering in Arabidopsis. JA promotes COI1-dependent degradation of JAZs, which in turn liberates transcriptional functions of the TOEs (TARGET of EAT1 and 2) to repress FT expression and mediate signaling cascades to delay flowering (Zhai et al., 2015). SOC1 is integrated with the GA pathway: for instance, the soc1 null mutant had reduced sensitivity to GA, and further SOC1 overexpression rescued the non-flowering phenotype of ga1-3 in Arabidopsis (Blazquez et al., 1998; Gocal et al., 2001). Even the genes responsive to SA were down-regulated in this study; it has previously been shown that SA regulates flowering time through photoperiod and autonomous pathways; however, it does not require functional flowering genes such as CO, FCA, and FLC (Martinez et al., 2004).
Fig. 7.
Proposed working model of GhAAI66 to induce the early flowering signaling cascade. (This figure is available in color at JXB online.)
The transcripts of ELF4 and its orthologous genes, GI, FT, and SOC1, were up-regulated in OE-GhAAI66 lines, whereas, previously, physical interaction proved ELF4 to be a regulator of GI nuclear distribution where GI binds the promoter of CO and directly activates the expression of FT (Sawa and Kay, 2011; Kim et al., 2013). Moreover, important flowering repressors including TEM1/2 and FLC were repressed in our transcriptomic data, and qRT–PCR analysis supported the previous findings that FLC protein interacts directly in vivo with SOC1 and FT (Helliwell et al., 2006), and prevents them from triggering a signal cascade of flower induction. For instance, overexpression of FLC completely blocked the activation of SOC by CO overexpression (Hepworth et al., 2002), whereas photoperiod deficiency in co and gi mutants was rescued when combined with the increased level of FLC generated by fca (Koornneef et al., 1998). Moreover, FT is negatively regulated by TEM1/2 and FLC (Zhai et al., 2015), and reports demonstrated that FT is the output of CO and SOC1 regulated through FT (Wigge et al., 2005; Yoo et al., 2005). Taken together, we concluded that GhAAI66 integrates multiple flower signaling pathways to induce early flowering in Arabidopsis.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Proposed names of putative AAI genes and their gene locus ID in A. thaliana, G. arboreum, G. hirsutum, G. raimondii, O. sativa, T. cacao, P. patens, S. moellendorffii, and Z. mays.
Table S2. Basic properties of GhAAI genes with locus ID, start and end point, strand, CDS, protein length, MW (molecular weight), pl (isoelectric point), GRAVY, as well as predicted subcellular localization.
Table S3. Gene duplication analysis in cotton.
Table S4. RNA-seq data of up-regulated genes in OE-GhAAI66 of three independent transgenic lines.
Table S5. RNA-seq data of down-regulated genes in OE-GhAAI66 of three independent transgenic lines.
Table S6. List of all primers used in this study for gene cloning and qPCR analysis.
Fig. S1. Conserved amino acid residue analysis among (A) Arabidopsis, (B) O. sativa, (C) G. hirsutum, (D) moss, and (E) fern AAI genes.
Fig. S2. Phylogenetic tree constructed using ME.
Fig. S3. The phylogenetic tree was generated using the NJ method for cotton including G. arboreum, G. hirsutum, and G. raimondii AAI genes in order to estimate the common ancestor hypothesis.
Fig. S4. Protein motif distribution and gene structure (exon/intron) analysis of GhAAI genes along with the phylogenetic tree inferred using the NJ method.
Fig. S5. Chromosomal location of GhAAI genes on different chromosomes.
Fig. S6. Expression pattern of GhAAI genes in different cotton tissues
Author contributions
ZY and FL designed the experiments and coordinated the project; GQ, LL, and ZL performed the experiments and analyzed the data; DY, KZ, and PH prepared the samples; and GQ, ZY, and FL wrote and revised the manuscript.
Acknowledgements
This project is supported by the Funds for Creative Research Groups of China (grant no. 31621005) and the Agricultural Science and Technology Innovation Program Cooperation and Innovation Mission (CAAS-XTCX2016). The authors declare that they have no competing interests.
References
- Altenhoff AM, Dessimoz C. 2009. Phylogenetic and functional assessment of orthologs inference projects and methods. PLoS Computational Biology 5, e1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biology 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelovici R, Lipka AE, Deason N, Gonzalez-Jorge S, Lin H, Cepela J, Buell R, Gore MA, Dellapenna D. 2013. Genome-wide analysis of branched-chain amino acid levels in Arabidopsis seeds. The Plant Cell 25, 4827–4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW. 2006. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Research 34, W369–W373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Hu GJ, Flagel LE, Salmon A, Bezanilla M, Paterson AH, Wang ZN, Wendel JF. 2011. Parallel up-regulation of the profilin gene family following independent domestication of diploid and allopolyploid cotton (Gossypium). Proceedings of the National Academy of Sciences, USA 108, 21152–21157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat A, Bagniewska-Zadworna A, Choi A, Plakkat U, DiLoreto DS, Yellanki P, Carlson JE. 2009. The cinnamyl alcohol dehydrogenase gene family in Populus: phylogeny, organization, and expression. BMC Plant Biology 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N, Doesseger B, Guyot R, et al. 2003. Whole-genome comparison of leucine-rich repeat extensins in Arabidopsis and rice. A conserved family of cell wall proteins form a vegetative and a reproductive clade. Plant Physiology 131, 1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäurle I, Dean C. 2006. The timing of developmental transitions in plants. Cell 125, 655–664. [DOI] [PubMed] [Google Scholar]
- Blazquez MA, Green R, Nilsson O, Sussman MR, Weigel D. 1998. Gibberellins promote flowering of arabidopsis by activating the LEAFY promoter. The Plant Cell 10, 791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss PK, Bastow RM, Mylne JS, Dean C. 2004. Multiple pathways in the decision to flower: enabling, promoting, and resetting. The Plant Cell 16(Suppl), S18–S31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon SB, Mitra A, Baumgarten A, Young ND, May G. 2004. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biology 4, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae K, Kieslich CA, Morikis D, Kim SC, Lord EM. 2009. A gain-of-function mutation of Arabidopsis lipid transfer protein 5 disturbs pollen tube tip growth and fertilization. The Plant Cell 21, 3902–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. 2008. Turning a hobby into a job: how duplicated genes find new functions. Nature reviews. Genetics 9, 938–950. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Research 14, 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng T, Yao H, Wang J, Wang J, Xue H, Zuo K. 2016. GhLTPG1, a cotton GPI-anchored lipid transfer protein, regulates the transport of phosphatidylinositol monophosphates and cotton fiber elongation. Scientific Reports 6, 26829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding G, Che P, Ilarslan H, Wurtele ES, Nikolau BJ. 2012. Genetic dissection of methylcrotonyl CoA carboxylase indicates a complex role for mitochondrial leucine catabolism during seed development and germination. The Plant Journal 70, 562–577. [DOI] [PubMed] [Google Scholar]
- Dossa K, Diouf D, Cissé N. 2016. Genome-wide investigation of hsf genes in sesame reveals their segmental duplication expansion and their active role in drought stress response. Frontiers in Plant Science 7, 1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frugoli JA, McPeek MA, Thomas TL, McClung CR. 1998. Intron loss and gain during evolution of the catalase gene family in angiosperms. Genetics 149, 355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaeta RT, Pires JC, Iniguez-Luy F, Leon E, Osborn TC. 2007. Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. The Plant Cell 19, 3403–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Britt RC Jr, Shan L, He P. 2011. Agrobacterium-mediated virus-induced gene silencing assay in cotton. Journal of Visualized Experiments 54, Doi: 10.3791/2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gipson AB, Morton KJ, Rhee RJ, et al. 2017. Disruptions in valine degradation affect seed development and germination in Arabidopsis. The Plant Journal 90, 1029–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocal GF, Sheldon CC, Gubler F, et al. 2001. GAMYB-like genes, flowering, and gibberellin signaling in Arabidopsis. Plant Physiology 127, 1682–1693. [PMC free article] [PubMed] [Google Scholar]
- Guo L, Yang H, Zhang X, Yang S. 2013. Lipid transfer protein 3 as a target of MYB96 mediates freezing and drought stress in Arabidopsis. Journal of Experimental Botany 64, 1755–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Wood CC, Robertson M, James Peacock W, Dennis ES. 2006. The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. The Plant Journal 46, 183–192. [DOI] [PubMed] [Google Scholar]
- Henderson IR, Shindo C, Dean C. 2003. The need for winter in the switch to flowering. Annual Review of Genetics 37, 371–392. [DOI] [PubMed] [Google Scholar]
- Hepworth SR, Valverde F, Ravenscroft D, Mouradov A, Coupland G. 2002. Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. The EMBO Journal 21, 4327–4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C-Y, Jenkins JN, Saha S, Ma D-P. 2005. Transcriptional regulation of the lipid transfer protein gene LTP3 in cotton fibers by a novel MYB protein. Plant Science 168, 167–181. [Google Scholar]
- Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G. 2015. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto M, Maekawa M, Saito A, Higo H, Higo K. 1998. Evolutionary relationship of plant catalase genes inferred from exon–intron structures: isozyme divergence after the separation of monocots and dicots. Theoretical and Applied Genetics 97, 9–19. [Google Scholar]
- Jacq A, Pernot C, Martinez Y, Domergue F, Payré B, Jamet E, Burlat V, Pacquit VB. 2017. The arabidopsis lipid transfer protein 2 (AtLTP2) is involved in cuticle–cell wall interface integrity and in etiolated hypocotyl permeability. Frontiers in Plant Science 8, 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia JT, Zhao PC, Cheng LQ, Yuan GX, Yang WG, Liu S, Chen SY, Qi DM, Liu GS, Li XX. 2018. MADS-box family genes in sheepgrass and their involvement in abiotic stress responses. BMC Plant Biology 18, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang HY, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. 1999. Activation tagging of the floral inducer FT. Science 286, 1962–1965. [DOI] [PubMed] [Google Scholar]
- Kim Y, Lim J, Yeom M, Kim H, Kim J, Wang L, Kim WY, Somers DE, Nam HG. 2013. ELF4 regulates GIGANTEA chromatin access through subnuclear sequestration. Cell Reports 3, 671–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. 1999. A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Blankestijn-de Vries H, Hanhart CJ, Peeters AJ. 1998. Genetic interactions among late-flowering mutants of Arabidopsis. Genetics 148, 885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Research 19, 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33, 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecharny A, Boudet N, Gy I, Aubourg S, Kreis M. 2003. Introns in, introns out in plant gene families: a genomic approach of the dynamics of gene structure. Journal of Structural and Functional Genomics 3, 111–116. [PubMed] [Google Scholar]
- Lee H, Suh SS, Park E, Cho E, Ahn JH, Kim SG, Lee JS, Kwon YM, Lee I. 2000. The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes & Development 14, 2366–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Lee I. 2010. Regulation and function of SOC1, a flowering pathway integrator. Journal of Experimental Botany 61, 2247–2254. [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. 2015. SMART: recent updates, new developments and status in 2015. Nucleic Acids Research 43, D257–D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Fan G, Lu C, et al. 2015. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nature Biotechnology 33, 524–530. [DOI] [PubMed] [Google Scholar]
- Li F, Fan G, Wang K, et al. 2014. Genome sequence of the cultivated cotton Gossypium arboreum. Nature Genetics 46, 567–572. [DOI] [PubMed] [Google Scholar]
- Li J, Fan SL, Song MZ, Pang CY, Wei HL, Li W, Ma JH, Wei JH, Jing JG, Yu SX. 2013. Cloning and characterization of a FLO/LFY ortholog in Gossypium hirsutum L. Plant Cell Reports 32, 1675–1686. [DOI] [PubMed] [Google Scholar]
- Li J, Yu D, Qanmber G, et al. 2019. GhKLCR1, a kinesin light chain-related gene, induces drought-stress sensitivity in Arabidopsis. Science China. Life Sciences 62, 63–75. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lu Y, Savage LJ, Larson MD, Wilkerson CG, Last RL. 2011. Chloroplast 2010: a database for large-scale phenotypic screening of Arabidopsis mutants. Plant Physiology 155, 1589–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez C, Pons E, Prats G, León J. 2004. Salicylic acid regulates flowering time and links defence responses and reproductive development. The Plant Journal 37, 209–217. [DOI] [PubMed] [Google Scholar]
- McGarry RC, Ayre BG. 2012. Geminivirus-mediated delivery of florigen promotes determinate growth in aerial organs and uncouples flowering from photoperiod in cotton. PLoS One 7, e36746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry RC, Prewitt S, Ayre BG. 2013. Overexpression of FT in cotton affects architecture but not floral organogenesis. Plant Signaling & Behavior 8, e23602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry RC, Prewitt SF, Culpepper S, Eshed Y, Lifschitz E, Ayre BG. 2016. Monopodial and sympodial branching architecture in cotton is differentially regulated by the Gossypium hirsutum SINGLE FLOWER TRUSS and SELF-PRUNING orthologs. New Phytologist 212, 244–258. [DOI] [PubMed] [Google Scholar]
- Mouradov A, Cremer F, Coupland G. 2002. Control of flowering time: interacting pathways as a basis for diversity. The Plant Cell 14(Suppl), S111–S130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson O, Lee I, Blázquez MA, Weigel D. 1998. Flowering-time genes modulate the response to LEAFY activity. Genetics 150, 403–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Wendel JF, Gundlach H, et al. 2012. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423–427. [DOI] [PubMed] [Google Scholar]
- Peng C, Uygun S, Shiu SH, Last RL. 2015. The impact of the branched-chain ketoacid dehydrogenase complex on amino acid homeostasis in arabidopsis. Plant Physiology 169, 1807–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prewitt SF, Ayre BG, McGarry RC. 2018. Cotton CENTRORADIALIS/TERMINAL FLOWER 1/SELF-PRUNING genes functionally diverged to differentially impact plant architecture. Journal of Experimental Botany 69, 5403–5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince VE, Pickett FB. 2002. Splitting pairs: the diverging fates of duplicated genes. Nature Reviews. Genetics 3, 827–837. [DOI] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G. 1995. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80, 847–857. [DOI] [PubMed] [Google Scholar]
- Qanmber G, Yu D, Li J, Wang L, Ma S, Lu L, Yang Z, Li F. 2018. Genome-wide identification and expression analysis of Gossypium RING-H2 finger E3 ligase genes revealed their roles in fiber development, and phytohormone and abiotic stress responses. Journal of Cotton Research 1, 1. [Google Scholar]
- Ratcliffe OJ, Amaya I, Vincent CA, Rothstein S, Carpenter R, Coen ES, Bradley DJ. 1998. A common mechanism controls the life cycle and architecture of plants. Development 125, 1609–1615. [DOI] [PubMed] [Google Scholar]
- Ratcliffe OJ, Riechmann JL. 2002. Arabidopsis transcription factors and the regulation of flowering time: a genomic perspective. Current Issues in Molecular Biology 4, 77–91. [PubMed] [Google Scholar]
- Ren Z, Yu D, Yang Z, et al. 2017. Genome-wide identification of the MIKC-Type MADS-Box gene family in Gossypium hirsutum L. unravels their roles in flowering. Frontiers in Plant Science 8, 384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SW, Gilbert W. 2005. Complex early genes. Proceedings of the National Academy of Sciences, USA 102, 1986–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SW, Gilbert W. 2006. The evolution of spliceosomal introns: patterns, puzzles and progress. Nature Reviews. Genetics 7, 211–221. [DOI] [PubMed] [Google Scholar]
- Roy SW, Penny D. 2007. A very high fraction of unique intron positions in the intron-rich diatom Thalassiosira pseudonana indicates widespread intron gain. Molecular Biology and Evolution 24, 1447–1457. [DOI] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G. 2000. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288, 1613–1616. [DOI] [PubMed] [Google Scholar]
- Sawa M, Kay SA. 2011. GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 108, 11698–11703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M, Parra S, Alcaraz LD, Guzmán P. 2006. The ATL gene family from Arabidopsis thaliana and Oryza sativa comprises a large number of putative ubiquitin ligases of the RING-H2 type. Journal of Molecular Evolution 62, 434–445. [DOI] [PubMed] [Google Scholar]
- Simpson GG, Dean C. 2002. Arabidopsis, the Rosetta stone of flowering time? Science 296, 285–289. [DOI] [PubMed] [Google Scholar]
- Sun TP, Gubler F. 2004. Molecular mechanism of gibberellin signaling in plants. Annual Review of Plant Biology 55, 197–223. [DOI] [PubMed] [Google Scholar]
- Sung S, Amasino RM. 2004. Vernalization and epigenetics: how plants remember winter. Current Opinion in Plant Biology 7, 4–10. [DOI] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Research 34, W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Guo Y, Wu C, Yang G, Li Y, Zheng C. 2008. Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice. BMC Genomics 9, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang H, Wang J, et al. 2011. The genome of the mesopolyploid crop species Brassica rapa. Nature Genetics 43, 1035–1039. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhang H, Yang J, Chen Y, Xu X, Mao X, Li C. 2013. Phylogenetic, expression, and bioinformatic analysis of the ABC1 gene family in Populus trichocarpa. ScientificWorldJournal 2013, 785070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel JF, Cronn RC. 2003. Polyploidy and the evolutionary history of cotton. Advances in Agronomy 78, 139–186. [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. 2005. Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309, 1056–1059. [DOI] [PubMed] [Google Scholar]
- Woodhouse MR, Schnable JC, Pedersen BS, Lyons E, Lisch D, Subramaniam S, Freeling M. 2010. Following tetraploidy in maize, a short deletion mechanism removed genes preferentially from one of the two homologs. PLoS Biology 8, e1000409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution 24, 1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang Z, Gong Q, Qin W, Yang Z, Cheng Y, Lu L, Ge X, Zhang C, Wu Z, Li F. 2017. Genome-wide analysis of WOX genes in upland cotton and their expression pattern under different stresses. BMC Plant Biology 17, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Gong Q, Wang L, et al. 2018. Genome-wide study of YABBY genes in upland cotton and their expression patterns under different stresses. Frontiers in Genetics 9, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Zhang C, Yang X, et al. 2014. PAG1, a cotton brassinosteroid catabolism gene, modulates fiber elongation. New Phytologist 203, 437–448. [DOI] [PubMed] [Google Scholar]
- Yanovsky MJ, Kay SA. 2003. Living by the calendar: how plants know when to flower. Nature Reviews. Molecular Cell Biology 4, 265–275. [DOI] [PubMed] [Google Scholar]
- Yi X, Du Z, Su Z. 2013. PlantGSEA: a gene set enrichment analysis toolkit for plant community. Nucleic Acids Research 41, W98–W103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin GJ, Xu HL, Xiao SY, Qin YJ, Li YX, Yan YM, Hu YK. 2013. The large soybean (Glycine max) WRKY TF family expanded by segmental duplication events and subsequent divergent selection among subgroups. BMC Plant Biology 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SK, Chung KS, Kim J, Lee JH, Hong SM, Yoo SJ, Yoo SY, Lee JS, Ahn JH. 2005. Constans activates suppressor of overexpression of constans 1 through flowering locus T to promote flowering in Arabidopsis. Plant Physiology 139, 770–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q, Zhang X, Wu F, Feng H, Deng L, Xu L, Zhang M, Wang Q, Li C. 2015. Transcriptional mechanism of jasmonate receptor COI1-mediated delay of flowering time in arabidopsis. The Plant Cell 27, 2814–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Liu J, Yang ZE, Chen EY, Zhang CJ, Zhang XY, Li FG. 2018. Genome-wide analysis of GRAS transcription factor gene family in Gossypium hirsutum L. BMC Genomics 19, 348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Hu Y, Jiang W, et al. 2015. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nature Biotechnology 33, 531–537. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.