Abstract
Autophagy plays a critical role in plants under biotic stress, including the response to pathogen infection. We investigated whether autophagy-related genes (ATGs) are involved in infection with Bamboo mosaic virus (BaMV), a single-stranded positive-sense RNA virus. Initially, we observed that BaMV infection in Nicotiana benthamiana leaves upregulated the expression of ATGs but did not trigger cell death. The induction of ATGs, which possibly triggers autophagy, increased rather than diminished BaMV accumulation in the leaves, as revealed by gene knockdown and transient expression experiments. Furthermore, the inhibitor 3-methyladenine blocked autophagosome formation and the autophagy inducer rapamycin, which negatively and positively affected BaMV accumulation, respectively. Pull-down experiments with an antibody against orange fluorescent protein (OFP)-NbATG8f, an autophagosome marker protein, showed that both plus- and minus-sense BaMV RNAs could associate with NbATG8f. Confocal microscopy revealed that ATG8f-enriched vesicles possibly derived from chloroplasts contained both the BaMV viral RNA and its replicase. Thus, BaMV infection may induce the expression of ATGs possibly via autophagy to selectively engulf a portion of viral RNA-containing chloroplast. Virus-induced vesicles enriched with ATG8f could provide an alternative site for viral RNA replication or a shelter from the host silencing mechanism.
Keywords: ATG5, autophagy, BaMV, chloroplast, chlorophagy, 3-MA, rapamycin, viral RNA replication
In cells of Nicotiana benthamiana infected with Bamboo mosaic virus, autophagy is hijacked by the virus to provide an alternative site for viral RNA replication or a shelter from the host silencing mechanism.
Introduction
Autophagy is a highly conserved intracellular self-degradation system in eukaryotes that differs from the ubiquitin–proteasomal pathway mechanism (Levine and Klionsky, 2004; Lin et al., 2010; Michaeli et al., 2016; Zientara-Rytter and Sirko, 2016). In addition to nutrient recycling, autophagy is involved in numerous physiological reactions including control of cell growth, starvation, anti-ageing, and defense mechanisms (Levine and Klionsky, 2004; Masclaux-Daubresse, 2016). Previous studies have explored the physiological mechanisms of autophagy in yeast, mammalian cells, and plants, but the detailed interactions between autophagy and pathogens in plants have only recently been clarified (Clavel et al., 2017; Wang et al., 2018). In general, under nutrient-poor conditions or pathogen attack, the autophagy-related gene (ATG)1/ATG13 kinase complex is activated to promote ATG9-mediated lipid delivery to the developed phagophore and to initiate the nucleation of the autophagosome with the addition of phosphatidyl inositol-3 phosphate. The key factor ATG8 is modified with phosphatidylethanolamine by the ATG12–ATG5/ATG16 complex and docked onto the expanding vesicle. Finally, the ATG8-containing vesicle engulfs the cargo, including damaged organelles, and is transported to the vacuole (Li and Vierstra, 2012; Masclaux-Daubresse et al., 2017).
The hypersensitive-response programmed cell death (HR-PCD) induced by the pathogens Pseudomonas syringae pv. tomato (Pst) DC3000 (AvrRps4) and Pst DC3000 (AvrRpm1) in Arabidopsis is suppressed in atg mutants (Hofius et al., 2009). In AtATG6-antisense and atg5 mutant plants, HR-PCD induced by Pst DC3000 (AvrRpm1) cannot be contained at the infection site, and it spreads to uninfected cells (Patel and Dinesh-Kumar, 2008). Similarly, ATG6/beclin1-silenced Nicotiana benthamiana plants carrying the N resistance gene are not able to restrict the HR-PCD response after infection with Tobacco mosaic virus (TMV), which results in unrestricted PCD of the uninfected tissue (Liu et al., 2005). These results indicate that autophagy plays a defense role against pathogen invasion by inducing HR-PCD. However, some viruses can benefit from the formation and maturation of autophagosomes when HR-PCD is negatively regulated after viral invasion, such as in infection by the Hepatitis C virus (Mizui et al., 2010), dengue virus, or Japanese encephalitis virus (Jin et al., 2013).
Besides regulating the HR-PCD defense mechanism, autophagy is also involved in some physiological functions. Transgenic plants with ATG knockdown, ATG18a-RNAi, have been shown to be defective in autophagosome formation and under increased oxidative stress when treated with H2O2 or the inducer of reactive oxygen species (ROS) methyl viologen (Xiong et al., 2007). ATG8-overexpression lines positively regulate their nutrient recycling, cell growth, and root-hair formation (Yano et al., 2007). The characteristics of knockout lines of Arabidopsis have shown that ATG6 is an essential gene, and heterozygous lines are defective in male gametophytes (Fujiki et al., 2007; Harrison-Lowe and Olsen, 2008; Patel and Dinesh-Kumar, 2008). In addition, autophagy is involved in chloroplast maturation; specifically, dark-treated leaves show decreased number and size of chloroplasts (Wada et al., 2009).
Chloroplasts in plant cells may play a defensive role by producing ROS that target invading pathogens (Li et al., 2016). Viruses have various close interactions with chloroplasts. Pathogens may target the ROS-producing proteins as a counter-defense mechanism, examples of which include Turnip mosaic virus (TuMV) (Wei et al., 2010) and Turnip yellow mosaic virus (TYMV) (Prod’homme et al., 2003) that associate with the chloroplast envelope during infection, pathogen-encoded proteins such as coat proteins of TMV (Reinero and Beachy, 1986), Cucumber necrosis virus (CNV) (Xiang et al., 2006) and Potato virus X (PVX) (Qiao et al., 2009) inside chloroplasts, and movement proteins of Potato mop-top virus (Cowan et al., 2012), Barley stripe mosaic virus (Torrance et al., 2006), and Alternanthera mosaic virus (AltMV) (Jang et al., 2013) that associate with the chloroplast membrane. However, the second layer of the host defense system would be triggered when chloroplasts are targeted by viral proteins. The destruction of chloroplasts after pathogen infection typically causes chlorosis symptoms and is proposed to be achieved by chlorophagy, a selective autophagy in chloroplast degradation (Dong and Chen, 2013).
Bamboo mosaic virus (BaMV) is a single-stranded positive-sense RNA virus belonging to Potexvirus of the family Alphaflexiviridae. The genome has five ORFs: ORF1 encodes the protein for viral RNA replication (Li et al., 1998, 2001a, 2001b; Huang et al., 2004; Meng and Lee, 2017); ORF2–ORF4 encode the proteins for cell-to-cell and systemic movement (Lin et al., 2004, 2006); and ORF5 encodes the capsid protein for encapsidation, symptom development, and viral movement (Lan et al., 2010; Hung et al., 2014a, 2014b). Host factors participating in the infection cycle of BaMV in N. benthamiana have been identified by using ultraviolet crosslinking (Lin et al., 2007; Prasanth et al., 2011; Huang et al., 2012), yeast two-hybrid (Lee et al., 2011), and cDNA-amplified fragment length polymorphism (Cheng et al., 2010) techniques. NbGSTU4 interacts with the 3′ UTR of BaMV RNA and enhances the minus-sense RNA synthesis at the early stage of BaMV replication by delivering glutathione to the replication complex to create a reduction state (Chen et al., 2013). NbHsp90 directly interacts with the 3′ UTR of BaMV and specifically regulates the synthesis of genomic RNA at the initial stage of BaMV infection (Huang et al., 2012). The chloroplast phosphoglycerate kinase interacts with the 3′ UTR of BaMV genomic RNA, targeting chloroplasts for RNA replication (Lin et al., 2007; Cheng et al., 2013; Huang et al., 2017).
Because BaMV targets chloroplasts, in this study we examined whether infection could trigger the chloroplast-associated pathogen response and induce autophagy as a viral defense. We found that the expression of ATGs was up-regulated after BaMV inoculation, and that the addition of inhibitors or knockdown of ATG expression could down-regulate BaMV replication. Thus, proteins encoded by these ATGs could play positive roles in facilitating BaMV RNA replication.
Materials and methods
The detection of ATGs
Total RNA was extracted from leaves or protoplasts using Tri-reagent (Clontech) and reverse-transcribed with primer (dT)25 by using the Im-PromII Reverse Transcription System (Promega). The ATGs (NbATG3, NbATG7, NbATG8f, NbATG12A, NbATG12B, and NbPI3K) were amplified using the primers listed in Supplementary Table S1 at JXB online. The expression of actin as an internal control.
Tobacco rattle virus (TRV)-based virus-induced gene silencing
The partial sequence of NbATG8f was PCR-amplified with specific primers for ATG8fF and ATG8fR and cloned into a TRV-based virus-induced gene silencing vector. The silencing efficiency was determined by qRT-PCR with the primer set ATG8fKDF and ATG8fKDR (Supplementary Table S1). Agrobacteria containing the plasmids TRV1, TRV2-PDS, TRV2-Luc (Luciferase), and TRV2-NbATG8f were cultured and mixed with TRV1 in a 1:1 ratio and infiltrated into N. benthamiana leaves as described previously (Chen et al., 2013). The BaMV, PVX, or CMV viral particle (500 ng) was inoculated onto the upper leaves of infiltrated leaves at ~10–14 d post agroinfiltration. The accumulation of coat protein was determined by western blot analysis at 5 d post-inoculation (dpi). NbATG8f-knockdown protoplasts isolated from the knockdown plants were inoculated with 2 µg BaMV viral RNA. The accumulation of BaMV coat protein and RNA was determined at 24 h post inoculation (hpi).
Total RNA extracted from BaMV-inoculated leaves or protoplasts was glyoxalated, separated on a 1% agarose gel, and transferred to nitrocellulose membranes as described by Lin et al. (1992). RNA positive and negative probes for BaMV were prepared from the plasmid pBL2.6 (Tsai et al., 1999) digested with HindIII to generate a 600-nt transcript complementary to the 3′-terminal region of the BaMV positive-sense genome, and plasmid pBaMV.S (Lin et al., 2005) digested with EcoRI to generate a 500-nt transcript complementary to the 3′-terminal region of the BaMV negative-sense genome.
Transient expression of NbATG8f and NbATG5
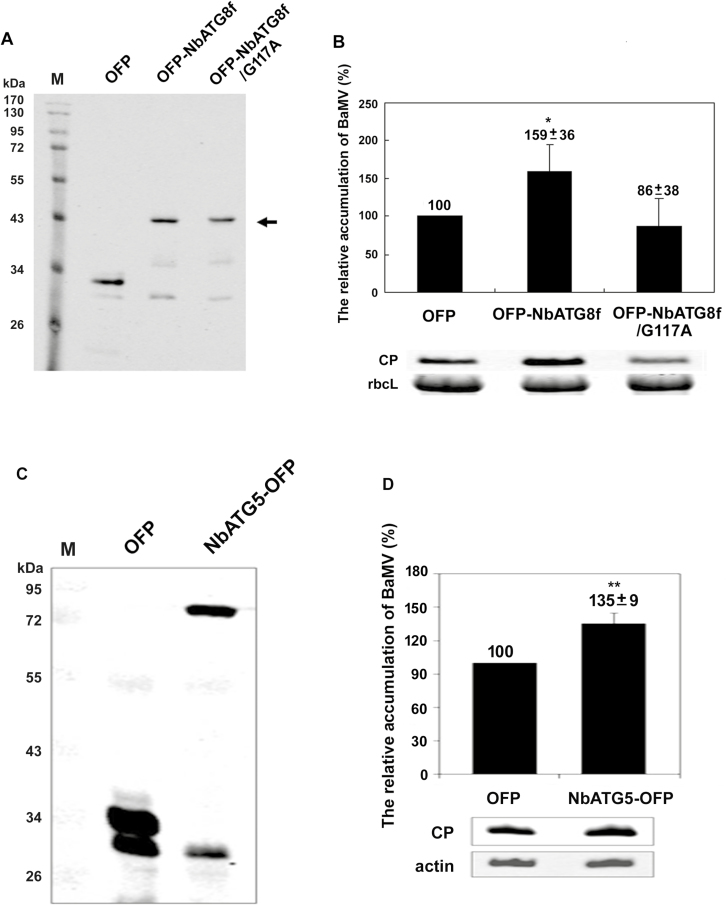
Full-length cDNA of NbATG8f and NbATG5 was PCR-amplified from total cDNA derived from N. benthamiana with the primer sets ATG8fcDNAF/ATG8fcDNAR and ATG5cDNAF /ATG5cDNAR, respectively (Supplementary Table S1). The mutant construct NbATG8f/G117A, which fails to associate with phosphatidylethanolamine (PE) to initiate the autophagosome formation, was PCR-amplified with ATG8fcDNAF and ATG8fcDNA-G117AR (5′-GGTACCCTACAGCTTGTTCAGGTCAGCG-3′). The BaMV coat protein was detected at 3 dpi in N. benthamiana leaves with transient expression of orange fluorescent protein (OFP)-NbATG8f and OFP-NbATG8f/G117A as a dominant negative control and OFP as a negative control. At 5 dpi, coat protein was detected in leaves with transient expression of NbATG5-OPF.
Detection of autophagosome-like structures
OFP-NbATG8f was transiently expressed by agro-infiltration in N. benthamiana leaves for 3 d, and infiltrated leaves were then inoculated with 500 ng BaMV, and treated with either 10 mM H2O2 as a positive control or inoculated with buffer (Mock) as a negative control. Images were obtained from the mock-, H2O2-, and BaMV-treated leaves at 1 d post-treatment by confocal laser-scanning microscopy. The number of granules obtained from the signal of OFP-NbATG8f was counted using the Image J software.
Treatment with an autophagy inhibitor or inducer
Protoplasts isolated from healthy N. benthamiana plants were inoculated with 2 µg BaMV, PVX, or CMV virion RNA (Tsai et al., 1999) and incubated with a medium (0.55 M mannitol-MES pH 5.7, 1 µM CuSO4, 10 mM MgSO4, 1 µM KI, 0.2 mM KPO4, 10 mM KNO3 pH 6.5, 10 mM CaCl2, 10 mg ml–1 gentamycin) containing the PI3K inhibitor 3-methyladenine (3-MA) 10 mM or the autophagy inducer rapamycin 100 nM. Accumulation of the viral coat protein and RNA was determined at 24 hpi.
Pull-down assays
Agrobacteria containing pBIN61/OFP (OFP only), pBIN61/OFP-NbATG8f, and pBIN61/OFP-NbATG8f/G117A were cultured and induced with 450 μM acetosyringone in 10 mM MgCl2 to a final concentration of OD600=1. Each of the constructs was mixed with agrobacteria containing pBIN61/HcPro in a 1: 1 ratio and infiltrated into N. benthamiana leaves. At 4 hpi, 500 ng BaMV viral particles were inoculated onto infiltrated leaves by rubbing on an abrasive suspension. Pull-down assays were performed at 5 dpi. The OFP and OFP-fusion proteins were extracted with buffer (20 mM Tris pH7.5, 2 mM MgCl2, 300 mM NaCl, 5 mM DTT, 2.5% PVPP, and 1× protease inhibitor) and purified by using a RFP-Trap_M kit (Chromotek). RNA was isolated from the pull-down extracts by using TriPure isolation reagent (Roche Diagnostics). cDNAs were synthesized by using oligo dT(25) with the ImProm-II Reverse Transcription System (Promega). BaMV RNA signals were detected by PCR after cDNA synthesis (reverse-transcribed with oligo dT primer) with the primer set BaMV-2002 5′-ATGTATCACGGAAATAAGAGTT-3′ and BaMV+1766 5′-CACATCCGGCACTTACCA-3′ (Chen et al., 2010). The RNA minus-sense BaMV was detected by PCR with the same set of primers after reverse-transcription by using BaMV+1766 primer. The pull-down proteins were examined by western blot analysis.
Localization with laser-scanning confocal microscopy
Agrobacteria containing pBIN61/OFP-NbATG8f and pBI-NLS-MS2-GFP were cultured and induced with 450 μM acetosyringone in 10 mM MgCl2 to a final concentration of OD600=1. All constructs were mixed with Agrobacteria containing pBIN61/HcPro in a 1:1:1 ratio and infiltrated into N. benthamiana leaves. The BaMV wild-type and BaMV/(MS2)8 transcripts (Cheng et al., 2013) were inoculated onto leaves that had been agro-infiltrated for 12 h. The protoplasts were isolated and observed at 4 dpi. Images were obtained under an Olympus Fluoview FV1000 laser-scanning confocal microscope with 488 nm and 543 nm laser excitation for GFP and OFP, respectively.
Immunostaining assays
Agrobacteria containing pBIN61/OFP (OFP only) or pBIN61/OFP-NbATG8f were cultured and induced with 450 μM acetosyringone in 10 mM MgCl2 to a final concentration of OD600=1. Each of the constructs was mixed with agrobacteria containing pBIN61/HcPro and pKn/BaMV/HA or the pKn/BaMV infectious clone in a 1:1:1 ratio and infiltrated into N. benthamiana leaves. Whole-mount immunofluorescence assays were performed at 4 dpi as described previously (Leivar et al., 2005; Kwon et al., 2013) with a few modifications. In brief, protoplasts were isolated and fixed with PMEG buffer (50 mM PIPES, pH 6.9, 5 mM MgSO4, 5 mM EGTA) containing 4% paraformaldehyde at 4 °C overnight. The fixed protoplasts were washed with PMEG buffer and then treated with PMEG buffer containing 0.5% Triton X-100 and 1% BSA for 5 min at room temperature. Cells were washed and incubated with anti-HA (mouse) and anti-OFP (rabbit) primary antibodies (10 ng µl–1) in 0.1 M phosphate buffered saline (PBS) buffer containing 3% BSA at 4 °C overnight. After washing with PBS, cells were incubated with Alexa Fluor 488-conjugated goat anti-mouse IgG (Alexa Fluor® 488, Invitrogen) and Alexa Fluor 555-conjugated goat anti-rabbit IgG (Alexa Fluor® 555, Invitrogen) at room temperature for 3 h. Images were obtained under a Olympus Fluoview FV1000 laser-scanning confocal microscope with 488 nm and 543 nm laser excitation for HA and OFP, respectively.
Results
ATGs are upregulated after BaMV inoculation in N. benthamiana
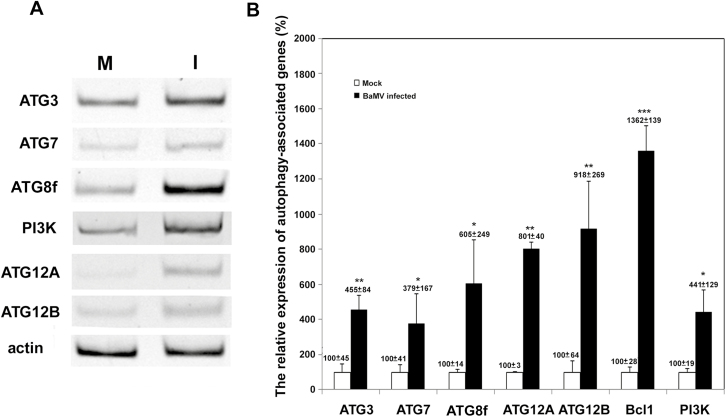
BaMV targets chloroplasts for replication and this possibly triggers the host defense and induces autophagy, or chlorophagy, which plays a role in chloroplast degradation against virus invasion (Dong and Chen, 2013). To investigate whether chloroplast damage occurs via the autophagy pathway (Wada et al., 2009), we examined whether it was activated after BaMV inoculation. First, we inspected the expression profile of some autophagosome formation-related ATGs after BaMV inoculation. The expression of ATGs (ATG3, ATG7, ATG8f, ATG12A, ATG12B) and phosphatidylinositol 3-kinases (PI3K/VPS34) was significantly upregulated at 3 dpi in N. benthamiana (Fig. 1A). To quantify the relative expression of these ATGs (normalized relative to actin) we used real-time quantitative RT-PCR. The expression of all these genes, including Bcl1 (Beclin 1/ATG6/VPS), was elevated 3- to 13-fold in BaMV-inoculated leaves as compared with mock-inoculated controls (Fig. 1B).
Fig. 1.
The expression of autophagy-related genes (ATGs) in inoculated leaves of Nicotiana benthamiana after BaMV inoculation. (A) RT-PCR of the mRNA expression of ATGs and actin (as the internal control) in mock control (M) and BaMV-inoculated (I) leaves at 3 d post-inoculation (dpi). (B) Real-time quantitative RT-PCR of the relative expression of ATGs quantified from three independent experiments with at least three individual plants for each experiment; the expression of mock-inoculated plants was set to 100%. Data are means (±SD). Significant differences were determined using Student’s t-test: *P<0.05; **P<0.01; ***P<0.001.
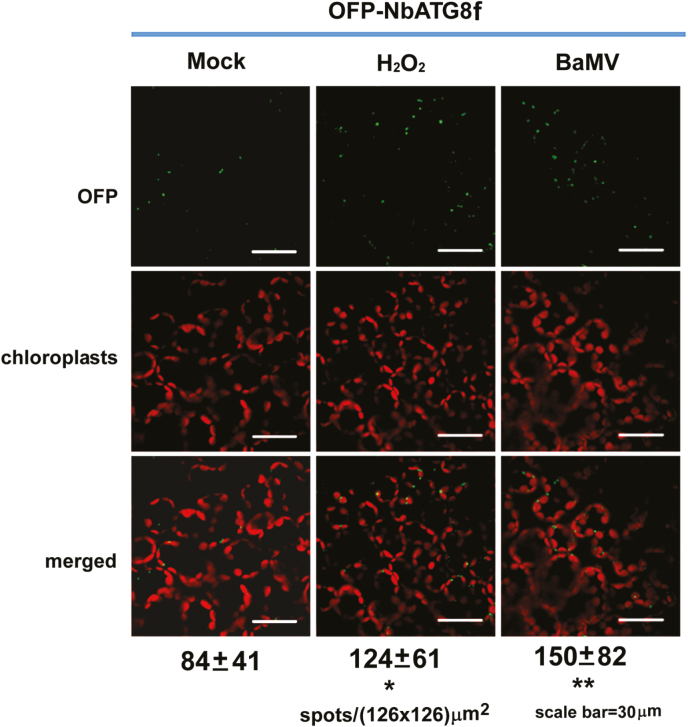
We then expressed OFP-labeled NbATG8f, the homolog of mammalian microtubule-associated proteins 1A/1B-light chain 3 (LC3s), commonly used as markers for autophagosome imaging, in N. benthamiana to visualize ATG8f-positive structures (Wang et al., 2013; Han et al., 2015). The number of fluorescent spots (representing granules derived from OFP-ATG8f clustering, possibly autophagosomes) in BaMV-inoculated cells was ~2-fold more than in mock-inoculated control cells (Fig. 2). Therefore, the autophagy pathway might be activated during BaMV infection.
Fig. 2.
ATG8f-enriched granules are induced after BaMV inoculation in Nicotiana benthamiana. The number of fluorescent vesicles (fluorescent spots in cells) was counted after orange fluorescent protein (OFP)-NbATG8f-expressing leaves were mock-treated (negative control), or treated with H2O2 (positive control) or BaMV inoculation. The autofluorescent signal is derived from chloroplasts. The numbers under the images are the mean (±SD) number of spots counted from 90 independent images of an area 126×126 μm2. Significant differences were determined using Student’s t-test: *P<0.05; **P<0.01. Scale bars are 30 μm.
ATGs play a positive role in BaMV replication
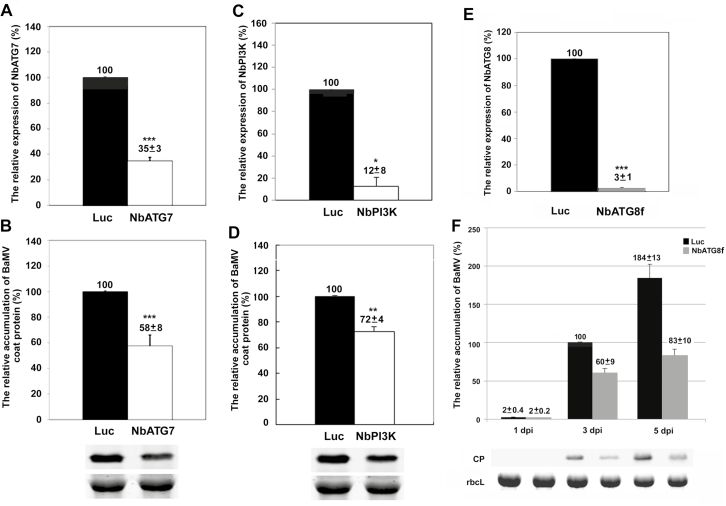
Because most of the ATGs were upregulated after BaMV inoculation, we then wondered whether some of these genes were involved in BaMV infection in N. benthamiana. TRV-based virus-induced gene silencing (VIGS) was used to knock-down the expression of ATGs and the accumulation of BaMV was evaluated. At 5 dpi, the BaMV coat protein levels in NbATG7- and NbPI3K-knockdown plants were reduced to 58% and 72% compared with Luc-knockdown plants, at which time the mRNA levels of NbATG7 and NbPI3K were reduced to 35% and 12%, respectively (Fig. 3A–D). Furthermore, when the NbATG8f level was reduced to 3% (Fig. 3E), the BaMV coat protein level was reduced to 60% and 45% at 3 dpi and 5 dpi, respectively, as compared with Luc-knockdown plants (Fig. 3F). However, PVX and CMV coat protein levels in NbATG8f-knockdown plants did not differ from those in Luc-knockdown plants at 3 dpi (Supplementary Fig. S1). Because it is the marker gene for the autophagy pathway, we focused on ATG8f thereafter.
Fig. 3.
The relative accumulation of BaMV coat protein in Luciferase (Luc)- and ATG-knockdown plants of Nicotiana benthamiana. Real-time RT-PCR analysis of the relative expression of (A) NbATG7, (C) NbPI3K, and (E) NbATG8f in control (Luc) and knockdown plants at 3 d post-inoculation (dpi). Western blot analysis of the relative accumulation of BaMV coat protein (CP) in inoculated leaves of (B) NbATG7- and (D) NbPI3K-knockdown plants at 3 dpi, and (F) NbATG8f-knockdown plants at 11–5 dpi. The Rubisco large subunit (rbcL) stained with Coomassie brilliant blue was a loading control for normalization. Luc-knockdown plants were the knockdown control, and the accumulation of BaMV CP in Luc-knockdown plants at 3 dpi was set to 100%. The numbers shown above the bars are means (±SE) derived from three independent experiments with at least three individual plants for each experiment. Significant differences were determined using Student’s t-test: *P<0.05; **P<0.01; ***P<0.001).
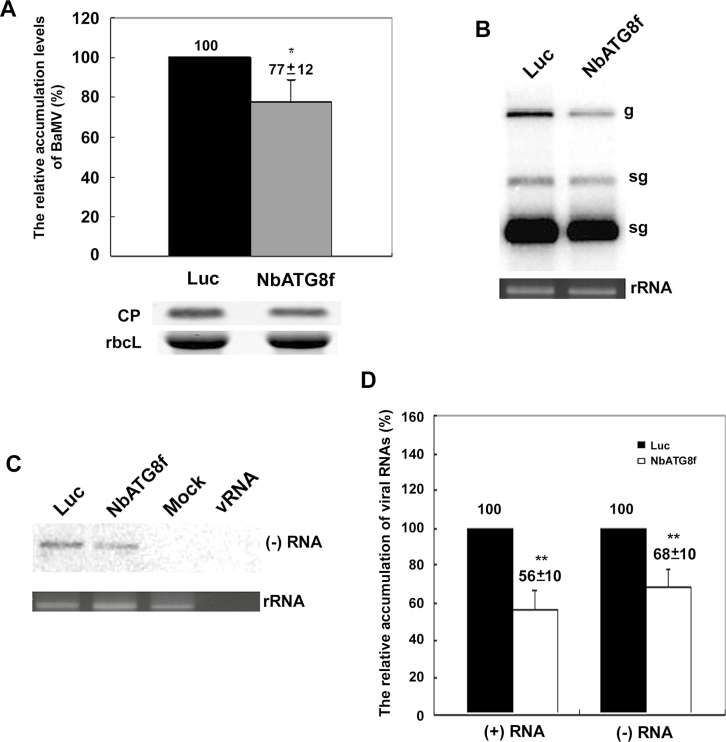
To exclude the possible effect of NbATG8f being involved in cell-to-cell movement of BaMV, we inoculated NbATG8f-knockdown protoplasts with BaMV RNA. The BaMV coat protein level in NbATG8f-knockdown protoplasts was reduced to of 77% of that of Luc-knockdown protoplasts at 24 hpi (Fig. 4A). Northern blot analysis indicated that the levels of BaMV RNA plus-sense (Fig. 4B) and RNA minus-sense (Fig. 4C) in NbATG8f-knockdown protoplasts were significantly reduced to 56% and 68% of control protoplast levels, respectively (Fig. 4D). Thus, the autophagy marker gene NbATG8 could play a positive role specifically in BaMV RNA replication.
Fig. 4.
The accumulation of BaMV in NbATG8f-knockdown protoplasts of Nicotiana benthamiana plants. The relative accumulation of (A) BaMV coat protein (CP), (B) plus-sense viral genomic RNA (g) and subgenomic RNAs (sg), and (C) minus-sense viral RNA in Luciferase (Luc-) and NbATG8f-knockdown protoplasts at 24 h post-inoculation (hpi) were quantified by western blot analysis for proteins and northern blot for RNA analyses. (D) Quantification of the results in (B, C). The numbers above the bars are means (±SE) derived from three independent experiments with at least three individual knockdown plants for protoplast preparation in each experiment. Significant differences were determined using Student’s t-test: *P<0.05; **P<0.01.
NbATG8f and NbATG5 facilitate BaMV replication
NbATG8f (122 amino acids; Supplementary Fig. S2) functions in phagophore expansion and in the completion of autophagosomes, in which autophagy-associated proteins are recruited to target autophagy receptors for selective degradation (Noda et al., 2010). To validate the results derived from the NbATG8f-knockdown experiment that NbATG8f could have a positive role in BaMV accumulation, we agro-infiltrated the construct OFP-NbATG8f into N. benthamiana (Fig. 5A) immediately after BaMV inoculation on the same leaves. The BaMV coat protein level in NbATG8f-transiently expressed leaves was increased to 159% of that of OFP-expressed control leaves (Fig. 5B) at 3 dpi, whereas the level in leaves with transient expression of the mutant construct NbATG8f/G117A (glycine 117 is used for the phosphatidylethanolamine linkage that is critical for membrane targeting; Woo et al., 2014) did not significantly differ from that in controls, although the accumulation was reduced to 86% (Fig. 5B). In addition to expressing NbATG8f in N. benthamiana leaves to show its assisting role in BaMV replication, we also expressed NbATG5 for further confirmation. The expression of NbATG5-OFP (Fig. 5C) also significantly increased BaMV accumulation to 135% of that of the control (Fig. 5D).
Fig. 5.
The accumulation of BaMV in Nicotiana benthamiana plants with transient expression of NbATG8f and NbATG5. Western blot analysis (A) of OFP-NbATG8f and its derivative and (C) NbATG5-OFP transient expression. Total proteins were probed with anti-OFP antibody. Relative accumulation of BaMV coat protein (CP) in plants with transient expression of (B) OFP, OFP-NbATG8f, and OFP-NbATG8f/G117A or (D) NbATG5-OFP. CP in OFP-expressed plant was set to 100%. The numbers shown above the bars are means (±SE) derived from three independent experiments with at least three individual plants for each experiment. Significant differences were determined using Student’s t-test: *P<0.05; **P<0.005.OFP, vector that expressed only OFP; OFP-NbATG8f, NbATG8f with OFP fused at its N-terminus; OFP-NbATG8f/G117A, mutant derived from OFP-NbATG8f; NbATG5-OFP, with OFP fused at its C-terminus. The rubisco large subunit (rbcL) stained with Coomassie brilliant blue (B) and β-actin (actin) detected with anti-actin antibody (D) were used as loading controls for normalization.
Autophagy plays a role in assisting BaMV replication
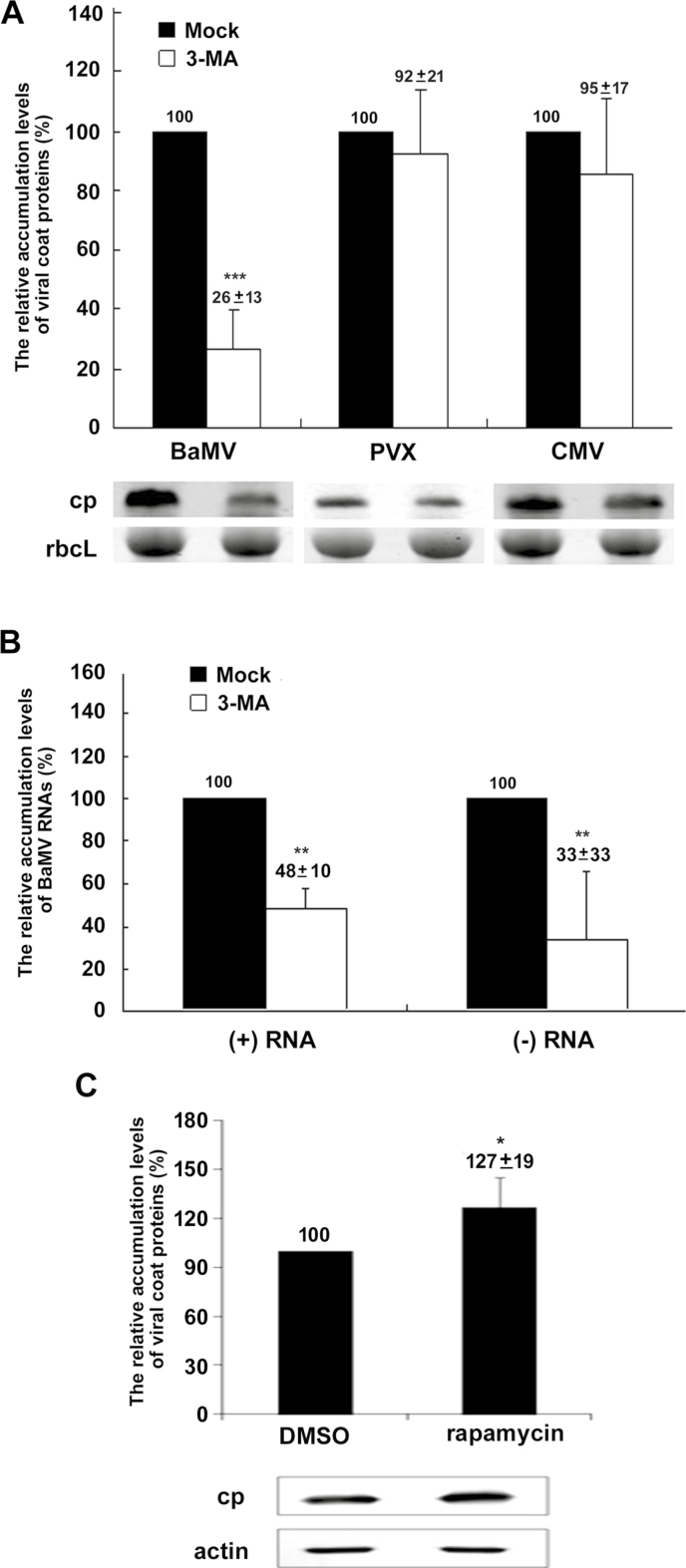
Because ATG8f is involved in the initiation step of autophagosome formation, and mutant NbATG8f/G117A fails in membrane targeting (Woo et al., 2014), we hypothesized that autophagosome formation might play a crucial role in facilitating the accumulation of BaMV. To test this hypothesis, we treated cells with the class-III PI3K specific inhibitor 3-methyladenine (3-MA) (Qin et al., 2012), which can halt vesicle trafficking, including autophagosome synthesis. The BaMV coat protein level was reduced to 26% after 3-MA treatment as compared with the control (Fig. 6A). However, PVX and CMV coat protein levels did not substantially differ that of the control after 3-MA treatment. Furthermore, the accumulation of plus- and minus-sense genomic RNA of BaMV was reduced significantly in 3-MA–treated protoplasts (Fig. 6B). Because 3-MA can specifically inhibit the function of class-III PI3K, which is involved in ATG8-dependent autophagosome expansion (Qin et al., 2012), the results of the linkage between NbATG8f-knockdown and 3-MA treatment implied that autophagosome maturation may be involved in BaMV accumulation. In addition to using the 3-MA inhibitor to suppress BaMV replication, rapamycin was used to induce autophagy in protoplasts in order to confirm the role of autophagy in supporting BaMV replication, and indeed, BaMV replication was increased to 127% of that of the control (Fig. 6C).
Fig. 6.
The effect of viral accumulation in Nicotiana benthamiana protoplasts treated with an autophagy inhibitor or an autophagy inducer. (A) Western blot analysis of the relative accumulation of BaMV, Potato virus X (PVX), and Cucumber mosaic virus (CMV) coat protein (CP) in mock controls and protoplasts treated with the inhibitor 3-methyladenine (3-MA). The Rubisco large subunit (rbcL) stained with Coomassie brilliant blue was a loading control for normalization. (B) The relative accumulation of BaMV plus- and minus-sense RNA in mock controls and 3-MA-treated protoplasts at 24 h post-inoculation (hpi). (C) Western blot analysis of the relative accumulation of BaMV CP in mock controls and protoplasts treated with the inducer rapamycin. β-actin detected by an anti-actin antibody was a loading control for normalization. The numbers above the bars are means (±SE) from at least three independent experiments. Significant differences were determined using Student’s t-test: *P<0.05; **P<0.01; ***P<0.001.
NbATG8f-enriched granules contain the virus replication complex
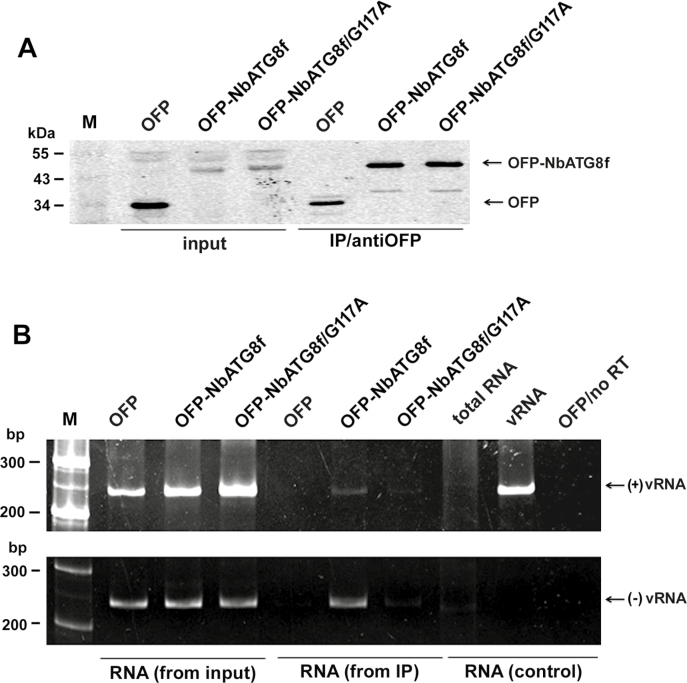
BaMV targets chloroplasts for replication (Cheng et al., 2013), which results in chloroplast damage via which BaMV RNA could be associated with NbATG8f-enriched granules if it induces autophagosome-like structures (i.e. possible chlorophagy). To determine whether NbATG8f-associated vesicles or granules (see Fig. 2) contained viral RNA, we analysed NbATG8f pull-down products. Plants were infiltrated with OFP-NbATG8f and inoculated with BaMV. Protein extracts were isolated from inoculated leaves at 5 dpi and incubated with anti-OFP magnetic beads to pull down OFP-fused proteins (Fig. 7A). RNA was then extracted from these pull-down products and analysed by RT-PCR. The OFP-NbATG8f-associated pull-down products (vesicles or the targeted organelles) contained both the plus- and minus-sense BaMV RNAs (Fig. 7B). Hence, NbATG8f could be associated with the BaMV replicative intermediate.
Fig. 7.
BaMV RNAs are associated with anti-NbATG8f immunoprecipitated products in Nicotiana benthamiana. OFP or OFP-fused proteins (OFP-NbATG8f and OFP-NbATG8f/G117A) were transiently expressed and leaves were inoculated with the BaMV virion. (A) Western blot analysis of proteins detected with anti-OFP antibody. (B) RT-PCR analysis of BaMV plus- and minus-sense RNAs indicated as (+)vRNA and (–)vRNA, respectively, at 5 d post-inoculation (dpi). The total extracts (protein in A and RNA in B) from inoculated leaves that were used as the control are indicated as ‘input’, and the pull-down products are indicated as ‘IP/antiOFP’. RNA extracted from healthy plants was used as a negative control and is indicated as ‘RNA (from input)’. RNA extracted from BaMV-inoculated OFP-expressed leaves that underwent PCR directly without reverse-transcription was used as another negative control and is indicated as ‘OFP/no RT’. Virion RNA (5 ng) indicated as ‘vRNA’ was detected by RT-PCR as a positive control for plus-sense RNA detection and as a negative control for minus-sense RNA detection with specific primers.
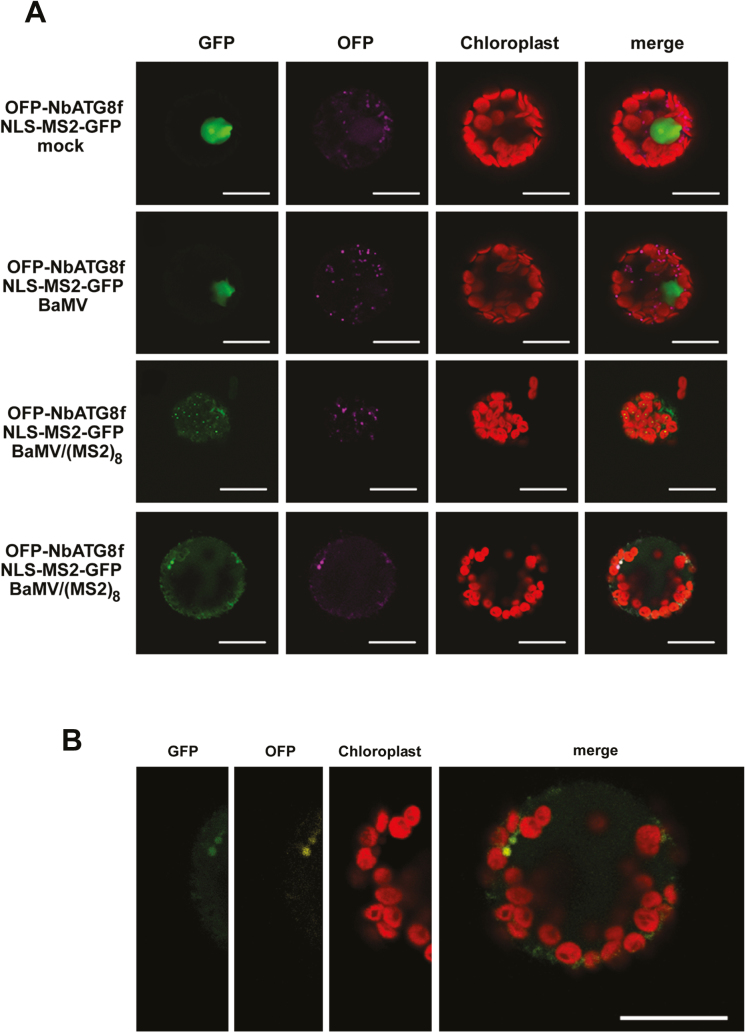
To visualize the viral RNA co-localized with the NbATG8f-associated vesicles (possibly autophagosomes) in live cells, we used the viral RNA-labeling system reported previously by Cheng et al. (2013). A chimera protein containing GFP, phage MS2 coat protein, and a nuclear-localization signal (NLS-MS2-GFP) was co-expressed with the autophagosome marker NbATG8f (OFP-NbATG8f) by agro-infiltration in N. benthamiana. The inoculated viral RNA transcript, BaMV/(MS2)8, contains eight copies of the MS2 coat-protein recognition sequence (19 nt per copy) on which the chimera protein (NLS-MS2-GFP) could interact with the traceable RNA molecules. Protoplasts were then isolated from these leaves at different times after inoculation with viral RNA [BaMV RNA as the control and BaMV/(MS2)8 as the sample] for confocal microcopy. Green fluorescence localized outside of the nucleus represented the modified viral RNA, BaMV/(MS2)8, with RNA molecules co-localized with chloroplasts; this signal was not observed in mock- or BaMV-inoculated protoplasts (Fig. 8A). The co-localization signal of BaMV RNA (green) and NbATG8f-enriched vesicles (magenta in Fig. 8A or yellow in Fig. 8B) was also associated with autofluorescence (possibly derived from chloroplasts). These image results suggested that a portion of the chloroplasts had split (or pinched) off to form NbATG8f-enriched vesicles, similar to the process of chlorophagy (Dong and Chen, 2013) (Fig. 8B). This supported the immunoprecipitation data (Fig. 7) that the NbATG8f-associated pull-down products contained the BaMV replicative intermediate.
Fig. 8.
The subcellular localization of NbAT8 and BaMV RNA in Nicotiana benthamiana leaves. (A) OFP-NbATG8f and the chimera protein containing GFP, MS2 coat protein (CP), and nucleus-localization signal (NLS-MS2-GFP) were transiently expressed in leaves. The agro-infiltrated leaves were then transfected with mock solution, BaMV, or BaMV/(MS2)8 as indicated. The bottom two rows are from different cells with the same treatment. (B) Enlargement of the bottom panel in (A). The signal for GFP is green; OFP is magenta in (A) and yellow in (B); the autofluorescence of chloroplasts is red. Scale bars are 20 µm.
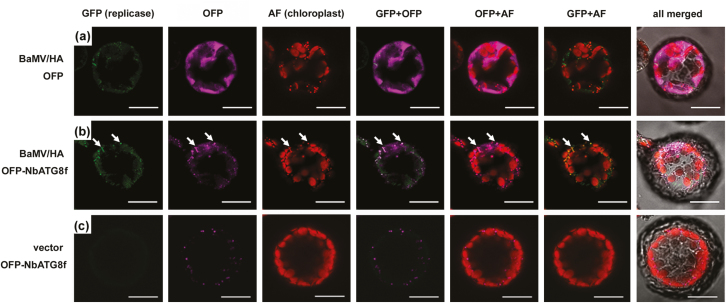
To determine whether the NbATG8f-enriched vesicles also contained the BaMV RNA replication complex, we inoculated modified BaMV containing the HA-tagged replicase (BaMV/HA) onto leaves with transient expression of OFP-NbATG8f. HA-tagged replicase could be detected by an anti-HA antibody followed by the Alexa Fluor 488 secondary antibody, and the OFP signal could be detected by an anti-OFP antibody followed by the Alexa Fluor 555 secondary antibody. BaMV replicase (Alexa Fluor 488) was localized in OFP-NbATG8f–enriched vesicles (Alexa Fluor 555) that also included the autofluorescence signal (Fig. 9). However, this phenomenon did not occur when the replicase was simply expressed by transient expression in OFP-NbATG8f-expressed cells (Supplementary Fig. S3). Taken together, these results suggested that the targeting of chloroplasts by BaMV RNA for replication possibly induces autophagy to attack virus-containing chloroplasts. Through this process, chlorophagy occurred and was hijacked to be an alternative replication site or a shelter away from the host silencing mechanism.
Fig. 9.
The subcellular localization of NbAT8 and BaMV replicase in Nicotiana benthamiana protoplasts. (a) OFP or (b, c) OFP-NbATG8f was transiently expressed in leaves which were then transfected with BaMV with HA-tagged replicase (BaMV/HA) or vector only, as indicated. Protoplasts were probed with an antibody against HA-tag for BaMV replicase and OFP for NbATG8f, then probed with Alexa Fluor 488-conjugated goat anti-mouse IgG for replicase (shown as GFP channel in green) and Alexa Fluor 555-conjugated goat anti-rabbit IgG for OFP (channel in magenta). The autofluorescence of chloroplast is in red. The co-localization of replicase and NbATG8f is indicated by arrows in (b). Scale bars are 20 µm.
Discussion
When plants undergo biotic or abiotic stresses, an anti-stress response is involved in activating the autophagy pathway to facilitate recovery. Here, we demonstrated that the expression of selected ATGs was up-regulated after BaMV inoculation in N. benthamiana leaves and the number of ATG8f-enriched granules (possibly the autophagosomes) in cells was increased after infection (Figs 1, 2). Knockdown (Figs 3, 4) and transient expression (Fig. 5) experiments and the addition of a PI3K-specific inhibitor to block NbATG8f-associated autophagosome formation (Fig. 6) indicated that the autophagy pathway might be involved in positively regulating BaMV accumulation in plants. In NbATG8f-knockdown protoplasts, the reduced accumulation of BaMV genomic plus-sense RNA and anti-genomic minus-sense RNA indicated that NbATG8f plays a crucial role in BaMV replication (Fig. 4). Furthermore, mutant NbATG8f/G117A, which fails to recruit phosphatidylethanolamine (Woo et al., 2014) and is unable to induce the formation of NbATG8f-associated autophagosome, did not enhance BaMV accumulation (Fig. 5). Although autophagy has been reported to assist in virus infection rather than serve primarily in defense for some animal RNA viruses (Richetta et al., 2013; Shinohara et al., 2013), here we demonstrate the first case among plant viruses that the ATGs (possibly via the formation of autophagosome-like structures) are involved in assisting in BaMV accumulation.
BaMV RNA targets chloroplasts for viral RNA replication (Cheng et al., 2013), which could induce the production of ROS and mark chloroplasts for degradation by autophagy (Wada et al., 2009; Ishida et al., 2014). We considered that symptom development, such as the chlorotic mosaic observed in N. benthamiana and necrosis in Chenopodium quinoa after BaMV inoculation, might result from the induction of autophagy targeting chloroplasts. Autophagy contributes to the degradation of chloroplasts, in that a granule-like structure is localized outside of the chloroplasts and sequestered by autophagosomes (Wang and Liu, 2013). In our study, we found granule-like NbATG8f-associated vesicles that were possibly derived from chloroplasts and contained BaMV RNA and replicase (Figs 7, 8, 9). Therefore, BaMV infection might exploit autophagosome formation and target the chloroplast to create conditions that are more favorable for RNA replication. Despite the mechanism of autophagy supporting BaMV accumulation via enhancement of viral RNA replication being unclear, NbATG8f-associated vesicles played a crucial role in facilitating this enhancement. Alternatively, ATG8f-enriched granules derived from BaMV-containing chloroplasts (the initial replication sites) could form stroma protein-based vesicles (Figs 8, 9), possibly representing a location for more efficient BaMV replication, with detectable replicase revealed by confocal microscopy (the alternative replication sites). The vesicles that formed (the autophagosomes) could be hijacked by BaMV for further replication while avoiding transport to vacuoles for degradation. Viral proteins presumably prevent the hijacked vesicles from being degraded in vacuoles or the autolysosome (Richards and Jackson, 2013), but they have not been identified in BaMV.
Positive-sense RNA viruses commonly target a specific organelle for replication (den Boon and Ahlquist, 2010; Laliberté and Sanfacon, 2010; Harak and Lohmann, 2015; Heinlein, 2015). BaMV targets chloroplasts; although the details of the intraplastidial location remain unresolved, the thylakoid membrane could be the site of BaMV RNA replication (Cheng et al., 2013). By contrast, the viral-encoded proteins of CMV and PVX localized in the chloroplasts, which might also trigger autophagy during infection; however, the replication sites of these two viruses are associated with the vacuole membrane and endoplasmic reticulum, respectively (Huh et al., 2011; Tilsner et al., 2012; Linnik et al., 2013), not the chloroplasts. ATG-knockdown experiments and the addition of the 3-MA inhibitor to halt the autophagy process (Supplementary Fig. S1, Fig. 6) revealed no significant difference in accumulation of these two viruses. The autophagosome-associated protein(s) such as NbATG8f and/or the membrane might be crucial for BaMV replication efficiency but not for that of PVX and CMV. However, selective autophagy could be initiated via the specific interaction between ATG8f and autophagy receptors that contain the ATG8f-interacting motif, the WXXL-like sequence (Noda et al., 2008, 2010). We observed three and one such sequence motifs in replicase and the triple gene block protein 1 (TGBp1) of BaMV, respectively, with only one in PVX replicase and none in CMV. The formation of autophagosome-like structures containing the BaMV replication complex derived from the chloroplast could involve selective interaction of NbATG8f with BaMV replicase and TGBp1 proteins.
Supplementary data
Supplementary data are available at JXB online.
Table S1. List of primers used in this study.
Fig. S1. The relative accumulation of PVX and CMV coat proteins in inoculated leaves of NbATG8f-knockdown plants.
Fig. S2. The amino acid sequence alignment of ATG8 from different species.
Fig. S3. The subcellular localization of NbAT8f and BaMV replicase transiently expressed in N. benthamiana protoplasts.
Acknowledgements
We are grateful to Professor Theo Dreher at Oregon State University for editing and making comments on the original manuscript. We appreciate the Cell Image Core Laboratory of the Graduate Institute of Biotechnology, National Chung Hsing University, for providing facilities and assistance. This work was financially supported by the Ministry of Science and Technology of Taiwan (MOST 102-2311-B-005-006) and in part by the Advanced Plant Biotechnology Center from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan.
References
- Chen IH, Chiu MH, Cheng SF, Hsu YH, Tsai CH. 2013. The glutathione transferase of Nicotiana benthamiana NbGSTU4 plays a role in regulating the early replication of Bamboo mosaic virus. New Phytologist 199, 749–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IH, Lin JW, Chen YJ, Wang ZC, Liang LF, Meng M, Hsu YH, Tsai CH. 2010. The 3´-terminal sequence of Bamboo mosaic virus minus-strand RNA interacts with RNA-dependent RNA polymerase and initiates plus-strand RNA synthesis. Molecular Plant Pathology 11, 203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SF, Huang YP, Chen LH, Hsu YH, Tsai CH. 2013. Chloroplast phosphoglycerate kinase is involved in the targeting of Bamboo mosaic virus to chloroplasts in Nicotiana benthamiana plants. Plant Physiology 163, 1598–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SF, Huang YP, Wu ZR, Hu CC, Hsu YH, Tsai CH. 2010. Identification of differentially expressed genes induced by Bamboo mosaic virus infection in Nicotiana benthamiana by cDNA-amplified fragment length polymorphism. BMC Plant Biology 10, 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavel M, Michaeli S, Genschik P. 2017. Autophagy: a double-edged sword to fight plant viruses. Trends in Plant Science 22, 646–648. [DOI] [PubMed] [Google Scholar]
- Cowan GH, Roberts AG, Chapman SN, Ziegler A, Savenkov EI, Torrance L. 2012. The Potato mop-top virus TGB2 protein and viral RNA associate with chloroplasts and viral infection induces inclusions in the plastids. Frontiers in Plant Science 3, 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Boon JA, Ahlquist P. 2010. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annual Review of Microbiology 64, 241–256. [DOI] [PubMed] [Google Scholar]
- Dong J, Chen W. 2013. The role of autophagy in chloroplast degradation and chlorophagy in immune defenses during Pst DC3000 (AvrRps4) infection. PLoS ONE 8, e73091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y, Yoshimoto K, Ohsumi Y. 2007. An Arabidopsis homolog of yeast ATG6/VPS30 is essential for pollen germination. Plant Physiology 143, 1132–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Wang Y, Zheng X, Jia Q, Zhao J, Bai F, Hong Y, Liu Y. 2015. Cytoplastic glyceraldehyde-3-phosphate dehydrogenases interact with ATG3 to negatively regulate autophagy and immunity in Nicotiana benthamiana. The Plant Cell 27, 1316–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harak C, Lohmann V. 2015. Ultrastructure of the replication sites of positive-strand RNA viruses. Virology 479-480, 418–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison-Lowe NJ, Olsen LJ. 2008. Autophagy protein 6 (ATG6) is required for pollen germination in Arabidopsis thaliana. Autophagy 4, 339–348. [DOI] [PubMed] [Google Scholar]
- Heinlein M. 2015. Plant virus replication and movement. Virology 479-480, 657–671. [DOI] [PubMed] [Google Scholar]
- Hofius D, Schultz-Larsen T, Joensen J, Tsitsigiannis DI, Petersen NH, Mattsson O, Jørgensen LB, Jones JD, Mundy J, Petersen M. 2009. Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 137, 773–783. [DOI] [PubMed] [Google Scholar]
- Huang YL, Han YT, Chang YT, Hsu YH, Meng M. 2004. Critical residues for GTP methylation and formation of the covalent m7GMP-enzyme intermediate in the capping enzyme domain of Bamboo mosaic virus. Journal of Virology 78, 1271–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YP, Chen IH, Tsai CH. 2017. Host factors in the infection cycle of Bamboo mosaic virus. Frontiers in Microbiology 8, 437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YW, Hu CC, Liou MR, Chang BY, Tsai CH, Meng M, Lin NS, Hsu YH. 2012. Hsp90 interacts specifically with viral RNA and differentially regulates replication initiation of Bamboo mosaic virus and associated satellite RNA. PLoS Pathogens 8, e1002726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh SU, Kim MJ, Ham BK, Paek KH. 2011. A zinc finger protein Tsip1 controls Cucumber mosaic virus infection by interacting with the replication complex on vacuolar membranes of the tobacco plant. New Phytologist 191, 746–762. [DOI] [PubMed] [Google Scholar]
- Hung CJ, Hu CC, Lin NS, Lee YC, Meng M, Tsai CH, Hsu YH. 2014a. Two key arginine residues in the coat protein of Bamboo mosaic virus differentially affect the accumulation of viral genomic and subgenomic RNAs. Molecular Plant Pathology 15, 196–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung CJ, Huang YW, Liou MR, Lee YC, Lin NS, Meng M, Tsai CH, Hu CC, Hsu YH. 2014b. Phosphorylation of coat protein by protein kinase CK2 regulates cell-to-cell movement of Bamboo mosaic virus through modulating RNA binding. Molecular Plant-Microbe Interactions 27, 1211–1225. [DOI] [PubMed] [Google Scholar]
- Ishida H, Izumi M, Wada S, Makino A. 2014. Roles of autophagy in chloroplast recycling. Biochimica et Biophysica Acta 1837, 512–521. [DOI] [PubMed] [Google Scholar]
- Jang C, Seo EY, Nam J, et al. 2013. Insights into Alternanthera mosaic virus TGB3 functions: interactions with Nicotiana benthamiana PsbO correlate with chloroplast vesiculation and veinal necrosis caused by TGB3 over-expression. Frontiers in Plant Science 4, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin R, Zhu W, Cao S, Chen R, Jin H, Liu Y, Wang S, Wang W, Xiao G. 2013. Japanese encephalitis virus activates autophagy as a viral immune evasion strategy. PLoS ONE 8, e52909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SI, Cho HJ, Kim SR, Park OK. 2013. The Rab GTPase RabG3b positively regulates autophagy and immunity-associated hypersensitive cell death in Arabidopsis. Plant Physiology 161, 1722–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laliberté JF, Sanfaçon H. 2010. Cellular remodeling during plant virus infection. Annual Review of Phytopathology 48, 69–91. [DOI] [PubMed] [Google Scholar]
- Lan P, Yeh WB, Tsai CW, Lin NS. 2010. A unique glycine-rich motif at the N-terminal region of Bamboo mosaic virus coat protein is required for symptom expression. Molecular Plant-Microbe Interactions 23, 903–914. [DOI] [PubMed] [Google Scholar]
- Lee CC, Ho YN, Hu RH, Yen YT, Wang ZC, Lee YC, Hsu YH, Meng M. 2011. The interaction between Bamboo mosaic virus replication protein and coat protein is critical for virus movement in plant hosts. Journal of Virology 85, 12022–12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, González VM, Castel S, Trelease RN, López-Iglesias C, Arró M, Boronat A, Campos N, Ferrer A, Fernàndez-Busquets X. 2005. Subcellular localization of Arabidopsis 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Plant Physiology 137, 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ. 2004. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Developmental Cell 6, 463–477. [DOI] [PubMed] [Google Scholar]
- Li F, Vierstra RD. 2012. Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends in Plant Science 17, 526–537. [DOI] [PubMed] [Google Scholar]
- Li Y, Cui H, Cui X, Wang A. 2016. The altered photosynthetic machinery during compatible virus infection. Current Opinion in Virology 17, 19–24. [DOI] [PubMed] [Google Scholar]
- Li YI, Chen YJ, Hsu YH, Meng M. 2001a. Characterization of the AdoMet-dependent guanylyltransferase activity that is associated with the N terminus of Bamboo mosaic virus replicase. Journal of Virology 75, 782–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YI, Cheng YM, Huang YL, Tsai CH, Hsu YH, Meng M. 1998. Identification and characterization of the Escherichia coli-expressed RNA-dependent RNA polymerase of Bamboo mosaic virus. Journal of Virology 72, 10093–10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YI, Shih TW, Hsu YH, Han YT, Huang YL, Meng M. 2001b. The helicase-like domain of plant potexvirus replicase participates in formation of RNA 5´ cap structure by exhibiting RNA 5´-triphosphatase activity. Journal of Virology 75, 12114–12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JW, Chiu HN, Chen IH, Chen TC, Hsu YH, Tsai CH. 2005. Structural and functional analysis of the cis-acting elements required for plus-strand RNA synthesis of Bamboo mosaic virus. Journal of Virology 79, 9046–9053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JW, Ding MP, Hsu YH, Tsai CH. 2007. Chloroplast phosphoglycerate kinase, a gluconeogenetic enzyme, is required for efficient accumulation of Bamboo mosaic virus. Nucleic Acids Research 35, 424–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin LT, Dawson PW, Richardson CD. 2010. Viral interactions with macroautophagy: a double-edged sword. Virology 402, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MK, Chang BY, Liao JT, Lin NS, Hsu YH. 2004. Arg-16 and Arg-21 in the N-terminal region of the triple-gene-block protein 1 of Bamboo mosaic virus are essential for virus movement. The Journal of General Virology 85, 251–259. [DOI] [PubMed] [Google Scholar]
- Lin MK, Hu CC, Lin NS, Chang BY, Hsu YH. 2006. Movement of potexviruses requires species-specific interactions among the cognate triple gene block proteins, as revealed by a trans-complementation assay based on the Bamboo mosaic virus satellite RNA-mediated expression system. The Journal of General Virology 87, 1357–1367. [DOI] [PubMed] [Google Scholar]
- Lin NS, Lin FZ, Huang TY, Hsu YH. 1992. Genome properties of Bamboo mosaic virus. Phytopathology 82, 731–734. [Google Scholar]
- Linnik O, Liesche J, Tilsner J, Oparka KJ. 2013. Unraveling the structure of viral replication complexes at super-resolution. Frontiers in Plant Science 4, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Czymmek K, Tallóczy Z, Levine B, Dinesh-Kumar SP. 2005. Autophagy regulates programmed cell death during the plant innate immune response. Cell 121, 567–577. [DOI] [PubMed] [Google Scholar]
- Masclaux-Daubresse C. 2016. Autophagy controls carbon, nitrogen, and redox homeostasis in plants. Autophagy 12, 896–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Chen Q, Havé M. 2017. Regulation of nutrient recycling via autophagy. Current Opinion in Plant Biology 39, 8–17. [DOI] [PubMed] [Google Scholar]
- Meng M, Lee CC. 2017. Function and structural organization of the replication protein of Bamboo mosaic virus. Frontiers in Microbiology 8, 522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaeli S, Galili G, Genschik P, Fernie AR, Avin-Wittenberg T. 2016. Autophagy in plants – what’s new on the menu? Trends in Plant Science 21, 134–144. [DOI] [PubMed] [Google Scholar]
- Mizui T, Yamashina S, Tanida I, et al. 2010. Inhibition of Hepatitis C virus replication by chloroquine targeting virus-associated autophagy. Journal of Gastroenterology 45, 195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda NN, Kumeta H, Nakatogawa H, Satoo K, Adachi W, Ishii J, Fujioka Y, Ohsumi Y, Inagaki F. 2008. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes to Cells 13, 1211–1218. [DOI] [PubMed] [Google Scholar]
- Noda NN, Ohsumi Y, Inagaki F. 2010. Atg8-family interacting motif crucial for selective autophagy. FEBS Letters 584, 1379–1385. [DOI] [PubMed] [Google Scholar]
- Patel S, Dinesh-Kumar SP. 2008. Arabidopsis ATG6 is required to limit the pathogen-associated cell death response. Autophagy 4, 20–27. [DOI] [PubMed] [Google Scholar]
- Prasanth KR, Huang YW, Liou MR, Wang RY, Hu CC, Tsai CH, Meng M, Lin NS, Hsu YH. 2011. Glyceraldehyde 3-phosphate dehydrogenase negatively regulates the replication of Bamboo mosaic virus and its associated satellite RNA. Journal of Virology 85, 8829–8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prod’homme D, Jakubiec A, Tournier V, Drugeon G, Jupin I. 2003. Targeting of the Turnip yellow mosaic virus 66K replication protein to the chloroplast envelope is mediated by the 140K protein. Journal of Virology 77, 9124–9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y, Li HF, Wong SM, Fan ZF. 2009. Plastocyanin transit peptide interacts with Potato virus X coat protein, while silencing of plastocyanin reduces coat protein accumulation in chloroplasts and symptom severity in host plants. Molecular Plant-Microbe Interactions 22, 1523–1534. [DOI] [PubMed] [Google Scholar]
- Qin T, Li J, Yuan M, Mao T. 2012. Characterization of the role of calcium in regulating the microtubule-destabilizing activity of MDP25. Plant Signaling & Behavior 7, 708–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinero A, Beachy RN. 1986. Association of TMV coat protein with chloroplast membranes in virus-infected leaves. Plant Molecular Biology 6, 291–301. [DOI] [PubMed] [Google Scholar]
- Richards AL, Jackson WT. 2013. How positive-strand RNA viruses benefit from autophagosome maturation. Journal of Virology 87, 9966–9972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richetta C, Grégoire IP, Verlhac P, Azocar O, Baguet J, Flacher M, Tangy F, Rabourdin-Combe C, Faure M. 2013. Sustained autophagy contributes to measles virus infectivity. PLoS Pathogens 9, e1003599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara Y, Imajo K, Yoneda M, et al. 2013. Unfolded protein response pathways regulate Hepatitis C virus replication via modulation of autophagy. Biochemical and Biophysical Research Communications 432, 326–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilsner J, Linnik O, Wright KM, Bell K, Roberts AG, Lacomme C, Santa Cruz S, Oparka KJ. 2012. The TGB1 movement protein of Potato virus X reorganizes actin and endomembranes into the X-body, a viral replication factory. Plant Physiology 158, 1359–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrance L, Cowan GH, Gillespie T, Ziegler A, Lacomme C. 2006. Barley stripe mosaic virus-encoded proteins triple-gene block 2 and γb localize to chloroplasts in virus-infected monocot and dicot plants, revealing hitherto-unknown roles in virus replication. The Journal of General Virology 87, 2403–2411. [DOI] [PubMed] [Google Scholar]
- Tsai CH, Cheng CP, Peng CW, Lin BY, Lin NS, Hsu YH. 1999. Sufficient length of a poly(A) tail for the formation of a potential pseudoknot is required for efficient replication of Bamboo mosaic potexvirus RNA. Journal of Virology 73, 2703–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada S, Ishida H, Izumi M, Yoshimoto K, Ohsumi Y, Mae T, Makino A. 2009. Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiology 149, 885–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Mugume Y, Bassham DC. 2018. New advances in autophagy in plants: regulation, selectivity and function. Seminars in Cell & Developmental Biology 80, 113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Liu Y. 2013. Autophagic degradation of leaf starch in plants. Autophagy 9, 1247–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yu B, Zhao J, et al. 2013. Autophagy contributes to leaf starch degradation. The Plant Cell 25, 1383–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T, Huang TS, McNeil J, Laliberté JF, Hong J, Nelson RS, Wang A. 2010. Sequential recruitment of the endoplasmic reticulum and chloroplasts for plant potyvirus replication. Journal of Virology 84, 799–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo J, Park E, Dinesh-Kumar SP. 2014. Differential processing of Arabidopsis ubiquitin-like Atg8 autophagy proteins by Atg4 cysteine proteases. Proceedings of the National Academy of Sciences, USA 111, 9325–9325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Kakani K, Reade R, Hui E, Rochon D. 2006. A 38-amino-acid sequence encompassing the arm domain of the Cucumber necrosis virus coat protein functions as a chloroplast transit Peptide in infected plants. Journal of Virology 80, 7952–7964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Contento AL, Nguyen PQ, Bassham DC. 2007. Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiology 143, 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K, Suzuki T, Moriyasu Y. 2007. Constitutive autophagy in plant root cells. Autophagy 3, 360–362. [DOI] [PubMed] [Google Scholar]
- Zientara-Rytter K, Sirko A. 2016. To deliver or to degrade – an interplay of the ubiquitin–proteasome system, autophagy and vesicular transport in plants. The FEBS Journal 283, 3534–3555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.