Abstract
Objective:
The objective of the study was to investigate Foot and Mouth Disease virus (FMDV) outbreak in cattle in the Sarankhola Upazila under Bagerhat district of Bangladesh with isolation, identification, and molecular characterization of FMDV during April 2018.
Materials and Methods:
This Upazila is located at southern border of Bangladesh and surrounded by mangrove forest Sundarban. The outbreak investigation team collected epidemiological data from outbreak location. In addition, the team collected a total of 30 (15 calves, 15 adult) tongue epithelial tissue samples from a clinically FMD-affected cattle. The confirmation of FMDV and its three serotypes (A, O, and Asia-1) was performed by Reverse Transcriptase Polymerase Chain Reaction (RT-PCR). An amplified product of the VP1 region of FMDV genome was sequenced by Sanger sequencing method after cultivation and reconfirmation of FMDV into the BHK21 cell line. Genetic variability was studied by constructing a phylogenetic tree.
Results:
The investigation survey was carried out in overall 8,393 (8,393/15,580; 53.89%) cases including 3,050 (3,050/8,393; 36.34%) cases in calf and 5,343 (5,343/8,393; 59.77%) cases in adult cattle. The overall case fatality rate (CFR) was recorded as 2.27% (354/15,580) with significantly highest CFR in the calf (71.46%; 253/354) compared to an adult. The collected all 30 samples found with FMDV positive and mixed infection of all samples with serotype Asia-1 and serotype O were observed. In BHK 21 cell line, the eight FMDV positive samples showed a typical cytopathic effect during the third passage. Finally, DNA sequence data of two isolates found closely related with the isolates of bordering country India and Myanmar.
Conclusion:
The investigation identified the risk factors involved in an outbreak of FMDV, namely, sharing the common paddy land after harvesting, no FMD vaccination, the interaction between cattle and wildlife, and cross bordering movement.
Keywords: FMDV, cattle, outbreak, sequence, Bangladesh
Introduction
Foot and mouth disease (FMD) is the most widespread viral disease of cloven-hoofed animal and locally called as “Khura Rog” in Bangladesh [1–3]. It is caused by Foot and mouth disease virus (FMDV), a virus under genus Aphthovirus and family Picornaviridae with single-stranded and positive-sense RNA genome. An icosahedral capsid layer surrounded the viral genome that composed of four structural proteins, namely, VP1, VP2, VP3, and VP4 with 60 copies of each protein. Totally seven immunologically distinct serotypes were identified including Types O, A, C, Asia 1, SAT 1-3 (South African territory) [4,5], and multiple subtypes circulating over the world [4].
In Bangladesh, serotype O was only circulated since 2000 [6] but the current study revealed that the three serotypes (A, O, and Asia-1) are being circulated in Bangladesh [7,8]. The prevalence of these three serotypes was 31% of both serotypes O and Asia-1, and 7% of serotype A, also found with mixed infection with serotype 7% O-Asia-1 and 2% serotype O-A [7]. Another researcher demonstrated that serotype O was responsible for 82% outbreak and its show dominance over serotype A and Asia-1 [9]. Three bordering countries of Bangladesh also circulated the same serotypes [10]. The largest border sharing country of Bangladesh is India where the prevalence of circulation serotypes O, A, and Asia-1 is 80%, 8%, and 12%, respectively [11].
FMDV is highly contagious and has a multi-route transmission capacity mostly direct contact with saliva, fomites, feeding, and watering places [12]. Also, it has transboundary potential that is responsible for the outbreak [13]. Livestock movements in inter-country are important factors for spreading the virus [14]. In Bangladesh, FMD prevalence was found in throughout the year but increases the incidence after two major festivals (Eid-ul-Fitarand and Eid-ul-Azha) [9]. It is the cause of increasing the animal movement throughout the country during this festival [15,16].
Now FMD is endemic in Bangladesh and outbreaks in every year [9,17]. It has a significant economic impact by reducing milk, meat production, and increasing calves mortality, also recovered animal losses in productivity [18,19]. Every year, the globally total impact of FMD is US$ 6.5 to 21 billion for endemic countries and >US$1.5 billion for FMD free countries [20].
In this study, an outbreak investigation was done and confirmation of FMDV infection by using clinical signs and Reverse Transcriptase Polymerase Chain Reaction (RT-PCR) of the tongue epithelial tissue sample. Then find out the genetic relationship of outbreak FMDV by phylogenetic analysis. Finally, addressed the risk factors that are responsible for this outbreak.
Materials and Methods
Ethical approval
All the protocols used in the study were approved by the Institutional Animal Experimentation Ethics Committee of the Bangladesh Livestock Research Institute, Savar, Dhaka.
Study area
In April 2018, the FMD outbreaks in cattle were suspected in three Unions (Dhansagar, Khontakata, and Royenda) of Sarankhola Upazila of Bagerhat district. Sarankhola Upazila located at the south border of Bangladesh and surrounded by world largest mangrove forest Sundarban and bank of Balaswar River that is directly connected with Bay of Bengal (Fig. 1). It is located geographically at 22.2917°N 89.7917°E with a total area of 756.61 km2;. The Upazila has a cattle population of total 15,580 and there is a conventional type of farming system. Where cattle are reared exclusively on an open field grazing pattern and general husbandry. Farmers have no experience in modern veterinary practices like vaccination and deworming technology. It has good access possibility to interact between wild animals and livestock due to the nearest Sarankhola Range of Sundarban.
Figure 1. Map showing the outbreak area in Sarankhola Upazila of Bagerhat district of Bangladesh. The map was created by using Geographic Information Systems (GIS) software ESRI ArcGIS version 10.6.1 [42].
Clinical sign and symptoms
The established clinical signs were observed to clinically suspect the FMD infection and following clinical signs were observed: primarily pyrexia up to 40°F, followed by the growth of vesicles on the whole oral cavity, including the tongue epithelium, gum, hard palate, dental pads, and muzzle, resulting in drooling of salivation. As vesicles developed on the teats of lactating cow, mastitis developed with other signs. Vesicles also developed on the inter-digital cleft and maggots grow when flies sited over it so that animal shows lameness. In the case of calf, high mortality with or without shows clinical symptoms.
Sample collection
Total of 30 tongue epithelial tissues of aphthous ulcers was collected from 30 (15 calves and 15 adults) clinically suspected cattle as 10 samples from each Union aseptically. The epithelial tissue samples were collected according to the guideline of OIE Terrestrial Manual [3]. Then the samples were shipped from the outbreak site to FMD Research Laboratory, Bangladesh Livestock Research Institute (BLRI), Savar, Dhaka in virus transfer media (VTM).
Preparation of VTM
The VTM prepared by Brain Heart Infusion Broth media with 1000,000 IU Penicillin G, Gentamicin, and Amphotericin B.
Epidemiological data collection
To consider the risk factors of outbreaks, a direct survey was conducted through pre-constructed questioner and data were collected from local people, farmers, and local veterinary service authority. Data were collected on the farming system, grazing pattern, housing, geographical location, animal interaction chances, wildlife and domestic animal relation, and animal marketing. Information on the population at risk, case, and case fatality rate also collected. The case fatality rate was calculated by the following formula:
Vaccination history
Farmers are not conscious of modern veterinary practices, vaccination, and deworming. So, there was no record of FMD vaccination in the outbreak area.
Detection of FMDV serotypes by RT-PCR test
An aliquot of epithelial tissue samples was ground by pastle and morter with phosphate-buffered saline (PBS) solution and centrifuged at 3,000 rpm for 10 min to obtain a 300 μl of supernatant fluid suspension. Then, the total genomic RNA was extracted by using QIAamp® Viral RNA kit (QIAGEN, Germany) following the manufacturer’s protocol. For the detection of FMDV, all extracted RNAs were subjected to RT-PCR test using QIAGEN One-Step RT-PCR Kit with 10 pM/sample universal primers. The list of primers used and PCR thermal profile are listed in Tables 1 and 2, respectively. Then the differentiation of serotypes Asia-1, A, and O by using serotype-specific primers. The corresponding amplification site of VP1 gene was visualized by stained 1 μg/ml ethidium bromide into 2% agarose gel in gel electrophoresis. For the test verification, lab positive control of each serotype was used in every step. The GenBank Accession number of lab-positive control was KP119763 (Type-A), KP119758 (Type-O), and KP119757 (Asia-1).
Table 1. List of oligonucleotide primers used for universal FMDV and serotyping of FMDV by RT-PCR.
| Serotype | Primer name | Primer sequence (5´ to 3´) | Location | PCR products (bp) | References |
|---|---|---|---|---|---|
| Universal | 1F | GCC TGG TCT TTC CAG GTC T | 5´UTR | 328 | [40] |
| 1R | CCA GTC CCC TTC TCA GAT C | 5´UTR | |||
| O | P38 | GCTGCCTACCTCCTTCAA | 1D | 402 | |
| P33 | AGCTTGTACCAGGGTTTGGC | 2B | |||
| Asia-1 | P74 | GACACCACTCAGGACCGCCG | 1D | 292 | |
| P33 | AGCTTGTACCAGGGTTTGGC | 2B | |||
| A | P110 | GT(G:A:T:C)ATTGACCT(G:A:T:C)ATGCA (G:A:T:C) AC (G:A:T:C) CAC | 1D | 732 | [41] |
| P33 | AGCTTGTACCAGGGTTTGGC | 2B |
Table 2. PCR conditions for RT-PCR for both universal FMDV and serotyping of FMDV.
| Sl. No. | Steps | Name of the primers | |
|---|---|---|---|
| 1F and 1R | P33, P38, P74, P110 | ||
| 1 | Reverse transcription | 50°C for 30 min | 50°C for 30 min |
| 2 | Initial PCR activation | 95°C for 15 min | 95°C for 15 min |
| 3 | Number of cycle | 30 | 35 |
| 4 | Denaturation | 94°C for 1 min | 94°C for 15 sec |
| 5 | Annealing | 55°C for 1 min | 55°C for 1 min |
| 6 | Extension | 72°C for 2 min | 72°C for 30 min |
| 7 | Final extension | 72°C for 7 min | 60°C for 6 min |
| 8 | Hold at | 4°C | 40C |
Cell culture for isolation of FMDV
A total of 12 samples were selected for propagating into BHK-21 cell line. Epithelial tissue samples were ground by sterile pastle and morter with an equal volume of sterilized PBS with antibiotics and then filtered through a syringe filter (0.2 μm). Then, 0.5 ml of filtered virus suspension was inoculated on the previously prepared BHK-21 monolayer cells containing 25 cm2 tissue culture flasks. Then incubated at 5% CO2 and 370C temperature for 48 h. The cultured cells are observed twice for cytopathic effects (CPEs). Then remove the virus from infected cells by freeze-thawing three times and centrifuged the supernatant fluid at 5,000 rpm for 5 min. Finally, verified the presence of FMDV in the supernatant fluid by RT-PCR test.
Sequencing and phylogeny analysis
The amplified region of FMDV genomic RNA by RT-PCR was sequenced on both forward and reverse strands as previously described by Knowles et al. [21]. The SeqMan software (DNAStar, Lasergene v.8) was used to assemble and verify the sequence data, and finally, nucleotide sequences were aligned by using BioEdit version 7.2.5 [22] and Clustal W [23]. Then reconstruct the alignments by Maximum Likelihood phylogenetic tree with TN93+G+I model existing in MEGA X [24]. The robustness of the phylogenetic tree topology was verified with 1,000 bootstrap replicates.
Statistics
The record of the outbreak investigation, sample size calculation, FMD case, case fatality rates, and prevalence was analyzed by using Epi Info™ version 7 (Epi Info™, CDC, USA).
Results
On the basis of clinical signs and physical examination, overall 8,393 (53.87%) cattle were found FMD positive cases. Where 3,050 (36.34%) cases were in calf and 5343 (59.77%) cases were in adult cattle. Location wise FMD cases were found in 2,524 (54.98%), 4,501 (70%), and 1,368 (30%) in Dhansagar, Khontakata, and Royenda Union, respectively. The highest 1,890 (41.99%) FMD cases were found in calves in Khontakata and 1,843 (73.02 %) in adult in Dhansagar Union (Table 3).
Table 3. The FMDV outbreak investigation data.
| Sl. No. | Location | Total cattle population | FMD cases | Case fatality rate | ||||
|---|---|---|---|---|---|---|---|---|
| Calves (1–24 months) | Adult | Total | Calves | Adult | Total | |||
| 1 | Dhansagar | 4590 | 681 (26.98%) | 1843 (73.02 %) | 2524 (54.98%) | 56 (74.66%) | 19 (25.33%) | 75 (2.97%) |
| 2 | Khontakata | 6430 | 1890 (41.99%) | 2611 (58%) | 4501 (70%) | 154 (68.44%) | 72 (32%) | 225 (5%) |
| 3 | Royenda | 4560 | 479 (35.01%) | 889 (64.98%) | 1368 (30%) | 43 (79.63%) | 11 (20.37%) | 54 (3.95%) |
| Total | 15580 | 3050 (36.34%) | 5343 (59.77%) | 8393 (53.87%) | 253 (71.46%) | 102 (28.81%) | 354 (2.27%) | |
| p | 0.000* | 0.001* | 0.000* | 0.001* | ||||
Significant at the 0.01 level.
In the outbreak area, the overall case fatality rates (CFR) on FMD were 354 (2.27%). But devastating CFR found in calves was 253 (71.46%). Total CFR in adult cattle was 102 (28.81%). The CFR was highest 43 (79.63%) in calves in Royenda and lowest 154 (68.44%) was in Khontakata Union. In the case of adult cattle, highest 72 (32%) and lowest 11 (20.37%) CFRs were in Khontakata and Royenda Union, respectively (Table 3).
A total of 30 randomly selected epithelial samples were collected from three locations of outbreak area for confirmatory diagnosis by RT-PCR test. At first, all the 30 samples were screened for FMDV by the universal primer of 5 UTR location of VP1 gene and found all samples are positive. Then differentiation of three serotypes (A, O, and Asia-1) made individually and found all 30 samples were positive for both serotypes Asia-1 and O but absent for serotype A (Table 4).
Table 4. Description of RT-PCR test results.
| Sample ID | Location | Type | Sample type | Results | ||||
|---|---|---|---|---|---|---|---|---|
| Universal | Asia 1 | O | A | Comments | ||||
| 1 | Dhansagar | Adult | Epithelium | + | + | + | - | Asia-1+O |
| 2 | Dhansagar | Calf | Epithelium | + | + | + | - | Asia-1+O |
| 3 | Dhansagar | Calf | Epithelium | + | + | + | - | Asia-1+O |
| 4 | Dhansagar | Adult | Epithelium | + | + | + | - | Asia-1+O |
| 5 | Dhansagar | Adult | Epithelium | + | + | + | - | Asia-1+O |
| 6 | Dhansagar | Calf | Epithelium | + | + | + | - | Asia-1+O |
| 7 | Dhansagar | Calf | Epithelium | + | + | + | - | Asia-1+O |
| 8 | Dhansagar | Adult | Epithelium | + | + | + | - | Asia-1+O |
| 9 | Dhansagar | Calf | Epithelium | + | + | + | - | Asia-1+O |
| 10 | Dhansagar | Adult | Epithelium | + | + | + | - | Asia-1+O |
| 11 | Khontakata | Calf | Epithelium | + | + | + | - | Asia-1+O |
| 12 | Khontakata | Calf | Epithelium | + | + | + | - | Asia-1+O |
| 13 | Khontakata | Calf | Epithelium | + | + | + | - | Asia-1+O |
| 14 | Khontakata | Adult | Epithelium | + | + | + | - | Asia-1+O |
| 15 | Khontakata | Adult | Epithelium | + | + | + | - | Asia-1+O |
| 16 | Khontakata | Calf | Epithelium | + | + | + | - | Asia-1+O |
| 17 | Khontakata | Adult | Epithelium | + | + | + | - | Asia-1+O |
| 18 | Khontakata | Adult | Epithelium | + | + | + | - | Asia-1+O |
| 19 | Khontakata | Adult | Epithelium | + | + | + | - | Asia-1+O |
| 20 | Khontakata | Calf | Epithelium | + | + | + | - | Asia-1+O |
| 21 | Royenda | Adult | Epithelium | + | + | + | - | Asia-1+O |
| 22 | Royenda | Calf | Epithelium | + | + | + | - | Asia-1+O |
| 23 | Royenda | Adult | Epithelium | + | + | + | - | Asia-1+O |
| 24 | Royenda | Calf | Epithelium | + | + | + | - | Asia-1+O |
| 25 | Royenda | Calf | Epithelium | + | + | + | - | Asia-1+O |
| 26 | Royenda | Adult | Epithelium | + | + | + | - | Asia-1+O |
| 28 | Royenda | Calf | Epithelium | + | + | + | - | Asia-1+O |
| 29 | Royenda | Adult | Epithelium | + | + | + | - | Asia-1+O |
| 30 | Royenda | Calf | Epithelium | + | + | + | - | Asia-1+O |
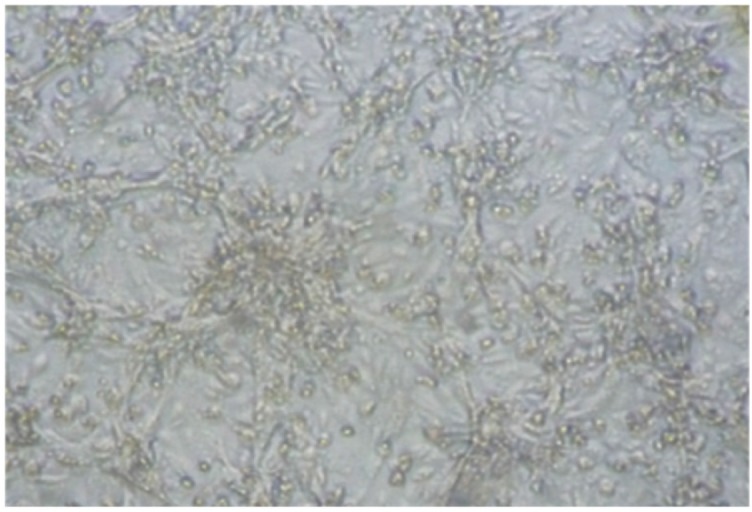
Eight out of 12 epithelial tissue samples were shown CPE during the third passage on BHK21cell culture (Fig. 2). The virus has also shown a positive result when RT-PCR was done from tissue culture fluid. Finally, these isolated three samples were considered for sequencing of the VP1 gene. The obtained sequences have been submitted in the NCBI GenBank under the accession numbers MK056240 (BLRI/BD/435.2), MH704868 (BD/BLRI/434.3), and MH699984 (BD/BLRI/435.4).
Figure 2. Syncytia formation of BHK-21 cells infected with Foot and Mouth Diseases Virus collected from clinical lesion, observed under 40× objective (Leica DMil, Germany).
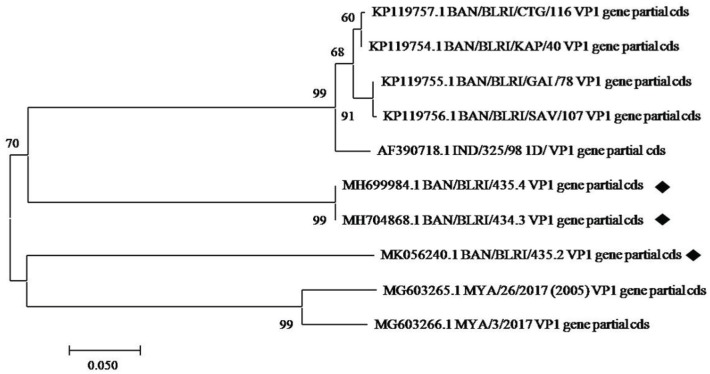
The phylogenetic tree was built by comparing the sequences with other sequences of Bangladesh and neighboring country collected from GenBank database (Fig. 3). All three isolates were found genetically closely related to isolates from the bordering country India and Myanmar. On the other hand, isolates BLRI/BD/435.2 (MK056240.1) and BD/BLRI/435.4 (MH699984.1) were closely related with isolate MG603265.1 (99%) from Myanmar and isolate AF390718.1 (70%) from India, respectively.
Figure 3. Neighbor-joining tree based on the partial virus protein VP1 coding sequence. The sequences generated for this study are marked with ♦ (filled diamond) symbol.
Discussion
The outbreak of FMD should be addressed at an early stage for effective control measures and prevent further transmission stated by Longjam et al.. The FMD virus can be transmitted very rapidly from the onset of shedding to surrounding areas due to its airborne transmission pattern [25].
The RT-PCR detected that all 30 clinical samples were tested for mixed infection with serotypes Asia-1 and Type O. Researchers demonstrated that the infection with multi serotypes of FMD virus increased the severity of infection as well as increased the mortality rates [26]. Serotypes A, Asia-1, and O are circulated in Bangladesh [6–8,27]. On the other hand, serotypes Asia-1 and O are more prevalent that is 31% and serotype A is less prevalent that is 7% [7]. Giasuddin et al. [7] recorded the highest (7%) prevalent in the mixed infection with serotypes Asia-1 and O than serotypes A and O in Bangladesh. On the other hand, Siddique et al. [9] revealed that serotype O was responsible for 82% outbreaks in Bangladesh and it is dominant over serotypes A and Asia-1.
According to the case report, it is confirmed that half (53.87%) of the cattle population of Sarankhola Upazila was infected with FMDV, resulting in massive outbreaks of FMDV. The same case was reported by Kouato et al. [28] as 54% case report of FMD outbreaks in Niger from September to October 2013.
The overall case fatality due to FMDV was 354 (2.27%) with no vaccination. The closely related CFR was demonstrated in Ethiopia at 2.8% by Rufael et al. [29] and 2.9%by Negusssie et al. [30], and 1.4% in South-East Asia during 2000–2010 reported by Madin [31].
In the case of calves, the CFR was 2.48 times higher than adult focused on the outbreak investigation. It indicated that calves are more vulnerable to FMDV due to the virus can invade into the myocardium of a left ventricle, resulting in necrosis of myocardium. This necrotic myocardium looks like a typical shape of striping like tiger skin, commonly known as ‘tiger heart diseases’ [32]. Sometimes calves can die without any clinical signs due to myocardial dysfunction [33]. The calves were kept intensively with mothers so they are in close contact with fomites, mangers, and other feeding materials [34]. The statement of CFR strongly agreed with Rufael et al. [29] and Negusssie et al. [30], they demonstrated 9.33 and 5.56 times more CFR in claves compared with the adult in Ethiopia. Radostits et al. [33] also concluded that the CFR in adult typically 2% and/or up to 20% in calves.
To find out the possible risk factors, questioner surveys were conducted, where the respondent local peoples reveled that FMD becomes epidemic during March–April months, due to after harvesting the paddy all cattle of surrounding villagers grazing freely in that paddy filed. So, cattle shared the common grazing land and watering points with neighboring villages that increase the chances of an outbreak, the observation in agreement with Cleland et al. [35]. The farmers were not aware of FMD vaccination, and they did not vaccinate their cattle, which is one of the contributing risk factors for FMDV outbreak. To prevent the FMDV in endemic area vaccination by circulating serotypes is an effective tool to combat the outbreak [36]. The bovine immunoglobulin G1 (IgG1) can be released via milk from vaccinated cows that have lifesaving importance to reduce early calves mortality against FMDV [37].
In consideration of the geographical location of Sarankhola Upazila, it has a potential risk of FMDV transmission from wildlife (ruminants, pigs) to cattle due to it is surrounded by Sundarban forest and close to Sarankhola Range of Sundarban forest [38].
A discussion with local villagers of outbreak area reveled that every week held a cattle market in Sarankhola Upazila. Where cattle interact with cattle of local sellers and traders those were from neighboring countries as cross-border cattle movement by water vehicles. The unsold cattle can carry the infection to the village that is the likely source of emerging serotypes of FMDV in Bangladesh [8]. FMDV has transboundary disease potential and involves in livestock trade pattern due to spreading through multiple routes and hosts [39,43].
From the above outbreak investigation, it is recommended that farmers should be confirmed regular vaccination with trivalent FMD vaccine of circulating serotypes and good health management due to the endemic area [9]. Farmers also have to sensitize about the potential risk of FMDV transmission of cattle from wildlife and should avoid sharing the common grazing land after harvesting paddy. To avoid transboundary transmission, unauthorized animal movement from neighboring countries should be stopped and strictly maintain quarantine protocols. It also recommended that unsold cattle should never be brought back to the farm and isolate the affected animal.
Conclusion
The investigation was conducted during the start of the outbreak of FMDV in Sarankhola Upazila of Bagerhat district, Bangladesh. The total case was found in 8393 and case fatality rate was 2.27%. The calves were more vulnerable than an adult and CFR of calves reached up to 71.46% that was highest CFR in any count. The investigation could not conclude the source of the outbreak but the risk factors and sequence analysis indicated that it could be from interaction with wildlife and cross-border animals of neighboring countries.
Acknowledgments
The research was funded by the development project of ‘Research on FMD and PPR in Bangladesh’ (Award no. 7080) of Ministry of Fisheries and Livestock, Bangladesh. The authors would like to thanks all members of the outbreak investigation team of Department of Livestock Services, Bangladesh.
Conflict of interests
The authors declare that they have no competing interests.
Authors’ contribution
Md Zulfekar Ali and Eusha Islam designed the experiment and conducted the actual experiments. Both the authors took part in the analysis of data and drafted the manuscript. All the authors read and finally approved the publication of this research work.
References
- [1].Kazi MK, Basant S, Sharma MC, Ratala DR, Akram M. Handbook on livestock and poultry diseases in SAARC countries. SAARC AC; Dhaka: 2007. Parasitic diseases; pp. 124–59. [Google Scholar]
- [2].Mohapatra JK, Das B, Rout M, Sreenivasa BP, Subramaniam S, Sanyal A, Pattnaik B. Alternate vaccine strain selection in the wake of emerging foot-and-mouth disease virus serotype A antigenic variants in India. Vaccine. 2018;36(23):3191–4. doi: 10.1016/j.vaccine.2018.04.090. https://doi.org/10.1016/j.vaccine.2018.04.090. [DOI] [PubMed] [Google Scholar]
- [3].Office International des Epizooties (OIE) Foot-and-mouth disease, in Manual of Standards for Diagnostic Tests and Vaccines for Terrestrial Animals 2010. [10 July 2011]. Available via www.oie.int/manual-of-diagnostic-tests-and-vaccines-for-terrestrial-animals.
- [4].Domingo E, Escarmís C, Baranowski E, Ruiz-Jarabo CM, Carrillo E, Núñez JI, et al. Evolution of foot-and-mouth disease virus. Virus Res. 2003;91(1):47–63. doi: 10.1016/s0168-1702(02)00259-9. https://doi.org/10.1016/S0168-1702(02)00259-9. [DOI] [PubMed] [Google Scholar]
- [5].Knowles NJ, Samuel AR. Molecular epidemiology of foot-and-mouth disease virus. Virus Res. 2003;91(1):65–80. doi: 10.1016/s0168-1702(02)00260-5. https://doi.org/10.1016/S0168-1702(02)00260-5. [DOI] [PubMed] [Google Scholar]
- [6].Loth L, Osmani MG, Kalam MA, Chakraborty RK, Wadsworth J, Knowles NJ, et al. Molecular characterization of foot and mouth disease virus: implications for disease control in Bangladesh. Transbound Emerg Dis. 2011;58(3):240–6. doi: 10.1111/j.1865-1682.2011.01206.x. https://doi.org/10.1111/j.1865-1682.2011.01206.x. [DOI] [PubMed] [Google Scholar]
- [7].Giasuddin M, Mahmud MS, Alam SMS, Samad MA, Islam MR, Ahasan MD, et al. Molecular epidemiology of foot-and-mouth disease viruses circulated in Bangladesh from 2011–2014. Br Microbiol Res J. 2016;16(4):1–13. https://doi.org/10.9734/BMRJ/2016/28040. [Google Scholar]
- [8].Nandi SP, Rahman MZ, Momtaz S, Sultana M, Hossain MA. Emergence and distribution of foot-and-mouth disease virus serotype A and O in Bangladesh. Transbound Emerg Dis. 2015;62(3):328–31. doi: 10.1111/tbed.12113. https://doi.org/10.1111/tbed.12113. [DOI] [PubMed] [Google Scholar]
- [9].Siddique MA, Ali MR, Alam ASMRU, Ullah H, Rahman A, Chakrabarty RP, et al. Emergence of two novel sublineages Ind2001 BD 1 and Ind2001 BD 2 of foot and mouth disease virus serotype O in Bangladesh. Transbound Emerg Dis. 2018;65(4):1009–23. doi: 10.1111/tbed.12834. https://doi.org/10.1111/tbed.12834. [DOI] [PubMed] [Google Scholar]
- [10].Mahapatra M, Upadhyay S, Aviso S, Babu A, Hutchings G, Parida S. Selection of vaccine strains for serotype O foot-and-mouth disease viruses (2007–2012) circulating in Southeast Asia, East Asia and Far East. Vaccine. 2017;35(51):7147–53. doi: 10.1016/j.vaccine.2017.10.099. https://doi.org/10.1016/j.vaccine.2017.10.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Subramaniam S, Pattnaik B, Sanyal A, Mohapatra JK, Pawar SS, Sharma GK, et al. Status of foot and mouth disease in India. Transbound Emerg Dis. 2013;60(3):197–203. doi: 10.1111/j.1865-1682.2012.01332.x. https://doi.org/10.1111/j.1865-1682.2012.01332.x. [DOI] [PubMed] [Google Scholar]
- [12].Mohr S, Deason M, Churakov M, Doherty T, Kao RR. Manipulation of contact network structure and the impact on foot-and-mouth disease transmission. Transbound Emerg Dis. 2018;157:8–18. doi: 10.1016/j.prevetmed.2018.05.006. https://doi.org/10.1016/j.prevetmed.2018.05.006. [DOI] [PubMed] [Google Scholar]
- [13].Dee SA, Bauermann FV, Niederwerder MC, Singrey A, Clement T, de Lima M, et al. Survival of viral pathogens in animal feed ingredients under transboundary shipping models. PLoS One. 2018;13(3):e0194509. doi: 10.1371/journal.pone.0194509. https://doi.org/10.1371/journal.pone.0194509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Abdul-Hamid NF, Hussein NM, Wadsworth J, Radford AD, Knowles NJ, King DP. Phylogeography of foot-and-mouth disease virus types O and A in Malaysia and surrounding countries. Infect Genet Evol. 2011;11:320–8. doi: 10.1016/j.meegid.2010.11.003. https://doi.org/10.1016/j.meegid.2010.11.003. [DOI] [PubMed] [Google Scholar]
- [15].Alam MA, Amin MR, Paul TK, Rizon MK. Clinically detection of foot and mouth disease at Kapasia Upazila under Gazipur district in Bangladesh. J Bangla Agri Uni. 2017;14(2):185–90. https://doi.org/10.3329/jbau.v14i2.32693. [Google Scholar]
- [16].Mannan MA, Siddique MP, Uddin MZ, Parvez MM. Prevalence of foot and mouth disease (FMD) in cattle at MeghnaUpazila in Comilla in Bangladesh. J Bangla Agri Uni. 2009;7(2):317–9. https://doi.org/10.3329/jbau.v7i2.4741. [Google Scholar]
- [17].Jamal SM, Belsham GJ. Foot-and-mouth disease: past, present and future. Vet Res. 2013;44(1):116. doi: 10.1186/1297-9716-44-116. https://doi.org/10.1186/1297-9716-44-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Brito BP, Rodriguez LL, Hammond JM, Pinto J, Perez AM. Review of the global distribution of foot-and-mouth disease virus from 2007 to 2014. Transbound Emerg Dis. 2017;64(2):316–32. doi: 10.1111/tbed.12373. https://doi.org/10.1111/tbed.12373. [DOI] [PubMed] [Google Scholar]
- [19].Onono JO, Wieland B, Rushton J. Constraints to cattle production in a semiarid pastoral system in Kenya. Trop Anim Health Prod. 2013;45:1415–22. doi: 10.1007/s11250-013-0379-2. https://doi.org/10.1007/s11250-013-0379-2. [DOI] [PubMed] [Google Scholar]
- [20].Knight-Jones TJD, Rushton J. The economic impacts of foot and mouth disease—what are they, how big are they and where do they occur? Prev Vet Med. 2013;112(3–4):161–73. doi: 10.1016/j.prevetmed.2013.07.013. https://doi.org/10.1016/j.prevetmed.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Knowles NJ, Wadsworth J, Bachanek-Bankowska K, King DP. VP1 sequencing protocol for foot and mouth disease virus molecular epidemiology. RevSci Tech Off Int Epiz. 2016;35(3):741–55. doi: 10.20506/rst.35.3.2565. https://doi.org/10.20506/rst.35.3.2565. [DOI] [PubMed] [Google Scholar]
- [22].Hall TA. BioEdit. A user friendly biological sequence alignment editor and analysis program for windows 98/98 NT. Nucl Acids Symp Ser. 1999;41:95–8. [Google Scholar]
- [23].Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. https://doi.org/10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–9. doi: 10.1093/molbev/msy096. https://doi.org/10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Nelson N, Paton DJ, Gubbins S, Colenutt C, Brown E, Hodgson S, et al. Predicting the ability of preclinical diagnosis to improve control of farm-to-farm foot-and-mouth disease transmission in cattle. J Clin Microbiol. 2017;55(6):1671–81. doi: 10.1128/JCM.00179-17. https://doi.org/10.1128/JCM.00179-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Valarcher J F, Knowles NJ, Zakharov V, Scherbakov A, Zhang Z, Shang YJ, et al. Multiple origins of foot-and-mouth disease virus serotype Asia 1 outbreaks, 2003–2007. Emerg Infect Dis. 2009;15(7):1046. doi: 10.3201/eid1507.081621. https://doi.org/10.3201/eid1507.081621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].VetsWeb. 100 cows die in two days FMD in Dimla, Bangladesh. 2009. [12 June 2010]. Available via http://www.vetsweb.com/news/100-cowsdie-in-two-days-fmd-in-dimla-bangladesh-381.html.
- [28].Kouato BS, Elliot FM, King DP, Hyera J, Knowles NJ, Ludi AB, et al. Outbreak investigations and molecular characterization of foot and mouth disease viruses circulating in southwest Niger. Transbound Emerg Dis. 2018;65(1):146–57. doi: 10.1111/tbed.12642. https://doi.org/10.1111/tbed.12642. [DOI] [PubMed] [Google Scholar]
- [29].Rufael T, Catley A, Bogale A, Sahle M, Shiferaw Y. Foot and mouth disease in the Borana pastoral system, southern Ethiopia and implications for livelihoods and international trade. Trop Anim Health Prod. 2008;40(1):29–38. doi: 10.1007/s11250-007-9049-6. https://doi.org/10.1007/s11250-007-9049-6. [DOI] [PubMed] [Google Scholar]
- [30].Negusssie H, Kyule MN, Yami M, Ayelet G. Outbreak investigations and genetic characterization of foot-and-mouth disease virus in Ethiopia in 2008/2009. Trop Anim Health Prod. 2011;43(1):235–43. doi: 10.1007/s11250-010-9683-2. https://doi.org/10.1007/s11250-010-9683-2. [DOI] [PubMed] [Google Scholar]
- [31].Madin B. An evaluation of Foot-and-Mouth Disease outbreak reporting in mainland South-East Asia from 2000 to 2010. Prev Vet Med. 2011;102(3):230–41. doi: 10.1016/j.prevetmed.2011.07.010. https://doi.org/10.1016/j. prevetmed.2011.07.010. [DOI] [PubMed] [Google Scholar]
- [32].Kitching RP. Clinical variation in foot and mouth disease: cattle. Rev Sci Tech Off Int Epiz. 2002;21(3):499–502. doi: 10.20506/rst.21.3.1343. https://doi.org/10.20506/rst.21.3.1343. [DOI] [PubMed] [Google Scholar]
- [33].Radostits OM, Gay CC, Blood DC, Hinchcliff KW. 9th. London, UK: W. B Saunders; 2000. Veterinary medicine: a textbook of the diseases of cattle, sheep, pigs, goats and horses. [Google Scholar]
- [34].Emami J, Rasouli N, McLaws M, Bartels CJM. Risk factors for infection with Foot-and-Mouth Disease virus in a cattle population vaccinated with a non-purified vaccine in Iran. Prev Vet Med. 2015;119(3–4):114–22. doi: 10.1016/j.prevetmed.2015.03.001. https://doi.org/10.1016/j. prevetmed.2015.03.001. [DOI] [PubMed] [Google Scholar]
- [35].Cleland PC, Baldock FC, Chamnanpood P, Gleeson LJ. Village level risk factors for foot-and-mouth disease in northern Thailand. Prev Vet Med. 1996;26(3–4):253–61. https://doi.org/10.1016/0167-5877(95)00552-8. [Google Scholar]
- [36].Novo SG, Malirat V, Maradei ED, Espinoza AM, Smitsaart E, Pedemonte AR, et al. Antigenic and immunogenic spectrum of foot-and-mouth disease vaccine strain O1 Campos against representative viruses of topotypes that circulated in Asia over the past decade. Vaccine. 2017;35(18):2303–7. doi: 10.1016/j.vaccine.2017.03.026. https://doi.org/10.1016/j. vaccine.2017.03.026. [DOI] [PubMed] [Google Scholar]
- [37].Armstrong RM, Mathew ES. Predicting herd protection against foot-and-mouth disease by testing individual and bulk tank milk samples. J Virol Methods. 2001;97(1–2):87–99. doi: 10.1016/s0166-0934(01)00342-1. https://doi.org/10.1016/S0166-0934(01)00342-1. [DOI] [PubMed] [Google Scholar]
- [38].Thomson GR, Bastos ADS. Foot-and- Mouth Disease. In: Coetzer JAW, Tustin RC, editors. Infectious Diseases of Livestock. 2nd. South Africa: Oxford University Press; 2004. pp. 1324–65. [Google Scholar]
- [39].Di Nardo A, Knowles NJ, Paton DJ. Combining livestock trade patterns with phylogenetics to help understand the spread of foot and mouth disease in sub-Saharan Africa, the Middle East and Southeast Asia. Rev Sci Tech OIE. 2011;30(1):63. doi: 10.20506/rst.30.1.2022. https://doi.org/10.20506/rst.30.1.2022. [DOI] [PubMed] [Google Scholar]
- [40].Vangrysperre W, De Clercq K. Rapid and sensitive polymerase chain reaction based detection and typing of foot-and-mouth disease virus in clinical samples and cell culture isolates, combined with a simultaneous differentiation with other genomically and/or symptomatically related viruses. Arch Virol. 1996;141(2):331–44. doi: 10.1007/BF01718403. https://doi.org/10.1007/BF01718403. [DOI] [PubMed] [Google Scholar]
- [41].Callens M, De Clercq K. Differentiation of the seven serotypes of foot-and-mouth disease virus by reverse transcriptase polymerase chain reaction. J Virol Meth. 1997;67(1):35–44. doi: 10.1016/s0166-0934(97)00074-8. https://doi.org/10.1016/S0166-0934(97)00074-8. [DOI] [PubMed] [Google Scholar]
- [42].ESRI ArcGIS for Desktop. Geographic Information Systems (GIS) software ESRI ArcGIS105 version 10.2.1. [26 May 2019];2018 http://www.esri.com/software/arcgis/arcgis-for-desktop.
- [43].Ali MZ, Sultana S, Bhuiyan ZA. Comparative efficacy of traditional medicine for the healing of aphthous ulcers due to foot and mouth disease in cattle. Am J Pharm Pharmacol. 2018;5(1):1–5. [Google Scholar]