Abstract
Objective:
The incidence of salmonellosis in humans and animals is still high due to the occurrence of virulence factors in Salmonella enterica which play a role in the process of infection in the host and the spread of disease and most of the S. enterica can infect humans and animals. The present study was aimed to identify Salmonella Enteritidis and detect virulence genes related to Salmonella pathogenicity islands (SPIs) and Salmonella plasmid virulence (Spv).
Materials and Methods:
A total of 27 S. Enteritidis archive isolates belonging to the National Veterinary Drug Assay Laboratory (NVDAL) were used in this study. The bacteria were collected in 2016 and 2017 from samples of the cloaca and fecal swabs from layer and broiler farms in five provinces of Java Island. Isolates were cultured in specific media, biochemical tests and Gram staining. Detection of S. Enteritidis and virulence genes was done by polymerase chain reaction (PCR) method.
Results:
Identification of serovar showed 100% (27/27) isolates were positive for the sdfI gene (304 bp). The result confirmed that all strains were S. Enteritidis. PCR based detection of virulence genes showed that 100% of isolates had virulence genes in SPI-1 to SPI-5, namely, invA, ssaQ, mgtC, spi4D, and pipA genes. All the isolates (27/27) were also positive to spvB gene-based PCR.
Conclusion:
All the isolates of S. Enteritidis in this study carry virulence genes related to SPI-1 to SPI-5 and plasmid virulence. The existence of virulent genes indicates that the S. Enteritidis strain examined in this study is highly virulent and poses a potential threat of worse disease outcome in humans and animals.
Keywords: Java Island, S. Enteritidis, virulence genes, SPIs, virulence plasmids
Introduction
Java Island is the center of poultry farming in Indonesia. More than 66% of the national population of layer and broiler chickens [1] are on Java Island. The large population of chickens allows the transmission of animal diseases, one of which is Salmonella infection. Cases of Salmonella infection continue to occur with a high percentage in both animals and humans. The European Surveillance System reported a total of 31 829 cases in 2015 and 3 709 incidents in early 2016 caused by S. Enteritidis infection from 18 countries in Europe [2]. The report of Salmonella enterica contamination in poultry products in several countries tends to be high, including S. Paratyphi B 76% and S. Heidelberg 23% in Columbia [3], S. Enteritidis 37.3% in Egypt [4], and S. Enteritidis 30% in Malaysia [5]. Salmonella enterica infections in humans mostly occur through foodborne transmissions. Humans can be infected by S. Enteritidis by consuming animal products (poultry, swine, fish, fish, crustaceans, and shellfish) and also indirectly related to the consumption of cheese, chocolate, vegetables, and juices [6]. Generally associated with gastrointestinal organs, S. enterica can cause invasive infections and bacteremia in young age and elderly people as well as in individuals with low immunity [7].
Salmonella enterica have many virulence factors that are necessary for the infection process to the host and the spread of the disease. The main virulence factor of S. enterica was determined in the chromosomal gene which was located in the Salmonella pathogenicity islands (SPI). SPI is needed for invasion and proliferation in host cells [8]. Some S. enterica serovars also have virulence plasmids called Salmonella plasmid virulence (Spv). This plasmid virulence has several virulence genes that cause suppression of host’s innate immune response [9]. Horizontal transfer of virulence genes can cause bacterial pathogenesis evolution. This evolution is the force that initiates the emergence of new pathogenic species, which can adapt to new hosts and are specific sites in the host. Most virulence genes can move between bacteria called pathogenicity island and are known as a quantum leap in the process of bacterial pathogenesis evolution [10]. In Salmonella bacteria, virulence gene clusters are located in specific areas of the chromosome called SPI. SPI act as virulence factors in the pathogenicity of Salmonella. SPI is characterized by an elemental composition that is different from the core genome. SPI is often associated with mobile genetic elements and tRNA genes, such as insertion element, phage genes, or transposons [11]. This difference indicates that there are sequences of SPI compilers obtained through horizontal transfers, although the origin of the sequence and the process of transferring sequences is still unclear [12].
Several studies on the isolation of Salmonella bacteria from animal products have been reported [13,14]. However, research on virulence genes in S. Enteritidis from samples of cloacal swabs and poultry feces is still lacking in Indonesia. This study aims to identify S. Enteritidis and detect virulence genes of S. Enteritidis originated from layer and broiler farms in Java Island.
Materials and Methods
Bacterial isolates and ethical approval of experimental animals
A total of 27 S. Enteritidis isolated from cloacal swabs and feces collected from the layer and broiler farms in the Province of West Java, Banten, Yogyakarta, Central Java, and East Java in 2016 and 2017 were used in the current study (Table 1). All isolates were belonging to NVDAL, West Java, Indonesia. No live animals were used in the present study. Therefore, no ethical approval was needed to conduct this study.
Table 1. Data of archival isolates of Salmonella Enteritidis bacteria (n = 27).
| No | Province | Districts | Farm | Isolate Code | Age | Year of sampling |
|---|---|---|---|---|---|---|
| 1 | Banten | Tangerang | Layer | SE/100/16L/Tangerang | 24 weeks | 2016 |
| 2 | Serang | Layer | SE/101/16L/Serang | 18 weeks | 2016 | |
| 3 | Serang | Layer | SE/105/16L/Serang | 18 weeks | 2016 | |
| 4 | West Java | Bogor | Broiler | SE/211/17B/Bogor | 13 days | 2017 |
| 5 | Bogor | Broiler | SE/236/17B/Bogor | 29 days | 2017 | |
| 6 | Bogor | Broiler | SE/237/17B/Bogor | 29 days | 2017 | |
| 7 | Ciamis | Broiler | SE/411/17B/Ciamis | 19 days | 2017 | |
| 8 | Ciamis | Broiler | SE/412/17B/Ciamis | 19 days | 2017 | |
| 9 | Ciamis | Broiler | SE/429/17B/Ciamis | 7 days | 2017 | |
| 10 | Ciamis | Broiler | SE/430/17B/Ciamis | 7 days | 2017 | |
| 11 | Ciamis | Broiler | SE/431/17B/Ciamis | 34 days | 2017 | |
| 12 | Ciamis | Broiler | SE/443/17B/Ciamis | 12 days | 2017 | |
| 13 | Central Java | Banyumas | Broiler | SE/157/17B/Banyumas | 15 days | 2017 |
| 14 | Kendal | Layer | SE/069/17L/Kendal | 28 weeks | 2017 | |
| 15 | Kendal | Broiler | SE/119/17B/Kendal | 35 days | 2017 | |
| 16 | Kendal | Broiler | SE/120/17B/Kendal | 32 days | 2017 | |
| 17 | Semarang | Layer | SE/155/16L/Semarang | 28 weeks | 2016 | |
| 18 | Semarang | Layer | SE/158/16L/Semarang | 28 weeks | 2016 | |
| 19 | Special Region of Yogyakarta | Bantul | Layer | SE/115/16L/Bantul | 1.5 years | 2016 |
| 20 | Bantul | Layer | SE/117/16L/Bantul | 1.5 years | 2016 | |
| 21 | Bantul | Layer | SE/118/16L/Bantul | 1.5 years | 2016 | |
| 22 | East Java | Kediri | Layer | SE/467/17L/Kediri | 20 months | 2017 |
| 23 | Kediri | Layer | SE/496/17L/Kediri | 12 months | 2017 | |
| 24 | Kediri | Layer | SE/502/17L/Kediri | 20 weeks | 2017 | |
| 25 | Kediri | Broiler | SE/508/17B/Kediri | 23 days | 2017 | |
| 26 | Kediri | Broiler | SE/511/17B/Kediri | 22 days | 2017 | |
| 27 | Kediri | Broiler | SE/513/17B/Kediri | 22 days | 2017 |
Identification of Salmonella Enteritidis and biochemical tests
Freeze dry bacteria were revived in Heart Infusion Broth (HIB) medium at 37°C for 24 h. The bacteria were then cultured on Xylose Lysine Deoxycholate (XLD) and Rapid Salmonella Agar (RSA) media which were incubated at 37°C for 24 h. Specific colonies of Salmonella on RSA media were cultured on the Heart Infusion agar (HIA) media for 24 h at 37°C. Then, the bacterial colonies were stained using Gram staining and were cultured on Triple Sugar Iron Agar (TSIA) media for 24 h at a temperature of 37°C. Salmonella bacteria cultured on TSIA media were subjected to be tested for biochemistry including indole test, Methyl Red-Voges Proskauer (MR-VP), lysine, and citrate. Indole test was done using Sulfide Indole Motility (SIM) media and incubated at 37°C for 24 h. After incubation, Kovac’s reagent indicator was given. The MR-VP test used MR-VP broth media and incubated at 37°C for 48 h. After incubation, the media were added with the reagent indicators [methyl red, Alpha naphtol, and potassium hydroxide (KOH)]. Lysine test was carried out using Lysine Decarboxylase Broth (LDB) media and incubated at 37°C for 48 h. Citrate test was done using Simmons Citrate Agar media and incubated at 37°C for 24 h [15].
Identification of Salmonella Enteritidis
Bacterial isolates positively as Salmonella in the biochemical test were enriched on HIB media for 24 h at 37°C. The isolates were purified by culturing on HIA media for 24 h at 37°C. Colonies grown on HIA media were taken 1–2 ose and dissolved in 100 μl PBS to extract DNA using the boiling method [16]. The samples were homogenized using vortex for 10–30 sec then heated at 100°C for 10 min. The samples were cooled for 2 min and centrifuged at 14000 rpm for 2 min, and the supernatant was taken as much as 50 μl as a DNA master stock.
Salmonella Enteritidis was identified by detecting sdfI gene on the amplicon length of 304 bp. The primary forward sequence of sdfI gene used was 5´-TGT GTT TTA TCT GAT GCA AGA GG-3´ while the primary reverse sequence of sdfI gene used was 5´-TGA ACT ACG TTC GTT CTT CTG G-3´ [17]. The amplification reagent of S. Enteritidis used in this study was the HotStarTaq® Master Mix Kit (Qiagen, Germany) with a total polymerase chain reaction (PCR) reagent volume of 25 μl consisting of 12.5 μl HotStarTaq master mix, 1 μl forward primer (20 μM), 1 μl reverse primer (20 μM), 5.5 μl dH2O, and 5μl DNA template. The PCR process begins with a pre-denaturation cycle of 95°C for 15 min, and lastly the next process was 30 cycles of denaturation at 94°C for 1 min, annealing at 57.5°C for 1 min, then 1 min of extension at 72°C, and a final extension at 72°C for 10 min. Visualization of PCR results was done using 1.5% agarose gel, SYBR safe staining, and 100-bp marker. The documentation was done using Gel Documentation Systems (Thermo Fisher Scientific, USA).
Detection of Virulence Genes in S. Enteritidis
Simplex PCR was performed targetinginvA (SPI-1), ssaQ (SPI-2), mgtC (SPI-3), spi4D (SPI-4), pipA (SPI-5), and spvB genes (plasmid virulence). The primer sequences, gene targets, amplicon sizes, and annealing temperatures are presented in Table 2.
Table 2. List of primer sequence virulence genes of S. Enteritidis.
| Gene target | Forward (F) and reverse (R) base pair sequence | Amplicon size (Annealing temperature) | Reference |
|---|---|---|---|
| invA (SPI-1) | (F) 5´-GTGAAATTATCGCCACGTTCGGGCAA-3´ (R) 5´-TCATCGCACCGTCAAAGGAACC-3´ |
284-bp (59°C) | [18] |
| ssaQ (SPI-2) | (F) 5´-GAATAGCGAATGAAGAGCGTCC-3´ (R) 5´-CATCGTGTTATCCTCTGTCAGC-3´ |
677-bp (60°C) | [19] |
| mgtC (SPI-3) | (F) 5´-TGACTATCAATGCTCCAGTGAAT-3´ (R) 5´-ATTTACTGGCCGCTATGCTGTTG-3´ |
655-bp (60°C) | [19] |
| spi4D (SPI-4) | (F) 5´-GAATAGAAGACAAAGCGATCATC-3´ (R) 5´-GCTTTGTCCACGCCTTTCATC-3´ |
1231-bp (60°C) | [19] |
| pipA (SPI-5) | (F) 5´- CTCTTGGATGATTTTCTTCTTTA-3´ (R) 5´-CTTATCTCAGGCGCGGGTGG-3´ |
406-bp (58°C) | [19] |
| spvB (Plasmid virulen) | (F) 5´-CTATCAGCCCCGCACGGAGAGCAGTTTTTA-3´ (R) 5´-GGAGGAGGCGGTGGCGGTGGCATCATA-3´ |
717-bp (60.6°C) | [20] |
PCR was performed in 25 μl reaction volume containing master mix reagents consisting of 12.5 μl HotStarTaq master mix (Qiagen, Germany), 1 μl forward primer (5 μM), 1 μl reverse primer (5 μM), 5.5μl dH2O, and 5μl DNA templates. The PCR process begins with a pre-denaturation cycle at 95°C for 15 min, followed by 30 cycles of 94°C for 1 min, annealing for 1 min (the primer and temperature used were shown in Table 2), and extension at 72°C for 1 min, and then a final extension at 72°C for 10 min. The PCR product was analyzed using 1.5% agarose (Thermo Fisher Scientific, USA) and visualized using Gel Documentation Systems (Thermo Fisher Scientific, USA) upon staining with SYBR safe (Thermo Fisher Scientific, USA).
Results and Discussion
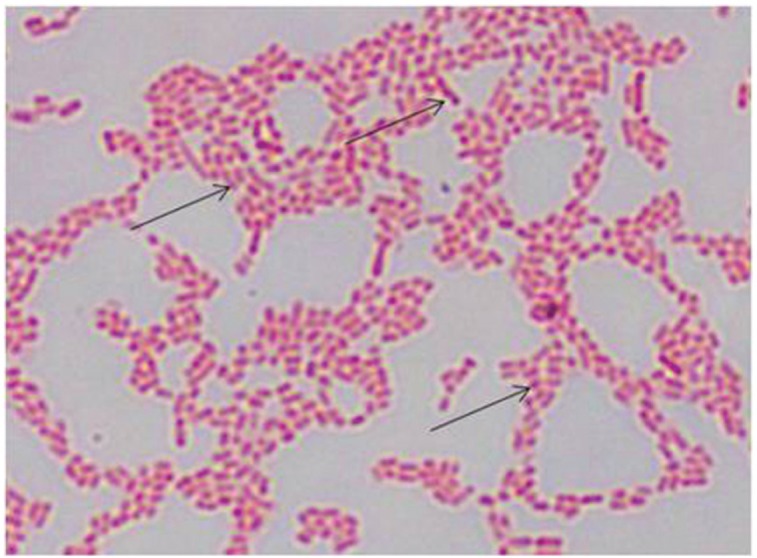
The results of the culture of S. Enteritidis isolates on XLD and RSA media showed that 100% (27/27) of isolates growing with typical colonies of Salmonella spp. Macroscopic observations of XLD showed pink colonies with a black center; there are also black colonies as a whole because H2S was produced (Fig. 1A). In RSA media, bacterial colonies are circular and the color is purple (Fig. 1B). Microscopic examination with Gram staining showed the stem morphology of S. Enteritidis bacteria and the color is red (Fig. 2).
Figure 1. Macroscopic observation of S. Enteritidis in XLD and RSA media. (A) S. Enteritidis colonies on XLD media. The arrows indicate S. Enteritidis colonies are pink with the black center because H2S was produced, there are also colonies that are full black because more H2S was produced. (B) S. Enteritidis colonies in RSA media. The arrows indicate the circular S. Enteritidis colonies and the color is purple.
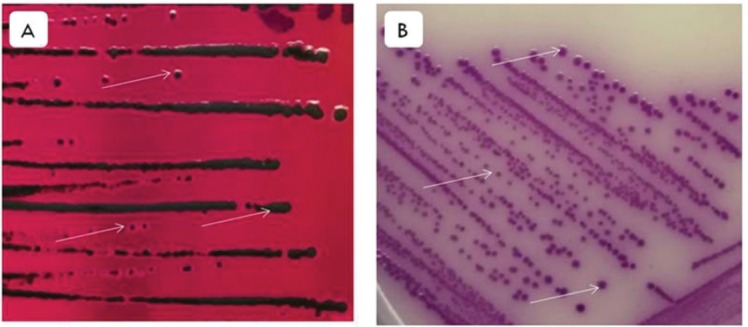
Figure 2. Microscopic observation of S. Enteritidis with Gram staining. The arrow indicates the morphology of the S. Enteritidis in the form of a stem and the color is red (1000×).
The results of culture on TSIA media showed that 100% (27/27) of the isolates reacted with the transformation to red in the TSIA slant media and yellow in the bottom part, H2S and gas bubbles were formed in the bottom part. Salmonella bacteria cannot ferment lactose and sucrose, so the TSIA media in the slant remains red. However, Salmonella bacteria can ferment glucose which produces pyruvic acid that causes the media on the bottom to become acidic and yellow because the red phenol indicator turns yellow [21]. The biochemical test showed that 100% (27/27) of S. Enteritidis isolates have a positive reaction in the MR, lysine, and citrate test. All isolates showed a negative reaction in the indole and VP test (Table 3).
Table 3. Results of biochemical tests on Salmonella Enteritidis (n = 27).
| Type of test | Result | Informationc | |
|---|---|---|---|
| Positive (+)a | Negative (-)b | ||
| Indole | 0 | 27 | The surface of the media is yellow; no pink ring is formed after the Kovac’s reagent was added |
| MRd | 27 | 0 | The methyl red indicator diffuses with the MR-VP media so that the color of the media is red |
| VPe | 0 | 27 | There is no red color on MR-VP media after alpha naphtol, and KOH were added |
| Lysine | 27 | 0 | The LDBf media returns to purple |
| Citrate | 27 | 0 | The development of Salmonella colonies shown with changes in the color of the media from green to blue |
The number of isolates was positive for biochemical tests,
the number of negative isolates against biochemical tests,
biochemical reactions that occurred,
MR =methyl red,
VP = Voges Proskauer,
LDB = Lysine Decarboxylase Broth.
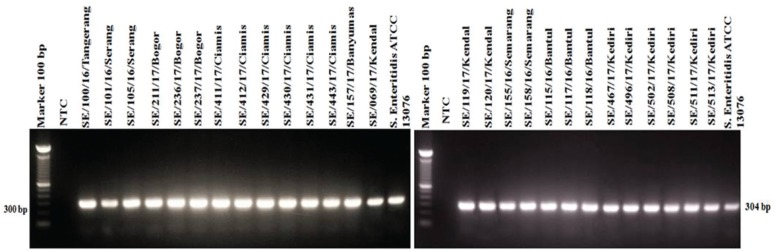
The results of detection of S. Enteritidis with the SdfI gene target showed all isolates and positive control of S. Enteritidis ATCC 13076 were positive for the sdfI gene with an amplicon length of 304 bp (Fig. 3). These results confirm that all isolates (27/27) of the archives were Salmonella Enteritidis.
Figure 3. Amplification of sdfI (304-bp) gene encoding S. Enteritidis. All or 100% (27/27) of isolates showed positive results against sdfI gene. NTC: non template control; S. Enteritidis ATCC 13076 as a positive control shows positive results against sdfI gene.
S. Enteritidis is the only serovar that has the unique fragment of sdfI gene. The unique fragments were isolated using the Suppression Subtractive Hybridization (SSH) method by multiplying fragments through matrix [17]. This unique fragment is not found in other bacteria so that its existence can be used as a diagnostic marker. The sdfI primer sequence specificity has been tested to 73 S. enterica non-Enteritidis isolates. The results showed sdfI primer did not amplify all S. enterica non-Enteritidis isolates. Other tests used 33 S. Enteritidis isolates from humans, pigs, cattle, turkeys, chickens, and poultry environmental samples. The result showed positive for sdfI primer in all 33 isolates [17].
Identification based on the results of bacterial culture on specific media, biochemical tests, Gram staining, and molecular identification showed that all isolates were S. Enteritidis. Rapid test and detection methods for the identification of S. enterica until the level of serovar are very important and fundamental to detect S. Enteritidis because of the high prevalence of salmonellosis and many serovars of S. enterica. The identification of S. Enteritidis with the sdfI gene target can be used because it provides a reliability benefit of an important test method with rapid, sensitive, and specific test results only for S. Enteritidis.
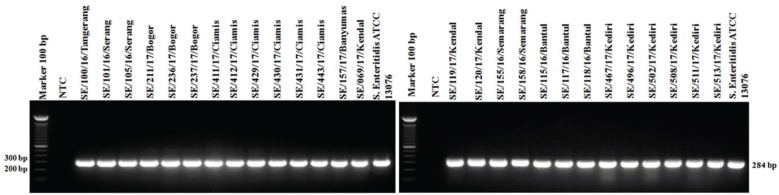
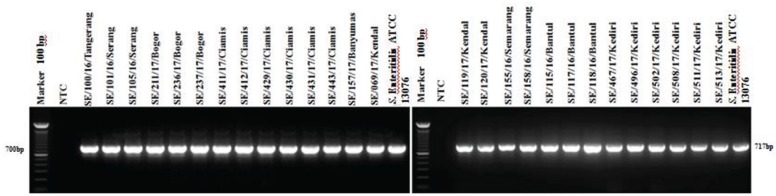
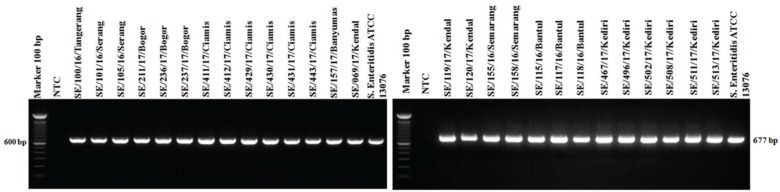
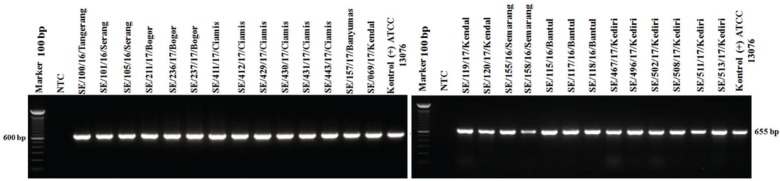
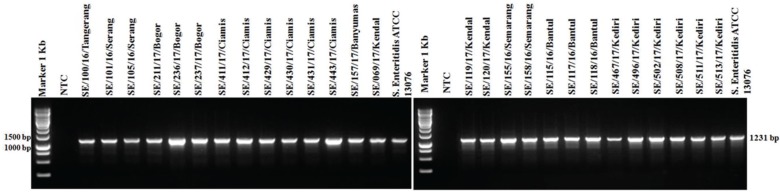
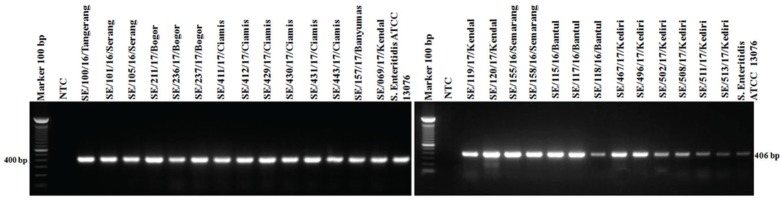
PCR targeting the virulence genes in SPI and SPV showed 100% isolates (27/27) of S. Enteritidis were positive to invA, ssaQ, mgtC, spi4D, pipA, and spvB genes (Figs. 4–9).
Figure 4. Amplification of invA gene on SPI-1 of S. Enteritidis. All isolates (27/27) showed positive invA results (284-bp). NTC: non-template control; positive control: S. Enteritidis ATCC 13076 showed positive invA results (284-bp).
Figure 9. Amplification of spvB gene on Spv of S. Enteritidis. All isolates (27/27) showed positive spvB results (717-bp). NTC: non-template control; positive control: S. Enteritidis ATCC 13076 proved positive of the spvB gene (717-bp).
There are a total of 17 SPI described in Salmonella [22]. SPI-1 to SPI-5 have been widely characterized and can be found in most S. enterica. Other SPIs were less widely distributed [12]. While the genes that make up each SPI are not known whether all serovars have the same SPI constituent gene. In S. Typhimurium [23] and S. Typhi [22], the SPI-1 to SPI-5 genes have been wholly identified. Amavisit et al. [24] reported that there were genetic variations between SPI-1, SPI-3, and SPI-5, while the genetic variation of SPI-4 and SPI-2 was not easily changed among 13 Salmonella serovars isolated from cattle, pigs, poultry, horses, environment, and humans [24].
Salmonella bacteria can be found in the digestive organs of both animals and humans. Besides being found in feces, eggs, and meat, Salmonella also can be found at any tools and part of the rooms in the abattoirs, such as knife, transport crate, chopping board, or even in the drain water and wash water, as well as in defeathering machines, drain swabs, and apron [25]. Salmonella can also be found in animal feed ingredients, soil, bedding, litter, and feces which are generally a source of Salmonella contamination on farms. Chickens can be exposed to and infected with Salmonella bacteria through vertical transmission. In addition, horizontal transmission from the environment of the genes can occur through equipment, transportation, feed, and vectors (insects, rodents, as well as humans) [26].
The mechanism of Salmonella infection and the role of SPI-2 and SPI-1 in bacterial infiltration and invasion of host epithelial cells, proliferation in mononuclear to systemic infections are as follows. Salmonella contained in the feed will enter the digestive tract, after passing gastric acid will go to the small intestine, large intestine, and cecum which has a low pH, and Salmonella can survive in a low pH environment. In the small intestine, Salmonella will attach to host epithelial cells, then SPI-1 will express the first Type 3 Secretion Systems (T3SS) or T3SS-1 which are multiprotein complexes that facilitate bacteria in the process of invasion and absorption through endothelial [22]. The T3SS-1 acts as a syringe molecule in the form of a channel to connect the bacterial cytoplasm with the membrane of the host cell. The T3SS-1 functions to transfer toxins and effector proteins of Salmonella encoded by SPI-1 into host intestinal cells. The number of effector proteins transferred is more than 20 structural proteins and regulating proteins [27]. One function of the effector protein makes the ruffling process on the host cell surface. Due to the wrinkled, supple, and curved surface of the host cell, Salmonella can easily enter the host cell membrane. Salmonella that have passed through the host cell membrane will bind to the host cell membrane to form a vacuole so that it is localized and known as Salmonella containing vacuole (SCV) [28].
Figure 5. Amplification of ssaQ gene on SPI-2 of S. Enteritidis. All isolates (27/27) showed positive results ssaQ (677-bp). NTC: non-template control; positive control: S. Enteritidis ATCC 13076 showed positive results of ssaQ gene (677-bp).
SPI-2 of Salmonella found in the SCV will express the second T3SS (T3SS-2). Through T2SS-2, the secretion of effector proteins has the function in the process of systemic infection and pathogenesis of infection inside the cell [26]. Other function of the effector proteins is their interaction with the motor protein and cytoskeleton in the formation of Salmonella induced filaments. T3SS-2 can modulate SCV movements to avoid fusion with lysosomes. SCV has a vital role in the process of survival as well as the proliferation of Salmonella bacteria in the macrophages and enterocytes [29].
After the Salmonella move through the intestinal epithelial cells, then enter the Peyer’s patches that will be presented as antigen-presenting cells as phagocytic and neutralization efforts [30]. Salmonella that can survive in mononuclear cells of Peyer’s patches will continue to grow and through the reticuloendothelial system will reach the liver and spleen to enter the bloodstream [31]. Salmonella will be excreted through feces which will contaminate water and become a source of transmission of Salmonella that can be spread through insects and other animals so that the bacterial life cycle will continue and increasingly spread [32].
All isolates S. Enteritidis (27 isolates) in this study carried virulence genes invA, ssaQ, and mgtC on the amplicon length 284, 677, and 655 bp, respectively. (Figs. 4–6). The central role of SPI-1 is to express the structure of T3SS and encode many effector proteins that are very important in the process of epithelial cell invasion and the invA virulence genes detected in the present study was reported to play a role in the process of invasion [22]. SPI-1 also plays a role in inducing inflammatory responses in the intestine [33]. Many genes are encoded by SPI-2 to form the secretion systems apparatus (ssa) is T3SS structure, the SPI-2 virulence gene detected in this study is the ssaQ gene which plays a role in the formation of the T3SS structure [25]. SPI-2 also has a gene that encodes effector proteins, namely, secretion system effector (Sse) and encodes specific chaperones of effector proteins called secretion system chaperone (Ssc). These three systems make Salmonella survive, multiply in phagocytic cells, and facilitate the spread of systemic infections in organs [27,34]. SPI-3 encoded the magnesium transport system that is regulated by the mgtB and mgtC genes. Availability of magnesium plays an essential role as a key signal for Salmonella inside the host cell [35]. The mgtC virulence gene detected in this study also plays a role in activating Na+, K+, and ATPase that regulates the potential membrane of Salmonella [36].
Figure 6. Amplification of mgtC gene on SPI-3 of S. Enteritidis. All isolates (27/27) showed mgtC positive results (655-bp). NTC: non-template control; positive control: S. Enteritidis ATCC 13076 showed positive of mgtC gene (655-bp).
Detection of virulence genes of spi4D on SPI-4 and pipA on SPI-5 in this study showed that 100% isolates were positive (Figs. 7 and 8). The role of the spi4D virulence gene in SPI-4 is to encode Type 1 Secretion Systems (T1SS), cognate substrate protein, and mediate adhesion [37], whereas the virulent gene of pipA encodes effector proteins for T3SS in SPI-2 [38].
Figure 7. Amplification of spi4D gene on SPI-4 of S. Enteritidis. All isolates (27/27) showed positive results of spi4D (1231-bp). NTC: non-template control; positive control: S. Enteritidis ATCC 13076 showed positive results spi4D (1231-bp).
Figure 8. Amplification of pipA gene on SPI-5 of S. Enteritidis. All isolates (27/27) showed positive pipA results (406-bp). NTC: non-template control; positive control: S. Enteritidis ATCC 13076 showed positive of pipA gene results (406-bp).
Plasmid containing the Salmonella plasmid virulence (spv) locus has an essential role in the bacterial multiplication in organs of the reticuloendothelial system, such as the spleen and the liver [39]. Virulent plasmids play a crucial role in septicemia caused by non-typhoid Salmonella serovar infections. In this study, all S. Enteritidis isolates (27 isolates) were positive for spvB (Fig. 9), and common results were found in study by Amini et al. [40] that the spv genes were prevalent in 90% (humans), 88.6% (poultry), and 100% (bovine) [40]. In contrast, Smith et al. [41] reported lower prevalence (5%) of spvB gene in Salmonella spp. from meat samples [41]. However, the distribution of virulence genes among Salmonella has been confirmed by other researchers irrespective of the hosts [42].
Detection of virulence genes of S. Enteritidis has been studied widely. Mezal et al. [43] conducted a study on 60 isolates of S. Enteritidis from animals and humans targeting virulence genes including invA and spvB [43]. The result showed that the invA gene was found to be 100% positive, but two isolates were negative for the spvB gene, whereas in this study, spvB genes were found in all S. Enteritidis isolates. In the study of Osman et al. [44] which detected several virulence genes including invA, ssaQ, and mgtC in three isolates of S. Enteritidis from imported ducks and two isolates of S. Enteritidis from domestic ducks. As a result, all isolates (5/5) were positive for the invA gene, but in the imported duck, there was one isolate negative for the mgtC gene and three isolates were negative of the ssaQ gene [44]. These results indicate that although in the same serovar, S. Enteritidis, the virulence genes they possess may differ because in this study all S. Enteritidis isolates had virulence genes (ssaQ and mgtC), whereas in the study of Osman et al. [44], there are several S. Enteritidis that do not have the ssaQ and mgtC genes. The differences in the occurrence of virulence genes may be due to the transfer of virulence genes obtained through horizontal transfer mechanisms.
Two key virulence properties possessed by S. enterica species including S. Enteritidis are the capability of attacking nonfagocyte cells such as enterocytes and phagocytic cells such as macrophages. Salmonella in phagocytic cells is facultative intracellular pathogens so that they can replicate in eukaryotic host cells [8]. The results of the study found that all S. Enteritidis isolates had six virulent genes, indicating they are capable of infecting and survive in the host’s body.
Conclusion
Isolates of S. Enteritidis from layer and broiler farms in Java Island carried virulence genes related to SPI-1 to SPI-5 and plasmids virulence. The existence of virulent S. Enteritidis in both layer and broiler farms can be a source of human and animal infection through contaminated eggs, meat, and the environment.
Acknowledgments
The authors gratefully acknowledge Dr. Sri Mukartini from National Veterinary Drug Assay Laboratory, West Java, Indonesia, for providing the facilities throughout the study. The funder of the study was The Agency of Agriculture Extension and Human Resources Development (AAEHRD) of the Ministry of Agriculture of the Indonesian Republic.
Conflict of interests
The authors declare that they have no conflict of interest.
Authors’ contribution
EA designed the study and the manuscript drafted under the supervision of AI and NLPIM. IR and I collected samples and compiled the resource materials. EA conducted the experimental and data analysis under the supervision of AI. EA performed the molecular test detection of virulence genes under the supervision of NLPIM. The final manuscript has been read and approved by all authors.
References
- [1].DJPKH. Direktorat Jenderal Peternakan dan Kesehatan Hewan. Jakarta, Indonesia: Kementerian Pertanian; 2017. Populasi Ayam Ras Pedaging dan Ras Petelur Menurut Provinsi. https://doi.org/10.18343/jipi.23.2.88. [Google Scholar]
- [2].EFSA. European Food Safety Authority, European Centre for Disease Prevention and Control. Stockholm, Sweden: EFSA; 2016. Multi-country outbreak of Salmonella Enteritidis phage typ8, MLVA type 2-9-7-3-2 and 2-9-6-3-2 infections. https://doi.org/10.2903/sp.efsa.2016.en-1110. [Google Scholar]
- [3].Donado-Godoy P, Byrne BA, Leon M, Castellanos R, Vanegas C, Coral A, et al. Prevalence, resistance patterns, and risk factors for antimicrobial resistance in bacteria from retail chicken meat in Colombia. J Food Prot. 2015;78:751–9. doi: 10.4315/0362-028X.JFP-14-349. https://doi.org/10.4315/0362-028x.jfp-14-349. [DOI] [PubMed] [Google Scholar]
- [4].Abd-Elghany SM, Sallam KI, Abd-Elkhalek A, Tamura T. Occurrence, genetic characterization and antimicrobial resistance of Salmonella isolated from chicken meat and giblets. Epidemiol Infect. 2015;143:997–1003. doi: 10.1017/S0950268814001708. https://doi.org/10.1017/s0950268814001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Thung TY, Mahyudin NA, Basri DF, Radzi CWJ, Nakaguchi Y, Nishibuchi M, et al. Prevalence and antibiotic resistance of Salmonella Enteritidis and Salmonella Typhimurium in raw chicken meat at retail markets in Malaysia. Poult Sci. 2016;95:1888–93. doi: 10.3382/ps/pew144. https://doi.org/10.3382/ps/pew144. [DOI] [PubMed] [Google Scholar]
- [6].EFSA and ECDC. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. European Food Safety Authority and European Centre for Disease Prevention and Control. EFSA J. 2018;16(12):1–262. doi: 10.2903/j.efsa.2018.5500. https://doi.org/10.2903/j.efsa.2018.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lokken KL, Walker GT, Tsolis RM. Disseminated infections with antibiotic-resistant non-typhoidal Salmonella strains: contributions of host and pathogen factors. Pathog Dis. 2016;74:ftw103. doi: 10.1093/femspd/ftw103. https://doi.org/10.1093/femspd/ftw103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hansen-Wester I, M. Hensel M. Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 2001;3:549–59. doi: 10.1016/s1286-4579(01)01411-3. https://doi.org/10.1016/s1286-4579(01)01411-3. [DOI] [PubMed] [Google Scholar]
- [9].Wu SY, Wang LD, Li JL, Xu GM, He ML, Li YY, Huang R. Salmonella spv locus suppresses host innate immune responses to bacterial infection. Fish Shellfish Immunol. 2016;58:387–96. doi: 10.1016/j.fsi.2016.09.042. https://doi.org/10.1016/j.fsi.2016.09.042. [DOI] [PubMed] [Google Scholar]
- [10].Hacker J, Kaper JB. Pathogenicity islands and the evolution of microbes. Ann Rev Microbiol. 2000;54:641–79. doi: 10.1146/annurev.micro.54.1.641. https://doi.org/10.1146/annurev.micro.54.1.641. [DOI] [PubMed] [Google Scholar]
- [11].Schmidt H, Hensel M. Pathogenicity islands in bacterial pathogenesis. Clin Microbiol Rev. 2004;17(1):14–56. doi: 10.1128/CMR.17.1.14-56.2004. https://doi.org/10.1128/cmr.17.1.14-56.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Parkhill J, Dougan G, James KD, Thomson NR, Pickard D, Wain J. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature. 2001;413:848–52. doi: 10.1038/35101607. https://doi.org/10.3410/f.1002051.19557. [DOI] [PubMed] [Google Scholar]
- [13].Restika KD. Bogor, Indonesia: Institut Pertanian Bogor; 2012. Keberadaan Salmonella pada daging ayam yang dijual di pasar tradisional di Kota Tangerang Selatan [skripsi] [Google Scholar]
- [14].Loisa, Lukman DW, Latif H. Resistensi Salmonella spp. terhadap beberapa antibiotik pada daging itik di Kabupaten Bogor yang dapat memengaruhi kesehatan konsumen. J Kedokteran Hewan. 2016;10(2):115–20. [Google Scholar]
- [15].[SNI] Standar Nasional Indonesia. Jakarta, Indonesia: Badan Standardisasi Nasional; 2008. Metode Pengujian Cemaran Mikroba dalam Daging, Telur dan Susu serta Hasil Olahannya. SNI No. 2897:2008. https://doi.org/10.1016/b978-0-12-416999-9.00007-1. [Google Scholar]
- [16].Barbosa C, Nogueira S, Gadanho M, Chaves S. Lisbon, Portugal: Elsevier Inc.; 2016. DNA extraction: finding the most suitable method. Molecular microbial diagnostic methods: pathways to implementation for the food and water industries; pp. 135–54. https://doi.org/10.1016/b978-0-12-416999-9.00007-1. [Google Scholar]
- [17].Agron PG, Walker RL, Kinde H, Sawyer SJ, Hayes DC, Wollard J, et al. Identification by subtractive hybridization of sequences specific for Salmonella enterica serotype Enteritidis. Appl Environ Microbiol. 2001;67(11):4984–91. doi: 10.1128/AEM.67.11.4984-4991.2001. https://doi.org/10.1128/aem.67.11.4984-4991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Oliveira SD, Santos LR, Schucha DMT, Silva AB, Salle CTP, Canal CW. Detection and identification of Salmonella from poultry-related samples by PCR. Vet Microbiol. 2002;87:25–35. doi: 10.1016/s0378-1135(02)00028-7. https://doi.org/10.1016/s0378-1135(02)00028-7. [DOI] [PubMed] [Google Scholar]
- [19].Soto SM, Rodríguez I, Rodicio MR, Vila J, Mendoza MC. Detection of virulence determinants in clinical strains of Salmonella enterica serovar Enteritidis and mapping on macrorestriction profiles. J Med Microbiol. 2006;55:365–73. doi: 10.1099/jmm.0.46257-0. 1 https://doi.org/10.1099/jmm.0.46257-0. [DOI] [PubMed] [Google Scholar]
- [20].Skyberg JA, Logue CM, Nolan LK. Virulence genotyping of Salmonella spp. with multiplex PCR. Avian Dis. 2006;50:77–81. doi: 10.1637/7417.1. https://doi.org/10.1637/7417.1. [DOI] [PubMed] [Google Scholar]
- [21].Percival Steven L, Williams David W. Amsterdam; Boston: Elsevier/Academic Press; 2014. Microbiology of waterborne diseases. Salmonella. Salmonella; pp. 209–22. https://doi.org/10.1016/b978-0-12-415846-7.00010-x. [Google Scholar]
- [22].Singh Y, Saxena A, Kumar R, Saxena MK. United Kingdom: Intech Open; 2018. Virulence system of Salmonella with special reference to Salmonella enterica; pp. 41–53. https://doi.org/10.5772/intechopen.77210. [Google Scholar]
- [23].Marcus SL, John H, Cheryl BG, Pfeifer B, Finlay B. Salmonella pathogenicity islands: big virulence in small packages. Microbes Infect. 2000;2(2):145–56. doi: 10.1016/s1286-4579(00)00273-2. https://doi.org/10.1016/s1286-4579(00)00273-2. [DOI] [PubMed] [Google Scholar]
- [24].Amavisit P, Lightfoot D, Browning GF, Markham PF. Variation between pathogenic serovars within Salmonella pathogenicity islands. J Bacteriol. 2003;185:3624–35. doi: 10.1128/JB.185.12.3624-3635.2003. https://doi.org/10.1128/jb.185.12.3624-3635.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Nidaullah H, Abirami N, Shamila-Syuhada AK, Chuah LO, Nurul H, Tan TP, et al. Prevalence of Salmonella in poultry processing environments in wet markets in Penang and Perlis, Malaysia. Vet World. 2017;10(3):286–92. doi: 10.14202/vetworld.2017.286-292. https://doi.org/10.14202/vetworld.2017.286-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].CCFH. Food safety risk profile for Salmonella species in broiler (young) chickens. Codex Committee on Food Hygiene Working Group on guidelines for control of Campylobacter and Salmonella species in broiler (young bird) chicken meat. 2007. https://doi.org/10.3358/shokueishi.22.1.
- [27].Kalergis AM, Bueno SM. New insights about excisable pathogenicity islands in Salmonella and their contribution to virulence. Microbes Infect. 2016;18(5):302–9. doi: 10.1016/j.micinf.2016.02.001. https://doi.org/10.1016/j. micinf.2016.02.001. [DOI] [PubMed] [Google Scholar]
- [28].LaRock DL, Chaudhary A, Miller SI. Salmonellae interactions with host processes. Nature Rev Microbiol. 2015;13:191–205. doi: 10.1038/nrmicro3420. https://doi.org/10.1038/nrmicro3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Meresse S, Unsworth KE, Habermann A, Griffiths G, Fang F, Martinez-lorenzo MJ, et al. Remodelling of the actin cytoskeletal is essential for replication of intravascular Salmonella. Cell Microbiol. 2001;3:567–57. doi: 10.1046/j.1462-5822.2001.00141.x. https://doi.org/10.1046/j.1462-5822.2001.00141.x. [DOI] [PubMed] [Google Scholar]
- [30].Denise MM, Anne M, Stanley F. Persistent bacterial infections: the interface of the pathogen and the host immune system. Nat Rev Microbiol. 2004;2:747–65. doi: 10.1038/nrmicro955. https://doi.org/10.1038/nrmicro955. [DOI] [PubMed] [Google Scholar]
- [31].House D, Bishop A, Parry CM, Dougan G, Wain J. Typhoid fever: pathogenesis and disease. Curr Opin Infect Dis. 2001;14:573–8. doi: 10.1097/00001432-200110000-00011. https://doi.org/10.1097/00001432-200110000-00011. [DOI] [PubMed] [Google Scholar]
- [32].Abulreesh HH. Vol. 2. United Kingdom: Intech Open; 2012. Salmonellae in the environment, in Salmonella— distribution, adaptation, control measures and molecular technologies; pp. 21–50. https://doi.org/10.5772/28201. [Google Scholar]
- [33].Coombes BK, Coburn BA, Potter AA, Gomis S, Mirakhur K, Li Y, Finlay BB. Analysis of the contribution of Salmonella pathogenicity islands 1 and 2 to enteric disease progression using a novel bovine ileal loop model and a murine model of infectious enterocolitis. Infect immun. 2005;73(11):7161–9. doi: 10.1128/IAI.73.11.7161-7169.2005. https://doi.org/10.1128/iai.73.11.7161-7169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Espinoza RA, Silva-Valenzuela CA, Amaya FA, Urrutia IM, Contreras I, Antiviago CA. Differential roles for pathogenicity islands SPI- 13 and SPI-8 in the interaction of Salmonella Enteritidis and Salmonella typhi with murine and human macrophages. Biol Res. 2017;50(5) doi: 10.1186/s40659-017-0109-8. https://doi.org/10.1186/s40659-017-0109-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Charles RC, Harris JB, Chase MR, Lebrun LM, Sheikh A, LaRocque RC, et al. Comparative proteomic analysis of the PhoP regulon in Salmonella enterica Serovar Typhi versus Typhimurium. PLoS One. 2009;4(9):6994. doi: 10.1371/journal.pone.0006994. https://doi.org/10.1371/journal.pone.0006994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gunzel DL, Kucharski M, Kehres DG, Romero MF, Maguire ME. The MgtC virulence factor of Salmonella enterica serovar Typhimurium activates Na(+), K(+)-ATPase. J Bacteriol. 2006;188:5586–94. doi: 10.1128/JB.00296-06. https://doi.org/10.1128/jb.00296-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Gerlach RG, Jackel D, Geymeier N, Hensel M. Salmonella Pathogenicity Island 4-Mediated Adhesion Is Coregulated with Invasion Genes in Salmonella enterica. Infect Immun. 2007;75(10):4697–709. doi: 10.1128/IAI.00228-07. https://doi.org/10.1128/iai.00228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Knodler LA, Celli J, Hardt WD, Vallance BA, Yip C, Finlay BB. Salmonella effectors within a single pathogenicity island are differentially expressed and translocated by separate type III secretion systems. Mol Microbiol. 2002;43:1089–103. doi: 10.1046/j.1365-2958.2002.02820.x. https://doi.org/10.1046/j.1365-2958.2002.02820.x. [DOI] [PubMed] [Google Scholar]
- [39].Van Asten AJ, van Dijk JE. Distribution of “classic” virulence factors among Salmonella spp. FEMS Immunol Med Microbiol. 2005;44:251–9. doi: 10.1016/j.femsim.2005.02.002. https://doi.org/10.1016/j.femsim.2005.02.002. [DOI] [PubMed] [Google Scholar]
- [40].Amini K, Salehi TZ, Nikbakht G, Ranjbar R, Amini J, Ashrafganjooei SB. Molecular detection of invA and spv virulence genes in Salmonella enteritidis isolated from humans and animals in Iran. Afr J Microbiol Res. 2010;4(21):2202–10. [Google Scholar]
- [41].Smith SI, Fowora MA, Tiba A, Anejo-Okopi J, Fingesi T, Adamu ME, et al. Molecular detection of some virulence genes in Salmonella Spp isolated from food samples in Lagos, Nigeria. Anim Vet Sci. 2015;3:22–7. https://doi.org/10.11648/j.avs.20150301.15. [Google Scholar]
- [42].Smith S, Opere B, Fowora M, Aderohunmu A, Ibrahim R, Omonigbehin E, et al. Molecular characterization of Salmonella spp directly from snack and food commonly sold in Lagos, Nigeria. Southeast Asian J Trop Med. 2012;43:718–23. [PubMed] [Google Scholar]
- [43].Mezal EH, Sabol A, Khan MA, Ali N, Stefanova R, Khan AA. Isolation and molecular characterization of Salmonella enterica serovar Enteritidis from poultry house and clinical samples during 2010. Food Microbiol. 2014;38:67–74. doi: 10.1016/j.fm.2013.08.003. https://doi.org/10.1016/j. fm.2013.08.003. [DOI] [PubMed] [Google Scholar]
- [44].Osman KM, Marouf SH, Zolnikov TR, Al Atfeehy N. Isolation and characterization of Salmonella enterica in day-old ducklings in Egypt. Pathog Global Health. 2014;108(1):37–48. doi: 10.1179/2047773213Y.0000000118. https://doi.org /10.1179/2047773213y.0000000118. [DOI] [PMC free article] [PubMed] [Google Scholar]