Abstract
Objectives:
The study was undertaken with the objectives to perform seromonitoring of Peste des Petits Ruminants (PPR) antibodies in goats vaccinated with PPR vaccine and molecular characterization of PPR virus (PPRV) from field cases in Bangladesh.
Materials and Methods:
Seromonitoring work was conducted in Char Kalibari, Mymensingh Sadar, Mymensingh. For this, a total of 50 goats were randomly selected and were divided into two groups; vaccinated (Group A; n = 25) and non-vaccinated (Group B; n = 25). The goats of both groups were again sub-divided into four age groups; (i) 0–6 months (n = 5), (ii) 6–12 months (n = 5), (iii) 12–24 months (n = 10), and (iv) >24 months (n = 5). Blood samples were collected on Day-0 and after 21 days of post-vaccination (DPV), and the sera were prepared. The sera were examined for the presence of antibodies against PPRV by competitive enzyme-linked immunosorbent assay. For molecular characterization, nasal swabs (n = 10) were collected from PPR infected goats in Jessore during PPR outbreak (February 2016). The causative agent, PPRV isolated from field cases were confirmed by N gene based on reverse transcription polymerase chain reaction (RT-PCR), followed by sequencing, phylogenetic analysis, and multiple sequence alignment analyses.
Results:
In the case of seromonitoring, the results revealed that before vaccination (at Day-0), overall, 44% (n = 22/50) goats were seropositive for PPRV. In Group A, 48% (n = 12/25) goats were seropositive, but after 21 DPV, 96% (n = 24/25) goats become seropositive. On the other hand, in Group B, 40% (n = 10/25) and 16% (n = 04/25) seropositive goats found at Day-0 and after 21 DPV, respectively, indicating that the antibody titer was increasing after vaccination and decreasing in convalescent goats. Out of 10 nasal swab samples, 40% (n = 4/10) was confirmed by RT-PCR targeting nucleocapsid (N gene). Phylogenetically, our isolate (KY039156/PPRV/BDG/Jes/2016) was similar to the other strains of PPRV under lineage IV. However, there was a unique amino acid substitution, where glycine (G) was recorded in place of arginine (R). The strain is closely related with other Chinese or Indian strains. The nucleotide sequence homology by NCBI BLAST search of the isolated strain ranged from 95% to 99% with other strains circulating in Bangladesh.
Conclusion:
The PPRV is prevailing in the Mymensingh and Jessore regions of Bangladesh. Effective control of PPR in goats may depend on vaccination with PPR vaccine. Molecular characterization of PPRV in Jessore reveals that the virus is differing from the strain prevalent in other regions of Bangladesh and the world.
Keywords: PPR virus, cELISA, Jessore, RT-PCR, Mymensingh, phylogenetic analysis, sequencing
Introduction
Peste des Petits Ruminants (PPR) is a viral disease of small ruminants, which is manifested by fever, ocular-nasal discharges, anorexia, necrotic stomatitis, fetid diarrhea, enteritis, and bronchopneumonia followed by either death or recovery [1]. The 4 to 24 months aged goats are more susceptible in the endemic areas [2]. The causal agent is PPR virus (PPRV), an envelope, a pleomorphic particle containing single-stranded RNA, approximately 16-kb long with negative polarity genome [3]. The genome of the virus codes for six structural (N, P, M, F, H, and L) and two nonstructural (C and V) proteins [4,5].
After the first report of PPR from Ivory Coast, the disease has been reported in the Middle East, the Arabian Peninsula, and most parts of Africa and Asia [1].
Among the South Asian subcontinent, PPRV under lineage IV was first reported in the southern part of India during 1987 [6]. Subsequently, frequent outbreaks of PPR have been recorded in South Asian countries like Pakistan, Bhutan, Nepal, Afghanistan, India, and Bangladesh [7]. Total goat population of Bangladesh is approximately 19.43 million [8]. In Bangladesh, the PPRV was first identified in 1993 during a severe PPR outbreak [2]. Now a day, the outbreak of PPR in goats occurs every year and considered as an endemic disease in Bangladesh [2]. The country faces a huge economic loss due to PPR outbreaks. High morbidity (100%) and mortality (50% to 90%) rates in goats caused by PPRV have been reported in Bangladesh [9]. Das et al. [10] also reported that PPR outbreaks caused 74.13% morbidity and 54.83% mortality in Black Bengal goats.
For the prevention and control of PPR, proper diagnosis is crucial. Agar gel immunodiffusion test, counter immunoelectrophoresis, indirect fluorescent antibody test, virus neutralization test, virus-specific monoclonal antibodies in an immunocapture enzyme-linked immunosorbent assay (ELISA), sandwich ELISA, and PCR techniques are generally used for identifying the PPR [4,11]. But more specifically, PPRV antibodies can be identified by competitive ELISA(c-ELISA) and serum neutralization test [12].
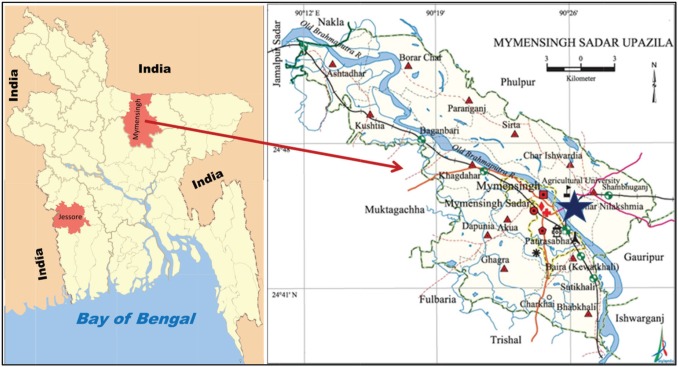
Due to its circulating in nature, the PPRV can easily be spread from one place to another. Sometimes genetic mutation occurs in its genomic structure [13]. For that reason, a locally produced vaccine can be a good choice to protect the animals susceptible to PPR viral infection. Thus, in Bangladesh, controlling this disease outbreak in small ruminants, regular seromonitoring, and molecular study are needed to evaluate the vaccine efficacy of the locally produced vaccine and genomic changes of PPRV in different regions of Bangladesh. Few studies in Bangladesh were previously reported on seromonitoring of antibodies against PPRV and molecular characterization of PPRV; for example, in the districts of Dhaka, Rajshahi, Gazipur, Netrokona, Narayanganj, and Mymensingh, there have been reported PPR outbreak investigation and seromonitoring activities [9,13–16]. However, seromonitoring of PPR in char region (Fig. 1) of Mymensingh Sadar (besides Brahmaputra River) has not done previously. Jessore region, a south-western district (Fig. 1) of Bangladesh, is yet to take under consideration for the investigation of genetic variability. This study was designed for the detection of PPRV-specific antibodies in serum of goats at the char area of Mymensingh by using monoclonal antibody-based c-ELISA and to characterize the circulating PPRV from Jessore region of Bangladesh.
Figure 1. Bangladesh Map showing research areas. Star in the Mymensingh Sadar Upazila was the site of seromonitoring, and Jessore was the place of sample collection for molecular characterization of PPRV.
Materials and Methods
Ethical approval
The experiment was approved by the Animal Welfare and Experimental Ethical Committee (AWEEC) of Bangladesh Agricultural University, Mymensingh [approval number: AWEEC/BAU/2018(16)].
Site selection and sample collection and transportation
For the detection and evaluation of PPRV-specific antibodies, Char Kalibari (Fig. 1) was selected as this area is an isolated area separated by the Brahmaputra River to the west. Total goat population of the Char Kalibari was 320 during this study. The village was divided into two experimental zones. The first zone populated with 205 goats considered as Group A and second area as Group B which populated with 115 goats. A total of 50 sera samples (25 sera from each group) were collected. The goats of each group were sub-divided into different age groups; (i) 0–6 months (n = 5), (ii) 6–12 months (n = 5), (iii) 12–24 months (n = 10), and (iv) >24 months (n = 5). After that, all the goats of Group A were vaccinated with PPR-Vac® vaccine manufactured by LRI (Livestock Research Institute), Dhaka, and the goats of Group B were considered as non-vaccinated control. After 21 days of vaccination, again 25 sera samples from both groups were collected. Blood samples were collected directly from the jugular vein of animals by the venipuncture method using sterile 5 ml syringe. The samples were placed in icebox without agitation until serum collection. The collected blood samples were placed without agitation for about 50–60 min. The serum samples were then collected into Eppendorf tube and marked specifically. These samples were then stored in the refrigerator at the Department of Microbiology and Hygiene, Bangladesh Agricultural University, Mymensingh, until transportation.
For the characterization of the PPRV, a total of 10 nasal swab samples PPR-suspected goats were collected from Jessore district (Fig. 1). Nasal secretions were taken by swab sticks from the PPR suspected goats. Swab sticks were then emerged into 2.5 ml microcentrifuge tube containing 1 ml viral transport medium. Then the upper part of the swab stick was broken up to the 2 marks of the microcentrifuge tube. Then the tubes were capped along with the broken swab sticks. The samples were stored at −70°C for further use.
Detection of PPRV-specific antibodies by c-ELISA
The serum samples were transported to the SAARC Regional Leading Diagnostic Laboratory (SAARC-RLDL) at the Animal Health Research Division, Bangladesh Livestock Research Institute (BLRI) Savar, Dhaka-1341, Bangladesh. The sera samples were then analyzed by c-ELISA by BDSL® cELISA kit of The Pirbright Institute (UK) by following the instructions of the manufacturer. The optical density (OD) values were taken by the BioTek® Power WaveTM 340 ELISA plate reader with interference filters of 492 nm machine. The obtained OD value was then calculated and analyzed to interpret the results. As per the instructions of the kit, the percentage inhibition (PI) value of antibody ≥50% indicates seropositive, whereas PI values <50% indicates the seronegative result.
RNA extraction, RT-PCR, and agarose gel electrophoresis
The viral RNA samples were extracted from nasal swab sample using Ambion® RNA extraction kit (USA) as per the manufacturer’s instructions. The one-step conventional reverse transcription polymerase chain reaction (RT-PCR) was conducted by using the Ambion® Kit (AgPath-IDTM One-step RT- PCR kit, USA) as per the manufacturer’s instructions. A 25 μl scale of reaction mixture consisted of 2× RT-PCR buffer 13 μl, forward and reverse primers (100 pmole/μl each) 0.5 μl, 2× RT-PCR enzyme mix 1.0 μl, template RNA 5 μl, and the rest 5 μl was nuclease-free water. The primers used for amplification of N gene of PPRV were NP3 (5ʹ-TCTCGGAAATCGCCTCACAGACTG-3ʹ) and NP4 (5ʹ-CCTCCTCCTGGTCCTCCAGAATCT-3ʹ) targeting an amplicon of 351-bp, as described by Couacy-Hymann et al. [17]. The oligonucleotide primers were obtained from Integrated DNA Technologies (Japan). The thermal profile for the RT-PCR reaction was followed by the conditions mentioned by Couacy-Hymann et al. [17]. Agarose gel electrophoresis was done for the visualization of the RT-PCR products after staining with ethidium bromide (10 mg/ml) (Sigma®, USA).
Sequencing of PPR viral N gene
RT-PCR products were sequenced directly using PCR primer specific for N gene. First, the RT-PCR products were purified with QIAquick PCR purification kit (QIAGEN, Germany) by following the manufacturer’s instructions. Then the cycle sequencing reaction was carried out with Big Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA) in a final reaction volume of 20 μl using 200 μl capacity thin walls PCR tube. Unincorporated dye terminators from extension products for sequencing were removed completely by Big Dye®XterminatorTM purification kit protocol. Capillary electrophoresis and data analysis were carried out from a commercial source on the ABI PRISM® 310 Genetic Analyzer (Applied Biosystem, USA). The nucleic acid sequences and their deduced amino acid sequences were aligned with other related sequences retrieved from the GenBank. Sequence editing, alignment, and phylogenetic tree construction were carried out with the software (CodonCode Aligner, USA; Mega5.2).
Phylogenetic and multiple sequence alignment analyses
The phylogenetic tree by the neighbor-joining method was constructed using partial sequences (85-aa) of the previously reported PPRV N gene sequences available in the GenBank by using the MEGA (Molecular Evolutionary Genetics Analysis) Ver. 6.0 [32]. The multiple sequence alignment was done using 55 other PPRV strains available in GenBank using MEGA Ver. 6.0.
Results
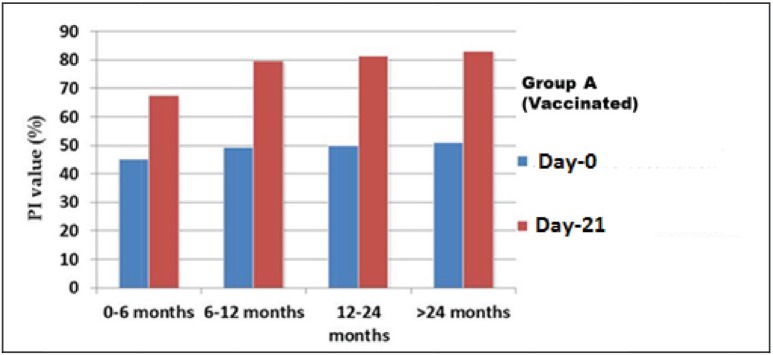
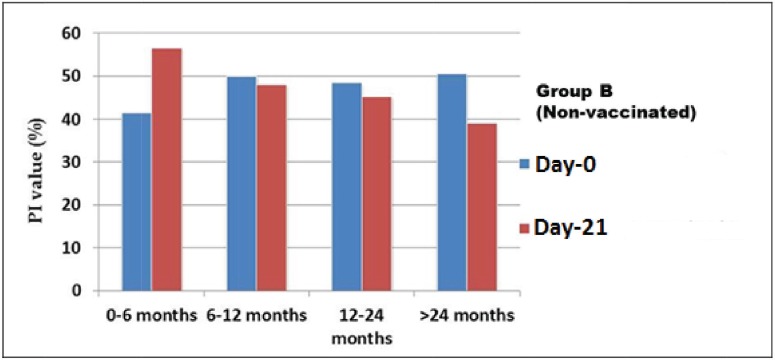
Based on the results of c-ELISA, the overall PPRV-specific antibody in the goats before vaccination was 44% (n = 22/50). Considering the goats of Group A (vaccinated), on Day-0, the prevalence was 48% (n = 12/25). On the other hand, 40% (n = 10/25) goats of Group B were seropositive on Day-0 (Fig. 2). At the pre-vaccination stage (Day-0), the average PI values of Group A of four different ages were 45.29%, 49.48%, 49.88%, and 51.0181%, respectively (Fig. 2). Similarly, on Day-0, the average PI values in Group B were 41.51%, 50.04%, 48.47%, and 50.55%, respectively (Fig. 3).
Figure 2. Antibody titer value of goats at pre-vaccination (Day-0) and post-vaccination (After 21 DPV)) stage (Group A). Here, the PI values in all categories goats are increased from Day-0 to 21 DPV. At Day-0 stage, the average PI values of all the groups were below 45% to 51% and after Day-21 of post-vaccination that was increased above 50% to above 80%.
Figure 3. Antibody titer value of goats at Day-0 and after 21 days without vaccination (Group B). Here, the average PI values of in Non-vaccinated flock decreased day-by-day except first group (0–6 months).
After 21 days of post-vaccination (DPV), 96% (n = 24/25) goats of vaccinated Group A were found seropositive (Fig. 2), whereas, only 16% (n = 04/25) goats of non-vaccinated Group B were seropositive (Fig. 3). The average PI values of the previously described four different aged groups (Group A) goats were 83.00%, 81.31%, 79.62%, and 67.58%, respectively (Fig. 2). Similarly, among the goats of Group B, the average PI value recorded were 56.56%, 48.20%, 45.36%, and 39.12%, respectively (Fig. 3).
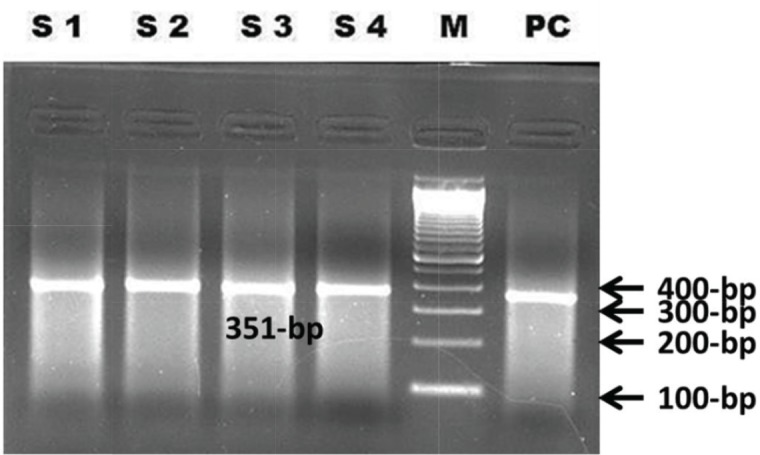
Among the nasal swab samples (n = 10), four were found RT-PCR positive (Fig. 4) targeting N gene showing an amplicon of 351-bp. Among the four amplified RT-PCR samples, one was sequenced directly using NP3 and NP4 primers (mentioned previously). Sequencing of the N gene was done in the Bangladesh Livestock Research Institute (BLRI), Savar, Dhaka. The nucleotide sequences obtained in this study (GenBank accession KY039156) were aligned with sequences of the other related PPRV strains retrieved from GenBank. The sequence revealed that our isolate had 95%–99% similarities with the previously reported PPRV isolates of Bangladesh.
Figure 4. N gene of PPRV amplification by RT-PCR; S1–S4 = Nasal swab samples, M = 100-bp Marker, PC = Positive control.
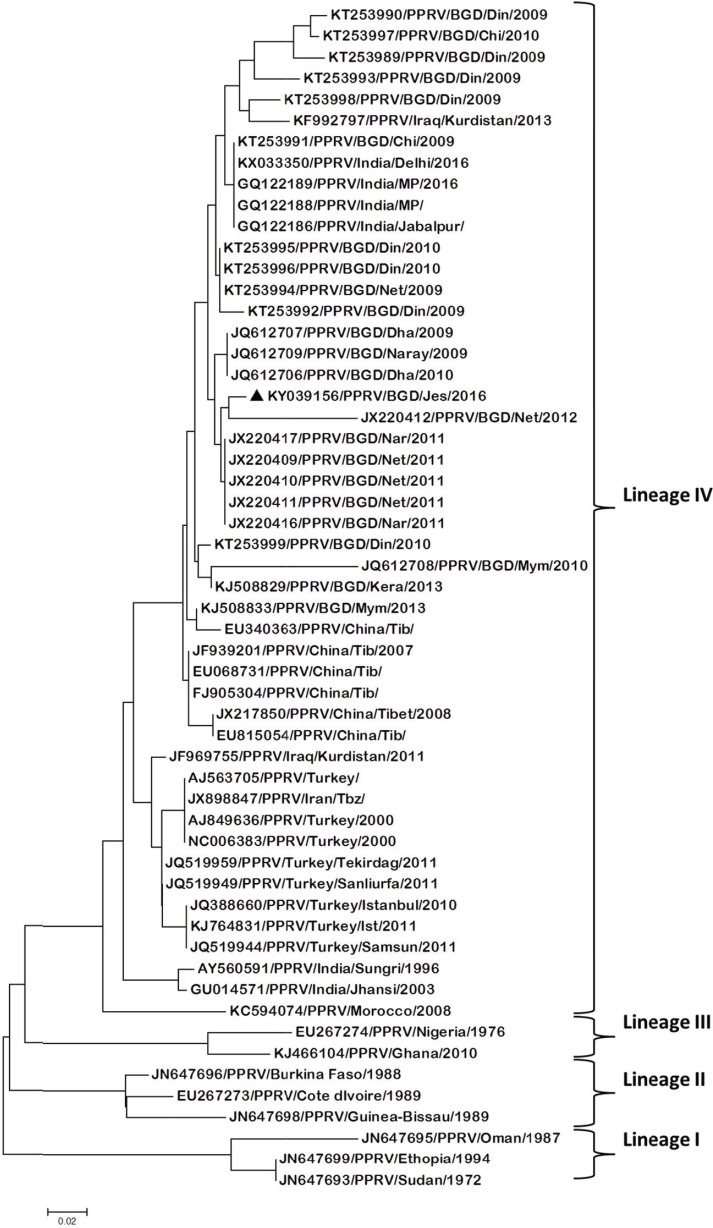
Phylogenetic analysis revealed that our isolate formed a sub-cluster with other Bangladesh isolates under lineage IV (Fig. 5). However, it is indicated that PPRV reported from African countries are mostly placed under lineage I to III. Multiple sequence alignment of the partial amino acid (N protein) sequence (85-aa) revealed that the PPRV isolate of this study changed its genomic structure. One amino acid (glycine; G) substitution has been found in place of arginine (R), which is considered as the usual amino acid at the same position (Fig. 6).
Figure 5. Phylogenetic relationship between PPR virus isolates based on the N gene sequence. The bootstrap test neighbor-joining method in MEGA 5 software (1000 replicates) was used to draw the tree. PPRV strain (KY039156/PPRV/BGD/Jes/2016) reported in this study is highlighted as a black triangle. Other sequences are retrieved from GenBank.
Figure 6. Multiple sequence alignment of the partial amino acid N protein sequences (85-aa) of the PPRV strains reported in Bangladesh so far along with other representative PPRV strains of Lineages I–IV. The column indicated by a star (*) has no difference among the strains. The columns having black background have difference in amino acid. The black arrow indicates the unique amino acid change of the strains reported in this study.
Discussion
PPR is now considered as an endemic disease and a threat toward the development of goat farming in Bangladesh [2,10,19]. Several studies had been reported on different aspects of PPRV in goats in Bangladesh; for example, pathological investigation [20], evaluation of antibiotic combined with hyperimmune serum therapy [21], and seroprevalence [16]. The present study investigated the seromonitoring of PPRV antibodies under the natural condition in vaccinated and unvaccinated goats. Birindwa et al. [22] reported that 64.7% unvaccinated goats were PPRV positive in Congo. It is crucial that effective implementation of control programs for PPR requires regular vaccination with effective vaccine and seromonitoring of immunity against PPRV. The present study was adopted for seromonitoring of PPR specific antibody in the vaccinated goats after field level vaccination in a selected area of Bangladesh and investigation of the genomic changes in field isolates of PPRV by molecular technique.
Based on seromonitoring, overall 44% goats were positive for PPR in this study. Recently, Rahman et al. [23] could identify RNA for PPRV in 38% goats by real-time reverse transcription polymerase chain reaction (rRT-PCR), which validated our study although the approach of identification was different. However, Islam et al. [14] reported an overall seroprevalence of 8.7% against PPR in Rajshahi, Gazipur and Sirajganj districts. This variation might be due to difference in geographical location.
In Mymensingh region, Banik et al. [7] found 76.60% average antibody titer at Day-21 of post-immunization in goat. Gowane et al. [24] reported that PI value at Day-0 of vaccination was 22.50%, and at 28 days of post-vaccination, it was 71.8%. They also found that 94.92% of the total animals showed protective titer on 28 days of post-vaccination. In Rajshahi and Sirajganj districts, the age group of 0–6 months, vaccinated samples had the highest seroprevalence (80.25%; n = 65/81) as compared to 12–24 (70.83%; n = 34/48) and >24 months (74.60%; n = 47/63) age groups of goats, respectively [14]. Yousuf et al. [25] reported the prevalence of PPR in different areas of Bangladesh; for example, Bogra (30%), Sirajganj (86.67%), Mymensingh (10%), and Rangpur (11.67%). In our study, we detected 44% prevalence of PPRV in goats in Mymensingh. This variation might be due to the difference in location within the region and season of sample collection.
Previously, in India, overall seroprevalence of PPRV antibody in goats was observed as 35% [3], whereas in Tanzania, Sudan, and the Republic of Congo, it was 49.50% [1], 59.15% [27], and 64.7% [22], respectively. A similar report from Sudan describing a prevalence of 45.6% PPRV antibodies by c-ELISA was reported by Salih et al. [28]. Thus, the difference in the seropositivity of PPRV antibody among different countries might be attributed to the difference in the agro-climatic condition of different countries.
In our experiment, we found 96% seropositive PI values in vaccinated goats through c-ELISA at 21 DPV, whereas Rahman et al. [9] found that 62% of the goats were seropositive after 21 DPV by using the same vaccine in Bangladesh. Similarly, Anderson and McKay [29] found that about 60%–70% animals were seropositive for PPRV. From the above findings, it proved that the animal gained high serum antibody level due to vaccination which made them seropositive, and the increasing trend of PI value (Fig. 2) indicated that PPR vaccine could produce immune response in goats for at least 21 days.
Recently, RT-PCR has emerged as a highly specific and sensitive test for molecular characterization of the PPRV. RT-PCR has become the most popular tool for diagnosis as well as molecular epidemiological studies [30]. RT-PCR targeting N gene was used for the confirmation of PPR. Libeau et al. [12] also described that among the structural proteins of PPRV, N protein is antigenically most conserved among Morbilliviruses and is highly immunogenic in spite of its internal location. So, in the present investigation, N gene was targeted for the detection of PPRV.
In this study, 10 nasal swab samples were collected and subjected to RT-PCR for molecular characterization. The primers NP3 and NP4 were used previously designed by Couacy-Hymann et al. [17]. Bhuiyan et al. [31] reported that the primers (NP3/NP4) against N gene were specific and sensitive to ensure efficient amplification and detection of PPRV in field samples. The primers produced an amplicon of 351-bp amplifying a region of N gene in four (n = 04) PPRV positive samples. These findings were closely related with the recent findings of Chowdhury et al. [32] in Bangladesh.
The deduced amino acid sequences of the present isolate (KY039156/PPRV/BDG/Jes/2016) were used to construct a phylogenetic tree with other 55 (n = 55) PPRV strains from 15 different countries (Bangladesh, India, China, Turkey, Kurdistan, Iran, Turkey, Morocco, Nigeria, Burkina-Faso, Cote d’ivoire, Guinea Bissau, Oman, Ethiopia, and Sudan) available in GenBank (Fig. 5). The tree shows that the strain of Jessore (KY039156/PPRV/BDG/Jes/2016) is grouped along with other reported strains under lineage IV. The phylogeny showed that the present isolate (KY039156/PPRV/BDG/Jes/2016) formed a sub-cluster under lineage IV, as reported by El Arbi et al. [33]. The PPRV strain isolated from Jessore, Bangladesh, depicts a common phylogenetic lineage IV (Fig. 5), and possible spread to/from India as a neighboring country through trans-boundary commerce.
From this study, it is revealed that the genomic similarity of the present strain (KY039156/PPRV/BDG/Jes/2016) was 95%–99% identical to each other of different PPRV strains of Bangladesh. Moreover, the sequence homology of the identified virus with rest of the viruses of China, India, Turkey, Iran, and Kurdistan was 97%–98%, 97%, 95%–96%, 95%, and 95%–97%, respectively. The strain isolated from Jessore, Bangladesh, revealed a unique amino acid substitution, where glycine (G) was found in place of arginine (R), isoleucine (I), and lysine (K) (Fig. 6), indicating that the strain is genetically different as compared to other strains reported from Bangladesh or other parts of the world. Rahman et al. [23] detected amino acid substitution of PPRV in a 59 amino acid window of N-protein (420 and 497). These close similarities of the variable part of the N gene suggest that the trans-boundary circulation of PPR virus among the countries might be the cause of the severe outbreak in Bangladesh. Low sample size for genetic characterization was the limitation of this study. Full-length N-gene analysis may reveal a more comprehensive understanding about the circulating PPRV in Bangladesh.
Conclusion
High antibody titer after vaccination of goats by PPR vaccine indicates that the goat responded well against the vaccine. The presence of PPRV is confirmed by RT-PCR in Jessore district of Bangladesh. Sequence homology and phylogenetic analysis reveal that the PPRV of this report is located within the same cluster under lineage IV of other Bangladeshi isolates. However, unique genetic variation has been detected in our strain. Thus, effective controlling of the PPR in goats can be depended on the use of effective vaccine and consideration of genetic variation. The current control strategies for PPRV in Bangladesh can be reevaluated considering regular seromonitoring and outbreak investigation of the disease throughout the country. This study provides baseline data, which may help in the development of an effective vaccine and controlling PPR in Bangladesh.
Acknowledgment
The chemical, reagents, and laboratory supports of the SAARC Regional Leading Diagnostic Laboratory (SAARC-RLDL) at the Animal Health Research Division, Bangladesh Livestock Research Institute (BLRI) Savar, Dhaka-1341, Bangladesh is accordingly acknowledged.
Conflict of Interest
The authors declare that they have no conflict of interest.
Authors’ contributions
SA and MAY designed the research work. SA, MAY, and MMI conducted the actual experiments. MYA, MAI, and MMM helped in conducting the research works and data analysis. MRI and KHMNHN supervised the research work. All the authors read and finally approved the manuscript for submission.
References
- [1].Balamurugan V, Krishnamoorthy P, Raju DSN, Rajak KK, Bhanuprakash V, Pandey AB, Gajendragad MR, Prabhudas K, Rahman H. Prevalence of Peste des petits ruminant virus antibodies in cattle, buffaloes, sheep and goats in India. Virus Dis. 2014;1:85–90. doi: 10.1007/s13337-013-0177-5. https://doi.org/10.1007/s13337-013-0177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Islam MR, Shamsuddin M, Rahman MA, Das PM, Dewan ML. An outbreak of Peste des Petits Ruminants in Black Bengal goats in Mymensingh, Bangladesh. Bangladesh Vet. 2001;18:14–9. https://doi.org/10.3329/bjm.v24i2.1260. [Google Scholar]
- [3].Barrett T, Banyard AC, Diallo A. Elsevier; Amsterdam, The Netherlands: 2005. Molecular biology of the Morbilliviruses virus plagues of large and small ruminants; pp. 31–67. [Google Scholar]
- [4].Bailey D, Banyard A, Dash P, Ozkul A, Barrett T. Full genome sequence of Peste des Petits Ruminants virus, a member of the Morbillivirus genus. Virus Res. 2005;110:119–24. doi: 10.1016/j.virusres.2005.01.013. https://doi.org/10.1016/j.virusres.2005.01.013. [DOI] [PubMed] [Google Scholar]
- [5].Mahapatra M, Sayalel K, Muniraju M. Spillover of Peste des Petits Ruminants virus from domestic to wild ruminants in the serengeti ecosystem, Tanzania. Emerg Infect Dis J. 2015;21:2230–34. doi: 10.3201/eid2112.150223. https://doi.org/10.3201/eid2112.150223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Shaila MS, Purushothaman V, Bhavasar D, Venugopal K, Venkatesan RA. Peste des petits ruminants of sheep in India. Vet Rec. 1989;125:602. [PubMed] [Google Scholar]
- [7].Banik SC, Podder SC, Samad MA, Islam MT. Sero-surveillance and immunization in sheep and goats against Peste des Petits Ruminants in Bangladesh. Bangladesh J Vet Med. 2008;6:185–90. https://doi.org/10.3329/bjvm.v6i2.2334. [Google Scholar]
- [8].BBS-Bangladesh Bureau of Statistics. Economic Review of 2013–2014, Government of the Peoples Republic of Bangladesh. Dhaka: 2015. Livestock population in Bangladesh. [Google Scholar]
- [9].Rahman MA, Shadmin I, Noor M, Parvin R, Chowdhury EH, Islam MR. Peste des Petits Ruminants virus infection of goats in Bangladesh: Pathological investigation, molecular detection and isolation of the virus. Bangladesh Vet. 2011;28:1–7. https://doi.org/10.3329/bvet.v28i1.8808. [Google Scholar]
- [10].Das KK, Shil NK, Islam MR. Sero-epidemiological investigation on Peste des Petits Ruminants in Black Bengal Goats. Bangladesh J Microbiol. 2007;24:143–5. https://doi.org/10.3329/bjm. v24i2.1260. [Google Scholar]
- [11].Sharma CS, Misraulia KS, Shukla PC, Bagherwal RK, Nidhi-Singh Studies on determination of Peste des Petits Ruminants antibodies in goats by c-ELISA in Madhya Pradesh. Indian J Field Vet. 2007;3:11–4. [Google Scholar]
- [12].Libeau G, Prehaud C, Lancelot R, Colas F, Guerre L, Bishop DHL, Diallo A. Development of a competitive ELISA for detecting antibodies to the Peste des Petits Ruminants virus using a recombinant nucleoprotein. Res Vet Sci. 1995;58:50–5. doi: 10.1016/0034-5288(95)90088-8. https://doi.org/10.1016/0034-5288(95)90088-8. [DOI] [PubMed] [Google Scholar]
- [13].Rahman MM, Parvin R, Bhuiyan AR, Giasuddin M, Chowdhury SMZH, Islam MR, et al. Genetic characterization of Peste des petits ruminants virus circulating in Bangladesh. Br J Virol. 2016;3(4):115–22. https://doi.org/10.17582/journal.bjv/2016.3.4.115.122. [Google Scholar]
- [14].Islam MM, Hasan MA, Yousuf MA, Islam UK, Shawan MMAK, Islam MR. Seroprevalence of Peste des Petits Ruminant Virus specific antibody in goats in different regions of Bangladesh. J Adv Vet Anim Res. 2016;3(2):127–33. http://doi.org/10.5455/javar.2016.c140. [Google Scholar]
- [15].Kabir ME, Hossain MM, Ershaduzzaman M, Yousuf MA, Islam MR. Sero-surveillance and sero-monitoring of locally produced PPR vaccine in the field and experimental level. Asian J Med Biol Res. 2016;2:33–7. https://doi.org/10.3329/ajmbr.v2i1.27566. [Google Scholar]
- [16].Razzaque MA, Rahman MB, Kafi MA, Islam MR, Khan MFR, Nazir KHMNH. Application of C-ELISA for detection of PPRV-specific antibodies in domestic ruminants in different areas of Mymensingh, Bangladesh. Mol Biol Biotechnol J. 2004;2:40–3. [Google Scholar]
- [17].Couacy-Hymann E, Roger F, Hurard C, Guillou JP, Libeau G, Diallo A. Rapid and sensitive detection of Peste des Petits Ruminants virus by a polymerase chain reaction assay. J Virol Methods. 2002;100:17–25. doi: 10.1016/s0166-0934(01)00386-x. https://doi.org/10.1016/s0166-0934(01)00386-x. [DOI] [PubMed] [Google Scholar]
- [18].Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. https://doi.org/10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Dhar P, Sreenivasa BP, Barrett T, Corteyn M, Singh RP, Bandyopadhyay SK. Recent epidemiology of Peste des Petits Ruminants virus (PPRV) Vet Microbiol. 2002;88:153–9. doi: 10.1016/s0378-1135(02)00102-5. https://doi.org/10.1016/s0378-1135(02)00102-5. [DOI] [PubMed] [Google Scholar]
- [20].Khan MR, Haider MG, Alam KJ, Hossain MG, Chowdhury SM, Hossain MM. Pathological investigation of Peste des Petits Ruminants (PPR) in goats. Bangladesh J Vet Med. 2012;3:134–8. https://doi.org/10.3329/bjvm.v3i2.11380. [Google Scholar]
- [21].Islam MR, Giasuddin M, Rahman MM, Kafi MA. Antibiotic combined hyperimmune serum therapy for Peste des Petits Ruminants infected goats. Bangladesh J Vet Med. 2003;1:49–51. https://doi.org/10.3329/bjvm.v1i1.1918. [Google Scholar]
- [22].Birindwa BA, George GC, Ntagereka BP, Christopher O, Lilly BC. Mixed infection of peste des-petits ruminants and Capripox in goats in South Kivu, Democratic Republic of Congo. J Adv Vet Anim Res. 2017;4(4):348–55. https://doi.org/10.5455/javar.2017.d233. [Google Scholar]
- [23].Rahman MZ, Haider N, Gurley ES, Ahmed S, Osmani MG, Hossain MB, et al. Epidemiology and genetic characterization of Peste des petits ruminants virus in Bangladesh. Vet Med Sci. 2018;(4):161–71. doi: 10.1002/vms3.98. https://doi.org/10.1002/vms3.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gowane GR, Akram N, Misra SS, Prakash V, Kumar A. Assessment of the antibody response to Peste des Petits Ruminants (PPR) disease vaccination in a flock of Sirohi goat kids. Small Rum Res. 2016;138:20–4. https://doi.org/10.1016/j.smallrumres.2016.03.031. [Google Scholar]
- [25].Yousuf MA, Rahman MM, Alauddin M, Rahman SMB, Islam SMS, Islam MR, et al. Sero-surveillance of peste des petits ruminant viral antibody in goats at different areas of Bangladesh. Asian J Med Biol Res. 2017;3(3):347–51. https://doi.org/10.3329/ajmbr. v3i3.34524. [Google Scholar]
- [26].Swai ES, Kapaga A, Kivaria F, Tinuga D, Joshua G, Sanka P. Prevalence and distribution of Peste des Petits Ruminants virus antibodies in various districts of Tanzania. Vet Res Commun. 2009;33:927–36. doi: 10.1007/s11259-009-9311-7. https://doi.org/10.1007/s11259-009-9311-7. [DOI] [PubMed] [Google Scholar]
- [27].Osman NA, Ali AS, Rahman ME, Fadol MA. Antibody seroprevalences against Peste des Petits Ruminants (PPR) virus in sheep and goats in Sudan. Trop Anim Health Prod. 2009;41:1449–53. doi: 10.1007/s11250-009-9333-8. https://doi.org/10.1007/s11250-009-9333-8. [DOI] [PubMed] [Google Scholar]
- [28].Salih HAM, Elfadil AAM, Saeed IK, Ali YH. Seroprevalence and risk factors of Peste des Petits Ruminants in sheep and goats in Sudan. J Adv Vet Anim Res. 2014;1(2):42–9. http://doi.org/10.5455/javar.2014.a12. [Google Scholar]
- [29].Anderson J, McKay JA. The detection of antibodies against PPR virus in cattle, sheep and goats and the possible implications to RP control programs. Epidemiol Infect. 1994;112:225–31. doi: 10.1017/s0950268800057599. https://doi.org/10.1017/s0950268800057599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Shaila MS, Shamaki D, Forsyth AM, Diallo A, Goatley L, Kitching RP, Barrett T. Geographic distribution and epidemiology of Peste des Petits Ruminants viruses. Virus Res. 1996;43:149–53. doi: 10.1016/0168-1702(96)01312-3. https://doi.org/10.1016/0168-1702(96)01312-3. [DOI] [PubMed] [Google Scholar]
- [31].Bhuiyan AR, Rahman MM, Begum JA, Islam MR, Chowdhury EH. Comparison of genes as target for molecular diagnosis of Peste des Petits Ruminants in goats. Bangladesh Vet. 2013;29:56–62. https://doi.org/10.3329/bvet.v29i2.14343. [Google Scholar]
- [32].Chowdhury EH, Bhiyan AR, Rahman MM, Siddique MSA, Islam MR. Natural Peste des Pestis Ruminants virus infection in Black Bengal goats: virological, pathological and immunohistochemical investigation. BMC Vet Res. 2014;10:263–4. doi: 10.1186/s12917-014-0263-y. https://doi.org/10.1186/s12917-014-0263-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].El Arbi AS, El Mamy AB, Salami H, Isselmou E, Kwiatek O, Libeau G, et al. Peste des petits ruminants virus, Mauritania. Emerg Infect Dis. 2014;20:333–6. doi: 10.3201/eid2002.131345. https://doi.org/10.3201/eid2002.131345. [DOI] [PMC free article] [PubMed] [Google Scholar]