Abstract
Background:
Germline mutations in BRCA2 have been linked to a higher risk of prostate cancer (PCa), and high frequency of BRCA1 and BRCA2 (BRCA1/2) gene alterations was recently reported in metastatic castration-resistant PCa specimens. Mutations in BRCA2 vary in racial and ethnic groups including African-American (AA) and Caucasian-American (CA) populations.
Methods:
BRCA1 and BRCA2 genes were sequenced (Ion AmpliSeq targeted sequencing) in archived blood DNA specimens in 1240 PCa patients, including 30% AA patients, in three different cohorts: localized early stage (T2) PCa (N = 935); advanced PCa (50% T3–4) (N = 189); and metastatic PCa (N = 116). The sequences were analyzed for known and novel mutations in BRCA1/2. Statistical analyses were performed to determine associations of the mutations with clinico-pathological parameters.
Results:
BRCA2 mutations with known pathogenic annotation were significantly more prevalent in men with advanced and metastatic PCa (3.1%) compared to patients with an organ-confined disease (0.7%). AA patients carried more frequently BRCA1/2 variants of unknown significance (VUS) when compared to Caucasian Americans (4.6 vs. 1.6%, respectively). Significantly, pathogenic BRCA2 mutations in men with localized early stage PCa increased the risk of distant metastasis.
Conclusions:
Germline variants of unknown significance in BRCA1/2 are more frequent in AA than CA PCa patients; however, the prevalence of pathogenic mutations were similar across the races. Patients carrying BRCA2 pathogenic mutations are more likely to progress to metastasis.
Subject terms: Cancer screening, Cancer genetics
Introduction
DNA damage repair genes (DDRGs) play a critical role in protecting genome integrity whereas the presence of germline mutations predispose to several cancer types (e.g., breast, ovarian). In prostate cancer (PCa), several recent studies described DDRG mutations that associated with aggressive disease. Exome and targeted sequencing-based findings for metastatic castration-resistant prostate cancer (mCRPC) revealed the importance of mutations and copy number changes in DDRG genes (including BRCA2, ATM, BRCA1, CHEK2, FANCA, RAD51B/D, PALB2, and MSH2) for disease progression [1–3]. Moreover, the IMPACT study demonstrated that germline BRCA1/2 mutations can be used to identify men with higher risk of developing PCa, and are associated with a more aggressive phenotype and poorer outcome [4, 5]. Thus, there is accumulating evidence that BRCA1/2 mutation carriers are at an increased risk for aggressive PCa. Because poly (ADP-ribose) polymerase (PARP) inhibitors can extend overall survival in mCRPC patients with DDRG mutations [6], the targeted identification of BRCA1/2 mutation carriers could improve survival in this group of patients.
Variants in BRCA1/2 are categorized into the following groups: pathogenic alleles that either inactivate the protein or are documented to confer risk for cancer, variants of unknown significance (VUS) that are as yet unclassified, likely benign and benign variants that are not known to confer cancer risk. However, this classification has been largely established for breast cancer patients and families; whether the same functional relationships exist for PCa is unclear. More studies are needed to establish if and how mutation frequencies in BRCA1 and BRCA2 vary in racial and ethnic groups including African-American (AA) and Caucasian-American (CA) populations [7]. It is important to note that AA men have the highest mortality rate from PCa [8] and their tumor biology remains understudied. Therefore, we included AA PCa patients into this study who were recruited either within the Department of Defense (DOD) Health Care System [9] or the National Cancer Institute (NCI) Maryland Case-Control Study [10], allowing for an evaluation of BRCA1/2 mutation frequencies in AA and CA men with PCa. An understanding for the role of racial/ethnic differences of BRCA1/2 variants in PCa is important and is also urgently needed because there is a persistent underrepresentation of samples from minorities in public databases (ClinVar, ENIGMA).
Materials and methods
Patient recruitment and characteristics
Germline DNA was obtained from 935 men (cohort 1) who were recruited at the Walter Reed National Medical Military Center (WRNMMC) (Supplementary Table 1). These patients were treated with radical prostatectomy between 1996 and 2012 for very low-, low-, and intermediate-risk PCa, according to National Comprehensive Cancer Network (NCCN) guidelines and were enrolled in the Center for Prostate Cancer Research (CPDR) longitudinal database and this study, under an Institutional Review Board (IRB)-approved protocol. Additional DNA samples were obtained from 189 participants in the NCI Maryland Prostate Cancer Case-Control study (cohort 2). This study recruited 976 AA and CA PCa patients between 2005 and 2016, as previously described [10]. Blood DNA samples were also obtained from participants in clinical trials for metastatic PCa at the National Cancer Institute (cohort 3, N = 116) (Supplementary Table 1). All studies were approved by IRBs of the individual institutions and written informed consent was obtained from all subjects.
Ion AmpliSeq targeted gene sequencing
BRCA1/2 genes were sequenced in archival blood DNA specimens (N = 1240) collected from PCa patients undergoing radical prostatectomy treatment at WRNMMC, from CPDR and from the NCI. Genomic DNA was extracted from peripheral blood lymphocytes using DNeasy Blood and Tissue DNA isolation kit. The DNA samples from the three studies were amplified and libraries prepared following the Ion AmpliseqTM Library Preparation protocol. Individual samples were barcoded, pooled, applied to chips, and sequenced on the Ion TorrentTM PGM Sequencer using the Ion PGMTM Template OT2 200 and Ion PGMTM Sequencing 200v2 kits. Mean read length after sequencing was 116 bp, and 94% of amplicons gave an average coverage of greater than 50 reads per sample. Resulting sequence reads were aligned to the human reference genome version hg19 using the TMAP aligner and single-nucleotide variants were called using both Genome Analysis Tool Kit (GATK) [11, 12] and Torrent Variant Caller (TSVC) as previously described [13]. The small insertions and deletions were called using the TSVC. All mutations, in Supplementary Table 1, were manually examined in Integrative Genomics Viewer (IGV) [14] to confirm an adequate number of mutant reads in both directions, and to eliminate false positives. The primer panel was previously validated against known mutations and in other studies [13]. We have previously compared the sensitivity of Ion Torrent sequencing and variant calling against Illumina and Complete genomics platforms and found them to be comparable [15]. The primer panel used here was previously shown to have over 99% coverage of the coding regions and splice sites of BRCA1 and 98% of BRCA2 [13]. Known mutations were used as positive controls and predicted mutations detected with this panel have been verified by Sanger sequencing in this and other studies [13]. The primer panel has been validated against a panel of 115 known variants and shown to have high sensitivity and specificity [16]. The sequences were analyzed for known and novel mutations in BRCA1/2, and variants less than 3% in frequency in the 1000 genomes database were selected for further analysis. Known variants were annotated with data from ClinVar to determine clinical relevance.
Statistical analyses
The chi-square test was used to compare the distribution of the clinico-pathological characteristics between CA and AA patients, as well as by BRCA1/2 mutation status. Unadjusted Kaplan–Meier estimate curves were used to evaluate prognostic significance. For all other data analysis, Student's t-test was used. P < 0.05 was considered to be statistically significant. All analyses were conducted using SAS software, version 9.3.
Results and discussion
To examine the frequency of BRCA1/2 germline variants and mutations in our cohort, we sequenced the two genes in early stage (T2) and advanced stage (T3–4 and metastatic) PCa patients and evaluated the impact of pathogenic BRCA1/2 mutations on PCa progression and distant metastasis in subjects unselected for family history, in both AA and CA patients.
To identify and determine the frequency of BRCA1/2 mutations in PCa, a total of 1240 germline DNA patient samples were amplified and sequenced (Supplementary Table 1). Variants in the BRCA1/2 genes were categorized as benign/likely benign (together with wild type), VUS, and pathogenic. Non-coding variants are shown in Supplementary Table 4. The population was comprised of mainly CA subjects 69.2% (855/1235) but enriched for AA patients (28.6% (353/1235)), while 2.6% (32/1235) were patients from different race/ethnic background (Asian, Hispanic) (Supplementary Table 1). Being of African descent was self-reported, providing an opportunity to compare the BRCA1/2 variant and mutational status between AA and CA patients. This comparison showed that AA patients had an increased frequency of germline VUS mutations in BRCA1 and BRCA2 genes compared to CA patients (4.6 vs. 1.6%, respectively); however, the prevalence of pathogenic mutations was similar (1.4 vs. 1.0%, respectively) (Table 1).
Table 1.
Association of BRCA1/2 germline mutations with race and tumor characteristics (combined three cohorts, N = 1240)
| Variable | WT/benign/likely benigna | VUSb | Pathogenicc | P value |
|---|---|---|---|---|
| Race | ||||
| AA | 412 (94.1) | 20 (4.6) | 6 (1.4) | 0.0063 |
| CA | 750 (97.4) | 12 (1.6) | 8 (1.0) | |
| Clinical stage | ||||
| Early stage (T2) | 984 (96.5) | 29 (2.8) | 7 (0.7) | 0.0206 |
| Advanced stage (T3–4/N1/M1/D3) | 182 (94.3) | 5 (2.6) | 6 (3.1) | |
| Biopsy Gleason sum | ||||
| 6 or less | 659 (96.6) | 19 (2.8) | 4 (0.6) | 0.0331 |
| 7 | 317 (95.5) | 11 (3.3) | 4 (1.2) | |
| 8–10 | 169 (95.5) | 2 (1.1) | 6 (3.4) | |
Numbers in parenthesis indicate % values
aWild type BRCA1/2 and benign/likely benign mutations in BRCA1/2
bVariants of unknown significance in BRCA1/2
cPathogenic mutations in BRCA1/2
Next, we analyzed the relationship between BRCA1/2 mutations and disease status. Our results show that there is a wide spectrum of germline mutations in BRCA1/2 and confirm that only a small percentage of men treated for localized PCa are carriers for BRCA1/2 mutations (7 of 1020 subjects carried pathogenic variants, 0.7%) [17]. However, as previously indicated, in patients with metastatic and advanced PCa, this percentage is significantly increased (over 4-fold) to 3.1% (6 of 193) (Table 1). In advanced PCa patients most of the pathogenic mutations were found in the BRCA2 gene (85.7%, 6 of 7 in BRCA2 vs. 14.3%, 1 of 7 in BRCA1). The Ashkenazi BRCA1 c.68_69delAG (185delAG) and BRCA2 c.5946delT (6174delT) founder mutations were each found in one metastatic PCa patient (Supplementary Table 2). All 19 pathogenic mutations in BRCA1 and BRCA2 are depicted in a schematic map format (Supplementary Fig. 1). An even higher frequency (6.0%, 9/150) of BRCA1/2 germline mutations was reported in a large multi-center study of CRPC [3]. Our data add to the increasing evidence that BRCA1/2 mutation carriers develop more aggressive PCa [18, 19].
The largest subset of the PCa patients (CPDR cohort, N = 935) in this study was treated by radical prostatectomy for localized PCa in an equal access DoD health care system (CPDR/WRNMMC). Mutations in BRCA1/2 were detected in 19% of PCa patients; however, most of them (over 15%) were categorized as benign or likely benign. Pathogenic or VUS mutations affected 35 of the 935 evaluable PCa patients (3.7%) (Supplementary Table 1). No significant association of BRCA1/2 mutations with ERG (erythroblast transformation-specific (ETS)-related gene) status of the tumor was apparent, although the number of pathogenic mutations was too low to conclude. Co-morbidity (other cancer) was not associated with BRCA1/2 mutations (Data not shown).
Most important, in this cohort with up to 20 years of follow-up time, pathogenic BRCA1/2 mutations associated with future distant metastasis and shorter time to metastasis (Kaplan–Meier analysis, p < 0.0001, Fig. 1), but had no significant association with biochemical recurrence (Supplementary Fig. 2). At the time of radical prostatectomy patients with higher pathologic T stage (pT3–4 vs pT2) had an increased percentage of pathogenic mutations, but this increase was not significant (Supplementary Table 3). A limitation of this study is that we did not use a method such as MLPA (multiplex ligation-dependent probe amplification) [20] to identify large insertions and deletions.
Fig. 1.
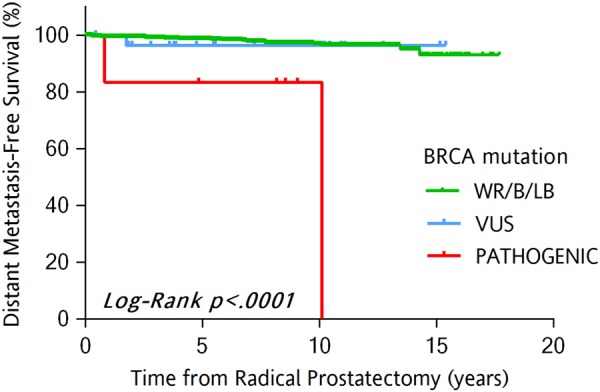
Kaplan–Meier distant metastasis-free survival curves across BRCA1/2 mutation status (cohort 1, N = 927). The figure depicts time to distant metastasis as survival estimate for the categories of patients based on BRCA1/2 mutation status. a Wild-type BRCA1/2 and benign/likely benign mutations in BRCA1/2; b variants of unknown significance in BRCA1/2; c pathogenic mutations in BRCA1/2; d metastasis
In summary, patients carrying pathogenic BRCA mutations, most frequently in BRCA2, confer a higher risk of more aggressive PCa phenotype, and a higher risk of metastasis with a shorter survival time. Increased frequency of VUS mutations in BRCA genes of AA patients, compared to CA patients, suggest that it may be important to evaluate these VUS mutations for potential pathogenic effects in PCa. Recent studies demonstrate that PARP inhibitors (inhibiting single-strand DNA break repair), such as Olaparib, can slow progression-free survival and extend overall survival in patients with BRCA1/2 mutations [21]. Therefore, testing for BRCA1/2 germline mutations in high-risk patients (familial history or tumor with poor prognostic clinico-pathologic features) may provide useful information for treatment stratification and for improving current nomograms.
Disclaimer
The views expressed in this article are those of the author and do not reflect the official policy of the Department of Army/Navy/Air Force, Department of Defense, or the U.S. Government. The identification of specific products, scientific instrumentation, or organization is considered an integral part of the scientific endeavor and does not constitute endorsement or implied endorsement on the part of the author, DoD, or any component agency.
Electronic supplementary material
Acknowledgements
This work was supported by the USUHS-CPDR fund (HU0001-10-2-0002) to ILR.
Funding
This work was supported by the USUHS-CPDR fund (HU0001-10-2-0002) to ILR.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41391-018-0114-1) contains supplementary material, which is available to authorized users.
References
- 1.McLeod DG, Srivastava S. Re: Inherited DNA-repair gene mutations in men with metastatic prostate cancer. Eur Urol. 2017;71:692. doi: 10.1016/j.eururo.2016.12.018. [DOI] [PubMed] [Google Scholar]
- 2.Pritchard CC, Mateo J, Walsh MF, De Sarkar N, Abida W, Beltran H, et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med. 2016;375:443–53. doi: 10.1056/NEJMoa1603144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;162:454. doi: 10.1016/j.cell.2015.06.053. [DOI] [PubMed] [Google Scholar]
- 4.Bancroft EK, Page EC, Castro E, Lilja H, Vickers A, Sjoberg D, et al. Targeted prostate cancer screening in BRCA1 and BRCA2 mutation carriers: results from the initial screening round of the IMPACT study. Eur Urol. 2014;66:489–99. doi: 10.1016/j.eururo.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castro E, Goh C, Olmos D, Saunders E, Leongamornlert D, Tymrakiewicz M, et al. Germline BRCA mutations are associated with higher risk of nodal involvement, distant metastasis, and poor survival outcomes in prostate cancer. J Clin Oncol. 2013;31:1748–57. doi: 10.1200/JCO.2012.43.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mateo J, Carreira S, Sandhu S, Miranda S, Mossop H, Perez-Lopez R, et al. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med. 2015;373:1697–708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Na R, Zheng SL, Han M, Yu H, Jiang D, Shah S, et al. Germline mutations in ATM and BRCA1/2 distinguish risk for lethal and indolent prostate cancer and are associated with early age at death. Eur Urol. 2017;71:740–7. doi: 10.1016/j.eururo.2016.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rebbeck TR. Prostate cancer genetics: variation by race, ethnicity, and geography. Semin Radiat Oncol. 2017;27:3–10. doi: 10.1016/j.semradonc.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brassell SA, Dobi A, Petrovics G, Srivastava S, McLeod D. The Center for Prostate Disease Research (CPDR): a multidisciplinary approach to translational research. Urol Oncol. 2009;27:562–9. doi: 10.1016/j.urolonc.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 10.Smith CJ, Dorsey TH, Tang W, Jordan SV, Loffredo CA, Ambs S. Aspirin use reduces the risk of aggressive prostate cancer and disease recurrence in African-American Men. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive. Oncology. 2017;26:845–53. doi: 10.1158/1055-9965.EPI-16-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dean M, Boland J, Yeager M, Im KM, Garland L, Rodriguez-Herrera M, et al. Addressing health disparities in Hispanic breast cancer: accurate and inexpensive sequencing of BRCA1 and BRCA2. Gigascience. 2015;4:50. doi: 10.1186/s13742-015-0088-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boland JF, Chung CC, Roberson D, Mitchell J, Zhang X, Im KM, et al. The new sequencer on the block: comparison of Life Technology's Proton sequencer to an Illumina HiSeq for whole-exome sequencing. Hum Genet. 2013;132:1153–63. doi: 10.1007/s00439-013-1321-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trujillano D, Weiss ME, Schneider J, Koster J, Papachristos EB, Saviouk V, et al. Next-generation sequencing of the BRCA1 and BRCA2 genes for the genetic diagnostics of hereditary breast and/or ovarian cancer. J Mol Diagn : JMD. 2015;17:162–70. doi: 10.1016/j.jmoldx.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 17.Kote-Jarai Z, Leongamornlert D, Saunders E, Tymrakiewicz M, Castro E, Mahmud N, et al. BRCA2 is a moderate penetrance gene contributing to young-onset prostate cancer: implications for genetic testing in prostate cancer patients. Br J Cancer. 2011;105:1230–4. doi: 10.1038/bjc.2011.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giri VN, Egleston B, Ruth K, Uzzo RG, Chen DY, Buyyounouski M, et al. Race, genetic West African ancestry, and prostate cancer prediction by prostate-specific antigen in prospectively screened high-risk men. Cancer Prev Res (Phila) 2009;2:244–50. doi: 10.1158/1940-6207.CAPR-08-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grossfeld GD, Latini DM, Downs T, Lubeck DP, Mehta SS, Carroll PR. Is ethnicity an independent predictor of prostate cancer recurrence after radical prostatectomy? J Urol. 2002;168:2510–5. doi: 10.1016/S0022-5347(05)64179-1. [DOI] [PubMed] [Google Scholar]
- 20.Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helleday T. PARP inhibitor receives FDA breakthrough therapy designation in castration resistant prostate cancer: beyond germline BRCA mutations. Ann Oncol. 2016;27:755–7. doi: 10.1093/annonc/mdw048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


