Abstract
Objectives
To comprehensively explore metabolic and genetic contributors to liver fat accumulation in overweight/obese children.
Methods
Two hundred thirty Italian children with obesity were investigated for metabolic parameters and genotyped for PNPLA3, TM6SF2, GCKR, and MBOAT7 gene variants. Percentage hepatic fat content (HFF%) was measured by nuclear magnetic resonance.
Results
HFF% was positively related with BMI, HOMAIR, metabolic syndrome, ALT, AST, γGT, and albumin. Carriers of [G] allele in PNPLA3, [T] allele in GCKR and [T] allele in TM6SF2 genes had significantly higher hepatic fat content than wild-type carriers. HFF% was explained for 8.7% by metabolic and for 16.1% by genetic factors and, a model including age, gender, BMI, HOMAIR, PNPLA3, GCKR, and TM6SF2 variants was the best predictor of HFF%, explaining 24.8% of its variation (P < 0.001). A weighted-genetic risk score combining PNPLA3, GCKR, and TM6SF2 risk alleles was associated with almost eightfold higher risk of NAFLD.
Conclusions
Our data highlighted the predominant role of genetic factors in determining the amount of liver fat content in children with obesity.
Introduction
Paralleling the worldwide epidemic of obesity in childhood, non-alcoholic fatty liver disease (NAFLD) has become the most common liver abnormality in children and adolescents.1 Indeed, it has been reported that NAFLD can be detected in up to 60% of overweight children.2–5 However, the fact that not all children with obesity develop NAFLD suggests that environmental and/or genetic factors may confer NAFLD susceptibility to each obese individual.
NAFLD predisposes to a broad spectrum of chronic liver diseases mostly occurring in adulthood, such as non-alcoholic steatohepatitis (NASH), cirrhosis, and hepatocellular carcinoma.6,7 Therefore, the identification of factors promoting increased liver fat content in children with obesity is clinically relevant as it may favor early preventive interventions in high-risk individuals. In addition, the recognition of these relationships at early stage may allow the better understanding of the steatogenic role of different factors since there is less potential for confounders (e.g., drug, alcohol consumption).
Several reports have recognized the leading role of insulin resistance in promoting hepatic fat content (HFF) in obesity, even though it has not been definitively established whether this condition may represent the cause or the consequence of the increased HFF in the liver.8,9 In addition, several susceptibility gene variants have been identified.10,11 In particular, the rs738409 C>G sequence variant in the Patatin-like Phospholipase domain-containing 3 (PNPLA3) gene, encoding for the I148M protein variation, has been identified as a major determinant of the inter-individual and ethnicity-related differences in HFF in adults.12,13 The mechanisms by which this substitution induces liver fat is related to an impaired hepatocellular triglycerides hydrolysis and increased lipogenesis associated to the 148M allele.14 The Transmembrane 6 Superfamily Member 2 (TM6SF2) gene has also been shown to increase NAFLD susceptibility causing an impaired mobilization of neutral lipids for very low-density lipoprotein assembly and secretion by the liver in rs58542926 C>T (E167K) carriers.15,16 Other pro-steatogenic variants have been identified in the Protein Phosphatase 1 Regulatory Subunit 3B (PPP1R3B), the Glucokinase Regulator (GCKR), Neurocan (NCAN), Lysophospholipase-Like 1 (LYPLAL1), and the Membrane Bound O-acyltransferase domain-containing 7 (MBOAT7-TMC4) genes, but their involvement in the pathogenesis of liver steatosis is less firmly established.17,18 We have recently reported that a genetic score based on PNPLA3, TM6SF2, GCKR, and MBOAT7 variants was highly predictive of NAFLD in adults.19 However, only some of these susceptibility genes have received direct confirmation in children.20–23
Thus, in this study we present a comprehensive evaluation of genetic and metabolic factors influencing HFF in a cohort of Italian children with obesity. As the sensitivity of ultrasound techniques is limited, we have used magnetic resonance imaging (MRI) to obtain an accurate quantitative assessment of the amount of liver fat.
Methods
Study subjects
This observational study included 230 overweight [body mass index (BMI) > 85th and < 95th percentile for age and gender] or with obesity (BMI ≥ 95th percentile for age and gender) children and adolescents (aged 6–16 years), who were consecutively admitted at the outpatient services of the Department of Pediatrics, Sapienza University of Rome,24,25 to receive a clinical evaluation for obesity. All children were Caucasians and ethnically homogeneous.
All study children had a complete physical examination as reported in detail elsewhere.25 The pubertal status was assessed according to Tanner.26 The degree of obesity was quantified using Cole’s least mean-square method, which normalizes the skewed distribution of BMI and expresses BMI as standard deviation (SD) score.24,25 Children with history of diabetes and endocrine or renal disease were excluded. Those with secondary causes of steatosis, including hepatic virus infections (hepatitis A-E and G, cytomegalovirus, and Epstein-Barr virus), autoimmune hepatitis, metabolic liver disease, α-1-antitrypsin deficiency, cystic fibrosis, Wilson’s disease, hemochromatosis, and celiac disease were also excluded.25 Metabolic syndrome (MetS) was diagnosed in the presence of any three of the following: (1) WC ≥ 90th age- and sex-specific percentiles according to reference curves for children aged 2–18 years;27,28 (2) elevated systolic and diastolic BP ≥ 90th percentile for age, gender, and height;29 (3) low high-density lipoprotein cholesterol (HDL) values (≤10th) according to reference curve for all ages/sexes;27,28 (4) elevated triglycerides (TG) values ≥ 90th sex and age-specific percentile according to reference curves for children aged 2–18 years,27,28 and (5) fasting glucose ≥ 5.6 mmol/L.27
The study protocol was reviewed and approved by the Ethics Committee of Policlinico Umberto I Hospital (EC approval #2464). Written informed consent was obtained from the next of kin, caretakers, or guardians on behalf of the children enrolled in this study, in accordance with principles of Helsinki Declaration.
Liver magnetic resonance imaging (MRI)
MRI data were acquired on a 3.0 T MR scanner with a 50 mT/m maximum gradient length and 200 T/m/s maximum slew rate (Discovery MR 750; GE Medical Systems, Milwaukee, WI) using an eight-element body torso-array coil system.30–32 All spectra were obtained in the stimulated echo acquisition mode, using a breath hold sequence with an acquisition time of ~24 s.32 HFF% was measured as previously described and validated.30–32 According to results of previous investigations, children showing HFF ≥ 5% were classified as having NAFLD.31,32
Laboratory measurements
Blood samples were taken from all study subjects after an overnight fast for estimation of glucose, insulin, total cholesterol (TC), HDL-C, TG, alanine aminotransferase (ALT), aspartate aminotransferase (AST), and gamma-glutamyl transferase (γGT).25 An oral glucose tolerance test (OGTT) was performed for all overweight/obese children as yet reported.30 Insulin sensitivity were estimated through the homeostasis model assessment (HOMAIR).30 We also classified subjects as having elevated ALT using cut off values established from NHANES data in boys (>25.8 U/L) and girls (>22.1 U/L).33 All analyses were conducted by COBAS 6000 (Roche Diagnostics).30 Insulin concentrations were measured by an electrochemiluminescent method.25
DNA genotyping
Genomic DNA was extracted from whole blood according standard procedures. The rs641738 C>G (I148M) (PNPLA3), rs58542926 C>T (E167K) (TM6SF2), rs1260326 C>T (L446P) (GCKR), and rs641738 C>T (G17E) (MBOAT7-TMC4) were considered for genotyping. They were selected because in a previous resequencing study carried out in an Italian adult cohort they emerged as significant genetic determinants of NAFLD.19 Genotyping were performed in duplicate by TaqMan 5′-Nucleotidase assay.19,34
Genetic risk score computation
The Genetic Risk Score (GRS) was calculated based on the four selected SNPs. As previously described, two methods were used: (1) a simple counting method (unweighted GRS) and (2) a weighted method (weighted GRS).19 The β-coefficients considered for weighted GRS calculation and each SNP were 0.2653 (rs738409), 0.2711 (rs58542926), 0.0649 (rs1260326), and 0.0575 (rs641738).35 The GRS was modeled as a continuous variable and then categorized into tertiles.19
Statistical analysis
Statistical analyses were performed by using the SPSS package (version 22.0), SPSS Inc., Chicago, IL. Data were reported as means and standard deviations for normally distributed variables, or as median and interquartile range for non-normally distributed variables. Differences between groups were evaluated by t-test or Mann–Whitney U-test, as appropriate. Proportions were compared by the χ2 test.
Genotype frequencies were assessed for Hardy–Weinberg equilibrium (HWE) using the goodness-of-fit χ2 test. It must be noted that because of the low frequency of the T allele (167K) in TM6SF2 gene (only one homozygous subject), all calculations were based on the dominant model of inheritance.
Predictors of HFF% were evaluated by linear regression analysis by adding once at time each independent variable. R2-value and β-coefficients were used to evaluate the percentage of variation of HFF explained by each variable and the degree of change in HFF% for every 1-unit of change of the independent variable. The association of PNPLA3, GCKR, TM6SF2, and MBOAT7 genotypes with HFF% was evaluated by a linear regression analysis adjusted for age, gender, BMI, pubertal stage, and HOMAIR. Finally, genetic and non-genetic variables, which emerged as significantly associated to HFF%, were then included into multivariate linear regression models to determine their independency in predicting HFF%. Age, gender, BMI, and HOMAIR were entered in the first model followed by PNPLA3, GCKR, and TM6SF2 variants (Enter method). R2-value, variation of R2, F-test, and P-value F-test were used to identify the best model able to predict HFF% variation and the significant amount of variance in HFF%. Collinear variables, evaluated by Pearson bivariate analysis, were excluded. Logistic regression analysis was used to examine the odds ratios (ORs) of NAFLD associated to metabolic and genetic variables and weighted GRS. ORs were adjusted for age, gender, BMI, and HOMAIR. Multiple comparisons were further adjusted by bootstrap correction based on 1000 bootstrap samples. Statistical significance was taken at a nominal P-value < 0.05 for all comparisons.
Results
Clinical characteristics of study population
Baseline characteristics are shown in Table 1. One hundred and thirty-one were boys. The mean age was 10.2 ± 3.0 years. Overall, 86% showed obesity as defined by sex- and gender-specific BMI values ≥ 95th percentile. Measures of adiposity, most notably BMI (P = 0.010) and WC (P = 0.004), were found to be higher in boys than girls. In the entire cohort 47.1% of children were classifiable as insulin resistant according to a previously reported criterion (HOMAIR ≥ 2.5).36 Systolic and diastolic BP as well as plasma lipids were within normal ranges and did not differ between genders, even after adjustment for age, BMI, and pubertal stage. Overall, 19.8% of boys and 14.1% of girls showed the characteristics of MetS (P = 0.25 for gender difference).
Table 1.
Clinical characteristics of the study population
| All | Boys | Girls | P | |
|---|---|---|---|---|
| N | 230 | 131 | 99 | |
| Age (years) | 10.2 ± 3.0 | 10.7 ± 3.1 | 9.5 ± 2.7 | 0.003 |
| Body composition | ||||
| BMI (kg/m2) | 25.4 (23.1–28.7) | 26.0 (23.4–29.5) | 24.6 (22.8–27.1) | 0.010 |
| BMI Z-score | 2.04 (1.8–2.3) | 2.1 (1.8–2.3) | 2.0 (1.8–2.2) | 0.36 |
| WC (cm) | 86.9 ± 11.6 | 88.8 ± 11.6 | 84.4 ± 11.1 | 0.004 |
| BMI ≥ 95th (n, %) | 196 (86) | 87 (83.7) | 109 (87.9) | 0.88 |
| Pubertal stage | 0.76 | |||
| Stage 1 | 88 (38.3) | 52 (39.7) | 36 (36.4) | 0.09 |
| Stage 2 | 59 (25.7) | 30 (22.9) | 29 (29.3) | 0.89 |
| Stage 3 | 33 (14.3) | 21 (16.0) | 12 (12.1) | 0.12 |
| Stage 4 | 21 (9.1) | 12 (9.2) | 9 (9.1) | 0.51 |
| Stage 5 | 28 (12.2) | 15 (11.5) | 13 (13.1) | 0.70 |
| MetS (n, %)a | 40 (17.4) | 26 (19.8) | 14 (14.1) | 0.26 |
| HFF (%) | 3.0 (1–10) | 5.0 (1–10) | 2.0 (1–7) | 0.042 |
| NAFLD (n, %)b | 105 (45.7) | 68 (51.9) | 37 (37.4) | 0.028 |
| Blood pressure | ||||
| Systolic (mmHg) | 111.5 (106–120) | 113.3 (106–120) | 111 (105–120) | 0.80 |
| Diastolic (mmHg) | 65 (60–70) | 65.0 (60.0–70.0) | 65.0 (60–70) | 0.95 |
| Plasma lipids | ||||
| TC (mg/dL) | 160.4 ± 31.9 | 161.4 ± 34.0 | 159.2 ± 29.04 | 0.60 |
| LDL-C (mg/dL) | 92.9 ± 27.6 | 93.2 ± 29.1 | 92.5 ± 25.6 | 0.80 |
| TG (mg/dL) | 75.0 (51–106) | 72.5 (51–106) | 75.0 (51.5–107) | 0.86 |
| HDL-C (mg/dL) | 50 ± 12.6 | 50.7 ± 13.4 | 52.1 ± 13.5 | 0.44 |
| APOB (mg/dL) | 0.46 (0.64–0.88) | 0.74 (0.63–0.85) | 0.79 (0.65–0.89) | 0.17 |
| APOAI (mg/dL) | 1.38 (1.25–1.54) | 1.39 (1.26–1.54) | 1.36 (1.23–1.54) | 0.33 |
| Measures of glucose homeostasis | ||||
| Blood glucose (mg/dL) | ||||
| Fasting | 83 (77–86) | 83.0 (79.0–86.0) | 81.0 (77.0–86.0) | 0.06 |
| 120 min | 95.2 ± 16.7 | 97.0 ± 17.7 | 92.5 ± 14.8 | 0.06 |
| Insulin (UI/L) | ||||
| Fasting | 11.8 (8–16.9) | 11.8 (7.8–17.1) | 11.8 (9.2–16.7) | 0.61 |
| 120 min | 42.4 (23.3–68.8) | 41.3 (26.6–72.6) | 42.7 (21.7–62.9) | 0.53 |
| HOMAIR | 2.4 (1.6–3.6) | 2.4 (1.5–3.6) | 2.3 (1.8–3.6) | 0.62 |
| Liver function tests | ||||
| ALT (UI/L) | 20.5 (16–28.2) | 22 (17–31) | 19 (15–24) | 0.005 |
| AST (UI/L) | 23 (20–27) | 24 (21–27) | 23 (20–26.5) | 0.28 |
| AST/ALT ratio | 1.10 (0.83–1.41) | 1.05 (0.76–1.35) | 1.17 (0.92–1.51) | 0.006 |
| GGT (UI/L) | 14 (11–18) | 14 (12–19) | 12 (10–16) | 0.002 |
| Albumin (g/L) | 49.0 (47–50.5) | 49.0 (47–51) | 48 (46–50) | 0.02 |
| Ferritin (μg/L) | 66.2 ± 35.8 | 68.7 ± 39.01 | 62.8 ± 30.9 | 0.35 |
| CRP (μg/L) | 1600 (800–3600) | 1400 (700–2850) | 1800 (900–4600) | 0.010 |
Data are expressed as percentage, mean (±SD) and median (25th–75th percentile range) as appropriate
APOA1 Apoliporotein A-I, APOB apoliporotein B, CRP C-reactive protein, LDL-C low-density lipoprotein cholesterol
aMetS was defined as reported in Material and methods
bNAFLD was defined as having hepatic fat content (HFF) ≥ 5% at MRI30–32
The distribution of HFF% values in the entire study cohort is shown in the Supplemental Figure S1 (online). Median HFF% was 3.0% (interquartile range 1–10%) and 45.7% of children showed HFF ≥ 5.0%, our threshold criteria for the definition of NAFLD. Compared to girls, a significantly higher proportion of boys showed NAFLD (P = 0.028), even though this difference disappeared when adjusted for age, BMI, and pubertal stage (Padj = 0.07). Boys showed significantly higher median ALT (P = 0.005) and γGT (P = 0.002) (Table 1).
Metabolic and genetic predictors of HFF
Supplemental Table S1 (online) shows the results of univariate linear regression analysis associating HFF% with demographic and biochemical variables. HFF% was positively related with BMI, glucose and insulin levels, MetS, ALT, AST, γGT, albumin and, negatively, with AST/ALT ratio.
To test whether genetic variants were associated with liver fat content, we first compared HFF% between genotypes. The distribution of genotyped SNPs was in HWE (all P > 0.05) and the minor allele frequency (MAF) for all variants was comparable to that reported in Europeans Non-Finnish.37 Overall, 122 (53.1%) subjects were carriers of G PNPLA3 allele, 163 (70.9%) of T MBOAT7 allele, 183 (79.5%) of T GCKR allele, and 32 (13.9%) of T TM6SF2 allele.
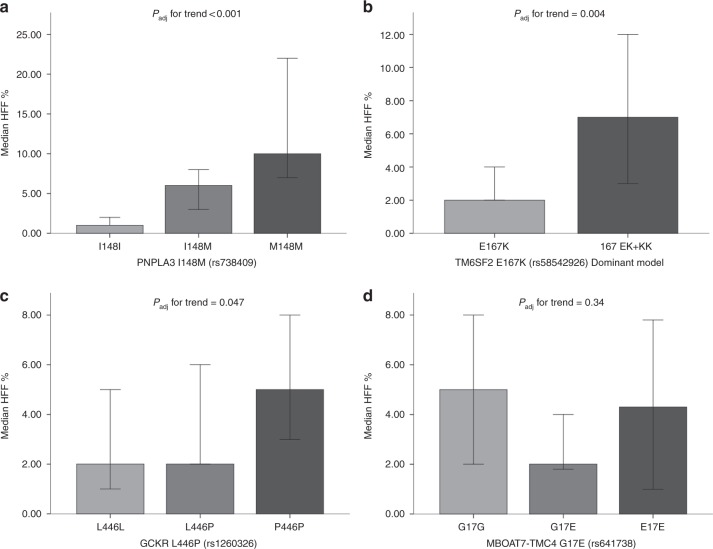
As reported in Fig. 1, carriers of G allele in PNPLA3, T allele in GCKR and T allele in TM6SF2 had higher HFF% than wild-type carriers, even after adjustment for age, gender, BMI, pubertal stage, and HOMAIR (Padj < 0.001, Padj < 0.004, and Padj = 0.047, respectively). We did not identify any association between HFF% and MBOAT7 genotypes. Noteworthy, each copy of G PNPLA3 allele determined a significant twofold increase in HFF% [median (interquartile range) = 6% (1.9–12.2) in CG vs. 10% (6.0–23.5) in GG (P = 0.019)].
Fig. 1.
Hepatic fat content according to genotypes. Linear regression analysis was used to evaluate associations between HFF% and a PNPLA3 I148M, b TM6SF2 E167K, c GCKR L446P and d MBOAT7-TMC4 G17E genotypes. *Padj for trend shows the significance level of association tested by linear regression analysis adjusted for age, gender, pubertal stage, BMI, and HOMAIR
Table 2 shows the contribution of genetic and non-genetic factors in predicting HFF. The model 2, which included age, gender, BMI, HOMAIR, PNPLA3, GCKR, and TM6SF2 gene variants (Enter method), showed the highest ability in predict HFF% explaining 24.8% of its variability (P < 0.001).
Table 2.
Multivariate models describing genetic and non-genetic predictors of hepatic fat content (HFF%) in children
| R2 | R2 variation | F-test | P-value F-test | β | 95% CI | P-value | |
|---|---|---|---|---|---|---|---|
| Model 1 | 0.087 | 0.087 | 5.220 | <0.001 | |||
| Age | 0.072 | −12.29–7.61 | 0.64 | ||||
| Gender | −0.085 | −4.47–0.94 | 0.19 | ||||
| BMI | 0.113 | −0.12–0.68 | 0.17 | ||||
| HOMAIR | 0.175 | 0.18–1.52 | 0.01 | ||||
| Model 2 | 0.248 | 0.161 | 10.141 | <0.001 | |||
| Age | −0.029 | −0.58–0.39 | 0.69 | ||||
| Gender | −0.091 | −4.38–0.57 | 0.13 | ||||
| BMI | 0.179 | 0.07–0.81 | 0.018 | ||||
| HOMAIR | 0.169 | 0.20–1.43 | 0.009 | ||||
| PNPLA3 variant (additive model) | 0.347 | 3.60–7.36 | 1 × 10−7 | ||||
| GCKR variant (additive model) | 0.100 | −0.30–3.31 | 0.10 | ||||
| TM6SF2 variant (dominant model) | 0.155 | 1.10–8.08 | 0.010 |
CI Confidence interval
Notably, PNPLA3 variant appeared to be the single best predictor of HFF% overcoming the potency of the other metabolic parameters. It is interesting to note that PNPLA3, GCKR, and TM6SF2 variants alone were able to explicate about 16.1% of HFF variation, a proportion higher than that due to metabolic factors alone (8.7%). The results did not change even after including in the model MetS diagnosis as covariate.
Predictors of NAFLD
Supplemental Table S2 (online) shows the baseline characteristics and genotypes of children with and without the presence of hepatic steatosis. Children with NAFLD showed significantly higher prevalence of obesity (Padj = 0.021) and MetS (P = 0.001) and about 54.3% of them had elevated ALT (Padj ≤ 0.001). The rs738409 in PNPLA3 and rs58542926 in TM6SF2, but not GCKR and MBOAT7 gene variants, emerged as significantly associated with NAFLD (Padj < 0.001 and Padj = 0.012). Notably, TM6SF2 167 EK + KK carriers had a threefold increased risk for hepatic steatosis (OR 3.1, 95% CI: 1.3–7.1; Padj = 0.008, data not shown) independently from age, gender, BMI, pubertal stage, and PNPLA3 I148M genotype. More importantly, homozygous PNPLA3 carriers (GG genotype) showed the highest risk of NAFLD (OR 14.9, 95% CI: 4.3–51.5; Padj < 0.001, data not shown).
When genetic factors were combined with demographic and metabolic variables (Table 3), the model including PNPLA3 and TM6SF2 variants and BMI had the highest discriminatory ability (all bootstrap Padj ≤ 0.013) in predicting the presence of NAFLD.
Table 3.
Genetic and non-genetic predictors of NAFLD in children
| Gene, SNP ID | β | OR (95%CI) | P-value | Padj-valuea |
|---|---|---|---|---|
|
PNPLA3, rs738409 Additive model |
1.27 | 3.56 (2.18–5.83) | <0.001 | 0.001 |
|
TM6SF2, rs58542926 Dominant model |
1.14 | 3.15 (1.29–7.68) | 0.011 | 0.013 |
| BMI (kg/m2) | 0.16 | 1.17 (1.08–1.27) | <0.001 | 0.011 |
In the model were included: age (years), gender (M/F), BMI (kg/m2), HOMAIR, rs738409 PNPLA3, rs1230326 GCKR, rs641738 MBOAT7 (additive models), and rs58542926 TM6SF2 (dominant model) (forward Wald ratio method). Only significant variables were reported
OR Odds ratio, CI confidence interval
aPadj-values were adjusted for multiple comparisons by using the bootstrap method
Comparing NAFLD children according to ALT levels, PNPLA3 rs738409 and BMI, but not TM6SF2 rs58542926 still emerged as predictors of NAFLD in those with elevated ALT (OR 6.0, 95% CI: 3.0–11.9; Padj < 0.001). Conversely, TM6SF2 167 EK + KK genotype was the best predictor of NAFLD in the presence of normal ALT conferring fivefold increased risk of fatty liver (Padj = 0.001), higher than that due to PNPLA3 I148M or M148M genotypes (OR 2.1, 95% CI: 1.13–4.00; Padj = 0.018).
Development of a genetic score for NAFLD
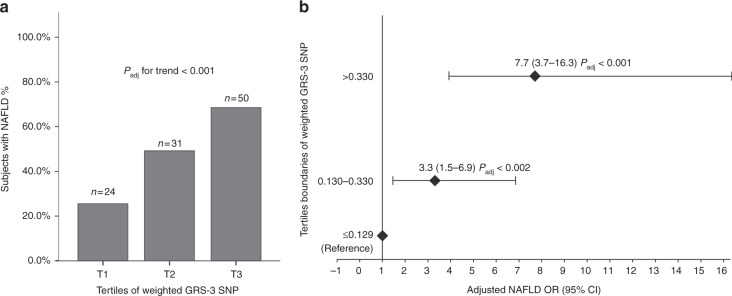
Finally, we evaluated the cumulative effect of the four SNPs on the risk of NAFLD and we found that a weighted 4-SNPs GRS conferred the highest risk of hepatic steatosis (OR 63.6, 95% CI: 13.4–292.9; Padj < 0.001). In order to explore the robustness of this association, we conducted analysis by excluding one genetic variant at a time and we found that the 3-SNPs GRS without-MBOAT7 variant was able to explain the highest risk of NAFLD (OR 83.5, 95% CI: 16.9–411.5; Padj < 0.001, bootstrap Padj = 0.001). Moreover, the prevalence of NAFLD among children significantly increased along with increasing tertiles of 3-SNPs GRS (Fig. 2a) and the adjusted risk of hepatic steatosis raised up to 7.7 for GRS values > 0.3302 (corresponding to the 3th tertile) (Padj < 0.001 Fig. 2b). The inclusion of MetS in the model did not change the power of weighted GRS in predicting fatty liver in children.
Fig. 2.
Association of weighted GRS 3-SNP with the risk of NAFLD. a Distribution of tertiles of weighted 3-SNP GRS in NAFLD patients; b NAFLD ORs adjusted for age, gender, BMI, and HOMAIR across tertiles of weighted 3-SNP GRS. Padj for trend was adjusted for age, gender, BMI, HOMAIR, and tertiles of weighted 3-SNPs GRS (χ2 Pearson followed by Stepwise regression analysis); NAFLD ORs were adjusted for age, gender, BMI, HOMAIR, and tertiles of weighted GRS (regression analysis, enter method)
Discussion
The present study was designed to weigh the impact of genetic and metabolic variables in liver fat accumulation in a cohort of children with obesity. Whereas increased body weight is a well-established determinant of fatty liver, the identification of factors interacting with obesity to produce a pro-steatogenic phenotype may have a clinical relevance.
Overall, we observed that genetic factors showed the strongest independent contribution to fatty liver and this effect was higher than that associated to insulin resistance, a typical obesity-linked metabolic abnormality. Indeed, we found that the variation of HFF was explained for 8.7% by metabolic factors and for 16.1% by the joint effect of PNPLA3, GCKR, and TM6SF2 variants. It is interesting to note that the rs738409 G allele in PNPLA3 showed the strongest effect with a standardized β-coefficient of 0.35, larger than that conferred by the increasing of 1 standard deviation of BMI (β = 0.18) and insulin resistance (β = 0.17).
In support of this data, we also found that children carrying the PNPLA3 G and/or TM6SF2 T alleles have more than threefold higher risk of NAFLD than non-carriers. In contrast, the presence of hepatic steatosis was not significantly associated with age, gender, pubertal status or HOMAIR, and only BMI and MetS emerged as independent metabolic predictor of NAFLD.
Obesity increases the risk of liver fat accumulation,4 but it is also evident that this measure alone is not sufficient for indicating increased HFF. Indeed, our data show that 87.9% of children with normal liver fat content were above the 95th percentile of BMI. It is also important to note that, in a recent study carried out in a cohort of 2042 children followed up over 30 years, Suomela et al.22 demonstrated that a combined risk score based on childhood BMI, insulin levels, birth weight and the genetic variants in PNPLA3 and TM6SF2 was superior to a model including only BMI and insulin values in predicting adult fatty liver.
In the present study, we have considered genetic variants in PNPLA3, TM6SF2, GCKR, and MBOAT7 genes for their recognized role in determining NAFLD in adults.19 However, as illustrated in Supplemental Table S3 (online), few studies have comprehensively evaluated these susceptibility genes in children. In our cohort PNPLA3 and TM6SF2 variants were strongly associated with HFF% as well as with NAFLD, while a much weaker association was detected with the common rs1260326 GCKR variant. This is in contrast with our recent observations where the GCKR T allele was associated with significantly higher risk of NAFLD in adults.19 Even though this result could be due to the small sample size, data shown in Supplemental Table S3 (online) highlight that the association of GCKR variant with NAFLD in children is controversial. To interpret this discrepancy, we could speculate that the weak pro-steatogenic effect of GCKR in young people could depend on the shorter time of exposure to environmental factors interacting with the GCKR gene variant. In fact, it has been reported that the rs1260326 variant, which encodes for the GCKR P446L protein, deregulates glucose storage and disposal thus promoting de novo lipogenesis.38 Because carbohydrates are the upstream substrates in the glucokinase-regulated pathway, the effect of GCKR variant on the NAFLD susceptibility may be mediated by the dietary intake of sugar.39 Therefore, a shorter exposure to sugar-enriched diet may account for the inconsistent results in the association between GCKR and fatty liver in childhood. Further investigations also considering the dietary carbohydrate intake are warranted to prove this hypothesis.
Moreover, we did not find any association between MBOAT7 gene variant and fatty liver accumulation. Three previous studies have explored this relation in obese children, but reporting inconsistent results.20,21,40 Two studies, where the effect of the rs641738 was explored in children evaluated by liver ultrasounds, did not find any independent association (see Supplementary Table S3, online). Conversely, Umano et al.40, which considered a different SNP in MBOAT7 gene, (rs626283) reported a positive association of this SNP with NMR-measured HFF%, but only in the subgroup of Caucasian children. Even though rs626283 is in strong LD with rs641738, it has been consistently associated with more advanced liver disease.41 It must be noted that compared to ours, children enrolled in the Umano’s study showed markedly higher indices of obesity (BMI 32.9 ± 7.5 kg/m2) and insulin resistance, thus leaving the possibility that they were presenting a more advanced liver damage. To this regard, the slightly higher levels of ALT reported in their homozygous carriers for rs626283 (ALT 30.2 ± 26.5 U/L) as compared to our homozygotes (ALT 23.0 ± 13.1 U/L) is highly suggestive of this possibility.
In line with previous studies in adults, we found that variants in the susceptibility genes act in an additive fashion in promoting liver steatosis in children with obesity. By using a weighted GRS, we showed that individuals carrying PNPLA3, GCKR, and TM6SF2 gene variants have about eightfold higher risk to develop fatty liver than non-carriers. There are few attempts in the literature to develop a GRS to illustrate the combined effect of genetic variants on liver fat accumulation in children. Walker et al.23 reported that a GRS comprising PNPLA3 and APOC3, but not the other susceptibility alleles, accounted for 12% of the variance in HFF and children with four risk alleles had threefold higher liver fat than non-variant carriers. The higher predictive power of our GRS is not easy to explain. The fact that Walker et al. included Hispanic children, which are very vulnerable to obesogenic environmental factors, might have limited the impact of genetic factors on the hepatic fat content variation. In fact, in the Walker’s cohort BMI and ALT showed the highest predictive power of HFF.23 Our findings indicate that a comprehensive weighted GRS significantly improves the risk classification of increased fat content also in children with obesity. Further confirmation in larger cohort of children with different ethnicity is needed. More importantly, since these genes have been also reported as modifiers of liver steatosis progression,42 their evaluation in combination with liver histology, measures of fibrogenesis or inflammatory biomarkers, is needed to better understand which genes play roles in the initial steps of steatogenesis as well as in the fibro-inflammatory evolution of NAFLD in children.
The present study has several strengths such as the careful metabolic evaluation of participants and the use of MRI to obtain the best accurate quantitative measurement of liver fat content. It has also limitations to be acknowledged. The cohort size is small, and we do not have longitudinal data, which might help in assessing the long-term effect of metabolic and genetic factors. Moreover, the lack of biochemical markers of fibrosis, as well as of measurement of visceral obesity or evaluation of other obesity-related factors [i.e., fructose-exceedingly rich diets or fructose intolerance3,43] did not permit a careful weighing of additional factors implicated in NAFLD or liver damage development.
In addition, we were not able to assess NAFLD by liver biopsy, but previous published studies have reported a high correlation between MRI-derived HFF% and histology in children.31,32 Moreover, in developing the weighted GRS, we have used β-coefficients obtained in adults19,35 which not necessarily may be applicable to a children population. Finally, an additional limitation of our study was the lack of replication in an independent sample. However, it must to be noted that we have considered well known genetic determinants of NAFLD not requiring replication.
In conclusion, we have shown that genetic factors have the greatest impact in determining the amount of liver fat content in children, higher than that attributable to metabolic alterations associated with obesity. Due to the large diffusion of obesity-related liver disease at all ages, there is a strong need to identify genotypes that correlate especially to pediatric NAFLD/NASH phenotype in order to decrease future fatty liver-related morbidity and develop a comprehensive risk factor panels finalized to the prediction of NAFLD risk at individual level. If definitely confirmed, this approach would provide a great help in designing early preventive intervention in high-risk children with obesity to prevent adult complication of NAFLD.
Supplementary information
Acknowledgements
This study was supported by grant from Sapienza University of Rome (2016).
Author contributions
A.D.C. performed genetic analysis, analyzed the data, generated figures, performed literature search, and prepared the manuscript; L.P. designed the study, collected the sample cohort, interpreted the data, and revised the manuscript; C.C. designed the study, interpreted the data, and revised the manuscript; F.M.P. collected the sample cohort and revised the manuscript; F.C. carried out the laboratory measurements and revised the manuscript; A.A. carried out the laboratory measurements and revised the manuscript; L.D.E. analyzed the data, prepared the manuscript, and performed literature search; M.D.M. carried out the MRI evaluation and revised the manuscript; MA designed the study, interpreted the data and prepared the manuscript. All authors edited and approved the final manuscript as written and take responsibility for its content.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41390-019-0303-1) contains supplementary material, which is available to authorized users.
References
- 1.Mencin AA, Lavine JE. Advances in pediatric nonalcoholic fatty liver disease. Pediatr. Clin. North Am. 2011;58:1375–1379. doi: 10.1016/j.pcl.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 2.Bugianesi E, et al. Low birthweight increases the likelihood of severe steatosis in pediatric non-alcoholic fatty liver disease. Am. J. Gastroenterol. 2017;112:1277–1286. doi: 10.1038/ajg.2017.140. [DOI] [PubMed] [Google Scholar]
- 3.Clemente MG, Mandato C, Poeta M, Vajro P. Pediatric non-alcoholic fatty liver disease: Recent solutions, unresolved issues, and future research directions. World J. Gastroenterol. 2016;22:8078–8093. doi: 10.3748/wjg.v22.i36.8078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson EL, et al. The prevalence of non-alcoholic fatty liver disease in children and adolescents: a systematic review and meta-analysis. PLoS ONE. 2015;10:e0140908. doi: 10.1371/journal.pone.0140908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mencin AA, Lavine JE. Non-alcoholic fatty liver disease in children. Curr. Opin. Clin. Nutr. Metab. Care. 2011;14:151–157. doi: 10.1097/MCO.0b013e328342baec. [DOI] [PubMed] [Google Scholar]
- 6.Chalasani N, et al. The diagnosis and management of non-alcoholic fatty liver disease: practice guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology and the American Gastroenterological Association. Hepatology. 2012;55:2005–2023. doi: 10.1002/hep.25762. [DOI] [PubMed] [Google Scholar]
- 7.Benedict M, Zhang X. Non-alcoholic fatty liver disease: an expanded review. World J. Hepatol. 2017;9:715–732. doi: 10.4254/wjh.v9.i16.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marchesini G, et al. Non-alcoholic fatty liver disease: a feature of the metabolic syndrome. Diabetes. 2001;50:1844–1850. doi: 10.2337/diabetes.50.8.1844. [DOI] [PubMed] [Google Scholar]
- 9.Gruben N, Shiri-Sverdlov R, Koonen DP, Hofker MH. Nonalcoholic fatty liver disease: A main driver of insulin resistance or a dangerous liaison? Biochim Biophys. Acta. 2014;1842:2329–2343. doi: 10.1016/j.bbadis.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 10.Nobili V, et al. A 360-degree overview of paediatric NAFLD: recent insights. J. Hepatol. 2013;58:1218–1229. doi: 10.1016/j.jhep.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Dongiovanni P, Valenti L. Genetics of nonalcoholic fatty liver disease. Metabolism. 2016;65:1026–1037. doi: 10.1016/j.metabol.2015.08.018. [DOI] [PubMed] [Google Scholar]
- 12.Romeo S, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sookoian S, Pirola CJ. Meta-analysis of the influence of I148M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatology. 2011;53:1883–1894. doi: 10.1002/hep.24283. [DOI] [PubMed] [Google Scholar]
- 14.Pirazzi C, et al. Patatin-like phospholipase domain-containing 3 (PNPLA3) I148M (rs738409) affects hepatic VLDL secretion in humans and in vitro. J. Hepatol. 2012;57:1276–1282. doi: 10.1016/j.jhep.2012.07.030. [DOI] [PubMed] [Google Scholar]
- 15.Kozlitina J, et al. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 2014;46:352–356. doi: 10.1038/ng.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahdessian H, et al. TM6SF2 is a regulator of liver fat metabolism influencing triglyceride secretion and hepatic lipid droplet content. Proc. Natl Acad. Sci. USA. 2014;111:8913–8918. doi: 10.1073/pnas.1323785111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Speliotes EK, et al. Genome-wide association analysis identifies variants associated with non- alcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7:e1001324. doi: 10.1371/journal.pgen.1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mancina RM, et al. The MBOAT7-TMC4 variant rs641738 increases risk of non-alcoholic fatty liver disease in individuals of European descent. Gastroenterology. 2016;150:1219–1230. doi: 10.1053/j.gastro.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Costanzo A, et al. Evaluation of polygenic determinants of non-alcoholic fatty liver disease (NAFLD) by a candidate genes resequencing strategy. Sci. Rep. 2018;27:3702. doi: 10.1038/s41598-018-21939-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin YC, Chang PF, Chang MH, Ni YH. Genetic determinants of hepatic steatosis and serum cytokeratin-18 fragment levels in Taiwanese children. Liver Int. 2018;38:1300–1307. doi: 10.1111/liv.13689. [DOI] [PubMed] [Google Scholar]
- 21.Di Sessa A, et al. The membrane-bound O-Acyltransferase7 rs641738 variant in pediatric nonalcoholic fatty liver disease. J. Pediatr. Gastroenterol. Nutr. 2018;67:69–74. doi: 10.1097/MPG.0000000000001979. [DOI] [PubMed] [Google Scholar]
- 22.Suomela E, et al. Childhood predictors of adult fatty liver. The cardiovascular risk in Young Finns Study. J. Hepatol. 2016;65:784–790. doi: 10.1016/j.jhep.2016.05.020. [DOI] [PubMed] [Google Scholar]
- 23.Walker RW, et al. Genetic and clinical markers of elevated liver fat content in overweight and obese Hispanic children. Obesity. 2013;21:E790–E797. doi: 10.1002/oby.20523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cole TJ, Bellizzi MC, Flegal KM, Dietz WH. Establishing a standard definition for child overweight and obesity worldwide: international survey. BMJ. 2000;320:1240–1243. doi: 10.1136/bmj.320.7244.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pacifico L, et al. Functional and morphological vascular changes in pediatric non-alcoholic fatty liver disease. Hepatology. 2010;52:1643–1651. doi: 10.1002/hep.23890. [DOI] [PubMed] [Google Scholar]
- 26.Marshall WA, Tanner JM. Variations in the pattern of pubertal changes in boys. Arch. Dis. Child. 1970;45:13–23. doi: 10.1136/adc.45.239.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pacifico L, et al. Association of serum triglyceride-to-HDL cholesterol ratio with carotid artery intima-media thickness, insulin resistance and nonalcoholic fatty liver disease in children and adolescents. Nutr. Metab. Cardiovasc Dis. 2014;24:737–743. doi: 10.1016/j.numecd.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 28.Cook S, Auinger P, Huang TT. Growth curves for cardio-metabolic risk factors in children and adolescents. J. Pediatr. 2009;155:S6.e15–26. doi: 10.1016/j.jpeds.2009.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.National High Blood Pressure Education Program Working Group on High Blood Pressure in Children and Adolescents. The fourth report on the diagnosis, evaluation, and treatment of high blood pressure in children and adolescents. Pediatrics. 2004;114(Suppl.2):555–576. [PubMed] [Google Scholar]
- 30.Pacifico L, et al. The impact of nonalcoholic fatty liver disease on renal function in children with overweight/obesity. Int J. Mol. Sci. 2016;17:1218. doi: 10.3390/ijms17081218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pacifico L, et al. T1-weighted dual-echo MRI for fat quantification in pediatric nonalcoholic fatty liver disease. World J. Gastroenterol. 2011;17:3012–3019. doi: 10.3748/wjg.v17.i25.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Di Martino M, et al. Comparison of magnetic resonance spectroscopy, proton density fat fraction and histological analysis in the quantification of liver steatosis in children and adolescents. World J. Gastroenterol. 2016;22:8812–8819. doi: 10.3748/wjg.v22.i39.8812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwimmer JB, et al. SAFETY study: alanine aminotransferase cutoff values are set too high for reliable detection of pediatric chronic liver disease. Gastroenterology. 2010;138:1357–1364. doi: 10.1053/j.gastro.2009.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Di Costanzo A, et al. Non-alcoholic fatty liver disease and subclinical atherosclerosis: a comparison of metabolically-versus genetically-driven excess fat hepatic storage. Atherosclerosis. 2017;257:232–239. doi: 10.1016/j.atherosclerosis.2016.12.018. [DOI] [PubMed] [Google Scholar]
- 35.Dongiovanni P, et al. Causal relationship of hepatic fat with liver damage and insulin resistance in nonalcoholic fatty liver. J. Intern Med. 2018;283:356–370. doi: 10.1111/joim.12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rijks J, et al. Glycaemic profiles of children with overweight and obesity in free-living conditions in association with cardiometabolic risk. Sci. Rep. 2016;6:31892. doi: 10.1038/srep31892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karczewski KJ, et al. The ExAC browser: displaying reference data information from over 60 000 exomes. Nucleic Acids Res. 2017;45(Database issue):D840–D845. doi: 10.1093/nar/gkw971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rees MG, et al. Cellular characterisation of the GCKR P446L variant associated with type 2 diabetes risk. Diabetologia. 2012;55:114–122. doi: 10.1007/s00125-011-2348-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin YC, Chang PF, Chang MH, Ni YH. Genetic variants in GCKR and PNPLA3 confer susceptibility to non-alcoholic fatty liver disease in obese individuals. Am. J. Clin. Nutr. 2014;99:869–874. doi: 10.3945/ajcn.113.079749. [DOI] [PubMed] [Google Scholar]
- 40.Umano GR, et al. The rs626283 variant in the MBOAT7 gene is associated with insulin resistance and fatty liver in caucasian obese youth. Am. J. Gastroenterol. 2018;113:376–383. doi: 10.1038/ajg.2018.1. [DOI] [PubMed] [Google Scholar]
- 41.Buch S, et al. A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcoholrelated cirrhosis. Nat. Genet. 2015;47:1443–1448. doi: 10.1038/ng.3417. [DOI] [PubMed] [Google Scholar]
- 42.Scott E, Anstee QM. Genetics of alcoholic liver disease and non-alcoholic steatohepatitis. Clin. Med. 2018;18(Suppl 2):s54–s59. doi: 10.7861/clinmedicine.18-2-s54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.D’Aniello R, et al. Emerging pathomechanisms involved in obesity. J. Pediatr. Gastroenterol. Nutr. 2015;60:113–119. doi: 10.1097/MPG.0000000000000559. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.