Abstract
The manufacture of large quantities of high-quality DNA is a major bottleneck in the production of viral vectors for gene therapy. Touchlight Genetics has developed a proprietary abiological technology that addresses the major issues in commercial DNA supply. The technology uses ‘rolling-circle’ amplification to produce large quantities of concatameric DNA that is then processed to create closed linear double-stranded DNA by enzymatic digestion. This novel form of DNA, Doggybone™ DNA (dbDNA™), is structurally distinct from plasmid DNA. Here we compare lentiviral vectors production from dbDNA™ and plasmid DNA. Lentiviral vectors were administered to neonatal mice via intracerebroventricular injection. Luciferase expression was quantified in conscious mice continually by whole-body bioluminescent imaging. We observed long-term luciferase expression using dbDNA™-derived vectors, which was comparable to plasmid-derived lentivirus vectors. Here we have demonstrated that functional lentiviral vectors can be produced using the novel dbDNA™ configuration for delivery in vitro and in vivo. Importantly, this could enable lentiviral vector packaging of complex DNA sequences that have previously been incompatible with bacterial propagation systems, as dbDNA™ technology could circumvent such restrictions through its phi29-based rolling-circle amplification.
Subject terms: Gene expression, Genetic vectors
Introduction
Lentiviral vectors provide important advantages in gene therapy applications, such as a large transgene-carrying capacity [1] and the ability to transduce dividing and non-dividing cells [2]. Despite the development of packaging and producer cell lines, lentivirus vectors are predominantly manufactured by transient transfection of HEK293T cells with plasmid DNA (pDNA) cassettes encoding the vector genome, HIV-1 structural proteins and trans-acting factors. These pDNAs are required at a sizeable manufacturing scale and represent a significant bottleneck to flexible clinical programmes.
There are several drawbacks associated with bio-production of lentivirus vector titres on a pDNA platform. GMP pDNA manufacture is costly, complex and the DNA product is ultimately contaminated with bacterial propagation elements that are unnecessary for virus production in mammalian cells. Furthermore, eukaryotic expression cassettes may occasionally contain gene sequences that produce toxic or problematic effects in bacteria, which limits their amplification.
Touchlight’s novel, synthetic in vitro amplification process is capable of producing GMP DNA to multi-gram scale in 2 weeks (WO 2010/086626 5 August 2010). The process uses rolling-circle amplification by phi29 polymerase to produce long concatameric repeats of the template DNA [3]. These concatamers are subsequently reduced to individual closed linear DNA molecules called Doggybone™ DNA (dbDNA™) through the action of the protelomerase enzyme TelN [3]. The process is high-fidelity (1/106–1/107), highly processive (approximately 70 kbp) and has the added benefit of being capable of amplifying complex DNA sequences, without loss of fidelity or yield [4]. The dbDNA™ is minimal, containing only the user-defined sequences of interest, with no antibiotic resistance gene or origin of replication. This has implications for the induction of immune stimulation effects [5] both in vitro [6] and in vivo [7]. Thus dbDNA™ potentially offers a safer, more clinically relevant DNA construct for use in virus manufacturing. We have previously observed that, unlike pDNA containing bacterial elements, dbDNA™ does not evoke recognition by TLR9 [7–10].
Here we investigated whether lentivirus vectors could be produced using dbDNA™. We demonstrate that lentiviral vectors can be produced using all combinations of dbDNA™ and pDNA in a three-component production system. Importantly, we show that vectors derived solely using dbDNA™ constructs produce comparable transgene expression to plasmid-derived lentivirus vectors in vitro and titre-matched vectors demonstrated similar transgene expression in vivo.
Results
Synthesis of dbDNA™ lentivirus vector payload
dbDNA™ was synthesised, as previously described [9], using a proTLx-K template plasmid derived from a lentivirus backbone containing codon-optimised firefly luciferase under transcriptional regulation of the spleen focus-forming virus promoter (SFFV), linked to green fluorescent protein (GFP) by a 2A self-cleaving peptide (pLNT-SFFV-Luc-2A-GFP, Supplementary Figure 1A) [11].
In brief, the lentivirus transgene backbone from pLNT-SFFV-Luc-2A-GFP (including the long terminal repeats) was cloned into the Touchlight proTLx-K template plasmid in between the protelomerase telRL sites. This construct was amplified by rolling circle replication using Bacillus subtilis phage phi29 DNA polymerase, generating long concatamer repeats. Escherichia coli phage N15 protelomerase TelN was used to cleave and join double-stranded sequences containing a single cassette. Initial amplification used small amounts of pDNA as a template, which, following its selective degradation with restriction enzymes and exonucleases, was fully degraded, leaving dbDNA™ only. Simple size-based separation removed dNTPs, primer and enzymes while facilitating buffer exchange. This process takes 4–5 days and is then followed by an external quality control, which is 1 working week.
In vivo transgene expression from payload dbDNATM
To determine whether the payload dbDNA™ construct was capable of mediating gene expression, we delivered it in vivo by hydrodynamic injection into the tail vein of adult mice as previously described [12].
The following constructs were administered; two plasmid constructs, pLNT-SFFV-Luc-2A-GFP and proTLx-K SFFV-Luc-2A-GFP, and the doggybone construct db-SFFV-Luc-2A-GFP (Supplementary Figure 1). The proTLx-K SFFV-Luc-2A-GFP construct is a matched control to the dbDNA™ construct, containing the telR/L sequences flanking the gene expression cassette.
Whole-body bioluminescence imaging revealed that luciferase expression from the dbDNA™ construct was significantly greater at each imaging time point than from the plasmid and proTLx-K payload constructs (Fig. 1).
Fig. 1.
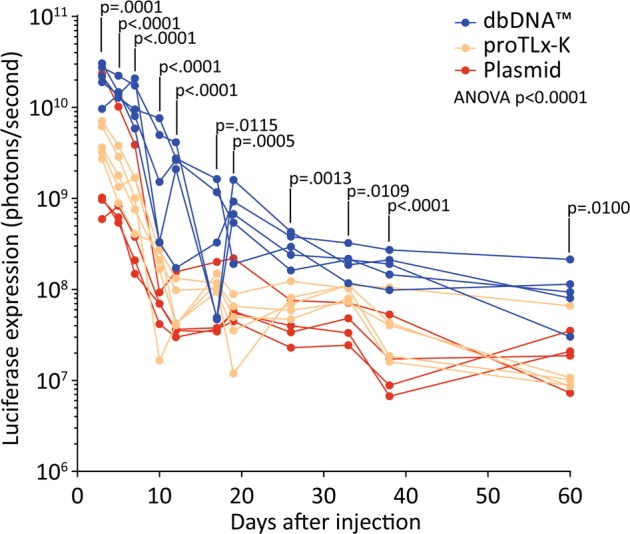
Luciferase expression quantified from Hydrodynamic injections of DNA. Plasmid, proTLx-K and dbDNA™ constructs were molarity matched and administered to 6-week-old mice by hydrodynamic injections via the tail vein (n = 5 for proTLx-K and dbDNA™ constructs and n = 4 for plasmid construct). The expression of luciferase was quantified post injection. Luciferase expression from dbDNA™ was consistently significantly higher than from plasmid DNA (repeated-measures analysis of variance with Dunnett’s post hoc test); luciferase expression from proTLx-K was similar to that from plasmid. Individual mice are plotted
Vector production using different combinations of plasmid and dbDNA™
Lentiviral vectors were produced using different combinations of dbDNA™ and pDNA encoding payload, envelope and packaging sequences to determine whether components required for vector production could be supplied by using the dbDNA™ constructs. The VSV-G envelope protein used in this vector has a broad tissue tropism [13].
Each lentiviral vector was quantified by three methods: p24 enzyme-linked immunosorbent assay (ELISA), fluorescence activated cell sorting (FACS) analysis, and quantitative polymerase chain reaction (qPCR). p24 ELISA reveals the total number of viral particles per ml, FACS analysis measures the number of transducing units per ml, and qPCR detects the reverse-transcribed viral genome copy number per ml. The plasmid templates (proTLx-K constructs) that were used to synthesise the dbDNA™ were also used to produce lentiviral vectors as a control (Supplementary Table 1B), as these constructs not only contain the same sequence as the dbDNA™ constructs but also have all the elements required for plasmid propagation in bacteria.
We demonstrate that lentiviral vectors could be produced using all combinations of dbDNA™ and comparable titres were obtained for each DNA format (six separate vector batches) (Fig. 2). Figure 3 shows the titres achieved in three separate experiments. The results revealed that both sets of constructs generated good titres of lentiviral vector. The titres of the dbDNA™ encoded vectors were significantly lower than the plasmid equivalents in the p24 assay (P < 0.001) but there was no difference by FACS or qPCR.
Fig. 2.
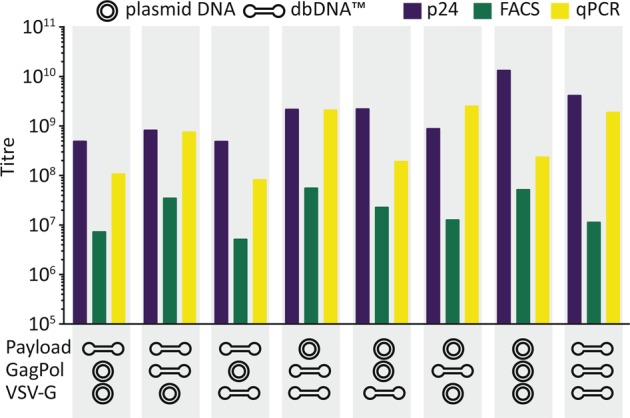
Summary of titration values for all lentiviral vectors produced. Lentiviral vectors using a combination of plasmid and dbDNA™ (payload, packaging and envelope DNA) constructs were titred by p24, fluorescence activated cell sorting and quantitative polymerase chain reaction analysis
Fig. 3.
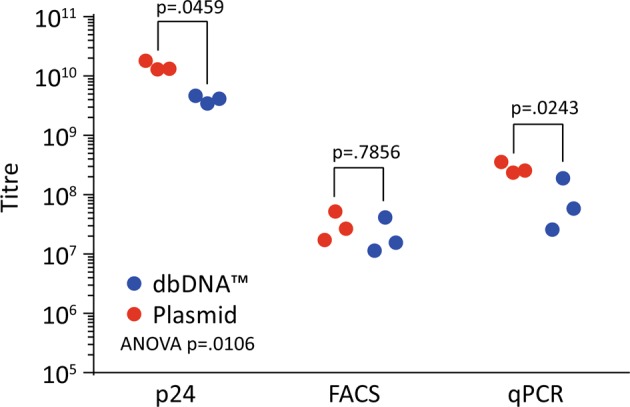
Comparison of titres between lentiviral vectors produced from plasmid and dbDNA™. Lentiviral vector produced from plasmid had a significantly higher titre than that produced from dbDNA™ when titred using p24 and quantitative polymerase chain reaction assay but not by fluorescence activated cell sorting (two-way analysis of variance with Sidak’s post hoc test)
In vitro comparison of transgene expression from pDNA or dbDNA™ vectors
To examine the transduction efficiencies of lentiviral vectors produced by either plasmid or dbDNA™, a dose escalation of each vector was applied to HEK 293T cells. Seventy-two hours after transduction, FACS analysis was conducted, revealing that plasmid and dbDNA™ lentiviral vectors produced a similar dose–response, as seen by plotting the percentage of GFP-positive cells vs the mean fluorescence intensity (MFI) (Fig. 4). However, comparing the lowest 3 doses, we observed that plasmid lentiviral vectors give rise to ~2-fold higher percentage of GFP-positive cells and MFI than dbDNA™ lentiviral vectors.
Fig. 4.
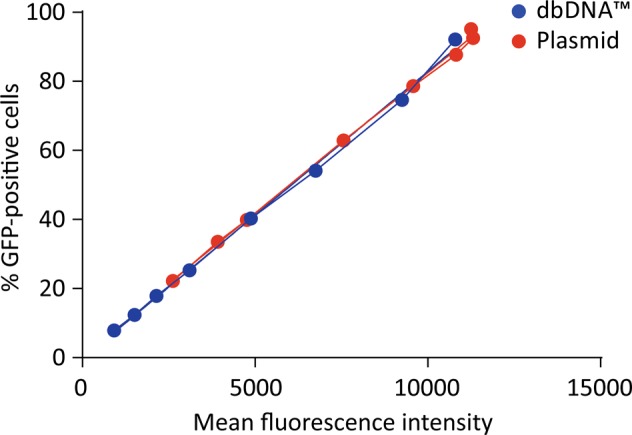
In vitro comparison of dbDNA™ and plasmid lentiviral vectors. Mean fluorescence intensity correlated equally with green fluorescent protein (GFP)-positive cells for vector generated by both dbDNA™ and plasmid vector
In vivo validation of titred matched plasmid vs dbDNA™ lentiviral vectors
Neonatal mice received a dose-matched intracerebroventricular injections of lentiviral vectors produced by either dbDNA™ or plasmid templates, with doses matched by either p24 titre (viral particles) or FACS titre (transducing units). The vectors were administered to P1 neonatal mice by intracranial injection, as described previously [14]. Luciferase expression was quantified by whole-body bioluminescence imaging over the course of development (Fig. 5a).
Fig. 5.
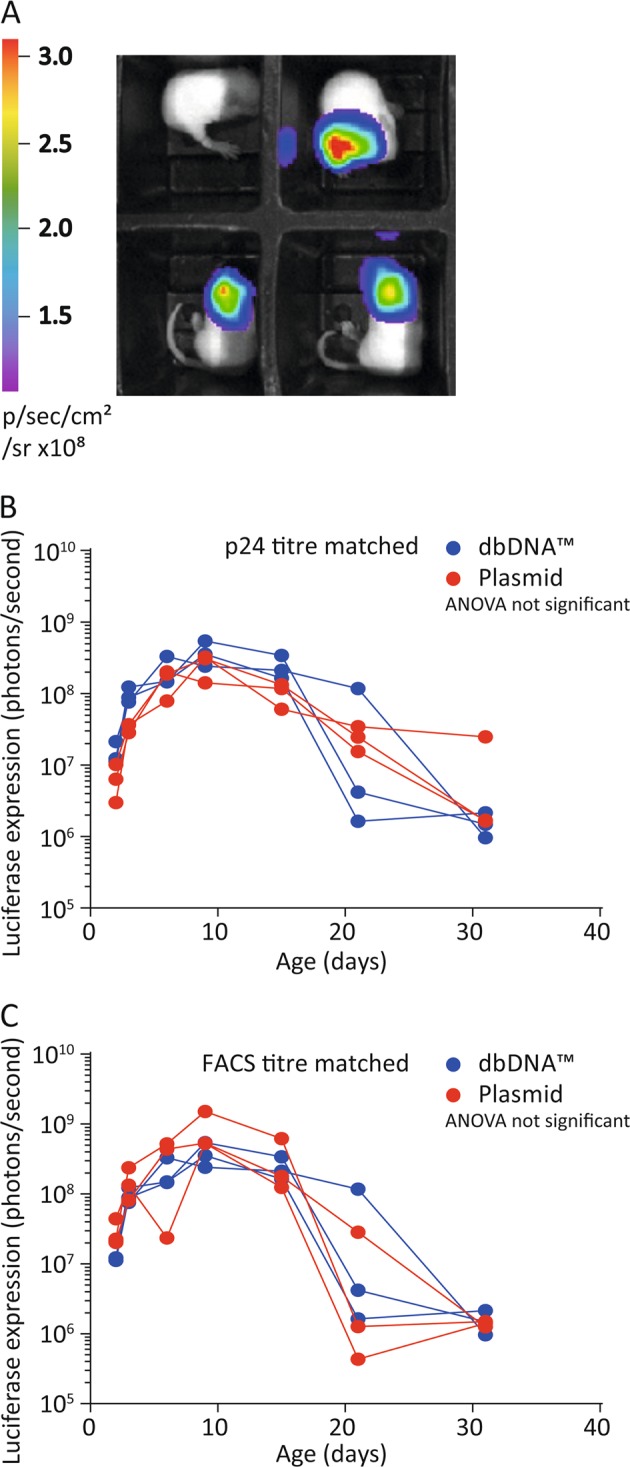
Whole-body bioluminescence imaging of titre-matched plasmid and dbDNA™ vector. a Plasmid and dbDNA™ lentiviral vector were titred matched by either p24 or fluorescence activated cell sorting (FACS) data. The vectors were administered to neonatal mice via intracranial administration (n = 3 for each vector). Whole-body bioluminescence images were taken every other day. There was no significant difference in bioluminescence following injection of either b p24 titre-matched vectors or c FACS titre-matched vectors. Bioluminescence from individual mice is plotted
There was no significant difference in luciferase expression between the two vectors in vivo with p24 matched virus, P = 0.43, 3 mice per group (Fig. 5b). We repeated this experiment with vector titres matched as per FACS titre where, again, we observed no significant difference in luciferase expression P = 0.33, 3 mice per group (Fig. 5c).
Discussion
In this report, we have described the ability of dbDNA™ constructs to generate functional lentiviral vectors with comparable in vivo transduction efficiency to those produced with plasmid constructs. From envelope, packaging and payload templates, it was possible to generate 1.5 mg/ml of payload, 2.6 mg/ml of envelope and 1.5 mg/ml of packaging DNA.
Hydrodynamic injection of pDNA was first reported by Liu et al. [15]. Here we observed significantly higher expression from dbDNA™ after a single injection, approximately 6-fold greater than pDNA 8 weeks after injection. This is consistent with the work of Lu and colleagues [16], where transcriptional silencing was observed with constructs containing >1 kb of DNA outside of the expression cassette. In our dbDNA™ constructs, 6.9 kb exists between the expression cassette and the 5’ and 3’ loops, respectively. An alternative explanation that TelN sequences confer persistence of expression can be disregarded, since proTLx-K plasmid does result in expression significantly higher than the original payload plasmid. Indeed, these data warrant further investigation into the mechanism for enhanced dbDNA™ expression in vivo, in future studies.
The primary aim of our experiment was to validate expression from the dbDNA™ payload construct. Importantly, our data demonstrate that production of functional lentiviral vectors using dbDNA™ can be achieved, with transgene expression levels comparable to those of plasmid-derived vectors.
We consequently tested these vectors in vivo by titre-matching the plasmid and dbDNA™ lentivirus based on p24 and FACS titration methods. The vectors were administered to neonatal mice via intracerebroventricular injections and luciferase expression was monitored over the course of development. These novel findings revealed a similar trend in expression from both plasmid and dbDNA™ titre-matched lentiviral vectors. Although vector generated from pDNA was significantly higher than that generated by dbDNA™, it is important to note that we did not optimise PEI:DNA ratios for dbDNA™.
The use of an enzymatic DNA amplification platform to produce constructs for lentivirus production has a number of potential key advantages, namely speed of production, cost of goods, elimination of bacterial sequences (such as origins of replication and antibiotic selection markers) and avoidance of carry-over of bacterial contaminants (such as lipopolysaccharide). However, a key advantage of dbDNA™ arises in its ability to amplify sequences that are difficult to propagate in bacteria, due to sequence toxicity or complexity. A number of therapeutically meaningful transgenes have been identified as being toxic to bacteria, including some sodium ion channels [17] and calcium channels [18]. The use of an enzymatic DNA amplification technology might therefore provide an important platform for the investigation and therapeutic development of candidates for diseases related to these transgenes. It would be useful to interrogate whether there is any difference in inflammatory and immune markers between plasmid- and dbDNA™-produced lentivirus vectors. We conducted the in vivo validation using neonatal mice, which, unlike adult mice, develop immune tolerance to vectorised transgenes [19, 20] and are immunologically ignorant to vector components [21]. It would be interesting to analyse the potential benefits for reduced inflammatory and immune markers in adult animals in future studies.
In summary, in this study we were able to show for the first time the production of lentiviral vectors using DNA constructs produced by enzymatic amplification. The in vitro and in vivo readouts were comparable with our traditional plasmid-produced lentiviral vectors, despite the utilisation of systems not optimised for the differing topology of dbDNA™ compared to plasmid. This platform could represent a significant advance for the development of gene therapies for previously intractable diseases limited by difficulty in manipulation of pDNA.
Material and methods
dbDNA™ production
Required sequences of the lentivirus payload, envelope and packaging plasmids were sub-cloned into the proTLx-K template plasmid inside the protelomerase telR/L sequences. Verification of correct clones was done by Sanger sequencing.
The template plasmids were denatured using 0.1 M NaOH for 5 min at 30 °C followed by quenching in reaction buffer (30 mM Tris–HCl pH 7.4, 5 mM (NH4)2SO4, 30 mM KCl, 7.5 mM MgCl2, 2 mM dithiothreitol) containing 50 μM custom primer (Metabion, Germany), 4 mM dNTPs (Bioline, London, UK), 200 units/ml Phi29 polymerase (Enzymatics, Manchester, UK) and 0.05 units/ml pyrophosphatase (Enzymatics). The reaction was incubated at 30 °C for 30 h and the resulting concatameric DNA processed by addition of 2 mM TelN protelomerase (Enzymatics). Additional processing was performed using 200 units/ml restriction enzyme suitable for plasmid backbone cleavage (template dependent) (Enzymatics or New England Biolabs, Hitchin, UK) and 200 units/ml exonuclease ExoIII (Enzymatics). The dbDNA™ was purified from the reaction components by addition of 500 mM NaCl/100 mM MgCl2 and precipitated using 3.9% polyethylene glycol (PEG) 8000 (Applichem, Bredbury, UK). dbDNA™ was pelleted (10 min, 4500 g) and re-suspended in 20 ml 500 mM NaCl/100 mM MgCl2. The 3.9% PEG 8000 precipitation was repeated with final resuspension in water prior to ethanol precipitation to remove residual salts.
Lentiviral vector production
HEK293T cells for viral production were seeded overnight in a T175-cm2 flask at 2 × 107 cells. Cells were transfected with 50 µg of payload plasmid or dbDNATM, 17.5 µg of pMD.G2 (VSV-G envelope plasmids, or VSV-G dbDNATM) and 32.5 µg of pCMVΔR8.74 (packaging plasmid and dbDNATM) with 1 µl of polyethylenimine (10 mM) (Sigma-Aldrich, Dorset, UK) in 12 ml of Optimem for 3 h. Media was replaced with fresh Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum. Forty-eight hours after transfection, vector supernatant was filter sterilised (0.22 µm) and concentrated by centrifugation at 5000 rpm for 20 h. The centrifuged viral vector pellet was resuspended with Optimem and stored at −80 °C.
Titration of vectors
HEK293T cells (2.5 × 105) were seeded into a 96-well plate. The detection of the enhanced GFP (eGFP) was undertaken using a BD Verse flow cytometer. eGFP fluorescence was excited using a 488 nm laser. Analysis of flow cytometry was conducted as previously described [22]. All flow cytometric data were analysed by the FlowJo software version 9.3.1 (Tree Star, Inc).
HEK293T cells (2.5 × 105) were plated into a 24-well plate and transduced with a range of volumes of concentrated lentivirus. Seventy-two hours after transduction, genomic DNA was extracted and the provirus titre calculated by qPCR, as described previously [23]. The viral capsid number was determined using a Lenti-XTM p24 Rapid Titer Kit (Takara, Saint-Germain-en-Laye, France). The capsid number was determined according to the kit manufacturer’s calculations and previously described in Vink et al. [22].
Animal procedures
Outbred CD1 mice used in this study were supplied by Charles Rivers. All animal experiments conducted within this study were in agreement with the United Kingdom Home Office guidelines, approved by the ethical review committee and following the institutional guidelines at University College London.
Hydrodynamic tail vein injections
Male CD1 mice of 6 weeks of age were used. Mice were placed in a chamber at 32 °C for 10 min. They were anaesthetised with isoflurane with 100% oxygen (Abbott Laboratories, London, UK). DNA amounts of 10 µg of plasmid and proTLx DNA and 7.8 µg of dbDNA™ (to ensure equal molarity of DNA) made up to 2 ml of phosphate-buffered saline were injected over approximately 5 s into adult mice.
Neonatal intracerebroventricular injections
For intracerebroventricular injections on the day of birth, mice were subjected to brief hypothermic anaesthesia followed by unilateral injections of concentrated lentiviral vector (5 µl in total) into the cerebral lateral ventricles using a 33-gauge Hamilton needle (Fisher Scientific, Loughborough), following co-ordinates provided by Kim et al. [24].
Whole-body bioluminescence imaging
Where appropriate, mice were anaesthetised with isoflurane with 100% oxygen and/or received an intraperitoneal injection of 15 mg/ml of D-luciferin (Gold Biotechnologies, St. Louis, USA). Mice were imaged after 5 min using a cooled charged-coupled device camera (IVIS machine, Perkin Elmer, Coventry, UK) between 1 s and 5 min. The regions of interest were measured using the Living Image Software (Perkin Elmer) and expressed as photons per second per centimetre squared per steradian (photons/second/cm2/sr).
Statistics
The exact sample sizes of each experimental group/condition are detailed in the respective legends of each figure. The number of replicates is stated in the respective section of Results. Where possible, two-way analysis of variance (ANOVA) with the appropriate post hoc test was performed, using GraphPad Prism. ANOVA has been shown to be robust to non-normal data. Individual plots are provided for each animal. Exact P values are stated except when the overall test is not significant. The number of replicates was the minimum possible to address our hypothesis of whether it was possible to produce lentivirus vectors using dbDNA™ and if this vector was equivalent, both in vitro and in vivo, to vector produced using pDNA. No power calculations were performed. All injected animals were included in the published analysis. Mice were selected randomly for injection. Imaging was not performed blind.
Supplementary information
Acknowledgements
RK and SNW received funding from MRC grants MR/P026494/1 and MR/R015325/1 and from SPARKS grant 17UCL01. JRC received funding from NIHR GOSH BRC grant 17BX23. This research was supported by the NIHR Great Ormond Street Hospital Biomedical Research Centre. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health. The authors wish to thank Touchlight employees, Anna Babula, Jon Extance, Karen Oliver, Ifeayinwa Orefo, Chris Ragbir and Shyam Thapa for the production of the dbDNA™ constructs used in this study.
Author contributions
SNW, RK, LJC and JPT developed experimental design. RK, JRC and KK contributed to data. RK, JRC, LJC, JPT and SNW contributed to manuscript.
Compliance with ethical standards
Conflict of interest
The study was funded in part by Touchlight Genetics, who were involved in all stages of the study conduct and analysis. RK was an employee at Touchlight and is now a Research Associate at UCL. KK, LJC and JPT are employees of Touchlight Genetics. SNW received funding for this research from Touchlight Genetics. JRC declares that he has no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41434-018-0056-1) contains supplementary material, which is available to authorized users.
References
- 1.Counsell JR, Asgarian Z, Meng J, Ferrer V, Vink CA, Howe SJ, et al. Lentiviral vectors can be used for full-length dystrophin gene therapy. Sci Rep. 2017;7:1–10. doi: 10.1038/s41598-016-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Naldini L, Blömer U, Gallay P, Ory D, Mulligan R, Gage FH, et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–7. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 3.Heinrich J, Schultz J, Bosse M, Ziegelin G, Lanka E, Moelling K. Linear closed mini DNA generated by the prokaryotic cleaving-joining enzyme TelN is functional in mammalian cells. J Mol Med. 2002;80:648–54. doi: 10.1007/s00109-002-0362-2. [DOI] [PubMed] [Google Scholar]
- 4.Garmendia C, Bernad A, Esteban JA, Blanco L, Salas M. The bacteriophage φ29 DNA polymerase, a proofreading enzyme. J Biol Chem. 1992;267:2594–9. [PubMed] [Google Scholar]
- 5.Krieg AM, Wagner H. Causing a commotion in the blood: immunotherapy progresses from bacteria to bacterial DNA. Immunol Today. 2000;21:521–6. doi: 10.1016/S0167-5699(00)01719-9. [DOI] [PubMed] [Google Scholar]
- 6.Messina JP, Gilkeson GS, Pisetsky DS. Stimulation of in vitro murine lymphocyte proliferation by bacterial DNA. J Immunol. 1991;147:1759–64. [PubMed]
- 7.Tokunaga T, Yamamoto H, Shimada S, Abe H, Fukuda T, Fujisawa Y, et al. Antitumor activity of deoxyribonucleic acid fraction from Mycobacterium bovis BCG. I. Isolation, physicochemical characterization, and antitumor activity. J Natl Cancer Inst. 1984;72:955–62. [PubMed] [Google Scholar]
- 8.Walters AA, Kinnear E, Shattock RJ, McDonald JU, Caproni LJ, Porter N, et al. Comparative analysis of enzymatically produced novel linear DNA constructs with plasmids for use as DNA vaccines. Gene Ther. 2014;21:645–52. doi: 10.1038/gt.2014.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scott VL, Patel A, Villarreal DO, Hensley SE, Ragwan E, Yan J, et al. Novel synthetic plasmid and DoggyboneTM DNA vaccines induce neutralizing antibodies and provide protection from lethal influenza challenge in mice. Hum Vaccin Immunother. 2015;11:1972–82. doi: 10.1080/21645515.2015.1022008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allen A, Wang C, Caproni LJ, Sugiyarto G, Harden E, Douglas LR, et al. Linear doggybone DNA vaccine induces similar immunological responses to conventional plasmid DNA independently of immune recognition by TLR9 in a pre-clinical model. Cancer Immunol Immunother. 2018;67:627–38. doi: 10.1007/s00262-017-2111-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buckley SMK, Delhove JMKM, Perocheau DP, Karda R, Rahim AA, Howe SJ, et al. In vivo bioimaging with tissue-specific transcription factor activated luciferase reporters. Sci Rep. 2015;5:11842. doi: 10.1038/srep11842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hibbitt Olivia C., Harbottle Richard P., Waddington Simon N., Bursill Christine A., Coutelle Charles, Channon Keith M., Wade-Martins Richard. Delivery and long-term expression of a 135 kbLDLR genomic DNA locusin vivo by hydrodynamic tail vein injection. The Journal of Gene Medicine. 2007;9(6):488–497. doi: 10.1002/jgm.1041. [DOI] [PubMed] [Google Scholar]
- 13.Kumar M, Bradow BP, Zimmerberg J. Large-scale production of pseudotyped lentiviral vectors using baculovirus GP64. Hum Gene Ther. 2003;14:67–77. doi: 10.1089/10430340360464723. [DOI] [PubMed] [Google Scholar]
- 14.Karda R, Perocheau DP, Suff N, Ng J, Delhove JMKM, Buckley SMK, et al. Continual conscious bioluminescent imaging in freely moving somatotransgenic mice. Sci Rep. 2017;7:6374. [DOI] [PMC free article] [PubMed]
- 15.Liu F, Song Y, Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6:1258–66. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- 16.Lu J, Zhang F, Fire AZ, Kay MA. Sequence-modified antibiotic resistance genes provide sustained plasmid-mediated transgene expression in mammals. Mol Ther. 2017;25:1187–98. doi: 10.1016/j.ymthe.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feldman DH, Lossin C. The Nav channel bench series: plasmid preparation. MethodsX. 2014;1:6–11. doi: 10.1016/j.mex.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mcmahon T, van Zijl PCM, Gilad AA. NIH Public Access. 2015;27:320–31.
- 19.Shi Y, Falahati R, Zhang J, Flebbe-Rehwaldt L, Gaensler KML. Role of antigen-specific regulatory CD4+CD25+ T cells in tolerance induction after neonatal IP administration of AAV-hF.IX. Gene Ther. 2013;20:987–96. doi: 10.1038/gt.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nivsarkar MS, Buckley SMK, Parker AL, Perocheau D, McKay TR, Rahim AA, et al. Evidence for contribution of CD4+CD25+ regulatory T cells in maintaining immune tolerance to human factor IX following perinatal adenovirus vector delivery. J Immunol Res. 2015;2015:397879. doi: 10.1155/2015/397879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waddington SN, Buckley SMK, Nivsarkar M, Jezzard S, Schneider H, Dahse T, et al. In utero gene transfer of human factor IX to fetal mice can induce postnatal tolerance of the exogenous clotting factor. Blood. 2003;101:1359–66. doi: 10.1182/blood-2002-03-0779. [DOI] [PubMed] [Google Scholar]
- 22.Vink Conrad A., Counsell John R., Perocheau Dany P., Karda Rajvinder, Buckley Suzanne M.K., Brugman Martijn H., Galla Melanie, Schambach Axel, McKay Tristan R., Waddington Simon N., Howe Steven J. Eliminating HIV-1 Packaging Sequences from Lentiviral Vector Proviruses Enhances Safety and Expedites Gene Transfer for Gene Therapy. Molecular Therapy. 2017;25(8):1790–1804. doi: 10.1016/j.ymthe.2017.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charrier S, Ferrand M, Zerbato M, Précigout G, Viornery A, Bucher-Laurent S, et al. Quantification of lentiviral vector copy numbers in individual hematopoietic colony-forming cells shows vector dose-dependent effects on the frequency and level of transduction. Gene Ther. 2011;18:479–87. doi: 10.1038/gt.2010.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim JY, Ash RT, Ceballos-Diaz C, Levites Y, Golde TE, Smirnakis SM, et al. Viral transduction of the neonatal brain delivers controllable genetic mosaicism for visualising and manipulating neuronal circuits in vivo. Eur J Neurosci. 2013;37:1203–20. doi: 10.1111/ejn.12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


