Abstract
Nitrogen (N) is an important element required for plant growth and development, which also affects plant yield and quality. Autophagy, a conserved pathway in eukaryotes, degrades and recycles cellular components, thus playing an important role in N remobilization. However, only a few autophagy genes related to N remobilization in rice (Oryza sativa) have been reported. Here, we identified a core autophagy gene in rice, OsATG8b, with increased expression levels under N starvation conditions. It was investigated the function of OsATG8b by generating three independent homozygous 35S-OsATG8b transgenic Arabidopsis thaliana lines. The overexpression of OsATG8b significantly enhanced autophagic flux in the transgenic Arabidopsis plants. It was also showed that over-expressing OsATG8b promoted growth and development of Arabidopsis, in which the rosette leaves were larger than those of the wild type (WT), and the yield increased significantly by 25.25%. In addition, the transgenic lines accumulated more N in seeds than in the rosette leaves. Further examination revealed that overexpression of OsATG8b could effectively alleviate the growth inhibition of transgenic Arabidopsis under nitrogen (N) stress. N partitioning studies revealed that nitrogen-harvest index (NHI) and nitrogen use efficiency (NUE) were significantly increased in the transgenic Arabidopsis, as well as the 15N-tracer experiments revealing that the remobilization of N to seeds in the OsATG8b-overexpressing transgenic Arabidopsis was high and more than WT. Based on our findings, we consider OsATG8b to be a great candidate gene to increase NUE and yield, especially under suboptimal field conditions.
Introduction
Nitrogen (N) is one of the most important nutrient elements for plants and is required for growth and development. N is also a vital component of amino acids, protein, nucleic acid, chlorophyll, and plant hormones [1]. In plants, especially cereal crops, the yield and quality of the grain depends not only on the absorbed N prior to flowering, but also on the mobilization of reused N from the mature leaves during senescence [2,3]. Plants are sedentary and cannot move to acquire nutrients and minerals as needed. Survival depends on their ability to consume the mineral nutrients available in the rhizosphere, metabolize, recycle, and conserve them efficiently during their lifespan [4,5]. The chloroplasts are the main source of C and N recycling in plants, which contribute to the 75–80% nitrogen of the leaf total protein in C3 plants [6]. Rubisco, a photosynthetic CO2 fixing enzyme, accounts for more than 50% of the soluble protein content in the leaves, and is the main source of nitrogen mobilization and reuse [7]. In the later stages of development, leaf protein (especially the chloroplast protein) is rapidly degraded as the output of nitrogen, as the released free amino acids and other compounds are transferred to the developing reproductive and storage organs, such as new leaves or seeds [2,8]. This mobilized nitrogen is an important source of nitrogen for seeds [9]. The circulation process for organic nitrogen transfer from aging tissues to seeds in plants is an important determinant of productivity and yield, especially under nitrogen deficiency stress [7]. Although the pathways and steps in protein turnover in aging leaves are not clear, plastid resident proteases, senescence-associated vacuoles (SAVs), and macroautophagy (hereafter referred to as autophagy) are considered to be three important routes [4,10].
Autophagy, which is a conserved vacuolar degradation pathway by which cells recycle components, including unwanted macromolecular substances or damaged organelles, plays a vital role in nutrient remobilization. Autophagy is continuously maintained at a basal level to maintain cell homeostasis under normal conditions. In response to stress or nutrient starvation, autophagy is enhanced to facilitate the degradation of increasing levels of toxic and damaged components, and nutrients mobilized from this recycled cell material is then used for the maintenance of cellular processes and adaptation to stress [11,12]. A pioneering study by Professor Yoshinori Ohsumi led to the discovery of the AuTophaGy-related genes (ATG genes) in yeast [13]. A total of 16 ATG genes were identified in the first yeast screening. Subsequently, many ATG genes have been characterized in different species, including mammals and plants [14,15]. In Arabidopsis (Arabidopsis thaliana), most ATG genes are transcriptionally up-regulated during leaf senescence and nutrient starvation [16,17]. Early senescence symptoms and hypersensitivity to C and N starvation were displayed in autophagy mutants [18], and a previous study revealed that the overaccumulation of salicylic acid (SA) is involved in the early senescence in atg mutants [19]. A study of Arabidopsis atg mutants provide the first evidence that N remobilization from the leaves to the seeds is controlled by autophagy [20]. In Arabidopsis, the wild type (WT) and atg mutants (atg18a RNAi, atg5 and atg9) were fed with 15NO3- during the vegetative growth stage, 15N remobilization sharply decreased in all the atg mutants compared to the WT under N-limited conditions, and the NUE decreased significantly to about 50% in the seeds. In the rice Osatg7-1 mutant, biomass and NUE during the vegetative growth stage were significantly reduced, and the mutant was unable to mobilize N in aging leaves [21]. In maize (Zea mays), 15N pulse-chase analysis revealed that the amount of recycled N was reduced by two-fold in the atg12 mutant compared with that of the WT [22].
Among the many plant ATG proteins, the abundance of ATG8 proteins at the cellular level can regulate the size of autophagosome. ATG8 is a ubiquitin-like protein conjugated to phosphatidylethanolamine (PE) on the autophagic membrane. ATG8 plays a central role in autophagy, which is involved in autophagic vesicle expansion [23,24] and is located on the autophagosome membrane [25]. The ATG8 is often used as a reliable marker of autophagic activity in plants and animals [12,26]. While ATG8 protein is unique in yeast, many isoforms exist in plants and are encoded by different genes. There are 7 identified ATG8 isoforms (OsATGa to OsATGg) in rice genome. OsATG8a was the first cloned rice ATG8 gene, interacted with OsATG4 [27]. OsATG8a, OsATG8b and OsATG8c share high levels of amino acid sequence similarity, while OsATG8d has high similarity to AtATG8i [28,29]. In addition, OsATG8a and OsATG8c had more closely phylogenetic relationships with ZmATG8a and ZmATG8c, respectively [30]. However, OsATG8e lacks supporting expressed sequence tag (EST) data, OsATG8f has no corresponding full-length cDNA, and OsATG8g has not been mapped to the rice genome [29]. Several studies indicate that overexpression of ATG8 promotes plant growth and increases tolerance to N starvation in plants. For example, transgenic Arabidopsis plants overexpressing AtAtg8f were slightly larger in size than the control plants, and flowered earlier than the control plants in a N-deficient medium [31]. Constitutive expression of apple MdATG8i and soybean GmATG8c in apple and soybean calli, respectively, not only led to better tolerance to N starvation, but also accelerated the growth of calli under optimal growth conditions. Overexpression of soybean and apple ATG8 genes, GmATG8c and MdATG8i, respectively, promoted growth and improved the yield in the transgenic Arabidopsis [30,32]. Similarly, Li et al observed that heterologous expression of foxtail millet SiATG8a in Arabidopsis and rice improved their performance under normal growth conditions and conferred tolerance to N starvation [33,34]. Recently, research of Masclaux-Daubresse’s group showed that overexpression of AtATG8s (AtATG8a, AtATG8e, AtATG8f, AtATG8g, respectively) in Arabidopsis increased autophagosome number and stimulated autophagic activity, increased N remobilization efficiency (NRE) only under full N conditions but did not affect yield and biomass [35].
In order to screen for potential yield and N remobilization genes in rice, the rice ATG8 gene OsATG8b (LOC_Os04g53240) was identified. OsATG8b has a 360-bp coding sequence (CDS) and encodes 120 amino acids. We then confirmed its function using 35S-OsATG8b in transgenic Arabidopsis plants. Compared to the WT, three independent transgenic lines were found to have improved vegetative growth, developed to the reproductive stage slightly earlier, were taller, and produced more siliques and seeds. Overexpression of OsATG8b therefore, not only led to better performance under normal growth conditions but also maintained growth under N starvation. The autophagic flux was increased in the transgenic lines. At the same time, the number of autophagosomes were significantly enhanced under N deficiency (N free with 0 mM nitrogen) with or without the addition of the autophagy inhibitor. Growth inhibition caused by nitrogen stress was effectively alleviated. N partitioning studies have shown that overexpression of OsATG8b increased N allocation to seeds and have enhanced NHI and NUE indicated by NHI/HI ratio [20], while the 15N allocation to seeds was significantly higher than that in the WT using 15N isotope tracer experiment. Therefore, our results indicate that OsATG8b may be an important candidate gene for rice with synergistically-enhanced NUE and better yield potential.
Results
OsATG8b displays N stress inducible expression in rice and the expression patterns of OsATG8b in transgenic Arabidopsis
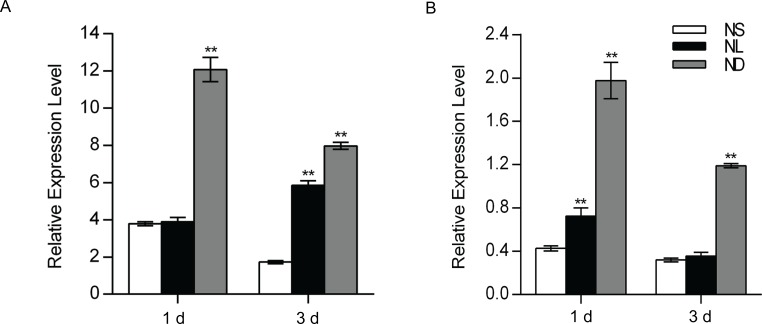
In order to analyze whether the expression of OsATG8b in rice is responsive to N limitation, we first examined the transcript levels of OsATG8b by real-time RT-PCR. When rice seedlings were placed under N stress for 1 day, the expression level of OsATG8b in leaves was not different in low N (NL) conditions when compared with that of the control, but was highly induced under N deficient (ND) conditions (Fig 1A). In addition, the levels of OsATG8b in the root increased with increasing degree of N stress (Fig 1B). After 3 days of N stress, the expression of OsATG8b in leaves increased with increasing degree of N stress but was highly induced only under ND conditions in the root (Fig 1). These results suggest that expression of OsATG8b in rice is induced by N stress.
Fig 1. Identification of OsATG8b as a N deficiency inducible gene of rice by real-time RT-PCR.
The rice seeds were sterilized and germinated at 28°C for 2 d, then seedlings cultured with half Hoagland’s N-sufficient solution for 14 days, and transferred to the same N-sufficient (NS, 3.5 mM N) solution as the control, low N (NL, 0.8 mM N) solution and the N-free (ND, 0 mM N) solution for 1 d and 3 d. (A) The expression of OsATG8b gene in leaves. (B) The expression of OsATG8b gene in roots. OsActin1 was used as an internal control. Values are means ± SD of three biological replicates, *P < 0.05, **P < 0.01 (t-test).
To examine the temporal and spatial expression patterns of OsATG8b, we performed histochemical analysis of the promoter-GUS fusion report systems. We transformed Arabidopsis with the ProOsATG8b-GUS construct, and histochemical staining indicated that GUS activity could be detected in the germinating seed, cotyledon, hypocotyl, the rosette leaf, root, as well as the mature inflorescence and silique in the transgenic seedlings (S1 Fig).
Overexpression of OsATG8b in transgenic Arabidopsis promotes growth and increases yield
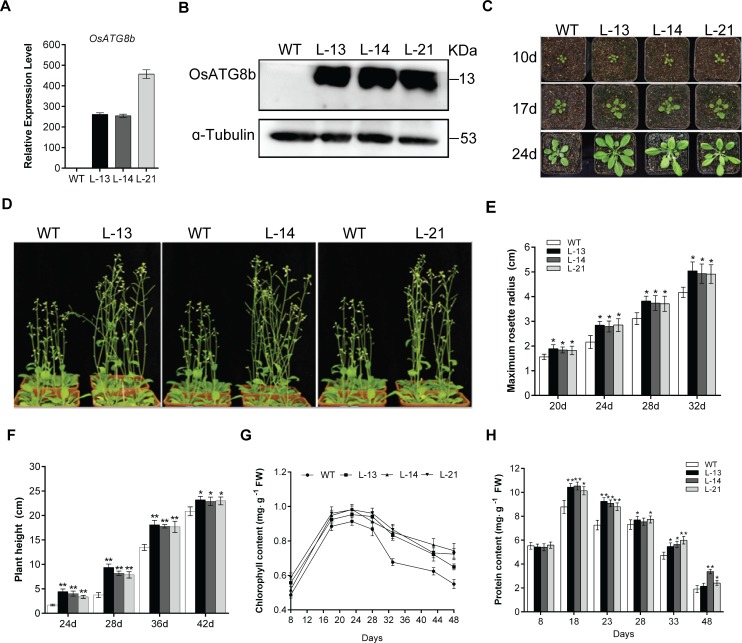
To fully understand the functional roles of OsATG8b, we transformed Arabidopsis plants with the 35S-OsATG8b binary expression vector. We obtained over 10 independent transgenic lines. In order to eliminate the influence of AtATGs, real-time RT-PCR was used to monitor the transcript levels of AtATGs. We found that the transcript levels of the endogenous AtATGs were unchanged and not affected in the transgenic lines (S2 Fig). The transcript and protein levels of OsATG8b were confirmed in three homozygous transgenic lines using PCR, real-time RT-PCR and western blotting (Figs 2A, 2B and 3). Lines 13, 14 and 21 (L-13, L-14, L-21) refer to the over-expressed lines. Phenotypic effects of OsATG8b were observed throughout the plant developmental stages. Under normal growth conditions, the rosette of the transgenic lines was significantly larger and grew rapidly than the WT (Fig 2C and 2E). Overexpression of OsATG8b accelerated plant development, bolting as flowering occurred 6 days earlier and also produced more total leaves in the transgenic lines when compared to that in the WT (Table 1), while the leaf number at bolting was not significant different with WT. On an average, plant height in the transgenic lines was significantly increased by up to 10.42% when compared to the WT plant height (Fig 2D and 2F). The chlorophyll content of the rosette leaves in the transgenic lines was consistently higher than those seen in the WT as the seedlings aged (Fig 2G). There was minimal or almost no difference in the soluble protein content in the rosettes of 8-day-old transgenic seedlings and the WT seedlings; however, from 8 days until plant maturity, the soluble protein content in the transgenic lines was consistently higher than those seen in the WT plants (Fig 2H). We further investigated the effect of OsATG8b on yield and found that the transgenic lines produced significantly higher number of siliques. As a result, the transgenic lines had a significantly higher thousand grain weight and total seed weight per plant than those in the WT (Table 2). These results show that constitutive expression of OsATG8b both promotes growth and increases yield in transgenic Arabidopsis.
Fig 2. Overexpression of OsATG8b promotes growth of transgenic Arabidopsis.
8-day-old seedlings were transferred to soil and cultivated under a long-day photoperiod. (A) Expression level of OsATG8b in 14-day-old seedlings of transgenic lines and WT. (B) Immunoblot analysis of the accumulation of OsATG8b in 14-day-old seedlings of the WT and 35S-OsATG8b transgenic lines (L-13, L-14, L-21) with a polyclonal anti-OsATG8b antibody. Equal protein loads were confirmed by immunoblot analysis with an α-tubulin antibody. (C) Panels from top to bottom show phenotypic observations of transgenic lines and WT at 10 days, 17 days and 24 days after transfer to soil, respectively. (D) Side view of transgenic lines and WT at 42 days after transfer to soil. (E)-(H) The maximum rosette radius, plant height, chlorophyll content and soluble protein content of transgenic lines and WT. Values are means ± SD of three biological replicates (n = 24), *P < 0.05, **P < 0.01 (t-test).
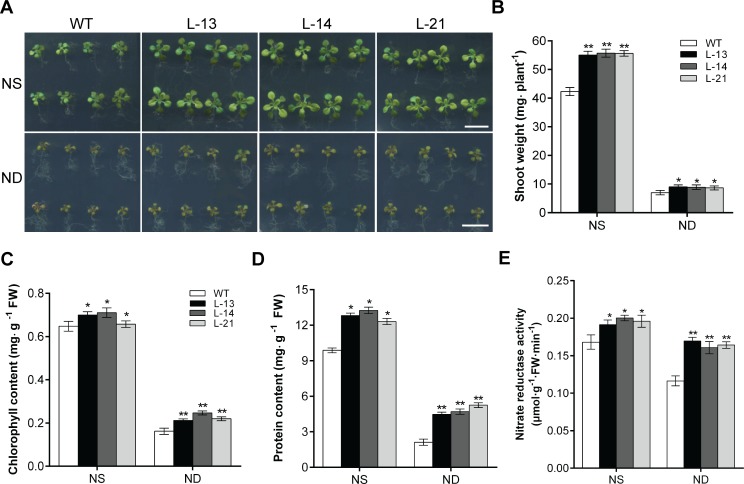
Fig 3. Overexpression of OsATG8b enhances tolerance to N deficiency in transgenic Arabidopsis.
(A) 7-day-old seedlings of transgenic lines and WT were transferred to ½ MS medium with sufficient (NS) or deficient (ND) N for 9 days. (B) The shoot weight of WT and transgenic Arabidopsis lines under NS or ND. (C) and (D) The chlorophyll content and soluble protein content in rosette leaves of transgenic lines and WT under NS or ND. (E) The nitrate reductase (NR) activity in rosette leaves of transgenic lines and WT under NS or ND. Bar = 1 cm. Values are means ± SD of three biological replicates (n = 16), *P < 0.05, **P < 0.01 (t-test).
Table 1. Total leaf number, Bolting and flowering times of WT and 35S-OsATG8b transgenic Arabidopsis.
| WT | L-13 | L-14 | L-21 | |
|---|---|---|---|---|
| Total leaf number | 14.12 ± 0.35 | 15.06 ± 0.47** | 15.19 ± 0.52** | 15.28 ± 0.58** |
| Bolting time (d) | 36.56 ± 1.58 | 30.78 ± 2.07** | 31.39 ± 1.91** | 31.06 ± 2.01** |
| Flowering time (d) | 42.67 ± 1.75 | 35.94 ± 1.98** | 36.61 ± 1.94** | 36.44 ± 2.01** |
Values are means ± SD of three biological replicates (n = 24)
**P < 0.01 (t-test), significant difference from the WT, the time to flowering and bolting were scored as days after sowing (DAS). The total leaf number was counted after the rosettes growth completed.
Table 2. Yield-related characteristics of WT and 35S-OsATG8b transgenic Arabidopsis.
| WT | L-13 | L-14 | L-21 | |
|---|---|---|---|---|
| Total number of siliques | 95.74 ± 3.86 | 139.26 ± 3.13** | 147.22 ± 3.62** | 140.74 ± 3.48** |
| Yield per plant (mg) | 132.34 ± 7.89 | 160.13 ± 6.02** | 172.77 ± 5.76** | 164.92 ± 5.13** |
| Thousand grain weight (mg) | 14.87 ± 0.23 | 16.36 ± 0.21** | 17.54 ± 0.41** | 16.31 ± 0.24** |
All seeds on a single-plant was harvested individually and the yield per plant was expressed as the weight of total seeds per plant, values are means ± SD of three biological replicates (n = 10)
**P < 0.01 (t-test).
Overexpression of OsATG8b in transgenic Arabidopsis enhances tolerance to N deficiency
To investigate the effect of OsATG8b overexpression on the plant responses to N stress, seven-day old transgenic seedlings and WT were transferred to ½ MS medium with sufficient or free N and incubated for 9 days. Changes to leaves and root were closely monitored. The transgenic lines overexpressing OsATG8b performed better than the WT, both under N-sufficient and N-free conditions. The transgenic lines had significantly larger rosettes (Fig 3A), as also the fresh weight of leaves in the transgenic lines was significantly higher both under N-sufficient and N-deficient conditions when compared to the WT (Fig 3B). The chlorophyll and soluble protein contents were also significantly higher than that of the WT (Fig 3C and 3D), and the enzymatic activity of nitrate reductase (NR) in transgenic Arabidopsis was obviously increased (Fig 3E), whether the supply of N was sufficient or deficient. These results indicate that the transgenic lines were able to tolerate N deficiency better and improve the N assimilation efficiency.
Overexpression of OsATG8b in transgenic Arabidopsis enhances the autophagic activity and flux
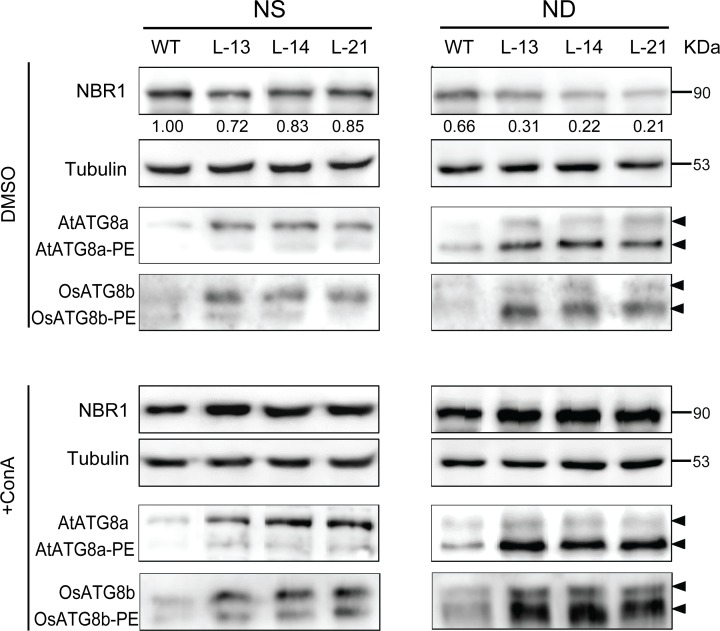
To investigate whether the autophagy activity is enhanced in the transgenic Arabidopsis overexpressing OsATG8b, seven-day-old seedlings of transgenic lines and WT were treated with ND for 12 h, and MDC staining was used to determine autophagic activity in the leaves. Concanamycin A (ConA) enables distinct observation of the autophagosome by increasing the internal pH in the vacuolar and resident hydrolase activity in an optimal environment [36,37]. The autophagy receptor NBR1 (Neighbor of BRCA1) protein of Arabidopsis is an autophagy substrate that degrades in the vacuole, and the degradation of NBR1 can be used as a measure of selective autophagic flux in plants [38,39]. As shown in Fig 4, the consumption of NBR1 protein was higher in transgenic lines, under NS condition, the expression level of NBR1 protein in transgenic lines were 15%-28% less than that of WT. While the degradation of NBR1 protein was 34% in WT under ND compared to NS condition, but it was as much as 41%-64% in transgenic lines (Fig 4, DMSO-ND). However, when applied with ConA to block the degradation of autophagic bodies and NBR1 protein, the accumulation of NBR1 protein was greatly increased (Fig 4, +ConA), especially under the ND treatment. The previous reports showed that the lipidation of ATG8 can be used to indicate the autophagic activity [40,41]. As the autophagy core protein ATG8a usually acts as the autophagic flux marker protein, and previous studies have used Arabidopsis anti-ATG8a antibody to exam the autophagic flux in various species (such as Arabidopsis, maize and tomato) [22,40–42]. The autophagy flux in our transgenic plants was monitored with both anti-AtATG8a and anti-OsATG8b antibodies. These results showed that overexpression of OsATG8b significantly increased the accumulation of both AtATG8a-PE and OsATG8b-PE in transgenic lines, thus the lipidation of OsATG8b was enhanced, especially under N deficiency condition (Fig 4, DMSO), and increased to a greater extent after ConA treatment (Fig 4, +ConA). The detection signal of lipidation used by anti-OsATG8b antibody was better than that of anti-AtATG8a antibody.
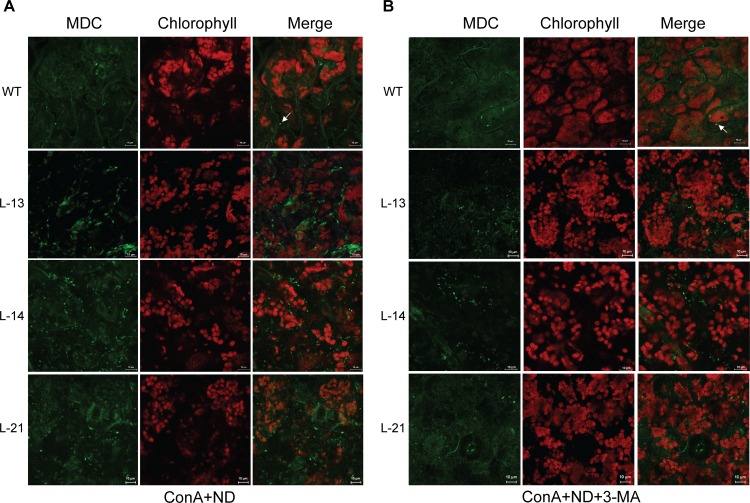
Fig 4. Overexpression of OsATG8b enhanced the autophagic flux in transgenic Arabidopsis.
9-day-old seedlings of transgenic lines and WT were transferred to NS or ND medium with 0.5 μM ConA or solvent control DMSO for 24h. Immunoblot analysis the accumulation of NBR1 with anti-Arabidopsis NBR1, near equal protein loads were confirmed by immunoblot analysis with an α-tubulin antibody. The membrane fraction was used to detect the level of lipidated (ATG8-PE) and free ATG8 with Arabidopsis anti-AtATG8a or an anti-OsATG8b antibody. The number under the blot indicated the intensity ratios of the NBR1 protein in each line after analyzed by Image J and normalized to the WT under NS condition (value set to 1).
Monodansylcadaverine (MDC), an acidophilic dye widely used in mammals and plants can be used as a probe to detect autophagosome [42]. To facilitate the observation of autophagic bodies, the H+-ATPase inhibitor ConA was added. The leaves of transgenic Arabidopsis and WT were excised and immediately observed by confocal microscopy. We observed significantly more MDC-positive autophagic structures in the mesophyll cells of the transgenic lines, and the fluorescence signal intensity was higher than that seen in the WT (Fig 5A). The autophagy inhibitor 3-methyladenine (3-MA), specifically inhibits PI3K activity [43,44]. The addition of 5 mM of 3-MA to the N-deficient treatment clearly suppressed the fluorescence signal of MDC-stained autophagosomes, but some fluorescent signal could still be observed in the transgenic lines (Fig 5B). The fluorescence ratio (positive signal/aera) in the WT was 30.87 under ND condition, and decreased to 13.07 after 3-MA treated, while the ratios in transgenic lines were 121.85 and 62.53. It was showed that the fluorescence ratios in the overexpressing lines were up to 3 or 4 times more than that in WT with or without 3-MA. All these results reveal that overexpression of OsATG8b could significantly increase the autophagosome number and autophagic flux in transgenic Arabidopsis. This can effectively improve the tolerance of transgenic Arabidopsis to N deficiency stress.
Fig 5. Overexpression of OsATG8b in Arabidopsis enhances the autophagic activity under N deficient condition.
(A) 7-day-old seedlings of transgenic lines and WT were transferred to in ND liquid medium with 1 μmol·L-1 ConA for 12h, MDC-stained autophagosomes in leaves were observed by confocal microscopy. (B) 7-day-old seedlings of transgenic lines and WT were transferred to ND liquid medium containing 1 μmol·L-1 ConA with 5 mM 3-MA for 12h, MDC-stained autophagosomes in leaves were observed by confocal microscopy.
Overexpression of OsATG8b in transgenic Arabidopsis modifies N use efficiency and N remobilization efficiency
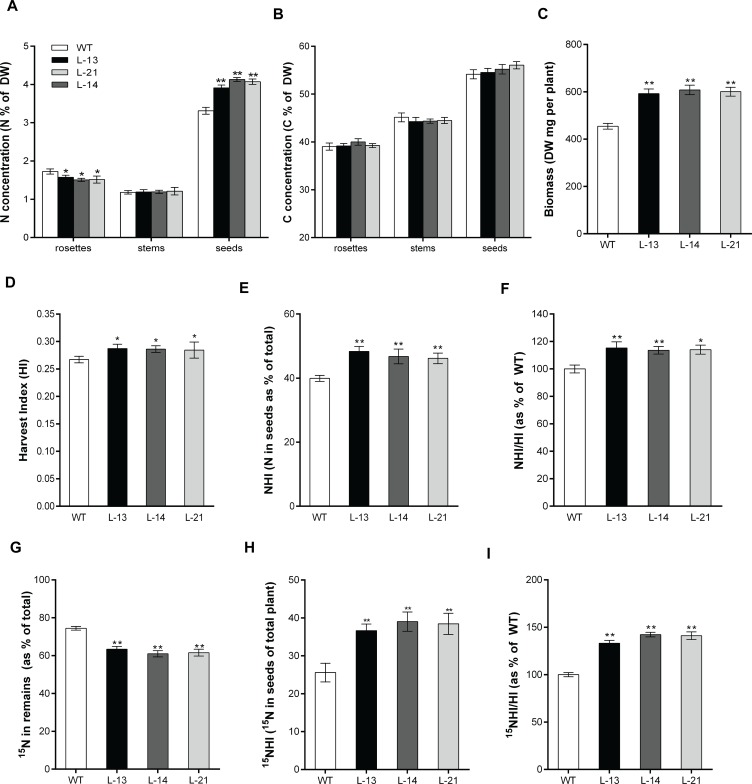
To determine whether the overexpression of OsATG8b modifies the N status at the whole plant level, the N concentration in rosettes, stem, and seeds was measured after harvesting. The N concentration (N%) in rosettes of the transgenic lines was much lower than that of the WT, but there was no difference in the stem samples. In contrast, the N% in seeds was significantly higher in the transgenic lines (Fig 6A). However, there was no significant difference in the C concentration (C%) between the transgenic lines and the WT (Fig 6B). Given the increased yield in transgenic lines, we further analyzed the biomass of plants. Relative to WT, the biomass of the transgenic lines increased by 25.95% to 26.89% (Fig 6C).
Fig 6. Evaluation of NUE and NRE in transgenic Arabidopsis.
(A) N concentration (N%, presented as mg per 100 mg DW) and (B) C concentration (C%, presented as mg per 100 mg DW) of the transgenic lines and WT were measured in rosettes, stems and seeds after harvesting. (C) Biomass accumulation as measured by dry weight of the whole plant. (D) HI was measured as the ratio of the dry weight of total seeds to the dry weight of remains. (E) NHI was measured as the partitioning total N in seeds. (F) NHI/HI ration was calculated as a percentage of WT and used to estimate NUE between the transgenic lines and WT. (G) Partitioning of 15N in remains of the transgenic lines and WT after harvesting. (H) 15NHI was measured as the partitioning total 15N in seeds. (I) 15NHI/HI ration was calculated as a percentage of WT and used to estimate NRE between the transgenic lines and WT. Values are means ± SD of three biological replicates (n = 12), *P < 0.05, **P < 0.01 (t-test).
We studied the effect of OsATG8b on plant yield, seed N filling, harvest index (HI) and N harvest index (NHI). While NHI is an important indicator of production, it also highlights the N allocation efficiency to seeds. As shown in Fig 6D and 6E, the HI and NHI were significantly higher in the transgenic lines compared to that in the WT plants. In order to determine the NUE, we used the NHI/HI ratio as an indicator of NUE variation [20]. The NHI/HI ratio in the transgenic lines was significantly higher than that of the WT (Fig 6F), suggesting that OsATG8b might be essential for both seed yield and NUE. These results clarify that overexpression of OsATG8b effectively redeploys N allocation from the leaves to seeds and increases the NUE of Arabidopsis plants.
To monitor N movements accurately, 15N-pulse chase experiments were performed at the bolting stage similarly as described in previous studies [20,22]. Samples of OsATG8b-overexpressing lines and WT were dissected into seed and remains (rosettes and stems), then traits for N utilization and remobilization were calculated. To determine the N remobilization efficiency (NRE), the partitioning of 15N in WT and transgenic lines were tracked. It was showed that overexpression of OsATG8b obviously reduced the partitioning of 15N into remains (Fig 6G). In contrast, the OsATG8b-overexpressing lines accumulated significantly more 15N in seeds relative to WT, as indicated by the higher 15NHI in transgenic lines (Fig 6H). The ratio of 15NHI to HI was next used to compare the NRE variation as described previously [20,22], and the 15NHI/HI were substantially increased in OsATG8b-overexpressing lines than in WT (Fig 6I), suggesting that through overexpression of OsATG8b could significantly improve the N remobilization into seeds, which resulted in favorably increased yield and NUE.
Discussion
Autophagy is involved in removal of damaged proteins and organelles, and response to biological and abiotic stresses [11]. Previous studies have shown that nutrition-limitation induces autophagy in plants [45,46]. Under C and N stress conditions, transcription of many plant ATG genes was significantly up-regulated similar to the high autophagy activity [28,29,33,47,48]. The transcriptional level of OsATG8b increased with N deficiency in rice seedlings (Fig 1), which is evidence that OsATG8b response to N stress. On the other hand, it would provide evidence that overexpression of OsATG8b could effectively increase tolerance to N stress (Fig 3A). The GUS activity of OsATG8b promoter was detected in the inflorescence, seedpod, and in particular was higher in the aging leaves, which indicated that OsATG8b gene may participate in seed formation in plants and also be involved in autophagic degradation during leaf senescence (S1A and S1E Fig).
With defective autophagy, the redundant N in the cells cannot be effectively recovered or utilized, leading to nutrient accumulation, wastage, cell death, and lower amino acid content, the atg deletion mutants showed accelerated senescence, decreased survival rate and other unfavorable phenotypes under nutrient stress conditions [11,20–22,49,50]. Consistent with this view, the transgenic Arabidopsis plants containing 35S-OsATG8b not only performed stronger and better than the WT under normal growth conditions, but also effectively overcome growth inhibition under N deficiency conditions (Fig 3A). In addition, we also found that the transgenic lines had increased chlorophyll content and soluble protein content in the leaves (Fig 2), so did the NR activity (Fig 3D). These results indicate that overexpression of OsATG8b leads to increased accumulation of available N in the ‘source’ of transgenic Arabidopsis. The overexpression of OsATG8b accelerated the switch from vegetative to reproductive growth in the transgenic Arabidopsis (Table 1), which was probably because of a healthy vegetative phase to have an extended reproductive phase resulting in increased number of siliques, and consequently, higher thousand grain weight and better yield (Table 2).
In the OsATG8b-overexpressing lines, the enhanced degradation of NBR1 and lipidation of ATG8, as well as the ConA-mediated increased accumulation of NBR1 and AtATG8a-PE/OsATG8b-PE, all these results revealing that the autophagic flux and activity significantly increased under ND condition (Fig 4-ND). Additionally, the MDC staining also supported this conclusion (Fig 5). Even under N sufficient condition, the nutrients still could be mobilized and reused from leaves to reproductive organs (seeds) to the maximum extent, as a result of enhanced autophagy activity (Fig 4-NS). The growth, development, and yield of transgenic Arabidopsis could be enhanced.
Autophagy is also as an important role in the remobilization and transfer of N to the grain [11,24]. Both NHI and seeds N concentration (N%Seeds) are major indicators of NUE and seed nutritional quality [7]. Overexpression of OsATG8b in transgenic Arabidopsis resulted in higher NHI, HI, NHI/HI, N%Seeds, and at the same time reduced N waste in their dry remains (Fig 6), indicating that OsATG8b plays a vital role in NUE. This could explain OsATG8b increases both grain production and the allocation of biomass to grain. On the other hand, 15NHI/HI ratio is used to measure the variations as Nitrogen Remobilization Efficiency (NRE). A significantly higher 15NHI/HI ratio observed in the transgenic lines compared to the WT (Fig 6I) implied that OsATG8b could also increase the allocation of N to seeds. The increased seed yield was at least partially caused by effective remobilization of assimilated N from the vegetative tissue to the developing seeds. Corresponding to the increased autophagy activity described above, since autophagy likely contributes to the nitrogen pool for seed production, we speculated that the transgenic Arabidopsis could increase N transported and accumulated during the vegetative growth stage, which in-turn could provide abundant nutrients for reproductive growth and higher yield even under N full condition.
A recent paper of Masclaux-Daubresse’s group has just reported that overexpression of four AtATG8 genes (AtATG8a, AtATG8e, AtATG8f, AtATG8g), respectively, in Arabidopsis could significantly increase the number of autophagosomes and the transcripts of other AtATG genes [35]. Under full N conditions, although N remobilization efficiency significantly increased relative to the control line by 15N isotope tracer test, the ATG8 overexpressors did not affect yield and biomass [35]. We two did similar but different studies during the same period. Although these similar conclusions were got in our research, there were also clear differences between us. It seemed that our OsATG8b gene was powerful in the development of plant. The overexpressing OsATG8b transgenic Arabidopsis not only unaffected the expression of other AtATGs (S2 Fig), but also significantly increased seed yield and thousand grains weight under normal growth conditions. Another point worth mentioned was the nitrogen absent (ND) condition of in our experiment was 0 mM N, it was a very very harsh stress condition. Our transgenic plants were still doing well and made a better state of growth, even restored the growth inhibition caused by N stress (Figs 2 and 3). Although rice and Arabidopsis are two type plants, there may be differences in gene function. In addition, the conclusions of we two groups could support or complement each other.
Taken together, our results suggest that the rice autophagy gene OsATG8b that can enhance NUE and improve crop yield, which has a positive role in the maintenance of plant fitness and adaptation to nutrient limitations. These results have also linked autophagy to plant nitrogen distribution/homeostasis, as well as increased yield.
Materials and methods
Plant materials and growth conditions
Rice (Japonica cv Shennong9816) seeds were sterilized and germinated at 28°C for 2 d, then grown hydroponically in a growth chamber (28°C/25°C and 10 h light/14 h dark). Arabidopsis seeds (Columbia-0) were surface-sterilized and stratified at 4°C for 2 days in the dark, then grow on ½ Murashige and Skoog (½ MS) medium (with 0.75% agar and 1% sucrose (w:v), pH = 5.7) in a growth chamber (22 ± 1°C and 16 h light/8 h dark). 8-day-old Arabidopsis seedlings were transferred to soil with sufficient nutrient and cultivated under the same conditions.
Total RNA extraction and gene expression analysis
To analysis OsATG8b expression responsive to N limitation in rice, rice seedlings cultured with half Hoagland’s solution (NS) as previous described [30] for 14 days, then transferred to the same solution as control, low N (NL) solution (with 0.6 mM KNO3 and 0.1 mM (NH4)2SO4, pH = 5.7) and the N-free (ND) solution (pH = 5.7), the lack of K+ was replaced with KCl. After 1 day and 3 days N limitation treatments, the leaves and roots were harvested separately for OsATG8b gene expression analysis. 10-day-old seedlings of transgenic lines and WT grown on NS were sampled for expression analysis of the endogenous AtATGs by real-time RT-PCR. Total RNA extraction was conducted using the Eastep Super Total RNA Extraction Kit (Promega) and first strand cDNA was synthesized with the PrimeScript RT Master Mix (TaKaRa). Real-time RT-PCR was performed as described previously [51] by using SYBR Premix Ex TaqII (TaKaRa) on an Applied Biosystems 7500 Real Time PCR System. The following standard thermal profile was used for all PCRs: 95°C for 30 s, 40 cycles of 95°C for 5 s and 60°C for 34 s. All reactions were done at least in triplicates. TIP41 was used as an internal control. The primers used for RT-PCR analysis were listed in S1 Table.
Binary vector construction and Arabidopsis genetic transformation
The complete coding region of OsATG8b was amplified by PCR using a pair of primers with full-length cDNA of rice as the template. The PCR product was then ligated into binary vector pCAMBIA1301, resulting in the 35S-OsATG8b fusion gene. For the ProOsATG8b-GUS construct, a 1988-bp promoter fragment of the OsATG8b was amplified from rice genomic DNA by PCR, and then ligated into binary vector pCAMBIA1301. All the construction was confirmed by DNA sequencing. The plant expression vector was transformed into Arabidopsis by Agrobacterium-mediated floral dipping method [52]. All the primers used were listed in S1 Table. T0 transgenic Arabidopsis seeds were screened on ½ MS medium containing 30 mg·L-1 hygromycin and seedlings with green true leaves were identified as transformants, 3 independents homozygous T3 lines were analyzed in this study.
Protein isolation and immunoblot analysis
Total proteins extraction was conducted using the Minute™ Total Protein Extraction Kit (Invent Biotechnologies, Inc). Total membrane protein extraction was conducted using the Minute™ Membrane Protein Isolation Kit for plants (Invent Biotechnologies, Inc). 14-day-old seedlings of transgenic lines and WT grown on NS were sampled to exam the protein level of OsATG8b, the full length OsATG8b was used to raise a polyclonal antibody (GenScript, Nanjing China). Protein concentration was determined with the BCA Protein Assay Kit (TaKaRa). For autophagic flux analysis, 9-day-old seedlings of transgenic lines and WT were transferred to NS or ND medium with 0.5 μM ConA or solvent control DMSO for 24h. All SDS-PAGE gels were prepared with 6 M urea, antibodies against Arabidopsis anti-ATG8a and anti-NBR1 (1:1000), anti-α-tubulin (1:1000) (Agirsera, Sweden) and anti-OsATG8b (1:1000) were used as primary antibodies; goat anti-rabbit (IgG) (Sigma-Aldrich) was used as the secondary antibody. Immunoblotting were done as described [28,30]. The protein band intensities of immunoblots was analyzed by Image J software. Each experiment was repeated for three times, and one representative result was shown.
Phenotypic analysis of OsATG8b transgenic Arabidopsis under optimum condition and N starvation
For the basal phenotypic analysis of transgenic lines and the WT under sufficient nutrient condition, 8-day-old (days after germination) seedlings were transferred to soil. The maximum rosette radius and the plant height were recorded every 3 days, the bolting and flowering times were recorded every day from Stage5.1 and Stage6.0 [53], respectively. The leaf number at bolting was counted when the inflorescence emergence at the Stage5.1, and the total leaf number was counted after the rosettes growth completed at about Stage6.0. Total number of siliques was counted when the plants growth to Stage6.9, the total seeds produced by a single-plant were harvested at the growth Stage9.7, and the yield per plant was expressed as the weight of total seeds per plant, and the thousand-grain weight were weighed. For N starvation treatments, 7-day-old seedlings of transgenic lines and WT were transferred to NS or ND for 9 days. The phenotypic changes were recorded by scanner (EPSON Perfection v33), the fresh weight of leaves was measured.
GUS histochemical staining
Seeds of ProOsATG8b-GUS transgenic Arabidopsis were screened on ½ MS medium containing 30 mg·L-1 hygromycin for temporal and spatial expression patterns analysis, then GUS histochemical staining was conducted as described [54].
MDC staining for detecting autophagosome activity
7-day-old seedlings of transgenic lines and WT were transferred to ND liquid medium containing 1 μmol·L-1 ConA with or without 5 mM 3-MA for 12h, then the leaves were excised and immediately vacuum infiltrated with 50 mM MDC (Sigma-Aldrich) for 10 min, MDC staining was conducted as described [48], and confocal images were acquired using an inverted Zeiss LSM 780 laser scanning microscope. Quantification of the MDC-stained autophagosomes in leaves was presented as the fluorescence ratio = positive signal/area. The number of positive signals was calculated from areas of 400 μm2, and more than 10 independent areas were used for the quantification.
Measurement of chlorophyll, soluble proteins, and nitrate reductase (NR) enzyme activity
Chlorophyll content was measured with Arnon method as describe [55]. Soluble protein content was determined with the Bradford method [56]. The NR enzyme activity was assayed as described previously [57].
Biomass, total N, C concentrations and indicator used to monitor NUE calculations
Plants were sampled by separation into rosettes (including cauline leaves), stems, and seeds at harvest, then all samples were kept in a dry oven at 80°C for 7 days, and dry mass were weighted as DWDr (dry weight of rosettes and stems) and DWSeeds (dry weight of total seeds). Then the dry mass of all these tissue organs were ground to powder, total N and C concentrations (N% and C%) were measured using an Elemental Analyzer (Elementa, Vario MAX C, Germany), which presented as N% (C%) = mg N (C)/100 mg DW. Biomass was calculated as DWDr+DWSeeds. Indicator used to monitor NUE were calculated as described [20]. The harvest index (HI) was calculated as DWSeeds/(DWSeeds+DWDr) ratio and was used as an important indicator for yield. The N harvest index (NHI) was used to evaluate grain N filling and was calculated as (N%Seeds×DWSeeds)/(N%Seeds×DWSeeds+N%Dr×DWDr). The ratio of NHI/HI was estimated as the percentage of WT to compare the NUE performance among genotypes.
15N-Labeling and NRE calculation
The 15N-Labeling experiment was conducted as Guiboileau et al (2012) [20] described on WT and 35S-OsATG8b transgenic lines at Stage5.1 (long time before flowering), the unlabeled watering solutions was replaced with one containing the same nutrient composition except that plants were supplied with 15NH4 and 15NO3 (10.6 atom% excess) for 3 d. Afterwards, the pots were rinsed thoroughly with distilled water to remove the remaining 15N, and unlabeled watering solutions were then applied until seed harvest. When the plants were grown to maturity, the plants were separated into remains [rosettes (including cauline leaves) and stems] and seeds. All samples were kept in a dry oven at 80°C for 7 days, then dry mass was weighted and ground to fine powder to assess N remobilization using an elemental analyzer/isotope ratio mass spectrometry system (Thermo Scientific, Flash2000/Delta V Advantage), and unlabeled plants were also collected to determine the natural 15N abundance. The 15N abundance of each sample was expressed as a percent of total N and calculated as atom% sample (A%sample), A%sample = 100×(15N)/(15N+14N). The natural abundance of 15N (Acontrol%) obtained from unlabeled samples were considered using the average value 0.365% to calculate 15N enrichment of labeled samples (E%), E% = A%sample-A%control. NRE in this experiment were estimated as described by Guiboileau et al (2012) [20], using the ratio of 15NHI/HI, and they were estimated as the percentage of WT to compare the NRE performance among genotypes. 15NHI was calculated as (E%Seeds×N%Seeds×DW Seeds)/[(E%Seeds×N%Seeds×DWSeeds)+(E%Dr×N%Dr×DWDr)].
Statistical analysis
The experiments were repeated three times, and data from one representative experiment were shown here. The statistically significant differences of all data were calculated based on Student's t-test at significance levels of *P < 0.05 and **P < 0.01.
Supporting information
(A) Seeds in germination; (B) 1-week-old seedlings; (C) 2-week-old seedlings; (D) 4-week-old of mature seedlings; (E) Early inflorescence; (F) Mature inflorescences and siliques. The left is transgenic Arabidopsis before staining, the right is that after staining. (A) and (B) Bars = 1 mm; (C) and (F) Bars = 5 mm.
(TIF)
(TIF)
PCR identification of genomic. DNA of transgenic lines. M, Molecular marker DL2000; 1–9, Independent transgenic lines; +, Positive vector containing 35S-OsATG8b plasmid; -, Negative control.
(TIF)
(DOCX)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This research was funded by the National Natural Science Foundation of China (grant number 31401298) and the National Key Research and Development Program of China (grant number 2018YFD0200200). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kraiser T, Gras DE, Gutiérrez AG, González B, Gutiérrez RA. A holistic view of nitrogen acquisition in plants. Journal of Experimental Botany. 2011; 62: 1455–1466. 10.1093/jxb/erq425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Havé M, Balliau T, Cottyn-Boitte B, Dérond E, Cueff G, Soulay F, et al. Increase of proteasome and papain-like cysteine protease activities in autophagy mutants: backup compensatory effect or pro cell-death effect? Journal of Experimental Botany. 2017; 69: 1369–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chardon F, Noël V, Masclaux-Daubresse C. Exploring NUE in crops and in Arabidopsis ideotypes to improve yield and seed quality. Journal of Experimental Botany. 2012; 63: 3401–3412. 10.1093/jxb/err353 [DOI] [PubMed] [Google Scholar]
- 4.Avila-Ospina L, Masclaux-Daubresse C. Autophagy, plant senescence, and nutrient recycling. Journal of Experimental Botany. 2014; 65: 3799–3811. 10.1093/jxb/eru039 [DOI] [PubMed] [Google Scholar]
- 5.Krapp A. Plant nitrogen assimilation and its regulation: a complex puzzle with missing pieces. Current Opinion in Plant Biology. 2015; 25: 115–122. 10.1016/j.pbi.2015.05.010 [DOI] [PubMed] [Google Scholar]
- 6.Makino A, Osmond B. Effects of Nitrogen Nutrition on Nitrogen Partitioning between Chloroplasts and Mitochondria in Pea and Wheat. Plant Physiology. 1991; 96: 355–362. 10.1104/pp.96.2.355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masclaux-Daubresse C, Daniel-Vedele F, Dechorgnat J, Chardon F, Gaufichon L, Suzuki A, et al. Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Annals of Botany. 2010; 105: 1141–1157. 10.1093/aob/mcq028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Masclaux-Daubresse C, Chardon F. Exploring nitrogen remobilization for seed filling using natural variation in Arabidopsis thaliana. Journal of Experimental Botany. 2011; 62: 2131–2142. 10.1093/jxb/erq405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patrick JW, Offler CE. Compartmentation of transport and transfer events in developing seeds. Journal of Experimental Botany. 2001; 52: 551–564. [PubMed] [Google Scholar]
- 10.Roberts IN, Caputo C, Criado MV, Funk C. Senescence-associated proteases in plants. Physiologia Plantarum. 2012; 145: 130–139. 10.1111/j.1399-3054.2012.01574.x [DOI] [PubMed] [Google Scholar]
- 11.Marshall RS, Vierstra RD. Autophagy: The Master of Bulk and Selective Recycling. Annual Review of Plant Biology. 2018; 69: 173–208. 10.1146/annurev-arplant-042817-040606 [DOI] [PubMed] [Google Scholar]
- 12.Yang X, Bassham DC. New Insight into the Mechanism and Function of Autophagy in Plant Cells. International Review of Cell & Molecular Biology. 2015; 320: 1–40. [DOI] [PubMed] [Google Scholar]
- 13.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. Febs Letters. 1993; 333: 169–174. 10.1016/0014-5793(93)80398-e [DOI] [PubMed] [Google Scholar]
- 14.Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Research. 2014; 24: 24–41. 10.1038/cr.2013.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yao Z, Delorme-Axford E, Backues SK, Klionsky DJ. Atg41/Icy2 regulates autophagosome formation. Autophagy. 2015; 11: 2288–2299. 10.1080/15548627.2015.1107692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rose TL, Bonneau L, Der C, Marty-Mazars D, Marty F. Starvation-induced expression of autophagy-related genes in Arabidopsis. Biology of the Cell. 2006; 98: 53–67. 10.1042/BC20040516 [DOI] [PubMed] [Google Scholar]
- 17.Thomas H. Senescence, ageing and death of the whole plant. New Phytologist. 2012; 197: 696–711. 10.1111/nph.12047 [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, Bassham DC. Autophagy: pathways for self-eating in plant cells. Annual Review of Plant Biology. 2012; 63: 215–237. 10.1146/annurev-arplant-042811-105441 [DOI] [PubMed] [Google Scholar]
- 19.Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, et al. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell. 2009; 21: 2914–2927. 10.1105/tpc.109.068635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guiboileau A, Yoshimoto K, Soulay F, Bataillé MP, Avice JC, Masclaux-Daubresse C, et al. Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytologist. 2012; 194: 732–740. 10.1111/j.1469-8137.2012.04084.x [DOI] [PubMed] [Google Scholar]
- 21.Wada S, Hayashida Y, Izumi M, Kurusu T, Hanamata S, Kanno K. Autophagy supports biomass production and nitrogen use efficiency at the vegetative stage in rice. Plant Physiology. 2015; 168: 60–73. 10.1104/pp.15.00242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li F, Chung T, Pennington JG, Federico ML, Kaeppler HF, Kaeppler SM, et al. Autophagic recycling plays a central role in maize nitrogen remobilization. Plant Cell. 2015; 27: 1389–1408. 10.1105/tpc.15.00158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie Z, Nair U, Klionsky DJ. An InCytes from MBC Selection: Atg8 Controls Phagophore Expansion during Autophagosome Formation. Molecular Biology of the Cell. 2008; 19: 3290 10.1091/mbc.E07-12-1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masclaux-Daubresse C, Chen Q, Havé M. Regulation of nutrient recycling via autophagy. Current Opinion in Plant Biology. 2017; 39: 8–17. 10.1016/j.pbi.2017.05.001 [DOI] [PubMed] [Google Scholar]
- 25.Kellner R, De la Concepcion JC, Maqbool A, Kamoun S, Dagdas YF. ATG8 Expansion: A Driver of Selective Autophagy Diversification? Trends in Plant Science. 2016; 22: 204–214. 10.1016/j.tplants.2016.11.015 [DOI] [PubMed] [Google Scholar]
- 26.Avin-Wittenberg T, Honig A, Galili G. Variations on a theme: plant autophagy in comparison to yeast and mammals. Protoplasma. 2012; 249: 285–299. 10.1007/s00709-011-0296-z [DOI] [PubMed] [Google Scholar]
- 27.Su W, Ma H, Liu C, Wu J, Yang J. Identification and characterization of two rice autophagy associated genes, OsAtg8 and OsAtg4. Molecular biology reports. 2006; 33: 273–278. 10.1007/s11033-006-9011-0 [DOI] [PubMed] [Google Scholar]
- 28.Chung T, Suttangkakul A, Vierstra RD. The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant Physiology. 2009; 149: 220–234. 10.1104/pp.108.126714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia K, Liu T, Ouyang J, Wang R, Fan T, Zhang M. Genome-Wide Identification, Classification, and Expression Analysis of Autophagy-Associated Gene Homologues in Rice (Oryza sativa L.). DNA Research. 2011; 18: 363–377. 10.1093/dnares/dsr024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xia T, Xiao D, Liu D, Chai W, Gong Q, Wang N. Heterologous expression of ATG8c from soybean confers tolerance to nitrogen deficiency and increases yield in Arabidopsis. PLoS ONE. 2012; 7: e37217 10.1371/journal.pone.0037217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slavikova S, Ufaz S, Avin-Wittenberg T, Levanony H, Galili G. An autophagy-associated Atg8 protein is involved in the responses of Arabidopsis seedlings to hormonal controls and abiotic stresses. Journal of Experimental Botany. 2008; 59: 4029–4043. 10.1093/jxb/ern244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang P, Sun X, Jia X, Wang N, Gong X, Ma F. Characterization of an autophagy-related gene MdATG8i from apple. Frontiers in plant science. 2016; 7: 720 10.3389/fpls.2016.00720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li W, Chen M, Zhong L, Liu JM, Xu ZS, Li L, et al. Overexpression of the autophagy-related gene SiATG8a from foxtail millet (Setaria italica L.) confers tolerance to both nitrogen starvation and drought stress in Arabidopsis. Biochemical & Biophysical Research Communications. 2015; 468: 800–806. [DOI] [PubMed] [Google Scholar]
- 34.Li W, Chen M, Wang E, Hu L, Hawkesford MJ, Zhong L, et al. Genome-wide analysis of autophagy-associated genes in foxtail millet (Setaria italica L.) and characterization of the function of SiATG8a in conferring tolerance to nitrogen starvation in rice. BMC Genomics. 2016; 17: 797 10.1186/s12864-016-3113-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Q, Soulay F, Saudemont B, Elmayan T, Marmagne A, Masclaux-Daubresse C. Overexpression of ATG8 in Arabidopsis stimulates autophagic activity and increases nitrogen remobilization efficiency and grain filling. Plant and Cell Physiology. 2019; 60: 343–352. 10.1093/pcp/pcy214 [DOI] [PubMed] [Google Scholar]
- 36.Ishida H, Yoshimoto K, Izumi M, Reisen D, Yano Y, Noda T, et al. Mobilization of rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiology. 2008; 148: 142–155. 10.1104/pp.108.122770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, et al. Processing of ATG8s, Ubiquitin-Like Proteins, and Their Deconjugation by ATG4s Are Essential for Plant Autophagy. Plant Cell. 2004; 16: 2967–2983. 10.1105/tpc.104.025395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hafrén A, Macia JL, Love AJ, Milner JJ, Drucker M, Hofius D. Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proceedings of the National Academy of Sciences. 2017; 114: E2026–E2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Svenning S, Lamark T, Krause K, Johansen T. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy. 2011; 7: 993–1010. 10.4161/auto.7.9.16389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klionsky DJ, Schulman BA. Dynamic regulation of macroautophagy by distinctive ubiquitin-like proteins. Nature structural & molecular biology. 2014; 21: 336–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu T, Zou L, Li Y, Yao X, Xu F, Deng X, et al. Mitochondrial alternative oxidase-dependent autophagy involved in ethylene-mediated drought tolerance in Solanum lycopersicum. Plant Biotechnology Journal. 2018; 16: 2063–2076. 10.1111/pbi.12939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Contento AL, Xiong Y, Bassham DC. Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP‐AtATG8e fusion protein. The Plant Journal. 2005; 42: 598–608. 10.1111/j.1365-313X.2005.02396.x [DOI] [PubMed] [Google Scholar]
- 43.Seglen PO, Gordon PB. 3-Methyladenine: Specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proceedings of the National Academy of Sciences of the United States of America. 1982; 79: 1889–1892. 10.1073/pnas.79.6.1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Inoue Y, Suzuki T, Hattori M, Yoshimoto K, Ohsumi Y, Moriyasu Y. AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells. Plant & Cell Physiology. 2006; 47: 1641–1652. [DOI] [PubMed] [Google Scholar]
- 45.Moriyasu Y, Ohsumi Y. Autophagy in tobacco suspension-cultured cells in response to sucrose starvation. Plant Physiology. 1996; 111: 1233–1241. 10.1104/pp.111.4.1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiology. 2005; 138: 2097–2110. 10.1104/pp.105.060673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun X, Jia X, Huo L, Che R, Gong X, Wang P, et al. MdATG18a overexpression improves tolerance to nitrogen deficiency and regulates anthocyanin accumulation through increased autophagy in transgenic apple. Plant Cell & Environment. 2018; 41: 469–480. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y, Yu B, Zhao J, Guo J, Li Y, Han S, et al. Autophagy contributes to leaf starch degradation. Plant Cell. 2013; 25: 1383–1399. 10.1105/tpc.112.108993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barros JA, Cavalcanti JHF, Medeiros DB, Nunes-Nesi A, Avin-Wittenberg T, Fernie AR, et al. Autophagy deficiency compromises alternative pathways of respiration following energy deprivation. Plant Physiology. 2017; 175: 62–76. 10.1104/pp.16.01576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ren C, Liu J, Gong Q. Functions of autophagy in plant carbon and nitrogen metabolism. Frontiers in plant science. 2014; 5: 301 10.3389/fpls.2014.00301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu D, Gong QQ, Ma YY, Li PL, Li JP, Yang SH, et al. cpSecA, a thylakoid protein translocase subunit, is essential for photosynthetic development in Arabidopsis. Journal of Experimental Botany. 2010; 61: 1655–1669. 10.1093/jxb/erq033 [DOI] [PubMed] [Google Scholar]
- 52.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998; 16: 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- 53.Boyes DC, Zayed AM, Ascenzi R, Mccaskill AJ, Hoffman NE, Davis KR, et al. Growth Stage-Based Phenotypic Analysis of Arabidopsis: A Model for High Throughput Functional Genomics in Plants. Plant Cell. 2001; 13: 1499–1510. 10.1105/TPC.010011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu F, Meng T, Li P, Yu Y, Cui Y, Wang Y, et al. A Soybean Dual-Specificity Kinase, GmSARK, and Its Arabidopsis Homolog, AtSARK, Regulate Leaf Senescence through Synergistic Actions of Auxin and Ethylene. Plant Physiology. 2011; 157: 2131–2153. 10.1104/pp.111.182899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arnon DI. Copper Enzymes in Isolated Chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiology. 1949; 24: 1–15. 10.1104/pp.24.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry. 1976; 72: 248–254. 10.1006/abio.1976.9999 [DOI] [PubMed] [Google Scholar]
- 57.Ferrario-Méry S, Valadier MH, Foyer CH. Overexpression of nitrate reductase in tobacco delays drought-induced decreases in nitrate reductase activity and mRNA. Plant Physiology. 1998; 117: 293–302. 10.1104/pp.117.1.293 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Seeds in germination; (B) 1-week-old seedlings; (C) 2-week-old seedlings; (D) 4-week-old of mature seedlings; (E) Early inflorescence; (F) Mature inflorescences and siliques. The left is transgenic Arabidopsis before staining, the right is that after staining. (A) and (B) Bars = 1 mm; (C) and (F) Bars = 5 mm.
(TIF)
(TIF)
PCR identification of genomic. DNA of transgenic lines. M, Molecular marker DL2000; 1–9, Independent transgenic lines; +, Positive vector containing 35S-OsATG8b plasmid; -, Negative control.
(TIF)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.