Abstract
A bacteriophage T4 DNA “synapsis model” proposes that the bacteriophage T4 terminase small subunit (TerS) apposes two pac site containing dsDNA homologs to gauge concatemer maturation adequate for packaging initiation. N-terminus, C-terminus, or both ends modified fusion Ter S proteins retain function. Replacements of the TerS gene in the T4 genome with fusion genes encoding larger (18 to 45 kDa) TerS-eGFP and TerS-mCherry fluorescent fusion proteins function without significant change in phenotype. Co-infection and co-expression by T4 phages encoding TerS-eGFP and TerS-mCherry shows in vivo FRET in infected bacteria comparable to that of the purified, denatured and then renatured, mixed fusion proteins in vitro. FRET of purified, denatured-renatured, mixed temperature sensitive and native TerS fusion proteins at low and high temperature in vitro shows that TerS ring-like oligomer formation is essential for function in vivo. Super-resolution STORM and PALM microscopy of intercalating dye YOYO-1 DNA and photoactivatable TerS-PAmCherry-C1 fusions support accumulation of TerS dimeric or multiple ring-like oligomer structures containing DNA and gp16-mCherry in vivo as well as in vitro to regulate pac site cutting.
Introduction:
Viral nucleic acid translocation by ATP driven motor proteins into preformed icosahedral proheads or procapsids is observed among numerous double stranded RNA and DNA bacteriophages as well as herpesviruses (Kainov et al., 2006). In large tailed dsDNA bacteriophages terminases drive DNA packaging to high final density (~500 mg/ml) through a channel located in a dodecameric portal structure situated at a unique vertex of the procapsid. Terminases also cut the replicated concatemeric head-to-tail joined genomic DNA to form the mature DNA ends within the mature virion, thus the name terminase (Black, 1989; Roy et al., 2012). Generally the ATP-driven packaging and cutting of the concatemer is carried out by a large terminase subunit (TerL) motor and nuclease component, whereas a small terminase (TerS) subunit is required for recognition and/or initiation cutting of the concatemeric DNA by TerL (Rao and Feiss, 2008). Following initiation cutting, the TerL nuclease carries out the second end, headful maturation cutting of the DNA genome through communication with the portal protein rather than TerS, to leave a single mature genome in the full head (Casjens et al., 1992; Tavares et al., 1992).
In phage T4 TerL (gp17, ~70 kDa) is the motor ATPase and nuclease protein whereas TerS (gp16, ~18 kDa) is required for initiating packaging of circular or concatemeric DNA (Fig. 1) (Black and Peng, 2006) (Rao and Black, 2010). T4 TerS is not required rather is inhibitory for packaging linear DNA in vitro despite TerS activation of TerL ATPase (Baumann and Black, 2003; Kondabagil et al., 2006; Leffers and Rao, 2000). This is consistent with the observation that TerS is not required for high velocity and high efficiency packaging of linear DNA in vitro by TerL that requires the high activity ATPase that results from multimerisation of TerL (Black and Peng, 2006; Fuller et al., 2007; Ray et al., 2010).
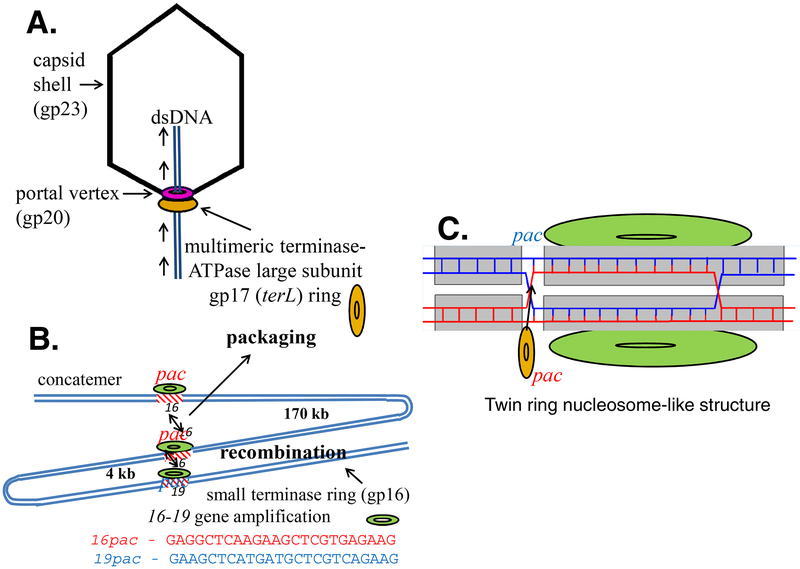
Fig. 1:
T4 packaging TerS synapsis model. A: The phage T4 TerS small subunit (gp16) hands off apposed pac site DNAs in the replicated concatemer (or circular DNA in vitro) to the large subunit TerL for initiation cutting and packaging. B: Synapsis of apposed homologous pac sequences in genes 16 and 19 can lead to gene amplification of genes 16 to 19 following recombination under selection for increased gene 17 expression. C:A twin ring nucleosome-like structure that pairs chromosome homologs is proposed to measure DNA replication adequate for packaging the genome by a strand swapped Holliday junction signal.
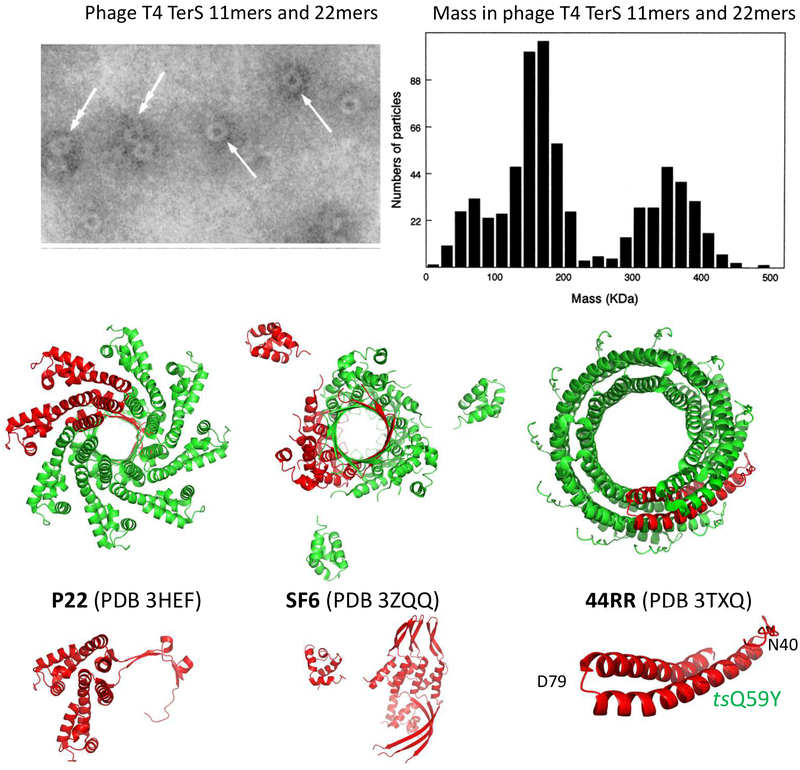
It has long been known that the terminase TerL is strikingly homologous among phages (Black, 1989), accordingly TerL genes act as superior informatics identifiers of tailed dsDNA phages (Serwer and Jiang, 2012). Crystallography shows the three dimensional structures of TerL proteins are highly conserved (Roy and Cingolani, 2012; Smits et al., 2009; Sun et al., 2008; Zhao et al., 2013). On the other hand, although crystallography studies of TerS from diverse pac site bacteriophages reveal a conserved subunit fold, TerS ring oligomers range from 8 to 12 subunits (Roy et al., 2012), (Zhao et al., 2010), (Maluf et al., 2006; Sun et al., 2012), (Buttner et al., 2012). For a view of some of the divergent size TerS ring structures see Fig. 2 (Lin et al., 1997; Roy et al., 2012).
Fig. 2:
Diverse pac site phage TerS structures from electron micrographs and crystallography. Pac site phage identities P22, SF6, and 44RR are given in the figure and the crystal structures are referenced in the text. The location of the phage T4 TerS gp16 ts mutation Q59Y used for temperature regulated renaturation determined by FRET (shown in Fig. 6) is displayed in the 44RR crystal structure (Sun et al., 2012). The top half electron micrograph figure and mass determination of T4 TerS protein only single and dimeric rings is from reference (Lin et al., 1997), as confirmed and refined by (van Duijn, 2010), the bottom half figure is from the protein data bank (PDB).
Importantly, enhanced transduction of phage T4 TerS pac site containing plasmids and prophages by a generalized T4 transducing phage derivative showed that T4 TerS recognizes and preferentially packages DNA containing a pac site DNA sequence located in its structural gene 16. Moreover, Southern blotting showed a pac site packaging fragment in mature T4 phage DNA derived from gene 16 pac to a unique BamH1 site located ~10 kb away from gene 16 in gene 10 (Lin and Black, 1998). In both these respects phage T4 is comparable to generalized transducing phages P1 and P22 that are known to contain pac sites. However, an additional feature of phage T4 is that recombination between the TerS gene 16 pac site and a homologous gene 19 pac-like site (see Fig. 1C) located ~ 4 kb from gene 16 can yield multiple intragenomic copies of TerL gene 17 (from 2 to more than 6 copies of TerL) to compensate for weak TerL expression. The gene 17 ~4kb gene amplifications that can be selected in phage T4 and in phage T4 gene derived plasmids require expression of a functional TerS protein. This TerL gene amplification provides genetic evidence for synapsis of the two homologous pac site containing duplex DNA sequences (16 and 19) by TerS to initiate recombination. Genetic analysis of this phenomenon led to a 16 to 16 pac site synapsis model for TerS function in DNA packaging as well as in initiating a specific 16 to 19 gene amplification recombination event (Black, 1995).
The purified untagged bacteriophage T4 TerS protein that lacks DNA forms both single and dimeric 11-mer side-by-side double rings that are shown in Fig. 2. Renaturation of the protein together with dsDNA showed enhanced DNA binding to the TerS coding sequence, supporting a pac site within the TerS structural gene. It was proposed that the side-by-side twin rings were nonplanar single and double lock washer-like helical structures reflecting a single type of subunit to subunit bonding interaction (Lin et al., 1997). More recent and precise values for the T4 TerS ring molecular weights prepared by the same purification procedure showed that the single and double rings are composed of 11-mers and dimers of 11-mers, respectively; and, importantly, are stable protein-only structures that are not held together by DNA judging by their masses (van Duijn, 2010). Thus, in addition to lacking the 12-mers that are found in addition to 11-mers by crystallography, these structures differ from the recently proposed TerS double ring structures suggested to form because they are held together by DNA (Sun et al., 2012).
Taken together the purified phage T4 TerS protein twin rings and sequence specific terminase gene amplifications fit a proposed “synapsis model” for TerS function (Fig. 1). This model proposed a mechanism allowing determination by TerS of sufficient DNA replication and concatemer maturation for TerL concatemer packaging initiation by hand off and cutting by the TerL nuclease (Black, 1995). Pairing by TerS of two identical pac sites in TerS or, less frequently, of the pac site in TerS with a homologous gene 19 pac-like site sequence ~4 kb downstream from TerS by T4 TerS, was proposed to explain amplifications of a gene 16 to 19 segment within the T4 chromosome. Under genetic selection two to more than six copies of head to tail joined gene 16 to 19 segment sequences were found within the genome (Wu and Black, 1987). Numerous gene 16 to 19 amplification mutations were readily isolated with the same end sequences under the same genetic selection scheme requiring enhanced TerL synthesis. The selection required a mutation that eliminated the Alt internal protein from within the capsid to allow packaging of significantly more DNA within the enlarged T4 capsid. This increased DNA capacity allowed viable addition of intragenomic gene amplifications to a headful of DNA. Other T4 recombination genes including UvsX are also required to yield the full intragenomic gene amplification following the TerS dependent initiation event. (Wu et al., 1991). The direct role of the TerS protein in this gene amplification was demonstrated in vivo using amber mutations in gene TerS expressed from plasmids. These plasmids showed a requirement for synthesis of the TerS protein to generate the recombinant sequence from the plasmid sequences (Wu et al., 1995) (Lin and Black, 1998). Recombination between the two apposed pac site containing duplex DNAs in vivo could increase the number of copies of gene TerL in the genome under strong genetic selection (poor suppression of gene TerL amber mutations) thereby allowing increased TerL synthesis (Fig. 1 B & C). It was proposed that this synapsed protein-DNA structure mimicked the twin ring protein structure characterized in vitro. Its standard packaging function in vivo would be to determine maturation of multiple copies of head to tail joined fully mature ~170 kb DNAs in the T4 DNA concatemer. By this mechanism pac to pac synapsis can control handing off of the DNA from TerS to TerL to initiate cutting and packaging initiation by the TerL nuclease-ATPase-translocase; thus this mechanism proposed a prokaryotic meiotic-like synapsis of chromosomes upon packaging maturation homologous to the eukaryotic synapsis in meiosis (Black, 1995).
Despite recent success in determining TerS crystal structures some of which are shown in Fig. 2, the structure most relevant for any phage TerS function in vivo, that of the phage TerS-DNA complex, remains to be established. In fact, very different TerS-DNA structures have been proposed, nucleosome-like wrapping of DNA around the TerS ring (Buttner et al., 2012) (Zhao et al., 2012) and through TerS ring channel DNA structures (Nemecek et al., 2008; Roy and Cingolani, 2012; Zhao et al., 2010). In addition TerS protein recognition of pac containing DNA sequences has been proposed to result from DNA binding motifs in the N-terminal, the C-terminal, or both regions of the TerS proteins (Gao and Rao, 2011; Nemecek et al., 2008; Roy et al., 2012; Zhao et al., 2010; Zhao et al., 2012), thus there is considerable controversy about the structure-function properties of TerS proteins. Although it has been shown experimentally that shuffling of TerS peptide components is possible without loss of function, possibly allowing the evolution of highly divergent TerS structures, it is not established that all of the ring crystal structures reported are biologically relevant since most have been purified as tag bearing polypeptides at very high expression levels (Leavitt et al., 2013) (Teschke, 2012). Evidence of a conserved ring structure is however found among all the pac site TerS phages despite differences in numbers of ring protomers; other properties such as ATP binding and TerL ATPase stimulation may also be conserved. The TerS rings are clearly not highly fixed structurally with respect to numbers of monomers in the ring, nor in the case of phage T4, numbers of protomers in a ring or of numbers of rings forming a stable oligomer.
In this work we show that both ends of the phage T4 TerS can be modified without loss of function, indeed both ends of TerS within the same molecule can be fused to peptides without loss of function. Remarkably, gene TerS can be replaced in the phage genome with fusion genes coding for much larger polypeptides (~18 kDa increased to ~45 kDa) coding for the fluorescent fusion proteins TerS-eGFP and TerS-mCherry. Low level T4 terminase late gene transcription-expression of the fusion protein genes by the weak single late gene TerS and TerL terminase gene promoter is sufficient for normal function. Also, the C-terminal fusion proteins fold to form active fluorophores while retaining full terminase function. Co-infection by two phages producing TerS fluorescent fusion proteins allows FRET analysis in vivo. This analysis shows that structures with monomer to monomer spacings characterized in the purified ring proteins are formed even at low native gene expression levels and in the presence of T4 concatemeric DNA. The TerS fluorescent gene fusion proteins do not produce detectable alteration of its essential DNA concatemer packaging function. Moreover, super-resolution localization fluorescence microscopy (Sauer and Heilemann, 2017), specifically STORM (Rust et al., 2006) was used for detection of organic fluorophores and PALM (Betzig et al., 2006) was realized with photoactivatable fluorescent proteins with localization precision in the range of 20 to 50nm. Accordingly, STORM and PALM were employed to examine the accumulation of TerS and spatially resolve the structures containing YOYO-1 DNA and photoactivatable TerS-PAmCherry-C1 fusions, respectively.
Results:
TerS-eGFP and TerS-mCherry provide full TerS function in vivo -
The phage T4 small terminase subunit TerS is strictly required in vivo for packaging. Amber mutants of the TerS structural gene 16 that are not suppressed fail to initiate DNA packaging and accumulate empty proheads in vivo (Rao and Black, 2010). Expression vectors for T4 TerS fluorescent fusion proteins TerS-eGFP and TerS-mCherry were constructed that synthesize an N-terminal 6X-His tag form of each fusion protein. A double amber mutant form of the TerS structural gene was used to show that when expressed in vivo both fusion proteins could complement the TerS gene 16 defect as efficiently as could expression of unmodified TerS (Table 1). Nearly wild type levels of mutant phages, as well as of intracellular fusion proteins upon purification, were produced upon IPTG induction of fusion protein synthesis.
Table 1:
Complementation of phage T4 TerS gene 16 double amN66-amN87 mutant by plasmid induced N-terminal 6X-His tag fusion proteins.
| Wild type/ Recombinant | Pfu/ml | |
|---|---|---|
| BL21-DE3 | CR63 | |
| 16-16-(amN66-amN87) phage | - | 3 × 1011 |
| T4 phage | 4 × 1012 | 5 × 1012 |
| Pl16 plasmid-UI | 3 × 108 | 7 × 1012 |
| Pl16 plasmid-I | 2 × 1010 | 2 × 1012 |
| gp16-GFP-UI | 8 × 108 | 6 × 1012 |
| gp16-GFP-I | 8 × 1010 | 8 × 1012 |
| gp16-mCherry-UI | 8 × 108 | 2 × 1012 |
| gp16-mCherry-I | 3 × 1010 | 8 × 1012 |
| pET28a plasmid | - | 2 × 1011 |
E. coli BL21DE3/ BL21DE3-gp16 plasmid were grown to OD-0.5 at 37°C, induction+/− with 0.5mM IPTG at 30°C, infection at OD 0.6; first infection 5 MOI, after 5 min second infection 5MOI, after 40 min phage-infected bacteria purified by pelleting at 6,000 rpm followed by CHCl3/DNase treatment and tittering of synthesized phages. E. coli CR63 amber supD was used as control for full T4 TerS expression. UI and I are uninduced and induced synthesis respectively of T4 TerS and TerS fusion proteins using IPTG
TerS-eGFP and TerS-mCherry form high molecular weight multimers in vivo -
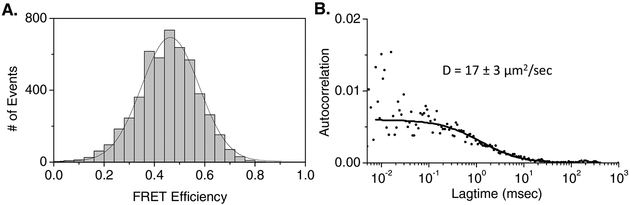
The TerS fusion proteins were found to be strongly fluorescent upon IPTG induction at low temperature. The concentrated fusion proteins, identified with a T4 TerS polyclonal antiserum (Lin et al., 1997), appeared fully soluble and highly pure by SDS-PAGE and Western blotting (Fig. 3A). The high level of purification of each fusion protein could be achieved by repeated and optimized His tag Ni2+-NTA column elution, followed by ammonium sulfate precipitation, and Superose 6 size exclusion chromatography (See Methods). Both fluorescent fusion proteins displayed highly similar properties during purification and following concentration by ammonium sulfate precipitation. Virtually all of each highly concentrated fluorescent fusion protein behaved as a mixture of high molecular weight multimers following the final column purification step, moving more slowly than, and together with, a 670 kDa thyroglobulin protein standard (Fig. 3B). When the ammonium sulfate precipitated and concentrated purified TerS-eGFP and TerS-mCherry proteins were mixed together and stored for several days there was no FRET transfer observed between the donor and acceptor fluorophores. However, when the purified proteins were denatured together in 6M urea, and then renatured by dialysis to eliminate the urea, ~50% FRET transfer was observed (Fig. 4A). The diffusion coefficient of the renatured protein complex was estimated to be 17 ± 3μm2/sec (Fig. 4B), consistent with the sizes determined by size sieving Superose 6 column determination. The diffusion coefficients determined by FCS for rhodamine, native GFP, and T4 proheads are ~280, ~50, and 4.4 μm2/sec respectively (Sabanayagam et al., 2007; Schwille et al., 1999) consistent with the determined values for the renatured complexes. Properties of the TerS fusion proteins appeared comparable to the unmodified native TerS, judging by low dissociation-reassociation of the multimers to monomers under physiological conditions; and capacity to reform ring-like structure following denaturation-renaturation from 6 M urea (Lin et al., 1997). Electron microscopic observations of the purified fluorescent fusion proteins showed large ring-like structures generally with a central hole.
Fig. 3.
A: SDS-PAGE of purified, concentrated TerS-eGFP and TerS-mCherry with Western blotting using gp16 antiserum for TerS identification and showing fluorescence of the purified concentrated fusion proteins. The sizes and purities of the boiled SDS buffer loaded protein are compared to rainbow marker size protein standards. B: Superose 6 size exclusion column chromatography profile of 50 μl of purified concentrated T4 TerS-eGFP (see Fig. 3A) plus 50 μl Biorad Gel filtration protein size standards with Typhoon assessed fluorescence (upper left inset). The identified four protein chromatography traces show 260nm, purple, 280nm green, 405nm orange, and 214nm gray wavelength absorptions of the size standards labeled in the figure. Size standards alone do not show the identified TerS-eGFP peak fluorescence or imidazole peak; fractions 3–31 are equal 0.2 ml volume column fractions; TerS-eGFP is found in fractions 3–13, ≥ 670 kDa. Left Y-axis provides the optical densities at the four wavelengths listed, top X-axis the fraction collector tube number.
Fig. 4:
Ring-like FRET in vitro of fluorescent TerS fusion proteins. A: 6X-His Tag Ni2+ and Superose 6B SEC purified TerS-eGFP and TerS-mCherry mixtures renatured together from 6 M urea show FRET of ~50%; B: diffusion (17 ± 3 μm2/sec) characteristic of >670 kDa multimeric ring complexes.
Single, double, triple, and even higher number rings lying side by side as originally observed for native TerS purified by the original procedure (See Fig. 2 from (Lin et al., 1997)) were revealed. Thus, the numbers of monomers in the fusion protein multimers and numbers of ring-like structures appeared to be variable and a general multimer structure of the fusion proteins could not be determined by cryo-EM or heavy metal staining.
TerS-eGFP and TerS-mCherry expression from fusion gene recombination into the phage genome -
The TerS gene was replaced in the phage genome with fusion genes coding for TerS-eGFP and TerS-mCherry. This required a partial synthesis of the overlap region between genes TerS and TerL (gene 17) to allow the fusions to be synthesized while the ribosome binding site of gene TerL was retained. These linked genes were recombined into the phage T4 genome in a single step using a double gene TerS-TerL amber to recombine in the mutant fusion gene segments; an rII deletion mutation also in the recombinant phage genome allowed the additional TerS fusion DNA to be added to the genome without functional deficit. PCR sequencing of the desired recombinant that lacked amber mutations showed that the gene fusion segment was correctly inserted. Moreover, Illumina sequencing showed that the eGFP and mCherry genes were present in the recombinant “pseudo-wild type” phages that lacked a detectable phenotype because of the apparently normal functioning of the TerS gene fusions. Evidently low-level expression of the fusion proteins by the weak single late T4 terminase gene promoter is sufficient for normal function.
When the genomic TerS-eGFP and TerS-mCherry phages were used to infect E. coli, fluorescence measurements readily detected both eGFP and mCherry fluorescence with the expected excitation and emission properties in infected bacteria. When the TerS-eGFP and TerS-mCherry phages were used to co-infect the same host at high multiplicity of infection (~10 mixed phage/bacterium), both fluorescent proteins were synthesized. FRET measurements showed that the fluorophores were in close proximity within the infected bacterium (Fig. 5 A & B). The FRET results are comparable when carried out on single infected bacteria and in the whole bacterial population (data not shown). The measurements of about 50% FRET transfer show the same average value as in the purified and renatured protein only ring-like structures, although significantly tighter FRET Gaussian distribution is seen in the presence of DNA in vivo, suggesting that the DNA-bound TerS is a more uniform and possibly more native structure (compare Figs. 4A and 5B). As a control, eGFP was strongly expressed from a plasmid in E. coli containing the eGFP gene that is under rhamnose induction control. Infection of this host bacterium with the TerS-mCherry phage did not display FRET transfer between the donor and acceptor fluorophores, although both fluorophores were detected, showing the requirement for the TerS portion of the donor for FRET energy transfer by multimer formation.
Fig. 5:
Ring-like FRET in vivo of fluorescent TerS fusion proteins: A: In vivo co-infection and co-expression scheme for T4 TerS-eGFP and TerS-mCherry gene containing T4 phages. B: A single bacterium co-infected with 5 TerS-eGFP + 5 TerS-mCherry phages that is attached to glass with polylysine displays ~50% FRET between normal phage production in the infected bacteria.
Ring-like oligomer formation of TerS is required for function -
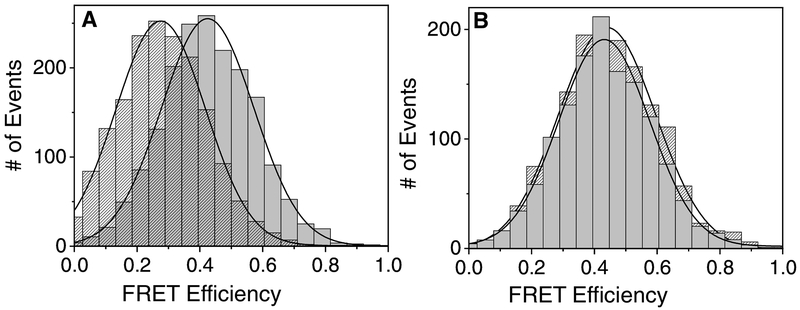
A TerS gene coding for a residue Q59Y mutation was constructed and found to be temperature sensitive (ts) by growth at 20 °C but not at 42 °C, unlike wild type T4 that grew well at both temperatures. In a T4-like 44RR crystal structure the Q59Y mutation is located in the alpha-helical interaction domains of the 12-mer structure, supporting disruption of the helix to helix forming ring structure under high temperature stress (Fig. 2(Sun et al., 2012)). Analysis of this mutation introduced into the TerS-mCherry plasmid or into the phage genome showed comparable temperature sensitive profiles. When analyzed by FRET, a mixture of urea renatured wild type TerS-eGFP and TerS resQ59Y-mCherry fusion proteins behaved very differently from the two wild type fusion proteins (Fig. 6A & B). FRET transfer from TerS-eGFP to TerS-mCherry was blocked at high but not low temperature only in the mutant containing mixture, thereby mimicking the effect of the ts mutation in the phage genome. Evidently the mutant TerS protein is unable to associate with wild type TerS protein to form ring-like oligomers at high temperature. This defect evidently causes loss of TerS function to initiate DNA packaging on the DNA concatemer in vivo.
Fig. 6:
Phage T4 TerS temperature sensitive and normal gene expression proteins purified as in Fig. 3, and mixed and renatured together in vitro at 20 deg and 42 deg.Left panel A: TerS-eGFP-wt + TerS tsQ59Y-mCherry at 20C (right, grey) and 42C (left, shaded). Right panel B: TerS-eGFP-wt + TerSmCherry -wt at 20C (grey) and 42C (shaded).
Super-resolution STORM and PALM optical microscopy supports multi-ring-like TerS oligomer formation in vivo -
Phage T4 infection degrades the E. coli DNA nucleoid and utilizes the nucleotides for viral DNA synthesis at ~ten-fold higher rate than in uninfected cells (Mathews and Allen, 1983). It is known that the DNA intercalating dye YOYO-1 is fluorescent essentially only when bound to DNA. This dye inactivates phage T4 development by selectively blocking DNA packaging (Dixit et al., 2012). STORM microscopy of YOYO-1 administered plasmid containing and T4 infected bacteria allowed the effect of the dye on DNA structure (Fig. 7 A & B) as well as on much (~10 fold) higher DNA concentration within the control infected versus uninfected cell to be measured (Fig. 8 E & F). PALM microscopy (Fig 8) of photoactivatable TerS-PACherry-C1 in the absence of YOYO-1 allowed direct visualization of the TerS-fusion protein expressed from a plasmid or from the T4 genome in the presence or absence of a functional TerL protein (gene TerL containing an amber mutation encoding amino acid residue 249). In the latter case the TerS-mCherry has no functional DNA packaging binding partner; initiation of DNA packaging and maturation cleavage are blocked and the TerS associated fluorescence appears more intense within the infected cell, apparently because the TerS self-association is retained without the possibility of handoff to TerL (Fig. 8 A versus 8 B). The difference is similar using YOYO-1 staining to inhibit DNA packaging using STORM microscopy to visualize the DNA associated dye (Fig. 8 C versus 8 D). Overall the results clearly support highly localized fluorescent sources within bacteria with significant intensity increases seen in the absence of the T4 TerL protein to block packaging. The size increase inferred by PALM measurements of the TerS-PACherry-C1 protein support the unmodified TerS twin ring structure previously shown in vitro (Lin et al., 1997) is also found in vivo. However, it appears that the fusion proteins have more variable and higher molecular weight forms than the unmodified TerS protein in vivo while retaining function.
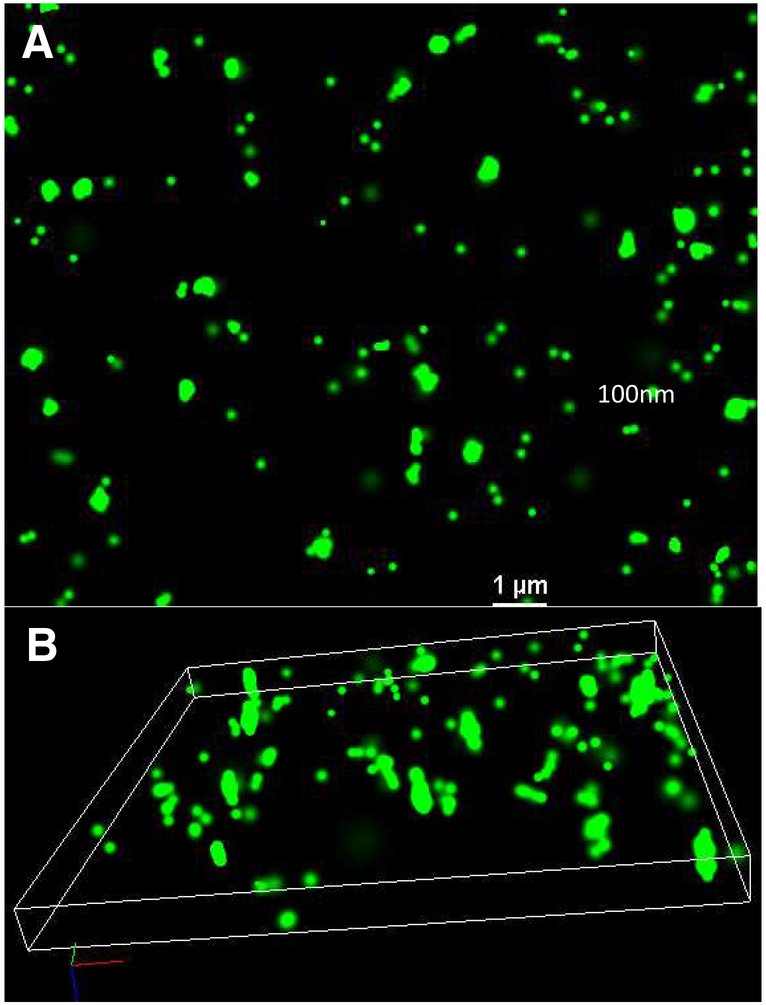
Fig. 7:
A: 2D STORM, and B: 3D STORM images of YOYO-1 stained T4 TerS-mCherry infected E. coli.
Fig. 8:
STORM and PALM super-resolution images with plasmids and T4 phages. A: T4 TerS-PAmCherry; B: T4 TerSPAmCherry-gene 17 amber; C: T4 TerS-mCherry YOY0–1; D: T4 TerS-mCherryY0Y0–1-gene 17amber; E: E. coli BL21 (DE3) pET28a induced TerS-mCherry infected with T4 TerS-mCherry plus Y0Y0–1; F: Uninfected E. coli BL21 (DE3) pET28a induced TerS-mCherry plus Y0Y0–1.
The figure 8 displays of fluorescence differences that are shown between TerS expression in the presence (Fig. 8 A & C) and absence (Fig. 8 B & D) of functional TerL result from genes incorporated into the phage genome. This analysis was possible by construction of the TerS-PACherry-C1 gene to allow PALM super-resolution optical microscopy with laser excitation of TerS-PACherry-C1. The images are of the TerS protein alone (PALM images (Fig. 8 A & B)) and as indirectly assessed with addition of Y0Y0–1 bound DNA where Y0Y0–1 has been shown to block packaging (STORM images (Fig. 8 C & D)). There are clearly enhanced intensities as well as more diffuse fluorescent images without the TerS gene 17 amber mutant TerL binding partner for both the PALM and STORM images. Since phage T4 TerS gp16 is apparently always found both in vitro and in vivo assembled in at least a single ring form, and never as a monomer (See Figs. 2 and 3 and references (Lin et al., 1997) and (van Duijn, 2010)), these results support the enhanced presence of two or more closely associated rings or ring-like oligomeric gp16 structures resulting in enhanced fluorescent intensity from the photo-activated TerS proteins in the absence of a TerL binding partner for hand-off.
Super-resolution microscopy resolution limits are challenged by structures located within E. coli. Since in the best case, the resolution of PALM in lateral (x-y) plane is ~50nm and for STORM, it is ~20nm, while in the axial (z-) plane, the resolution is ~100nm (for PALM) and ~50nm (for STORM), we could not expect to see the actual structure of TerS single (~8 nm diameter native rings (Lin et al., 1997)), double, or even more numerous ring-like oligomers by super-resolution in the ~1000 by ~4000 nm infected E. coli. Instead we see in Figure 8 ~50 to ~100 nm fluorescence spots, with enhanced and more diffuse fluorescence with twinned or more TerS rings-like oligomers when compared with the single rings resulting from handoff to functional packaging active TerL.
Discussion:
Substantial experimental evidence suggests that native non-tagged and non-DNA bound phage T4 TerS proteins form both protein-only 11-mer single rings and protein-only 22-mer dimeric side-by-side twin rings in vitro (Lin et al., 1997; van Duijn, 2010). As shown here, T4 TerS self-association is strong in vitro and in vivo even in fluorescent fusion protein forms. However TerS association with its other expected binding partners (TerL, portal protein, and gp49 endonuclease VII resolvase) are too weak to allow detection, including by high molecular weight mass spectroscopy (Lin et al., 1997; Golz and Kemper, 1999; van Duijn, 2010). The high frequency and stability of the double ring form as well as the variety of multimers found in TerS ring structures among pac site phages is most consistent with the proposal that the double ring form is likely to be a nonplanar lock washer like structure where the ring ends have not joined. A fraction of the single rings could have a comparable structure. Quite possibly such structures would be excluded from protein crystals. In support of this interpretation, in addition to the T4 TerS twin rings, Liang Tang has found that the purified phage Sf6 TerS is also found in a double ring form in the absence of DNA (personal communication).
The seminal headful cutting and terminal redundancy proposal of Streisinger et al. is difficult to distinguish conceptually as well as mechanistically from pac site cutting since most of the same elements are maintained (Sun et al., 2012) (Fokine and Rossmann, 2014) (Streisinger et al., 1967). Pac site phages invariably carry out headful packaging whereas DNA pac site cutting specificity is variable. In phage T4 direct evidence for a gene 16 to BamH1 mature DNA pac segment was found by Southern blotting. Moreover direct evidence by transduction and site directed mutagenesis for a gene TerS 16 situated pac site came from a transducing phage variant of T4GT7, as is generally found to be the case among pac site phages such as P22 or P1 phages that can also be generalized transducing phages (Lin and Black, 1998). As previously discussed, five pac site directed single base mutational changes in the gene 16 pac site that reduce the gene amplifications are not lethal to the phage, suggesting back up mechanisms (e.g. continued use of the mutated gene 16 pac site, use of the homologous pac sequence in gene 19 that leads to the 16 to 19 amplifications, use of other more distant genomic pac-like sites, or of other back up terminase assisting DNA trimming proteins). It may also be the case, as proposed, that binding of the small terminase to DNA is favored by pairing two homologous sequence DNAs. As analyzed in numerous pac site phages, only the first, in what may be a long series of processive packaging events, is expected to display any pac site sequence specificity, and even the initial DNA cutting directed by the pac site may be at some distance to a specific pac sequence whose boundaries may not be precisely defined. It is known that cutting of DNA around a pac site can be quite imprecise, as shown for phage Sf6 where cutting occurs over 2 kb around the pac site and leaves a fuzzy mature pac segment (Leavitt et al., 2013), or in the case of Sapi1 phage pac cutting can be redirected to more distant sites in the Sapi1 DNA (Bento et al., 2014). Specific TerS pac site containing DNA binding has been difficult to demonstrate in vitro in a number of pac site phages and has been observed to be relatively sequence unrelated when studied in vitro e.g. in phage Sf6 or SPP1 (Djacem et al., 2017; Zhao et al., 2012).
Our experiments reported here show that TerS gp16 forms multimers in vivo with spacings comparable to those found in the purified protein rings. These multimers appear to be the predominant active form of the protein in vivo found in association with concatemeric DNA. The spacings do not appear to be altered by association with DNA in vivo. More work must be done to determine the structure of both double and single ring forms of the protein in association with DNA to establish a detailed mechanism for the double ring form of TerS. Analysis of a ts mutation in gp16 TerS supports ring formation as necessary for TerS function in vivo. Super-resolution microscopy supports the formation and action of a double or multimeric TerS ring-like complex in vivo. It is remarkable that substantial protein fusions to either end of the T4 TerS molecule leave its function intact, especially the large C-terminal GFP and mCherry fusions. Evidently these fusion proteins still allow gp16 to hand off the concatemeric DNA to TerL with adequate efficiency to initiate normal cutting and DNA packaging. A double or multimeric ring structure with DNA wound around both rings supports a nucleosome-like DNA-protein structure rather than a protein ring threaded through with DNA. Moreover it is likely in view of the generation of gene recombinants by TerS that the formation of a Holliday junction four stranded DNA structure from two apposed pac site duplex DNAs is done in large part by DNA base pairing in addition to DNA protein recognition (See Fig. 1C). This structure would explain TerS generation of recombinants at pac sites as well as being essential for packaging initiation cutting when promoted by the TerS protein at pac site(s) in collaboration with the TerL nuclease initiation cutting. It is also consistent with the very general recognition features of Pac sequences found among Pac site phages (Lin and Black, 1998; Zhao et al., 2012; Djacem et al., 2017). More experimental work is required to establish the requirement for a Holiday junction DNA structure in TerS DNA recognition.
Materials and Methods
Fluorescence, FRET, and temperature controlled FRET analysis of TerS in vitro and in vivo
Expression and fluorescence of eGFP and mCherry in E. coli BE was measured by growth in M9 minimal medium containing glycerol for low fluorescence background. The nonfusion GFP was induced from plasmid pJOE5751.1 maintained by addition of ampicillin, a generous gift of J. Altenbuchner, University of Stuttgart, by rhamnose addition (Wegerer et al., 2008). FRET and FCS measurements were performed with an excitation wavelength at 470nm in a confocal microscope with single molecule detection sensitivity as reported earlier (Ray et al., 2010). Briefly, fluorescence responses from the donor and the acceptor molecules were separated by a 50/50 beam splitter and detected by two avalanche photodiode detectors (APD) using the method of time-correlated single photon counting and the Time-Tagged Time-Resolved (TTTR) mode. High quality bandpass (Chroma) filters were used for recording donor (500–550 nm) and acceptor (590–650 nm) fluorescence in two separate detection channels. The collected single photon data was binned by 1 msec bin in each channel (donor or acceptor) which resulted in intensity-time traces and count-rate histograms. Threshold values in each channel were used to identify the single molecule bursts from the corresponding background signal level.
Fluorescence bursts were recorded simultaneously in donor and acceptor channels and FRET efficiencies were calculated using E = IA/(IA + γID) where ID and IA are the sums of donor counts and acceptor counts for each burst, taking into account the possible difference in the detection efficiency (γ) in two separate channels (Schuler et al., 2002). The donor-to-acceptor distance (r) in terms of efficiency of energy transfer (E) and Forster Distance (R0) is given by r = R0[1/E − 1]1/6. For temperature-controlled FRET measurements, the samples were placed in a custom-made temperature-controlled stage-top chamber by means of water circulation in water tight channels where the temperatures were set at 20±1°C or 42±1°C.
Super-resolution imaging was carried out using the Nikon N-STORM microscope (Nikon Instruments Inc., Melville, NY) equipped with 100X CFI Apo TIRF oil-immersion objective (1.49 NA) (Mengistu et al., 2015). Single molecule fitting and Gaussian images were rendered using the N-STORM software NIS Elements. The localization precision for 3D dSTORM was determined to be 20 nm and 50 nm in-plane and axial-resolution respectively. Intercalating dye YOYO-1 stained T4 TerS infected E. coli was imaged by excitation with a 473nm laser and emission was recorded in the 500 to 550 nm spectral region by the dSTORM set-up. The oxygen-scavenging imaging buffer was 14mg glucose oxidase and 50μL of 17mg/mL catalase (Sigma) in 200μL Component A (10mM Tris, 50mM NaCl); Component B (50mM Tris-HCl, 10mM NaCl, 10% glucose); and 1M cysteamine (MEA). PALM microscopy was performed with photoactivatable 16-PAmCherryC1 upon continuous excitation with 561nm laser which is near the excitation maximum of the PAmCherry. A second laser at 405nm was used for activating the PAmCherry. Emission was recorded in the 580 to 640 nm spectral range. Imaging was performed in the total internal reflection fluorescence (TIRF) microscopy set-up coupled with an Andor EMCCD camera capable of detecting single photons.
Cloning, expression, and purification of TerS(gp16)-mCherry and TerS-eGFP from adjacent overlapping genes 16 (TerS) and 17 (TerL).
Gp16 was first cloned in pET28a vector using Forward primer (5’ GGAATTGCATATGATGGAAGGTCTTGATATAAACAAACTT 3’) with start codon ATG and NdeI site and Reverse primer (CCGGAATTCATCGGTTGTTCCATTTATCACCTT) with EcoRI site but no stop codon. mCherry or eGFP were cloned at the C-terminus of gp16 using mCherry primers (Forward primer with an EcoRI site, a 3 aminoacid linker GSG and no start codon 5’ CCGGAATTCGGCAGCGGCGTGAGCAAGGGCGAGGAGGAT 3’ and reverse primer with NotI site and a stop codon TAA 5’ ATAAGAATGCGGCCGCCTACTTGTACAGCTCGTCCATGCC 3’) or eGFP primers (Forward primer with EcoRI site, a 3 amino acid linker GSG and no start site 5’CCGGAATTCGGCAGCGGCGCGGCTAGCAAAGGAGAAGAACTT 3’and reverse primer with NotI site and a stop codon 5’ ATAAGAATGCGGCCGCTTATCGACGGTATCGATAAGCTTG 3’). The 6X-His tagged proteins were purified using Nickel -NTA- columns, followed by ammonium sulfate precipitations -gp16- eGFP (20%) and gp16-mcherry (35%)-the percentage saturation solid ammonium sulfate is slowly added to the sample with gentle shaking by hand at room temperature. Resuspended ammonium sulfate precipitates were completely dissolved in buffer without ammonium sulfate and spun at high speed (17000 rpm for 30 min). Proteins were dissolved in a small volume of buffer and left at room temperature for 30 min followed by gel size exclusion chromatography and dialysis. Resuspension buffer consists of pH 7 Sodium phosphate buffer (50mM), 300mM NaCl, protease inhibitor cocktail, and 5mM Imidazole, followed by washing in the same buffer with 10, 20, 50, and 100 mM Imidazole, for final his tag column elution in 300mM Imidazole. Purified proteins were analyzed by SDS and Typhoon imager. For Typhoon image and identification of the gp16 fusion proteins on Superose 6 chromatography fluorescence identified most of the fluorescence in the fractions 5–7 followed by lesser amounts in fraction 8–13 as shown in Fig. 4 (upper left) run with and without size standards (not shown). For fluorescence Typhoon image no boiling and no dye was added to samples run on 10% SDS-PAGE gel by loading in glycerol only as mCherry protein is proteolyzed by harsh treatment like SDS and boiling.
Rescue and Complementation assays
For rescue test of BL21 (DE3) cells carrying gp16-mCherry-pET28a, gp16-eGFP-pET28a, or gp16-pET28a plasmids, bacterial cultures were grown and gene 16 double amber phage was used to infect the cells on LB-agar plates. The progeny plaques were sequenced. For complementation assays 16 double amber mutant phage was used to infect cells producing IPTG induced levels of the gp16, gp16-mCherry, or gp16-eGFP fusion proteins. In all of the infections into induced cells expressing high levels of fusion protein, the yield of amber mutant phages is significantly higher than of the WT phage yield, suggesting that the phage are produced by complementation rather than from genetic recombination between the infecting amber mutant and the WT gene on the resident plasmid in the infected cells (See Table 1).
Construction of recombinant gp16-mcherry/eGFP-gp17 phages
gp17 gene carrying the ribosome binding site and part of gp16 was amplified using forward primer with NotI site 5’ ATAAGAAT GCGGCCGCAAGCTCGTGAGAAGG TGATAAATGGAA 3’and reverse primer with XhoI site 5’ CCGCTCGAGTTATACCATTGACATACCATGAGATACTGG 3’ and was cloned at the C-terminus of gp16-mCherry/eGFP fusion in pET28a vector. Phages carrying recombinant gp16-mCherry/eGFP-gp17 genes were selected by infecting recombinant plasmid carrying cells with 16amber17amber rII phage. Phages were sequenced using gp16, mcherry/eGFP or gp17 primers. Western blotting was carried using gp16 antiserum (Lin et al., 1997). Briefly, E. coli BE were infected with the recombinant phages or T4 or 13am-16am-17am phages (5 MOI) and just after 20 min of infections cells were pelleted and saved at −80°C and used for sample preparation after chloroform, DNAse treatment and boiling in loading dye for 10 min.
Construction of photoactivatable 16-PAmCherryC1 for PALM microscopy in vitro and in vivo
The same primers used for mCherry cloning were used with plasmid containing the gene for PAmCherryC1 (Clontech) (Subach et al., 2009). This was used to introduce the analogous construct described above into the same plasmids. An amber mutation in phage T4 TerL gene 17 was introduced into the T4 encoding the PAmCherryC1 by selection of the gene 17 amber recombinant that can confer 9 amino acridine resistance.
Significance:
Bacteriophage T4 DNA packaging is shown to involve a eukaryotic meiotic-like synapsis of two homologous pac site DNAs to gauge adequate DNA concatemer synthesis for cutting and packaging of a full genome’s amount of DNA. Single molecule fluorescence and super-resolution optical microscopy of fully functional terminase small subunit fusion proteins in vitro and in vivo support a TerS twin ring model for the synapsis. TerS gene pac site to gene 19 pac site gene amplifications can be selected in phage T4 only with functional TerS protein. This provides genetic evidence for synapsis of the two homologous pac site containing duplex DNA segments by TerS double rings. The results can also explain the divergent and highly variable TerS protein monomer and multimer crystal structures found among bacteriophages: Dimeric protein rings binding two homologous dsDNA duplexes based on many TerS monomer structures can supply the same function.
Acknowledgments
LWB and ABD were funded by NIH R01 grants AI011676 and GM118766, KR by NIH R01 GM117836. We thank Naiqian Cheng for cryo-electron microscopy and negative stain electron microscopy of Superose 6 chromatography purified TerS-mCherry proteins. We thank Greg Snyder IHV UMB for assistance in making Fig. 2 from PDB data.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baumann RG, Black LW, 2003. Isolation and characterization of T4 bacteriophage gp17 terminase, a large subunit multimer with enhanced ATPase activity. J.Biol.Chem. 278, 4618–4627. [DOI] [PubMed] [Google Scholar]
- Bento JC, Lane KD, Read EK, Cerca N, Christie GE, 2014. Sequence determinants for DNA packaging specificity in the S. aureus pathogenicity island SaPI1. Plasmid 71, 8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF, 2006. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645. [DOI] [PubMed] [Google Scholar]
- Black LW, 1989. DNA packaging in dsDNA bacteriophages. Annu Rev Microbiol 43, 267–292. [DOI] [PubMed] [Google Scholar]
- Black LW, 1995. DNA packaging and cutting by phage terminases: control in phage T4 by a synaptic mechanism. Bioessays 17, 1025–1030. [DOI] [PubMed] [Google Scholar]
- Black LW, Peng G, 2006. Mechanistic coupling of bacteriophage T4 DNA packaging to components of the replication-dependent late transcription machinery. J Biol Chem 281, 25635–25643. [DOI] [PubMed] [Google Scholar]
- Buttner CR, Chechik M, Ortiz-Lombardia M, Smits C, Ebong IO, Chechik V, Jeschke G, Dykeman E, Benini S, Robinson CV, Alonso JC, Antson AA, 2012. Structural basis for DNA recognition and loading into a viral packaging motor. Proc Natl Acad Sci U S A 109, 811–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S, Wyckoff E, Hayden M, Sampson L, Eppler K, Randall S, Moreno ET, Serwer P, 1992. Bacteriophage P22 portal protein is part of the gauge that regulates packing density of intravirion DNA. J Mol Biol 224, 1055–1074. [DOI] [PubMed] [Google Scholar]
- Dixit AB, Ray K, Black LW, 2012. Compression of the DNA substrate by a viral packaging motor is supported by removal of intercalating dye during translocation. Proc Natl Acad Sci U S A 109, 20419–20424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djacem K, Tavares P, Oliveira L, 2017. Bacteriophage SPP1 pac Cleavage: A Precise Cut without Sequence Specificity Requirement. J Mol Biol 429, 1381–1395. [DOI] [PubMed] [Google Scholar]
- Fokine A, Rossmann MG, 2014. Molecular architecture of tailed double-stranded DNA phages. Bacteriophage 4, e28281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller DN, Raymer DM, Kottadiel VI, Rao VB, Smith DE, 2007. Single phage T4 DNA packaging motors exhibit large force generation, high velocity, and dynamic variability. Proc Natl Acad Sci U S A 104, 16868–16873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Rao VB, 2011. Specificity of interactions among the DNA-packaging machine components of T4-related bacteriophages. J Biol Chem 286, 3944–3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golz S, Kemper B, 1999. Association of holliday-structure resolving endonuclease VII with gp20 from the packaging machine of phage T4. J Mol Biol 285, 1131–1144. [DOI] [PubMed] [Google Scholar]
- Kainov DE, Tuma R, Mancini EJ, 2006. Hexameric molecular motors: P4 packaging ATPase unravels the mechanism. Cell Mol Life Sci 63, 1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondabagil KR, Zhang Z, Rao VB, 2006. The DNA translocating ATPase of bacteriophage T4 packaging motor. J Mol Biol 363, 786–799. [DOI] [PubMed] [Google Scholar]
- Leavitt JC, Gilcrease EB, Wilson K, Casjens SR, 2013. Function and horizontal transfer of the small terminase subunit of the tailed bacteriophage Sf6 DNA packaging nanomotor. Virology 440, 117–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffers G, Rao VB, 2000. Biochemical characterization of an ATPase activity associated with the large packaging subunit gp17 from bacteriophage T4. J.Biol.Chem. 275, 37127–37136. [DOI] [PubMed] [Google Scholar]
- Lin H, Black LW, 1998. DNA requirements in vivo for phage T4 packaging. Virology 242, 118–127. [DOI] [PubMed] [Google Scholar]
- Lin H, Simon MN, Black LW, 1997. Purification and characterization of the small subunit of phage T4 terminase, gp16, required for DNA packaging. J Biol Chem 272, 3495–3501. [DOI] [PubMed] [Google Scholar]
- Maluf NK, Gaussier H, Bogner E, Feiss M, Catalano CE, 2006. Assembly of bacteriophage lambda terminase into a viral DNA maturation and packaging machine. Biochemistry 45, 15259–15268. [DOI] [PubMed] [Google Scholar]
- Mathews CK, Allen JR, 1983. Enzymes and Proteins of DNA Metabolism, Bacteriophage T4: ASM Press, p. 59. [Google Scholar]
- Mengistu M, Ray K, Lewis GK, DeVico AL, 2015. Antigenic properties of the human immunodeficiency virus envelope glycoprotein gp120 on virions bound to target cells. PLoS Pathog 11, e1004772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemecek D, Lander GC, Johnson JE, Casjens SR, Thomas GJ Jr., 2008. Assembly architecture and DNA binding of the bacteriophage P22 terminase small subunit. J Mol Biol 383, 494–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao VB, Black LW, 2010. Structure and assembly of bacteriophage T4 head. Virol J 7, 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao VB, Feiss M, 2008. The Bacteriophage DNA Packaging Motor. Annu Rev Genet. [DOI] [PubMed] [Google Scholar]
- Ray K, Sabanayagam CR, Lakowicz JR, Black LW, 2010. DNA crunching by a viral packaging motor: Compression of a procapsid-portal stalled Y-DNA substrate. Virology 398, 224–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Bhardwaj A, Datta P, Lander GC, Cingolani G, 2012. Small terminase couples viral DNA binding to genome-packaging ATPase activity. Structure 20, 1403–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Cingolani G, 2012. Structure of p22 headful packaging nuclease. J Biol Chem 287, 28196–28205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust MJ, Bates M, Zhuang X, 2006. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat Methods 3, 793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabanayagam CR, Oram M, Lakowicz JR, Black LW, 2007. Viral DNA packaging studied by fluorescence correlation spectroscopy. Biophys J 93, L17–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer M, Heilemann M, 2017. Single-Molecule Localization Microscopy in Eukaryotes. Chem Rev 117, 7478–7509. [DOI] [PubMed] [Google Scholar]
- Schuler B, Lipman EA, Eaton WA, 2002. Probing the free-energy surface for protein folding with single-molecule fluorescence spectroscopy. Nature 419, 743–747. [DOI] [PubMed] [Google Scholar]
- Schwille P, Haupts U, Maiti S, Webb WW, 1999. Molecular dynamics in living cells observed by fluorescence correlation spectroscopy with one- and two-photon excitation. Biophys J 77, 2251–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serwer P, Jiang W, 2012. Dualities in the analysis of phage DNA packaging motors. Bacteriophage 2, 239–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits C, Chechik M, Kovalevskiy OV, Shevtsov MB, Foster AW, Alonso JC, Antson AA, 2009. Structural basis for the nuclease activity of a bacteriophage large terminase. EMBO Rep 10, 592–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streisinger G, Emrich J, Stahl MM, 1967. Chromosome structure in phage t4, iii. Terminal redundancy and length determination. Proc Natl Acad Sci U S A 57, 292–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subach FV, Patterson GH, Manley S, Gillette JM, Lippincott-Schwartz J, Verkhusha VV, 2009. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nature methods 6, 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Gao S, Kondabagil K, Xiang Y, Rossmann MG, Rao VB, 2012Structure and function of the small terminase component of the DNA packaging machine in T4-like bacteriophages. Proc Natl Acad Sci U S A 109, 817–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Kondabagil K, Draper B, Alam TI, Bowman VD, Zhang Z, Hegde S, Fokine A, Rossmann MG, Rao VB, 2008. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell 135, 1251–1262. [DOI] [PubMed] [Google Scholar]
- Tavares P, Santos MA, Lurz R, Morelli G, de Lencastre H, Trautner TA, 1992. Identification of a gene in Bacillus subtilis bacteriophage SPP1 determining the amount of packaged DNA. J Mol Biol 225, 81–92. [DOI] [PubMed] [Google Scholar]
- Teschke CM, 2012. Themes and variations of viral small terminase proteins. Structure 20, 1291–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duijn E, 2010. Current limitations in native mass spectrometry based structural biology. J Am Soc Mass Spectrom 21, 971–978. [DOI] [PubMed] [Google Scholar]
- Wegerer A, Sun T, Altenbuchner J, 2008. Optimization of an E. coli L-rhamnose-inducible expression vector: test of various genetic module combinations. BMC Biotechnol 8, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Lin H, Black LW, 1995. Bacteriophage T4 gene 17 amplification mutants: evidence for initiation by the T4 terminase subunit gp16. J Mol Biol 247, 523–528. [PubMed] [Google Scholar]
- Wu DG, Black LW, 1987. Gene amplification mechanism for the hyperproduction of T4 bacteriophage gene 17 and 18 proteins. J Mol Biol 195, 769–783. [DOI] [PubMed] [Google Scholar]
- Wu DG, Wu CH, Black LW, 1991. Reiterated gene amplifications at specific short homology sequences in phage T4 produce Hp17 mutants. J Mol Biol 218, 705–721. [DOI] [PubMed] [Google Scholar]
- Zhao H, Christensen TE, Kamau YN, Tang L, 2013. Structures of the phage Sf6 large terminase provide new insights into DNA translocation and cleavage. Proc Natl Acad Sci U S A 110, 8075–8080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Finch CJ, Sequeira RD, Johnson BA, Johnson JE, Casjens SR, Tang L, 2010. Crystal structure of the DNA-recognition component of the bacterial virus Sf6 genome-packaging machine. Proc Natl Acad Sci U S A 107, 1971–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Kamau YN, Christensen TE, Tang L, 2012. Structural and functional studies of the phage Sf6 terminase small subunit reveal a DNA-spooling device facilitated by structural plasticity. J Mol Biol 423, 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]