Abstract
Cancers are caused by mutations to genes that regulate cell normal functions. The capability to rapid and reliable detection of specific target gene variations can facilitate early disease detection and diagnosis, and also enables personalized treatment of cancer. Most of the currently available methods for DNA mutation detection are time-consuming and/or require the use of labels or sophisticated instruments. In this work, we reported a label-free enzymatic reaction-based nanopore sensing strategy to detect DNA mutations, including base substitution, deletion, and insertion. The method was rapid and highly sensitive with a detection limit of 4.8 nM in a 10-minute electrical recording. Furthermore, the nanopore assay could differentiate among perfect-match, one-mismatch, and two-mismatches. In addition, simulated serum samples were successfully analyzed. Our developed nanopore-based DNA mutation detection strategy should find useful application in genetic diagnosis.
Graphical Abstract
Introduction
Mutations to genes that regulate cell normal functions can cause serious genetic disorders and even cancers. The capability of rapid and accurate detection of specific target gene sequences and base variations is of paramount importance since such a genetic diagnostic technology not only benefits early disease detection and diagnosis, but also enables personalized treatment, thus improving outcomes. Thus far, three major approaches have been developed for gene mutation detection, including direct sequencing (e.g., PCR), DNA hybridization, and restriction enzyme digestion methods.1–6 Among them, taking advantage of a complementary DNA or PNA probe to detect the presence of a specific target nucleic acid sequence is one of the most popular strategies used by the current optical-, electrochemical-, piezoelectric-, or mass-based nanobiotechnologies to detect gene mutations.7–14 However, most of these methods are time consuming, and/or require the use of labels or expensive instruments. Therefore, development of improved mutation detection techniques is still highly desirable.
Nanopore stochastic sensing is an emerging label-free technique for measuring single molecules.15–19 By monitoring the ionic current modulations produced by the interaction between analyte molecules and a nano-scale sized pore, nanopore sensing technology has successfully been utilized for various applications, including environmental protection,20, 21 homeland security and bio-defense,22, 23 pharmaceutical screening,24–26 and medical diagnosis.27,28 At present, there are two major types of nanopore technology: biological protein pore29–34 and synthetic solid-state nanopore35,36. Protein pores generally provide a better resolution and selectivity to analyte detection than synthetic nanopores but have a reputation for being fragile. In contrast, solid-state nanopores, which can have flexible pore diameters & lengths, are stable and could tolerate a variety of extreme conditions, and are ideal for field deployable applications.17,18 In addition to the ionic current-based detection strategy, fluorescence- and surface-enhanced raman (SER)- based nanopore sensing techniques have also successfully been developed.37,38 Recently, molecular dynamics simulations showed that plasmonic nanopores coupling with SER detection offered the possibility for DNA sequencing.39 At the moment, it is hard to gauge the long-term successfulness of these novel concepts due to the lack of experimental data thus far. However, just like other variations of solid-state nanopores, one of the key challenges to their sensitive detection of small molecules and even to achieve single-base resolution for DNA sequencing is to introduce new surface functions inside the nanopore, preferably at a specific position. Furthermore, reducing background noise can also improve their sensing resolution. In earlier studies, we reported a sensitive and selective α-hemolysin (αHL) nanopore sensing method for the detection of anthrax lethal factor by using a complementary single-stranded DNA as a molecular probe.40 The similar nucleic acid hybridization strategies were also utilized by the Kang group and the Gu group for the successful detection of HBV DNA and cancer biomarker microRNA.41,42 Note that, in those sensing systems, the constrictions of the nanopores were slightly larger than the diameter of ssDNA but smaller than that of dsDNA, so that ssDNA could rapidly translocate through the nanopore, while dsDNA needed to be unzipped into a form of ssDNA before translocation, thus producing significantly longer residence time events than ssDNA. Although single base resolution has been demonstrated in these nanopore nucleic acid sensors, early studies also demonstrated that the residence time of the dsDNA events increased significantly with the increase in the DNA length, and was also affected by the DNA sequence (e.g., GC content).43–45 Therefore, the hybridization-based nanopore nucleic acid assay is generally limited to the detection of rather short sequences (~10 base) of DNA/RNA without sacrificing single-base resolution.46
In this work, by taking advantage of single-strand specific nuclease, we developed a nanopore enzymatic sensing strategy for rapid detection of DNA mutations. Our method overcame the length limitation of the well-documented hybridization-based nanopore nucleic acid assay, and could be utilized as a generic nucleic acid detection method for analyzing DNA/RNA biomarkers (usually 18 – 22 nucleotides in length). Single-strand specific nuclease, which acts characteristically on single-stranded nucleic acids or single-stranded regions in double-stranded nucleic acids, are extensively employed in DNA mutation detection.47 A variety of nucleases such as S1, P1, mung bean nuclease, and Surveyor Nuclease have been identified thus far. Surveyor Nuclease was used as a model nuclease in this work to proof-of-concept demonstrate our new nanopore strategy for DNA mutation analysis due to its several unique properties.48 First, this enzyme shows accurate detection in not only bacterial genomic DNA but also human gene.49 Second, unlike other single-strand specific nucleases, which have the optimum reaction pH around 4–5, Surveyor Nuclease works most efficiently at ~ pH 7, avoiding the depurination of DNA in acidity.50 Third, S1, mung bean, and some other single-strand specific nucleases occasionally could not recognize some single-base mismatches,51 while Surveyor Nuclease activates on each mismatch site although the cleavage efficiency varies with the sequence of the mismatch.52
Methods
Materials.
Surveyor Nuclease kit and DNA polymers with standard purification (desalting) were ordered from Integrated DNA Technologies (Coralville, IA). All the other chemicals, including sodium chloride, Trizma base, hydrochloric acid, pentane, hexadecane, HPLC-grade water, and DNase, RNase free water, were purchased from Sigma-Aldrich (St. Louis, MO). 1,2-diphytanoylphosphatidylcholine was bought from Avanti Polar Lipids (Alabaster, AL). Rabbit blood was obtained from HemoStat Laboratories (Dixon, CA).
Bilayer experiment and data analysis.
The procedure for single channel recordings have been described previously.27 Briefly, a Teflon film (Goodfellow Malvern, PA) with a 150-μm diameter orifice separated two Teflon chamber compartments. Planar bilayer was formed according to the Montal-Muller method. Unless otherwise noted, the experiments were performed at 24 ± 1 °C using the wild-type αHL protein nanopore under symmetrical buffer conditions with the two chamber compartments filled with a solution consisting of 1 M NaCl, and 10 mM Tris (pH 7.5). Both the αHL protein and DNA polymers were added to the cis chamber compartment. The applied potential was +120 mV, unless otherwise noted. Ionic currents were recorded with Axopatch 200B amplifier (Molecular Devices, Sunnyvale, CA), filtered with a four-pole low-pass Bessel filter at 5 kHz, and then digitized with a Digidata 1440A converter (Molecular Devices) at a sampling frequency of 10 kHz. An average of 450 events was recorded in each of the single channel recording experiments. The event blockage amplitude, residence time, and number of occurrences (i.e., event counts) were obtained by using Clampfit 10.5 software (Molecular Devices).
DNA hybridization and surveyor nuclease digestion.
DNA sample pretreatment procedure is illustrated in a flowchart (Supporting Information, Fig. S1). Briefly, 0.5 μL 1 mM single-stranded DNA samples and 0.5 μL 1 mM of their corresponding hybridization ssDNA probes were mixed and incubated at 95 °C for 5 min, and then cooled to room temperature. Nuclease digestion was carried out by adding 6 μL DNase, RNase free water, 10 μL Surveyor Nuclease, 10 μL Surveyor Enhancer, and 3 μL 0.15 M MgCl2 to the hybridized dsDNA, and incubated at 42 °C for two hours. After cooling to room temperature, the mixture solutions were added to the cis chamber compartment for single-channel recording.
Simulated serum sample analysis.
Serum was prepared by collecting the supernatant after centrifugation (2000 rpm) of rabbit blood at 4 °C for 10 min, and was stored at −80 °C. 2 μL serum, 5 μL 100 μM LF (LF1) DNA, 5 μL 100 μM BP DNA, and 8 μL DNase, RNase free water were mixed and incubated at 95 °C for 5 min. Then, the samples were cooled to room temperature, and followed by nuclease digestion and single-channel recording as described in the previous section.
Result and discussion
Principle for nanopore detection of DNA mutations
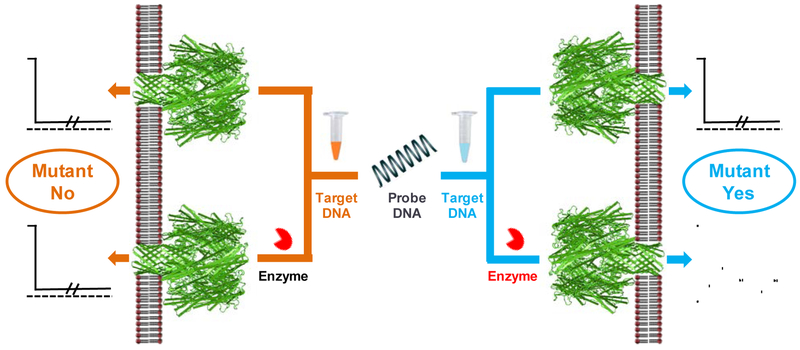
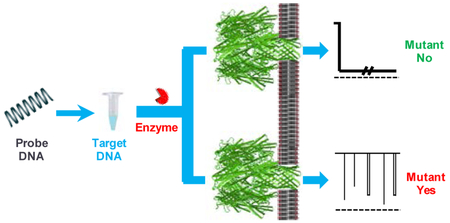
Nanopore detection of DNA mutations is accomplished by monitoring the hybridization mixture of a ssDNA sample and a ssDNA probe in the absence and in the presence of a nuclease. As showed in Scheme 1, in the event that the hybridization between the DNA analyte and the DNA probe produces completely-matched dsDNA, the event signature of the DNA mixture sample would not change significantly in the absence / presence of the nuclease: the nanopore is always blocked for quite a long time. In contrast, if the hybridization produces dsDNA with mismatches, the long-lived DNA events (in the absence of the nuclease) would become less frequent or even disappear after addition of the nuclease to the DNA sample; furthermore, new types of events with smaller residence time could possibly be observed due to the shorter fragments produced by the enzymatic cleavage of the dsDNA substrate.
Scheme 1.
Nanopore detection of DNA mutations. If the sample contains mutant DNA, its hybridization with the DNA probe would produce dsDNA with mismatches; and hence, the event signature of the hybridized dsDNA in the nanopore would change significantly in the absence / presence of the nuclease. In contrast, if the sample contains wild-type DNA, its hybridization with the DNA probe would produce completely-matched dsDNA. Thus, addition of the nuclease to the hybridized DNA sample would not affect the event signature.
Base-base substitution
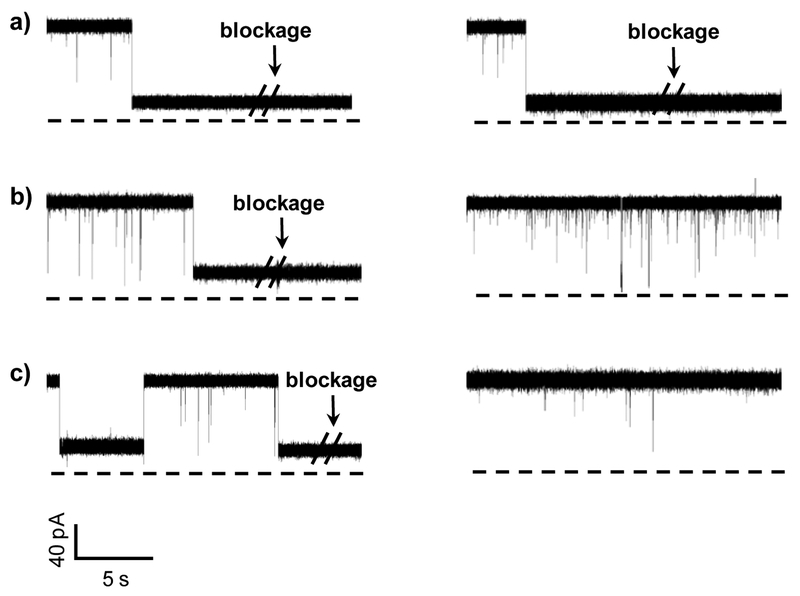
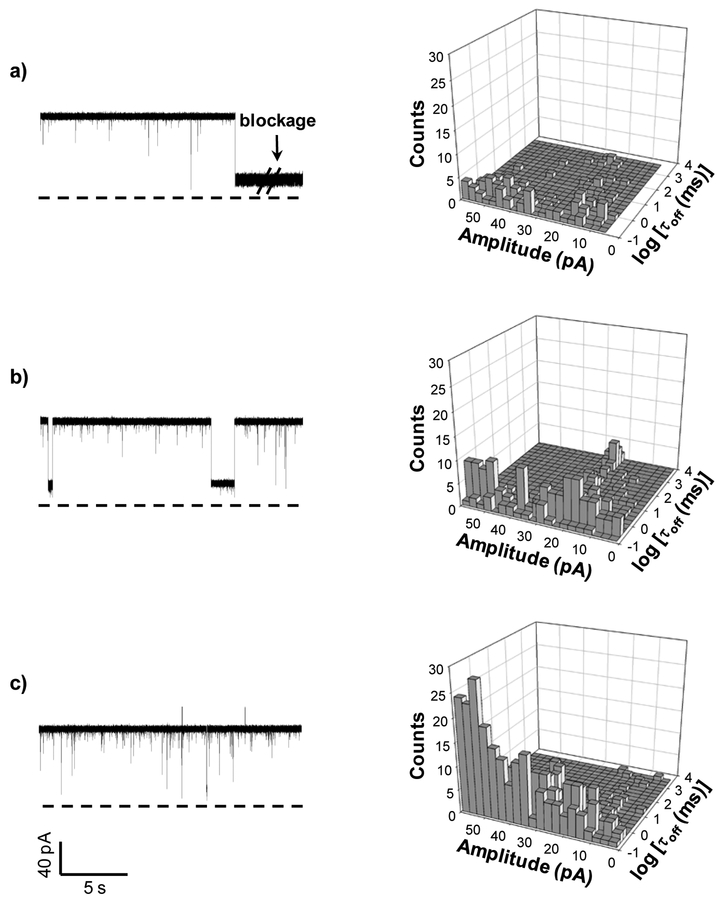
Initial experiments were performed in an electrolyte buffer solution containing 1 M NaCl and 10 mM Tris (pH7.5) using the wild-type αHL protein pore as the sensing element. Three 20-mer single-stranded DNA samples (LF, LF1, LF2) were used as the target analytes with their sequences summarized in Table 1. Note that these three ssDNA samples had similar sequences and were able to hybridize with the 20-mer probe DNA (BP) to form perfectly-matched dsDNA, dsDNA with one mismatch, and dsDNA with two mismatches, respectively. The experimental results were summarized in Fig. 1. In the case of the LF DNA sample, which could hybridize with the probe DNA to form completely-matched dsDNA, two types of blockage events were observed after addition of the LF-BP mixture sample to the nanopore (Fig. 1a). One type of events showed small residence time (< 1 ms) and a wide range of current blockage amplitudes (from ~38.2% to 89.7% of full channel blockage), which are believed to be attributed to the brief residency of DNA polymers in the vestibule or their collision with the opening of the αHL pore.53 The possibility that those events were due to the translocation of unhybridized (free) ssDNA through the pore was not supported by our control experiment (Supporting Information, Fig. S2), where ssDNA samples produced events with significantly different characteristics (especially event distribution and blockage amplitude) from those of the LF-BP mixture. The other type of events presented a narrow range of current blockage amplitudes (a mean of 80.2 ± 2.0 % of full channel blockage) but with a large spread of durations (ranging from hundreds of milliseconds to seconds or even longer), which were caused by the tangling of dsDNA with/near the constriction region of the channel.50 For convenience, the long-lived events with residence time more than 10 s were called permanent block, and were excluded in our data analysis. Note that the mixture sample often permanently blocked the nanopore, and we had to flip the applied potential polarity to make the channel reopen, indicating that the perfectly-matched dsDNA could hardly be unzipped under our experimental condition. The same phenomenon was observed in the experiment with the mixture sample consisting of LF, BP, and Surveyor Nuclease, suggesting that the nuclease had no effect on the perfectly-matched dsDNA. It is worth mentioning that, due to the size difference between ssDNA and dsDNA (the constriction of the αHL nanopore was slightly larger than the diameter of ssDNA but smaller than that of dsDNA), ssDNA and dsDNA produce significantly different residence time events in the nano-channel. Furthermore, our experiments (Supporting Information, Fig. S3) showed that, with an increase in the DNA length, an increased event residence time difference between ssDNA and dsDNA was observed. Our finding was in agreement with the previous observation that with an increase in the DNA length, the event mean residence time of ssDNA linearly increased, while that of dsDNA rose exponentially.ref As to the one-mismatch dsDNA sample (i.e. the mixture of LF1 and BP), similar to the completely-matched dsDNA, it often permanently blocked the nanopore (with an amplitude of 78.7 ± 0.9 % of full channel blockage) in the absence of the nuclease. However, in sharp contrast, in the presence of the nuclease, those long duration (seconds) events disappeared; instead, a new type of events having a mean residence time of 2.7 ± 0.3 ms and a mean residual current of 32.2 ± 0.5 pA appeared (Fig. 1b), suggesting that the nuclease was able to cut the dsDNA into shorter fragments, which could be unzipped and translocated through the nanopore. As an important and interesting side point, we noticed a significant (~ 4.5 folds) increase in the number of short-lived (< 1 ms) events after the nuclease was added to the LF1 and BP mixture (Fig. 1a and Supporting Information, Fig. S4a). It is not unreasonable considering that the total number of DNA molecules increased after nuclease cleavage of the BP-LF1 dsDNA; further, earlier studies have shown that the event frequency for biomolecular interaction with the nanopore was strongly affected by the length of the biomolecule.54 Similar to the observation we made with the one-mismatch dsDNA, the two-mismatch dsDNA (i.e., BP-LF2) also often permanently blocked the nanopore. After addition of the nuclease to the LF2 and BP mixture, the long duration double stranded DNA events (with a mean residual current of 28.9 ± 0.4 pA, i.e., 71.9 ± 0.4 % of full channel blockage) disappeared, and a new type of events having a mean residence time of 1.41 ± 0.12 ms and a mean residual current of 26.8 ± 1.4 pA was identified (Fig. 1c, and Supporting Information, Fig. S5). The results suggested that the two-mismatched dsDNA sample was cleaved into short fragments by the nuclease, thus being rapidly unzipped and translocated through the nanopore. However, interestingly, we noticed that, unlike the one-mismatch dsDNA, the number of short-lived events (< 1 ms) of the two-mismatch dsDNA didn’t change significantly in the absence/presence of the nuclease. One likely interpretation is that the short 7 bp dsDNA fragment (sequence: 5’-GGATTATG-3’ / 3’-CCTAATA-5’), which resulted from the nuclease cleavage of the BP-LF2, passed through the nanopore so rapidly that most of their events were missed by the patch-clamp instrument (with a ~ 200 μs resolution under our experimental conditions). This interpretation was confirmed by direct measurement of current blockages using single standards of the cleavage fragments of the one-mismatch and two-mismatch dsDNA. As shown in the supporting information, Fig. S6, the 7-bp dsDNA fragment (sequence: 5’-GGATTATG-3’ / 3’-CCTAATA-5’), i.e., one of the major cleavage products of the two-mismatch dsDNA produced significantly less frequent events than the 9-bp dsDNA (sequence: 5’-AAATATTGA-3’/3’-ATTTATAACT-5’). It is worth mentioning that, the observation and discussion we made above with nanopore analysis of BP-LF, BP-LF1, and BP-LF2, as well as their nuclease digestion products were also supported by the results of similar experiments in which dsDNA samples were analyzed by agarose gel electrophoresis. As shown in the Supporting Information, Fig. S7, no new bands were observed after addition of Surveyor Nuclease to BP-LF. In contrast, in the presence of the nuclease, BP-LF1 and BP-LF2 showed a new band of ~ 10 bp, indicating they were being digested by Surveyor Nuclease. Note that, bands of 2 bp and 7 bp were not observed for the digestion products of BP-LF2, suggesting that the resolution of agarose gel electrophoresis was not sufficient to resolve these two fragments.
Table 1.
The sequences of single-stranded DNAs used in this work
| Target DNA | Probe DNA | ||
|---|---|---|---|
| Nucleic acids | Sequence | Nucleic acids | Sequence |
| LF | 5’-GGATTATrGTrAAATATTGA-3’ | BP | 5’-TCAATATTTAACAATA ATCC-3’ |
| LF-1 | 5’-GGATTATTGTGAAATATTGA-3’ | BP | 5’-TCAATATTTAACAATA ATCC-3’ |
| LF-2 | 5’-GGATTATGGTGAAATATTG A-3’ | BP | 5’-TCAATATTTAACAATA ATCC-3’ |
| TMS | 5’-CTAATGCTAATCGTGATAGGGG-3’ | TP | 5’-CCCCTATCACGATTAGCATTAAAAAAAAAA-3’ |
| BDS | 5’-TTAATGCTAATTGATAGGGG-3’ | TP | 5’-CCCCTATCACGATTAGCATTAAAAAAAAAA-3’ |
*The mismatched or inserted bases are highlighted.
Figure 1.
Typical trace segments of (a) the completely-matched dsDNA; (b) one-mismatch dsDNA; and (c) two-mismatch dsDNA in the (Left) absence and (Right) presence of the nuclease. The experiments were performed at +120 mV with the wild-type αHL protein pore in 1 M NaCl solution buffered with 10 mM Tri•HCl (pH 7.5).
To investigate the effect of the applied voltage bias on nanopore detection of DNA mutations, the two dsDNA samples with one- and two- mismatches were further examined at +140 mV and +160 mV, respectively. The results were summarized in Supporting Information, Figs. S4 and S5. Similar to our observation made at +120 mV, the long duration (including permanent block) dsDNA events disappeared after the DNA samples were incubated with Surveyor Nuclease. In addition, we noticed that, in the absence of the nuclease, the ratio of the number of permanent block events over the number of long duration (from hundreds of milliseconds to 10 s) events decreased with an increase in the applied potential bias for both DNA samples. Specifically, as the voltage increased from +120 mV to +160 mV, the number of long duration events increased by ~ 4 folds and ~ 8 folds for the BP-LF1 and BP-LF2 samples, respectively, suggesting that high voltage could facilitate dsDNA unzipping. This is similar to previous reports53 in our laboratory. Although both linear and exponential (non-linear) correlations between the applied potential and the number of DNA events have been reported54–56, our experimental results (Supporting Information, Fig. S8) favored their exponential relationship, suggesting that dsDNA’s entrance into the αHL nanopore was the rate limiting step57,58. It should be mentioned that, the principle for nanopore detection of DNA mismatches is based on the disappearance of long-lived events due to the nuclease cleavage of the dsDNA substrate. Since a better event contrast could be obtained at an increased applied potential bias, voltage could be utilized as an important parameter to improve the nanopore sensor sensitivity in the detection of DNA mismatches.
Taken together, the combined results demonstrated that Surveyor Nuclease had no effect on the completely-matched dsDNA but could cleave both one-mismatch and two-mismatch dsDNA into shorter fragments, thus producing new types of events in the nanopore. Therefore, the nanopore sensor was indeed able to rapidly differentiate completely-matched dsDNA from mismatched dsDNA. In addition, by taking advantage of the blockage amplitudes (78.7% vs. 71.9% of full channel blockage) of the long-lived events of BP-LF1 and BP-LF2, we could readily tell the difference between one-mismatch dsDNA and two-mismatch dsDNA. One likely reason why one mismatch difference between the two dsDNA produced events with significantly different blockage amplitudes might be attributed to the different orientations in which the two dsDNA polymers entered the nanopore. It has been well documented that the event amplitude, residence time, and frequency were dependant on the orientation of the nucleic acids when they entered the nano-cavity.34
Terminal base-base substitution mismatch detection
Although various base-base substitution mismatch detection methods7–14 have been reported, developing a technique which is capable of terminal base-base substitution mismatch detection remains a challenging task. In the previous section, a ssDNA probe which could hybridize with the target ssDNA to form blunt-ended dsDNA was employed to detect base-base substitution mismatches. However, if the mismatch occurs at the terminal location, this blunt-ended dsDNA approach might not be successful. Since after the nuclease cleavage, one of the produced fragments was only one base pair shorter than the substrate dsDNA so that they may be difficult to be differentiated by the nanopore sensor. On the other hand, the other DNA fragment is too small to be detected by the nanopore due to its rapid translocation through the nano-channel (note that Surveyor Nuclease cleaves specifically any mismatch site in the DNA double strands at 3’-carbon side.52). To demonstrate the potential application of our nanopore sensor for detecting terminal base-base substitution mismatch, another strategy which uses a ssDNA probe to hybridize with the target DNA to form dsDNA with overhang was designed. Specifically, a 32-mer ssDNA molecule with a poly(A) tail (TP) was employed to analyze a 22-mer cancer biomarker DNA (TMS) using the mutant (M113F)7 αHL nanopore, which was obtained by mutating the amino acid residue methionine at position 113 of the wild-type αHL protein to phenylalanine. One criterion about whether a mutant or a wild-type αHL protein nanopore should be used for dsDNA analysis is the GC content of the target dsDNA. As observed in our previous study503 wild-type αHL pore was inefficient to unzip GC base pairs, so that it was generally only useful for analyzing dsDNA containing no or very low GC content or with short length. In contrast, the (M113F)7 αHL nanopore could facilitate unzipping of double stranded DNA, which led to a reduced probability for DNA to permanently block the nanochannel, and allowed investigating a variety of dsDNA molecules, including those with high GC content. However, although the (M113F)7 αHL nanopore could be utilized to investigate the base substitution system (i.e., LF, LF1, and LF2) in the previous section, its performance and resolution to differentiate the three DNA sequences (i.e., full match, one-mismatch, and two-mismatch) was not as good as that of the wild-type αHL pore due to the rapid unzipping of these dsDNA molecules and hence the produced small residence time events in the (M113F)7 αHL nanopore. It is worth mentioning that, another advantage of utilizing the (M113F)7 αHL nanopore instead of the wild-type αHL pore is that single-stranded DNA molecules showed more frequent events with a larger mean residence time in this engineered nanopore so that the sensor had a better resolution to ssDNA detection.59 Note that, in this terminal base-base substitution mismatch investigation, the translocation of the 10 base-long poly(A) tail produced after nuclease cleavage of the substrate dsDNA (TP-TMS) was utilized to monitor the nuclease cleavage events. Our experimental results (Fig. 2) showed that, in the absence of Surveyor Nuclease, although the TP-TMS mixture sample sometimes permanently blocked the nanopore, we did observe much more frequent long duration events than that of BP-LF1 in the wild-type αHL nanopore. These long-lived events could be further divided into two types: one type had a mean residence time of 179 ± 15 ms and a residual current of 6.0 ± 0.5 pA (i.e., 94 ± 0.5 % of full channel blockage), while the other presented a mean residence time of 42 ± 4 ms and a residual current of 9.2 ± 0.3 pA (91 ± 0.3 % of full channel blockage). These two types of long residence time events might be attributed to the two different orientations in which the dsDNA entered the nanopore, as reported by previous research.40 In contrast, in the presence of the nuclease, these long duration and large block amplitude events disappeared; instead, a new type of events with small residence time (1.00 ± 0.10 ms) but large amplitude (92 ± 0.8 % of full channel blockage) were observed (Fig. 2). Clearly, these new events were attributed to the nuclease cleavage of TP-TMS dsDNA.
Figure 2.
Nanopore detection of terminal base mismatch. (a) Without and (b) with Surveyor Nuclease. Amplitude in Fig. 2 was blockage residual current. The experiments were performed at +140 mV with the mutant (M113F)7 αHL pore in 1 M NaCl solution buffered with 10 mM Tri•HCl (pH 7.5).
DNA base insertion detection
As common as base substitution during DNA replication, base deletion and base insertion are two other kinds of mutations. To demonstrate that our nanopore sensing platform could not only be utilized to detect base substitution, but also is able to distinguish DNA base insertion or base deletion from completely-matched dsDNA, we further studied the interaction between a mixture of two ssDNA molecules (BDS with a sequence of 5’-TTAATGCTAATTGATAGGGG-3’ and TP with a sequence of 5’-CCCCTATCACGATTAGCATTAAAAAAAAAA-3’) and the (M113F)7 αHL nanopore in the absence/presence of Surveyor Nuclease. Note that these two ssDNA molecules could form a 20-bp completely matched dsDNA with two extra bases (which were highlighted) inserted in the middle region. The experimental results were summarized in Fig. 3. Similar to the observation we made in the nuclease digestion of TP-TMS in the previous “terminal base-base substitution mismatch detection” section, in the absence of the nuclease, in addition to the short-lived events, two types of long duration events were clearly identified. One type of events had a mean residence time of 83.2 ± 9 ms and a residual current of 4.6 ± 0.3 pA, while the other showed a mean residence time of 770 ± 58 ms and a residual current of 30.1 ± 0.6 pA. Again, these events should be attributed to the two different orientations in which the dsDNA entered the nanopore. In contrast, in the presence of the nuclease, the long duration events disappeared, and a new type of events with much smaller residence time (τoff = 13.6 ± 0.9 ms) could be observed, suggesting that the DNA mixture sample could be cleaved by the nuclease, so that the two ssDNA molecules were not completely matched.
Figure 3.
Nanopore detection of DNA insertion/deletion. (a) Without; and (b) with Surveyor Nuclease. Amplitude in Fig. 3 was blockage residual current. The experiments were performed at +140 mV with the mutant (M113F)7 αHL protein pore in 1 M NaCl solution buffered with 10 mM Tri•HCl (pH 7.5).
Detection limit of the base-base substitution detection
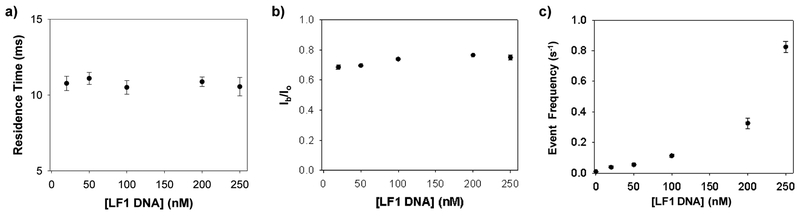
As a proof-of-principle purpose, detection of point mutation (one mismatch) based on the formation of blunt-ended dsDNA was utilized as a model system in this investigation. Under the commonly used symmetric electrolyte condition with 1 M NaCl in both the cis and trans compartments of the nanopore sensing chamber, LF1 could be detected at as low as ~ 50 nM (data not shown). To improve the sensitivity of the nanopore sensor for analysis of DNA biomarkers in human serum / blood (note that their concentrations in healthy people are normally in the range from few nanomolar to tens of nanomolar60,61), nanopore detection of LF1 was further carried out in a salt gradient. It has been well documented that using a salt gradient instead of a symmetric electrolyte buffer condition could significantly increase the event frequency for the translocation of nucleic acid molecules through a nanopore, thus improving the detection limit of the nanopore sensor.62 Briefly, the cis chamber compartment was filled with an electrolyte buffer solution consisting of 0.5 M NaCl and 5 mM Tris-HCl (pH 7.5), while a solution of 3 M NaCl and 10 mM Tris-HCl (pH 7.5) was added to the trans compartment. The concentration of BP DNA was 250 nM, while the concentrations of LF1 ssDNA ranged from 20 nM to 250 nM. The mixture solutions of LF1 DNA and BP DNA were incubated in the presence of 10 μL Surveyor nuclease, 10 μL Surveyor enhancer, and 3 μL MgCl2 for 2h at 42 °C before added to the protein nanopore for electrical recording at +120 mV. Our experimental results (Figs. 4a and 4b) showed that both the mean residence time and blockage amplitude of the new type of events (i.e., attributed to the BP-LF1 cleavage products) were unvaried with the changing concentration of the LF1 DNA. Therefore, residence time and amplitude could be used as signatures for identifying LF1. Our experiments (Fig. 4c) also demonstrated that the frequency of the new events increased exponentially with the increase in the concentration of the LF1 ssDNA, suggesting that, in addition to diffusion and electrophoretic effect, the interaction between the dsDNA molecules and the αHL nanopore might play a significant role in the DNA capture rate63. The detection limit of this sensor system (defined as the concentration corresponding to three times the standard deviation of a blank signal) in a 10-minute recording period was ~4.8 nM. Although the sensitivity of the nanopore sensor operated under our investigated experimental conditions was not very impressive, it is expected that the detection limit for point mutation could be significantly improved by using a larger salt gradient (e.g., 0.15 M NaCl (cis) / 3 M NaCl (trans)) and with a greater applied voltage (e.g., + 180 mV), as documented in our previous studies.40,64
Figure 4.
Effect of DNA concentration on the characteristics of current blockage events. Plot of (a) residence time, (b) blockage amplitude, and (c) event frequency as a function of LF1 DNA concentration, showing that both the event mean residence time and amplitude were unvaried with the changing concentration of added DNA, while the event frequency increased with increasing DNA concentration. Ib/Io in Figure 4b is normalized blockage current, which was obtained by dividing the average blockage amplitude of an event by the average open channel current. The experiments were performed with the wild-type αHL protein pore at +120 mV in the presence of 250 nM BP DNA. An asymmetric buffer condition (with 3 M NaCl and 10 mM Tris-HCl (pH 7.5) in the trans compartment and 0.5 M NaCl and 10 mM Tris-HCl (pH 7.5) in the cis compartment) was used. The events with residence time less than 3 ms were not included in the data analysis to minimize the potential interference from the short-lived events attributed to the brief residency of DNA molecules in the vestibule or their collision with the opening of the αHL pore.
Detection of base-base substitutions in serum
To proof-of-concept demonstrate the potential application of the developed nanopore sensing platform as a useful tool for clinical analysis of DNA mutations, simulated serum samples, which were prepared based on the base-base substitution system as described in the previous section, were examined. Briefly, the mixture of LF DNA and BP DNA as well as the mixture of LF1 DNA and BP DNA were spiked into the rabbit serum, which were then analyzed by the wild-type αHL protein nanopore sensor in the absence and in the presence of Surveyor Nuclease at +120 mV. The experimental results were summarized in Fig. 5 and Supporting Information, Fig. S9. Similar to the observation we made in the previous sections (i.e., without serum), the completely-matched dsDNA / serum mixture sample blocked the nanopore most of the time both in the absence and in the presence of the nuclease (Fig. S9), while the one-mismatch dsDNA / serum mixture sample often permanently blocked the channel in the absence of the enzyme but produced short duration events after addition of the nuclease to the mixture sample (note that the possibility that these frequent long duration events were attributed to the serum was ruled out by the control experiment, in which the serum only blocked the nanopore occasionally). Furthermore, interestingly, we noticed that, long residence time events were still able to be observed in the one-mismatch dsDNA / serum mixture sample even when 10 μL nuclease was added to the solution (in comparison, with 10 μL nuclease, all of these long-lived events disappeared in the absence of serum). One possible interpretation is that the nuclease might have a reduced activity in the serum medium. This interpretation is supported by another experiment, in which all of the long duration events disappeared, and significantly more frequent new type of short duration events could be observed when the amount of added nuclease was increased to 20 μL (Fig. 5).
Figure 5.
Nanopore analysis of one-mismatch dsDNA in serum. (a) 0; (b) 10 μL; and (c) 20 μL Surveyor Nuclease. The experiments were performed at +120 mV with the wild-type αHL protein pore in 1 M NaCl solution buffered with 10 mM Tri•HCl (pH 7.5).
Conclusions
In summary, by monitoring the interaction between a nanopore and a DNA mixture sample (containing a DNA analyte and a DNA probe) in the absence and in the presence of a nuclease, a highly sensitive nanopore biosensor for DNA mutation detection was developed. Our method took advantage of the ability of nuclease to cleave mismatched DNA into short fragments, allowing analysis of longer DNA sequences than the well-documented hybridization-based nanopore nucleic acid assay did. Unlike various traditional detection techniques, our nanopore sensor was rapid (10 min assay), inexpensive, and does not require the use of labels. Furthermore, due to the excellent mismatch recognition capability of the nuclease, our method could be utilized to detect various types of dsDNA mutations, including base substitution, deletion, and insertion. It should be noted that, with the wild-type αHL protein as the sensing element and using a ssDNA probe to hybridize with the target ssDNA to form blunt-ended dsDNA, our nanopore sensor can analyze DNA with ~20 bases in length. Longer DNA samples can be investigated if an engineered αHL protein nanopore and/or a ssDNA probe which can react with the target ssDNA to form overhang dsDNA is used.53 It should be noted that, our developed enzymatic reaction-based nanopore DNA mismatch detection strategy can be coupled with other genome-targeting technologies such as CRISPR/Cas9 systems,65,66 which produce RNA-guided site-specific DNA cleavage, to investigate a variety of other genetic related diseases. For example, in spite of the DNA length limitation (~20 bases), our nanopore sensor could be utilized to investigate Huntington disease, which consists of an abnormal expansion of a CAG repeat (e.g., 36 or more repeats) in the genetic code. When analyzing such a ssDNA sample (> 100 bases), we can divide the entire gene into many segments of ~20 bases long and design their corresponding ssDNA probes, with their hybridization mixtures analyzed by the nanopore sensor sequentially or using an array of nanopores simultaneously.67
Supplementary Material
ACKNOWLEDGMENT
This work was financially supported by the National Institutes of Health (2R15GM110632–02) and National Science Foundation (1708596).
Footnotes
ASSOCIATE CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Additional figures, including procedure flowchart of DNA sample pretreatment, translocation of ssDNA in the wild-type αHL pore, effect of DNA length on the event residence time, effects of the applied potential bias on nanopore detection of one-mismatch dsDNA and two-mismatch dsDNA, 3-D plots of event counts vs. blockage amplitude vs. residence time of three dsDNA cleavage fragments, analysis of dsDNA digestion products by agarose gel electrophoresis and nanopore analysis, effect of applied potential on the frequency of dsDNA events, and nanopore analysis of complementary-match dsDNA in serum.
Notes
The authors declare no competing financial interests.
References
- 1.Botezatu IV; Panchuk IO; Stroganova AM; Senderovich AI; Kondratova VN; Shelepov VP; Lichtenstein AV TaqMan Probes as Blocking Agents for Enriched PCR Amplification and DNA Melting Analysis of Mutant Genes. BioTechniques, 2017, 62, 62–68. [DOI] [PubMed] [Google Scholar]
- 2.Sho S; Court CM; Kim S; Braxton DR; Hou S; Muthusamy VR; Watson RR; Sedarat A; Tseng HR; Tomlinson JS Digital PCR Improves Mutation Analysis in Pancreas Fine Needle Aspiration Biopsy Specimens. PloS One, 2017, 12: e0170897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Su X; Li L; Wang S; Hao D; Wang L; Yu C Single-Molecule Counting of Point Mutations by Transient DNA Binding. Sci. Rep, 2017, 7, 43824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stancescu M; Fedotova TA; Hooyberghs J; Balaeff A; Kolpashchikov DM Nonequilibrium Hybridization Enables Discrimination of a Point Mutation within 5–40 °C. J. Am. Chem. Soc, 2016, 138, 13465–13468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu Y; Wu T; Johnson-Buck A; Li L; Su X A Two-Layer Assay for Single-Nucleotide Variants Utilizing Strand Displacement and Selective Digestion. Biosens. Bioelectron, 2016, 82, 248–254. [DOI] [PubMed] [Google Scholar]
- 6.Cai Z; Chen Y; Lin C; Wu Y; Yang CJ; Wang Y; Chen X A Dual-signal Amplification Method for the DNA Detection Based on Exonuclease III. Biosens. Bioelectron, 2014, 61, 370–373. [DOI] [PubMed] [Google Scholar]
- 7.Toren P; Ozgur E; Bayindir M Real-Time and Selective Detection of Single Nucleotide DNA Mutations Using Surface Engineered Microtoroids. Anal. Chem, 2015, 87, 10920–10926. [DOI] [PubMed] [Google Scholar]
- 8.Wu WH; Zhu KD Proposition of a Silica Nanoparticle-Enhanced Hybrid Spin-Microcantilever Sensor Using Nonlinear Optics for Detection of DNA in Liquid. Sensors, 2015, 15, 24848–24861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MacConaghy KI; Chadly DM; Stoykovich MP; Kaar JL Label-Free Detection of Missense Mutations and Methylation Differences in the p53 Gene Using Optically Diffracting Hydrogels. The Analyst, 2015, 140, 6354–6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song Y; Zhang Y; Wang TH Single Quantum Dot Analysis Enables Multiplexed Point Mutation Detection by Gap Ligase Chain Reaction. Small Weinh. Bergstr. Ger, 2013, 9, 1096–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye M; Zhang Y; Li H; Zhang Y; Tan P; Tang H; Yao S A Novel Method for the Detection of Point Mutation in DNA Using Single-Base-Coded CdS Nanopores. Biosens. Bioelectron, 2009, 24, 2339–2345. [DOI] [PubMed] [Google Scholar]
- 12.Pang L; Li J; Jiang J; Shen G; Yu R DNA Point Mutation Detection Based on DNA Ligase Reaction and Nano-Au Amplification: A Piezoelectric Approach. Anal. Biochem, 2006, 358, 99–103. [DOI] [PubMed] [Google Scholar]
- 13.Benvidi A; Firouzabadi AD; Moshtaghiun SM; Mazloum-Ardakani M; Tezerjani MD Ultrasensitive DNA Sensor Based on Gold Nanoparticles/Reduced Graphene Oxide/Glassy Carbon Eelectrode. Anal. Biochem, 2015, 484, 24–30. [DOI] [PubMed] [Google Scholar]
- 14.Skotadis E; Voutyras K; Chatzipetrou M; Tsekenis G; Patsiouras L; Madianos L; Chatzandroulis S; Zergioti I; Tsoukalas D Label-Free DNA Biosensor Based on Resistance Change of Platinum Nanoparticles Assemblies. Biosens. Bioelectron, 2016, 81, 388–394. [DOI] [PubMed] [Google Scholar]
- 15.Liu L; Wu HC DNA-Based Nanopore Sensing. Angew. Chem. Int. Ed Engl, 2016, 55, 15216–15222. [DOI] [PubMed] [Google Scholar]
- 16.Schneider G, F.; Dekker. C. DNA Sequence with Nanopores. Nat Biotechnol, 2012, 30, 326–328. [DOI] [PubMed] [Google Scholar]
- 17.Wang G; Wang L; Han Y; Zhou S; Guan X Nanopore Stochastic Detection: Diversity, Sensitivity, and Beyond. Acc. Chem. Res, 2013, 46, 2867–2877. [DOI] [PubMed] [Google Scholar]
- 18.Haque F; Li J; Wu HC; Liang XJ; Guo P Solid-State and Biological Nanopore for Real-Time Sensing of Single Chemical and Sequencing of DNA. Nano Today, 2013, 8, 56–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu LQ; Wanunu M; Wang MX; McReynolds L; Wang Y Detection of miRNAs with a Nanopore Single-Molecule Counter. Expert Rev. Mol. Diagn, 2012, 12, 573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mereuta L; Schiopu I;Asandei A; Park Y; Hahm KS; Luchian T Protein Nanopore-Based, Single-Molecule Exploration of Copper Binding to an Antimicrobial-derived, Histidine-Containing Chimera Peptide. Langmuir ACS J. Surf. Colloids, 2012, 28, 17079–17091. [DOI] [PubMed] [Google Scholar]
- 21.Perera RT; Fleming AM; Johnson RP; Burrows CJ; White HS Detection of Benzo[α]pyrene-Guanine Adducts in Single-Stranded DNA Using the α-Hemolysin Nanopore. Nanotechnology, 2015, 26, 074002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han Y; Zhou S; Wang L; Guan X Nanopore Back Titration Analysis of Dipicolinic Acid. Electrophoresis, 2015, 36, 467–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guan X; Gu LQ; Cheley S; Braha O; Bayley H Stochastic Sensing of TNT with a Genetically Engineered Pore. Chembiochem Eur. J. Chem. Biol, 2005, 6, 1875–1881. [DOI] [PubMed] [Google Scholar]
- 24.Gao C; Ding S; Tan Q; Gu LQ Method of Creating a Nanopore-Terminated Probe for Single-Molecule Enantiomer Discrimination. Anal. Chem, 2009, 81, 80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X; Price NE; Fang X; Yang Z; Gu LQ; Gates KS Characterization of Interstrand DNA-DNA Cross-Links Using the α-Hemolysin Protein Nanopore. ACS Nano, 2015, 9, 11812–11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tavassoly O; Kakish J; Nokhrin S; Dmitriev O; Lee JS The Use of Nanopore Analysis for Discovering Drugs Which Bind to α-Synuclein for Treatment of Parkinson’s Disease. Eur. J. Med. Chem, 2014, 88, 42–54. [DOI] [PubMed] [Google Scholar]
- 27.Wang G; Wang L; Han Y; Zhou S; Guan X Nanopore Detection of Copper Ions Using a Polyhistidine Probe. Biosens. Bioelectron, 2014, 53, 453–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou S Wang L; Chen X; Guan X Label-Free Nanopore Single-Molecule Measurement of Trypsin Activity. ACS Sens, 2016, 1, 607–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu LQ; Cheley S; Bayley H Capture of a Single Molecule in a Nanocavity. Science 2001, 291, 636–640. [DOI] [PubMed] [Google Scholar]
- 30.Capone R; Blake S; Restrepo MR; Yang J; Mayer M Designing Nanosensors Based on Charged Derivatives of Gramicidin A. J. Am. Chem. Soc, 2007, 129, 9737–9745. [DOI] [PubMed] [Google Scholar]
- 31.Manrao EA; Derrington IM; Laszlo AH; Langford KW; Hopper MK; Gillgren N; Pavlenok M; Niederweis M; Gundlach JH Reading DNA at Single-Nucleotide Resolution with a Mutant Mspa Nanopore and phi29 DNA Polymerase. Nat. Biotechnol 2012, 30, 349–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ying YL; Wang HY; Sutherland TC; Long YT Monitoring of an ATP-Binding Aptamer and its Conformational Changes using an α-hemolysin Nanopore. Small. 2011, 7, 87–94. [DOI] [PubMed] [Google Scholar]
- 33.Zhang X; Zhang JJ; Ying YL; Tian H; Long YT Molecule Analysis of Light-Regulated RNA:Spiropyran Interactions. Chem. Sci 2014, 5, 2642–2646. [Google Scholar]
- 34.Meng FN; Li ZY; Ying YL; Liu SC; Zhang J; Long YT Structural Stability of the Photo-Responsive DNA Duplexes Containing one Azobenzene via a Confined Pore. Chem. Commun 2017, 53, 9462–9465. [DOI] [PubMed] [Google Scholar]
- 35.Li J; Stein D; McMullan C; Branton D; Aziz MJ; Golovchenko JA Ion-Beam Sculpting at Nanometre Length Scales. Nature 2001, 412, 166–169. [DOI] [PubMed] [Google Scholar]
- 36.Heins EA; Siwy ZS; Baker LA; Martin CR Detecting Single Porphyrin Molecules in a Conically Shaped Synthetic Nanopore. Nano Lett. 2005, 5, 1824–1829. [DOI] [PubMed] [Google Scholar]
- 37.Cecchini MP; Wiener A; Turek VA; Chon H; Lee S; Ivanov AP; McComb DW; Choo J; Albrecht T; Maier SA; Edel JB Rapid Ultrasensitive Single Particle Surface-enhanced Raman Spectroscopy using Metallic Nanopores. Nano Lett. 2013, 13, 4602–4609. [DOI] [PubMed] [Google Scholar]
- 38.McNally B; Singer A; Yu Z; Sun Y; Weng Z; Meller A Optical Recognition of Converted DNA Nucleotides for Single-molecule DNA Sequencing using Nanopore Arrays. Nano Lett. 2010, 10, 2237–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Belkin M; Chao SH; Jonsson MP; Dekker C; Aksimentiev A Plasmonic Nanopores for Trapping, Controlling Displacement, and Sequencing of DNA. ACS Nano 2015, 9, 10598–10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang L; Han Y; Zhou S; Wang G; Guan X Nanopore Biosensor for Label-Free and Real-Time Detection of Anthrax Lethal Factor. ACS Appl. Mater. Interfaces, 2014, 6, 7334–7339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao F; Zhang Y; Wei Y; Kang X A Rapid and Sensitive Detection of HBV DNA Using a Nanopore Sensor. Chem. Commun. Camb. Engl, 2014, 50, 13853–13856. [DOI] [PubMed] [Google Scholar]
- 42.Zhang X; Wang Y; Fricke BL; Gu LQ Programming Nanopore Ion Flow for Encoded Multiplex MicroRNA Detection. ACS Nano, 2014, 8, 3444–3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carson S; Wanunu M Challenges in DNA Motion Control and Sequence Readout Using Nanopore Devices. Nanotechnology, 2015, 26, 074004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ying YL; Zhang J; Gao R; Long YT Nanopore-Based Sequencing and Detection of Nucleic Acids. Angew. Chem. Int. Ed Engl, 2013, 52, 13154–13161. [DOI] [PubMed] [Google Scholar]
- 45.Venkates BM; Bashir R Nanopore Sensors for Nucleic Acid Analysis. Nat. Nanotechnol, 2011, 6, 615–624. [DOI] [PubMed] [Google Scholar]
- 46.Tian K; He Z; Wang Y; Chen SJ; Gu LQ Designing a Polycationic Probe for Simultaneous Enrichment and Detection of MicroRNAs in a Nanopore. ACS Nano 2013, 7, 3962–3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gite SU; Shankar V Single-Strand-Specific Nucleases. Crit. Rev. Microbiol, 1995, 21, 101–122. [DOI] [PubMed] [Google Scholar]
- 48.Qiu P; Shandilya H; D’Alessio JM; O’Connor K; Durocher J; Gerard GF Mutation Detection Using Surveyor Nuclease. BioTechniques, 2004, 36, 702–707. [DOI] [PubMed] [Google Scholar]
- 49.Pilch J; Asman M; Jamroz E; Kajor M; Kotrys-Puchalska E; Goss M; Krzak M; Witecka J; Gmiński J; Sieroń AL Surveyor Nuclease Detection of Mutations and Polymorphisms of mtDNA in Children. J. Pediatr. Neurol, 2010, 43, 325–330. [DOI] [PubMed] [Google Scholar]
- 50.Desai NA; Shankar V Single-Strand-Specific Nucleases. FEMS Microbiol. Rev, 2003, 26, 457–491. [DOI] [PubMed] [Google Scholar]
- 51.Till BJ; Burtner C; Comai L; Henikoff S Mismatch Cleavage by Single-Strand Specific Nucleases. Nucleic Acids Res, 2004, 32, 2632–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oleykowski CA; Bronson Mullins CR; Godwin AK; Yeung AT Mutation Detection Using a Novel Plant Endonuclease. Nucleic Acids Res, 1998, 26, 4597–4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu A; Zhao Q; Krishantha DMM; Guan X Unzipping of Double-Stranded DNA in Engineered α-Hemolysin Probes. J. Phys. Chem. Lett, 2011, 2, 1372–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bell NA; Muthukumar M; Keyser UF. Translocation Frequency of Double-Stranded DNA through a Solid-State Nanopore. Phys Rev E. 2016, 93, 022401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maglia G; Restrepo MR; Mikhailova E; Bayley H Enhanced translocation of single DNA molecules through alpha-hemolysin nanopores by manipulation of internal charge. Proc. Natl. Acad. Sci. U S A. 2008, 105, 19720–19725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Q; Wu H; Wu L; Xie X; Kong J; Ye X; Liu L Voltage-driven translocation of DNA through a high throughput conical solid-state nanopore. PLoS One 2012, 7, e46014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meller A; Branton D Single Molecule Measurements of DNA Transport through a Nanopore. Electrophoresis 2002, 23, 2583–2591. [DOI] [PubMed] [Google Scholar]
- 58.Henrickson SE; Misakian M; Robertson B; Kasianowicz JJ Driven DNA Transport into an Asymmetric Nanometer-Scale Pore. Phys. Rev. Lett 2000, 85, 3057–3060. [DOI] [PubMed] [Google Scholar]
- 59.Zhao Q; Jayawardhana DA; Wang; D.; Guan, X. Study of Peptide Transport through Engineered Protein Channels. J. Phys. Chem. B, 2009, 113, 3572–3578. [DOI] [PubMed] [Google Scholar]
- 60.Schwarzenbach H; Pantel K Circulating DNA as Biomarker in Breast Cancer. Breast Cancer Res. 2015, 17, 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Elshimali YI; Khaddour H; Sarkissyan M; Wu Y; Vadgama JV The Clinical Utilization of Circulating Cell Free DNA (CCFDNA) in Blood of Cancer Patients. Int. J. Mol. Sci 2013, 14, 18925–18958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wanunu M; Morrison W; Rabin Y; Grosberg AY; Meller A Electrostatic Focusing of Unlabelled DNA into Nanoscale Pores Using a Salt Gradient. Nat. Nanotechnol, 2010, 5, 160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sen YH; Karnik R Investigating the Translocation of λ-DNA Molecules through PDMS Nanopores. Anal. Bioanal. Chem 2009, 394, 437–446. [DOI] [PubMed] [Google Scholar]
- 64.Roozbahani GM; Chen X; Zhang Y; Xie R; Ma R; Li D; Li H; Guan X Peptide-Mediated Nanopore Detection of Uranyl Ions in Aqueous Media. ACS Sens. 2017, 2, 703–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cong L; Ran FA; Cox D; Lin S; Barretto R; Habib N; Hsu PD; Wu X; Jiang W; Marraffini LA; Zhang F Multiplex Genome Engineering using CRISPR/Cas Systems. Science 2013, 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mali P; Yang L; Esvelt KM; Aach J; Guell M; DiCarlo JE; Norville JE; Church GM RNA-guided Human Genome Engineering via Cas9. Science 2013, 339, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao Q; Wang D; Jayawardhana DA; Guan X Stochastic Sensing of Biomolecules in a Nanopore Sensor Array. Nanotechnology 2008, 19, 505504. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.