Long-term persistence, not ongoing virus replication, is primarily responsible for maintaining HIV during antiretroviral therapy.
Abstract
HIV persistence during combination antiretroviral therapy (cART) is the principal obstacle to cure. Mechanisms responsible for persistence remain uncertain; infections may be maintained by persistence and clonal expansion of infected cells or by ongoing replication in anatomic locations with poor antiretroviral penetration. These mechanisms require different strategies for eradication, and determining their contributions to HIV persistence is essential. We used phylogenetic approaches to investigate, at the DNA level, HIV populations in blood, lymphoid, and other infected tissues obtained at colonoscopy or autopsy in individuals who were on cART for 8 to 16 years. We found no evidence of ongoing replication or compartmentalization of HIV; we did detect clonal expansion of infected cells that were present before cART. Long-term persistence, and not ongoing replication, is primarily responsible for maintaining HIV. HIV-infected cells present when cART is initiated represent the only identifiable source of persistence and is the appropriate focus for eradication.
INTRODUCTION
Combination antiretroviral therapy (cART) suppresses, but does not eradicate, HIV infections. Reports of successful HIV cure after bone marrow transplant and reports of long-term suppression of HIV following discontinuation of cART in some individuals show that both eradication and functional control of HIV are possible (1–5). The fact that these are rare events that were not observed in others undergoing bone marrow transplant (6–8) means that improving the outcomes for the majority of treated individuals will require a better understanding of the sources and mechanisms of HIV persistence during therapy. Two distinct mechanisms have been proposed to explain how the population of cells containing replication-competent HIV, denoted the HIV reservoir, are maintained in individuals undergoing successful cART: long-term persistence and clonal expansion of HIV-infected cells, and, alternatively, ongoing cycles of active HIV infection that continuously replenish the HIV reservoir. We and others reported, in those on cART, that HIV RNA in virus populations present in plasma or DNA populations obtained from peripheral blood mononuclear cells (PBMCs) do not diverge from samples obtained prior to therapy, suggesting that the persistence of long-lived infected cells, and not ongoing replication, is primarily responsible for maintenance of the reservoir (9–17). In addition, we and others demonstrated that clonal expansion of HIV-infected cells plays a major role in generating the HIV DNA populations that are present in those undergoing cART (18–22). In contrast, recent reports that analyzed samples from HIV-infected individuals, including those from lymph node, gut-associated lymphoid tissue (GALT), and cerebrospinal fluid suggested that certain anatomic locations could act as sanctuaries for HIV replication, at least in some individuals (23–26). Lorenzo-Redondo et al. (24) recently reported finding genetic changes in HIV proviruses [proviruses are the integrated form of retrovirus DNA (27)] in the 6 months following the initiation of cART, leading to the generation of populations of HIV proviruses in tissues that were distinct from the population of HIV proviruses in PBMCs. That report and others (23, 25, 28, 29) suggested that HIV replication that was restricted to tissue sanctuary sites resulted in the generation of markedly different populations of HIV proviruses in blood and tissues after prolonged cART. In contrast, if the reservoir consists of cells (or their descendants) that were infected before cART was initiated, there will be no additional genetic changes in the populations of proviruses during cART. These two proposed mechanisms of HIV persistence would necessitate fundamentally different HIV eradication strategies: Ongoing HIV replication in sanctuary sites would require improved cART. In contrast, if cART completely blocks HIV replication, the problem that needs to be addressed is the survival and clonal expansion of cells that were infected before cART was initiated.
We conducted a critical test of the question of whether ongoing HIV replication persists in sanctuary locations by analyzing HIV populations in blood and tissues from individuals after prolonged cART (8 to 16 years). In untreated individuals, HIV replication is rapid and error prone (27), leading to the accumulation of genetic changes that are readily detectable in the proviruses. Here, we found that infected cells persist and clonally expand, but there was no detectable evidence of HIV molecular evolution nor was there any evidence of compartmentalization of specific HIV proviruses in blood or tissues in six individuals who were studied during effective cART for prolonged periods, cumulatively totaling over 60 patient-years of follow-up.
To compare HIV RNA and DNA populations in blood cells and tissues, we first identified individuals from whom samples were available for analysis after prolonged cART (table S1). We chose to study individuals who initiated cART early after infection (AVBIO2-15, AVBIO2-04, and HAMB-1; table S1 and see the Supplementary Materials for clinical characteristics) and had tissue samples from colonoscopy (AVBIO2-15, AVBIO2-04, ileum, and colon) or autopsy (HAMB-1) after years of suppressive cART. The HIV DNA and RNA populations in individuals who initiate cART early after infection are genetically homogeneous, facilitating the identification of any genetic changes that arise during long-term cART. In addition, we also studied chronically infected individuals from whom we obtained samples from autopsy (HAMB-2) or routine research colonoscopy (AVBIO2-35 and AVBIO2-37; see the Supplementary Materials for clinical characteristics). All of the participants in this study were male with a median age of 40 years at diagnosis who had undergone cART for a median of 17.8 years (range, 8 to 22.7 years) at the time tissue samples were obtained.
To determine whether there was detectable molecular evolution of HIV in tissues during cART, we analyzed single HIV RNA and DNA genomes recovered from total PBMCs and tissues to determine whether detectable genetic changes occurred during cART. We first determined the levels of HIV RNA in plasma and HIV DNA in PBMCs and tissues. HIV RNA was present in plasma, and the level declined after cART was initiated in all individuals (Figs. 1 to 3). HIV DNA was detectable in PBMCs and tissue samples in all participants after prolonged cART (table S2). Consistent with previous reports (30, 31), individuals who initiated cART early after infection (AVBIO2-15, AVBIO2-04, and HAMB-1) generally had lower levels of cell-associated HIV DNA (median, 54; range, <1 to 90 HIV DNA copies/106 cells) in their tissues compared with HIV DNA levels in their PBMCs (median, 195.3; range, 97 to 270.1 HIV DNA copies/106 PBMCs). The highest levels of HIV DNA were present in lymphoid tissue, including GALT and lymph node, while the lowest levels of HIV were detected in the central nervous system (table S2). Individuals with AIDS who initiated cART during chronic HIV infection after years of infection and progressive immunodeficiency had higher levels of HIV DNA, which could be detected in all tissues, including the frontal and occipital lobes of the central nervous system (PBMCs: median, 779; tissues: median, 96 HIV DNA copies/106 cells; table S2). The relatively small number of HIV proviruses in tissues from the individuals who were treated early in infection precluded the use of next generation sequencing approaches for the analysis of the HIV DNA sequences (32, 33). To ask whether there was detectable genetic change over time, we obtained single-genome sequences (SGSs) of the pro-pol region of HIV; we and others have previously demonstrated that, in the absence of cART, genetic changes reproducibly accumulate in this region of the genome over periods of 1 to 3 years (34, 35). We also investigated HIV RNA sequences derived from virions present in the plasma prior to cART. In some cases, we were also able to recover HIV RNA sequences from the low level (<50 c/ml) of virus present in blood during cART.
Fig. 1. No evidence of HIV molecular evolution in the colon or ileum during suppressive ART in individuals undergoing initiated therapy early after HIV infection (AVBIO2-15 and AVBIO2-04).
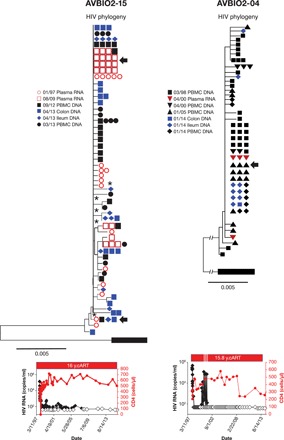
HIV-infected study participants underwent antiretroviral therapy within months of HIV infection (see the Supplementary Materials), and response to cART is indicated in plots of CD4 and viral RNA levels. HIV was sampled from plasma and PBMCs prior to and during cART, and at the time of routine colonoscopy after prolonged ART. AVBIO2-04 underwent several treatment interruptions, as indicated by short increases in viral RNA. HIV pro-pol sequences were obtained and then aligned; neighbor-joining phylogenetic trees were constructed; and bootstrap support >75 is indicated (*). Hypermutated sequences were removed, and identical sequences were grouped for ease of presentation. ycART, years of cART.
Two individuals who were treated early underwent colonoscopy (Fig. 1, AVBIO2-15 and AVBIO2-04) after 8 to 16 years of cART, and one individual (HAMB-1) underwent autopsy after 8 years of cART. Pretherapy HIV populations were analyzed using sequences of HIV RNA from plasma (AVBIO2-15) and PBMC-derived HIV DNA (AVBIO2-04). For HAMB-1, PBMC-derived DNA samples were obtained at years 6 and 8 of cART, and HIV DNA sequences were determined from tissues obtained at autopsy. Baseline populations from the study participants who initiated cART early after the initial infection had low genetic diversity (Table 1), with sequences displaying 0.04 to 0.46% average pairwise difference. This result is similar to prior reports (36, 37) that showed that there was limited genetic diversity in HIV genomic RNA sequences obtained from individuals treated early after infection. We performed phylogenetic analysis of these HIV sequences (Figs. 1 to 3). As expected, pretherapy samples from AVBIO2-15, AVBIO2-04, and HAMB-1 yielded numerous identical pro-pol sequences, consistent with infection by one or perhaps two HIV variants (36, 37). These variants persisted after prolonged cART, as demonstrated by the sequences of HIV RNA derived from plasma, from HIV DNA derived from PBMCs, and from all of the tissues we sampled at colonoscopy and autopsy, including colon, ileum, lung, lymph node, spleen, and jejunum (Figs. 1 to 3, arrow). We used root-to-tip phylogenetic analysis (Fig. 4), an approach that quantifies genetic changes that have accumulated in the populations of HIV RNA and DNA sequences (11). Using a consensus HIV sequence obtained from pretherapy plasma HIV (AVBIO2-15) or PBMC-derived HIV (AVBIO2-04) as the baseline root, we calculated the genetic distance to sequences obtained at subsequent time points. If there were genetic changes, then the genetic distances from this baseline sequence to the sequences obtained later (root to tip) would increase with time. As shown in Fig. 4, there was no increase in the root-to-tip distances for HIV DNA sequences from PBMC, colon, or ileum samples obtained following prolonged cART. Similarly, for HAMB-1, using a PBMC-derived consensus HIV sequence obtained after 6 years of therapy as the baseline root, we found no increase in root-to-tip distances in any of the HIV DNA sequences obtained from the PBMC samples taken before autopsy or from any of the tissue samples taken at autopsy (Fig. 4). These data show that there is no evidence of sequence evolution, which implies that there is little or no ongoing replication in any of the locations we sampled. Instead, it is likely that the HIV DNA sequences are from cells infected prior to the initiation of cART or are from cells infected prior to cART that have clonally expanded (18, 19, 21). The low genetic diversity of the HIV DNA in individuals treated early in infection means that many of the sequences are so similar that we cannot use sequence differences to infer clonal expansion of the infected cells. In some cells, HIV DNA may undergo extensive G-to-A changes during replication as the result of the APOBEC (apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like) innate immune defense system, yielding hypermutated proviruses; patterns of hypermutation are specific for individual cells, and recovering multiple sequences of proviral DNA with identical hypermutation patterns is evidence of clonal expansion. Analysis of hypermutants revealed that there was clonal expansion of some of the HIV-infected cells in these individuals (fig. S1). For instance, we identified a hypermutant sequence present in PBMCs in AVBIO2-04 prior to cART and subsequently identified 42 copies of the identical hypermutant in both ileum- and colon-derived tissues obtained 16 years later (fig. S1), demonstrating that there are HIV-infected cells that can persist and proliferate during therapy in individuals who initiated cART shortly after infection, including in areas of GALT with distinct immune functions, including ileum with lymphoid follicles and an inductive immune function and colon with predominantly effector immune functions.
Table 1. No evidence of molecular evolution during effective cART.
PID, participant ID.
| PID |
ART duration (years) |
Source |
Total SGSs recovered |
Hypermutants |
Total SGS hypermutants |
Average pairwise difference (%) |
Mean root-to-tip distance |
| AVBIO2-15 | 0.0 | Plasma | 17 | 0 | 17 | 0.2 | 0.001 |
| 11.7 | Plasma | 21 | 0 | 21 | 0.11 | 0.001 | |
| 14.8 | PBMC | 17 | 0 | 17 | 0.18 | 0.001 | |
| 15.3 | PBMC | 14 | 1 | 13 | 0.22 | 0.001 | |
| 15.3 | Colon | 19 | 1 | 18 | 0.29 | 0.001 | |
| 15.3 | Ileum | 12 | 0 | 12 | 0.24 | 0.001 | |
| AVBIO2-04 | 0.0 | PBMC | 30 | 2 | 28 | 0.12 | 0.001 |
| 20 | PBMC | 6 | 1 | 5 | 0.0005 | 0.0005 | |
| 2.1 | Plasma | 4 | 0 | 4 | 0.0004 | 0.0002 | |
| 6.8 | PBMC | 17 | 1 | 16 | 0.1 | 0.001 | |
| 15.9 | PBMC | 14 | 6 | 8 | 0.04 | 0.0002 | |
| 18.9 | Ileum and colon | 41 | 31 | 10 | 0.08 | 0.0004 | |
| HAMB-1 | 6.0 | PBMC | 20 | 1 | 19 | 0.28 | 0.0015 |
| 6.2 | PBMC | 29 | 5 | 24 | 0.34 | 0.0016 | |
| 6.6 | PBMC | 30 | 5 | 25 | 0.29 | 0.0018 | |
| 7.8 | PBMC | 43 | 18 | 25 | 0.26 | 0.0015 | |
| 8.0 | Spleen | 38 | 10 | 28 | 0.26 | 0.0019 | |
| 8.0 | Lymph node | 30 | 4 | 26 | 0.32 | 0.0018 | |
| 8.0 | Kidney and lung | 8 | 2 | 6 | 0.46 | 0.0024 | |
| 8.0 | Testes | 2 | 1 | 1 | – | – | |
| 8.0 | Colon | 5 | 5 | 0 | – | – | |
| 8.0 | Ileum | 23 | 2 | 21 | 0.25 | 0.0014 | |
| 8.0 | Jejunum | 12 | 2 | 10 | 0.17 | 0.0009 | |
| HAMB-2 | 19.1 + 19.2 | PBMC | 46 | 6 | 40 | 1.5 | 0.009 |
| 22.7 | Spleen | 21 | 0 | 22 | 1.7 | 0.01 | |
| 22.7 | Lymph node-1 | 20 | 1 | 19 | 1.6 | 0.011 | |
| 22.7 | Lymph node-2 | 30 | 3 | 27 | 1.8 | 0.011 | |
| 22.7 | Lung | 33 | 1 | 32 | 1.8 | 0.011 | |
| 22.7 | Testes | 5 | 0 | 5 | 1.6 | 0.009 | |
| Frontal | |||||||
| 22.7 | +Occipital lobe | 6 | 0 | 6 | 1.2 | 0.007 | |
| AVBIO2-35 | 0.0 | Plasma | 23 | 0 | 23 | 1 | 0.006 |
| 0.0 | PBMC | 4 | 0 | 4 | 1.2 | 0.007 | |
| 8.3 | Plasma | 45 | 0 | 45 | 1 | 0.021 | |
| 19.4 | PBMC | 20 | 0 | 20 | 1.4 | 0.017 | |
| 19.7 | Plasma | 9 | 0 | 9 | 0.9 | 0.021 | |
| 19.7 | PBMC | 33 | 2 | 31 | 1.5 | 0.016 | |
| 19.7 | Colon | 22 | 2 | 20 | 1.1 | 0.02 | |
| 19.7 | Ileum | 14 | 0 | 14 | 1.1 | 0.021 | |
| AVBIO2-37 | 0.0 | Plasma | 17 | 0 | 17 | 1.7 | 0.01 |
| 0.0 | PBMC | 20 | 0 | 20 | 1.8 | 0.012 | |
| 3.3 | Plasma | 28 | 0 | 28 | 1.1 | 0.026 | |
| 19.4 | Plasma | 17 | 0 | 17 | 1.6 | 0.015 | |
| 19.4 | PBMC | 27 | 1 | 26 | 2.5 | 0.018 | |
| 19.7 | Colon | 19 | 1 | 18 | 2.7 | 0.02 | |
| 19.7 | Ileum | 30 | 1 | 29 | 2.6 | 0.019 | |
| 19.7 | Plasma | 17 | 0 | 17 | 1.6 | 0.015 | |
| 19.7 | PBMC | 26 | 0 | 26 | 2.5 | 0.017 | |
| 20.0 | Plasma | 39 | 4 | 35 | 1.3 | 0.014 |
Fig. 4. No genetic divergence of HIV during cART.
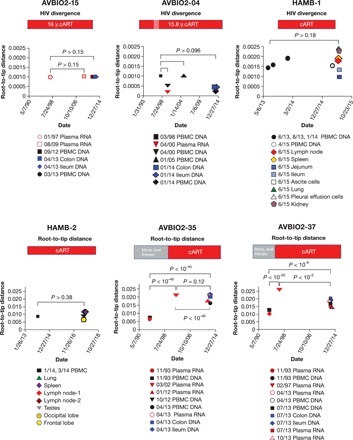
HIV sequences were subjected to root-to-tip analysis to investigate genetic divergence during cART. To calculate root-to-tip distances, a consensus HIV sequence was constructed for each individual from the earliest available time point, and root-to-tip distances were determined for HIV sequences from each time point as described in Materials and Methods. No divergence was detected during cART for AVBIO2-15, AVBIO2-04, HAMB-1, and HAMB-2. For AVBIO2-35 and AVBIO2-37, genetic divergence was detected during suboptimal therapy; upon initiating cART with suppression of viremia, no further divergence was detected. A decrease in root-to-tip distances was detected comparing samples obtained in plasma prior to cART and HIV DNA in samples obtained during suppressive cART because of the persistence of HIV DNA sequences from the pre-cART period.
It is possible that individuals like the study participants AVBIO2-15, AVBIO2-04, and HAMB-1 who were treated early had a well-preserved immune system, which might explain why there was little or no ongoing replication in the tissues in this group once cART was initiated. Thus, we also investigated individuals who had AIDS when they initiated cART to determine whether there was evidence of detectable changes in the HIV DNA sequences present in samples from participants with progressive immunodeficiency (HAMB-2, AVBIO2-35, and AVBIO2-37; Figs. 2 and 3 and Table 1). We studied one individual who underwent autopsy (HAMB-2) and two individuals who underwent routine colonoscopy (AVBIO2-35 and AVBIO2-37) after prolonged cART. In contrast to the limited genetic diversity detected in individuals who were treated early in infection, study participants initiating therapy during chronic HIV infection had genetically diverse HIV populations (HAMB-2, AVBIO2-35, and AVBIO2-37; Table 1), with average pairwise differences (1.0 to 3.4%) that were 8- to 10-fold higher than those detected in AVBIO2-15, AVBIO2-04, and HAMB-1, reflecting the HIV mutations that accumulated over the years of active replication. HIV genetic diversity was present in all tissues in HAMB-2 at autopsy, including lymph nodes, spleen, lung, testes, and the frontal and occipital lobes of the brain. As expected, this resulted in phylogenetic trees of HIV sequences with complex branching (Figs. 2 and 3, HAMB-2, AVBIO2-35, and AVBIO2-37). Identical HIV sequences suggestive of probable clones were recovered from PBMCs and multiple tissues at once, including the brain (Fig. 2, HAMB-2, black arrows), indicating probable clones traffic widely through infected individuals.
Fig. 2. No evidence of HIV molecular evolution in diverse tissues obtained at autopsy (HAMB-1 and HAMB-2).
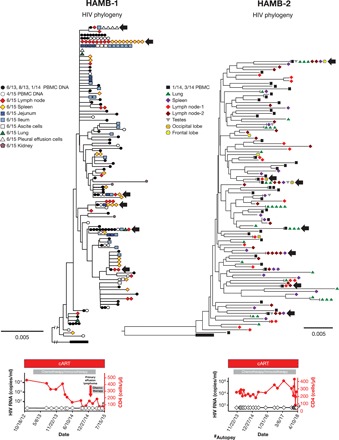
PBMCs were obtained from individuals after years of cART, and tissue samples were taken at autopsy after these individuals expired. HIV pro-pol sequences were obtained and then aligned; neighbor-joining phylogenetic trees were constructed). Hypermutated sequences were removed, and root-to-tip distances were determined as in Fig. 1. HAMB-1 initiated cART early after infection, and HAMB-2 initiated ART during chronic infection (see the Supplementary Materials). HIV DNA sequences obtained at the time of autopsy did not diverge from HIV DNA sequences recovered from PBMCs years previously. HIV DNA sequences that were identical to HIV DNA sequences from the PBMCs were found in all the tissues sampled, including the lung, spleen, multiple lymph nodes, testes, and the frontal and occipital lobes of the brain taken at autopsy; as a result, there were numerous groups with identical sequences present from multiple anatomic locations and PBMCs (black arrows).
Longitudinal analysis of samples taken from two of these individuals (AVBIO235 and AVBIO2-37) who had undergone mono- and dual therapy prior to receiving effective cART provided opportunities to demonstrate that we could detect HIV sequence evolution during suboptimal antiretroviral therapy. Prior to effective cART, phylogenetic analysis showed that there were new and distinct nodes for branches in the HIV phylogenetic trees, and increasing branch lengths, both of which demonstrated the accumulation of genetic changes in the population. As a result, root-to-tip distances increased during mono- or dual therapy (Fig. 4, AVBIO2-35 and AVBIO2-37). AVBIO2-35 was sampled while on mono- and dual therapy with persistent viremia, and subsequently after years of effective cART. As shown in Fig. 4, there were significant (3.3-fold) increases in root-to-tip distances during the 8.5-year period of nonsuppressive therapy, and drug resistance mutations accumulated during nonsuppressive therapy (Fig. 4). After switching to effective cART, however, there were no additional increases in root-to-tip distances (Fig. 4, AVBIO2-35 and AVBIO2-37), and no additional resistance mutations were detected in the proviral DNA in blood or tissues. Combining data from all individuals, no genetic change in HIV was detected sampling over 60 person-years of suppressive cART.
Phylogenetic analysis of sequences from the chronically infected individuals also revealed that the HIV DNA and RNA sequences obtained from plasma, PBMCs, or tissues during the course of suppressive cART were highly related, with few sequences having branches with significant bootstrap support (Fig. 2, HAMB-2; Fig. 3, AVBIO2-35 and AVBIO2-37), demonstrating that these sequences remained well mixed over prolonged periods, with multiple examples of identical sequences derived from different anatomic sources.
Fig. 3. Evidence of HIV molecular evolution during suboptimal therapy, but not during cART.
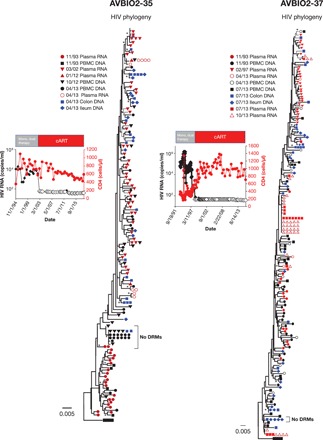
HIV RNA was obtained from plasma, and HIV DNA was obtained from PBMCs and tissues obtained at colonoscopy from individuals (AVBIO2-35 and AVBIO2-37) who underwent suboptimal therapy prior to suppressive cART as indicated; samples were obtained prior to any therapy, during suboptimal ART, and after effective combination therapy was initiated as indicated. Single-genome sequencing was performed; hypermutated sequences were removed as in Figs. 1 and 2. Groups of identical sequences with no drug resistance mutations (DRMs; bracket) were detected after prolonged suppression on cART in PBMCs and in tissues that were similar to sequences obtained prior to initiating any ART.
The sequential emergence of the drug resistance mutations during suboptimal therapy provided useful markers that could be used to determine the relative ages of the proviruses. For example, AVBIO2-35 underwent prolonged suboptimal antiretroviral therapy beginning in 1993, and analysis of plasma-derived HIV viral RNA populations in 2002 showed that all the SGSs obtained from plasma samples had acquired numerous mutations, including D67N and Y188C in reverse transcriptase (RT). The individual underwent suppressive therapy, and, at the time colonoscopy samples were obtained in 2013, after 11 years of effective cART, most of the proviruses contained these two resistance mutations, but no new resistance mutations were found. We also identified, however, a series of SGSs from proviruses with the wild-type D67 and Y188 sequence in samples from the colon and PBMCs (Fig. 3, AVBIO2-35, bracket), strongly suggesting that the cells that carried the wild-type RT sequences were infected prior to the emergence of the viruses that contained the drug resistance mutations. Similarly, AVBIO2-37 (Fig. 3) initiated suboptimal therapy in 1993; by 1997, every RNA sequence from plasma-derived HIV contained numerous drug resistance mutations. There were proviruses with the same resistance mutations in the HIV DNA sequences from colon and ileum samples, and there were also a series of identical proviruses present whose sequences were similar to pretherapy HIV and had no drug resistance mutations. The cells containing these proviruses, which were probably clonally expanded, which were present in tissues, would have been present for over 20 years (Fig. 3, AVBIO2-37, bracket). In these two individuals (Fig. 3, AVBIO2-35 and AVBIO2-37), we detected an overall decrease in root-to-tip distance. This can be explained by clonal expansion and persistence of a subset of the infected cells with no resistance mutations (Fig. 3).
These data show that, if therapy is suboptimal, detectable genetic changes in HIV proviruses accumulated over time. After the introduction of effective cART, however, no additional genetic changes were detected, and cells that were infected prior to suppressive therapy persisted during effective cART.
We performed additional population genetics analyses of nucleotide variation in plasma-, PBMC-, and tissue-derived HIV RNA and DNA (see the Supplementary Materials) to look for any evidence of genetic changes, including changes occurring at positions conferring predicted immunological escape from cytotoxic T lymphocytes (CTLs). We found (table S2), as expected, that HIV populations in recently infected individuals were highly uniform prior to therapy (36, 37); after cART, no shift in the overall population during therapy occurred (fig. S2, HAMB-1). In contrast, population shifts were detected in individuals while undergoing suboptimal cART, but no additional shift occurred after effective cART was introduced (fig. S2, AVBIO 2-37). In addition to the overall population analysis, we also investigated whether individual genetic changes had occurred in HIV over prolonged periods; we found no new immune escape variants emerged during cART (see the Supplementary Materials, population genetics analysis). In contrast, new variants did emerge and persist during suboptimal antiretroviral therapy (AVBIO2-35 and AVBIO2-37) or when cART was interrupted, even for brief periods (see the Supplementary Materials).
Determining the source(s) of HIV persistence during cART is central to developing curative and functional control strategies for HIV infection (38–42). Reservoirs of long-lived cells have been identified as long-lived reservoirs of CD4+ lymphocytes infected with replication-competent HIV, at least some of which undergo clonal expansion (20, 43, 44). “Shock and kill” strategies have been proposed as a way to activate and eliminate latently infected cells (40, 45, 46). In contrast, a number of studies have reported ongoing HIV replication as the result of substantially reduced levels of antiretroviral drug concentrations in tissues, including lymphoid cells in the lymph node, gut mucosa, and GALT, preventing complete inhibition of active viral replication; low drug levels have not been a universal finding in all tissues, including GALT (47). In settings where low drug levels persist, modeling studies predict ongoing cycles of active HIV replication in resident lymphocytes potentially confined in these tissues. Modeling studies have reported that ART levels that are sufficiently low will permit full HIV replication without selection of drug-resistant mutations. In such circumstances, active replication with error-prone RT-mediated HIV replication will lead to accumulation of genetic change in HIV genomes in these sanctuary sites and may not be detected in populations of peripheral blood lymphocytes, which will contain cells trafficking from a variety of sources where replication is not taking place. In contrast, the tissue-derived cells will be enriched for cells with evidence of ongoing replication (23–25). If HIV continues to replicate during cART, more effective cART would be required to prevent new infections and accelerate the decay of the HIV reservoir. In a critical test of the ongoing replication model, we detected no evidence of genetic changes in HIV (DNA) sequences from blood or tissue after many years of cART. In chronically infected individuals sampled over time during cART (HAMB-2, AVBIO2-35, and AVBIO2-37), HIV DNA and RNA sequences in tissues, plasma, and PBMCs remained highly related, with identical variants present in distinct locations, indicating a well-mixed population; well-mixed populations of HIV have been previously reported in GALT and peripheral blood (48). Here, we additionally find that potential sanctuary sites such as the brain, lung, and lymph node share identical HIV sequences (Fig. 2, HAMB-2). In individuals with acquired drug resistance who subsequently underwent suppressive cART with the addition of new agents, no new resistance mutations accumulated in tissue sources, even though the effective therapy included antivirals that were not fully effective (AVBIO2-35 and AVBIO2-37).
The study reported here did not detect evidence of ongoing cycles of HIV replication; prior work reporting ongoing replication during cART (24) analyzed next generation sequences obtained from blood and lymph node from individuals after 3 to 6 months of cART. In a detailed analysis, Rosenbloom and co-workers (49) demonstrated that the evidence of molecular evolution during this relatively short period of cART may be explained by the rapid loss of small subpopulations of HIV-infected cells that are eliminated shortly after cART is introduced. In addition, Kearney and co-workers noted the next generation sequence datasets used in (24) had limitations arising from sampling issues and amplification bias (50). It is impossible to formally rule out infrequent replication events using phylogenetic techniques, and we were, in fact, able to detect genetic changes that occurred during relatively short (circa 4 weeks) periods of cART interruption (see the Supplementary Materials, AVBIO2-04). During continuous therapy, however, we did not detect evidence of ongoing replication during a total of over 60 person-years of cART.
Approaches to quantify and characterize HIV populations will be necessary to define HIV eradication or functional control. The data reported here and elsewhere (11–13, 15, 22, 51–53) indicate that cure efforts should focus on determining the fundamental characteristics of HIV populations present during cART other than ongoing replication. We need to better define the fraction of proviruses that are replication competent, their genetic diversity, the degree of clonal expansion of the infected cells, and what controls the proportion of proviruses expressing HIV. Characterizing the genetic diversity of the proviruses that form the reservoir would include identifying the possible presence of CTL and antibody escape mutations, which could emerge and would require specific elimination. Determining the extent of clonal expansion of the infected cells that carry infectious proviruses is critical. The presence of clones, including those containing infectious HIV, is substantial and will require extensive and potent efforts for elimination or functional control. Determining the proportion of all proviruses, both defective and replication competent, that are expressing HIV is important because both intact and defective proviruses may contribute to ongoing inflammation and HIV pathogenesis (51, 54, 55). Developing new approaches for the eradication or functional control of HIV is daunting, but characterizing the genetic diversity and extent of clonal expansion of infected cells and understanding what controls the expression of these proviruses will define the magnitude of the challenge.
MATERIALS AND METHODS
Study participants
Samples were obtained from six individuals enrolled in studies of HIV infection and treatment approved by the National Institute of Allergy and Infectious Diseases (protocol 97-I-0082, 95-I-0027, or 08-I-0221) or the National Cancer Institute (NCI) (protocol 01-C0038, 16-C- 0047A, or 16-C-0066) Intramural Institutional Review Boards (FWA00005897), and conducted at the National Institutes of Health (NIH) Clinical Center in Bethesda, Maryland (table S1). Study participants provided written consent for research studies. Clinical information was abstracted from medical records at the NIH Clinical Center; plasma and PBMCs were obtained during protocol visits by phlebotomy, cytopheresis, or both. Routine CD4 cell counts and HIV viral RNA determinations were obtained as previously described (27). Samples from the colon and ileum were obtained through research colonoscopy (protocol 95-I-0027). Autopsies were performed within 14 to 72 hours after expiration, and dissected samples were stored at −70oC.
HIV sequence analysis
HIV sequence analysis DNA was prepared from PBMC and tissue samples, and HIV DNA copy numbers were determined using real-time polymerase chain reaction assays as previously described; in parallel, total cell equivalents were determined for each sample by quantifying CCR5 gene DNA copy number (45). In plasma samples, HIV RNA was extracted as previously described (10, 27). SGSs of HIV p6-RT were obtained and then aligned, and neighbor-joining phylogenetic analyses were performed [MEGA (46)]. Hypermutants were identified using HYPERMUT (www.hiv.lanl.gov/content/sequence/HYPERMUT/hypermut.html); for ease of visualization of phylogenetic trees, hypermutants were removed from phylogenetic trees in Figs. 1 to 3. In individuals who acquired drug resistance mutations during the course of therapy, phylogenetic trees were also constructed with drug resistance mutations removed; no differences in phylogenetic relationships were detected, and only minor changes were detected in branch lengths. Genetic diversity (average pairwise difference) was determined (46). To investigate the distribution of pairwise differences, we determined the nucleotide differences between each pair of sequences for the SGS within time points and between time points. The distribution of the intra– and inter–time point pairwise differences was compared in histogram analysis using Microsoft Excel programs. To determine root-to-tip distances, we first constructed a consensus root sequence that was obtained from sequences obtained from the earliest time point sampled (www.hiv.lanl.gov/content/sequence/CONSENSUS/consensus.html); for participants AVBIO2-15, AVBIO2-04, and HAMB-1, this consensus represented early HIV infection, and for individuals with chronic HIV infection (HAMB-2, AVBIO2-35, and AVBIO2-37), this consensus represented the earliest sampling date. While this consensus may not represent the root of all subsequent sequences, it is a sample predating all subsequent samples. The root-to-tip distance was calculated using the nucleotide distance [p distance, MEGA (46)] from this consensus root to each single genome sequence obtained at each time point; differences in root-to-tip distances between time points were determined by Student’s t test and by determining slope using the maximum likelihood approach (14), which yielded consistent results. Sequences are deposited in GenBank with accession numbers: MN461570- MN462555.
Population genetics characteristics
Population genetics characteristics (numbers of polymorphic sites, numbers of singleton sites) were determined using DnaSP (DNA Sequence Polymorphism) (47). To investigate whether the HIV population was evolving neutrally or violated one or more neutral population characteristics (e.g., random mating, constant population size, absence of selection or recombination), we used the Tajima’s D statistic in DnaSP (48), which calculates the differences between genetic diversity (average pairwise difference) and numbers of segregating sites (polymorphisms). Resistance mutation profiles were determined using the Stanford HIV database (https://hivdb.stanford.edu).
Supplementary Material
Acknowledgments
We are grateful to the volunteers and families who participated in this study. We thank S. Hughes, J. Kovacs, V. Pathak, W.-S. Hu, M. A. Polis, C. Lane, H. Imamichi, J. Coffin, and E. Freed for the helpful suggestions. Funding: This study was supported, in part, by an NIH Bench to Bedside Award (F.M.) and federal funds from the NCI, NIH (contract HHSN261200800001E; R.G. and B.F.) and the NIH Intramural Research Program Support (ZIA BC011700, ZIA BC011466, and ZIA BC010885). The content of this publication does not necessarily reflect the views or policies of the U.S. Department of Health and Human Services nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. Author contributions: Study conception and design: G.B., F.R.S., S.A.W., R.Y., T.U., and F.M. Acquisition of data: G.B., F.R.S., S.A.W., E.M.A., M.G., P.R., C.L., R.G., B.F., S.K., S.W., S.He., D.E.K., J.H., S.Hi., J.B., and C.R. Analysis and interpretation of data: G.B., F.R.S., S.A.W., E.M.A., W.S., M.F.K., M.J.B., and Z.G. Drafting of manuscript: G.B., F.R.S., S.A.W., T.U., and F.M. Critical revision: G.B., F.R.S., S.A.W., T.U., and F.M. Competing interests: R.Y. and T.U. are inventors on a patent related to this work filed by Celgene Corporation (no. 10,001,483 B2; filed 19 June 2018). The authors declare that they have no other competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/9/eaav2045/DC1
Supplementary Text
Fig. S1. Persistence and clonal expansion over 16 years.
Fig. S2. Histogram analysis of genetic diversity during cART.
Table S1. Demographic and clinical characteristics of study participants.
Table S2. HIV DNA recovery from tissue samples and PBMC.
Table S3. Population genetics analyses of HIV sequences.
REFERENCES AND NOTES
- 1.Hütter G., Nowak D., Mossner M., Ganepola S., Müßig A., Allers K., Schneider T., Hofmann J., Kütcherer C., Blau O., Hofmann I. W. K., Thiel E., Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N. Engl. J. Med. 360, 692–698 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Namazi G., Fajnzylber J. M., Aga E., Bosch R. J., Acosta E. P., Sharaf R., Hartogensis W., Jacobson J. M., Connick E., Volberding P., Skiest D., Margolis D., Sneller M. C., Little S. J., Gianella S., Smith D. M., Kuritzkes D. R., Gulick R. M., Mellors J. W., Mehraj V., Gandhi R. T., Mitsuyasu R., Schooley R. T., Henry K., Tebas P., Deeks S. G., Chun T.-W., Collier A. C., Routy J.-P., Hecht F. M., Walker B. D., Li J. Z., The control of HIV after antiretroviral medication pause (CHAMP) study: Post-treatment controllers identified from 14 clinical studies. J. Infect. Dis. 218, 1954–1963 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sáez-Cirión A., Bacchus C., Hocqueloux L., Avettand-Fenoel V., Girault I., Lecuroux C., Potard V., Versmisse P., Melard A., Prazuck T., Descours B., Guergnon J., Viard J.-P., Boufassa F., Lambotte O., Goujard C., Meyer L., Costagliola D., Venet A., Pancino G., Autran B., Rouzioux C.; ANRS VISCONTI Study Group , Post-treatment HIV-1 controllers with a long-term virological remission after the interruption of early initiated antiretroviral therapy ANRS VISCONTI Study. PLOS Pathog. 9, e1003211 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yukl S. A., Boritz E., Busch M., Bentsen C., Chun T.-W., Douek D., Eisele E., Haase A., Ho Y.-C., Hütter G., Justement J. S., Keating S., Lee T.-H., Li P., Murray D., Palmer S., Pilcher C., Pillai S., Price R. W., Rothenberger M., Schacker T., Siliciano J., Siliciano R., Sinclair E., Strain M., Wong J., Richman D., Deeks S. G., Challenges in detecting HIV persistence during potentially curative interventions: A study of the Berlin patient. PLOS Pathog. 9, e1003347 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.A. K. Gupta. R. K. McCoy, L. Mok, H. P. Peppa, D. Lee, H. Nastouli, E. Lambert, J. Pace, M. Frater, J. Edwards, S. Olavarria, E. Gabriel, I. paper presented at the Conference on Retroviruses and Opportunistic Infections, Seattle, WA, 2019. [Google Scholar]
- 6.Henrich T. J., Hanhauser E., Marty F. M., Sirignano M. N., Keating S., Lee T.-H., Robles Y. P., Davis B. T., Li J. Z., Heisey A., Hill A. L., Busch M. P., Armand P., Soiffer R. J., Altfed M., Kuritzkes D. R., Antiretroviral-free HIV-1 remission and viral rebound after allogeneic stem cell transplantation: Report of 2 cases. Ann. Intern. Med. 161, 319–327 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henrich T. J., Hu Z., Li J. Z., Sciaranghella G., Busch M. P., Keating S. M., Gallien S., Lin N. H., Giguel F. F., Lavoie L., Ho V. T., Armand P., Soiffer R. J., Sagar M., LaCasce A. S., Kuritzkes D. R., Long-term reduction in peripheral blood HIV type 1 reservoirs following reduced-intensity conditioning allogeneic stem cell transplantation. J. Infect. Dis. 207, 1694–1702 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koelsch K. K., Rasmussen T., Hey-Nguyen W., Pearson C., Xu Y., Bailey M., Marks K., Sasson S., Taylor M., Tantau R., Obeid S., Milner B., Morrissey O., Pinto A., Suzuki K., Busch M., Keating S., Kaiser P., Yukl S., Wong J., Hiener B., Palmer S., Zaunders J., Post J., Chan D., Avery S., Milliken S., Kelleher A., Lewin S., Cooper D., Impact of allogeneic hematopoietic stem cell transplantation on the HIV reservoir and immune response in 3 HIV-infected individuals. J. Acquir. Immune Defic. Syndr. 75, 328–337 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hosmane N. N., Kwon K. J., Bruner K. M., Capoferri A. A., Beg S., Rosenbloom D. I., Keele B. F., Ho Y.-C., Siliciano J. D., Siliciano R. F., Proliferation of latently infected CD4+ T cells carrying replication-competent HIV-1: Potential role in latent reservoir dynamics. J. Exp. Med. 214, 959–972 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Josefsson L., von Stockenstrom S., Faria N. R., Sinclair E., Bacchetti P., Killian M., Epling L., Tan A., Ho T., Lemey P., Shao W., Hunt P. W., Somsouk M., Wylie W., Douek D. C., Loeb L., Custer J., Hoh R., Poole L., Deeks S. G., Hecht F., Palmer S., The HIV-1 reservoir in eight patients on long-term suppressive antiretroviral therapy is stable with few genetic changes over time. Proc. Natl. Acad. Sci. U.S.A. 110, E4987–E4996 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kearney M. F., Spindler J., Shao W., Yu S., Anderson E. M., O’Shea A., Rehm C., Poethke C., Kovacs N., Mellors J. W., Coffin J. M., Maldarelli F., Lack of detectable HIV-1 molecular evolution during suppressive antiretroviral therapy. PLOS Pathog. 10, e1004010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z., Gurule E. E., Brennan T. P., Gerold J. M., Kwon K. J., Hosmane N. N., Kumar M. R., Beg S. A., Capoferri A. A., Ray S. C., Ho Y.-C., Hill A. L., Siliciano J. D., Siliciano R. F., Expanded cellular clones carrying replication-competent HIV-1 persist, wax, and wane. Proc. Natl. Acad. Sci. U.S.A. 115, E2575–E2584 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Persaud D., Pierson T., Ruff C., Finzi D., Chadwick K. R., Margolick J. B., Ruff A., Hutton N., Ray S., Siliciano R. F., A stable latent reservoir for HIV-1 in resting CD4+ T lymphocytes in infected children. J. Clin. Invest. 105, 995–1003 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruff C. T., Ray S. C., Kwon P., Zinn R., Pendleton A., Hutton N., Ashworth R., Gange S., Quinn T. C., Siliciano R. F., Persaud D., Persistence of wild-type virus and lack of temporal structure in the latent reservoir for human immunodeficiency virus type 1 in pediatric patients with extensive antiretroviral exposure. J. Virol. 76, 9481–9492 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Zyl G. U., Katusiime M. G., Wiegand A., McManus W. R., Bale M. J., Halvas E. K., Luke B., Boltz V. F., Spindler J., Laughton B., Engelbrecht S., Coffin J. M., Cotton M. F., Shao W., Mellors J. W., Kearney M. F., No evidence of HIV replication in children on antiretroviral therapy. J. Clin. Invest. 127, 3827–3834 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mok H. P., Norton N. J., Hirst J. C., Fun A., Bandara M., Wills M. R., Lever A. M. L., No evidence of ongoing evolution in replication competent latent HIV-1 in a patient followed up for two years. Sci. Rep. 8, 2639 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brodin J., Zanini F., Thebo L., Lanz C., Bratt G., Neher R. A., Albert J., Establishment and stability of the latent HIV-1 DNA reservoir. eLife 5, e18889 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohn L. B., Silva I. T., Oliveira T. Y., Rosales R. A., Parrish E. H., Learn G. H., Hahn B. H., Czartoski J. L., McElrath M. J., Lehmann C., Klein F., Caskey M., Walker B. D., Siliciano J. D., Siliciano R. F., Jankovic M., Nussenzweig M. C., HIV-1 integration landscape during latent and active infection. Cell 160, 420–432 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maldarelli F., Wu X., Su L., Simonetti F. R., Shao W., Hill S., Spindler J., Ferris A. L., Mellors J. W., Kearney M. F., Coffin J. M., Hughes S. H., Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 345, 179–183 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simonetti F. R., Sobolewski M. D., Fyne E., Shao W., Spindler J., Hattori J., Anderson E. M., Watters S. A., Hill S., Wu X., Wells D., Su L., Luke B. T., Halvas E. K., Besson G., Penrose K. J., Yang Z., Kwan R. W., Van Waes C., Uldrick T., Citrin D. E., Kovacs J., Polis M. A., Rehm C. A., Gorelick R., Piatak M., Keele B. F., Kearney M. F., Coffin J. M., Hughes S. H., Mellors J. W., Maldarelli F., Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc. Natl. Acad. Sci. U.S.A. 113, 1883–1888 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagner T. A., McLaughlin S., Garg K., Cheung C. Y., Larsen B. B., Styrchak S., Huang H. C., Edlefsen P. T., Mullins J. I., Frenkel L. M., HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science 345, 570–573 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reeves D. B., et al. , A majority of HIV persistence during antiretroviral therapy is due to infected cell proliferation. Nat. Commun. 9, 4811 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fletcher C. V., Staskus K., Wietgrefe S. W., Rothenberger M., Reilly C., Chipman J. G., Beilman G. J., Khoruts A., Thorkelson A., Schmidt T. E., Anderson J., Perkey K., Stevenson M., Perelson A. S., Douek D. C., Haase A. T., Schacker T. W., Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc. Natl. Acad. Sci. U.S.A. 111, 2307–2312 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lorenzo-Redondo R., Fryer H. R., Bedford T., Kim E.-Y., Archer J., Pond S. L. K., Chung Y.-S., Penugonda S., Chipman J., Fletcher C. V., Schacker T. W., Malim M. H., Rambaut A., Haase A. T., McLean A. R., Wolinsky S. M., Persistent HIV-1 replication maintains the tissue reservoir during therapy. Nature 530, 51–56 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nolan D. J., Rose R., Rodriguez P. H., Salemi M., Singer E. J., Lamers S. L., McGrath M. S., The spleen is an HIV-1 sanctuary during combined antiretroviral therapy. AIDS Res. Hum. Retroviruses 34, 123–125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joseph S. B., Kincer L. P., Bowman N. M., Evans C., Vinikoor M. J., Lippincott C. K., Gisslén M., Spudich S., Menezes P., Robertson K., Archin N., Kashuba A., Eron J. J., Price R. W., Swanstrom R., HIV-1 RNA detected in the cns after years of suppressive antiretroviral therapy can originate from a replicating cns reservoir or clonally expanded cells. Clin. Infect. Dis. 2018, ciy1066 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu W.-S., Hughes S. H., HIV-1 reverse transcription. Cold Spring Harb. Perspect. Med. 2, a006882 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rose R., Lamers S. L., Nolan D. J., Maidji E., Faria N. R., Pybus O. G., Dollar J. J., Maruniak S. A., McAvoy A. C., Salemi M., Stoddart C. A., Singer E. J., McGrath M. S., HIV maintains an evolving and dispersed population in multiple tissues during suppressive combined antiretroviral therapy in individuals with cancer. J. Virol. 90, 8984–8993 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez-Picado J., Zurakowski R., Buzón M. J., Stevenson M., Episomal HIV-1 DNA and its relationship to other markers of HIV-1 persistence. Retrovirology 15, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ananworanich J., Chomont N., Eller L. A., Kroon E., Tovanabutra S., Bose M., Nau M., Fletcher J. L. K., Tipsuk S., Vandergeeten C., O’Connell R. J., Pinyakorn S., Michael N., Phanuphak N., Robb M. L.; RV217 and RV254/SEARCH010 study groups , HIV DNA set point is rapidly established in acute hiv infection and dramatically reduced by early ART. EBioMedicine 11, 68–72 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crowell T. A., Fletcher J. L., Sereti I., Pinyakorn S., Dewar R., Krebs S. J., Chomchey N., Rerknimitr R., Schuetz A., Michael N. L., Phanuphak N., Chomont N., Ananworanich J.; RV254/SEARCH010 Study Group , Initiation of antiretroviral therapy before detection of colonic infiltration by HIV reduces viral reservoirs, inflammation and immune activation. J. Int. AIDS Soc. 19, 21163 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boltz V. F., Rausch J., Shao W., Hattori J., Luke B., Maldarelli F., Mellors J. W., Kearney M. F., Coffin J. M., Ultrasensitive single-genome sequencing: Accurate, targeted, next generation sequencing of HIV-1 RNA. Retrovirology 13, 87 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emery A., Zhou S., Pollom E., Swanstrom R., Characterizing HIV-1 splicing by using next-generation sequencing. J. Virol. 91, e02515-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maldarelli F., Kearney M., Palmer S., Stephens R., Mican J., Polis M. A., Davey R. T., Kovacs J., Shao W., Rock-Kress D., Metcalf J. A., Rehm C., Greer S. E., Lucey D. L., Danley K., Alter H., Mellors J. W., Coffin J. M., HIV populations are large and accumulate high genetic diversity in a nonlinear fashion. J. Virol. 87, 10313–10323 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shankarappa R., Gupta P., Learn G. H. Jr., Rodrigo A. G., Rinaldo C. R. Jr., Gorry M. C., Mullins J. I., Nara P. L., Ehrlich G. D., Evolution of human immunodeficiency virus type 1 envelope sequences in infected individuals with differing disease progression profiles. Virology 241, 251–259 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Kearney M., Maldarelli F., Shao W., Margolick J. B., Daar E. S., Mellors J. W., Rao V., Coffin J. M., Palmer S., Human immunodeficiency virus type 1 population genetics and adaptation in newly infected individuals. J. Virol. 83, 2715–2727 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keele B. F., Giorgi E. E., Salazar-Gonzalez J. F., Decker J. M., Pham K. T., Salazar M. G., Sun C., Grayson T., Wang S., Li H., Wei X., Jiang C., Kirchherr J. L., Gao F., Anderson J. A., Ping L.-H., Swanstrom R., Tomaras G. D., Blattner W. A., Goepfert P. A., Kilby J. M., Saag M. S., Delwart E. L., Busch M. P., Cohen M. S., Montefiori D. C., Haynes B. F., Gaschen B., Athreya G. S., Lee H. Y., Wood N., Seoighe C., Perelson A. S., Bhattacharya T., Korber B. T., Hahn B. H., Shaw G. M., Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U.S.A. 105, 7552–7557 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barton K., Winckelmann A., Palmer S., HIV-1 reservoirs during suppressive therapy. Trends Microbiol. 24, 345–355 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chun T.-W., Moir S., Fauci A. S., HIV reservoirs as obstacles and opportunities for an HIV cure. Nat. Immunol. 16, 584–589 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Shan L., Siliciano R. F., From reactivation of latent HIV-1 to elimination of the latent reservoir: The presence of multiple barriers to viral eradication. Bioessays 35, 544–552 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spivak A. M., Planelles V., Novel latency reversal agents for HIV-1 cure. Annu. Rev. Med. 69, 421–436 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong J. K., Yukl S. A., Tissue reservoirs of HIV. Curr. Opin. HIV AIDS 11, 362–370 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bui J. K., Sobolewski M. D., Keele B. F., Spindler J., Musick A., Wiegand A., Luke B. T., Shao W., Hughes S. H., Coffin J. M., Kearney M. F., Mellors J. W., Proviruses with identical sequences comprise a large fraction of the replication-competent HIV reservoir. PLOS Pathog. 13, e1006283 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lorenzi J. C., Cohen Y. Z., Cohn L. B., Kreider E. F., Barton J. P., Learn G. H., Oliveira T., Lavine C. L., Horwitz J. A., Settler A., Jankovic M., Seaman M. S., Chakraborty A. K., Hahn B. H., Caskey M., Nussenzweig M. C., Paired quantitative and qualitative assessment of the replication-competent HIV-1 reservoir and comparison with integrated proviral DNA. Proc. Natl. Acad. Sci. U.S.A. 113, E7908–e7916 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kwon K. J., Siliciano R. F., HIV persistence: Clonal expansion of cells in the latent reservoir. J. Clin. Invest. 127, 2536–2538 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pitman M. C., Lau J. S. Y., McMahon J. H., Lewin S. R., Barriers and strategies to achieve a cure for HIV. Lancet HIV 5, e317–e328 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Asmuth D. M., Thompson C. G., Chun T.-W., Ma Z.-M., Mann S., Sainz T., Serrano-Villar S., Utay N. S., Garcia J. C., Troia-Cancio P., Pollard R. B., Miller C. J., Landay A., Kashuba A. D., Tissue pharmacologic and virologic determinants of duodenal and rectal gastrointestinal-associated lymphoid tissue immune reconstitution in HIV-infected patients initiating antiretroviral therapy. J. Infect. Dis. 216, 813–818 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Imamichi H., Degray G., Dewar R. L., Mannon P., Yao M., Chairez C., Sereti I., Kovacs J. A., Lack of compartmentalization of HIV-1 quasispecies between the gut and peripheral blood compartments. J. Infect. Dis. 204, 309–314 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosebloom D. I. S., Hill A. L., Laskey S. B., Siliciano R. F., Re-evaluating evolution in the HIV reservoir. Nature 551, E6–E9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kearney M. F., Wiegand A., Shao W., McManus W. R., Bale M. J., Luke B., Maldarelli F., Mellors J. W., Coffin J. M., Ongoing HIV replication during ART reconsidered. Open Forum Infect. Dis. 4, ofx173 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bruner K. M., Murray A. J., Pollack R. A., Soliman M. G., Laskey S. B., Capoferri A. A., Lai J., Strain M. C., Lada S. M., Hoh R., Ho Y. C., Richman D. D., Deeks S. G., Siliciano J. D., Siliciano R. F., Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat. Med. 22, 1043–1049 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ho Y. C., Shan L., Hosmane N. N., Wang J., Laskey S. B., Rosenbloom D. I., Lai J., Blankson J. N., Siliciano J. D., Siliciano R. F., Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155, 540–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.von Stockenstrom S., Odevall L., Lee E., Sinclair E., Bacchetti P., Killian M., Epling L., Shao W., Hoh R., Ho T., Faria N. R., Lemey P., Albert J., Hunt P., Loeb L., Pilcher C., Poole L., Hatano H., Somsouk M., Douek D., Boritz E., Deeks S. G., Hecht F. M., Palmer S., Longitudinal genetic characterization reveals that cell proliferation maintains a persistent HIV type 1 DNA pool during effective HIV therapy. J. Infect. Dis. 212, 596–607 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Imamichi H., Dewar R. L., Adelsberger J. W., Rehm C. A., O’Doherty U., Paxinos E. E., Fauci A. S., Lane H. C., Defective HIV-1 proviruses produce novel protein-coding RNA species in HIV-infected patients on combination antiretroviral therapy. Proc. Natl. Acad. Sci. U.S.A. 113, 8783–8788 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Imamichi H., Natarajan V., Adelsberger J. W., Rehm C. A., Lempicki R. A., Das B., Hazen A., Imamichi T., Lane H. C., Lifespan of effector memory CD4+ T cells determined by replication-incompetent integrated HIV-1 provirus. AIDS 28, 1091–1099 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/9/eaav2045/DC1
Supplementary Text
Fig. S1. Persistence and clonal expansion over 16 years.
Fig. S2. Histogram analysis of genetic diversity during cART.
Table S1. Demographic and clinical characteristics of study participants.
Table S2. HIV DNA recovery from tissue samples and PBMC.
Table S3. Population genetics analyses of HIV sequences.


