Several genes lost in the ancestors of whales and dolphins likely contributed to adapting to a fully aquatic environment.
Abstract
The transition from land to water in whales and dolphins (cetaceans) was accompanied by remarkable adaptations. To reveal genomic changes that occurred during this transition, we screened for protein-coding genes that were inactivated in the ancestral cetacean lineage. We found 85 gene losses. Some of these were likely beneficial for cetaceans, for example, by reducing the risk of thrombus formation during diving (F12 and KLKB1), erroneous DNA damage repair (POLM), and oxidative stress–induced lung inflammation (MAP3K19). Additional gene losses may reflect other diving-related adaptations, such as enhanced vasoconstriction during the diving response (mediated by SLC6A18) and altered pulmonary surfactant composition (SEC14L3), while loss of SLC4A9 relates to a reduced need for saliva. Last, loss of melatonin synthesis and receptor genes (AANAT, ASMT, and MTNR1A/B) may have been a precondition for adopting unihemispheric sleep. Our findings suggest that some genes lost in ancestral cetaceans were likely involved in adapting to a fully aquatic lifestyle.
INTRODUCTION
The ancestors of modern cetaceans (whales, dolphins, and porpoises) transitioned from a terrestrial to a fully aquatic lifestyle during the Eocene about 50 million years ago (1). This process constitutes one of the most marked macroevolutionary transitions in mammalian history and was accompanied by profound anatomical, physiological, and behavioral transformations that allowed cetaceans to adapt and thrive in the novel habitat (2). Remarkable changes in cetacean anatomy include streamlined bodies and loss of body hair to reduce drag during swimming, a much thicker skin that lacks sweat and sebaceous glands and has enhanced physical barrier properties, a thick layer of blubber for insulation, the loss of hindlimbs after propulsion by the tail flukes evolved, and reduced olfactory and gustatory systems, which became less important in water (3). To efficiently store and conserve oxygen for prolonged breath-hold diving, cetaceans developed a variety of adaptations. These adaptations include increased oxygen stores that result from large blood volumes and elevated concentrations of hemoglobin, myoglobin, and neuroglobin in blood, muscle, and brain tissue, respectively; a high-performance respiratory system that allows rapid turnover of gases at the surface; and a flexible ribcage that allows the lung to collapse at high ambient pressure (4).
Comparative analysis of cetacean genomes has provided important insights into the genomic determinants of cetacean traits and aquatic specializations. Several studies revealed patterns of positive selection in genes with roles in the nervous system, osmoregulation, oxygen transport, blood circulation, or bone microstructure (5–7). An adaptive increase in myoglobin surface charge likely permitted a high concentration of this oxygen transport and storage protein in cetacean muscles (8). In addition to patterns of positive selection, the loss (inactivation) of protein-coding genes is associated with derived cetacean traits. For example, cetaceans have lost a large number of olfactory receptors, taste receptors, and hair keratin genes (9–12). Furthermore, all or individual cetacean lineages lost the ketone body–synthesizing enzyme HMGCS2 (13), the nonshivering thermogenesis gene UCP1 (14), the protease KLK8 that plays distinct roles in the skin and hippocampus (15), and short wave– and long wave–sensitive opsin genes (16). During evolution, gene loss not only can be a consequence of relaxed selection on a function that became obsolete but also can be a mechanism for adaptation (17). For example, the loss of the erythrocyte-expressed AMPD3 gene in the sperm whale, one of the longest- and deepest-diving cetacean species, is likely beneficial by enhancing oxygen transport (18). The loss of the elastin-degrading protease MMP12 may have contributed to “explosive exhalation,” and the loss of several epidermal genes (GSDMA, DSG4, DSC1, and TGM5) likely contributed to hair loss and the remodeling of the cetacean epidermal morphology (18).
Because of an extensive series of intermediate fossils, the shift from a terrestrial to a fully aquatic environment is one of the best-characterized macroevolutionary transitions in mammalian evolution (5). However, the important genomic changes that occurred during this transformation remain incompletely understood. Because recent work has shown that the loss of ancestral protein-coding genes is an important evolutionary force (17–19), we conducted a systematic screen for genes that were inactivated on the stem Cetacea branch, i.e., after the split between Cetacea and Hippopotamidae but before the split between Odontoceti (toothed whales) and Mysticeti (baleen whales). This revealed a number of gene losses that are associated with the evolution of adaptations to a fully aquatic environment.
RESULTS AND DISCUSSION
Screen for coding genes that were inactivated in the cetacean stem lineage
To investigate the contribution of gene inactivation to the evolution of adaptations to a fully aquatic environment in cetaceans, we systematically searched for protein-coding genes that were inactivated in the cetacean stem lineage (a flowchart of the screen is shown in fig. S1). Briefly, we considered 19,769 genes annotated in the human genome and searched for gene-inactivating mutations throughout a phylogeny of 62 mammalian species, comprising four cetaceans, two pinnipeds, a manatee, and 55 terrestrial mammals (table S1). To detect gene-inactivating mutations, we used a comparative approach that makes use of genome alignments to search for mutations that disrupt the protein’s reading frame (stop codon mutations, frameshifting insertions or deletions, and deletions of entire exons) and mutations that disrupt splice sites (18). Excluding members of the large olfactory receptor and keratin-associated gene families, whose losses have been studied in detail before [for example, in (9, 11, 12)], we identified 236 genes that do not have an intact ortholog in cetaceans and are inactivated in at most 3 of the 55 terrestrial mammal species. Of these 236 genes, 110 exhibit inactivating mutations that are shared between the two extant cetacean clades, odontocetes and mysticetes (Fig. 1A). Odontocetes were represented in our screen by the bottlenose dolphin, killer whale, and sperm whale (20–22), and mysticetes were represented by the common minke whale (6). The most parsimonious hypothesis for inactivating mutations shared between odontocetes and mysticetes is that they occurred before the split of these two clades in the common ancestral branch of Cetacea. To precisely identify genes that were inactivated during the transition from land to water in the cetacean stem lineage, we made use of the recently sequenced genome of the common hippopotamus (23), a semi-aquatic mammal that, along with the pygmy hippopotamus, is the closest living relative to cetaceans, and considered only genes with no detected inactivating mutations in the hippopotamus. This resulted in a set of 85 lost genes that exhibit shared inactivating mutations in odontocetes and mysticetes, 62 (73%) of which have not been reported before (table S2).
Fig. 1. Key coagulation factors that promote thrombosis were lost in the cetacean stem lineage.
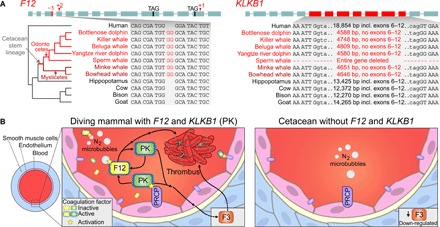
(A) F12 (coagulation factor XII) and KLKB1 (kallikrein B1) were lost in the cetacean stem lineage, consistent with previous findings (6, 22, 25). Boxes illustrate coding exons superimposed with those gene-inactivating mutations that are shared among odontocetes and mysticetes (both lineages are labeled in the phylogenetic tree) and thus likely occurred before the split of these lineages. The inset shows one representative inactivating mutation. Shared breakpoints imply that the deletion of KLKB1 coding exons 6 to 12 occurred in the cetacean stem lineage (intronic bases adjacent to exons 5 and 13 are in lowercase letters). All inactivating mutations in both genes are shown in figs. S4 and S5. (B) Left: F12 encodes a zymogen that autoactivates by contact with a variety of surfaces, which likely include nitrogen microbubbles that form during breath-hold diving (27, 29). KLKB1 encodes another zymogen that can be activated to plasma kallikrein (PK) by either activated F12 or by the endothelial membrane–associated endopeptidase prolylcarboxypeptidase (PRCP) (26). PK, in turn, can activate F12. Both activated F12 and PK proteases promote thrombosis formation (26). Right: Gene knockouts in mice suggest that loss of F12 and KLKB1 has no major effect on wound sealing but protects from thrombus formation via different mechanisms. While loss of KLKB1 protects from thrombosis by reducing the expression of F3 (coagulation or tissue factor III) (30), loss of F12 prevents activation on nitrogen microbubbles during diving. Because a vasoconstriction-induced reduction in blood vessel diameters and nitrogen microbubble formation increase the risk of thrombosis for frequent divers, the loss of both genes was likely beneficial for cetaceans.
For these 85 genes, we performed additional analyses to confirm evolutionary loss in the cetacean stem lineage. First, inactivating mutations shared between the four cetaceans used in the genomic screen imply that other species that descended from their common ancestor should share these mutations. We tested this by aligning the genomes of two additional odontocetes [Yangtze River dolphin (11) and beluga whale (24)] and an additional mysticete [bowhead whale (25)]. Manually inspecting the gene loci in these additional species confirmed the presence of shared inactivating mutations. Second, the manual inspection of genome alignments also revealed no evidence for an undetected functional copy of these genes in cetaceans. Together, these analyses show that these genes were inactivated on the stem Cetacea branch, i.e., after the split between Cetacea and Hippopotamidae but before the split between Odontoceti and Mysticeti.
We intersected the 85 genes with functional annotations of their human and mouse orthologs (table S2) and performed a literature search. This revealed a number of genes that we hypothesize to be related to aquatic adaptations [F12 (coagulation factor XII), KLKB1 (kallikrein B1), POLM (DNA polymerase mu), MAP3K19 (mitogen-activated protein kinase 19), SEC14L3 (SEC14-like lipid binding 3), SLC6A18 (solute carrier family 6 member 18), SLC4A9 (solute carrier family 4 member 9), and AANAT (aralkylamine N-acetyltransferase)] by being involved in thrombosis, repair of oxidative DNA damage, oxidative stress–induced lung inflammation, renal amino acid transport, saliva secretion, and melatonin synthesis. For these eight genes, we further verified that they have an intact reading frame not only in the common hippopotamus but also in the pygmy hippopotamus, the only other extant species in the family Hippopotamidae. Furthermore, we validated the correctness of all inactivating mutations with raw DNA sequencing reads that were used to assemble the cetacean genomes. We found that the vast majority of inactivating mutations (248 of 251; 98.8%) are confirmed by DNA sequencing reads (fig. S2 and table S3). We further estimated that the remnants of the coding regions of genes evolve under relaxed selection in cetaceans (highly significant for all genes except MAP3K19 with P = 0.08; table S4). Last, we analyzed available expression data of the bottlenose dolphin and minke whale, which revealed that the remnants of these genes either are not expressed anymore or do not produce full-length and properly spliced transcripts (fig. S3). With the exception of POLM and AANAT, which are also lost in the pangolin, these genes are either exclusively lost in the cetacean stem lineage (F12, KLKB1, MAP3K19, SEC14L3, and SLC6A18) or convergently lost in the aquatic manatee (SLC4A9 and AANAT). In the following, we describe how the loss of these eight genes likely relates to adaptations to a fully aquatic environment.
Loss of coagulation-associated factors and reduced thrombus formation
Diving results in a systemic response, consisting of a decrease in heart rate (bradycardia) and reduced peripheral blood flow, which is achieved by contraction of endothelial smooth muscle cells (peripheral vasoconstriction) (4). A frequent vasoconstriction-induced reduction in blood vessel diameter during diving increases the risk of thrombus (blood clot) formation. Our screen detected two blood coagulation-associated factors, F12 and KLKB1, that are specifically lost in cetaceans and no other analyzed mammal. Several shared inactivating mutations show that both genes were lost in the cetacean stem lineage (Fig. 1A and figs. S4 and S5). While the loss of these genes in various cetacean species was noted before (6, 22, 25), the mechanisms by which these two gene losses likely protect from thrombus formation during diving have not been described.
F12 initiates thrombus formation via the contact activation system (CAS) (26). F12 encodes a zymogen that autoactivates upon encountering a variety of foreign or biological surfaces (27). The activated zymogen functions as a serine protease that engages in a reciprocal activation cycle with the serine protease encoded by KLKB1, resulting in platelet activation and the formation of a blood clot (26). Consistently, knockout or knockdown of F12 protects various mammals from induced thrombosis but, importantly, did not impair wound sealing after blood vessel injury (hemostasis) (28). Eliminating CAS-based coagulation by inactivating F12 may have been especially advantageous for cetaceans, as nitrogen microbubbles, which readily form in the blood upon repeated breath-hold diving, may act as foreign F12-activating surfaces entailing harmful thrombus formation (Fig. 1B) (29).
The KLKB1-encoded zymogen prekallikrein is activated by proteolytic cleavage to form the serine protease plasma kallikrein (PK). Similar to the knockout of F12, the knockout of KLKB1 in mice also granted protection from induced thrombosis while only slightly prolonging wound sealing (28). Thrombosis protection in KLKB1 knockout mice is mediated by a CAS-independent mechanism (30). KLKB1 knockout leads to reduced levels of bradykinin, the main target of PK, which, in turn, leads to reduced expression of coagulation factor III (also called tissue factor, F3) (30), a key initiator of the blood coagulation cascade. The reduction of coagulation factor III alone is sufficient to reduce the risk of thrombosis. In addition to being activated by F12, prekallikrein can be activated by the endothelial membrane–associated endopeptidase prolylcarboxypeptidase (PRCP) (31). Evidently, this type of activation should happen more frequently in a diving cetacean, where constricted blood vessels increase the proximity of prekallikrein and PRCP (Fig. 1B). Moreover, the activity of PRCP is pH dependent and peaks in slightly acidified plasma (31), a condition found in diving cetaceans.
In summary, all cetaceans are deficient in two key factors that promote thrombosis but largely do not affect wound sealing. In support of the hypothesis that wound sealing mechanisms remain intact, we found that the key coagulation factors facilitating hemostasis upon tissue damage (encoded by the genes F2, F3, F7, and F10) are intact in the cetaceans and other mammals included in our screen. The risk of thrombus-induced occlusion of blood vessels is higher for frequent divers, as smaller blood vessel diameters and nitrogen microbubble formation during diving both increase the likelihood of F12 or prekallikrein activation (Fig. 1B). Because inactivating F12 or KLKB1 reduces the risk of thrombus formation via different and likely additive mechanisms, both gene losses were potentially advantageous for stem cetaceans. Consistent with this, previous studies found that several genes involved in blood clotting evolved under positive selection in cetaceans (21).
Loss of a DNA repair gene and improved tolerance of oxidative DNA damage
The pronounced peripheral vasoconstriction evoked by the diving response restricts blood supply to peripheral tissues of the diving mammal, causing an oxygen shortage (ischemia). Restoration of blood flow (reperfusion) to these tissues causes the production of reactive oxygen species (ROS), which can damage DNA. Diving mammals are better adapted to tolerate frequent ischemia/reperfusion-induced ROS generation by having high levels of antioxidants (32). In addition to these increased antioxidant levels, we detected the inactivation of POLM in the cetacean stem lineage (Fig. 2A and fig. S6). POLM lacks inactivation mutations in any other mammal with the exception of the Chinese pangolin, a burrowing mammal that inhabits higher elevations.
Fig. 2. Loss of an error-prone DNA repair polymerase could have improved tolerance of oxidative DNA damage in cetaceans.
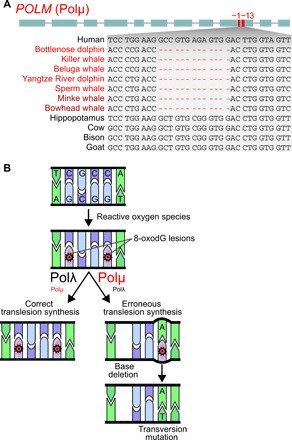
(A) POLM (DNA polymerase mu) was lost in the cetacean stem lineage, as shown by shared gene-inactivating mutations. Visualization as in Fig. 1A. All inactivating mutations are shown in fig. S6. (B) ROS (reactive oxygen species) induce DNA damage, which includes oxidation of guanine (8-oxodG) as one the most frequent lesions. POLM encodes the DNA repair polymerase Polμ, which often does not perform a correct translesion synthesis (left) but instead introduces errors (right). In particular, Polμ typically deletes bases (35) or erroneously incorporates deoxy-adenosine opposite to 8-oxodG (instead of the correct deoxy-cytosine), which results in a C:G to A:T transversion mutation (34). In contrast to Polμ, another DNA repair polymerase Polλ is much less error prone (36). Loss of POLM in cetaceans may have reduced the mutagenic potential of diving-induced oxidative stress by increasing the utilization of the more precise Polλ and accurate homology-directed DNA repair.
Loss of POLM has implications for improved tolerance of oxidative DNA lesions. POLM encodes the DNA polymerase Polμ, which plays an integral role in DNA damage repair (33). The most severe type of DNA damage caused by ROS is a DNA double-strand break. One mechanism to repair double-strand breaks is nonhomologous end joining (NHEJ), a process that ligates DNA strands without requiring a homologous template and resynthesizes missing DNA bases by DNA polymerases. Polμ is able to direct synthesis across a variety of broken DNA backbone types, including ends that lack any complementarity (33). This high flexibility comes at the cost of making Polμ more error prone than Polλ, the second DNA polymerase that participates in NHEJ (33). One of the most frequent types of DNA damage caused by ROS is the oxidation of guanine, creating 8-oxo-7-hydrodeoxyguanosine (8-oxodG) (34). 8-oxodG is highly mutagenic, as the bypassing Polμ resolves this lesion either by deleting bases (35) or by creating a transversion mutation (Fig. 2B) (34). In contrast to Polμ, Polλ performs translesion synthesis with a much lower error rate (36). Outside the context of DNA double-strand breaks, Polβ is the main polymerase facilitating 8-oxodG repair (37). Our screen detected no inactivating mutations in cetaceans (and other mammals) in the genes encoding Polλ (POLL) and Polβ (POLB), suggesting that other DNA repair polymerases remain functional.
However, in a regime of frequent oxidative stress, as experienced by diving cetaceans, the error-prone DNA repair polymerase Polμ likely constitutes a mutagenic risk factor. Inactivation of Polμ in the cetacean stem lineage may have enhanced the fidelity of bypassing 8-oxodG lesions and repairing double-stranded breaks by increased utilization of the more precise Polλ, which is supported by mouse experiments. Compared with wild-type mice, POLM knockout mice showed significantly reduced mutagenic 8-oxodG translesion synthesis and exhibited a higher endurance when challenged with severe oxidative stress (38, 39). POLM knockout mice also displayed improved learning abilities and greater liver regenerative capacity at high age (38, 39) but were found to suffer from reduced hematopoiesis and impaired adaptive immunity (40). Consequently, the benefits of losing POLM may only predominate under frequent exposure to oxidative stress, where its loss reduces the mutagenic potential of ROS, which readily form during repeated ischemia/reperfusion processes in diving mammals.
Loss of lung-related genes and a high-performance respiratory system
During diving, the cetacean lung collapses and reinflates during ascent. While lung collapse would represent a severe clinical problem for humans, it serves to reduce both buoyancy and the risk of developing decompression sickness in cetaceans (41). Our screen revealed two genes that are exclusively lost in cetaceans and have specific expression patterns in the lung, MAP3K19 and SEC14L3 (Fig. 3, A and B, and figs. S7 and S8).
Fig. 3. Loss of lung-related and renal transporter genes in the cetacean stem lineage.
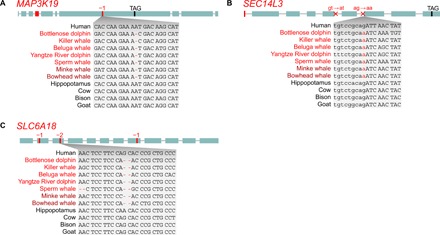
(A and B) The loss of MAP3K19 (mitogen-activated protein kinase 19) and SEC14L3 (SEC14-like lipid binding 3), which are specifically expressed in cell types of the lung, may relate to the high-performance respiratory system of cetaceans. (C) The loss of the renal amino acid transporter SLC6A18 (solute carrier family 6 member 18) offers an explanation for the low plasma arginine levels in cetaceans and may have contributed to stronger vasoconstriction during the diving response. Visualization as in Fig. 1A. A shared donor (gt ➔ at) and acceptor (ag ➔ aa) splice site disrupting mutation is indicated in (B). All inactivating mutations are shown in figs. S7 to S9.
MAP3K19 is expressed in bronchial epithelial cells, type II pneumocytes, and pulmonary macrophages (42). Overexpression of MAP3K19 was detected in pulmonary macrophages of human patients suffering from idiopathic pulmonary fibrosis (42). This disease is believed to be caused by aberrant wound healing in response to injuries of the lung epithelium, leading to the abnormal accumulation of fibroblasts (fibrosis), excessive collagen secretion, and severely impaired lung function (42). Consistent with a fibrosis-promoting function of MAP3K19, inhibition of MAP3K19 in mice protects from induced pulmonary fibrosis by significantly reducing fibrosis and collagen deposition (42). In a similar manner, MAP3K19 loss may also have a protective effect in cetaceans where repeated lung collapse/reinflation events during deep dives cause shear forces that could increase the incidence of pulmonary microinjuries.
Furthermore, overexpression of MAP3K19 was also detected in human patients suffering from chronic obstructive pulmonary disease (COPD) (43), a disease associated with cigarette smoking–induced oxidative stress. MAP3K19 is up-regulated in cells in response to oxidative and other types of environmental stress and promotes the expression of pro-inflammatory chemokines (43). Further supporting a role of MAP3K19 in the pathogenesis of COPD, inhibition of MAP3K19 in mouse COPD models strongly reduced pulmonary inflammation and airway destruction (43). A hallmark of COPD is a reduction of alveolar elasticity caused by elastin degradation, which contributes to an incomplete emptying of the lung. Cetaceans exhibit the opposite phenotype and have extensive elastic tissue in their lungs (41), which contributes to “explosive exhalation,” a breathing adaptation that allows renewal of ~90% of the air in the lung in a single breath (3). Therefore, similar to the previously described loss of the elastin-degrading and COPD-overexpressed MMP12 in aquatic mammals (18), the loss of MAP3K19 may also be involved in the evolution of this breathing adaptation. More generally, the frequent oxidative stress faced by diving cetaceans, especially upon reoxygenation of the reinflated hypoxic lung, would increase the risk for MAP3K19-mediated chronic pulmonary inflammation and compromised respiratory function, which could have contributed to MAP3K19 loss.
The second lung-expressed gene SEC14L3 is expressed in airway ciliated cells and in alveolar type II cells that secrete pulmonary surfactant, the lipid-protein complex that prevents alveoli collapse (44, 45). Similar to other surfactant-associated genes, SEC14L3 expression is highly induced in the lungs before birth (45). SEC14L3 functions as a sensor of liposomal lipid-packing defects and may affect surfactant composition (45). Alterations in surfactant composition may be relevant for cetaceans and other diving mammals. A study in seals suggested that pulmonary surfactants with anti-adhesive properties are important for diving mammals by facilitating alveolar reinflation after collapse (46). Because cetacean surfactants have not been characterized, it remains to be investigated whether the cetacean-specific loss of the surfactant-related SEC14L3 is associated with changes in the composition and anti-adhesive properties of cetacean surfactants.
Loss of a renal transporter gene and enhanced vasoconstriction during the diving response
Our screen revealed the cetacean-specific loss of SLC6A18 (Fig. 3C and fig. S9), which encodes a renal amino acid transporter that participates in reabsorption of arginine and other amino acids in the kidney proximal tubules. Knockout of SLC6A18 in mice resulted in reduced plasma arginine levels (47). Thus, the loss of SLC6A18 and its renal arginine reabsorbing activity provides one possible explanation for why cetaceans exhibit considerably lower plasma arginine levels in comparison to mice (48). In addition, SLC6A18 knockout in mice resulted in stress-induced hypertension (47), a condition that involves vasoconstriction. This hypertension phenotype likely arises because lower arginine levels reduce the main substrate for the production of nitric oxide, a highly diffusible vasodilating substance (47). Consistently, SLC6A18 inactivation caused persistent hypertension in a different mouse strain that is more susceptible to perturbations of nitric oxide production (49). This raises the possibility that the evolutionary loss of SLC6A18 in the cetacean stem lineage may have contributed to an increased diving capacity by indirectly enhancing vasoconstriction during the diving response.
Loss of an ion transporter gene and feeding in an aquatic environment
Saliva plays a role in lubricating the oral mucosa, in providing starch-degrading enzymes, and in the perception of taste. All these functions became less important in an aquatic environment, where the abundance of water sufficiently lubricates food and dilutes salivary digestive enzymes. In addition, the hyperosmotic marine environment necessitates strict housekeeping of freshwater resources in marine species (50); thus, freshwater loss via saliva secretion may be detrimental. We found that SLC4A9, a gene participating in saliva secretion, was lost in the cetacean stem lineage (Fig. 4A and fig. S10). Moreover, we found a convergent inactivation of this gene in the manatee, representing the only other fully aquatic mammalian lineage (fig. S10).
Fig. 4. Loss of a pleiotropic ion transporter in cetaceans relates to the dispensability of saliva secretion.
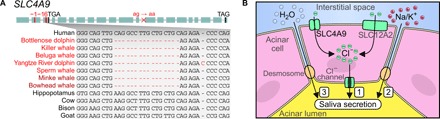
(A) Several shared inactivating mutations indicate that SLC4A9 (solute carrier family 4 member 9) was lost in the cetacean stem lineage. Visualization as in Fig. 1A. All inactivating mutations are shown in fig. S10. (B) Simplified illustration of saliva secretion. SLC4A9 encodes an ion transporter. (1) In the submandibular salivary gland, SLC4A9 participates in creating a transepithelial chloride anion flux into the acinar lumen, together with another transporter SLC12A2 and chloride channels (52). (2) This first evokes a passive movement of cations across the tight junctions into the acinar lumen. (3) The resulting osmotic gradient induces a flow of water, which constitutes fluid secretion into the acinar lumen, the initial site of saliva secretion. SLC4A9 knockout in mice leads to a 35% reduction in saliva secretion (52). The remaining saliva secretion potential in SLC4A9 knockout mice is maintained by SLC12A2 (52). However, SLC12A2 lacks inactivating mutations in cetaceans; these mutations lead to severe phenotypes in humans and mice (55), suggesting that gene essentiality maintained this gene in cetaceans. In addition to saliva secretion, SLC4A9 is also involved in transepithelial sodium ion flux in the kidney (not shown here) and participates in sodium chloride reabsorption (56), a process that is less important in hyperosmotic marine environments.
SLC4A9 encodes an electroneutral ion exchange protein (51), which is expressed in the submandibular salivary gland. SLC4A9 is restricted to the basolateral membrane of acinar cells, where it participates in saliva secretion (Fig. 4B) (52). SLC4A9 knockout mice displayed a 35% reduction in saliva secreted from the submandibular gland (52). This suggests that loss of SLC4A9 in cetaceans could contribute to a reduction of saliva secretion, which is in agreement with morphological observations that salivary glands are absent or atrophied in cetaceans (53). A second gene decisively contributing to saliva secretion is SLC12A2. Knockout of this gene in mice reduces saliva secretion by more than 60% (54). However, SLC12A2 is involved in a multitude of other physiological processes, and mutations in SLC12A2 entail severely detrimental phenotypes (55). Pleiotropy therefore likely explains why SLC12A2 lacks inactivating mutations in cetaceans and all other analyzed mammals.
In addition to the submandibular salivary gland, SLC4A9 is also expressed at the basolateral membrane of β-intercalated cells of the kidney, where it contributes to sodium chloride reabsorption (56). For species living in a hyperosmotic environment, where they incidentally ingest seawater with their prey, salt reabsorption by the kidney is probably less important (or even harmful) relative to efficient salt excretion. Thus, the loss of the salt reabsorbing factor SLC4A9 may contribute to the high urinary concentrations of sodium and chloride in cetaceans as compared to cows (57). In summary, the pleiotropic SLC4A9 gene was likely lost because both of its physiological processes, secretion of saliva and salt reabsorption, became dispensable in marine aquatic environments.
Loss of melatonin biosynthesis/reception and the evolution unihemispheric sleep
Commitment to a fully aquatic lifestyle also required distinct behavioral adaptations in stem cetaceans. Specifically, prolonged periods of sleep are obstructed by the needs to surface regularly to breathe and to constantly produce heat in the thermally challenging environment of the ocean. Cetaceans are the only mammals thought to sleep exclusively unihemispherically, a type of sleep that allows one brain hemisphere to sleep while the awake hemisphere coordinates movement for surfacing and heat generation (58). Our screen uncovered that AANAT was lost in the stem cetacean lineage (Fig. 5A and fig. S11). AANAT is a key gene required for synthesis of melatonin, the sleep hormone that influences wakefulness and circadian rhythms. Because genes that are functionally linked in a pathway tend to be co-eliminated (17, 59), we inspected ASMT (acetylserotonin O-methyltransferase), encoding the second enzyme required for melatonin synthesis, and MTNR1A and MTNR1B (melatonin receptors 1A and 1B), encoding the two membrane-bound melatonin receptors. Supporting a pattern of co-elimination of melatonin-related genes, we found that all three genes were lost in all analyzed cetaceans, with MTNR1B being inactivated in the cetacean stem lineage (Fig. 5A and fig. S12), while ASMT and MTNR1A were probably inactivated independently after the split of odontocetes and mysticetes (figs. S13 and S14). Thus, cetaceans have lost all genes required for melatonin biosynthesis and reception (Fig. 5B). In line with these findings, cetaceans exhibit low levels of circulating melatonin, which does not follow a circadian pattern (60, 61). Because dietary melatonin is readily transported into the blood stream, our finding that the melatonin-synthesizing enzymes AANAT and ASMT are lost in cetaceans further indicates that previously measured melatonin levels in cetaceans are not endogenous, but rather of dietary origin. Furthermore, the loss of the ASMT gene suggests that the previously reported immunohistochemistry signal of ASMT protein in the retina, Harderian gland, and gut of bottlenose dolphin (61) may be attributed to antibody cross-reactivity.
Fig. 5. Complete loss of melatonin synthesis and reception may have been a precondition to exclusively adopt unihemispheric sleep in cetaceans.
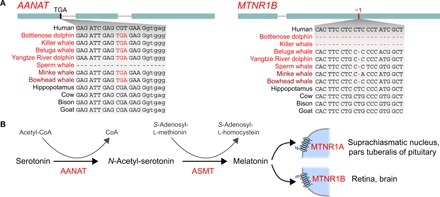
(A) Shared inactivating mutations indicate that AANAT (aralkylamine N-acetyltransferase), the first enzyme required to synthesize melatonin, and MTNR1B, one of the two melatonin receptors, were lost in the cetacean stem lineage. Subsequently, the second enzyme ASMT (acetylserotonin O-methyltransferase) and the second receptor MTNR1A were probably independently lost in cetaceans after the split of odontocetes and mysticetes; however, overlapping deletions of the last ASMT coding exon and MTNR1A exon 2 do not exclude the possibility of ancestral gene losses. Visualization as in Fig. 1A. All inactivating mutations are shown in figs. S11 to S14. (B) Pathway to synthesize melatonin from serotonin and the main sites of expression of the two melatonin transmembrane receptors.
Melatonin is synthesized in the pineal gland in the absence of light (i.e., at night) by serial action of the enzymes AANAT and ASMT and thereby relays information on daytime and season. Polymorphisms in AANAT or ASMT affect sleep patterns in humans (62). Furthermore, knockout of AANAT in zebrafish decreased the length of sleep bouts, causing an ~50% reduction in nightly sleep time (63). It has been suggested that melatonin influences sleep-wake cycles mainly by binding the receptors encoded by MTNR1A and MTNR1B on cells of the suprachiasmatic nucleus. Accordingly, elimination of these two receptors significantly increased the time spent awake in mutant mice (64). Furthermore, a polymorphism in the promoter region of MTNR1A was linked to insomnia symptoms (65). In addition to influencing sleep, melatonin has also been shown to regulate core body temperature in a circadian manner, and high circulating melatonin levels evoke a reduction of core body temperature through increased distal heat loss (66).
Therefore, the potential benefits of abolishing melatonin production and reception for cetaceans were likely twofold. First, by helping to decouple sleep-wake patterns from daytime, the loss of circadian melatonin production may have been a precondition to adopt unihemispheric sleep as the exclusive sleep pattern. Consistently, sleep in several cetacean species was observed to be equally distributed between day- and nighttime and is thought to be primarily influenced by prey availability (58). Second, mechanisms that reduce core body temperature appear detrimental for species inhabiting a thermally challenging environment.
When we examined the melatonin biosynthesis/reception genes in the manatee, we found inactivating mutations in three of the four genes (AANAT, ASMT, and MTNR1B; figs. S11 to S13). Similar to unihemispheric sleep, manatees also display considerable interhemispheric asymmetry during slow-wave sleep (58). In addition, manatees seem to lack a pineal gland. The pangolin, the only terrestrial mammal in our dataset that exhibits an inactivated AANAT gene, also lacks a pineal gland.
The results of our genomic analysis also have implications for the conflicting results on the morphological presence of the pineal gland in cetaceans. This gland has been reported to be absent or rudimentary in several cetaceans (but its absence can sometimes be variable between different individuals), while other species such as beluga, harbour porpoise, and sperm whale appear to have a fully developed pineal gland (61). Even if a pineal gland is present in some cetacean individuals or species, inactivating mutations in melatonin synthesis and receptor genes in all cetaceans, including beluga and sperm whale, preclude a role for this gland in melatonin-mediated circadian rhythms.
Loss of genes involved in immune system, muscle function, metabolism, and development
Despite the fact that cetacean phenotypes have been extensively studied, our genomic screen for genes lost in the cetacean stem lineage detected several gene losses that imply changes in particular phenotypes, which have not been well characterized. For example, we found losses of genes involved in defense to infectious agents such as bacteria and viruses (TRIM14 and TREM1; figs. S15 and S16 and table S2). Furthermore, while mammals generally have four genes encoding peptidoglycan recognition proteins, which are receptors important for antimicrobial function and for maintaining a healthy gut microbiome, cetaceans have lost three of these four genes (PGLYRP1/3/4; figs. S17 to S19 and table S2). While the loss of these genes highlights differences in the cetacean immune system, it is not clear whether these losses are potentially related to different pathogens encountered in a fully aquatic environment, changes in gut microbiome composition in these obligate carnivores, or other reasons. Another example is MSS51, a gene that is predominantly expressed in fast glycolytic fibers of the skeletal muscle. Inactivation of MSS51 in muscle cell lines directs muscle energy metabolism toward beta-oxidation of fatty acids. MSS51 was lost in the cetacean stem lineage (fig. S20 and table S2), suggesting that muscle metabolism may be largely fueled by fatty acids, which would be consistent with a high intramuscular lipid content in cetaceans. Cetaceans also lost ACSM3 (fig. S21 and table S2), a gene involved in oxidation of the short-chain fatty acid butyrate, but it is not clear whether this loss relates to their carbohydrate-poor diet. The loss of ADH4 (fig. S22 and table S2), a gene that metabolizes retinol and other substrates, suggests differences in vitamin A metabolism. Last, the cetacean loss of SPINK7 (fig. S23 and table S2), a gene involved in esophageal epithelium development, could be linked to the specific ontogeny of the cetacean esophagus, which is homologous to a ruminant’s forestomach. Overall, this highlights the need for further studies to investigate how the loss of these genes may affect immunity, metabolism, and development in cetaceans.
Loss of less well-characterized genes in cetaceans
Last, we detected losses of genes that have no experimentally characterized function (table S2). Some of these genes have tissue-specific expression patterns, exemplified by FABP12 (fig. S24 and table S2), a member of the fatty acid–binding protein family that is expressed in retina and testis of rats; ASIC5 (fig. S25 and table S2), an orphan acid-sensing ion channel specifically expressed in interneuron subtypes of the vestibulocerebellum that regulates balance and eye movement; or C10orf82 (fig. S26 and table S2), which is specifically expressed in the human testis. Natural losses of these uncharacterized genes provide intriguing candidates for future functional studies, which may help to relate evolutionary gene losses to particular cetacean phenotypes.
Convergent gene losses in other semi-aquatic and aquatic mammals
Several of the 85 genes lost on the stem Cetacea branch are also convergently inactivated in the fully aquatic manatee (including SLC4A9 and AANAT) or in semi-aquatic pinnipeds (table S2). We further tested whether these three lineages of (semi-) aquatic mammals have convergently lost more genes than their closest terrestrial relatives in our phylogenetic tree. To this end, we determined the number of genes that are convergently inactivated in at least two of the (semi-) aquatic mammals (represented by killer whale, Pacific walrus, and manatee) but intact in all their respective terrestrial sister species (represented by cow, polar bear, and elephant). For comparison, we determined the number of genes that are convergently inactivated in at least two of these three terrestrial mammals but intact in all their respective (semi-) aquatic sister mammals. Indeed, we found 20 genes that are convergently inactivated in at least two (semi-) aquatic mammals, whereas only two genes are convergently inactivated between at least two of their respective terrestrial sister species (fig. S27 and table S5). This finding is in contrast to a previous study showing that there are not more convergent amino acid substitutions among (semi-) aquatic mammals than there are among their terrestrial sister species (21), which might be related to the fact that the loss of a gene is generally a rarer and more radical genomic change than the substitution of an amino acid.
Summary
By conducting a systematic screen for coding genes that were inactivated in the cetacean stem lineage, we found 85 gene losses, 62 (73%) of which have not been reported before. Many of these gene losses were likely neutral, and their loss happened because of relaxed selection to maintain their function. This “use it or lose it” principle may also apply to pleiotropic genes that are involved in more than one process. An example is the loss of the pleiotropic SLC4A9 gene, which was likely permitted in cetaceans because both of its functions (saliva secretion and renal salt reabsorption) became less important in marine environments. Together with KLK8, a pleiotropic gene with epidermal and hippocampal functions that is convergently lost in cetaceans and manatees (15), this adds to a rather small list of known pleiotropic gene losses (67).
In addition to likely neutral gene losses, some of the genes that were for the most part specifically lost in the cetacean stem lineage could have contributed to adapting to an aquatic environment, particularly in relation to the challenges of diving. The loss of F12 and KLKB1 likely reduced the risk of thrombus formation during diving. The loss of POLM likely reduced the mutagenic potential of ROS by indirectly enhancing the fidelity of oxidative DNA damage repair. The loss of MAP3K19 protects from pulmonary fibrosis and from lung inflammation induced by oxidative stress. Because ischemia followed by reperfusion during diving generates ROS, losing these two genes may have contributed to better tolerating frequent diving-induced oxidative stress. SLC6A18 loss could be involved in reduced plasma arginine levels and thus could indirectly enhance the vasoconstriction capacity during diving by reducing the substrate for synthesis of the potent vasodilator nitric oxide. Last, the composition of pulmonary surfactant is important to allow lung reinflation after deep diving–induced alveolar collapse, which makes it interesting to investigate whether the loss of SEC14L3 affects composition and functional properties of surfactants in cetaceans.
In conclusion, our findings suggest that gene losses in cetaceans not only are associated with aquatic specializations but could have been involved in adapting to a fully aquatic environment, which further supports that loss of ancestral genes can be a mechanism for phenotypic adaptation (17, 18). More generally, our study highlights important genomic changes that occurred during the transition from land to water in the cetacean lineage and thus helps to understand the molecular determinants of their remarkable adaptations.
MATERIALS AND METHODS
Detecting genes lost in cetaceans during the transition from land to water
We first searched for genes that exhibit inactivating mutations in the bottlenose dolphin, killer whale, sperm whale, and common minke whale using data generated by a previously developed gene loss detection approach (18). Briefly, this approach used the human Ensembl (www.ensembl.org) version 90 gene annotation and a genome alignment with the human hg38 assembly as the reference (68) (all analyzed assemblies with their accession numbers are listed in table S1) to detect stop codon mutations, frameshifting insertions or deletions, deletions of entire exons or genes, and mutations that disrupt splice sites (18). The approach performs a series of filter steps to remove artifacts related to genome assembly or alignment and evolutionary exon-intron structure changes in conserved genes. These steps comprise excluding those deletions that overlap assembly gaps in a query genome, re-aligning all coding exons with CESAR (Codon Exon Structure Aware Realigner) to detect evolutionary splice site shifts and to avoid spurious frameshifts due to alignment ambiguities (69), and excluding alignments to paralogs or processed pseudogenes. Last, the approach considers all principal or alternative APPRIS isoforms (http://appris.bioinfo.cnio.es) of a gene and outputs data for the isoform with the smallest number of inactivating mutations.
To identify genes lost on the stem Cetacea branch, our genomic screen comprised the following steps (summarized in fig. S1). Starting with 19,769 genes annotated in the human genome, we considered the 18,363 genes, which are present in the genome assemblies of at least half (31) of the 62 placental mammals. Next, we extracted the 2472 genes that were not classified as intact in any of the four analyzed cetaceans. To obtain candidate genes whose loss may be involved in aquatic adaptations, we excluded all genes that were inactivated in more than 5% (3 of 55) of the terrestrial mammals, resulting in 350 genes. These genes included 114 genes belonging to the keratin-associated and olfactory receptor gene families. Because genome alignments have lower accuracy in aligning members of these large gene families and because losses of keratin-associated and olfactory receptor genes have been studied in detail previously (9, 11, 12), we focused on the 236 remaining genes.
For these 236 genes, we manually investigated whether they were inactivated before the split of odontocetes and mysticetes. To this end, we first extracted those genes with stop codon, frameshift, or splice site mutations that are shared between species from both clades. Second, we classified genes that are partially or completely deleted in odontocetes and mysticetes into two groups, those that have shared deletion breakpoints between at least one toothed and baleen whale and those where the deletion breakpoints are not shared between both lineages. To assess the deletion breakpoints up- and downstream of the deleted genes, we manually inspected the pairwise genome alignment chains (70) between human and cetaceans in the University of California, Santa Cruz (UCSC) genome browser. We only included genes that exhibit shared breakpoints (such as KLKB1; see Fig. 1A) because the most parsimonious explanation is a single deletion event in the cetacean stem lineage. However, it should be noted that a single ancestral deletion event may have been obscured by subsequent decay of the breakpoint regions in individual lineages; thus, gene deletions without shared breakpoints, which are not included in this study, may have also occurred in the cetacean stem lineage. Likewise, we also excluded genes that exhibit smaller (stop codon, frameshift, and splice site) inactivating mutations in one clade and are deleted in the other clade, although the deletion may have happened after a single gene inactivation event occurred in the cetacean stem lineage. This analysis resulted in 110 genes that were inactivated before the split of odontocetes and mysticetes.
Excluding genes with inactivating mutations in the hippopotamus lineage
To detect those genes that were inactivated during the transition from land to water in the cetacean stem lineage, we next excluded all genes that have inactivating mutations in the hippopotamus lineage. To this end, we first aligned the genome of the common hippopotamus (23) to the human hg38 assembly using lastz (parameters K = 2400, L = 3000, and default scoring matrix), axtChain, chainCleaner, and chainNet (all with default parameters) as done before (13) and used our approach to detect inactivating mutations in the common hippopotamus. We obtained 85 genes that were inactivated in the cetacean stem lineage, meaning after the split between Cetacea and Hippopotamidae.
For the 11 genes discussed in detail (F12, KLKB1, POLM, MAP3K19, SEC14L3, SLC6A18, SLC4A9, AANAT, ASMT, MTNR1A, and MTNR1B), we further corroborated the lack of inactivating mutations in the hippopotamus lineage. To this end, we made use of unassembled sequencing reads of the pygmy hippopotamus (Choeropsis liberiensis). We mapped these reads to exonic sequences of the common hippopotamus, including ~60–base pair (bp) flanking intron on each side using Geneious 11.1.5 (www.geneious.com). In cases where an exon was not present in the common hippopotamus because of an assembly gap, we used the orthologous exon from cow and BLASTed the Sequence Read Archive (SRA) of the common hippopotamus (SRR5663647) to recover and assemble the missing exons. Manual inspection confirmed that all of the intact exons in the common hippopotamus also lack inactivating mutations in the pygmy hippopotamus.
Validating gene loss in additional cetacean species
For all 85 genes listed in table S2, we investigated whether shared inactivating mutations are present in the genomes of three additional cetaceans that were not part of the whole-genome alignment (68), the Yangtze River dolphin, beluga whale, and bowhead whale (11, 24, 25). To this end, we aligned these genomes to the hg38 assembly as described above and manually confirmed the presence of shared inactivation mutations. Furthermore, we used our alignment chains, which are sensitive enough to capture many gene duplications that happened before the divergence of mammals, to confirm that these 85 lost genes do not have another intact copy elsewhere in a cetacean genome due to a more recent gene duplication event.
Validation of inactivating mutations with raw sequencing reads
For the 11 genes discussed in detail (F12, KLKB1, POLM, MAP3K19, SEC14L3, SLC6A18, SLC4A9, AANAT, ASMT, MTNR1A, and MTNR1B), we validated the correctness of all 251 inactivating mutations that are present in the bottlenose dolphin, killer whale, sperm whale, common minke whale, and manatee genome using raw sequencing reads. To this end, we extracted 100-bp sequences covering the respective mutations and used BLAST to retrieve alignments to the species’ raw sequencing reads deposited on the SRA (experiment IDs are listed in table S3). We then determined the number of reads that support or do not support each mutation (table S3).
Analysis of available RNA-seq data
For these 11 genes, we further investigated whether they still produce spliced transcripts using available RNA sequencing (RNA-seq) data of cetaceans. We downloaded RNA-seq data of the bottlenose dolphin skin and blood (PRJNA385781, SRA study: SRP106690) and the minke whale brain, heart, kidney, liver, lung, and muscle (PRJNA72723, SRA study: SRP025154) (6) from the National Center for Biotechnology Information (NCBI) SRA. We processed all SRA read files with fastq-dump using parameters for removing technical reads (skip technical), filtering (read-filter = pass) and removing tags (clip), splitting paired-end reads into according files (split files), keeping read identifiers (readids), and formatting data into base space (dumpbase). Reads were then mapped to the genome assembly of the bottlenose dolphin (turTru3) and the minke whale (balAcu1) using STAR [Spliced Transcripts Alignment to a Reference (version 2.4.2a); https://github.com/alexdobin/STAR/releases/tag/STAR_2.4.2a). For bottlenose dolphin, genome indexes were generated with default parameters. For minke whale, we adjusted the number of bins for the indexes according to the number of total bases and scaffolds in the assembly (genomeChrBinNbits = 18). The minke whale RNA-seq data consist of paired-end reads, whereas the dolphin RNA-seq data consist of single-end reads. During mapping, we specified input files according to paired- or single-end read data. All runs were mapped separately using parameters for removing reads that map to many different locations (outFilterMultimapNmax = 20) and limiting the number of allowed mismatches for mapped reads (outFilterMismatchNoverLmax = 0.04). We used bedtools (https://bedtools.readthedocs.io/en/latest/) and bedGraphToBigWig (UCSC genome browser source code) to visualize the read coverage in the UCSC genome browser. The dolphin RNA-seq data contain 10 runs from blood samples and 25 runs from skin samples. We combined all runs from blood samples and all runs from skin samples using bigWigMerge. The read coverage across the seven candidate gene losses was then inspected to assess whether remnants of the lost genes are still expressed and properly spliced in dolphin and minke whale.
Convergent gene losses between aquatic or semi-aquatic mammals
We used our gene loss detection approach to investigate which of the genes that are lost in the cetacean stem lineage are also convergently lost in other aquatic (manatee) or semi-aquatic (pinnipeds) mammalian lineages. Because pinnipeds were only represented by the Weddell seal (family Phocidae) and the walrus, we downloaded the genome of the Antarctic fur seal (family Otariidae) (https://datadryad.org/resource/doi:10.5061/dryad.8kn8c.2), aligned it to the hg38 assembly as described above, and applied our gene loss detection pipeline. All genes convergently lost between cetaceans and manatees and/or pinnipeds are indicated in table S2.
We further screened for genes that are convergently lost between any aquatic or semi-aquatic mammalian lineages and compared the prevalence of these convergent gene losses to convergent losses between their terrestrial sister species. To this end, we first extracted from our dataset those genes that are classified as lost in one representative of the three (semi-) aquatic lineages but that are classified as intact in their respective terrestrial sister species, focusing on genes not belonging to keratin-associated and olfactory receptor gene families. Specifically, we extracted genes lost in the killer whale but not in the cow, genes lost in the Pacific walrus but not in the polar bear, and genes lost in the manatee but not in the elephant. Then, we asked how many of these genes are convergently lost in at least two (semi-) aquatic mammals (table S5). We compared this number to the number of convergent losses detected when swapping the three (semi-) aquatic and three terrestrial mammals. These data are visualized as Venn diagrams in fig. S27.
Supplementary Material
Acknowledgments
We thank the genomics community for sequencing and assembling the genomes and the UCSC genome browser group for providing software and genome annotations. We also thank J. G. Roscito for help with gene annotations, G. Amato (NYZS) for providing a tissue sample for pygmy hippo genome sequencing, M. Collin for help with laboratory work, and the Computer Service Facilities of the MPI-CBG and MPI-PKS for support. Funding: This work was supported by the Max Planck Society, the German Research Foundation (HI1423/3-1), the Leibniz Association (SAW-2016-SGN-2), and the National Science Foundation (USA) grant (DEB-1457735). Author contributions: M.Hi., J.G., and M.S.S. conceived the study. M.Hu. performed the gene loss screen and analyzed the data. N.H. analyzed the data. M.S.S., J.G., V.S., and M.Hi. contributed to the data analysis and interpretation. M.Hu. and M.Hi. drafted the manuscript. All authors critically revised the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Specifically, all analyzed genome assemblies (table S1) are publicly available on the UCSC genome browser and from NCBI. The list of 85 genes lost in the cetacean stem lineage, together with functional annotations, is provided in table S2. The list of genes convergently inactivated in two or more (semi-) aquatic mammals is provided in table S5. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/9/eaaw6671/DC1
Fig. S1. Workflow for identifying gene losses that are lost in the cetacean stem lineage.
Fig. S2. Raw DNA sequencing read validation of shared inactivating mutations.
Fig. S3. Expression of the remnants of genes lost in cetaceans.
Fig. S4. Inactivating mutations in F12 in cetaceans.
Fig. S5. Inactivating mutations in KLKB1.
Fig. S6. Inactivating mutations in POLM.
Fig. S7. Inactivating mutations in MAP3K19.
Fig. S8. Inactivating mutations in SEC14L3.
Fig. S9. Inactivating mutations in SLC6A18.
Fig. S10. Inactivating mutations in SLC4A9.
Fig. S11. Inactivating mutations in AANAT.
Fig. S12. Inactivating mutations in MTNR1B.
Fig. S13. Inactivating mutations in ASMT.
Fig. S14. Inactivating mutations in MTNR1A.
Fig. S15. Inactivating mutations in TRIM14.
Fig. S16. Inactivating mutations in TREM1.
Fig. S17. Inactivating mutations in PGLYRP1.
Fig. S18. Inactivating mutations in PGLYRP3.
Fig. S19. Inactivating mutations in PGLYRP4.
Fig. S20. Inactivating mutations in MSS51.
Fig. S21. Inactivating mutations in ACSM3.
Fig. S22. Inactivating mutations in ADH4.
Fig. S23. Inactivating mutations in SPINK7.
Fig. S24. Inactivating mutations in FABP12.
Fig. S25. Inactivating mutations in ASIC5.
Fig. S26. Inactivating mutations in C10orf82.
Fig. S27. Convergent gene losses between any of the three aquatic or semi-aquatic mammalian lineages.
Table S1. Species and genome assemblies used in this study.
Table S2. Genes lost in the cetacean stem lineage.
Table S3. Validation of all smaller inactivating mutations with raw DNA sequencing reads.
Table S4. Analysis of relaxed selection.
Table S5. Convergently inactivated genes between (semi-) aquatic mammalian clades, represented by killer whale, manatee, and Pacific walrus.
REFERENCES AND NOTES
- 1.Thewissen J. G. M., Cooper L. N., Clementz M. T., Bajpai S., Tiwari B. N., Whales originated from aquatic artiodactyls in the Eocene epoch of India. Nature 450, 1190–1194 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Gatesy J., Geisler J. H., Chang J., Buell C., Berta A., Meredith R. W., Springer M. S., McGowen M. R., A phylogenetic blueprint for a modern whale. Mol. Phylogenet. Evol. 66, 479–506 (2013). [DOI] [PubMed] [Google Scholar]
- 3.A. Berta, J. L. Sumich, K. M. Kovacs, Marine Mammals: Evolutionary Biology (Academic Press, 2005). [Google Scholar]
- 4.Kooyman G. L., Ponganis P. J., The physiological basis of diving to depth: Birds and mammals. Annu. Rev. Physiol. 60, 19–32 (1998). [DOI] [PubMed] [Google Scholar]
- 5.McGowen M. R., Gatesy J., Wildman D. E., Molecular evolution tracks macroevolutionary transitions in Cetacea. Trends Ecol. Evol. 29, 336–346 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Yim H.-S., Cho Y. S., Guang X., Kang S. G., Jeong J.-Y., Cha S.-S., Oh H.-M., Lee J.-H., Yang E. C., Kwon K. K., Kim Y. J., Kim T. W., Kim W., Jeon J. H., Kim S.-J., Choi D. H., Jho S., Kim H.-M., Ko J., Kim H., Shin Y.-A., Jung H.-J., Zheng Y., Wang Z., Chen Y., Chen M., Jiang A., Li E., Zhang S., Hou H., Kim T. H., Yu L., Liu S., Ahn K., Cooper J., Park S.-G., Hong C. P., Jin W., Kim H.-S., Park C., Lee K., Chun S., Morin P. A., O'Brien S. J., Lee H., Kimura J., Moon D. Y., Manica A., Edwards J., Kim B. C., Kim S., Wang J., Bhak J., Lee H. S., Lee J.-H., Minke whale genome and aquatic adaptation in cetaceans. Nat. Genet. 46, 88–92 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou X., Sun D., Guang X., Ma S., Fang X., Mariotti M., Nielsen R., Gladyshev V. N., Yang G., Molecular footprints of aquatic adaptation including bone mass changes in cetaceans. Genome Biol. Evol. 10, 967–975 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mirceta S., Signore A. V., Burns J. M., Cossins A. R., Campbell K. L., Berenbrink M., Evolution of mammalian diving capacity traced by myoglobin net surface charge. Science 340, 1234192 (2013). [DOI] [PubMed] [Google Scholar]
- 9.McGowen M. R., Clark C., Gatesy J., The vestigial olfactory receptor subgenome of odontocete whales: Phylogenetic congruence between gene-tree reconciliation and supermatrix methods. Syst. Biol. 57, 574–590 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Feng P., Zheng J., Rossiter S. J., Wang D., Zhao H., Massive losses of taste receptor genes in toothed and baleen whales. Genome Biol. Evol. 6, 1254–1265 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou X., Sun F., Xu S., Fan G., Zhu K., Liu X., Chen Y., Shi C., Yang Y., Huang Z., Chen J., Hou H., Guo X., Chen W., Chen Y., Wang X., Lv T., Yang D., Zhou J., Huang B., Wang Z., Zhao W., Tian R., Xiong Z., Xu J., Liang X., Chen B., Liu W., Wang J., Pan S., Fang X., Li M., Wei F., Xu X., Zhou K., Wang J., Yang G., Baiji genomes reveal low genetic variability and new insights into secondary aquatic adaptations. Nat. Commun. 4, 2708 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nery M. F., Arroyo J. I., Opazo J. C., Increased rate of hair keratin gene loss in the cetacean lineage. BMC Genomics 15, 869 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jebb D., Hiller M., Recurrent loss of HMGCS2 shows that ketogenesis is not essential for the evolution of large mammalian brains. eLife 7, e38906 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaudry M. J., Jastroch M., Treberg J. R., Hofreiter M., Paijmans J. L. A., Starrett J., Wales N., Signore A. V., Springer M. S., Campbell K. L., Inactivation of thermogenic UCP1 as a historical contingency in multiple placental mammal clades. Sci. Adv. 3, e1602878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hecker N., Sharma V., Hiller M., Transition to an aquatic habitat permitted the repeated loss of the pleiotropic KLK8 gene in mammals. Genome Biol. Evol. 9, 3179–3188 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meredith R. W., Gatesy J., Emerling C. A., York V. M., Springer M. S., Rod monochromacy and the coevolution of cetacean retinal opsins. PLOS Genet. 9, e1003432 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Albalat R., Cañestro C., Evolution by gene loss. Nat. Rev. Genet. 17, 379–391 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Sharma V., Hecker N., Roscito J. G., Foerster L., Langer B. E., Hiller M., A genomics approach reveals insights into the importance of gene losses for mammalian adaptations. Nat. Commun. 9, 1215 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hecker N., Sharma V., Hiller M., Convergent gene losses illuminate metabolic and physiological changes in herbivores and carnivores. Proc. Natl. Acad. Sci. U.S.A. 116, 3036–3041 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindblad-Toh K., Garber M., Zuk O., Lin M. F., Parker B. J., Washietl S., Kheradpour P., Ernst J., Jordan G., Mauceli E., Ward L. D., Lowe C. B., Holloway A. K., Clamp M., Gnerre S., Alföldi J., Beal K., Chang J., Clawson H., Cuff J., Palma F. D., Fitzgerald S., Flicek P., Guttman M., Hubisz M. J., Jaffe D. B., Jungreis I., Kent W. J., Kostka D., Lara M., Martins A. L., Massingham T., Moltke I., Raney B. J., Rasmussen M. D., Robinson J., Stark A., Vilella A. J., Wen J., Xie X., Zody M. C.; Broad Institute Sequencing Platform; Whole Genome Assembly Team, Worley K. C., Kovar C. L., Muzny D. M., Gibbs R. A.; Baylor College of Medicine Human Genome Sequencing Center Sequencing Team, Warren W. C., Mardis E. R., Weinstock G. M., Wilson R. K.; Genome Institute at Washington University, Birney E., Margulies E. H., Herrero J., Green E. D., Haussler D., Siepel A., Goldman N., Pollard K. S., Pedersen J. S., Lander E. S., Kellis M., A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478, 476–482 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foote A. D., Liu Y., Thomas G. W. C., Vinař T., Alföldi J., Deng J., Dugan S., van Elk C. E., Hunter M. E., Joshi V., Khan Z., Kovar C., Lee S. L., Lindblad-Toh K., Mancia A., Nielsen R., Qin X., Qu J., Raney B. J., Vijay N., Wolf J. B. W., Hahn M. W., Muzny D. M., Worley K. C., Gilbert M. T. P., Gibbs R. A., Convergent evolution of the genomes of marine mammals. Nat. Genet. 47, 272–275 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warren W. C., Kuderna L., Alexander A., Catchen J., Pérez-Silva J. G., López-Otín C., Quesada V., Minx P., Tomlinson C., Montague M. J., Farias F. H. G., Walter R. B., Marques-Bonet T., Glenn T., Kieran T. J., Wise S. S., Wise J. P. Jr., Waterhouse R. M., Wise J. P. Sr., The novel evolution of the sperm whale genome. Genome Biol. Evol. 9, 3260–3264 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Árnason U., Lammers F., Kumar V., Nilsson M. A., Janke A., Whole-genome sequencing of the blue whale and other rorquals finds signatures for introgressive gene flow. Sci. Adv. 4, eaap9873 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones S. J. M., Taylor G. A., Chan S., Warren R. L., Hammond S. A., Bilobram S., Mordecai G., Suttle C. A., Miller K. M., Schulze A., Chan A. M., Jones S. J., Tse K., Li I., Cheung D., Mungall K. L., Choo C., Ally A., Dhalla N., Tam A. K. Y., Troussard A., Kirk H., Pandoh P., Paulino D., Coope R. J. N., Mungall A. J., Moore R., Zhao Y., Birol I., Ma Y., Marra M., Haulena M., The genome of the beluga whale (Delphinapterus leucas). Genes (Basel) 8, E378 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keane M., Semeiks J., Webb A. E., Li Y. I., Quesada V., Craig T., Madsen L. B., van Dam S., Brawand D., Marques P. I., Michalak P., Kang L., Bhak J., Yim H.-S., Grishin N. V., Nielsen N. H., Heide-Jørgensen M. P., Oziolor E. M., Matson C. W., Church G. M., Stuart G. W., Patton J. C., George J. C., Suydam R., Larsen K., López-Otín C., O’Connell M. J., Bickham J. W., Thomsen B., de Magalhães J. P., Insights into the evolution of longevity from the bowhead whale genome. Cell Rep. 10, 112–122 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmaier A. H., The contact activation and kallikrein/kinin systems: Pathophysiologic and physiologic activities. J. Thromb. Haemost. 14, 28–39 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Long A. T., Kenne E., Jung R., Fuchs T. A., Renné T., Contact system revisited: An interface between inflammation, coagulation, and innate immunity. J. Thromb. Haemost. 14, 427–437 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Kokoye Y., Ivanov I., Cheng Q., Matafonov A., Dickeson S. K., Mason S., Sexton D. J., Renné T., McCrae K., Feener E. P., Gailani D., A comparison of the effects of factor XII deficiency and prekallikrein deficiency on thrombus formation. Thromb. Res. 140, 118–124 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Radziwon P., Olszański R., Korsak J., Siermontowski P., Dąbrowiecki Z., Factor XII–A limitation for divers? Polish Hyperbaric Res. 52, 7–12 (2015). [Google Scholar]
- 30.Stavrou E. X., Fang C., Merkulova A., Alhalabi O., Grobe N., Antoniak S., Mackman N., Schmaier A. H., Reduced thrombosis in Klkb1−/− mice is mediated by increased Mas receptor, prostacyclin, Sirt1, and KLF4 and decreased tissue factor. Blood 125, 710–719 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shariat-Madar Z., Mahdi F., Schmaier A. H., Recombinant prolylcarboxypeptidase activates plasma prekallikrein. Blood 103, 4554–4561 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Wilhelm Filho D., Sell F., Ribeiro L., Ghislandi M., Carrasquedo F., Fraga C. G., Wallauer J. P., Simões-Lopes P. C., Uhart M. M., Comparison between the antioxidant status of terrestrial and diving mammals. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 133, 885–892 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Pryor J. M., Waters C. A., Aza A., Asagoshi K., Strom C., Mieczkowski P. A., Blanco L., Ramsden D. A., Essential role for polymerase specialization in cellular nonhomologous end joining. Proc. Natl. Acad. Sci. U.S.A. 112, E4537–E4545 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shibutani S., Takeshita M., Grollman A. P., Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature 349, 431–434 (1991). [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y., Wu X., Guo D., Rechkoblit O., Taylor J.-S., Geacintov N. E., Wang Z., Lesion bypass activities of human DNA polymerase μ. J. Biol. Chem. 277, 44582–44587 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Maga G., Villani G., Crespan E., Wimmer U., Ferrari E., Bertocci B., Hübscher U., 8-oxo-guanine bypass by human DNA polymerases in the presence of auxiliary proteins. Nature 447, 606–608 (2007). [DOI] [PubMed] [Google Scholar]
- 37.van Loon B., Markkanen E., Hübscher U., Oxygen as a friend and enemy: How to combat the mutational potential of 8-oxo-guanine. DNA Repair (Amst) 9, 604–616 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Lucas D., Delgado-García J. M., Escudero B., Albo C., Aza A., Acín-Pérez R., Torres Y., Moreno P., Enríquez J. A., Samper E., Blanco L., Fairén A., Bernad A., Gruart A., Increased learning and brain long-term potentiation in aged mice lacking DNA polymerase μ. PLOS ONE 8, e53243 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Escudero B., Lucas D., Albo C., Dhup S., Bacher J. W., Sánchez-Muñoz A., Fernández M., Rivera-Torres J., Carmona R. M., Fuster E., Carreiro C., Bernad R., González M. A., Andrés V., Blanco L., Roche E., Fabregat I., Samper E., Bernad A., Polμ deficiency increases resistance to oxidative damage and delays liver aging. PLOS ONE 9, e93074 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lucas D., Escudero B., Ligos J. M., Segovia J. C., Estrada J. C., Terrados G., Blanco L., Samper E., Bernad A., Altered hematopoiesis in mice lacking DNA polymerase μ is due to inefficient double-strand break repair. PLOS Genet. 5, e1000389 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piscitelli M. A., Raverty S. A., Lillie M. A., Shadwick R. E., A review of cetacean lung morphology and mechanics. J. Morphol. 274, 1425–1440 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Boehme S. A., Franz-Bacon K., DiTirro D. N., Ly T. W., Bacon K. B., MAP3K19 is a novel regulator of TGF-β signaling that impacts bleomycin-induced lung injury and pulmonary fibrosis. PLOS ONE 11, e0154874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boehme S. A., Franz-Bacon K., Ludka J., DiTirro D. N., Ly T. W., Bacon K. B., MAP3K19 is overexpressed in COPD and is a central mediator of cigarette smoke-induced pulmonary inflammation and lower airway destruction. PLOS ONE 11, e0167169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shan L., Noritake S., Fujiwara M., Asano S., Yoshida-Noro C., Noro N., Yamashita K., Kawakami T., Sec14l3 is specifically expressed in mouse airway ciliated cells. Inflammation 35, 702–712 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Hishikawa D., Shindou H., Harayama T., Ogasawara R., Suwabe A., Shimizu T., Identification of Sec14-like 3 as a novel lipid-packing sensor in the lung. FASEB J. 27, 5131–5140 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Miller N. J., Postle A. D., Orgeig S., Koster G., Daniels C. B., The composition of pulmonary surfactant from diving mammals. Respir. Physiol. Neurobiol. 152, 152–168 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Singer D., Camargo S. M. R., Huggel K., Romeo E., Danilczyk U., Kuba K., Chesnov S., Caron M. G., Penninger J. M., Verrey F., Orphan transporter SLC6A18 is renal neutral amino acid transporter B0AT3. J. Biol. Chem. 284, 19953–19960 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miyaji K., Nagao K., Bannai M., Asakawa H., Kohyama K., Ohtsu D., Terasawa F., Ito S., Iwao H., Ohtani N., Ohta M., Characteristic metabolism of free amino acids in cetacean plasma: Cluster analysis and comparison with mice. PLOS ONE 5, e13808 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quan H., Athirakul K., Wetsel W. C., Torres G. E., Stevens R., Chen Y. T., Coffman T. M., Caron M. G., Hypertension and impaired glycine handling in mice lacking the orphan transporter XT2. Mol. Cell. Biol. 24, 4166–4173 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ortiz R. M., Osmoregulation in marine mammals. J. Exp. Biol. 204, 1831–1844 (2001). [DOI] [PubMed] [Google Scholar]
- 51.Peña-Münzenmayer G., George A. T., Shull G. E., Melvin J. E., Catalán M. A., Ae4 (Slc4a9) is an electroneutral monovalent cation-dependent Cl−/HCO3− exchanger. J. Gen. Physiol. 147, 423–436 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pena-Munzenmayer G., Catalán M. A., Kondo Y., Jaramillo Y., Liu F., Shull G. E., Melvin J. E., Ae4 (Slc4a9) anion exchanger drives Cl− uptake-dependent fluid secretion by mouse submandibular gland acinar cells. J. Biol. Chem. 290, 10677–10688 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.B. Cozzi, S. Huggenberger, H. A. Oelschläger, Anatomy of Dolphins (Elsevier Science, 2016). [Google Scholar]
- 54.Evans R. L., Park K., Turner R. J., Watson G. E., Nguyen H.-V., Dennett M. R., Hand A. R., Flagella M., Shull G. E., Melvin J. E., Severe impairment of salivation in Na+/K+/2Cl− cotransporter (NKCC1)-deficient mice. J. Biol. Chem. 275, 26720–26726 (2000). [DOI] [PubMed] [Google Scholar]
- 55.Flagella M., Clarke L. L., Miller M. L., Erway L. C., Giannella R. A., Andringa A., Gawenis L. R., Kramer J., Duffy J. J., Doetschman T., Lorenz J. N., Yamoah E. N., Cardell E. L., Shull G. E., Mice lacking the basolateral Na-K-2Cl cotransporter have impaired epithelial chloride secretion and are profoundly deaf. J. Biol. Chem. 274, 26946–26955 (1999). [DOI] [PubMed] [Google Scholar]
- 56.Chambrey R., Kurth I., Peti-Peterdi J., Houillier P., Purkerson J. M., Leviel F., Hentschke M., Zdebik A. A., Schwartz G. J., Hübner C. A., Eladari D., Renal intercalated cells are rather energized by a proton than a sodium pump. Proc. Natl. Acad. Sci. U.S.A. 110, 7928–7933 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Birukawa N., Ando H., Goto M., Kanda N., Pastene L. A., Nakatsuji H., Hata H., Urano A., Plasma and urine levels of electrolytes, urea and steroid hormones involved in osmoregulation of cetaceans. Zoolog. Sci. 22, 1245–1257 (2005). [DOI] [PubMed] [Google Scholar]
- 58.Lyamin O. I., Manger P. R., Ridgway S. H., Mukhametov L. M., Siegel J. M., Cetacean sleep: An unusual form of mammalian sleep. Neurosci. Biobehav. Rev. 32, 1451–1484 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martí-Solans J., Belyaeva O. V., Torres-Aguila N. P., Kedishvili N. Y., Albalat R., Cañestro C., Coelimination and survival in gene network evolution: Dismantling the RA-signaling in a chordate. Mol. Biol. Evol. 33, 2401–2416 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Funasaka N., Yoshioka M., Suzuki M., Ueda K., Miyahara H., Uchida S., Seasonal difference of diurnal variations in serum melatonin, cortisol, testosterone, and rectal temperature in indo-pacific bottlenose dolphins (Tursiops aduncus). Aquatic Mammals 37, 433–442 (2011). [Google Scholar]
- 61.Panin M., Gabai G., Ballarin C., Peruffo A., Cozzi B., Evidence of melatonin secretion in cetaceans: Plasma concentration and extrapineal HIOMT-like presence in the bottlenose dolphin Tursiops truncatus. Gen. Comp. Endocrinol. 177, 238–245 (2012). [DOI] [PubMed] [Google Scholar]
- 62.Hohjoh H., Takasu M., Shishikura K., Takahashi Y., Honda Y., Tokunaga K., Significant association of the arylalkylamine N-acetyltransferase (AA-NAT) gene with delayed sleep phase syndrome. Neurogenetics 4, 151–153 (2003). [DOI] [PubMed] [Google Scholar]
- 63.Gandhi A. V., Mosser E. A., Oikonomou G., Prober D. A., Melatonin is required for the circadian regulation of sleep. Neuron 85, 1193–1199 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Comai S., Ochoa-Sanchez R., Gobbi G., Sleep–wake characterization of double MT1/MT2 receptor knockout mice and comparison with MT1 and MT2 receptor knockout mice. Behav. Brain Res. 243, 231–238 (2013). [DOI] [PubMed] [Google Scholar]
- 65.Park H. J., Park J. K., Kim S. K., Cho A.-R., Kim J. W., Yim S.-V., Chung J.-H., Association of polymorphism in the promoter of the melatonin receptor 1A gene with schizophrenia and with insomnia symptoms in schizophrenia patients. J. Mol. Neurosci. 45, 304–308 (2011). [DOI] [PubMed] [Google Scholar]
- 66.Kräuchi K., Cajochen C., Pache M., Flammer J., Wirz-Justice A., Thermoregulatory effects of melatonin in relation to sleepiness. Chronobiol. Int. 23, 475–484 (2006). [DOI] [PubMed] [Google Scholar]
- 67.Sharma V., Hiller M., Loss of enzymes in the bile acid synthesis pathway explains differences in bile composition among mammals. Genome Biol. Evol. 10, 3211–3217 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sharma V., Hiller M., Increased alignment sensitivity improves the usage of genome alignments for comparative gene annotation. Nucleic Acids Res. 45, 8369–8377 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sharma V., Schwede P., Hiller M., CESAR 2.0 substantially improves speed and accuracy of comparative gene annotation. Bioinformatics 33, 3985–3987 (2017). [DOI] [PubMed] [Google Scholar]
- 70.Kent W. J., Baertsch R., Hinrichs A., Miller W., Haussler D., Evolution's cauldron: Duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl. Acad. Sci. U.S.A. 100, 11484–11489 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kodama Y., Shumway M., Leinonen R.; International Nucleotide Sequence Database Collaboration , The Sequence Read Archive: Explosive growth of sequencing data. Nucleic Acids Res. 40, D54–D56 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Haeussler M., Zweig A. S., Tyner C., Speir M. L., Rosenbloom K. R., Raney B. J., Lee C. M., Lee B. T., Hinrichs A. S., Gonzalez J. N., Gibson D., Diekhans M., Clawson H., Casper J., Barber G. P., Haussler D., Kuhn R. M., Kent W. J., The UCSC Genome Browser database: 2019 update. Nucleic Acids Res. 47, D853–D858 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sharma V., Elghafari A., Hiller M., Coding exon-structure aware realigner (CESAR) utilizes genome alignments for accurate comparative gene annotation. Nucleic Acids Res. 44, e103 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kinsella R. J., Kähäri A., Haider S., Zamora J., Proctor G., Spudich G., Almeida-King J., Staines D., Derwent P., Kerhornou A., Kersey P., Flicek P., Ensembl BioMarts: A hub for data retrieval across taxonomic space. Database 2011, bar030 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dziarski R., Kashyap D. R., Gupta D., Mammalian peptidoglycan recognition proteins kill bacteria by activating two-component systems and modulate microbiome and inflammation. Microb. Drug Resist. 18, 280–285 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dziarski R., Gupta D., How innate immunity proteins kill bacteria and why they are not prone to resistance. Curr. Genet. 64, 125–129 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moyer A. L., Wagner K. R., Mammalian Mss51 is a skeletal muscle-specific gene modulating cellular metabolism. J. Neuromuscul. Dis. 2, 371–385 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Velten B. P., Dillaman R. M., Kinsey S. T., McLellan W. A., Pabst D. A., Novel locomotor muscle design in extreme deep-diving whales. J. Exp. Biol. 216, 1862–1871 (2013). [DOI] [PubMed] [Google Scholar]
- 79.Fujino T., Takei Y. A., Sone H., Ioka R. X., Kamataki A., Magoori K., Takahashi S., Sakai J., Yamamoto T. T., Molecular identification and characterization of two medium-chain acyl-CoA synthetases, MACS1 and the Sa gene product. J. Biol. Chem. 276, 35961–35966 (2001). [DOI] [PubMed] [Google Scholar]
- 80.Molotkov A., Deltour L., Foglio M. H., Cuenca A. E., Duester G., Distinct retinoid metabolic functions for alcohol dehydrogenase genes Adh1 and Adh4 in protection against vitamin A toxicity or deficiency revealed in double null mutant mice. J. Biol. Chem. 277, 13804–13811 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.W. F. Perrin, B. Würsig, J. G. M. Thewissen, Encyclopedia of Marine Mammals (Academic Press, 2008). [Google Scholar]
- 82.Smathers R. L., Petersen D. R., The human fatty acid-binding protein family: Evolutionary divergences and functions. Hum. Genomics 5, 170–191 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Boiko N., Kucher V., Wang B., Stockand J. D., Restrictive expression of acid-sensing ion channel 5 (asic5) in unipolar brush cells of the vestibulocerebellum. PLOS ONE 9, e91326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.The GTEx Consortium , The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 348, 648–660 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/9/eaaw6671/DC1
Fig. S1. Workflow for identifying gene losses that are lost in the cetacean stem lineage.
Fig. S2. Raw DNA sequencing read validation of shared inactivating mutations.
Fig. S3. Expression of the remnants of genes lost in cetaceans.
Fig. S4. Inactivating mutations in F12 in cetaceans.
Fig. S5. Inactivating mutations in KLKB1.
Fig. S6. Inactivating mutations in POLM.
Fig. S7. Inactivating mutations in MAP3K19.
Fig. S8. Inactivating mutations in SEC14L3.
Fig. S9. Inactivating mutations in SLC6A18.
Fig. S10. Inactivating mutations in SLC4A9.
Fig. S11. Inactivating mutations in AANAT.
Fig. S12. Inactivating mutations in MTNR1B.
Fig. S13. Inactivating mutations in ASMT.
Fig. S14. Inactivating mutations in MTNR1A.
Fig. S15. Inactivating mutations in TRIM14.
Fig. S16. Inactivating mutations in TREM1.
Fig. S17. Inactivating mutations in PGLYRP1.
Fig. S18. Inactivating mutations in PGLYRP3.
Fig. S19. Inactivating mutations in PGLYRP4.
Fig. S20. Inactivating mutations in MSS51.
Fig. S21. Inactivating mutations in ACSM3.
Fig. S22. Inactivating mutations in ADH4.
Fig. S23. Inactivating mutations in SPINK7.
Fig. S24. Inactivating mutations in FABP12.
Fig. S25. Inactivating mutations in ASIC5.
Fig. S26. Inactivating mutations in C10orf82.
Fig. S27. Convergent gene losses between any of the three aquatic or semi-aquatic mammalian lineages.
Table S1. Species and genome assemblies used in this study.
Table S2. Genes lost in the cetacean stem lineage.
Table S3. Validation of all smaller inactivating mutations with raw DNA sequencing reads.
Table S4. Analysis of relaxed selection.
Table S5. Convergently inactivated genes between (semi-) aquatic mammalian clades, represented by killer whale, manatee, and Pacific walrus.


