Blood culture bottles containing antibiotic binding resins are routinely used to minimize artificial sterilization in the presence of antibiotics. However, the resin binding kinetics can differ between antibiotics and concentrations.
KEYWORDS: Pseudomonas aeruginosa, Escherichia coli, Klebsiella pneumoniae; bacteremia; antimicrobial stewardship; carbapenem; cephalosporin; piperacillin; levofloxacin
ABSTRACT
Blood culture bottles containing antibiotic binding resins are routinely used to minimize artificial sterilization in the presence of antibiotics. However, the resin binding kinetics can differ between antibiotics and concentrations. This study assessed the impact of clinically meaningful peak, midpoint, and trough concentrations of meropenem, imipenem, cefepime, cefazolin, levofloxacin, and piperacillin-tazobactam on the recovery of Pseudomonas aeruginosa, Escherichia coli, and Klebsiella pneumoniae from resin-containing BacT/Alert FA Plus and Bactec Aerobic/F blood culture bottles. P. aeruginosa-inoculated bottles alarmed positive in 4/20 (20%), 16/20 (80%), and 20/20 (100%) of those with peak, midpoint, and trough concentrations of antipseudomonal agents, respectively (P ≤ 0.001). E. coli was recovered from 8/24 (33%), 11/24 (46%), and 14/24 (58%) of bottles with peak, midpoint, and trough concentrations, respectively (P = 0.221). K. pneumoniae was recovered from 8/16 (50%) at all concentrations of the studied antibiotics (P = 1.0). BacT/Alert and Bactec bottles inoculated with antibiotics and P. aeruginosa had similar times to detection (TTD) (P = 0.352); however, antibiotic-containing BacT/Alert bottles had a shorter TTD compared with antibiotic-containing Bactec bottles for E. coli (P = 0.026) and K. pneumoniae (P ≤ 0.001). Pathogen recovery in BacT/Alert FA Plus and Bactec Aerobic/F blood culture bottles containing antibiotic binding resins was greatly reduced in the presence of antibiotics, especially at higher concentrations. These data support the practice of drawing blood cultures immediately before an antibiotic dose to maximize the chances of pathogen recovery.
INTRODUCTION
Bloodstream infections (BSIs) remain a significant health care concern in the United States and globally. It is estimated that the United States experiences 536,000 to 628,000 BSI episodes and 72,000 to 85,000 deaths from BSIs per year, making it the seventh most common cause of death and the leading cause of death caused by infection (1). BSIs also pose a large financial burden on patients and the health care system. Depending on the patient’s level of care, it can cost anywhere between $10,000 and $75,000 per case (2, 3).
Because timely identification of the causative organism and initiation of appropriate antimicrobial are vital when treating BSIs, blood cultures play a central role in the diagnosis. Furthermore, the lack of antimicrobial susceptibility testing when a pathogen is not available may also result in prolonged, inappropriate courses of broad-spectrum antimicrobials. The Surviving Sepsis Campaign guidelines recommend that blood cultures be obtained before initiating antimicrobial therapy due to the risk of culture sterilization, which can occur only minutes after an initial dose (4). Unfortunately, as many as 50 to 80% of hospitalized patients have blood cultures collected after receiving an antibiotic (5). To minimize this risk, blood culture bottles containing proprietary binding resins that inactivate residual concentrations are available. However, these bottles and their binding resins perform differently based on the specific organisms and antimicrobials being combined (6–10). Lovern and colleagues observed differences in binding affinities for antibiotics across several classes, including β-lactams, fluoroquinolones, and glycopeptides, and these differences translated into the ability for BSI organisms to grow and be identified (11).
In previous in vitro sterilization blood culture studies, only a single antibiotic concentration was tested; moreover, this was usually a peak concentration. The actual concentration tested is critical with respect to timing of blood cultures when patients have already received a dose. For example, Grupper and colleagues observed that Pseudomonas aeruginosa isolates were only consistently recovered from BacT/Alert FA Plus and Bactec Plus Aerobic/F bottles in the presence of trough concentrations of meropenem, ceftolozane-tazobactam, and ceftazidime-avibactam (10). Higher concentrations (e.g., peak or midpoint) sterilized the bottles. Therefore, if it not possible to collect blood cultures before antibiotics are administered, they should be obtained immediately before a patient’s upcoming dose (i.e., trough or lowest concentration of antibiotic present) (10). To our knowledge, no data confirming that this theory applies to Enterobacteriaceae are available. The objective of this study was to further investigate the antibiotic binding and microbiological recovery abilities of BacT/Alert FA Plus and Bactec Plus Aerobic/F blood culture bottles by introducing P. aeruginosa, Escherichia coli, and Klebsiella pneumoniae to blood containing a variety of antibiotics at clinically relevant concentrations.
MATERIALS AND METHODS
Antibiotics.
Commercially available formulations of meropenem (Fresenius Kabi USA, Inc., Lake Zurich, IL), imipenem (Fresenius Kabi USA, Inc., Lake Zurich, IL), cefepime (WG Critical Care, LLC, Paramus, NJ), cefazolin (Sagent Pharmaceuticals, Schaumburg, IL), levofloxacin (Aurobindo Pharma Ltd., Telangana, India), and piperacillin-tazobactam (Fresenius Kabi USA, Inc., Lake Zurich, IL) were purchased through Cardinal Health (Dublin, OH). The agents were reconstituted per their package inserts.
Bacteria.
Three well-established quality control Gram-negative bacteria were utilized in this study. With the exception of cefazolin against K. pneumoniae, isolates were selected to be susceptible to the antibiotics, because previous observations with highly resistant isolates demonstrated growth regardless of concentration present (9). P. aeruginosa ATCC 27853, E. coli ATCC 25922, and K. pneumoniae ATCC 13883 were acquired from American Type Culture Collection (ATCC; Manassas, VA). The isolates were frozen on skim milk and stored at –80°C for the duration of the study. Prior to each experiment, they were subcultured twice on 5% blood Trypticase soy agar plates and incubated at 37°C overnight. MICs were determined in triplicate using Clinical and Laboratory Standards Institute broth microdilution methodology for the following combinations: MICs for all antibiotics were obtained for E. coli, while meropenem, imipenem, cefepime, levofloxacin, and piperacillin-tazobactam MICs were obtained against P. aeruginosa, and meropenem, cefazolin, cefepime, and piperacillin-tazobactam MICs were determined for K. pneumoniae.
Blood culture bottles.
BacT/Alert FA Plus Aerobic bottles were provided by bioMérieux, Inc. (Durham, NC) and tested in the bioMérieux BacT/Alert Virtuo detection system in the Center for Anti-Infective Research and Development (CAIRD) laboratory. Bactec Plus Aerobic/F bottles (Becton, Dickinson and Company, Franklin Lakes, NJ) were purchased from the manufacturer and tested on the BD Bactec FX detection system used in the Hartford Hospital clinical microbiology department.
Blood culture bottle preparation.
After obtaining written informed consent, fresh whole blood was collected from healthy adult volunteers into tubes containing sodium polyanethole sulfonate, an anticoagulant commonly found in blood culture bottles, and pooled on the first day of each experiment. The study antibiotics were then supplemented into the pooled blood to achieve the mean peak, midpoint, or trough plasma concentrations seen in regimens for critically ill patients (Table 1) (12–17). Four blood culture bottles (2 BacT/Alert FA Plus Aerobic and 2 Bactec Plus Aerobic/F bottles) were prepared for each organism/antibiotic/concentration combination.
TABLE 1.
Targeted total plasma concentrations for antibiotic regimens in critically ill patientsa
| Antibiotic | Dosing regimenb | Concentration (μg/ml) at: |
||
|---|---|---|---|---|
| Peak | Midpoint | Trough | ||
| Meropenem | 2 g q8h (3-h infusion) | 40 | 20 | 5 |
| Imipenem | 500 mg q6h (0.5-h infusion) | 25 | 3 | 1 |
| Levofloxacin | 750 mg q24h (1.5-h infusion) | 13 | 3 | 1.5 |
| Cefazolin | 1 g q8h (0.5-h infusion) | 108 | 13 | 3 |
| Cefepime | 2 g q8h (0.5-h infusion) | 105 | 33 | 21 |
| Piperacillin-tazobactam | 4.5g (4 g piperacillin-0.5 g tazobactam) q6h (0.5-h infusion) | 177 | 42 | 12 |
After incubation, bacteria were emulsified in sterile 0.9% sodium chloride to a McFarland density of 1 and diluted to obtain 7 to 30 CFU per 0.5 ml. Blood culture bottles were then inoculated with this bacterial suspension to simulate the minimum CFU detectable by the BacT/Alert Virtuo and Bactec FX blood culture systems (10, 11). Once inoculated, 10 ml of antibiotic-containing blood was added to each bottle. For controls, two BacT/Alert bottles and two Bactec bottles were inoculated and injected with whole blood that did not contain any antibiotics. One bottle per manufacturer/organism/antibiotic/concentration combination was loaded into its respective blood culture detection system within 1 h of preparation and incubated until positive growth was detected, up to a maximum of 120 h. Similarly, the duplicate bottles from each manufacturer were placed on an agitator and incubated at 37°C in the CAIRD laboratory.
Blood culture bottle sampling.
CAIRD blood culture bottles were sampled for antibiotic concentrations at 0, 4, and 12 h after inoculation, and for bacterial CFU at 0, 4, 12, 24, 48, 72, 96, and 120 h after inoculation. Samples for antibiotic concentration determination were frozen at –80°C until assayed. CFU were quantified by performing manual colony counts of serially diluted plates with a lower limit of detection of 1.7 log10 CFU/ml. The Virtuo and FX detection system bottles were sampled at 0 h for both antibiotic concentration and bacterial CFU. When a bottle alarmed positive, it was again sampled for CFU within 6 h of the alert, while noting the time to detection (TTD) from the detection system’s log. Bottles that did not alarm by 120 h were sampled at that time to confirm the absence of the study organism.
Data analyses.
TTD was normalized to the loading time into each detection systems. Bottles negative at 120 h were listed as “no growth.” Mean CFU/ml at each sampling time point for replicate bottles was determined. Chi-square or Fisher’s exact tests were used to compare the proportion of bottles that alarmed positive for each drug/organism combination at each concentration as well as to compare between BacT/Alert and Bactec bottles. The mean CFU per bottle and TTD between manufacturers were compared using Student’s t test or the Mann-Whitney rank sum test, depending on the Gaussian distribution of the data. All analyses were conducted using Sigma Plot version 14 (Systat Inc., San Jose, CA). An a priori P of <0 0.05 was considered statistically significant.
Concentrations of meropenem, cefepime, cefazolin, and the piperacillin component of piperacillin-tazobactam in both whole blood and blood culture bottle samples were assayed using validated high-performance liquid chromatography (HPLC) assays developed by CAIRD. In summary, all assays consisted of a Waters 626 gradient pump (Waters Associates, Milford, MA) and 717 plus autosampler (Waters) with a programmable UV detector (model 526; ESA Inc., Chelmsford, MA). The autosampler was cooled to 10°C. Columns were maintained at room temperature. The EZChrom Elite chromatography data system (Scientific Software Inc., Pleasanton, CA) was used to quantify the peak heights. Each sample was injected into the system once for determination of concentration. For meropenem, the lower limit of detection was 0.25 μg/ml for both matrices; the intraday coefficient of variation (CV) for low and high check samples was <5%. For cefepime, the lower limit of detection was 0.5 μg/ml in both matrices; the intraday CV for low and high check samples in whole blood was 3.65% and 2.85%, respectively; CV in blood culture bottles was 6.09% and 6.73%, respectively. For cefazolin, the lower limit of detection was 0.5 μg/ml in both matrices; the CV for low and high check samples in whole blood was 5.47% and 3.38%, respectively; CV in blood culture bottles was 1.58% and 3.57%, respectively. Finally, the lower limit of detection for piperacillin was 2 μg/ml in both matrices; the CV for low and high check samples was <5%. Assayed concentrations were plotted graphically to compare the rates of antibiotic binding relative to the antibiotic MICs for the organism. Zero-hour concentrations in bottles were calculated as the total concentration in whole blood divided by 2 to account for 50% hematocrit and then divided by the dilution of volume from blood culture media (4-fold in both detection systems).
RESULTS
MICs.
Antibiotic MICs against the three Gram-negative organisms are provided in Table 2. All organisms were susceptible to the antibiotics included in the study, except for K. pneumoniae ATCC 13883 to cefazolin, which was intermediate at an MIC of 4 μg/ml.
TABLE 2.
Modal MICs and susceptibility categorization of study isolates and antibiotics
| Isolate | Antibiotic MIC (μg/ml) (susceptibility) fora
: |
|||||
|---|---|---|---|---|---|---|
| Meropenem | Imipenem | Levofloxacin | Cefazolin | Cefepime | Piperacillin-tazobactam | |
| P. aeruginosa ATCC 27853 | 0.25 (S) | 1 (S) | 1 (S) | NT | 1 (S) | 4/4 (S) |
| E. coli ATCC 25922 | ≤0.063 (S) | 0.25 (S) | ≤0.063 (S) | 2 (S) | ≤0.063 (S) | 4/4 (S) |
| K. pneumoniae ATCC 13883 | 0.25 (S) | NT | NT | 4 (I) | 0.125 (S) | 4/4 (S) |
For P. aeruginosa, CLSI susceptibility breakpoints are ≤2 μg/ml (meropenem and imipenem), ≤1 μg/ml (levofloxacin), ≤8 μg/ml (cefepime), and ≤16/4 μg/ml (piperacillin-tazobactam). For Enterobacteriaceae, CLSI susceptibility breakpoints are ≤1 μg/ml (meropenem and imipenem), ≤0.5 μg/ml (levofloxacin), ≤2 μg/ml (cefazolin), ≤2 μg/ml (cefepime), and ≤16/4 μg/ml (piperacillin-tazobactam). S, susceptible; I, intermediate; NT, not tested.
Baseline CFU and target antibiotic concentrations.
For P. aeruginosa, E. coli, and K. pneumoniae experiments, the starting inocula obtained in bottles were 12.8 ± 4.0 CFU/bottle, 5.5 ± 1.8 CFU/bottle, and 6.6 ± 2.7 CFU/bottle, respectively. Observed concentrations of antibiotics in whole blood were nearly identical to the target concentrations for critically ill patients (Table 3).
TABLE 3.
Observed antibiotic concentrations in whole blood before inoculation into blood culture bottlesa
| Antibiotic | Mean observed ± SD (target) (μg/ml) fora
: |
||
|---|---|---|---|
| Peak | Midpoint | Trough | |
| Meropenem | 21.5 ± 2.9 (20) | 11.2 ± 3.2 (10) | 4.9 (2.5) |
| Imipenem | NT | NT | NT |
| Levofloxacin | NT | NT | NT |
| Cefazolin | 60.2 ± 3.4 (54) | 7.9 ± 0.4 (6.5) | 1.8 ± 0.2 (1.5) |
| Cefepime | 49.0 ± 1.3 (52.5) | 12.4 ± 0.3 (16.5) | 9.0 ± 0.9 (10.5) |
| Piperacillin | 76.5 ± 2.75 (88.5) | 20.5 ± 2.2 (21) | 6.0 ± 0.9 (6) |
Data are presented as mean ± standard deviation for all antibiotic concentrations (μg/ml) except the meropenem trough where a single observation was tested. Target whole-blood concentration was determined as 1/2 target plasma concentration based on the assumption of 50% hematocrit of blood from the healthy volunteers.
P. aeruginosa growth.
Control bottles without antibiotic grew to 8.71 ± 0.32 log10 CFU/ml in the BacT/Alert bottles and 8.71 ± 0.24 log10 CFU/ml in the Bactec bottles. For bottles containing antibiotics, growth was observed in 4/20 (20%), 16/20 (80%), and 20/20 (100%) of bottles containing peak, midpoint, and trough concentrations, respectively, of meropenem, imipenem, cefepime, levofloxacin, and piperacillin-tazobactam (P ≤ 0.001) (Table 4). Compared by bottle, both BacT/Alert and Bactec bottles saw growth in 2/10 (20%), 8/10 (80%), and 10/10 (100%) of bottles with peak, midpoint, and trough concentrations, respectively (P ≤ 0.001). The only bottles with peak concentrations that grew P. aeruginosa were those containing piperacillin-tazobactam. At midpoint concentrations, all bottles except for those containing meropenem alarmed positive. Finally, both systems detected growth across all bottles when only trough concentrations were introduced.
TABLE 4.
Blood culture bottle positivity and time to detection observations for P. aeruginosa, E. coli, and K. pneumoniae in BacT/Alert FA Plus Aerobic and Bactec Plus Aerobic/F blood culture bottles
| Antibiotic | Category | Mean TTD ± SD (h) fora: |
|||||
|---|---|---|---|---|---|---|---|
| BacT/Alert FA Plus Aerobic |
Bactec Plus Aerobic/F |
||||||
| P. aeruginosa | E. coli | K. pneumoniae | P. aeruginosa | E. coli | K. pneumoniae | ||
| Control | NAb | 15.6 ± 1.0 | 10.6 ± 0.2 | 11.3 (11.0–11.4) | 17.4 ± 0.4 | 11.8 ± 0.4 | 12.5 (12.2–12.6) |
| Meropenem | Peak | NG | NG | NG | NG | NG | NG |
| Midpoint | NG | NG | NG | NG | NG | NG | |
| Trough | 21.2 | NG | NG | 18.8 | NG | NG | |
| Imipenem | Peak | NG | NG | Not done | NG | NG | Not done |
| Midpoint | 17.1 | 29.5 ± 0.1 | Not done | 18.5 | NG | Not done | |
| Trough | 16.8 | 13.0 | Not done | 17.7 | NG | Not done | |
| Levofloxacin | Peak | NG | NG | Not done | NG | NG | Not done |
| Midpoint | 17.2 ± 2.0 | 13.6 ± 0.1 | Not done | 18.9 ± 1.3 | NG | Not done | |
| Trough | 17.3 ± 1.0 | 12.2 ± 1.2 | Not done | 19.0 | 14.8 ± 1.4 | Not done | |
| Cefazolin | Peak | Not done | 10.3 | 11.7 ± 0.2 | Not done | 11.8 ± 0.6 | 12.7 ± 0.4 |
| Midpoint | Not done | 11.3 | 11.5 ± 0.1 | Not done | 11.2 | 12.7 ± 0.2 | |
| Trough | Not done | 10.6 | 11.3 ± 0.0 | Not done | 11.8 | 13.6 ± 0.6 | |
| Cefepime | Peak | NG | NG | NG | NG | NG | NG |
| Midpoint | 20.1 | NG | NG | 21.8 ± 3.1 | NG | NG | |
| Trough | 18.9 ± 4.7 | NG | NG | 17.5 ± 0.9 | NG | NG | |
| Piperacillin-tazobactam | Peak | 15.5 | 10.9 | 11.2 ± 0.2 | 16.7 | 11.8 | 12.3 ± 0.2 |
| Midpoint | 15.7 | 10.8 | 11.3 | 17.0 | 11.7 | 12.3 | |
| Trough | 15.2 | 10.5 | 11.0 | 16.7 | 11.8 | 12.2 | |
Data are provided as mean TTD (h) ± standard deviations for those with multiple observations except for K. pneumoniae control data, which are provided as median TTD (h) (interquartile range) due to non-Gaussian distribution of data. TTD, time to detection; NG, no growth.
NA, not applicable.
E. coli growth.
Over the course of each 120-h experiment, control bottles grew to 8.94 ± 0.14 log10 CFU/ml and 8.94 ± 0.22 log10 CFU/ml in the BacT/Alert and Bactec bottles, respectively. E. coli growth was detected in 8/24 (33%), 11/24 (46%), and 14/24 (58%) of bottles with peak, midpoint, and trough concentrations of the studied antibiotics, respectively (P = 0.221) (Table 4). Neither system displayed a statistically significant difference between the proportions of detected bottles at any concentration, but the Virtuo system detected E. coli in the presence of certain antibiotic concentrations, including the midpoint and trough concentrations of imipenem and the midpoint of levofloxacin, when the FX system did not. Meropenem and cefepime bottles caused in vitro sterilization across all concentrations in both systems, while all cefazolin and piperacillin-tazobactam bottles alarmed positive.
K. pneumoniae growth.
At 120 h, control bottles had grown to 8.45 ± 0.57 log10 CFU/ml in the BacT/Alert bottles and 8.18 ± 1.08 log10 CFU/ml in the Bactec bottles. K. pneumoniae growth was detected in 8/16 (50%) bottles at each concentration (P = 1.0). The same antibiotic/concentration bottles alarmed positive between the two detection systems. Similarly to the bottles inoculated with E. coli, none of the bottles containing meropenem or cefepime concentrations alarmed positive with K. pneumoniae, while all cefazolin and piperacillin-tazobactam bottles grew.
Time to detection.
A summary of the observed TTDs of the BacT/Alert and Bactec bottles is presented in Table 4. For bottles inoculated with P. aeruginosa, antibiotic-free controls were detected at a mean of 15.6 ± 1.0 h versus 17.4 ± 0.4 h (P = 0.046) in the BacT/Alert and Bactec bottles, respectively. TTD for E. coli control bottles was 10.6 ± 0.2 h versus 11.8 ± 0.4 (P ≤ 0.001) hours in the BacT/Alert and Bactec bottles, respectively. Finally, TTD for K. pneumoniae control bottles was also shorter for BacT/Alert bottles (median, 11.3 [interquartile range (IQR), 11.0 to 11.4] versus 12.5 [IQR, 12.2 to 12.6] hours; P = 0.003).
For blood culture bottles containing antibiotics, growth, if observed, was usually detected within the first 24 h. One exception was the observation of E. coli in a BacT/Alert bottle containing the imipenem midpoint concentration, which alarmed positive at 29.5 h, though the corresponding Bactec bottle remained negative for the duration of the experiment. Differences in TTD were isolate dependent for antibiotic-containing bottles that alarmed in both systems. For example, BacT/Alert and Bactec bottles inoculated with antibiotics and P. aeruginosa had similar TTD (17.5 ± 2.0 versus 18.3 ± 1.5 h, P = 0.352). However, BacT/Alert bottles inoculated with antibiotics and E. coli had a shorter TTD compared with Bactec bottles (10.8 [IQR, 10.5 to 11.3] versus 11.8 [IQR, 11.7 to 11.8] hours; P = 0.026). K. pneumoniae-containing BacT/Alert bottles also had a shorter TTD compared with the Bactec bottles (11.3 ± 0.2 versus 12.6 ± 0.5 h; P ≤ 0.001).
Antibiotic concentrations.
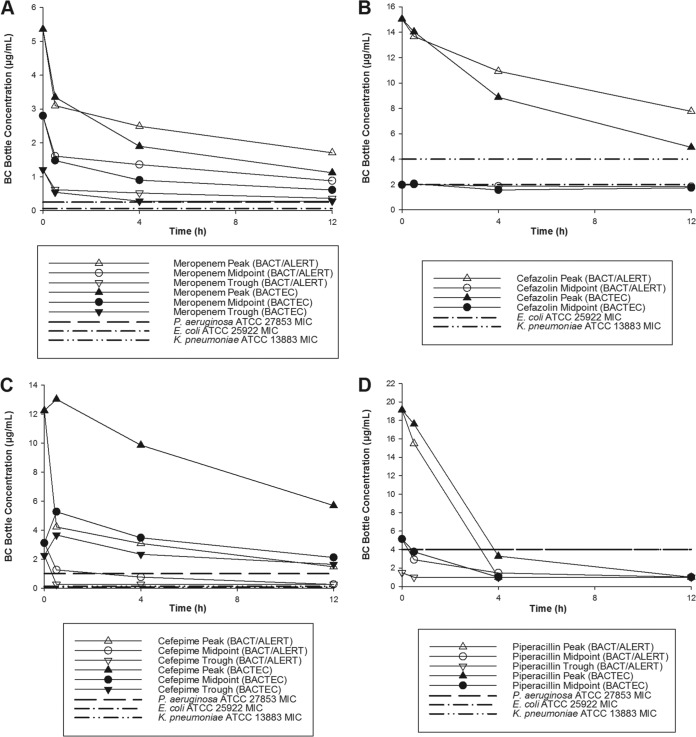
Observed antibiotic concentrations at the start of the experiment (0 h) and then within 0.5, 4, and 12 h of inoculation are plotted in Fig. 1 for meropenem, cefepime, cefazolin, and the piperacillin component of piperacillin-tazobactam. The mean percent reduction in antibiotic concentration at each time point is provided in Table 5. For meropenem, both BacT/Alert FA Plus and Bactec Plus Aerobic/F blood culture bottles equally reduced peak, midpoint, and trough concentrations by 42 to 56% over the first 0.5 h; afterwards, very little meropenem was further reduced over 12 h. For cefepime, BacT/Alert bottles reduced peak, midpoint, and trough concentrations by 60 to 73% over 0.5 h, while Bactec bottles had no meaningful reductions in concentrations. By 4 h, cefepime concentrations were reduced by 75 to 76% and 16 to 44% in BacT/Alert and Bactec bottles, respectively. Neither BacT/Alert nor Bactec bottles caused significant reductions in cefazolin concentrations until 12 h. Finally, both bottles resulted in minimal and variable reductions of piperacillin concentrations over the first 0.5 h; however, by 4 h, the majority of piperacillin was inactivated in both BacT/Alert and Bactec bottles.
FIG 1.
Meropenem (A), cefazolin (B), cefepime (C), and piperacillin (D) concentrations over the initial 12 h after inoculating blood culture bottles with antibiotic and MICs against study isolates.
TABLE 5.
Percent reduction in meropenem, cefepime, cefazolin, and piperacillin concentrations over 12 h in BacT/Alert FA Plus Aerobic and Bactec Plus Aerobic/F blood culture bottles
| Antibiotic | Category (observed 0-h concentration [μg/ml])a | Mean % reduction of observed zero concentration by test over: |
|||||
|---|---|---|---|---|---|---|---|
| 0.5 h |
4 h |
12 h |
|||||
| BacT/Alert | Bactec Plus | BacT/Alert | Bactec Plus | BacT/Alert | Bactec Plus | ||
| Meropenemb | P (5.36) | 42 | 38 | 54 | 65 | 68 | 79 |
| M (2.80) | 43 | 47 | 52 | 68 | 69 | 78 | |
| T (1.21) | 49 | 56 | 58 | 77 | 70 | 77 | |
| Cefazolinc | P (15.04) | 8 | 7 | 27 | 41 | 48 | 67 |
| M (1.97) | +2d | +4d | 5 | 20 | 7 | 12 | |
| T (0.46) | BDLg | BDL | BDL | BDL | BDL | BDL | |
| Cefepimee | P (12.24) | 66 | +2d | 75 | 44 | 88 | 64 |
| M (3.11) | 60 | +33d | 76 | 23 | BDL | 50 | |
| T (2.25) | BDL | +3d | BDL | 16 | BDL | 46 | |
| Piperacillinf | P (19.13) | 19 | 8 | BDL | 83 | BDL | BDL |
| M (5.14) | 43 | 27 | BDL | BDL | BDL | BDL | |
| T (1.49) | BDL | BDL | BDL | BDL | BDL | BDL | |
Zero-hour concentration in blood culture bottles was 50% of the observed concentration in whole blood, divided by 4 to account for dilution in each bottle (10 ml whole blood inoculated with antibiotic and 30 ml blood culture media). P, peak; M, midpoint; T, trough.
The lower limit of detection for meropenem was 0.25 μg/ml.
The lower limit of detection for cefazolin was 0.5 μg/ml.
Positive numbers denote that the observed concentration was higher than that at 0 h, suggesting no binding reductions in the first 0.5 h.
The lower limit of detection for cefepime was 0.5 μg/ml.
The lower limit of detection for piperacillin was 2 μg/ml.
BDL, below detectable limit.
DISCUSSION
Prompt pathogen identification and subsequent antibiotic susceptibility testing best facilitates antibiotic selection when treating bloodstream infections and is best accomplished by obtaining untreated (i.e., antibiotic-free) blood. Scheer and colleagues observed that detection rates of blood cultures from adult intensive care unit patients who had received antibiotics were approximately half of those obtained from patients who had not yet started therapy (18). While it is ideal and continually recommended to collect blood cultures prior to administration of antibiotics, this is not feasible in many clinical situations. As a result, additional interventions aimed at increasing pathogen identification are necessary. The use of blood culture bottles that contain proprietary antibiotic binding resins to “bind” or “inactivate” residual antibiotic concentrations in blood is a practical approach for hospital laboratories. While these binding resins successfully bind many antibiotics, there are some drug classes, including carbapenems and cephalosporins, for which variable binding performance has been reported. It is therefore important to understand how clinically meaningful antibiotic concentrations in blood culture bottles may affect the growth and identification of susceptible Gram-negative bacteria.
This study expands on the prior observations made by Grupper and colleagues, who investigated P. aeruginosa growth in blood culture bottles exposed to meropenem, ceftolozane-tazobactam, and ceftazidime-avibactam using peak, midpoint, and trough concentrations observed from patients (10). In that study, only trough concentrations of the tested antibiotics consistently resulted in P. aeruginosa growth. Here, two additional pansusceptible Enterobacteriaceae species, E. coli and K. pneumoniae, were included to determine if the in vitro sterilization observed with P. aeruginosa also transpires in these more susceptible (i.e., lower-MIC) organisms. Recovery results were generally consistent with the prior study. In fact, the repeated combination of meropenem against P. aeruginosa ATCC 27853 yielded the same detection results from both manufacturers’ bottles. For all antibiotics tested against P. aeruginosa in this study (meropenem, imipenem, cefepime, and piperacillin-tazobactam), both BacT/Alert and Bactec blood culture bottles containing antibiotic binding resins only consistently alarmed positive at simulated trough concentrations. Midpoint concentrations for imipenem alarmed positive in both manufacturers’ bottles, while bottles with piperacillin-tazobactam alarmed positive at all simulated concentrations tested (peak, midpoint, and trough). Therefore, the findings in the current study align quite well with the conclusions of Grupper and colleagues (10) in that collection of blood cultures right before the scheduled dose, when antibiotic concentrations are predicted to be the lowest, should be a best practice for obtaining blood cultures for patients already receiving antibiotics. Interestingly, Menchinelli and colleagues recently reported P. aeruginosa recovery at all tested concentrations of meropenem, which were higher than the ones used here, whereas this study only had trough bottles alarm positive (19). This difference in observations may be attributed to the higher target used for the starting inoculums (50 to 100 CFU per 0.5 ml versus 7 to 30 CFU per 0.5 ml) (19).
While the above conclusions are well supported for P. aeruginosa, the results against E. coli and K. pneumoniae were not as clear. These Enterobacteriaceae species routinely had MICs that were several dilutions lower than that of P. aeruginosa (Table 2). As a result, growth even at trough concentrations was variable. Only 58% of E. coli-containing bottles and 50% of K. pneumoniae-containing bottles alarmed positive when inoculated with trough concentrations. Notable exceptions included meropenem and cefepime, both of which remained negative at all concentrations when inoculated with Enterobacteriaceae, and in contrast, cefazolin and piperacillin-tazobactam, which alarmed positive at all tested concentrations. Again, Menchinelli and colleagues observed better recovery rates among their levofloxacin bottles. In addition to the higher inoculum, the lower levofloxacin peak concentration used in their study may have contributed to these differences (19). The following differences between manufacturers were observed: BacT/Alert bottles alarmed positive for E. coli paired with midpoint and trough concentrations of imipenem and for the midpoint concentration of levofloxacin, while the corresponding Bactec bottles remained negative. These observations demonstrate the impact of specific antibiotics, concentrations, and binding kinetics of resins on organism recovery. Therefore, while the recommendation for blood culture collection just prior to the next antibiotic dose may still be the best practice in general to maximize pathogen identification, there still remains a risk of in vitro sterilization of bottles containing very susceptible Enterobacteriaceae, particularly if patients received potent antibiotics, such as meropenem or cefepime. This may be an advantageous, strategic scenario for the use of direct-from-blood molecular tests that, in theory, should be unaffected by the presence of residual antibiotic concentrations (20). Further research in this area is needed. Another possible contributing factor for the differences between manufacturers is the loading method. The BacT/Alert system employs a conveyor belt that carries the bottles into the body of the machine and an automatic robotic arm for loading, while the Bactec system requires the machine shelves to be slid out and the bottles to be loaded in manually. Because of this, the BacT/Alert system would be able to maintain a more consistent temperature, potentially providing a more fertile environment.
The majority of the above observations can be explained by looking at the residual concentrations of the assayed antibiotics over the initial 12 h of the experiment relative to the MIC (Fig. 1). For example, both bottle systems reduced meropenem concentrations significantly over the first 0.5 h, but very little afterward, leaving residual peak and midpoint concentrations above the MICs for all tested organisms over 12 h. A similar observation was made for cefepime in the BacT/Alert bottles, where concentrations were reduced by 60 to 73% over the initial 0.5 h, but little afterwards. In contrast, Bactec bottles resulted in small reductions in cefepime concentration, if any, over the initial 0.5 h, but more substantial declines over the ensuing 12 h. Regardless, neither bottle system was efficacious at reducing cefepime concentrations to below the MICs of the selected susceptible isolates, thereby causing in vitro sterilization in all experiments except the bottles inoculated with trough concentrations and P. aeruginosa. It is yet unclear how quickly residual antibiotic concentrations must be reduced to below the MIC to allow isolate growth to sufficient thresholds to trigger alarm. Lovern and colleagues (11) measured concentrations for a number of Gram-positive and Gram-negative antibiotics in BacT/Alert FA Plus and Bactec Plus Aerobic/F bottles (i.e., the same bottles studied here) and found more rapid and overall better binding kinetics in the Virtuo system for all of the β-lactams tested, as well as for levofloxacin. However, the kinetic studies were done in phosphate buffer solution instead of whole blood, and no bacteria were inoculated into the bottles. While a more rapid reduction appears beneficial, very little binding was observed for piperacillin after the initial 0.5 h, and it was not until the measurement at 4 h that peak concentrations were reduced to below the MIC. Despite this observation, all tested organisms grew in the presence of all piperacillin concentrations. It should be noted that no samples were collected between 0.5 and 4 h; therefore, it is likely that piperacillin concentrations declined below the MIC well before 4 h. For cefazolin, very little binding of the peak concentrations was observed over the 12-h measurement in both bottles, yet, like piperacillin-containing bottles, Enterobacteriaceae were identified at all cefazolin concentrations. This is likely best explained by the high protein binding of cefazolin (∼90%) (15) combined with the poor potency of this antibiotic against the selected isolates for testing; MICs were 2 μg/ml (susceptible) for E. coli and 4 μg/ml (intermediate) for K. pneumoniae. When corrected for unbound concentrations, which are active against bacteria, even free peak cefazolin concentrations would have been below these MICs by 0.5 h.
A major strength of this study was the utilization of clinically relevant antibiotic concentrations over the dosing interval to define timing scenarios where collection can occur to maximize recovery. Lovern and colleagues (11) previously investigated cefepime-E. coli ATCC 25922, cefepime-P. aeruginosa ATCC 27853, levofloxacin-E. coli ATCC 25922, piperacillin-E. coli ATCC 25922, and piperacillin-P. aeruginosa ATCC 27853. However, the isolates were only exposed to single concentrations of each antibiotic, which may affect interpretation. For example, the selected levofloxacin concentration (5.7 μg/ml) from their study was similar to the midpoint (3 μg/ml) concentrations targeted here. Notably, differences in favor of BacT/Alert were observed between simulated midpoint levofloxacin concentrations against the E. coli strain in our study, which was consistent with the results of Lovern and colleagues. However, the simulated peak concentrations in our study resulted in in vitro sterilization of all bottles in both systems, while trough concentrations resulted in uniform recovery, offering very different conclusions from the previous study.
TTD was also compared between the BacT/Alert and Bactec bottles in their respective systems. For all isolates tested in the absence of any antibiotic concentrations (i.e., control bottles), TTD was faster for the BacT/Alert bottles in the Virtuo system, which is consistent with previous studies (21). These observations carried over to the antibiotic-containing bottles, where TTD was significantly shorter for BacT/Alert bottles when inoculated with E. coli and K. pneumoniae. However, no difference in TTD was observed for antibiotic-containing bottles inoculated with P. aeruginosa. The lack of difference may have been due to the slower growth curves of the selected P. aeruginosa strain. Even without antibiotics present in the bottles, TTD for P. aeruginosa was approximately 5 h longer than that of E. coli or K. pneumoniae isolates. Moreover, when bottles did alarm positive in the presence of antibiotics, the TTD appeared to be very similar for the same species compared to that of control bottles containing no antibiotics.
There are several limitations in this study, some of which remain the same as those in Grupper and colleagues’ study, which have already been reported (10). The fresh whole blood utilized in this experiment was donated by healthy volunteers and was not sourced from clinically ill patients; furthermore, target drug concentrations were obtained by inoculating the whole blood, as opposed to via administering the drug to the actual patient and sampling the actual attained concentration. In doing so, better accuracy and control of the target concentrations is maintained, but we must assume that plasma concentrations are approximately 50% of whole blood based on the value of a normal hematocrit in a healthy adult. Second, only susceptible isolates were selected for inclusion. Resistant isolates are more likely to grow because antibiotic concentrations will likely be below the MIC in both bottles with binding resins (10). Finally, the simulated antibiotic concentrations were that of aggressive high-dose regimens, which was done to create the worst-case scenario. Lower concentrations obtained with reduced doses may lend to increase recovery in the clinical setting, which is encouraging.
In conclusion, most clinically meaningful concentrations of meropenem, imipenem, cefepime, and levofloxacin increase the likelihood of artificial sterilization of BacT/Alert FA Plus Aerobic and Bactec Plus Aerobic/F blood culture bottles containing antibiotic binding resins specifically designed to mitigate this issue. Cefazolin and piperacillin-tazobactam were the only antibiotics that consistently allowed for detectable growth of all tested isolates at all concentrations. In order to maximize the chances of recovery for patients who have previously received these antibiotics, we conclude that blood cultures should be drawn immediately prior to the upcoming dose.
ACKNOWLEDGMENTS
This study was funded by bioMérieux, Inc., Durham, NC, USA.
We report no conflicts of interest.
We acknowledge Elizabeth Cyr, Lee Steere, Michelle Insignares, Debora Santini, Courtney Bouchard, Elias Mullane, Kimelyn Greenwood, Sara Giovagnoli, Janice Cunningham, Nicole DeRosa, Lauren McLellan, Alissa Padgett, Christina Sutherland, Jennifer Tabor-Rennie, Tomefa Asempa, James Kidd, Lindsay Avery, and Safa Abuhussain from the Center for Anti-Infective Research and Development, and Yanice Maldonado from the Department of Microbiology, Hartford Hospital, for their assistance with the conduct of this study.
REFERENCES
- 1.Goto M, Al-Hasan MN. 2013. Overall burden of bloodstream infection and nosocomial bloodstream infection in North America and Europe. Clin Microbiol Infect 19:501–509. doi: 10.1111/1469-0691.12195. [DOI] [PubMed] [Google Scholar]
- 2.Kilgore M, Brossette S. 2008. Cost of bloodstream infections. Am J Infect Control 36:S172.e1–3. doi: 10.1016/j.ajic.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Hollenbeak CS. 2011. The cost of catheter-related bloodstream infections: implications for the value of prevention. J Infus Nurs 34:309–313. doi: 10.1097/NAN.0b013e3182285e43. [DOI] [PubMed] [Google Scholar]
- 4.Rhodes A, Evans LE, Alhazzani W, Levy MM, Antonelli M, Ferrer R, Kumar A, Sevransky JE, Sprung CL, Nunnally ME, Rochwerg B, Rubenfeld GD, Angus DC, Annane D, Beale RJ, Bellinghan GJ, Bernard GR, Chiche JD, Coopersmith C, De Backer DP, French CJ, Fujishima S, Gerlach H, Hidalgo JL, Hollenberg SM, Jones AE, Karnad DR, Kleinpell RM, Koh Y, Lisboa TC, Machado FR, Marini JJ, Marshall JC, Mazuski JE, McIntyre LA, McLean AS, Mehta S, Moreno RP, Myburgh J, Navalesi P, Nishida O, Osborn TM, Perner A, Plunkett CM, Ranieri M, Schorr CA, Seckel MA, Seymour CW, Shieh L, Shukri KA, Simpson SQ, Singer M, Thompson BT, Townsend SR, Van der Poll T, Vincent JL, Wiersinga WJ, Zimmerman JL, Dellinger RP. 2017. Surviving Sepsis Campaign: international guidelines for management of sepsis and septic shock: 2016. Crit Care Med 45:486–552. doi: 10.1097/CCM.0000000000002255. [DOI] [PubMed] [Google Scholar]
- 5.Zadroga R, Williams DN, Gottschall R, Hanson K, Nordberg V, Deike M, Kuskowski M, Carlson L, Nicolau DP, Sutherland C, Hansen GT. 2013. Comparison of 2 blood culture media shows significant differences in bacterial recovery for patients on antimicrobial therapy. Clin Infect Dis 56:790–797. doi: 10.1093/cid/cis1021. [DOI] [PubMed] [Google Scholar]
- 6.Flayhart D, Borek AP, Wakefield T, Dick J, Carroll KC. 2007. Comparison of BACTEC PLUS blood culture media to BacT/Alert FA blood culture media for detection of bacterial pathogens in samples containing therapeutic levels of antibiotics. J Clin Microbiol 45:816–821. doi: 10.1128/JCM.02064-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spaargaren J, van Boven CP, Voorn GP. 1998. Effectiveness of resins in neutralizing antibiotic activities in Bactec Plus Aerobic/F culture medium. J Clin Microbiol 36:3731–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vigano EF, Vasconi E, Agrappi C, Clerici P, Melloni P. 2004. Use of simulated blood cultures for antibiotic effect on time to detection of the two blood culture systems BacT/ALERT and BACTEC 9240. New Microbiol 27:235–248. [PubMed] [Google Scholar]
- 9.Mitteregger D, Barousch W, Nehr M, Kundi M, Zeitlinger M, Makristathis A, Hirschl AM. 2013. Neutralization of antimicrobial substances in new BacT/Alert FA and FN Plus blood culture bottles. J Clin Microbiol 51:1534–1540. doi: 10.1128/JCM.00103-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grupper M, Nicolau DP, Aslanzadeh J, Tanner LK, Kuti JL. 2017. effects of clinically meaningful concentrations of antipseudomonal beta-lactams on time to detection and organism growth in blood culture bottles. J Clin Microbiol 55:3502–3512. doi: 10.1128/JCM.01241-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lovern D, Katzin B, Johnson K, Broadwell D, Miller E, Gates A, Deol P, Doing K, van Belkum A, Marshall C, Mathias E, Dunne WM Jr. 2016. Antimicrobial binding and growth kinetics in BacT/ALERT(R) FA Plus and BACTEC(R) Aerobic/F Plus blood culture media. Eur J Clin Microbiol Infect Dis 35:2033–2036. doi: 10.1007/s10096-016-2759-9. [DOI] [PubMed] [Google Scholar]
- 12.Crandon JL, Ariano RE, Zelenitsky SA, Nicasio AM, Kuti JL, Nicolau DP. 2011. Optimization of meropenem dosage in the critically ill population based on renal function. Intensive Care Med 37:632–638. doi: 10.1007/s00134-010-2105-0. [DOI] [PubMed] [Google Scholar]
- 13.Sakka SG, Glauner AK, Bulitta JB, Kinzig-Schippers M, Pfister W, Drusano GL, Sorgel F. 2007. Population pharmacokinetics and pharmacodynamics of continuous versus short-term infusion of imipenem-cilastatin in critically ill patients in a randomized, controlled trial. Antimicrob Agents Chemother 51:3304–3310. doi: 10.1128/AAC.01318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drusano GL, Preston SL, Fowler C, Corrado M, Weisinger B, Kahn J. 2004. Relationship between fluoroquinolone area under the curve: minimum inhibitory concentration ratio and the probability of eradication of the infecting pathogen, in patients with nosocomial pneumonia. J Infect Dis 189:1590–1597. doi: 10.1086/383320. [DOI] [PubMed] [Google Scholar]
- 15.Smyth RD, Pfeffer M, Glick A, Van Harken DR, Hottendorf GH. 1979. Clinical pharmacokinetics and safety of high doses of ceforanide (BL-S786R) and cefazolin. Antimicrob Agents Chemother 16:615–621. doi: 10.1128/aac.16.5.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicasio AM, Ariano RE, Zelenitsky SA, Kim A, Crandon JL, Kuti JL, Nicolau DP. 2009. Population pharmacokinetics of high-dose, prolonged-infusion cefepime in adult critically ill patients with ventilator-associated pneumonia. Antimicrob Agents Chemother 53:1476–1481. doi: 10.1128/AAC.01141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felton TW, Roberts JA, Lodise TP, Van Guilder M, Boselli E, Neely MN, Hope WW. 2014. Individualization of piperacillin dosing for critically ill patients: dosing software to optimize antimicrobial therapy. Antimicrob Agents Chemother 58:4094–4102. doi: 10.1128/AAC.02664-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scheer CS, Fuchs C, Grundling M, Vollmer M, Bast J, Bohnert JA, Zimmermann K, Hahnenkamp K, Rehberg S, Kuhn SO. 2019. Impact of antibiotic administration on blood culture positivity at the beginning of sepsis: a prospective clinical cohort study. Clin Microbiol Infect 25:326–331. doi: 10.1016/j.cmi.2018.05.016. [DOI] [PubMed] [Google Scholar]
- 19.Menchinelli G, Liotti FM, Giordano L, De Angelis G, Sanguinetti M, Spanu T, Posteraro B. 2019. Efficient inactivation of clinically relevant antimicrobial drug concentrations by BacT/Alert or Bactec resin-containing media in simulated adult blood cultures. Antimicrob Agents Chemother 63. doi: 10.1128/AAC.00420-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Angelis G, Posteraro B, De Carolis E, Menchinelli G, Franceschi F, Tumbarello M, De Pascale G, Spanu T, Sanguinetti M. 2018. T2Bacteria magnetic resonance assay for the rapid detection of ESKAPEc pathogens directly in whole blood. J Antimicrob Chemother 73:iv20–iv26. doi: 10.1093/jac/dky049. [DOI] [PubMed] [Google Scholar]
- 21.Menchinelli G, Liotti FM, Fiori B, De Angelis G, D’Inzeo T, Giordano L, Posteraro B, Sabatucci M, Sanguinetti M, Spanu T. 2019. In vitro evaluation of BACT/ALERT(R) VIRTUO(R), BACT/ALERT 3D(R), and BACTEC FX automated blood culture systems for detection of microbial pathogens using simulated human blood samples. Front Microbiol 10:221. doi: 10.3389/fmicb.2019.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]