Abstract
Phosphorylation at aspartic acid residues represents an abundant and critical post-translational modification (PTM) in prokaryotes. In contrast to most characterized PTMs, such as phosphorylation at serine or threonine, the phosphoaspartate moiety is intrinsically labile, and therefore incompatible with common proteomic profiling methods. Herein, we report a nucleophilic, desthiobiotin-containing hydroxylamine (DBHA) chemical probe that covalently labels modified aspartic acid residues in native proteomes. DBHA treatment coupled with LC-MS/MS analysis enabled detection of known phosphoaspartate modifications, as well as novel aspartic acid sites in the E. coli proteome. Coupled with isotopic labelling, DBHA-dependent proteomic profiling also permitted global quantification of changes in endogenous protein modification status, as demonstrated with the detection of increased E. coli OmpR phosphorylation, but not abundance, in response to changes in osmolarity.
Keywords: chemoproteomics, phosphoaspartate modification, response regulator, two-component signaling
Post-translational modifications (PTMs) have evolved as a selective and reversible means to alter protein function and thus propagate signals within and between cells. These chemical tags are exceptionally diverse in structure and function and are known to target a majority of proteins in many organisms, including mammals.[1, 2] While the prevalence and perceived importance of PTMs is conserved across all forms of life, the relative abundance of specific PTMs is extremely variable. Humans, for example, encode more than 500 protein kinases that catalytically phosphorylate the alcohol functionality in serine, threonine, and tyrosine residues.[3, 4] Protein phosphorylation is recognized as a major communication mechanism in mammalian cells, a fact that is bolstered by high association with disease when phosphorylation networks are deregulated.[5] An important technical consideration when reflecting upon the vast number of phosphorylation marks that are known in mammalian systems is the fact that systematic, quantitative analysis of these PTMs is made possible through several technologies, including detection with phospho-specific antibodies, and global identification with LC-MS/MS proteomic approaches.[6, 7] Combined, these methods have identified thousands of phosphorylation sites in diverse cell types and organisms. The comprehensive set of tools available to study protein phosphorylation, which is invoked in this study as a prototypical PTM, hinges on a single aspect of the modification itself: chemical stability to biochemical and proteomic workflows.
Prokaryotes also employ phosphorylation as a major form of biochemical communication, but in contrast to eukaryotes, the number of serine/threonine phosphorylation sites appears to be orders of magnitude lower.[8] Instead, these lower organisms rely more on two-component signalling pathways that involve phosphorylation of histidine “receiver” residues, which ultimately phosphorylate aspartic acids to form an acylphosphate modification.[9–11] The resulting phosphoaspartate residue (pD) is generally thought to cause changes in protein structure, which in the case of many characterized response regulator (RR) transcription factors can promote or inhibit DNA binding and subsequent downstream signalling. While thought to represent the major form of signal transduction, the widespread discovery and dynamic quantification of these modifications has not been possible owing to the intrinsic lability of the pD group itself. As a result, pD sites have almost exclusively been studied one-by-one with the use of 32P-ATP as a means to radiolabel the active protein for visualization by gel electrophoresis.[12] Even in these settings, however, protein purification is often required and the endogenous environment cannot be probed. Therefore, new methods for the study of these chemically labile yet functionally important modifications are necessary. Herein, we report a chemoproteomic approach to rapidly trap the electrophilic acylphosphate group unique to pD sites within native bacterial proteomes (Figure 1a). We demonstrate that a desthiobiotin-containing hydroxylamine (DBHA) probe selectively enriches and site-specifically identifies both known and novel electrophilic aspartic acid sites throughout the E. coli proteome. Furthermore, we demonstrate the potential to measure signalling dynamics in the osmolarity sensing OmpR/OmpF two-component signalling pathway by combining isotopic proteome labelling and pD-site enrichment from native proteomes.
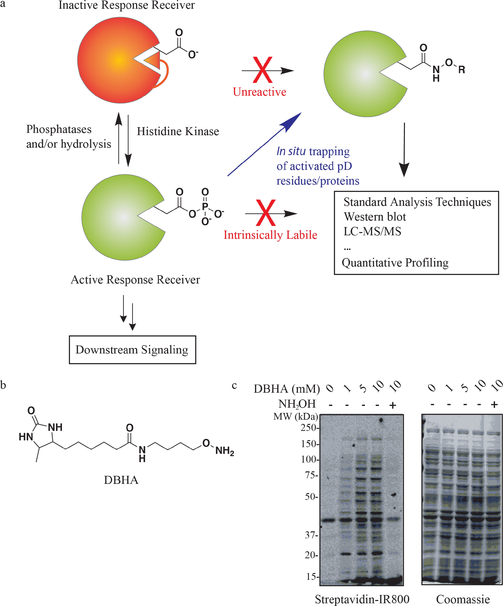
Figure 1.
a) Schematic of dynamic phosphorylation at aspartic acids, as well as labelling with a prototypical nucleophilic probe in situ, permitting detection and quantification. b) Chemical structure of the desthiobiotin-linked hydroxylamine (DBHA) probe. c) Western blot visualization of desthiobiotin-labelled proteins demonstrates dose-dependent and hydroxylamine competitive labelling of E. coli proteins in DBHA-treated lysates.
We previously reported that intrinsically labile metabolites, such as the glycolytic metabolite 1,3-bisphosphoglycerate, could be efficiently trapped and converted into a stable hydroxamic acid derivative upon treatment with hydroxylamine in situ.[13] Based on the success of this approach for acylphosphate-containing metabolites, and precedence for mapping ADP-ribosylation,[14] we reasoned that a hydroxylamine-based chemical probe may be useful for capturing pD sites in the proteome. Such a probe would encompass two main components: 1) An O-alkyl hydroxylamine warhead capable of forming a stable intermediate with pD sites in native proteins under mild conditions and 2) a retrieval tag that permits direct enrichment and elution of modified proteins or peptides for LC-MS/MS or other quantitative detection methods (Figure 1a and the Supporting Information, Figure S1). We first attempted to use a minimal probe, O-propargyl hydroxylamine, which would label modified aspartates with a small alkyne tag that could be used for detection or enrichment following [3+2] Huisgen click chemistry with secondary retrieval/visualization tags.[15] This approach is similar to a recent report that employed an alkyne-containing hydrazine probe in mammalian cells.[16] Our preliminary experiments using O-propargyl hydroxylamine suffered from poor retrieval of tagged species as measured by either in-gel fluorescence or LC-MS/MS, which we attribute to decreased efficiency that comes with additional steps involved in click chemistry sample processing. Therefore, we sought to develop a probe that could be directly retrieved and processed for a variety of proteomic detection methods. We synthesized a desthiobiotin-containing O-alkyl hydroxylamine (DBHA) probe that could bypass the need for subsequent chemical modification prior to detection or enrichment (Figure 1b). Desthiobiotin was specifically used in place of biotin to permit efficient release of enriched peptides for LC-MS/MS detection, which can be a major bottleneck in chemical proteomic workflows.[17]
We first tested whether incubation of DBHA across assay conditions would covalently label native proteins in K12 E. coli. We found that DBHA labelling was optimal under the denaturing conditions of 6m urea, which likely improves probe access to pD modification sites. Proteome incubation with DBHA and western blot detection of desthiobiotin labelled proteins revealed dose-dependent labelling of the E. coli proteome across a wide range of concentrations (Figure 1c). These labelling events were inhibited by pretreatment of proteome with soluble hydroxylamine or acidic conditions (pH 2), confirming that the proteins and sites being visualized were dependent upon intact reactivity (Figure 1c and Figure S2). Determining the site of labelling is particularly important for probes such as DBHA because they could, in principle, react with numerous modifications on proteins. For example, hydrazine-probes have previously been shown to modify glycosylated proteins in their open-chain form,[18] as well as capture pyruvoyl and glyoxylyl modifications in mammalian cells.[16] Despite the potential for interaction with other electrophilic moieties that are present on proteins, our probe and workflow should differentiate these events through the detection of the DBHA-modified aspartic acids directly on tryptic peptides with high resolution LC-MS/MS and site-of-labelling analysis (see below; Figure S1). To identify the proteins and specific sites being modified by DBHA, we developed an efficient workflow formodified protein enrichment and processing by LC-MS/MS analysis (Figure S3). DBHA treatments were performed at 10 mM and 100 mM concentrations in order to maximally capture both low and high abundance sites within the E. coli proteome. Modified proteins were directly enriched with streptavidin–agarose immunoprecipitation, washed to remove non-labelled proteins, and processed for on-bead tryptic digestion. Importantly, bulk tryptic peptides from enriched proteins could be collected in the digest eluent, followed by elution of DBHA-modified peptides. The combination of these two pools enables high-confidence detection of target proteins (that is, from many detected tryptic peptides per protein) as well as site-specific resolution of DBHA modification sites. DBHA-labelled sites were filtered based on stringent mass tolerance, decoy database false-discovery rates, and MS2 statistics. Beyond these initial filters, we required that a putative modification site be observed in four or more biological replicates to be included in this dataset. Finally, in order to identify the specific DBHA-modified residue in each detected tryptic peptide we analyzed the “hits” that passed our initial specificity filters above using the LuciPHor modification site localization algorithm.[19] Application of this analysis pipeline to LC-MS/MS data from the 10 mM (n=eight replicates) and 100 mM (n=six replicates) DBHA datasets permitted detection of 138 high confidence pAsp sites from 98 proteins (site global false-localization rate in LuciPHor at <1%; Table S1).
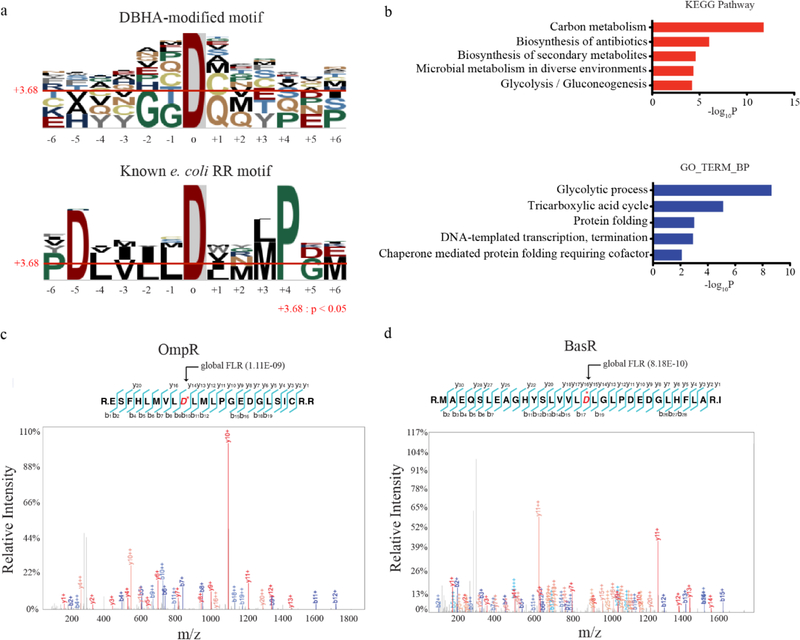
The identification of over 100 modified aspartic acids in the E. coli. proteome was surprising since there are circa 20–30 predicted RR’s present in this and other typical prokaryotes.[20] Analysis of the local sequence context of DBHA modified aspartic acids revealed a modest motif preference for small, hydrophilic residues around the DBHA-modified aspartic acid, as well as preference for proline at the +4 position (Figure 2a). This overall profile was distinct from the motif surrounding known response regulator pD sites in E. coli. However, a conserved preference for proline at the +4 position was also present, confirming partial similarities between the two motifs (Figure 2a). Bioinformatic analysis of modified proteins with DAVID and KEGG pathway analysis revealed a dominant signature of proteins involved in metabolism, protein folding, and transcriptional regulation (Figure 2b and Figure S4). These sites were found in proteins spanning the entire range of relative abundance in E. coli, from metabolic proteins like Pgk (estimated at ca. 100 000 copies/cell) at one extreme, to transcription factors like BasR (estimated at ca. 20 copies/cell) on the other (Table S1 and Figure S5).[20] Among the lower abundance proteins identified were response regulators and related two component signalling proteins with known pD sites. For example, the response regulators OmpR and BasR were identified harboring DBHA-modifications at D55 and D51, respectively (Figure 2c,d). These sites are known phosphorylation sites that regulate OmpR and BasR DNA-binding activity.[21, 22] Additional DBHA-modification sites included known phosphohistidine-containing proteins, like Asp38 in Crr. Intriguingly, the DBHA modification site in Crr was found to be highly proximal to a known phosphohistidine site, perhaps indicating that dynamic relay between pH and pD sites occurs within this and other proteins (Figure S6).[23] Taken together, this dataset validated DBHA-labelling and enrichment as a method to detect both known and potentially novel phosphoaspartate sites in native proteins from whole prokaryotic proteome.
Figure 2.
a) Enriched sequence motifs derived from DBHA-modified aspartic acid sites (top) compared to the motif generated by known response regulator pD sites in E. coli (bottom). b) Gene-ontology biological process categories (GOTERM BP; bottom) and KEGG pathways (top) enriched among DBHA-modified proteins. Representative, statistically significant categories are shown, with a complete list in Figure S4. c,d) MS/MS spectra of DBHA-modified tryptic peptides at two known pD sites in the response regulator proteins c) OmpR and d) BasR. Observed b- and y-ions are labelled on each spectrum and highlighted on the tryptic peptide. The known pD sites at D55 and D51 in OmpR and BasR, respectively, are highlighted and starred (*) in red.
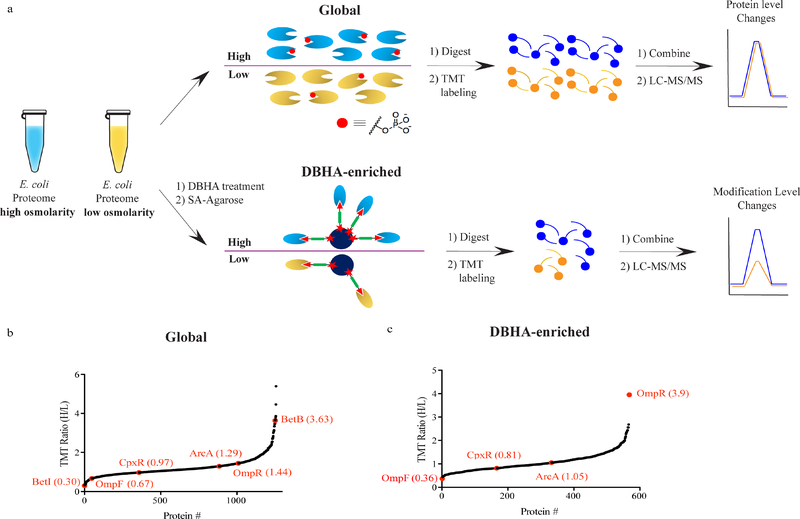
DBHA-enrichment of proteins through the labelling of pD sites presents an opportunity to directly interrogate changes in phosphorylated proteins in parallel to, and distinct from, monitoring global changes in protein abundance. To determine whether DBHA-profiling could identify physiologically relevant changes in two-component signalling, we interrogated the well-characterized EnvZ/OmpR pathway in E. coli. This pathway facilitates sensing of extracellular osmolarity by the histidine kinase and sensor protein EnvZ, which phosphorylates the response regulator OmpR at D55 in response to increased osmolarity. Phosphorylated OmpR exhibits markedly increased DNA-binding activity and differential regulation of transcription of the membrane porin proteins OmpF and OmpC (Figure S7).[24,25]To track this activation directly in cells, E. coli growing in standard LB media were switched to either 20% sucrose-containing NB (high osmolarity) or NB alone (mock) and grown for 12 h. A minor fraction of each bulk proteome was trypsinized, isotopically labelled with unique tandem-mass tag (TMT) channels,[26] and pooled to compare protein abundance changes resulting from high osmolarity (Figure 3a). In parallel, the bulk of the native proteome from each condition was labelled with DBHA probe, enriched on streptavidinagarose resin, and typrisinized on-bead. The resulting tryptic peptides were labelled with unique TMT channels and pooled for LC-MS/MS quantitation of phosphorylation changes in response to high osmolarity (Figure 3a). Abundance changes for more than 1200 proteins were quantified between conditions (Figure 3b and Table S2). OmpR protein levels were increased 1.4-fold in response to high osmolarity, which matches literature precedent showing that OmpR levels remain relatively constant or slightly increase in response to increased osmolarity (Figure 3b).[27] This abundance profile also detected significantly decreased levels of OmpF protein, consistent with repression by active OmpR. Significant changes in the levels of other proteins involved in osmoregulation were also apparent in the global profile, including the transcriptional regulator BetI (downregulated in high osmolarity) and the target of its repression, BetB (upregulated in high osmolarity; Figure 3b).[28, 29] Quantification of DBHA-enriched proteins, in contrast, identified OmpR as the most affected protein, with nearly 4-fold higher levels of DBHA-enriched OmpR present in the cells grown in 20% sucrose (Figure 3c and Table S2). Other known response regulators that harbor pD sites, including CpxR and ArcA, were detected and quantified in the enriched profile but were not affected by osmolarity at either the protein abundance (Figure 3b) or pD level (Figure 3c). Combined, these data validate that DBHA enrichment coupled to quantitative proteomics enables global profiling of response regulator phosphorylation, as opposed to abundance, in response to physiological signals in E. coli.
Figure 3.
a) Proteomic workflow to quantify global changes in protein levels (top) and pD or other DBHA-reactive modifications (bottom) by LC-MS/MS. Lysates from high and low osmolarity samples of E. coli are split for protein level quantification (top) or treated with DBHA probe (bottom). This workflow thus generates two isotopically labelled tryptic peptide pools that provide quantitative information on bulk protein level changes (top) and DBHA-sensitive modification levels (bottom). b) Global quantification of protein level changes in response to high osmolarity in E. coli. Proteins are ranked from most down-regulated (left) to up-regulated (right) under high osmolarity conditions. Proteins discussed in the text are labelled in red with their quantitative ratio listed. c) Quantification of DBHA-enriched proteins in response to high osmolarity in E. coli, thus reporting on specific changes in modification status, in contrast to changes in abundance alone. The osmoregulatory response regulatory OmpR was the most affected protein in the dataset. All data points in (b) and (c) represent the mean ratios from n=4 (two technical of two biological) replicate runs.
In summary, we have developed a novel chemical proteomic workflow that offers several advantages over current approaches to interrogate phosphoaspartate sites, and likely other electrophilic modifications, in native proteomes. First, peptide-based enrichment and LC-MS/MS profiling provides unequivocal identification of modified residues. This not only enables quantitation of pD signaling dynamics at specific sites but also allows for exploratory mapping in unannotated proteomes and organisms. In line with this attribute, the dataset herein contained many high confidence, reproducible DBHA-modified residues that are not previously annotated as phosphoaspartate sites. These sites may represent heretofore unrecognized phosphoaspartate sites that are introduced through enzymatic or nonenzymatic means, as has been observed for other intrinsically reactive metabolic intermediates like acetylphosphate,[30–32] which is used in vitro to chemically phosphorylate Asp sites in response regulators.[33] Beyond phosphoaspartate, other modifications could contribute to the profile observed, including methylesterification, ADP-ribosylation, and potentially other less-characterized PTMs that occur at aspartic acids in prokaryotes. Despite this potential for overlap, we view the labelling of additional electrophilic modifications as a feature not a flaw of DBHA-profiling because the use quantitative LC-MS/MS provides site-specific resolution necessary for follow up analysis at a site-by-site level. The dataset produced herein represents a starting point for these future efforts. Finally, we have demonstrated the ability to detect physiologically relevant changes in phosphoaspartic acid levels, distinct from protein level differences, in the EnvZ/OmpR osmoregulatory pathway. This work is the first to enable global, quantitative measurement of phosphoaspartate modification dynamics in native proteome, which should be broadly applicable to other organisms and biologic perturbations, providing an unprecedented view of this elusive, yet critical, signalling mechanism.
Supplementary Material
Acknowledgements
We thank M. Rust and G. Pattanayak for discussions surrounding the manuscript, and R. Park for conversations about proteomic analyses. We are grateful for financial support of this work from the following: Kwanjeong Educational Fellowship (to G.L.); NIH 2T32GM008720–16 (to J.E.M.); NIH R00CA175399, DP2GM128199–01, and 2R01CA093577–11 (to R.E.M.); the Damon Runyon Cancer Research Foundation (DFS08–14 to R.E.M.), and The University of Chicago.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supporting information and the ORCID identification number(s) forthe author(s) of this article can be found under: https://doi.org/10.1002/anie.201809059.
Reference
- [1].Walsh C, Posttranslational modification of proteins: expanding natureQs inventory (Roberts and Co. Publishers, Englewood, Colo: ), 2006, pp. xxi. [Google Scholar]
- [2].Olsen JV, Mann M, Mol. Cell. Proteomics 2013, 12, 3444–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Cohen P, Trends Biochem. Sci 2000, 25, 596–601. [DOI] [PubMed] [Google Scholar]
- [4].Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S, Science 2002, 298, 1912–1934. [DOI] [PubMed] [Google Scholar]
- [5].Brognard J, Hunter T, Curr. Opin. Genet. Dev 2011, 21, 4–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Mann M, Jensen ON, Nat. Biotechnol 2003, 21, 255–261. [DOI] [PubMed] [Google Scholar]
- [7].Kee JM, Oslund RC, Perlman DH, Muir TW, Nat. Chem. Biol 2013, 9, 416–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Stock AM, Robinson VL, Goudreau PN, Annu. Rev. Biochem. 2000, 69, 183–215. [DOI] [PubMed] [Google Scholar]
- [9].Stock JB, Ninfa AJ, Stock AM, Microbiol. Rev 1989, 53, 450–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Potel CM, Lin MH, Heck AJR, Lemeer S, Nat. Methods 2018, 15, 187–190. [DOI] [PubMed] [Google Scholar]
- [11].Attwood PV, Besant PG, Piggott MJ, Amino Acids 2011, 40, 1035–1051. [DOI] [PubMed] [Google Scholar]
- [12].Igo MM, Ninfa AJ, Silhavy TJ, Genes Dev. 1989, 3, 598–605. [DOI] [PubMed] [Google Scholar]
- [13].Chang JW, Lee G, Coukos JS, Moellering RE, Anal. Chem 2016, 88, 6658–6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhang Y, Wang J, Ding M, Yu Y, Nat. Methods 2013, 10, 981–984. [DOI] [PubMed] [Google Scholar]
- [15].Speers AE, Cravatt BF, Chem. Biol 2004, 11, 535–546. [DOI] [PubMed] [Google Scholar]
- [16].Matthews ML, He L, Horning BD, Olson EJ, Correia BE, Yates JR III, Dawson PE, Cravatt BF, Nat. Chem 2017, 9, 234–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hirsch JD, Eslamizar L, Filanoski BJ, Malekzadeh N, Haugland RP, Beechem JM, Haugland RP, Anal. Biochem 2002, 308, 343 357. [DOI] [PubMed] [Google Scholar]
- [18].Zhang H, Li XJ, Martin DB, Aebersold R, Nat. Biotechnol 2003, 21, 660–666. [DOI] [PubMed] [Google Scholar]
- [19].Fermin D, Walmsley SJ, Gingras AC, Choi HW, Nesvizhskii AI, Mol. Cell. Proteomics 2013, 12, 3409–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Schmidt A, Kochanowski K, Vedelaar S, Ahrne E, Volkmer B, Callipo L, Knoops K, Bauer M, Aebersold R, Heinemann M, Nat. Biotechnol 2016, 34, 104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yamamoto K, Hirao K, Oshima T, Aiba H, Utsumi R, Ishihama A, J. Biol. Chem 2005, 280, 1448–1456. [DOI] [PubMed] [Google Scholar]
- [22].Delgado J, Forst S, Harlocker S, Inouye M, Mol. Microbiol 1993, 10, 1037–1047. [DOI] [PubMed] [Google Scholar]
- [23].Presper KA, Wong CY, Liu L, Meadow ND, Roseman S, Proc. Natl. Acad. Sci. USA 1989, 86, 4052–4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Jo YL, Nara F, Ichihara S, Mizuno T, Mizushima S, J. Biol. Chem 1986, 261, 15252–15256. [PubMed] [Google Scholar]
- [25].Aiba H, Nakasai F, Mizushima S, Mizuno T, J. Biol. Chem 1989, 264, 14090–14094. [PubMed] [Google Scholar]
- [26].Dayon L, Hainard A, Licker V, Turck N, Kuhn K, Hochstrasser DF, Burkhard PR, Sanchez JC, Anal. Chem 2008, 80, 2921–2931. [DOI] [PubMed] [Google Scholar]
- [27].Cai SJ, Inouye M, J. Biol. Chem 2002, 277, 24155–24161. [DOI] [PubMed] [Google Scholar]
- [28].Rkenes TP, Lamark T, Strom AR, J. Bacteriol 1996, 178, 1663–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Styrvold OB, Falkenberg P, Landfald B, Eshoo MW, Bjornsen T, Strom AR, J. Bacteriol 1986, 165, 856–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Moellering RE, Cravatt BF, Science 2013, 341, 549–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Weinert BT, Iesmantavicius V, Wagner SA, Scholz C, Gummesson B, Bell P, Nystrom T, Choudhary C, Mol. Cell 2013, 51, 265–272. [DOI] [PubMed] [Google Scholar]
- [32].Harmel R, Fiedler D, Nat. Chem. Biol 2018, 14, 244–252. [DOI] [PubMed] [Google Scholar]
- [33].Rajeev L, Luning EG, Dehal PS, Price MN, Arkin AP, Mukhopadhyay A, Genome Biol. 2011, 12, R99. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.