Abstract
BALB/cJ mice exhibit considerable phenotypic differences with other BALB/c substrains. Some of these traits involve the liver, including persistent postnatal expression of genes that are normally expressed only in the fetal liver and reduced expression of major urinary proteins. These traits are due to a mutation that dramatically reduces expression of the gene encoding the transcription factor Zinc fingers and homeoboxes 2 (Zhx2). BALB/cJ mice also exhibit reduced serum lipid levels and resistance to atherosclerosis compared to other mouse strains when placed on a high fat diet. This trait is also due, at least in part, to the Zhx2 mutation. Microarray analysis identified many genes affecting lipid homeostasis, including Lipoprotein lipase, that are dysregulated in BALB/cJ liver. This led us to investigate whether hepatic lipid levels would be different between BALB/cJ and BALB/c mice when placed on a normal chow or a high fat chow diet. On the high fat chow, BALB/cJ mice had increased weight gain, increased liver/body weight ratio, elevated hepatic lipid accumulation and markers of liver damage when compared to BALB/c mice. These traits in BALB/cJ mice were only partially reversed by a hepatocyte-specific Zhx2 transgene. These data indicate that Zhx2 reduces liver lipid levels and is hepatoprotective in mice on a high fat diet, but the partial rescue by the Zhx2 transgene suggests a contribution by both parenchymal and non-parenchymal cells. A model to account for the cardiovascular and liver phenotype in mice with reduced Zhx2 levels is provided.
Keywords: Zhx2, BALB/cJ, high fat diet, liver damage
Introduction
The highly related BALB/c and BALB/cJ substrains separated in 1937 from a line of albino mice established in the early 1900’s (Potter, 1985). BALB/cJ mice exhibit considerable physiological and behavioral differences compared to BALB/c and other BALB substrains (Potter, 1985). Several notable phenotypes unique to the BALB/cJ substrain include reduced levels of major urinary proteins (Mups), persistent postnatal expression of hepatic genes [including alpha-fetoprotein (AFP)] normally expressed only in the fetal liver and higher aggression in adult males (Dow et al., 2011; Duncan et al., 1988; Olsson et al., 1977). Genetic analysis indicates that postnatal AFP expression and reduced Mup levels are transmitted in an autosomal recessive manner, suggesting that each is controlled by a single locus (Duncan et al., 1988; Olsson et al., 1977).
Elevated serum AFP protein levels in adult BALB/cJ mice was first observed by Rhouslatti and colleagues in 1977 (Olsson et al., 1977). Subsequent studies indicated that this was due to elevated AFP mRNA levels in the adult liver and was governed by a locus on Chromosome 15 called alpha-fetoprotein regulator 1 (Afr1)(Belayew and Tilghman, 1982; Blankenhorn et al., 1988; Peyton et al., 2000). The imprinted H19 gene and X-linked glypican 3 (Gpc3) gene, both normally expressed in the fetal liver and silenced at birth, are also targets of Afr1 based on their continued expression in adult BALB/cJ liver (Morford et al., 2007; Pachnis et al., 1984). We used positional cloning to identify the Zinc fingers and homeoboxes 2 (Zhx2) gene as being responsible for the Afr1 phenotype (Perincheri et al., 2005). Zhx2 encodes an 838 amino acid protein that is predicted to contain two C2-H2 zinc fingers and four homeodomains, suggesting a role in nucleic acid binding and gene regulation. Zhx2 is a member of a small gene family that includes Zhx1 and Zhx3 (Spear et al., 2006). The hypomorphic BALB/cJ Zhx2 allele (Zhx2Afr1) contains a mouse endogenous retroviral element in the first intron that dramatically reduces, but does not eliminate, Zhx2 mRNA levels; the low levels of Zhx2 mRNA that are expressed can encode normal Zhx2 protein (Perincheri et al., 2008). This mutation is not found in the BALB/c or BALB/cByJ Zhx2 genes. Expression of a hepatocyte-specific Zhx2 transgene (TTR-Zhx2; in which the Zhx2 coding region was linked to a hepatocyte-specific Transthyretin enhancer-promoter cassette) in BALB/cJ mice resulted in normal AFP, H19 and Gpc3 repression, providing direct evidence that Zhx2 regulates postnatal silencing of these target genes (Perincheri et al., 2005). We have found that the dramatic reduction in Mups, which are synthesized primarily in the liver, in BALB/cJ mice is also due to the Zhx2 mutation (Jiang et al., 2017).
Another unique BALB/cJ trait is their resistance to atherosclerosis and reduced serum lipid levels compared to other mouse strains, including MRL/lpr mice, when placed on a high fat diet (Gu et al., 1999). A Quantitative Trait Locus (QTL) analysis using BALB/cJ mice and MRL/lpr mice identified several loci associated with hyperlipidemia and atherosclerosis. One QTL called Hyperlipidemia 2 (Hyplip2) was localized to the region on chromosome 15 where Zhx2 resides (Wang et al., 2004). Further studies demonstrated that BALB/cJ mice expressing the TTR-Zhx2 transgene had increased serum triglyceride and cholesterol levels compared to non-transgenic BALB/cJ littermates, demonstrating that Zhx2 is a novel regulator of serum lipid levels and responsible for the Hyplip2 phenotype (Gargalovic et al., 2010).
Several hepatic proteins that regulate serum lipids, including Lipoprotein lipase (Lpl), are altered in the adult liver when Zhx2 levels are reduced (Gargalovic et al., 2010). This led us to test the hypothesis that hepatic lipids would also be affected by changes in Zhx2 expression in mice on a normal or high fat diets. BALB/cJ and BALB/c mice were placed on a normal chow or a high fat chow diet for eight weeks. BALB/cJ mice on the high fat chow had increased weight gain, increased liver/body weight ratio, elevated hepatic lipid accumulation and markers of liver damage when compared to BALB/c mice on the same diet. Similar trends were observed when diet studies were performed in BALB/cJ mice that did or did not contain the hepatocyte-specific TTR-Zhx2 transgene, although the effects were not as robust as in the BALB/cJ:BALB/c comparison. These data indicate that Zhx2 is hepatoprotective in mice on a high fat diet and suggests that both parenchymal (i.e., hepatocytes) and non-parenchymal cells (i.e., Kupffer cells, stellate cells, endothelial cells) contribute to this Zhx2-mediated protection. A model to account for the cardiovascular and liver phenotype in mice with reduced Zhx2 levels is provided.
Methods and Materials
Mice and diet study.
All mice were housed in the University of Kentucky Division of Laboratory Animal Research (DLAR) facility and kept according to Institutional Animal Care and Use Committee (IACUC)-approved protocol and received food and water ad libitum. BALB/cJ and BALB/c mice were obtained from The Jackson Lab and Harlan, respectively. BALB/cJ mice containing TTR-Zhx2-Flag transgene, which is comprised of a transthyretin enhancer/promoter cassette fused to a full-length mouse Zhx2 cDNA and is expressed specifically in hepatocytes at physiological levels, were described previously (Perincheri et al., 2005). For the diet studies, six-week old female mice were placed on either standard chow with 6.8% fat or high fat “Paigen” chow (Harlan Teklad TD 90221) with 15.8% of fat for eight weeks; each cohort contained five mice. Mice were weighed weekly and at the end of the study, mice were fasted for four hours and then euthanized. Blood was collected in a serum separator tube (Becton-Dickinson Co.) and centrifuged at 13,000 x g for 1 min to separate red blood cells from serum, which was snap-frozen and stored at −80°C. Livers were removed, weighed and liver pieces were embedded in OCT and snap frozen, placed in formalin and then embedded in paraffin, or snap frozen and stored at −80°C.
RNA extraction and Real-Time-quantitative PCR (RT-qPCR).
RNA was prepared from ~100 mg of liver using Trizol (Invitrogen) as described (Clinkenbeard et al., 2012). One µg of RNA was converted into cDNA using the qscript kit (Quanta Biosciences) according to the manufacturer’s instructions and run in the reverse transcriptase protocol in an iCycler (BioRad). qPCR was carried out using 2.5 µl of 1:10 diluted cDNA with 10 µL of Sybr Green (Quanta Biosciences), 5 µL of primer mix and 2.5 µL of water and analyzed with Bio-rad iCycler. All CT (MyIQ) levels were normalized against ribosomal protein L30 using the ΔΔCt method (Livak and Schmittgen, 2001). The following primers were used: AFP (ATCAGTGTCTGCTGGCACGCA and GGCTGGGGCATACATGAAGGGG), Lipoprotein lipase (Lpl: TGGCTACACCAAGCTGGTGGGA and GGTGAACGTTGTCTAGGGGGTAGT), Lipocalin 2 (Lcn2: CTACAATGTCACCTCCATCCTG and AGCTCTGTACCTGAGGATACC), ribosomal protein L30 (L30: ATGGTGGCCGCAAAGAAGACGAA and CCTCAAAGCTGGACAGTTGTTGGCA).
Oil Red O staining.
Frozen OCT livers were cryosectioned at 10 µm thickness and placed on slides. Sections were fixed in ice-cold formalin, rinsed extensively in water, equilibrated in propylene glycol, and then placed in 0.5% Oil Red O (Sigma) in propylene glycol at 60°C for 8 min. Sections were then placed in 85% propylene glycol for 5 min, rinse with water, and incubated in haematoxylin for 1 min to counterstain the nuclei blue. After a 3 min rinse in water, VECTAshield (Vector Laboratories) was added and slides were coverslipped. Images were captured using a Ziess Upright Microscope and AxioVision software.
H&E staining and liver pathology.
Formalin-fixed, paraffin-embedded liver sections were stained with Hematoxylin & Eosin and analyzed in a “blinded” manner by Eun Lee, MD, a liver pathologist at the University of Kentucky. Livers from the different groups on the normal chow had no identifiable differences.
Alanine Aminotranserase (ALT) measurement.
One µl of serum and 9 µl of 0.9% Saline were incubated with 1 ml of prepared ALT solution using an ALT kit (Pointe Scientific) according to manufacturer’s instructions. Samples were incubated at 37°C and kinetic measurements were taken every minute for a total of 3 readings on the Biomate 3 spectrophotometer (Thermo-Scientific). Differences in the absorbance readings were calculated to determine serum ALT concentrations (IU/L).
Lipid extraction and Cholesterol analysis.
Total lipid from whole liver was extracted as previously described with minor modifications (Folch et al., 1957). Fifty milligrams of each liver was mixed with 3 mL of 2:1 chloroform:methanol and heated to 55°C for 1–2 hours. Extracted lipid in the supernatant was removed and dried under in a 55°C water bath. The resulting lipid was purified by adding 6 mL of 2:1 chloroform:methanol and 1.2mL 0.05% and vortexed, allowing for the phases to separate for 10 minutes at room temperature. Purified lipid was mixed with 2 mL of 1% Triton X100 in chloroform and dried under in a 55°C water bath. Total lipid was suspended in 1mL of distilled water and heated to 55°C for lipid to dissolve fully. Cholesterol levels were measured in total purified lipid using the Pointe Scientific cholesterol reagent (Pointe Scientific). Fifty µl of lipid solution was mixed with 150 µl of cholesterol reagent in a 96-well plate and incubated for 1hr at room temperature. Absorbance of each sample was read at 595nm. Each sample was run in duplicate and normalized to the wet weight of the liver sample.
Statistical analysis.
All values within a group were averaged and plotted as mean +/− standard deviation. The p-values were calculated between two groups using student’s t-test. Diet study groups were analyzed with ANOVA followed by Tukey’s test. To determine significant interaction between strain and type of diet, two-way ANOVA analysis was performed. A p-value less than 0.05 was considered significant.
Results
BALB/cJ mice have increased body weight and liver:body weight ratio on a high-fat chow compared to BALB/c mice.
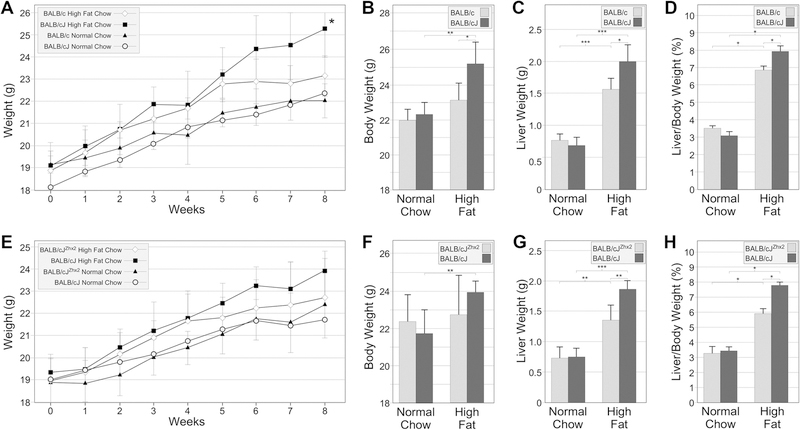
Based on the hypomorphic Zhx2 mutation in BALB/cJ mice and the role of Zhx2 in controlling serum lipid levels in mice, we investigated whether changes in Zhx2 levels impacted the liver in mice on a high fat diet. In the first study, six-week old female BALB/c and BALB/cJ mice were place on regular chow or high fat chow and weighed weekly for eight weeks. All four cohorts gained weight during the 8-week study. No significant differences were observed between BALB/cJ and BALB/c maintained on a regular chow (Fig. 1A, 1B). Weight gain was greater in both substrains on a high fat chow compared to those on a regular chow, although this difference was significant only in BALB/cJ mice (Fig 1A, B). By week 8, BALB/cJ mice had significantly greater weight gain than BALB/c mice on the high fat chow, indicating a significant interaction between strain and diet on weight gain. (Fig. 1A, B). At the end of 8 weeks, all mice were sacrificed and the dissected livers were weighed prior to processing. Livers of all mice on the high fat chow had a much whiter color than those on a normal chow, indicative of substantial lipid accumulation (not shown). Liver weight in both substrains was greater on a high fat chow than on normal chow (Fig 1C). Whereas there was no difference in liver weight in both substrains on a normal chow, the liver weight in BALB/cJ mice was significantly greater than the liver weight of BALB/c mice on a high fat chow (Fig. 1C). Liver weight normalized to body weight showed that both cohorts on high fat chow had significantly increased liver:body weight ratios compared to those on normal chow (Fig. 1D). A significant interaction was found between strain and diet type since this ratio in BALB/cJ mice was greater than in BALB/c.
Figure 1. BALB/cJ mice exhibit greater overall body weight and liver:body weight ratio than BALB/c mice on a high fat chow.
Female BALB/cJ and BALB/c mice (Figs. 1A–G) and BALB/cJ mice with or without the TTR-Zhx2 transgene (Figs. 1E–H) were maintained on a normal chow or high fat chow for 8 weeks, at which time mice were killed and livers and serum were collected. A, B. There was no difference in overall body weight BALB/cJ and BALB/c mice on normal chow but BALB/cJ mice had significantly higher body weight than BALB/c mice after 8 weeks on high fat chow. BALB/cJ mice but not BALB/c mice had increased body weight on a high fat chow compared to those on a normal chow. C. Liver weight increased in both BALB/c and BALB/cJ on a high fat chow compared to those on normal chow and to a greater extent in BALB/cJ mice than in BALB/c mice. D. The liver:body weight ratio in both substrains on high fat chow was greater than those on normal chow. The liver:body weight ratio of BALB/cJ mice was greater than BALB/c mice on a high fat chow for 8 weeks. E, F. There was no significant difference in overall body weight in BALB/cJ with or without the TTR-Zhx2 transgene on either normal chow or high fat chow, although high fat chow significantly increased body weight in BALB/cJ. G. Liver weight increased in transgenic and non-transgenic mice on a high fat chow compared to those on normal chow and significantly more in non-transgenic mice compared to BALB/cJZhx2 littermates. H. The liver:body weight ratio in BALB/c mice with or without the TTR-Zhx2 transgene on high fat chow was greater than those on a normal chow. The liver:body weight ratio of BALB/cJ mice without the TTR-Zhx2 transgene was greater than BALB/cJ mice with the transgene on a high fat chow for 8 weeks. *p<0.05, **p<0.01, ***p<0.001
A hepatocyte-specific Zhx2 transgene significantly reduced liver-body weight ratio but not overall body weight in BALB/cJ on high-fat chow.
Since the hepatocyte-specific TTR-Zhx2 transgene was previously shown to alter serum lipid levels in BALB/cJ mice (Gargalovic et al., 2010), we tested whether this transgene would influence high fat chow-induced weight gain and liver/body weight ratio. Six week-old female BALB/cJ mice with or without the TTR-Zhx2 transgene (BALB/cJ Zhx2 and BALB/cJ, respectively), were maintained on a regular or high fat chow as described above. Over the eight-week period, weight gain was greater in mice on a high fat chow and BALB/cJ gained more than BALB/cJZhx2 mice (Fig. 1E, F). The trend was similar to what was seen between BALB/c and BALB/cJ mice, but the difference in transgenic and non-transgenic cohorts on the high fat chow did not reach significance although body weight gain was significantly greater in non-transgenic BALB/cJ than in BALB/cJZhx2 mice on a high fat chow (Fig. 1F). Liver weight was the same in both transgenic and non-transgenic mice on normal chow and, in both groups liver weight increased significantly on a high fat chow (Fig. 1G). This liver weight gain on high fat chow was greater in BALB/cJ mice than in BALB/cJZhx2 mice (Fig. 1G). Both cohorts on high fat chow had significantly increased liver:body weight ratios compared to littermates on normal chow (Fig. 1H). Similarly to what was observed with the BALB/c:BALB/cJ comparison, this ratio was greater in BALB/cJ than in BALB/cJZhx2 littermates, demonstrating a significant interaction between diet and Zhx2 expression. The common difference between BALB/cJ and BALB/c or BALB/cJZhx2 suggests that decreased hepatocyte Zhx2 expression is directly responsible for increased liver weight gain in mice on high fat chow.
Lipid accumulation, histological alterations and serum ALT levels are greater in BALB/cJ mice than BALB/c on a high fat chow and are not rescued by the TTR-Zhx2 transgene.
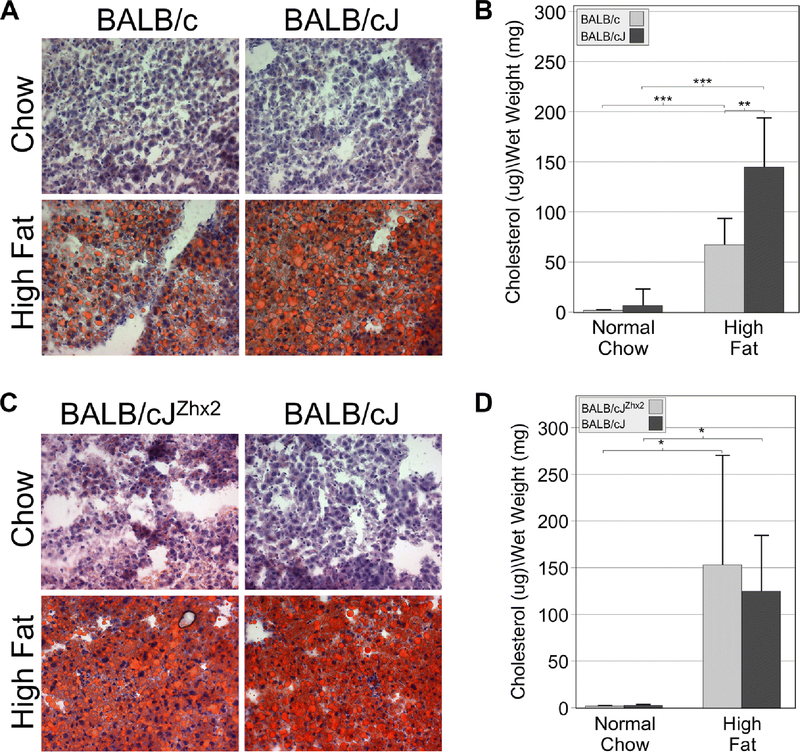
The increased liver:body weight ratio in BALB/cJ mice on high-fat chow led us to investigate potential differences in lipid accumulation and liver morphology in both diet studies. Frozen livers sections from mice after the eight-week diet study were stained with Oil Red O. In the BALB/c:BALB/cJ comparison, minimal Oil Red O staining was observed in frozen liver sections of mice on a normal chow (Fig. 2A, upper panels). Staining was increased in both groups on a high fat chow, but was greater in BALB/cJ than in BALB/c liver (Fig. 2A, lower panels). This data is supported by quantitation of liver cholesterol levels (Fig. 2B). Livers from both substrains of mice on high fat chow had much higher cholesterol levels than mice on normal chow and cholesterol levels were significantly greater in BALB/cJ mice than BALB/c mice. However, a different pattern was observed in the comparison between BALB/cJ and BALB/cJZhx2 mice. While both cohorts exhibited higher Oil Red O staining on a high fat chow, these levels were not obviously different between transgenic and non-transgenic mice (Fig. 2C). Consistent with this staining, overall cholesterol levels in BALB/cJ and BALB/cJZhx2 livers from high fat chow-fed mice were elevated compared to chow-fed mice, but were not significantly different from each other (Fig. 2D).
Figure 2. BALB/cJ mice have higher liver lipid accumulation than BALB/c mice on a high fat chow.
A. Oil Red O staining of frozen liver sections was low in both BALB/cJ and BALB/c mice on a normal chow. Oil Red O staining was elevated in both substrains on a high fat chow, but BALB/cJ mice had higher staining than BALB/c mice. B. Cholesterol levels were higher in both substrains on a high fat chow compared to those on a normal chow but were higher in BALB/cJ mice than BALB/c mice on a high fat chow. C. Oil Red O staining was greater in BALB/cJ mice with or without the TTR-Zhx2 transgene on a high fat chow compared to those on a normal chow, but the presence of the TTR-Zhx2 transgene did not dramatically change Oil Red O staining in mice on a high fat chow. D. Cholesterol levels were higher in BALB/cJ mice with or without the TTR-Zhx2 transgene on a high fat chow compared to those on a normal chow but were not significantly different between both groups on a high fat chow. *p<0.05; **p<0.01; ***p<0.001
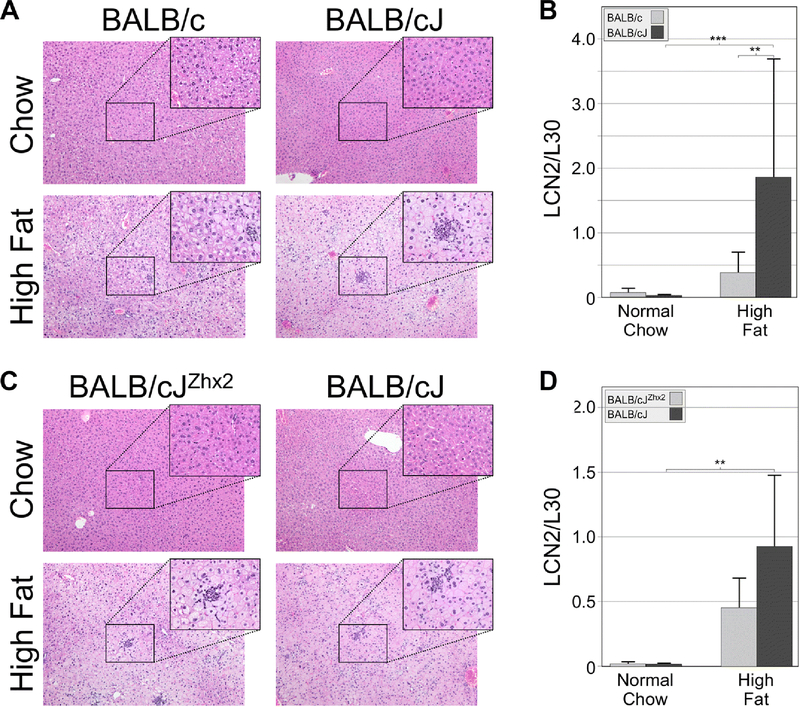
To explore further diet-induced changes in liver morphology, formalin-fixed, paraffin-embedded liver sections were stained with H&E. Livers from BALB/c and BALB/cJ on the normal chow looked normal and had no obvious differences. Livers from mice on the high fat chow had evidence of hepatocyte ballooning and neutrophil infiltration, with these changes being greater in BALB/cJ than in BALB/c mice (Fig. 3A). To quantitate neutrophil infiltration, RT-qPCR was used to measure mRNA levels of Lipocalin 2 (Lcn2), a neutrophil marker (Ariza et al., 2016). Lcn2 mRNA levels were higher in both substrains on a high fat chow, but were significantly greater in BALB/cJ than in BALB/c mice (Fig. 3B). In contrast, H&E staining revealed similar levels of increased hepatocyte ballooning and neutrophil infiltration in livers of BALB/cJ and BALB/cJZhx2 mice on a high fat chow (Fig. 3C). Furthermore, while hepatic Lcn2 mRNA levels were somewhat lower in TTR-Zhx2 transgenic mice, this difference did not reach significance (Fig. 3D).
Figure 3. BALB/cJ livers exhibit greater hepatocyte ballooning and neutrophil infiltration than livers of BALB/c mice on a high fat chow.
A. H&E staining indicated that livers of BALB/cJ and BALB/c on a normal chow showed no abnormalities. When placed on a high fat chow, hepatocyte ballooning and neutrophil infiltration was greater in BALB/cJ than in BALB/c liver. B. Levels of LCN2 mRNA, a neutrophil marker, were low in both BALB/cJ and BALB/c mice on a normal chow and increased significantly in both substrains on a high fat chow; this high fat-induced increase was greater in BALB/cJ mice than in BALB/c mice. C. H&E staining indicated normal liver morphology in BALB/cJ mice with or without the TTR-Zhx2 transgene on a normal chow and no obvious difference in the degree of hepatocyte ballooning and neutrophil infiltration was observed between transgenic and non-transgenic mice on a high fat chow. D. LCN2 mRNA levels increased in BALB/cJ mice with or without the TTR-Zhx2 transgene on a high fat chow; although this increase was greater in non-transgenic animals, this difference did not reach significance. In both A and C, sections are shown at 10X; boxed regions are also shown at 40X; clusters of neutrophils are evident at 40X magnification. **p<0.1; ***p<0.01
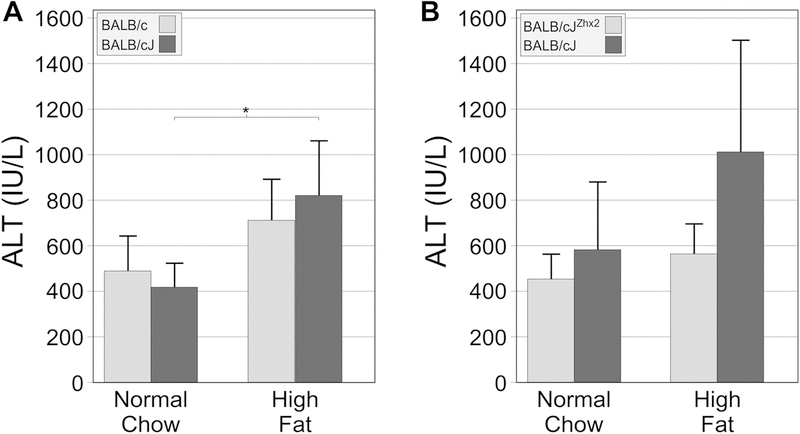
Because the liver histology of mice on the high fat chow showed evidence of damage and to determine whether increased liver damage was occurring in BALB/cJ mice, serum levels of alanine aminotransferase (ALT, a marker of hepatocyte injury) were measured at the end of the study (Fig. 4). BALB/c mice on a high fat chow exhibited ~1.5-fold increase in serum ALT levels compared to those on normal chow. In BALB/cJ mice, serum ALT levels were higher and showed a significant, roughly 2-fold increase in the high fat chow cohort compared to the chow-fed mice (Fig. 4A). While the high fat chow increased serum ALT levels in BALB/cJ mice with the TTR-Zhx2 transgene more than those without the transgene (1.74- and 1.25-fold, respectively), in neither group was this increase significant (Fig. 4B).
Figure 4. The high fat chow significantly increased serum ALT levels in BALB/cJ mice but not in BALB/c mice.
Serum ALT levels were analyzed at the end of the eight-week study. A. Serum ALT levels significantly increased only in BALB/cJ mice receiving the high fat chow. B. Serum ALT levels modestly increased in both BALB/cJZhx2 and BALB/cJ mice on high fat chow compared to those on normal chow, but in neither case was this increase significant. *p<0.05 vs same genotype on normal chow.
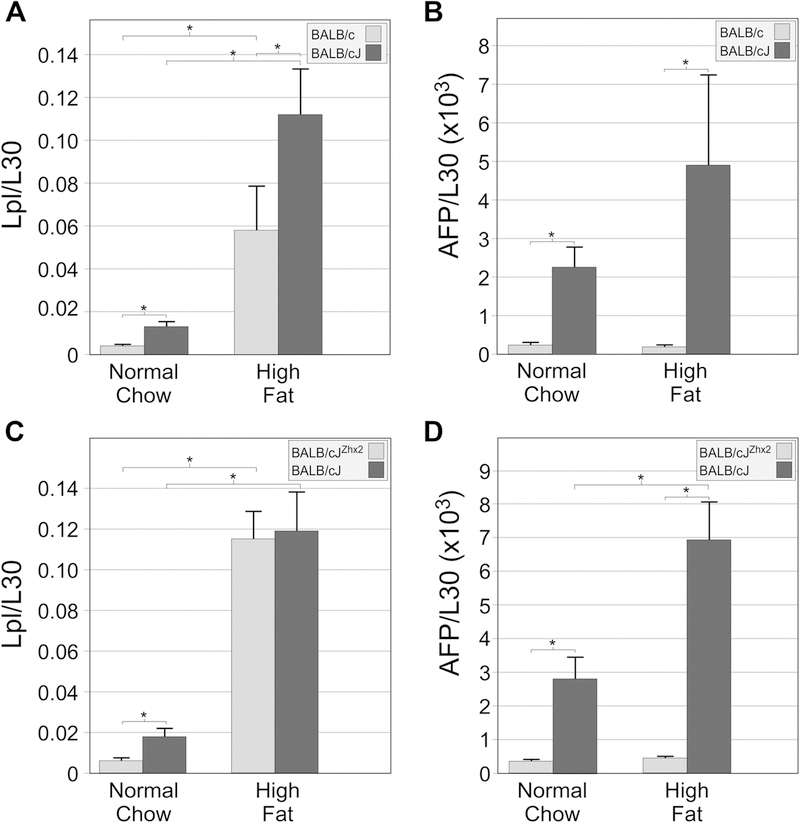
Hepatocyte-specific Zhx2 transgene does not affect elevated Lpl expression induced by high fat chow.
Lpl, which hydrolyzes triglycerides on lipoproteins and facilitates their uptake into cells, is normally not expressed in the adult liver (Kirchgessner et al., 1989). However, Lpl mRNA levels are elevated in BALB/cJ livers, indicating that it, like AFP, H19 and Gpc3, is also normally repressed by Zhx2 in the adult liver (Gargalovic et al., 2010). Since Lpl could contribute to the changes in hepatic lipid levels in mice on the high fat chow, liver Lpl mRNA levels were measured in both sets of diet study mice. In the BALB/c:BALB/cJ comparison, BALB/cJ mice on normal chow had higher Lpl mRNA levels than BALB/c mice (Fig. 5A), consistent with previous studies. Lpl mRNA levels increased significantly in both substrains on a high fat chow, although levels were significantly lower in BALB/c mice than in BALB/cJ mice (Fig. 5A). A similar analysis was performed in BALB/cJ and BALB/cJZhx2 mice. On normal chow, Lpl mRNA levels were lower in transgenic mice, consistent with the TTR-Zhx2 transgene repressing basal expression of Lpl (Fig. 5C). However, on a high fat chow, Lpl mRNA levels were increased to roughly the same level in both groups of mice, irrespective of the presence of the hepatocyte-specific Zhx2 transgene (Fig. 5C). As a control to show that the TTR-Zhx2 transgene was functioning as expected in the high fat chow fed mice, AFP mRNA levels were also measured. Like Lpl, AFP mRNA levels were higher in BALB/cJ mice than in either BALB/c or BALB/cJZhx2 mice on normal chow (Fig. 5B, D). AFP mRNA levels increased over 2-fold in BALB/cJ mice on a high fat chow in both studies, likely due to increased liver damage in these mice. However, in contrast to Lpl, AFP mRNA levels in both BALB/c and BALB/cZhx2 mice on a high fat chow did not increase but were essentially the same as those mice on normal chow, indicating that the TTR-Zhx2 transgene was still repressing AFP gene expression (Fig. 5B, D). These results suggest that a portion of the diet-induced increase in Lpl expression must come from non-hepatocyte sources.
Figure 5. Liver Lpl mRNA levels increase in mice on a high fat chow; this increase is greater in BALB/cJ than in BALB/c mice but are the same in BALB/cJ mice with or without the TTR-Zhx2 transgene.
RNA was analyzed by RT-qPCR; Lpl and AFP mRNA levels were normalized to L30. A. Liver Lpl mRNA levels are higher in BALB/cJ than in BALB/c mice on a normal chow. These levels increase in both substrains on a high fat chow, but increase to a greater extent in BALB/cJ mice than in BALB/c mice. B. Liver AFP levels are higher in BALB/cJ than in BALB/c mice on both the normal chow and high fat chow. C. Liver Lpl mRNA levels are higher in BALB/cJ than in BALB/cJZhx2 mice on a normal chow. These levels increase in both cohorts on a high fat chow but are not significantly different between BALB/cJ and BALB/cJZhx2 mice. D. Liver AFP mRNA levels are higher in BALB/cJ than in BALB/cJZhx2 mice on a normal chow and high fat chow. Whereas BALB/cJZhx2 mice showed no change in AFP mRNA on high fat chow, liver AFP mRNA in BALB/cJ mice on the high fat chowed showed a further induction over normal chow. *p<0.05
Discussion
Several traits distinguish BALB/cJ mice from other mice, including lower serum lipid levels when placed on a diet that is high in fat (Potter, 1985; Wang et al., 2004). This phenotype is due to a natural hypomorphic mutation in the BALB/cJ Zhx2 allele that dramatically reduces Zhx2 levels (Gargalovic et al., 2010). While previous diet studies have focused on the role of Zhx2 in altering serum lipid levels and associated cardiovascular traits, studies described here investigated the impact of Zhx2 on the liver in response to a high fat diet. These studies used BALB/cJ mice compared to the highly related BALB/c substrain which has a wild-type Zhx2 allele, as well as BALB/cJ mice with or without a hepatocyte-specific TTR-Zhx2 transgene. We show that BALB/cJ mice on a high fat chow have increased body weight and liver/body weight ratios, as well as greater hepatic lipid accumulation, hepatocyte ballooning, neutrophil accumulation and liver damage, as measured by serum ALT levels, when compared to BALB/c mice. Thus, in contrast to the cardioprotective effects of the Zhx2 mutation, our data indicate that reduced Zhx2 levels lead to elevated liver damage in BALB/cJ mice on a high fat chow.
Previous adult liver microarray analysis using BALB/cJ mice and a Zhx2+ BALB/cJ congenic strain identified numerous genes that were affected, some positively and others negatively, by dramatically reduced Zhx2 expression (Gargalovic et al., 2010). Many of these hepatic genes could influence the physiological response to a high fat diet. One gene of particular interest is Lpl, which promotes the cellular uptake of cholesterol and free fatty acids and is normally not expressed in the adult liver. However, similarly to AFP, Lpl is highly expressed in the fetal liver, silenced postnatally, and continues to be expressed in the adult BALB/cJ liver. We found that hepatic cholesterol levels (Fig. 2B) and Lpl mRNA levels (Fig. 5A) are roughly 2-fold higher in BALB/cJ mice than in BALB/c mice on the high fat chow, consistent with the possibility that elevated Lpl expression contributes to increased hepatic lipid levels. Evidence for a direct role of Lpl in liver lipid accumulation comes from several previous studies. Overexpression of a hepatocyte-specific Lpl transgene in Lpl knock-out mice resulted in dramatically increased liver lipid accumulation (Merkel et al., 1998). In a tree shrew model of NAFLD that increased liver Lpl expression, pharmacological inhibition of Lpl reduced both hepatic lipid accumulation and inflammation (Zhang et al., 2015). While our data support a role for Lpl in lipid accumulation, it is unlikely that misregulated Lpl accounts for the entire liver phenotype in BALB/cJ mice on high fat chow. We have found that Mup mRNA levels are dramatically reduced in the absence of Zhx2 (Jiang et al., 2017), and several Mups have been found to influence lipid levels (Zhou et al., 2009). Several Cyp enzymes and Elongation of very long chain fatty acid 3 (Elovl3) are also dramatically reduced in Zhx2-deficient livers (Creasy et al., 2016)(K.T. Creasy, E.C. M.L.P. and B.T.S., manuscript in preparation).
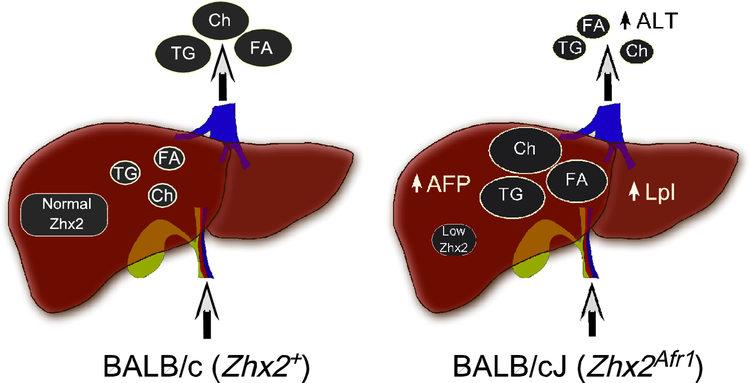
A model to explain how Zhx2-mediated changes in hepatic gene expression could account for the seemingly discordant cardiovascular and hepatic diet-induced traits in mice on high fat chow is shown in Fig. 6. In BALB/c mice fed a high fat chow, normal Zhx2 levels result in efficient lipid clearance from the liver, resulting in elevated serum lipids and subsequent increase in atherosclerotic lesions. In contrast, reduced Zhx2 levels in BALB/cJ mice alters the expression of Zhx2 target genes, including Lpl, that are involved in lipid uptake, transport and/or metabolism. While this increased hepatic lipid accumulation is accompanied by lower serum lipid levels, which reduces the extent of cardiovascular injury, it also causes increased hepatocyte damage as judged by hepatocyte ballooning, neutrophil infiltration, and serum ALT levels. It should be noted that several human GWAS studies have found an association between Zhx2 and cardiovascular disease (Bis et al., 2011; Li et al., 2015), although to date there are no human data linking Zhx2 and liver disease.
Figure 6. Model to account for the cardiovascular and liver phenotypes in BALB/cJ and BALB/c mice on a high fat diet.
The Zhx2 gene contains a hypomorphic mutation in BALB/cJ mice (Zhx2Afr1) but not in the highly related BALB/c substrain (Zhx2+). When placed on a high fat chow in the presence of the wild-type Zhx2 allele (BALB/c), hepatic lipid levels remain relatively low but serum lipid levels are elevated, contributing to the increased atherosclerosis. When BALB/cJ mice are maintained on a high fat chow, lipids are redistributed so that hepatic lipid levels are increased and serum lipid levels are lower. This increase in hepatic lipids may be due, in part, to elevated Lpl levels. Higher hepatic lipid levels enhance liver damage as determine by increased hepatocyte ballooning, neutrophil infiltration and serum ALT levels, whereas reduced serum lipid levels reduce atherosclerosis.
If hepatocytes are solely responsible for the diet-induced increased lipid accumulation and liver damage in BALB/cJ compared to BALB/c mice, the TTR-Zhx2 transgene should have reversed all aspects of the liver phenotype in BALB/cJ mice fed a high fat chow. However, the TTR-Zhx2 transgene, whose expression is hepatocyte-specific (Perincheri et al., 2005; Wu et al., 1996), only partially corrected the liver phenotype in BALB/cJ mice. The endogenous Zhx2 gene is expressed in both hepatocytes and non-parenchymal cells of the adult liver (Jiang et al., 2017), and some Zhx2 targets, including Lpl, are expressed in both hepatocytes and non-parenchymal cells in the absence of Zhx2 (data not shown). In fact, the equally high expression of Lpl in BALB/cJ mice on a high fat chow with or without the hepatocyte-specific TTR-Zhx2 transgene (Fig. 5C) is consistent with the high Lpl expression coming from the non-parenchymal cells. In mice on high fat chow, immunohistochemical staining confirmed Lpl expression in non-parenchymal cells and hepatocytes although this latter expression is restricted to pericentral hepatocytes (data not shown), consistent with previous single-cell RNA seq studies (Bahar Halpern et al., 2017). Lpl expression in non-parenchymal cells, such as Kupffer cells, would not be affected by the TTR-Zhx2 transgene and could also explain the similarly high lipid/cholesterol levels in BALB/cJ and BALB/cJZhx2 mice on high fat chow (Fig. 2C, D). We therefore believe that changes in Zhx2 target gene expression in both hepatocytes and non-hepatocytes account for the increased liver damage in BALB/cJ mice on the high fat chow. Since Zhx2 is ubiquitously expressed and BALB/cJ mice have reduced Zhx2 expression is all tissues (Perincheri et al., 2005)(data not shown), it is also possible that altered expression of Zhx2 target genes in other tissues could affect the differences seen on a high fat chow. We have generated mice with a floxed Zhx2 allele on a C57BL/6 background (Creasy et al., 2016; Jiang et al., 2017) and further studies using these mice will help identify the contributions different tissues and/or hepatic cell subpopulations make to the liver phenotype seen in BALB/cJ mice on the high fat chow.
Increased and persistent liver damage can lead to steatohepatitis, fibrosis and end stage liver disease, cirrhosis and hepatocellular carcinoma (HCC) (Aravalli et al., 2013; Baffy et al., 2012; Bruix et al., 2004). Our data demonstrating increased high fat diet-induced liver damage in BALB/cJ mice raises the possibility that Zhx2 and its target genes may contribute to liver disease. Several other studies support this possibility. Liver fibrosis caused by chronic treatment with the hepatoxin Carbon tetrachloride (CCl4) is greater in BALB/cJ mice compared to other strains (Shi et al., 1997). A QTL analysis identified several loci that are responsible for this phenotype, include hepatic fibrosis 1 (hFib1) (Hillebrandt et al., 2002). Furthermore, studies suggest that Zhx2 functions as an HCC tumor suppressor in liver cell culture and xenograft tumor studies (Song et al., 2018; Yue et al., 2012). Taken together, these data indicate that Zhx2 has an important hepatoprotective role in multiple aspects of liver disease. Additional studies will be needed to further elucidate the role of Zhx2 in maintaining liver health in response to injury. Since Zhx2 is a member of a small gene family that also includes Zhx1 and Zhx3 (Kawata et al., 2003; Spear et al., 2006; Yamada et al., 2002; Yamada et al., 2003), it will also be interesting to investigate whether these two related genes also influence lipid homeostasis and/or liver function.
Acknowledgements:
We thank Sean Thatcher, Ryan Temel and Eun Lee, University of Kentucky, for their assistance. This study was supported by Public Health Service Grants DK059866 and DK074816 from the National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Conflict of Interest Statement: On behalf of all authors, the corresponding author states that there is no conflict of interest.
REFERENCES:
- Aravalli RN, Cressman EN, and Steer CJ (2013). Cellular and molecular mechanisms of hepatocellular carcinoma: an update. Archives of Toxicology 87:227–247. [DOI] [PubMed] [Google Scholar]
- Ariza X, Graupera I, Coll M, Sola E, Barreto R, Garcia E, Moreira R, Elia C, Morales-Ruiz M, Llopis M, et al. (2016). Neutrophil gelatinase-associated lipocalin is a biomarker of acute-on-chronic liver failure and prognosis in cirrhosis. J Hepatol 65:57–65. [DOI] [PubMed] [Google Scholar]
- Baffy G, Brunt EM, and Caldwell SH (2012). Hepatocellular carcinoma in non-alcoholic fatty liver disease: an emerging menace. J Hepatol 56:1384–1391. [DOI] [PubMed] [Google Scholar]
- Bahar Halpern K, Shenhav R, Matcovitch-Natan O, Toth B, Lemze D, Golan M, Massasa EE, Baydatch S, Landen S, Moor AE, et al. (2017). Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 542:352–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belayew A, and Tilghman SM (1982). Genetic analysis of α-fetoprotein synthesis in mice. Mol Cell Biol 2:1427–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bis JC, Kavousi M, Franceschini N, Isaacs A, Abecasis GR, Schminke U, Post WS, Smith AV, Cupples LA, Markus HS, et al. (2011). Meta-analysis of genome-wide association studies from the CHARGE consortium identifies common variants associated with carotid intima media thickness and plaque. Nat Genet 43:940–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenhorn EP, Duncan R, Huppi C, and Potter M (1988). Chromosomal location of the regulator of mouse α-fetoprotein, afr-1. Genetics 119:687–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruix J, Boix L, Sala M, and Llovet JM (2004). Focus on hepatocellular carcinoma. Cancer Cell 5:215–219. [DOI] [PubMed] [Google Scholar]
- Clinkenbeard EL, Butler JE, and Spear BT (2012). Pericentral activity of alpha-fetoprotein enhancer 3 and glutamine synthetase upstream enhancer in the adult liver are regulated by beta-catenin in mice. Hepatology 56:1892–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creasy KT, Jiang J, Ren H, Peterson ML, and Spear BT (2016). Zinc Fingers and Homeoboxes 2 (Zhx2) Regulates Sexually Dimorphic Cyp Gene Expression in the Adult Mouse Liver. Gene Expression 17:7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow HC, Kreibich AS, Kaercher KA, Sankoorikal GM, Pauley ED, Lohoff FW, Ferraro TN, Li H, and Brodkin ES (2011). Genetic dissection of intermale aggressive behavior in BALB/cJ and A/J mice. Genes, Brain, and Behavior 10:57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan R, Matthai R, Huppi K, Roderick T, and Potter M (1988). Genes that modify expression of major urinary proteins in mice. Mol Cell Biol 8:2705–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch J, Lees M, and Sloane Stanley GH (1957). A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem 226:497–509. [PubMed] [Google Scholar]
- Gargalovic PS, Erbilgin A, Kohannim O, Pagnon J, Wang X, Castellani L, Leboeuf R, Peterson ML, Spear BT, and Lusis AJ (2010). Quantitative trait locus mapping and identification of Zhx2 as a novel regulator of plasma lipid metabolism. Circ Cardiovasc Genet 3:60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L, Johnson MW, and Lusis AJ (1999). Quantitative trait locus analysis of plasma lipoprotein levels in an autoimmune mouse model : interactions between lipoprotein metabolism, autoimmune disease, and atherogenesis. Arterioscler Thromb Vasc Biol 19:442–453. [DOI] [PubMed] [Google Scholar]
- Hillebrandt S, Goos C, Matern S, and Lammert F (2002). Genome-wide analysis of hepatic fibrosis in inbred mice identifies the susceptibility locus Hfib1 on chromosome 15. Gastroenterology 123:2041–2051. [DOI] [PubMed] [Google Scholar]
- Jiang J, Creasy KT, Purnell J, Peterson ML, and Spear BT (2017). Zhx2 (zinc fingers and homeoboxes 2) regulates major urinary protein gene expression in the mouse liver. J Biol Chem 292:6765–6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawata H, Yamada K, Shou Z, Mizutani T, Yazawa T, Yoshino M, Sekiguchi T, Kajitani T, and Miyamoto K (2003). Zinc-fingers and homeoboxes (ZHX) 2, a novel member of the ZHX family, functions as a transcriptional repressor. Biochem J 373:747–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchgessner TG, LeBoeuf RC, Langner CA, Zollman S, Chang CH, Taylor BA, Schotz MC, Gordon JI, and Lusis AJ (1989). Genetic and developmental regulation of the lipoprotein lipase gene: loci both distal and proximal to the lipoprotein lipase structural gene control enzyme expression. J Biol Chem 264:1473–1482. [PubMed] [Google Scholar]
- Li C, Chen W, Jiang F, Simino J, Srinivasan SR, Berenson GS, and Mei H (2015). Genetic association and gene-smoking interaction study of carotid intima-media thickness at five GWAS-indicated genes: the Bogalusa Heart Study. Gene 562:226–231. [DOI] [PubMed] [Google Scholar]
- Livak KJ, and Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408. [DOI] [PubMed] [Google Scholar]
- Merkel M, Weinstock PH, Chajek-Shaul T, Radner H, Yin B, Breslow JL, and Goldberg IJ (1998). Lipoprotein lipase expression exclusively in liver. A mouse model for metabolism in the neonatal period and during cachexia. J Clin Invest 102:893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morford LA, Davis C, Jin L, Dobierzewska A, Peterson ML, and Spear BT (2007). The oncofetal gene glypican 3 is regulated in the postnatal liver by zinc fingers and homeoboxes 2 and in the regenerating liver by alpha-fetoprotein regulator 2. Hepatology 46:1541–1547. [DOI] [PubMed] [Google Scholar]
- Olsson M, Lindahl G, and Roushlahti E (1977). Genetic control of alpha-fetoprotein synthesis in the mouse. J Exp Med 145:819–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachnis V, Belayew A, and Tilghman SM (1984). Locus unlinked to α-fetoprotein under the control of the murine raf and Rif genes. Proc Natl Acad Sci USA 81:5523–5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perincheri S, Dingle RW, Peterson ML, and Spear BT (2005). Hereditary persistence of alpha-fetoprotein and H19 expression in liver of BALB/cJ mice is due to a retrovirus insertion in the Zhx2 gene. Proc Natl Acad Sci, USA 102:396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perincheri S, Peyton DK, Glenn M, Peterson ML, and Spear BT (2008). Characterization of the ETnII-alpha endogenous retroviral element in the BALB/cJ Zhx2 ( Afr1 ) allele. Mammalian Genome 19:26–31. [DOI] [PubMed] [Google Scholar]
- Peyton DK, Huang M-C, Giglia MA, Hughes NK, and Spear BT (2000). The alpha-fetoprotein promoter is the target of Afr1-mediated postnatal repression. Genomics 63, 173–180. [DOI] [PubMed] [Google Scholar]
- Potter M (1985). History of the BALB/c family. Curr Top Microbiol Immunol 122:1–5. [DOI] [PubMed] [Google Scholar]
- Shi Z, Wakil AE, and Rockey DC (1997). Strain-specific differences in mouse hepatic wound healing are mediated by divergent T helper cytokine responses. Proc Natl Acad Sci USA 94: 10663–10668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Tan S, Wu Z, Xu L, Wang Z, Xu Y, Wang T, Gao C, Gong Y, Liang X, et al. (2018). HBV suppresses ZHX2 expression to promote proliferation of HCC through miR-155 activation. Int J Cancer 143:3120–3130. [DOI] [PubMed] [Google Scholar]
- Spear BT, Jin L, Ramasamy S, and Dobierzewska A (2006). Transcriptional control in the mammalian liver: liver development, perinatal repression, and zonal gene regulation. Cell Mol Life Sci 63:2922–2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Gargalovic P, Wong J, Gu JL, Wu X, Qi H, Wen P, Xi L, Tan B, Gogliotti R, et al. (2004). Hyplip2, a new gene for combined hyperlipidemia and increased atherosclerosis. Arterioscler Thromb Vasc Biol 24:1928–1934. [DOI] [PubMed] [Google Scholar]
- Wu H, Wade M, Krall L, Grisham J, Xiong Y, and Van Dyke T (1996). Targeted in vivo expression of the cyclin-dependent kinase inhibitor p21 halts hepatocyte cell-cycle progression, postnatal liver development and regeneration. Genes and Development 10:245–260. [DOI] [PubMed] [Google Scholar]
- Yamada K, Kawata H, Matsuura K, Shou Z, Hirano S, Mizutani T, Yazawa T, Yoshino M, Sekiguchi T, Kajitani T, et al. (2002). Functional analysis and the molecular dissection of zinc-fingers and homeoboxes 1 (ZHX1). Biochem Biophys Res Comm 297:368–374. [DOI] [PubMed] [Google Scholar]
- Yamada K, Kawata H, Shou Z, Hirano S, Mizutani T, Yazawa T, Sekiguchi T, Yoshino M, Kajitani T, and Miyamoto K (2003). Analysis of zinc-fingers and homeoboxes (ZHX)-1-interacting proteins: molecular cloning and characterization of a member of the ZHX family, ZHX3. Biochem J 373:167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue X, Zhang Z, Liang X, Gao L, Zhang X, Zhao D, Liu X, Ma H, Guo M, Spear BT, et al. (2012). Zinc fingers and homeoboxes 2 inhibits hepatocellular carcinoma cell proliferation and represses expression of Cyclins A and E. Gastroenterology 142:1559–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhang Z, Li Y, Liao S, Wu X, Chang Q, and Liang B (2015). Cholesterol induces lipoprotein lipase expression in a tree shrew (Tupaia belangeri chinensis) model of non-alcoholic fatty liver disease. Sci Rep 5:15970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Jiang L, and Rui L (2009). Identification of MUP1 as a regulator for glucose and lipid metabolism in mice. J Biol Chem 284:11152–11159. [DOI] [PMC free article] [PubMed] [Google Scholar]