Abstract
In order to arise and progress, cancers need to evade immune elimination. Consequently, progressing tumors are often MHC-I low and express immune inhibitory molecules, such as PD-L1, which allows them to avoid the main anti-tumor host defense, CD8+ T cells. The molecular mechanisms that led to these alterations were incompletely understood. Here, we identify loss of the transcription factor IRF2 as a frequent underlying mechanism that leads to a tumor immune evasion phenotype in both humans and mice. We identified IRF2 in a CRISPR-based forward genetic screen for genes that controlled MHC-I antigen presentation in HeLa cells. We then found that many primary human cancers, including lung, colon, breast, prostate and others, frequently downregulated IRF2. Although generally known as a transcriptional repressor, we found that IRF2 was a transcriptional activator of many key components of the MHC I pathway, including immunoproteasomes, TAP and ERAP1, whose transcriptional control was previously poorly understood. Upon loss of IRF2, cytosol-to-ER peptide transport and N-terminal peptide trimming become rate limiting for antigen presentation. In addition, we found that IRF2 is a repressor of PD-L1. Thus, by downregulating a single non-essential gene, tumors become harder to see (reduced antigen presentation), more inhibitory (increased checkpoint inhibitor), and less susceptible to being killed by CD8+ T cells. Importantly, we found that the loss of antigen presentation caused by IRF2 downregulation could be reversed by interferon-stimulated induction of the transcription factor IRF1. The implication of these findings for tumor progression and immunotherapy are discussed.
Introduction
The importance of adaptive immunity in preventing cancer was revealed through studies in which immunodeficient animals, such as those lacking IFNγ, perforin, or RAG-2, were found to have a marked increase in spontaneous and mutagen-induced tumors (1–3). In addition, tumors derived from such immunodeficient animals grew when transplanted into other immunodeficient hosts but were rejected when placed into immunocompetent hosts (3–5), providing further evidence that the immune system recognized such tumors and could reject them. In contrast, many tumors arising in immunocompetent animals grew after being transplanted into immunocompetent hosts (3–5), thereby showing that cancers that arise and successfully progress in the face of the immune system have undergone immunoediting to escape from immune control. This immunoediting process is thought to be why many cancers express low levels of MHC-I and upregulate certain inhibitory molecules (6). The underlying molecular mechanisms responsible for these changes are poorly understood but have obvious potential impact on tumor progression and immunotherapy (7, 8).
The major histocompatibility complex class I (MHC-I) presentation pathway is critical for immune recognition and elimination of tumors by CD8+ T cells. In this process, a fraction of peptides that are generated by proteasomal degradation of cellular proteins are transported by the TAP transporter into the endoplasmic reticulum (ER), wherein they can be further trimmed by the aminopeptidase, ERAP1 (9–11). Subsequently, peptides of the correct length and sequence bind to MHC-I molecules and these complexes are then transported to the cell surface for display to CD8+ T cells. This allows activated CD8+ T cells to identify and kill cells that are presenting tumor-specific peptides (e.g., from mutant proteins) on their MHC-I (12).
We performed an unbiased, forward-genetic screen in human cervical carcinoma HeLa H1 cells to identify genes whose loss downregulated the MHC-I pathway. In this screen, the second strongest hit, second only to β2-microglobulin (the MHC-I light chain), was IRF2, an interferon regulatory transcription factor that had not been previously recognized to positively regulate this pathway. Here we show that under basal conditions, not only does IRF2 positively regulate the MHC-I pathway by transcriptionally activating genes necessary for peptide transport and processing, but it also transcriptionally represses the expression of PD-L1, an important immune checkpoint molecule. These results, together with our findings that many human cancers have downregulated IRF2 expression and that tumor cells lacking IRF2 are more difficult to kill, demonstrate that loss of IRF2 by cancers is a common immune evasion mechanism and this has obvious therapeutic implications.
Materials and Methods
Cells
DC3.2 is a J2 virus-immortalized dendritic cell line (13). A particular DC3.2 clone (with Renilla luciferase) was used for all experiments in this study as this clone has very strong cross-presentation and MHC class II presentation, as compared to other clones. RF33.70 is a T cell hybridoma that recognizes the ovalbumin (OVA) peptide OVA257–264 in the context of H2-Kb (14). MF2.2D9 is a T cell hybridoma that recognizes OVA258–276 in the context of I-Ab (13). RF33.70 and MF2.2D9 were transduced with lentivirus containing NFAT-luciferase. NIH-3T3 cells were stably transfected with the mouse H2-Kb molecule. A549 and MCF7 were kindly provided by Leslie Shaw (UMass), and the D53m and H50m mouse MCA-induced sarcoma lines were kindly provided by Robert Schreiber (Wash U, St. Louis). The MCA-induced sarcoma lines were grown in R10 media and all other cell lines were grown in RPMI 1640 (Gibco) supplemented with 10% FBS (Hyclone), 1% NEAA (Gibco), 1% HEPES (Gibco), 1% Antibiotic-Antimycotic (Gibco), and 5 × 10−5 M 2-ME (Sigma). The MCF7 growth media was also supplemented with 10μg/mL insulin. Antibiotic selection for CRISPR-Cas9 knockout cells was done for two weeks in media containing 5μg/mL blasticidin (Invivogen). All cells were grown in a 10% CO2 atmosphere at 37°C.
Plasmids
The LentiCRISPRv2 plus blasticidin selection plasmid (15, 16) was acquired from Addgene (83480) and, unmodified, is the same as the “no sgRNA” plasmid. The plasmids used to target mouse B2m, TAP1, TAP2, ERAP1, IRF2, IRF1 or to target human IRF2 were constructed by inserting the following sgRNA sequences, respectively, into the LentiCRISPRv2 plasmid: Mouse B2m: 5’-AGTATACTCACGCCACCCAC-3’; Mouse TAP1: 5’-ACTAATGGACTCGCACACGT-3’; Mouse TAP2: 5’-ATTACACGACCCGAATAGCG-3’; Mouse ERAP1: 5’-TGCAGCATCCAGAGCATAAT-3’; Mouse IRF2: 5’-TCCGAACGACCTTCCAAGAA-3’; Mouse IRF1: 5’-CTCATCCGCATTCGAGTGAT-3’; Human IRF2: 5’-TGCATGCGGCTAGACATGGG-3’. To insert these sgRNA sequences into LentiCRISPRv2, two primers were created for each sgRNA sequence: (1) a forward primer wherein CACCG preceded the aforementioned sgRNA sequence; and (2) a reverse primer where the reverse complement of the aforementioned sgRNA sequence was flanked by AAAC at the 5’ end and C at the 3’ end. Then, these 100μM primer sets were annealed and diluted 1:50. 3μg of LentiCRISPRv2 plasmid was digested for 3hrs at 55°C with BsmBI (NEB) and removal of the 2kb filler sequence was confirmed by gel electrophoresis. The larger molecular weight band was gel extracted and quick ligated with the diluted annealed oligos according to the manufacturer’s instructions (NEB). Stable competent E. coli (NEB) were then transformed with 2μL of the quick ligation product according to the manufacturer’s instructions and grown overnight at 30°C on LB+Ampicillin (100μg/mL) plates. Plasmids were isolated (Clontech) from individual colonies and sequenced (Genewiz) using the primer hU6-F: 5’-GAGGGCCTATTTCCCATGATT-3’ to confirm proper insertion of the sgRNA into LentiCRISPRv2. In addition, sgRNAs were checked for high indel efficiencies in transduced cells by TIDE analysis (17).
Rescue plasmids were constructed by inserting mouse IRF2, TAP2, or ERAP1 cDNA or human IRF2 cDNA into the constitutive expression vector, pCDH-CMV (Addgene), with a modified multiple cloning site. Overlapping PCRs were run using the primers below on either mouse cDNA from DC3.2 cells or human cDNA from HeLa H1 cells to create IRF2 cDNA sequences containing 6 synonymous mutations within the IRF2 sgRNA target site. The mouse IRF2 K78R sequence was constructed by further mutating A to G at nucleotide 233. The wild-type TAP2 and ERAP1 cDNA sequences were of C57BL/6 origin. All plasmids were sequenced to confirm correct sequences and reading frames. The primer sets for cloning the rescue/overexpression constructs were as follows: Human IRF2 AgeI forward: 5’-GACTACCGGTATGCCGGTGGAAAGGATGCGCATG-3’; human IRF2 sgRNA mut reverse: 5’-GCCGTGCCTCGCTGCGTGCATCCAGGGGATCTGAAAAATCTTCTTTTCCTTG-3’; human IRF2 sgRNA mut forward: 5’-GATGCACGCAGCGAGGCACGGCTGGGATGTGGAAAAAGATGCACCACTCTTTAGAAA-3’; human IRF2 MluI reverse: 5’-GATCACGCGTTTAACAGCTCTTGACGCGGGCCTGG-3’; mouse IRF2 AgeI forward: 5’-GATCACCGGTATGCCGGTGGAACGGATGCGAATG-3’; mouse IRF2 sgRNA mut reverse: 5’-CCTTTTTTCGAGGGGCGCTCTGATAAGGGCAGCATCCGGTAGACTCTGAAGGCG-3’; mouse IRF2 sgRNA mut forward: 5’- CTTATCAGAGCGCCCCTCGAAAAAAGGAAAGAAACCAAAGACAGAAAAAGAAGAGAG-3’; and mouse IRF2 MluI reverse: 5’-GATCACGCGTTTAACAGCTCTTGACACGGGCCTGG-3’.
Cell surface staining
Where indicated, mouse cells were blocked with 2.4G2 and stained for surface MHC class I levels with anti-Kb-APC (eBioscience, AF6–88.5.5.3), MHC class II levels with anti-IA/IE-PECy7 (BioLegend, M5/114.15.2), PD-L1 levels with anti-PD-L1-PE (BioLegend, 10F.9G2), or with isotype controls (eBioscience mouse IgG2a-APC eBM2a, eBioscience rat IgG2b κ-PE eB149) at 1:200 dilutions. Where indicated, human cells were stained for surface MHC class I levels with W6/32. W6/32 staining was performed either by two-step labeling with W6/32 hybridoma supernatant followed by 1:500 donkey-anti-mouse Alexa 647 (Life Technologies) or by one-step labeling with 1:200 FITC-conjugated W6/32 (eBioscience). Where indicated, human cells were stained for surface PD-L1 levels with 1:200 rabbit anti-PD-L1 (Abcam, 28–8), followed by 1:500 donkey-anti-rabbit Alexa 647 (Life Technologies). Normalized MFI was computed by dividing the geometric MFI of each knockout cell line by the geometric MFI of the WT (no sgRNA) cell line.
T cell hybridoma Ag presentation
Cross-presentation and MHC class II presentation were measured by co-culturing DC3.2 lines in the presence of the indicated concentrations of OVA-coated iron-oxide beads (Polysciences) and RF33.70-Luc CD8+ T cells or MF2.2D9-Luc CD4+ T cells, respectively, for 16–18 hours. Then, One-Glo luciferase substrate (Promega) was added and luciferase activity quantified by an EnVision plate reader (Perkin Elmer). Rescue experiments were performed by adding the OVA-beads and RF33.70-Luc cells 48hrs post-transduction of the DC3.2 lines. Data from a representative cross-presentation and MHC class II presentation experiment (of N=4) are shown where points represent mean ± SD of technical duplicates. Normalized CD8+ T cell activation was calculated for rescue experiments by dividing the CD8+ T cell activation (RLU of luciferase) at each point by the CD8+ T cell activation of the DC3.2 no sgRNA line transduced with EV (WT+EV).
siRNA transfections
104 HEK293T or HeLa H1 cells were transfected with 10nM Silencer Select siRNA (Invitrogen) and 0.3μL Lipofectamine RNAiMAX (Invitrogen) per well in flat-bottom 96-well plates. Individual siRNAs used were negative control #1 (4390843) human β2m (s1854), human TAP1 (s13778), human IRF2 #1 (s7506), and human IRF2 #2 (s7504). After 72 hours, adherent cells were trypsinized, washed in PBS supplemented with 2% FBS, and stained with the surface MHC I pan-HLA-A/B/C antibody, W6/32. Normalized MFI was computed by dividing the geometric MFI of each experimental siRNA by the geometric MFI of the negative control siRNA.
Minigene transfections
104 3T3-Kb cells were transfected with 100ng of various pTracer-CMV2 plasmids (Invitrogen) containing SIINFEKL precursors (18) and 0.4uL Lipofectamine 2000 (Invitrogen) in flat-bottom 96-well plates. After 72 hours, cells were stained with 25-D1.16 (specific for the combination of SIINFEKL and H-2Kb) (19), followed by donkey-anti-mouse Alexa 647 (Life Technologies) and analyzed by flow cytometry. Transfected cells were identified by GFP expression and the MFI of 25-D1.16 staining was measured on the gated transfected cells. Transfection efficiency, based on GFP expression, ranged from 5–20%, depending on the vector. The normalized MFI for each experiment (with technical duplicates) was calculated by dividing the MFI of each knockout line by the MFI of the wild-type (“no sgRNA”) line.
RNA-Seq
RNA was extracted using the RNeasy kit (Qiagen) after 2 hours of stimulating the DC3.2 lines with 5,000U/mL mouse IFNα, 2ng/mL mouse IFNγ, or media alone. A standard library preparation protocol was used with 50ng of total RNA as starting material. Libraries were checked for appropriate fragment size traces by Bioanalyzer (Agilent) and concentrations were determined to achieve similar sequencing depth per library. Libraries were run on NextSeq 500/550 high-output and mid-output kits (Illumina) and all libraries had at least 107 reads with single index paired-end sequencing. Trimmomatic-0.32 (20) was used to remove 5’ or 3’ stretches of bases having an average quality of less than 20 in a window size of 10. Only reads longer than 36 bases were kept for further analysis. RSEM v1.2.28 (21) was used to estimate gene expression, with parameters -p 4 --bowtie-e 70 --bowtie-chunkmbs 100 --strand-specific. Gene quantification was run on the transcriptome (RefSeq v69 downloaded from UCSC Table Browser (22). Genes with more than 15 TPM in any time point were considered expressed, and genes that did not achieve this threshold were removed from further analysis. Batch effects were observed between samples from different replicates. We used the log transformed TPM normalized expression values as input to ComBat (package sva version 3.18.0) (23, 24) with default parameters and a model that specified different replicates as batches. Corrected TPM values were transformed back to read counts using the expected size of each transcript informed by RSEM. We only considered genes with at least 15 TPMs in at least one replicate at any time point. The expressed gene list was filtered to include only genes with homologs as defined by the previous step. We used the batch corrected counts per gene to identify differentially expressed genes by at least 2 fold between unstimulated cells (time 0) and 2 hours following stimulation with IFNα or IFNγ and whose change in expression was significant (p-adjusted < 0.05) according to the package DESeq2 (v1.10.1) (25) in R (v3.5.1). Due to the large transcriptional changes observed in this system, we turned off the fold change shrinkage in DESeq2 with betaPrior=FALSE and we added a pseudocount of 32 to all timepoints to avoid spurious large fold change estimates from lowly abundant genes. RNA-Seq data has been deposited to GEO (accession number GSE133089; www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE133089).
Human lung specimens
27 non-small cell lung cancers were acquired from UMass Pathology archive of formalin-fixed and paraffin-embedded patient samples. PD-L1 expression was determined at the time of diagnosis by immunohistochemistry (22C3 pharmDx assay; Agilent) by a surgical pathologist. RNA was extracted from the archival material and analyzed by RT-PCR (see below).
RT-PCR
RNA was extracted from cell lines using the RNeasy kit (Qiagen) or from formalin-fixed paraffin-embedded non-small cell lung cancers using the RNeasy FFPE kit (Qiagen) and reverse transcribed to cDNA using EcoDry pre-mix random hexamers (Clontech). 50ng cDNA were used per well (done in triplicate) with indicated TaqMan probes (Applied Biosystems), according to the manufacturer’s instructions. The TaqMan probes used are as follows: mouse β-actin - 4352933, mouse β2m - Mm00437762_m1, mouse H2-K1 - Mm01612247_m1, mouse H2-Ab1 - Mm00439216_m1, mouse Tap1 - Mm00443188_m1, mouse Tap2 - Mm01277033_m1, mouse Erap1 - Mm00472842_m1, mouse ERp57 - Mm00433130_m1, mouse tapasin - Mm00493417_m1, mouse calnexin - Mm00500330_m1, mouse calreticulin - Mm00482936_m1, mouse Tapbpr - Mm00520408_m1, mouse Irap - Mm00555903_m1, mouse Psme1- Mm00650858_g1, mouse Psme2 - Mm01702833_g1, mouse Psmb8 - Mm00440207_m1, mouse Psmb9 - Mm00479004_m1, mouse Psmb10 - Mm00479052_g1, mouse PD-L1 - Mm03048248_m1, human GAPDH - Hs02758991_g1, human IRF2 - Hs01082884_m1, human TAP2 - Hs00241060_m1, and human ERAP1 - Hs00429970_m1. mRNA expression levels in the IRF2-knockout DC3.2 were compared to those in the wild-type DC3.2 by first normalizing to the mRNA expression of β-actin (mouse) in each sample (2^-ΔΔCt). Statistical analysis was done by comparing the expression of a given gene to that of H2-Ab1. Relative mRNA expression levels in each human lung tumor were measured by first normalizing to the mRNA expression of GAPDH (human) in each specimen (2^-ΔCt). Results are displayed after further normalization to one of the tumors. Statistical analysis was done using two-tailed Mann-Whitney U-tests and linear regression models with R2 for goodness of fit.
Chromatin immunoprecipitations
ChIP procedure generally followed the Thermo Fisher ChIP protocol. In short, 107 DC3.2-no sgRNA cells were stimulated for 2hrs with 2ng/mL IFNγ or media alone, harvested and fixed with 1% formaldehyde, quenched with glycine, and washed in cell lysis buffer then nuclear lysis buffer. Chromatin was sheared by sonication for 20min and fragment size (~300bp) was determined by gel electrophoresis. Immunoprecipitations of the sheared chromatin were done using 2μg of primary antibody – normal rabbit IgG (Santa Cruz sc-2027), rabbit anti-IRF1 (Abcam ab186384), or rabbit anti-IRF2 (Invitrogen B-80 H53L46) – by incubating for 1hr at RT then overnight at 4°C. The next day, 25uL of pre-washed Protein A/G beads (Pierce) were added to each of the samples and incubated at RT for 30min then 90min at 4°C. After washing sequentially with low-salt buffer, high-salt buffer, LiCl buffer, and TE buffer, the DNA was eluted from the beads. All immunoprecipitated samples were treated with RNase A (Qiagen) and Proteinase K (Qiagen) and then column purified (Clontech). ChIP-qPCR was performed in triplicate wells using SYBR Green (Bio Rad) and unique primer sets (below) flanking the IRF1/2-binding site within the gene’s promoter (26–28), according to the manufacturer’s instructions. Data shown as fold enrichment (2^-ΔCt) over the normal rabbit IgG control IP (N=2). The primer sets for ChIP-qPCR were as follows: mouse TAP2 forward: 5’-CAAATTGACAGGCGCCATCT-3’; mouse TAP2 reverse: 5’-GCTTCTTCTCAAACTGGATCTCC-3’; mouse ERAP1 forward: 5’-CTTAGGCTTGCTCTCTTTTAGCG-3’; mouse ERAP1 reverse: 5’-GACTCCTGCTCCCGATCCTC-3’; mouse PD-L1 forward: 5’-CAAGAAAGCTAATGCAGGTTTCAC-3’; and mouse PD-L1 reverse: 5’-CCTGCGGATGACTTTAGAGTC-3’.
Western blotting
Whole cell lysates were prepared in RIPA buffer with protease inhibitor (Pierce), protein concentrations were determined by BCA assay (Pierce), and 10μg of denatured samples were run on 10% reducing gels (Genscript). After transfer, PVDF membranes (Millipore) were blocked with TBS-Tween 1x + 5% milk and then blotted with rabbit anti-IRF2 (Abcam ab124744) or rabbit anti-IRF1 (Abcam ab186384) in TBS-Tween 1x + 2% milk overnight at 4°C. The following day, membranes were washed 3x with TBS-Tween 1x, goat-anti-rabbit HRP (Millipore) was added for 1hr at RT, membranes were washed 3x, and HRP substrate (Millipore) was added. Following exposure, membranes were stripped (Millipore), blocked, and re-blotted with mouse anti-β-actin (Santa Cruz sc-47778) in TBS-Tween 1x + 2% milk overnight at 4°C. The following day, membranes were prepared as above except anti-mouse HRP (Pierce) was used instead.
In vitro cytotoxicity assays
OT-I CD8+ T cells were pre-activated by co-culturing them for 4–5 days with irradiated, SIINFEKL-pulsed wild-type LPS-stimulated B cell blasts in T cell media containing 30ng/mL IL-2. After this time, OT-I were checked for CD8 expression and upregulation of CD27 and CD44. WT or IRF2-null RMA cells were counted, sub-divided into respective tubes, SIINFEKL-pulsed at the indicated concentrations or kept un-pulsed, and CFSE-labeled as high (1μM) for cells pulsed with SIINFEKL and CFSE-labeled as low (0.1μM) for un-pulsed cells. Pulsed and un-pulsed cells were re-counted, mixed 1:1, and 105 cells total were plated per well in U-bottom 96-well plates. 5 × 104 OT-I were added to the respective wells and incubated at 37°C for 4hrs. Live RMA cells were gated by flow cytometry and CFSE levels analyzed. Specific killing was determined for each RMA line by calculating 100 * (1 – [(% Targets / % Bystanders) / (% Control Targets / % Control Bystanders)]).
Results
Regulation of MHC-I presentation by IRF2
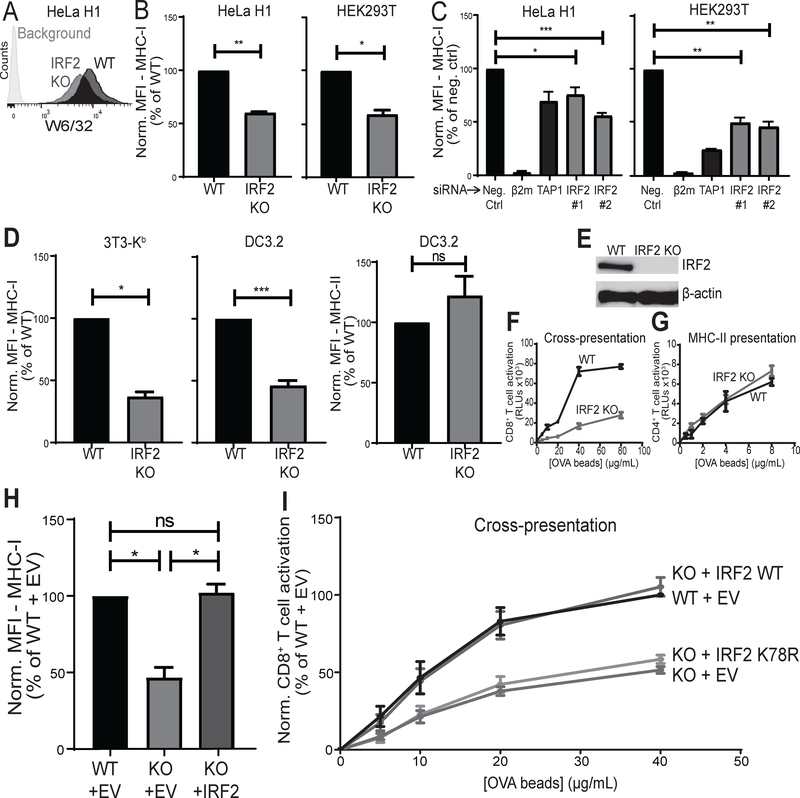
When HeLa H1 cervical carcinoma cells were immunoselected for MHC-I low variants in a genome-wide CRISPR-Cas9 screen, IRF2 scored as the second most targeted gene, with six out of six independent IRF2 guide RNAs hitting. To validate this hit, we tested whether IRF2 affects surface MHC-I levels in human cells by knocking out IRF2. HeLa H1 cells and HEK293T kidney cells were transduced with vectors expressing either Cas9 and a sgRNA targeting human IRF2 or Cas9 alone and surface MHC-I levels were checked by flow cytometry (Fig. 1A). The IRF2-knockout HeLa H1 and HEK293T had significantly lower surface MHC-I levels than their wild-type controls (Fig. 1B). To further validate this finding with another technique, we silenced IRF2 expression with two independent siRNAs and found that both siRNAs similarly decreased surface MHC-I levels in HeLa H1 and HEK293T (Fig. 1C), confirming the phenotype. The magnitude of the decrease in MHC-I was similar to that observed when the expression of the TAP transporter was silenced (Fig. 1C). To determine whether IRF2 also affects surface MHC-I levels in mouse cells, we transduced NIH-3T3 fibroblasts stably transfected with H2-Kb (3T3-Kb) and DC3.2 dendritic cells (13) with vectors expressing either Cas9 and a sgRNA targeting mouse IRF2 or Cas9 alone. The IRF2-knockout (IRF2-KO) mouse fibroblasts and DC cells also had significantly reduced surface MHC-I levels (Fig. 1D). Disruption of the IRF2 gene was validated by TIDE analysis (17) and loss of IRF2 expression was confirmed by western blot (Fig. 1E). DC3.2 cells also express MHC-II molecules and we found no change in surface MHC-II levels in the IRF2-KO cells (Fig. 1D), which demonstrates that IRF2 is selectively affecting the MHC-I pathway; further evidence supporting this conclusion will be described below.
Figure 1. IRF2 positively regulates MHC-I presentation under basal conditions.
(A) Representative histograms of surface MHC I levels by W6/32 staining in HeLa H1 lines stably transduced with the LentiCRISPRv2 constructs; “IRF2 KO” (IRF2 sgRNA) or “WT” control (no sgRNA). Background is with secondary antibody staining only; (B) Normalized MFI of surface MHC I levels on HeLa H1 (left) or HEK293T (right) stable knockout lines; (C) normalized MFI of surface MHC I levels on HeLa H1 (left) or HEK293T (right) cells after 72hrs silencing with 10nM indicated siRNA; (D) Normalized MFI of surface MHC I levels by AF6 staining on 3T3-Kb stable knockout lines (left), surface MHC I levels on DC3.2 stable knockout lines (middle), or surface MHC II levels on DC3.2 stable knockout lines (right); (E) Representative western blot in DC3.2 lines “IRF2 KO” (IRF2 sgRNA) or “WT” control (no sgRNA) for protein expression of IRF2 or β-actin as control; (F) Representative cross-presentation experiment of OVA-beads by DC3.2 lines to RF33.70-Luc CD8+ T cell hybridoma; (G) Representative MHC-II presentation experiment of OVA-beads by DC3.2 lines to MF2.2D9-Luc CD4+ T cell hybridoma; (H) Normalized MFI of surface MHC I levels on DC3.2 lines after transducing with pCDH expressing empty vector (EV) or wild-type IRF2 containing six synonymous mutations within the IRF2 sgRNA target site (IRF2); (I) Cross-presentation after transducing the DC3.2 lines with pCDH expressing empty vector (EV), wild-type IRF2 containing six synonymous mutations within the IRF2 sgRNA target site (IRF2 WT), or mutant IRF2 (IRF2 K78R) which also contains the same six synonymous mutations as IRF2 WT. (B, C, D, H, I) Bars represent mean + SEM (N≥3). Statistical analysis by two-tailed ratio paired t-tests; (F, G) Points represent mean ± SD of technical duplicates. *p<0.05, **p<0.01, ***p<0.001, ns=not significant.
To investigate the functional consequence of this reduction in MHC-I levels, we evaluated the importance of IRF2 for MHC-I cross-presentation (i.e., the presentation of peptides derived from exogenous antigen on MHC-I). The IRF2-KO DCs cross-presented more poorly than their wild-type controls (Fig. 1F), demonstrating that IRF2 positively regulates MHC-I antigen presentation. In the same experiments, MHC-II presentation was unaffected, again showing selectivity in IRF2 effects (Fig. 1G). Lastly, to confirm that IRF2 is responsible for these differences, we overexpressed IRF2 in the IRF2-KO DC3.2 line and found that it completely restored surface MHC-I levels (Fig. 1H) and the ability of these cells to cross-present antigen (Fig. 1I). Evidence that loss of IRF2 also compromises the MHC-I presentation of endogenous cellular antigens will be described below.
To determine whether IRF2 was exerting its function as a transcription factor, we introduced a lysine to arginine point mutation at position 78 which prevents acetylation at this site and thereby prevents IRF2 from binding its DNA target sequences (29). Overexpressing this mutant IRF2 did not restore function in the IRF2-KO DC3.2 cells (Fig. 1I). Therefore, IRF2 is necessary for optimal transcriptional regulation of MHC-I antigen presentation pathways.
Transcriptional control of MHC-I pathway components by IRF2
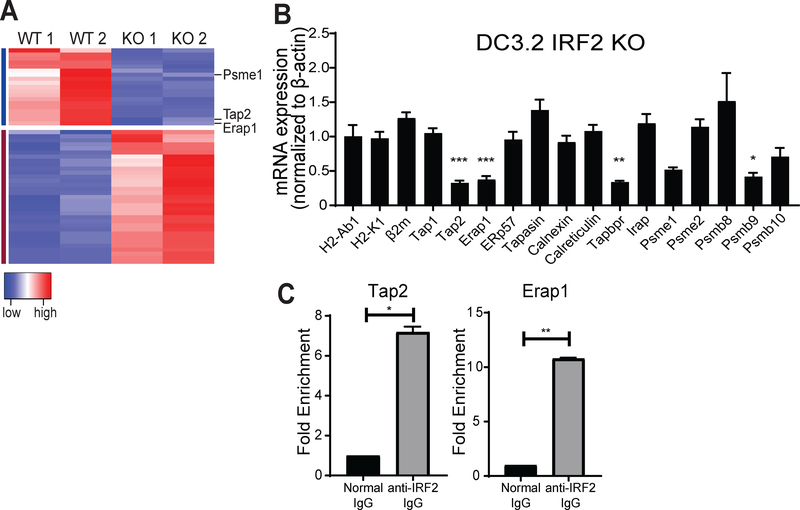
Since IRF2 is functioning as a transcription factor, we next sought to determine what genes are regulated by IRF2 and could be responsible for producing the low MHC-I phenotype observed in the IRF2-KO cells. RNA-seq was performed on the wild-type and IRF2-KO DC3.2 lines (Fig. 2A, Table S1). Surprisingly, relatively few genes were differentially expressed by >2-fold (19 decreased and 33 increased). Of these 52 differentially expressed genes, we identified TAP2, ERAP1, and the immunoproteasome subunit PSME1 as potential contributors to the decreased MHC-I levels. To further confirm this result, we performed qPCR in these cell lines to check expression of all MHC-I pathway genes (Fig. 2B), which showed that the mRNA levels of TAP2, ERAP1, TAPBPR, and PSME9 (another immunoproteasome subunit) were significantly downregulated in the IRF2-KO DC3.2; PSME1 levels were reduced but this decrease did not achieve statistical significance. Interestingly, the mRNA levels of the MHC heavy chain (H2-K1) and MHC light chain (Beta2-microglobulin; β2m) were unaffected (Fig. 2B), indicating that IRF2 is not required for synthesis of the MHC-I heterodimer. ChIP-qPCR experiments confirmed that IRF2 regulates TAP2 and ERAP1 mRNA expression by directly binding to their promoters (Fig. 2C). To test whether functional IRF2 is needed for TAP2 and ERAP1 mRNA expression, we transduced wild-type IRF2 or the IRF2-K78R mutant in the DC3.2 IRF2-KO cells and found that the TAP2 and ERAP1 mRNA levels were increased in the cells expressing wild-type but not mutant IRF2 (Fig. S1).
Figure 2. IRF2-mediated transcriptional regulation of MHC-I pathway under basal conditions.
(A) Heatmap of genes (52) differentially expressed by >2-fold between the DC3.2 no sgRNA (“WT”) and IRF2 sgRNA (“KO”) lines. Columns represent independent duplicate RNA-seq runs in these two lines. Red → blue indicates high → low expression for a given row (gene). For clarity, only three of the downregulated genes in the KO line (Psme1, Tap2, and Erap1) are shown, and all are listed in Table S1; (B) mRNA expression levels by qPCR in DC3.2 IRF2 KO relative to those in DC3.2 WT; normalized to the mRNA expression of mouse β-actin in each sample (2^-ΔΔCt). Values >1 indicate higher expression in DC3.2 IRF2 KO and values <1 indicate lower expression in the DC3.2 IRF2 KO. Bars represent mean + SEM mRNA expression (N≥3). Statistical analysis by two-tailed unpaired t-tests by comparing the expression of a given gene to that of H2-Ab1 (control); (C) ChIP-qPCR in DC3.2 WT for Tap2 (left) or Erap1 (right) DNA with rabbit anti-IRF2 IgG or normal rabbit IgG (control). Bars represent mean + SEM fold enrichment (2^-ΔCt) over the normal rabbit IgG control IP (N=2). Statistical analysis by two-tailed ratio paired t-tests. *p<0.05, **p<0.01, ***p<0.001, ns=not significant.
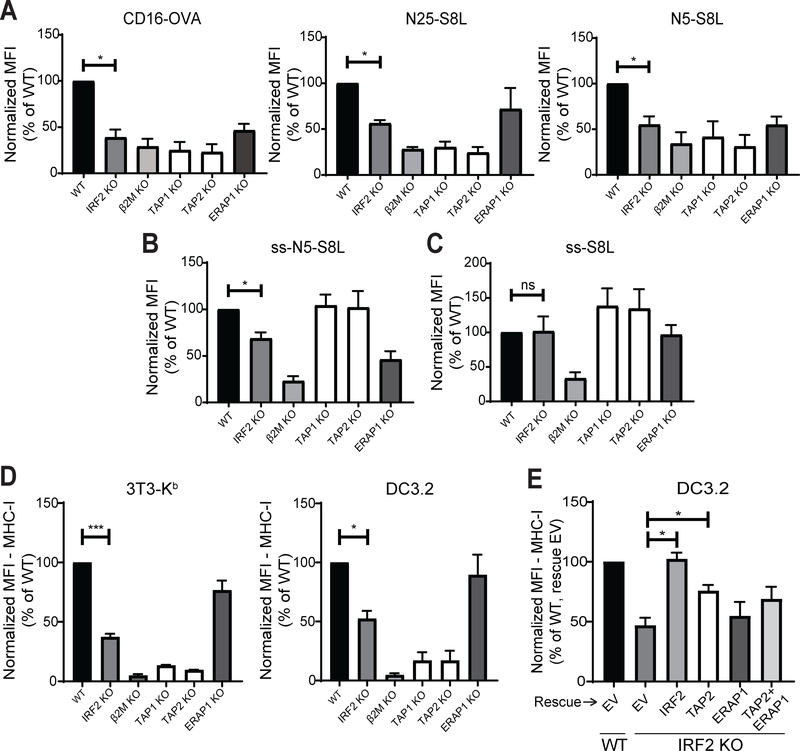
To more closely examine the functional effects of IRF2 on peptide transport and trimming, 3T3-Kb IRF2-KO cells were transfected with various pTracer plasmids, each of which contained a GFP reporter and a minigene encoding a peptide that could be processed down to the mature epitope (i.e., SIINFEKL/S8L). The cells were surface stained with the antibody 25-D1.16, which recognizes H2-Kb-S8L complexes (19), and transfected cells (GFP-positive) were analyzed. In each case, β2m-KO cells were included to show MHC I-dependence and the maximum inhibition of S8L presentation, and TAP1-, TAP2-, and ERAP1-KO cells were included as positive or negative controls, depending on the minigene examined. IRF2-KO cells presented fewer H2-Kb-S8L complexes than wild-type cells when given the TAP-dependent, ERAP1-dependent antigens CD16-OVA (full-length ovalbumin protein) (30), N25-S8L (a S8L precursor extended by 25 amino acids on the N-terminus), or N5-S8L (a S8L precursor extended by 5 amino acids on the N-terminus) (Fig. 3A). We also tested the presentation of a version of the precursor peptide with 5 extra N-terminal residues that was targeted into the ER by a co-linear signal sequence (ss-N5-S8L). Since the signal sequence allows this peptide to enter the ER through SEC61 instead of TAP, its presentation is TAP-independent but still dependent on ERAP1 to remove the extra N-terminal residues (Fig. 3B). IRF2-KO cells also presented fewer H2-Kb-S8L complexes than wild-type cells when transfected with ss-N5-S8L (Fig 3B). However, IRF2-deficient cells were equally capable of presenting H2-Kb-S8L complexes when given a TAP-independent, ERAP1-independent peptide (S8L with no extra N-terminal residues that was targeted into the ER via a co-linear signal sequence; ss-S8L) (Fig. 3C), demonstrating that while IRF2 affects the transport and processing of MHC-I epitopes, it does not affect the ability of such peptides to be loaded onto MHC-I.
Figure 3. IRF2 affects antigen transport and processing.
(A-C) H-2Kb presentation of SIINFEKL derived from (A) the TAP-dependent, ERAP1-dependent antigens CD16-OVA (left), N25-S8L (middle), or N5-S8L (right); (B) the TAP-independent, ERAP1-dependent antigen ss-N5-S8L; and (C) the TAP-independent, ERAP1-independent antigen ss-S8L on 3T3-Kb knockout lines. 25-D1.16 staining analyzed on transfected (GFP+) cells; (D) Normalized MFI of surface MHC I levels on 3T3-Kb (left) or DC3.2 (right) knockout lines; (E) Normalized MFI of surface MHC I levels on DC3.2 lines 48hrs after transduction with pCDH expression vectors containing empty vector (EV), IRF2, TAP2, ERAP1, or dual transduction of TAP2 and ERAP1. (A-E) Bars represent the mean + SEM of the normalized MFI from independent experiments (N≥3). Statistical analysis by two-tailed ratio paired t-tests. *p<0.05, ***p<0.001, ns=not significant.
We also compared the MHC-I phenotype in IRF2-knockout 3T3-Kb to that observed in the same cells with knockouts of TAP2 or ERAP1. The magnitude of the reduction in MHC-I levels on IRF2-KO cells was in between that of the TAP2- and ERAP1-knockout cells (Fig. 3D), which is consistent with our findings that IRF2 positively regulates TAP2 and ERAP1 but their expression is not entirely lost in IRF2-KO cells. Lastly, we performed rescue experiments wherein we overexpressed TAP2 and/or ERAP1 in the IRF2-knockout DC3.2 and checked surface MHC-I levels two days after transduction (Fig. 3E). Although the double-rescue partially restored MHC-I levels, it was not complete, suggesting that other genes regulated by IRF2 also contribute to surface MHC-I expression.
IRF2 and PD-L1
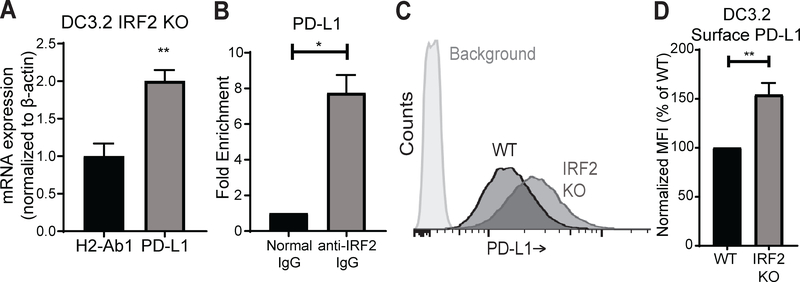
One of the upregulated genes in the IRF2-KO cells was Cd274 (Fig. S1), also known as programmed death-ligand 1 (PD-L1), which is often upregulated in certain cancers (e.g., non-small cell lung cancer) and functions as a checkpoint inhibitor to suppress antigen-specific CD8+ T cell effector function (31, 32). To validate our RNA-seq finding with an independent technique, we performed qPCR on the IRF2-KO and wild-type DC3.2 lines and found that the IRF2-KO cells expressed approximately twice as much PD-L1 mRNA as the wild-type controls (Fig. 4A). Additionally, ChIP-qPCR revealed that IRF2 regulates PD-L1 mRNA expression by directly binding the PD-L1 promoter (Fig. 4B). Therefore, IRF2 acts as a transcriptional repressor of PD-L1 in these cells. To evaluate the extent to which a roughly 2-fold increase in PD-L1 mRNA translates to surface PD-L1 expression, we analyzed these cells by flow cytometry (Fig. 4C). This analysis reveals that the surface PD-L1 levels also increased by roughly 50% in the IRF2-KO cells (Fig. 4D). Taken together, these results demonstrate that IRF2 plays a role in repressing PD-L1 expression under basal conditions.
Figure 4. IRF2 represses PD-L1 expression under basal conditions.
(A) H2-Ab1 and PD-L1 mRNA expression levels by qPCR in DC3.2 IRF2 KO relative to those in DC3.2 WT; normalized to the mRNA expression of mouse β-actin in each sample (2^-ΔΔCt). Values >1 indicate higher expression in DC3.2 IRF2-KO. Bars represent mean + SEM mRNA expression (N=3). Statistical analysis by two-tailed unpaired t-test by comparing the expression of PD-L1 to that of H2-Ab1 (control); (B) ChIP-qPCR of DC3.2 WT for PD-L1 DNA with rabbit anti-IRF2 IgG or normal rabbit IgG (control); bars represent mean + SEM fold enrichment (2^-ΔCt) over the normal rabbit IgG control IP (N=2). Statistical analysis by two-tailed ratio paired t-test; (C) Representative histograms of surface PD-L1 levels by 10F.9G2 staining in DC3.2 lines stably transduced with the LentiCRISPRv2 constructs; “IRF2 KO” (IRF2 sgRNA) or “WT” control (no sgRNA). Background = isotype control staining; (D) Normalized MFI of surface PD-L1 levels on DC3.2 lines; bars represent mean + SEM (N=6). Statistical analysis by two-tailed ratio paired t-test. *p<0.05, **p<0.01.
Contributions of IRF1 vs. IRF2
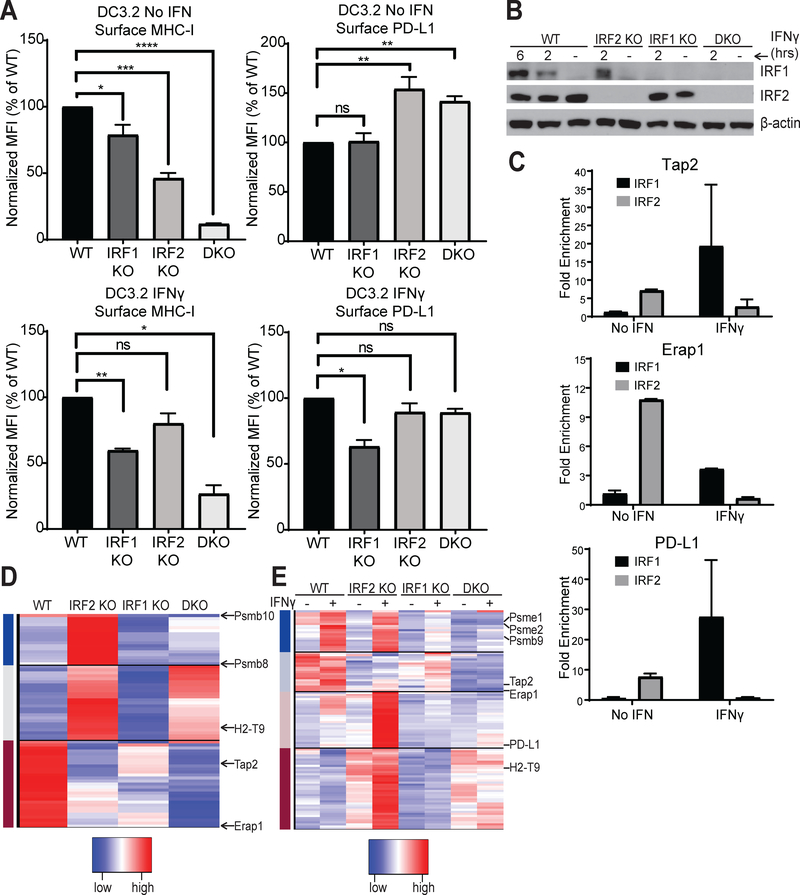
Because IRF2 is an interferon regulatory transcription factor, we wanted to see how interferon induction would affect IRF2’s regulation of the MHC-I pathway and PD-L1 expression. Interestingly, stimulation with either IFNγ or IFNα restored the surface MHC-I expression in the IRF2-KO DC3.2 (Fig. 5A, Fig. S2). This is an important finding because it indicates that impairment of the MHC-I pathway from loss of IRF2 is reversible.
Figure 5. Contributions of IRF1 vs. IRF2.
(A) Normalized MFI of surface MHC I and PD-L1 levels on the indicated single or double (IRF1+IRF2, DKO) DC3.2 knockout lines after overnight incubation with either media alone or 2ng/mL IFNγ; bars represent mean + SEM (N≥3); (B) Western blot for IRF1 and IRF2 (and β-actin as loading control) in the DC3.2 knockout lines in the absence or presence of 2ng/mL IFNγ for the durations indicated; (C) ChIP-qPCR of DC3.2 WT cells stimulated with ± 2ng/mL IFNγ for two hours for Tap2 (top), Erap1 (middle), or PD-L1 (bottom) DNA with rabbit anti-IRF2 IgG, rabbit anti-IRF1 IgG, or normal rabbit IgG (control). Bars represent mean + SEM fold enrichment (2^-ΔCt) over the normal rabbit IgG control IP (N=2); (D, E) Heatmap of genes differentially expressed between the DC knockout lines (D) at baseline or (E) after stimulation with ± 2ng/mL IFNγ for 2hrs. For clarity, only a few genes are shown, and all are listed in Table S1. Statistical analysis by two-tailed ratio paired t-tests. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, ns=not significant.
IRF1 and IRF2 recognize the same IFN-stimulated response element (ISRE) (33–35) and, whereas IRF2 is constitutively expressed and minimally affected by interferon induction, IRF1 is significantly upregulated in response to interferon (Fig. 5B, Fig. S2) (36–38). Such upregulation of IRF1 causes IRF1 to compete with IRF2 for binding to the TAP2, ERAP1, and PD-L1 promoters, ultimately displacing it from them (Fig. 5C). Consistent with the literature that IRF1 positively regulates both MHC-I and PD-L1 expression under IFN-stimulated conditions (39–41), we found that knocking out IRF1 decreased both surface MHC-I and PD-L1 levels after IFNγ stimulation (Fig. 5A). Interestingly, when we examined the dual effects of IRF1 and IRF2 on surface MHC-I and PD-L1 levels using single or double knockout DC3.2 lines (Fig. 5A, Fig. S2), we found that: (1) IRF1/2 double knockouts (DKO) have a larger reduction in surface MHC-I than is observed in either single knockout; and (2) IRF2-knockouts have a larger effect than the IRF1-knockouts on both MHC-I and PD-L1 expression under basal conditions. This suggested that, although these two IRFs recognize the same ISRE, the subset of genes they each primarily regulate differs and that a cell’s dependence on IRF2 vs. IRF1 for any given gene may vary depending on the cues (e.g., interferon) which that cell receives from its environment. To better characterize these expression changes more globally, we performed RNA-seq on the single and double knockout DC3.2 lines under basal conditions (Fig. 5D) or after adding IFNγ (Fig. 5E) or IFNα (Fig. S2). From this analysis, several genes known to be important for antigen presentation and immune cell function were identified (Table S1). Furthermore, this analysis showed that genes influenced by IRF1 and IRF2 could be grouped into multiple classes. Under basal conditions, there was a subset of antigen presentation-related genes whose expression was activated by IRF2 (e.g., TAP2, ERAP1), others that were repressed by IRF2 and remained so in the double knockouts (e.g., H2-T9), and yet others that were repressed by IRF2 but did not remain so in the double knockouts (e.g., PSMB8, PSMB10) (Fig. 5D). After stimulating with IFNγ, some genes were primarily activated by IRF1 (e.g., PSME1, PSME2, PSMB9), others were primarily repressed by IRF2 (e.g., PD-L1), and yet others were activated by both IRF1 and IRF2 (e.g., TAP2, ERAP1) (Fig. 5E). Collectively, these studies demonstrate that while some genes are acted on antagonistically by IRF1 and IRF2, other genes are regulated synergistically by these two transcription factors and that the relative contributions of IRF1 vs. IRF2 in mediating these expression changes varies depending on the inflammatory state of the cell.
IRF2 in cancer
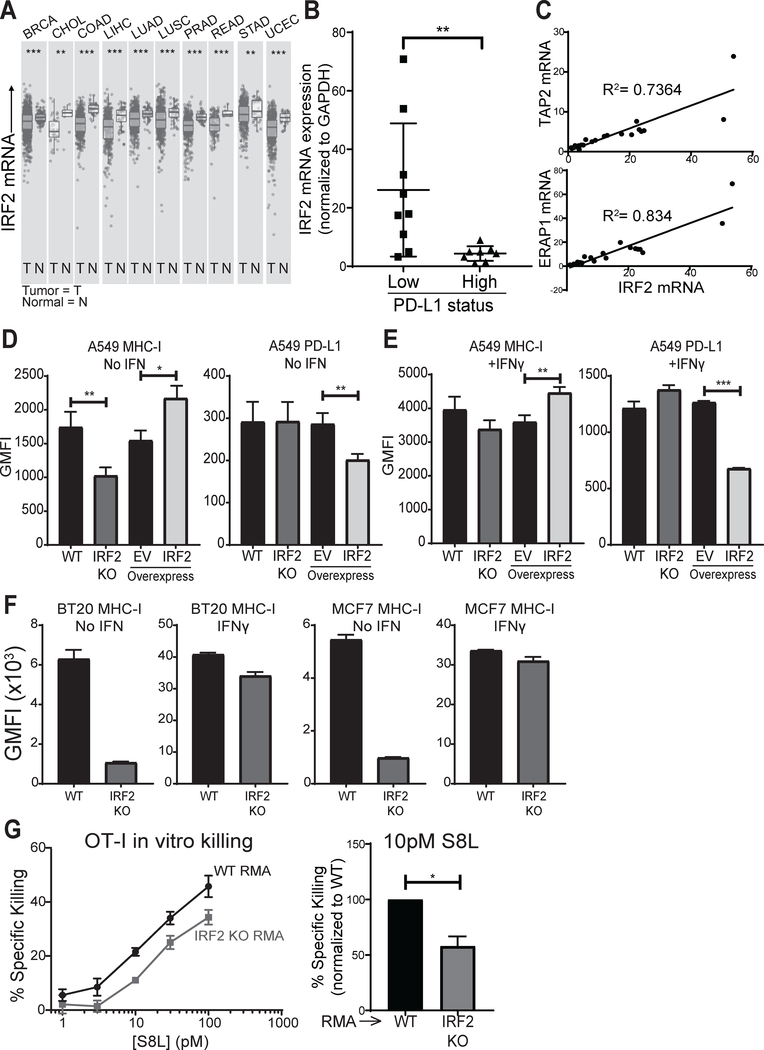
Given that experimentally-induced loss of IRF2 both compromises MHC-I presentation and increases PD-L1 expression, it was of interest to see how often IRF2 is downregulated in primary cancers. We used the TIMER bioinformatics tool (42) to mine publicly available databases for IRF2 expression in primary human cancers. Remarkably, IRF2 was downregulated in several kinds of human cancers and the overall reductions were highly statistically significant (Fig. 6A). For each of the IRF2-low cancers, which included invasive breast carcinoma, cholangiocarcinoma, colon adenocarcinoma, liver hepatocellular carcinoma, lung adenocarcinoma and squamous cell carcinoma (non-small cell lung cancers), prostate, rectum and stomach adenocarcinomas, and uterine corpus endometrial carcinoma, a subset of patients had very low levels of IRF2. We chose one of the IRF2-low cancers, non-small cell lung cancer (NSCLC), to determine whether IRF2 levels were functionally limiting in primary cancers. At our institution, NSCLCs are screened for PD-L1 expression by immunohistochemistry (IHC) at the time of diagnosis, which enabled us to randomly select tumors spanning a spectrum of PD-L1 expression. We extracted RNA from archival patient biopsy material and quantified expression of IRF2, TAP2, and ERAP1 by qPCR. In these lung cancers, IRF2 mRNA levels and PD-L1 IHC status were significantly inversely correlated (Fig. 6B). Additionally, consistent with our cell line findings, TAP2 and ERAP1 mRNA levels positively and significantly correlated with IRF2 mRNA levels (Fig. 6C). To formally test cause and effect for these correlations, we analyzed a human NSCLC cell line, A549, that is IRF2-low relative to other NSCLCs tested in the NCI-60 panel (43). A549 cells are also MHC-I-low and PD-L1-positive (44, 45). Eliminating the residual IRF2 in A549 cells by CRISPR-Cas9-mediated knockout further decreased surface MHC-I but did not further increase surface PD-L1 (Fig. 6D). In contrast, restoring IRF2 by transfection repressed surface PD-L1 expression and increased surface MHC-I expression (Fig. 6D). Stimulation of A549 with IFNγ augmented both surface MHC-I and PD-L1 expression, as expected, but transfection of IRF2 still had the same pattern of effects as without IFN (repressing PD-L1 and further increasing MHC-I expression) (Fig. 6E). To further generalize these findings to other cancers, we analyzed two human breast cancers (chosen because breast cancers are often IRF2 low (Fig. 6A) as well as two mouse sarcomas (chosen because sarcomas are known to undergo immunoediting (3) and a mouse lymphoma (chosen for use in CD8+ T cell killing assays) and found similar results (Fig. 6F, Fig. S3).
Figure 6. IRF2 and cancer.
(A) Differential IRF2 expression in tumor and normal tissue from patients with the indicated cancer types (TCGA abbreviations), as queried from TIMER (42); (B) IRF2 mRNA expression in patient NSCLC specimens scored as PD-L1 low (1–50%) or high (>50%) by immunohistochemistry. IRF2 mRNA expression was normalized to GAPDH mRNA expression in each lung specimen and then IRF2 expression across specimens was compared by calculating fold changes over the lowest IRF2-expressing specimen, which was set equal to 1. Mann-Whitney U test, **p<0.01; (C) TAP2 (top) and ERAP1 (bottom) mRNA correlations with IRF2 mRNA in NSCLC specimens. Linear regression models shown with R2 for goodness of fit; (D-E) Geometric MFI of surface MHC-I (left) and PD-L1 (right) on (D) unstimulated or (E) overnight IFNγ-stimulated A549 lines knocked out for IRF2 (“IRF2 KO”) or control (“WT”) or overexpressing IRF2 or empty vector (EV) as control. Bars represent mean + SEM (N=3). Two-tailed ratio paired t-tests, *p<0.05, **p<0.01, ***p<0.001; (F) Geometric MFI of surface MHC-I on unstimulated or IFNγ-stimulated human breast carcinoma (BT20 and MCF7) lines knocked out for IRF2 (“IRF2 KO”) or control (“WT”). Bars represent mean + SEM (N=3). Two-tailed ratio paired t-tests, **p<0.01, ***p<0.001; (G) Pre-activated OT-I effectors were cultured with pairs of wild-type (“WT RMA”) or IRF2 KO (“IRF2 KO RMA”) cells that were S8L-pulsed or not and labeled with different amounts of the dye CFSE. After 4 hours, specific killing was quantified by flow cytometry. (Left) % specific killing dose-titration curve for one representative experiment using an effector to target ratio of 1:2 and [S8L] indicated, points show mean ± SD of technical triplicates; (Right) % specific killing of IRF2 KO relative to that of WT RMA at 10pM S8L. Bars represent mean + SEM (N=4). Two-tailed ratio paired t-test, *p<0.05.s
The effects of IRF2 loss on antigen presentation and checkpoint inhibition are predicted to make it harder for CD8+ T cells to kill IRF2-low cells. To test this and quantify the magnitude of the effect, we analyzed mouse wild-type vs. IRF2-KO lymphoma (RMA) cell lines. RMA cells were chosen because they are known to be very good targets for cytotoxic T cell killing assays (and are, therefore, a stringent test) and express H2-Kb, which allows the use of potent CD8+ T cell effectors from the H2-Kb-S8L-specific TCR transgenic OT-I model. Pre-activated OT-I effectors were cultured with pairs of wild-type or IRF2-KO cells that were S8L-pulsed or not and labeled with different amounts of the dye CFSE. After 4 hours, specific killing was quantified by flow cytometry and the dose-response curve for the IRF2-KO cells was shifted up ~3-fold (which equates to a decreased killing efficiency of ~67%) in the IRF2-KO RMA, as compared to the wild-type RMA (Fig. 6G), demonstrating that tumor cells lacking IRF2 are harder for CD8+ T cells to eliminate. Taken together, these findings show that IRF2 downregulation leads to immune evasion and that there are several types of human cancers which may use this escape mechanism.
Discussion
Our findings that IRF2 both positively regulates the MHC class I pathway and negatively regulates PD-L1 expression have implications for cancer progression and immunotherapy. Cancers need to evade the immune system and, if they are immunogenic, require editing to escape and progress (6). One of the ways that murine tumors can evade immune elimination is by downregulating the MHC-I pathway (3). Similar immunoediting of this pathway occurs in humans as it has been found that tumors, which were predominantly MHC-I positive at early stages, subsequently become homogeneously MHC-I deficient (46, 47) and progressing cancers are frequently MHC-I deficient (48, 49). Another way tumors can evade immune elimination is by expressing checkpoint inhibitors. Indeed, many human cancers upregulate PD-L1 and those that do tend to be more aggressive and have fewer T cells present in the tumor (50). Blocking PD-L1 or its receptor (PD-1) can lead to tumor rejection in both mice and humans, proving that this is an important immune evasion mechanism (51, 52). The molecular mechanisms that allow cancers to exploit these immune evasion strategies are incompletely understood but are important to elucidate for understanding pathogenesis, how they affect prognosis and immunotherapy, and how they might be reversed to improve therapy.
Our studies have uncovered an immune evasion mechanism that is relatively frequent. Many cancers of diverse origins (NSCLC, breast, colorectal, liver, stomach, prostate, uterine) downregulate IRF2 expression, which is a molecule that is not essential for viability in fully differentiated cells. In silico analysis through cBioPortal (53, 54) revealed that IRF2 is rarely mutated in cancers and, therefore, the loss of expression must be epigenetic. As a result of losing IRF2, tumor cells decrease their surface MHC-I expression and increase their surface PD-L1 expression. Thus, loss of a single gene can generate a “double whammy” for the immune system as it enables tumor cells lacking IRF2 to become both harder to identify (loss of MHC-I antigen presentation) and better able to suppress T cell-mediated elimination (increased checkpoint inhibition). As shown in our in vitro cytotoxicity assay, the loss of IRF2 renders such tumor cells more difficult for CD8+ T cells to kill.
Analysis of IRF2-knockout cells revealed that PD-L1 was highly upregulated under basal conditions. It is well-established that IRF1 can promote IFNγ-inducible activation of PD-L1 (40, 41). Recently, Dorand et al. found that hyperphosphorylation of IRF2BP2 (an IRF2-binding protein) could lead to decreased PD-L1 expression after IFNγ stimulation (55). Additionally, Wu et al. recently discovered that a loss of IRF2BP2 can lead to enhanced IRF2 binding to the PD-L1 promoter and that IFNγ stimulation releases IRF2 from the PD-L1 promoter (56). However, the role of IRF2 on PD-L1 expression had not been directly examined. Our ChIP-qPCR data shows that IFNγ stimulation leads to both increased IRF1 binding and decreased IRF2 binding to the PD-L1 promoter. In addition, we show here for the first time that, under basal conditions, a loss of IRF2 significantly increases PD-L1 mRNA and surface expression and IRF2 overexpression produces the opposite effects. Collectively, these results establish the repressive role of IRF2 on PD-L1 expression in the absence and presence of IFNγ and reveal that unstimulated cells can upregulate PD-L1 expression if upstream regulatory factors, such as IRF2, are defective/absent.
The transcriptional control of many MHC class I pathway components, particularly under basal conditions, has not been well defined (57). Here, we identified IRF2 as a novel positive regulator of MHC-I antigen presentation and show that, under basal conditions, it is important for both classical MHC-I presentation and cross-presentation. IRF2 binds the promoters of TAP2 and ERAP1 and transcriptionally activates their expression. In the absence of IRF2, cells express less TAP2 and ERAP1 and, due to the consequent defects in antigen transport and processing, present fewer peptide-MHC-I complexes at the cell surface. Additionally, IRF2 regulates the expression of many other genes, as demonstrated by others (58, 59) and shown here, such as immunoproteasome subunits, which likely also influence the MHC-I presentation in IRF2-deficient cells (60). Interestingly, in one of these earlier studies, IRF2 was reported to bind to the promoter of TAP1 as assessed by electrophoretic mobility shift assays (59); however, we found no evidence that IRF2 was needed for transcription of endogenous TAP1 by either RNA-seq or qPCR. Most likely, this discrepancy is due to the nature of the different assays (e.g. extract binding to exogenous probes vs. endogenous promoter activity).
In the DC line where we focused most of our studies and in other cell lines described in the literature, IRF2 is constitutively expressed, relatively stable with a half-life of 8 hours, and minimally affected by IFN (36–38). However, in these same sets of cells, IRF1 is minimally expressed under basal conditions but strongly induced in the presence of IFN and more short-lived than IRF2, with a half-life of only 0.5 hours (36). Due to these differences between IRF2 and IRF1, even though they both recognize and bind to the same ISRE (33–35), we hypothesized that the relative contributions of IRF2 and IRF1 to certain signaling pathways varies depending on the inflammatory state of the cell. Although IRF1 and IRF2 were originally characterized as an activator and repressor of IFN-α/β expression, respectively (35), several reports since then have highlighted that IRF1 and IRF2 do not always act as such (37, 61, 62). Yet, to the best of our knowledge, there have been no global differential expression analyses in IRF1- and/or IRF2-lacking cells of the same cell type to better understand the synergistic vs. antagonistic roles these transcription factors perform. Here, we conducted RNA-seq on IRF1- and/or IRF2-knockout DCs in the absence or presence of IFN to help fill this void. Globally, we found that the expression profile of the double knockout DCs differed from that of either IRF single knockout and that the relative contributions of IRF1 vs. IRF2, in terms of their ability to positively or negatively regulate certain genes, varied depending on the inflammatory state of the cell. Transcriptional control of genes can be highly cell-type dependent and therefore, it is possible that in other cells, the contribution of IRF2 on surface MHC-I and PD-L1 expression may vary. However, our concordant observations in multiple mouse and human cell lines and primary tumors suggest that the number of cell types in which IRF2 regulates these genes in this manner is quite large.
Immune evasion due to tumor PD-L1 upregulation can be reversed by blocking the PD-L1/PD-1 interaction, which is the basis of targeted checkpoint blockade immunotherapy. However, any natural or invigorated (from checkpoint blockade) CD8+ T cell response to kill tumors that have downregulated their MHC-I expression will continue to be impaired. In this context, it is of considerable interest and potential importance that the downregulation of the MHC-I pathway from the loss of IRF2 is reversible. When IRF2-deficient cells were treated with IFN, MHC-I levels were restored, likely because of induction of IRF1 and possibly some other transcriptional activators. These findings suggest that interferons (which are FDA-approved for other indications) or interferon-inducing agents could be used to restore MHC-I antigen presentation in IRF2-low tumors. This would be predicted to enhance the effects of immunotherapies, such as checkpoint blockade, that are ultimately dependent on T cell receptor recognition of tumor MHC-I presentation. Currently, checkpoint therapy is effective in only some patients and, based on the mechanisms we have uncovered, it is conceivable that reversing the IRF2 defects might increase the number of patients that can benefit. In addition, because checkpoint blockade is an extremely expensive therapy and one that can have serious side effects, there is a need for good biomarkers to identify those patients that would be more likely to benefit from this type of therapy. Therefore, it will be of interest to examine in future studies whether expression of IRF2 and its downstream target genes (e.g., TAP2 and ERAP1) could be used as biomarkers to help identify checkpoint blockade-responsive patients.
Supplementary Material
Key Points.
Many primary human cancers frequently downregulate the transcription factor IRF2
IRF2 loss causes an immune evasion phenotype of low MHC-I and high PD-L1
The MHC-I defect in IRF2-low cells can be restored upon interferon stimulation
Acknowledgments
The authors thank Kate Fitzgerald for helpful discussions, Janice BelleIsle for technical assistance, and Leslie Shaw and Robert Schreiber for sharing cells and reagents.
Grant support
Financial support provided by NIH grants R01 AI114495 (KLR), T32 AI095213 (BAK), and T32 GM107000 (BAK). This work was funded by an Investigators in the Pathogenesis of Infectious Disease grant from the Burroughs Wellcome Fund to ALB. ALB is grateful to the Bill and Melinda Gates Foundation and to Gilead Sciences Inc. for their support.
References
- 1.Kaplan DH, Shankaran V, Dighe AS, Stockert E, Aguet M, Old LJ, and Schreiber RD. 1998. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc Natl Acad Sci U S A 95: 7556–7561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Street SE, Cretney E, and Smyth MJ. 2001. Perforin and interferon-gamma activities independently control tumor initiation, growth, and metastasis. Blood 97: 192–197. [DOI] [PubMed] [Google Scholar]
- 3.Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, and Schreiber RD. 2001. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature 410: 1107–1111. [DOI] [PubMed] [Google Scholar]
- 4.Svane IM, Engel AM, Nielsen MB, Ljunggren HG, Rygaard J, and Werdelin O. 1996. Chemically induced sarcomas from nude mice are more immunogenic than similar sarcomas from congenic normal mice. Eur J Immunol 26: 1844–1850. [DOI] [PubMed] [Google Scholar]
- 5.Engel AM, Svane IM, Rygaard J, and Werdelin O. 1997. MCA sarcomas induced in scid mice are more immunogenic than MCA sarcomas induced in congenic, immunocompetent mice. Scand J Immunol 45: 463–470. [DOI] [PubMed] [Google Scholar]
- 6.Mittal D, Gubin MM, Schreiber RD, and Smyth MJ. 2014. New insights into cancer immunoediting and its three component phases--elimination, equilibrium and escape. Curr Opin Immunol 27: 16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sabbatino F, Villani V, Yearley JH, Deshpande V, Cai L, Konstantinidis IT, Moon C, Nota S, Wang Y, Al-Sukaini A, Zhu AX, Goyal L, Ting DT, Bardeesy N, Hong TS, Fernandez-del Castillo C, Tanabe KK, Lillemoe KD, Ferrone S, and Ferrone CR. 2016. PD-L1 and HLA Class I Antigen Expression and Clinical Course of the Disease in Intrahepatic Cholangiocarcinoma. Clin Cancer Res 22: 470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gettinger S, Choi J, Hastings K, Truini A, Datar I, Sowell R, Wurtz A, Dong W, Cai G, Melnick MA, Du VY, Schlessinger J, Goldberg SB, Chiang A, Sanmamed MF, Melero I, Agorreta J, Montuenga LM, Lifton R, Ferrone S, Kavathas P, Rimm DL, Kaech SM, Schalper K, Herbst RS, and Politi K. 2017. Impaired HLA Class I Antigen Processing and Presentation as a Mechanism of Acquired Resistance to Immune Checkpoint Inhibitors in Lung Cancer. Cancer Discov 7: 1420–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abele R, and Tampe R. 2004. The ABCs of immunology: structure and function of TAP, the transporter associated with antigen processing. Physiology (Bethesda) 19: 216–224. [DOI] [PubMed] [Google Scholar]
- 10.Saric T, Chang SC, Hattori A, York IA, Markant S, Rock KL, Tsujimoto M, and Goldberg AL. 2002. An IFN-gamma-induced aminopeptidase in the ER, ERAP1, trims precursors to MHC class I-presented peptides. Nat Immunol 3: 1169–1176. [DOI] [PubMed] [Google Scholar]
- 11.Serwold T, Gonzalez F, Kim J, Jacob R, and Shastri N. 2002. ERAAP customizes peptides for MHC class I molecules in the endoplasmic reticulum. Nature 419: 480–483. [DOI] [PubMed] [Google Scholar]
- 12.Zamora AE, Crawford JC, and Thomas PG. 2018. Hitting the Target: How T Cells Detect and Eliminate Tumors. J Immunol 200: 392–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen Z, Reznikoff G, Dranoff G, and Rock KL. 1997. Cloned dendritic cells can present exogenous antigens on both MHC class I and class II molecules. J Immunol 158: 2723–2730. [PubMed] [Google Scholar]
- 14.Rock KL, Rothstein L, and Gamble S. 1990. Generation of class I MHC-restricted T-T hybridomas. J Immunol 145: 804–811. [PubMed] [Google Scholar]
- 15.Sanjana NE, Shalem O, and Zhang F. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11: 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, and Zhang F. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343: 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brinkman EK, Chen T, Amendola M, and van Steensel B. 2014. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res 42: e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hearn A, York IA, and Rock KL. 2009. The specificity of trimming of MHC class I-presented peptides in the endoplasmic reticulum. J Immunol 183: 5526–5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Porgador A, Yewdell JW, Deng Y, Bennink JR, and Germain RN. 1997. Localization, quantitation, and in situ detection of specific peptide-MHC class I complexes using a monoclonal antibody. Immunity 6: 715–726. [DOI] [PubMed] [Google Scholar]
- 20.Bolger AM, Lohse M, and Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li B, and Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pruitt KD, Tatusova T, Brown GR, and Maglott DR. 2012. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res 40: D130–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson WE, Li C, and Rabinovic A. 2007. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8: 118–127. [DOI] [PubMed] [Google Scholar]
- 24.Leek JT, Johnson WE, Parker HS, Jaffe AE, and Storey JD. 2012. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28: 882–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Love MI, Huber W, and Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mould AW, Morgan MA, Nelson AC, Bikoff EK, and Robertson EJ. 2015. Blimp1/Prdm1 Functions in Opposition to Irf1 to Maintain Neonatal Tolerance during Postnatal Intestinal Maturation. PLoS Genet 11: e1005375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doody GM, Stephenson S, McManamy C, and Tooze RM. 2007. PRDM1/BLIMP-1 modulates IFN-gamma-dependent control of the MHC class I antigen-processing and peptide-loading pathway. J Immunol 179: 7614–7623. [DOI] [PubMed] [Google Scholar]
- 28.Lu C, Redd PS, Lee JR, Savage N, and Liu K. 2016. The expression profiles and regulation of PD-L1 in tumor-induced myeloid-derived suppressor cells. Oncoimmunology 5: e1247135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masumi A, Yamakawa Y, Fukazawa H, Ozato K, and Komuro K. 2003. Interferon regulatory factor-2 regulates cell growth through its acetylation. J Biol Chem 278: 25401–25407. [DOI] [PubMed] [Google Scholar]
- 30.Fernandes DM, Vidard L, and Rock KL. 2000. Characterization of MHC class II-presented peptides generated from an antigen targeted to different endocytic compartments. Eur J Immunol 30: 2333–2343. [DOI] [PubMed] [Google Scholar]
- 31.Dong H, Strome SE, Salomao DR, Tamura H, Hirano F, Flies DB, Roche PC, Lu J, Zhu G, Tamada K, Lennon VA, Celis E, and Chen L. 2002. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med 8: 793–800. [DOI] [PubMed] [Google Scholar]
- 32.Yu H, Boyle TA, Zhou C, Rimm DL, and Hirsch FR. 2016. PD-L1 Expression in Lung Cancer. J Thorac Oncol 11: 964–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanaka N, Kawakami T, and Taniguchi T. 1993. Recognition DNA sequences of interferon regulatory factor 1 (IRF-1) and IRF-2, regulators of cell growth and the interferon system. Mol Cell Biol 13: 4531–4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Darnell JE Jr., Kerr IM, and Stark GR. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264: 1415–1421. [DOI] [PubMed] [Google Scholar]
- 35.Harada H, Fujita T, Miyamoto M, Kimura Y, Maruyama M, Furia A, Miyata T, and Taniguchi T. 1989. Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell 58: 729–739. [DOI] [PubMed] [Google Scholar]
- 36.Watanabe N, Sakakibara J, Hovanessian AG, Taniguchi T, and Fujita T. 1991. Activation of IFN-beta element by IRF-1 requires a posttranslational event in addition to IRF-1 synthesis. Nucleic Acids Res 19: 4421–4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ren G, Cui K, Zhang Z, and Zhao K. 2015. Division of labor between IRF1 and IRF2 in regulating different stages of transcriptional activation in cellular antiviral activities. Cell Biosci 5: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oshima S, Nakamura T, Namiki S, Okada E, Tsuchiya K, Okamoto R, Yamazaki M, Yokota T, Aida M, Yamaguchi Y, Kanai T, Handa H, and Watanabe M. 2004. Interferon regulatory factor 1 (IRF-1) and IRF-2 distinctively up-regulate gene expression and production of interleukin-7 in human intestinal epithelial cells. Mol Cell Biol 24: 6298–6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hobart M, Ramassar V, Goes N, Urmson J, and Halloran PF. 1997. IFN regulatory factor-1 plays a central role in the regulation of the expression of class I and II MHC genes in vivo. J Immunol 158: 4260–4269. [PubMed] [Google Scholar]
- 40.Lee SJ, Jang BC, Lee SW, Yang YI, Suh SI, Park YM, Oh S, Shin JG, Yao S, Chen L, and Choi IH. 2006. Interferon regulatory factor-1 is prerequisite to the constitutive expression and IFN-gamma-induced upregulation of B7-H1 (CD274). FEBS Lett 580: 755–762. [DOI] [PubMed] [Google Scholar]
- 41.Garcia-Diaz A, Shin DS, Moreno BH, Saco J, Escuin-Ordinas H, Rodriguez GA, Zaretsky JM, Sun L, Hugo W, Wang X, Parisi G, Saus CP, Torrejon DY, Graeber TG, Comin-Anduix B, Hu-Lieskovan S, Damoiseaux R, Lo RS, and Ribas A. 2017. Interferon Receptor Signaling Pathways Regulating PD-L1 and PD-L2 Expression. Cell Rep 19: 1189–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, Jiang P, Shen H, Aster JC, Rodig S, Signoretti S, Liu JS, and Liu XS. 2016. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol 17: 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rajapakse VN, Luna A, Yamade M, Loman L, Varma S, Sunshine M, Iorio F, Sousa FG, Elloumi F, Aladjem MI, Thomas A, Sander C, Kohn KW, Benes CH, Garnett M, Reinhold WC, and Pommier Y. 2018. CellMinerCDB for Integrative Cross-Database Genomics and Pharmacogenomics Analyses of Cancer Cell Lines. iScience 10: 247–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teo J, Mirenska A, Tan M, Lee Y, Oh J, Hong LZ, Wnek R, Yap YS, Shih SJ, AA SB, Chin CL, and Skibinski DA. 2017. A preliminary study for the assessment of PD-L1 and PD-L2 on circulating tumor cells by microfluidic-based chipcytometry. Future Sci OA 3: FSO244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Redondo M, Ruiz-Cabello F, Concha A, Cabrera T, Perez-Ayala M, Oliva MR, and Garrido F. 1991. Altered HLA class I expression in non-small cell lung cancer is independent of c-myc activation. Cancer Res 51: 2463–2468. [PubMed] [Google Scholar]
- 46.Carretero R, Romero JM, Ruiz-Cabello F, Maleno I, Rodriguez F, Camacho FM, Real LM, Garrido F, and Cabrera T. 2008. Analysis of HLA class I expression in progressing and regressing metastatic melanoma lesions after immunotherapy. Immunogenetics 60: 439–447. [DOI] [PubMed] [Google Scholar]
- 47.del Campo AB, Kyte JA, Carretero J, Zinchencko S, Mendez R, Gonzalez-Aseguinolaza G, Ruiz-Cabello F, Aamdal S, Gaudernack G, Garrido F, and Aptsiauri N. 2014. Immune escape of cancer cells with beta2-microglobulin loss over the course of metastatic melanoma. Int J Cancer 134: 102–113. [DOI] [PubMed] [Google Scholar]
- 48.Marincola FM, Jaffee EM, Hicklin DJ, and Ferrone S. 2000. Escape of human solid tumors from T-cell recognition: molecular mechanisms and functional significance. Adv Immunol 74: 181–273. [DOI] [PubMed] [Google Scholar]
- 49.Hicklin DJ, Marincola FM, and Ferrone S. 1999. HLA class I antigen downregulation in human cancers: T-cell immunotherapy revives an old story. Mol Med Today 5: 178–186. [DOI] [PubMed] [Google Scholar]
- 50.Zou W, and Chen L. 2008. Inhibitory B7-family molecules in the tumour microenvironment. Nat Rev Immunol 8: 467–477. [DOI] [PubMed] [Google Scholar]
- 51.Pardoll DM 2012. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 12: 252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang X, Bao Z, Zhang X, Li F, Lai T, Cao C, Chen Z, Li W, Shen H, and Ying S. 2017. Effectiveness and safety of PD-1/PD-L1 inhibitors in the treatment of solid tumors: a systematic review and meta-analysis. Oncotarget 8: 59901–59914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, and Schultz N. 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2: 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, and Schultz N. 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dorand RD, Nthale J, Myers JT, Barkauskas DS, Avril S, Chirieleison SM, Pareek TK, Abbott DW, Stearns DS, Letterio JJ, Huang AY, and Petrosiute A. 2016. Cdk5 disruption attenuates tumor PD-L1 expression and promotes antitumor immunity. Science 353: 399–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu A, Wu Q, Deng Y, Liu Y, Lu J, Liu L, Li X, Liao C, Zhao B, and Song H. 2019. Loss of VGLL4 suppresses tumor PD-L1 expression and immune evasion. EMBO J 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jongsma MLM, Guarda G, and Spaapen RM. 2017. The regulatory network behind MHC class I expression. Mol Immunol. [DOI] [PubMed] [Google Scholar]
- 58.Fairfax BP, Humburg P, Makino S, Naranbhai V, Wong D, Lau E, Jostins L, Plant K, Andrews R, McGee C, and Knight JC. 2014. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science 343: 1246949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rouyez MC, Lestingi M, Charon M, Fichelson S, Buzyn A, and Dusanter-Fourt I. 2005. IFN regulatory factor-2 cooperates with STAT1 to regulate transporter associated with antigen processing-1 promoter activity. J Immunol 174: 3948–3958. [DOI] [PubMed] [Google Scholar]
- 60.Kincaid EZ, Che JW, York I, Escobar H, Reyes-Vargas E, Delgado JC, Welsh RM, Karow ML, Murphy AJ, Valenzuela DM, Yancopoulos GD, and Rock KL. 2011. Mice completely lacking immunoproteasomes show major changes in antigen presentation. Nat Immunol 13: 129–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jesse TL, LaChance R, Iademarco MF, and Dean DC. 1998. Interferon regulatory factor-2 is a transcriptional activator in muscle where It regulates expression of vascular cell adhesion molecule-1. J Cell Biol 140: 1265–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vaughan PS, Aziz F, van Wijnen AJ, Wu S, Harada H, Taniguchi T, Soprano KJ, Stein JL, and Stein GS. 1995. Activation of a cell-cycle-regulated histone gene by the oncogenic transcription factor IRF-2. Nature 377: 362–365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.