Abstract
The ability to manipulate and fuse nano-compartmentalized volumes addresses a demand for spatiotemporal control in the field of synthetic biology, for example in the bottom-up construction of (bio)chemical nanoreactors and for the interrogation of enzymatic reactions in confined space. Herein, we mix entrapped sub-attoliter volumes of liposomes (~135 nm diameter) via lipid bilayer fusion, facilitated by the hybridization of membrane-anchored lipidated oligonucleotides. We report on an improved synthesis of the membrane-anchor phosphoramidites that allows for a flexible choice of lipophilic moiety. Lipid-nucleic acid conjugates (LiNAs) with and without triethylene glycol spacers between anchor and the 17 nt binding sequence were synthesized and their fusogenic potential evaluated. A fluorescence-based content mixing assay was employed for kinetic monitoring of fusion of the bulk liposome populations at different temperatures. Data obtained at 50 °C indicated a quantitative conversion of the limiting liposome population into fused liposomes and an unprecedently high initial fusion rate was observed. For most conditions and designs only low leakage during fusion was observed. These results consolidate LiNA-mediated membrane fusion as a robust platform for programming compartmentalized chemical and enzymatic reactions.
Subject terms: Membrane biophysics, Membrane structure and assembly, Origin of life
Introduction
Conjugation of oligonucleotides with neutral lipids (lipid-nucleic acid conjugates, LiNAs) has proven a viable strategy for functionalization of artificial lipid membranes1,2. Synthetic lipid-based membranes and liposomes represent useful model systems for studying membrane docking and fusion, and serve as platforms for nano-scaffolded (bio)chemical processes, e.g. reactions catalyzed by immobilized or compartmentalized enzymes, including a variety of other applications revolving around efficient spatiotemporal manipulation of sub-attoliter compartments.
In recent years, techniques involving LiNAs for controlled aggregation3–6 and fusion7–10 of phospholipid-based liposomes have been developed, but only recently, substantial content mixing (CM) between LiNA-encoded liposome populations has been reported (i.e. ≥20% of encapsulated volume)11,12. To use DNA as a tool for programming multiple rounds of fusion, efficiencies above 20% are needed to achieve detectable fusion beyond the first round. Recently, the feature of programmability in LiNA-mediated fusion was demonstrated in experiments where liposomes were fused over three sequentially dependent rounds, based on the design of the nucleobase sequences of the LiNA fusogens. An estimated bulk fusion yield of 60-80% per round was reported13.
The most well-described proteins that drive biological fusion are the soluble N-ethylmaleimide-sensitive-factor attachment receptors (SNAREs). SNAREs force biological membranes into contact through the interaction of membrane-anchored subunits. Compared to the LiNAs mentioned above, they also rely on transmembrane domains (TMDs) and a conserved linker-domain for efficient fusion14–16. SNARE-model systems such as SNARE sub-domains (SNAREpins)17 and SNARE-derived lipidated coiled-coil peptides18,19 were also shown to induce fusion and content mixing of liposomes. Using nucleic acid hybridization as a recognition unit provides means to make liposome fusion programmable, making use of Watson-Crick base pairing to achieve specific recognition and fusion between multiple populations of liposomes. In Biology, SNAREs rely on a variety of co-factors for specificity, and coiled-coil forming peptides lack the intrinsic programmability of nucleic acids16,20.
Further model systems include peptide nucleic acids (PNA) conjugated to the SNARE TMD-peptides, which were used to study the role of TMDs in fusion pore opening20–22. Recently PNAs with aliphatic membrane anchors were employed23, serving as a useful uncharged alternative to LiNAs. Many of the earlier fusion systems have been reviewed in the literature showing either low levels of content mixing or very substantial leakage for designs with higher levels of content mixing24,25.
Compared to SNARE-derived lipidated coiled-coil peptides, only a low degree of functionalization of LiNAs is needed to facilitate efficient content mixing between liposomes (~0.5 to 5 mol% versus 0.01 to 0.1 mol% of total lipid for lipopeptides and LiNAs, respectively)18,19. Still, it was estimated that the number of LiNA duplexes per liposome required to open a fusion pore is around 10–209,13, while native SNAREs are effective at 1-2 complexes per vesicle26.
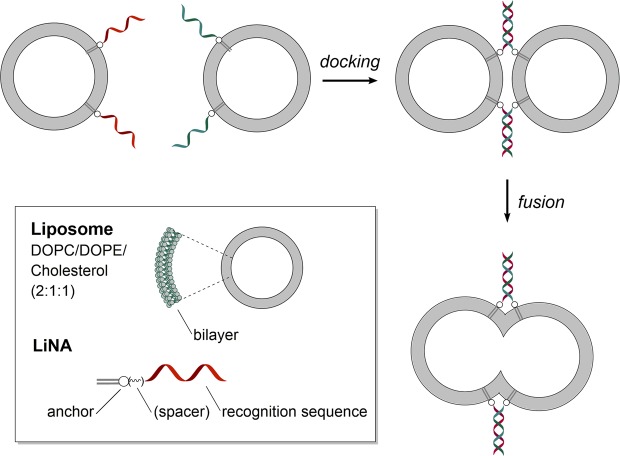
Regarding the mechanism of membrane fusion, SNARE-proteins act via coiled-coil formation between different α-helical subunits, and via local destabilization of the cell membrane induced via the TMDs and linker-domains of the SNARE-complex20–22. Concerning LiNAs or other SNARE-mimics lacking a TMD, we earlier proposed the hypothesis12 that the main driver for fusion are Brownian collisions between bilayers in tight apposition (docked state, Fig. 1). Spontaneous fusion requires intermembrane distances of ≤2 nm, at which the hydration shell of the lipid headgroups begins to be disturbed27,28. Notably, the rate of spontaneous membrane fusion, as measured by François-Martin et al.28,29, could simply be amplified in a docked state forcing intermembrane distances of a few nanometers across an extended contact area.
Figure 1.
LiNA-mediated fusion of liposomes. Functionalized liposomes with complementary LiNAs dock via hybridization of LiNA strands leading to membrane fusion (drawing not to scale).
Applications of controlled and/or programmable liposome fusion have already been reported. Direct fusion to cell-membranes present a promising strategy for bypassing endosomal pathways during drug delivery. Lipidated SNARE-peptides have been used to fuse liposomal vehicles to live HeLa cells in culture (loaded with small molecules30 or oligonucleotides31) or even to endothelial cells in the blood stream of live zebrafish embryos32. Just recently, Sun et al. reported on DNA-mediated fusion to deliver horseradish peroxidase directly to the cytosol of L1210 and HeLa cells as a model for protein delivery33.
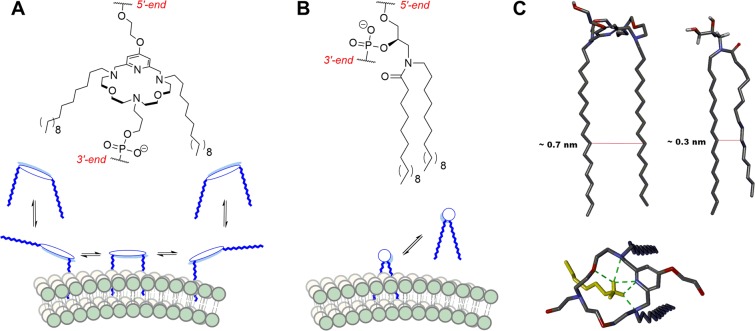
In our efforts to develop a robust platform for sequence-programmable membrane fusion to be applied in systems biochemistry and as nanoreactors, we have previously studied the effect of different anchor moieties on fusion efficiency, i.e. two linear (palmityl), branched (phytanyl) or planar (cholesteryl) moieties were attached to the same N-atom of a 3-amino-1,2-propanediol scaffold34. In the present study, we used a macrocyclic polyaza crown ether scaffold to give an approximate 0.7 nm spacing between the two aliphatic chains (C16, palmityl) serving as anchors (see Fig. 2). When attached to the crown-ether scaffold, the lipophilic chains can act as two separate membrane anchor moieties, which was proposed to improve anchoring of the oligonucleotides compared to the case where both C16 chains are attached to the same N-atom. Furthermore, crown-ethers are known for their ability to bind cations. Aza-crown ethers differ from their oxo counterparts by their selectivity shift towards NH4+ over K+ and Na+ ions, when containing more than two N-atoms. For triaza crown ethers the affinity for binding ammonium ions was highest among the measured monovalent cations, in solution35 and as part of an oligonucleotide backbone36, respectively. As lipid mixtures used for membrane fusion models typically contain phosphatidylethanolamine (PE) lipids, the crown-ether may further enhance anchoring by binding to PE headgroups (present as zwitter-ions at neutral pH, with the ammonium moiety closest to the membrane surface)36. Here, we report that LiNAs carrying this macrocyclic non-nucleosidic building block efficiently mediate content mixing between two liposome populations engrafted with complementary LiNA strands.
Figure 2.
Anchor design. Different anchor scaffold structures and proposed modes of anchoring (A) Anchor based on aza-crown ether scaffold has a 6-bond spacing over a rigid aromatic system giving the two lipophilic chains an approximate spacing of 0.7 nm and allowing each chain to interact with a different subset of lipids in the bilayer. (B) In the anchor based on 3-amino-1,2-propanediol scaffold both chains are attached to the same atom, favoring intramolecular interactions of the two chains, effectively resulting in a bigger lipophilic moiety which is expected to interact with the membrane in a concerted manner (C) Molecular representation of average lipid chain distances (red lines) for relaxed palmityl chains in aza crown ether and amino propanediol membrane anchors (hydrogens not shown for clarity) and top view of the aza crown ether macrocycle with hydrogen bonded (green dotted lines) ammonium head group of aminoethanol lipids from the lipid bilayer.
Results and Discussion
Synthesis of double-chain anchor building blocks for solid-phase DNA synthesis
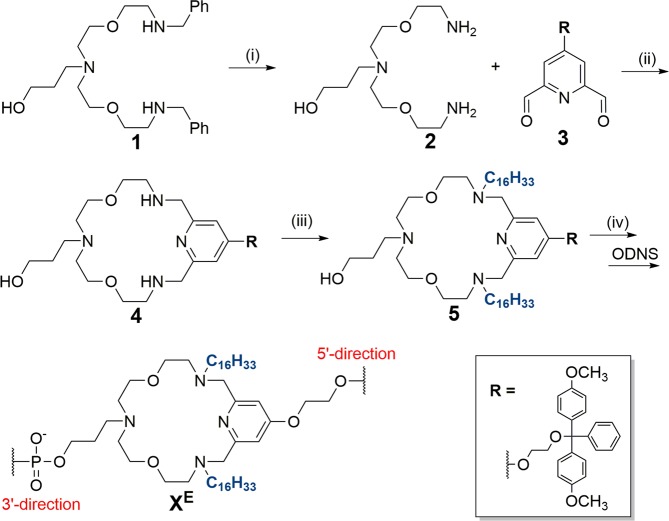
We have previously reported on a lipid functionalized macrocyclic anchor building block that was inserted into DNA oligomers to facilitate DNA-mediated assembly of liposomes3. Rohr et al. described an early-stage insertion of lipid moieties into the polyaza crown ether scaffold, i.e., at the level of compound 1 (Fig. 3). Herein, we report on an improved synthetic strategy that allows for later-stage lipid anchor functionalization. The syntheses of benzylated diamine 1 and dialdehyde 3 have been described previously37. Hence, starting from 1 (see Fig. 3), the final phosphoramidite 5a was obtained over four steps. First, treating 1 with palladium hydroxide in methanol at room temperature provided the deprotected diamine 2. Next, reacting 2 with dialdehyde 3 in a standard reductive amination under dry conditions afforded the desired cyclized polyaza crown ether 4, which was subsequently condensed with hexadecanal to give 5. Finally, phosphitylation of 5 provided the desired lipid anchor building block 5a primed for standard automated solid-phase oligonucleotide synthesis of LiNAs 1–4 (Fig. 4).
Figure 3.
Synthesis and reaction conditions for compound 4-5a. Synthesis of 1 and 3 are described in ref.37. (i) Pd(OH)2, MeOH, rt (2, 90%). (ii) 1: DMTr protected dialdehyde, CaCl2, anhydrous MeOH, 3Å mol. sieves, reflux; 2: NaBH4, 5 °C → rt (4, 59%). (iii) 1: Hexadecanal, 2: NaBH(OAc)3, anhydrous DCE, 5 °C → rt, 3Å mol. sieves (5, 50%). (iv) 2-Cyanoethyl-N,N-diisopropylchlorophosphoramidite, anhydrous DCE, 5 °C → rt (5a, 80%). 5a incorporated of into oligonucleotides = XE.
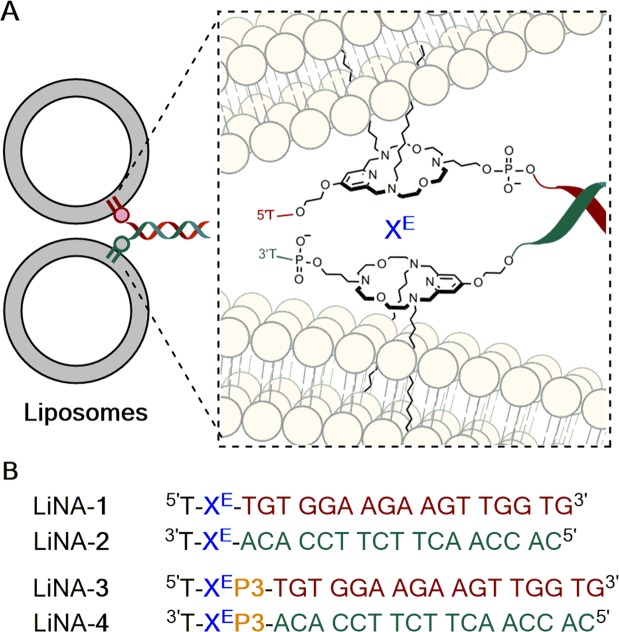
Figure 4.
LiNAs for liposome fusion. (A) Schematic of liposome docking via hybridization of LiNA strands. The inset shows anchor structure and connectivity (drawing not to scale). (B) LiNA sequences with or without triethylene glycol phosphate spacer unit (P3). Duplexes between LiNAs complementary reference DNAs, i.e. the 17-nt zipper-sequence alone, have a Tm of around 57 ± 1 °C (see Supplementary information, Table S2).
Design, synthesis and characterization of LiNA fusogens
Previous studies used a design of DNA oligomers that are able to fold up into a zipper-like geometry (Fig. 4) giving rise to a close distance between bilayers upon hybridization8,10,34. In this study we synthesized two pairs of modified DNA oligonucleotides that hybridize in the same manner. The first pair contained a single T overhang and one insertion of the 5a building block within the phosphodiester backbone (designated XE) at the 5′ or 3′ end, respectively (LiNA-1 and LiNA-2). The single nucleotide overhang was introduced as in previous studies3,4 to prevent the self-aggregation of LiNA molecules into micelles in aqueous phase, driven by hydrophobic interactions between the anchors38. In presence of the extra nucleotide, two negatively charged phosphate groups flank the anchor building block electrostatically and sterically disfavoring micelle formation (absence of foaming of aqueous LiNA stock solutions). The second set contained a triethylene glycol phosphate spacer (P3, see Fig. 4) inserted in between the anchor XE and the DNA-based zipper unit adding approximately 3 nm (1.5 nm for each spacer in the duplex, similar to the ones described for coiled-coil fusogens by Daudey et al.)39 to the maximal separation between docked bilayers (LiNA-3 and LiNA-4). Previously, using LiNAs based on the 3-amino-1,2-propanediol backbone, the presence of a P3 spacer increased fusion efficiency12. In these LiNAs the distance between the 5′-OH and the N-atom bearing the aliphatic anchors was only four bonds, compared to the nine bonds in the present design (XE anchor). We hypothesized that relative to LiNA-1 and LiNA-2, which do not contain spacer P3, less strain-release would be expected upon liposomal fusion mediated by LiNA-3 and LiNA-4. In this study, these two pairs serve to evaluate the effect of linker size and flexibility on LiNA-mediated fusion of liposomes.
The oligonucleotides listed in Fig. 4B were purified on a reverse phase HPLC as previously described34. Circular dichroism spectroscopy revealed that both pairs (LiNA-1/2 and LiNA-3/4, Fig. 4B) formed standard B-type DNA duplexes (see the Supplementary Information). Thermal denaturation and circular dichroism experiments of the complementary strands confirmed that each XE or XE-P3-modified DNA could bind to unmodified complementary sequences (Supplementary Information, Table S2 and Figure S1 and S2). The Tm of such duplexes (56 to 58 °C) were comparable to the unmodified reference duplex (55 °C), however, duplexes of LiNA-1/2 and LiNA-3/4 exhibited an increased Tm (77 ± 1 °C). This increase was attributed to the hydrophobic interactions between aligned alkyl chains. In the presence of liposomes, the lipid anchors are already embedded into the nonpolar environment of different phospholipid bilayers and cannot exhibit the abovementioned stabilization. When carrying out the fusion experiments at 50 °C, we argue that this is approx. 6-8 °C below the actual Tm of the system. No fusion above the Tm of the LiNA/DNA duplexes was observed at 60 °C (data not shown).
LiNA-Liposome binding using Surface Plasmon Resonance (SPR)
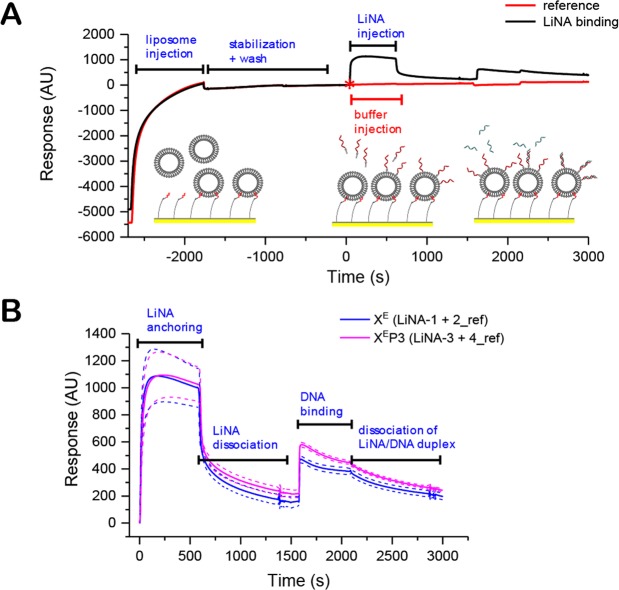
The spontaneous anchoring of LiNAs into liposome bilayers was assayed using surface plasmon resonance, a method that generates a signal based on the bulk of material present a few hundred nanometers above a gold surface. Liposomes were immobilized onto a sensor chip coated with an alkyl-modified dextran matrix (Biacore L1 sensor chip) in the instrument flow cell (Fig. 5A)40. LiNAs were injected slowly (contact time 0-600 s, 2 µl/min, Fig. 5B) giving rise to a fast binding response, saturating the surface of the immobilized liposomes within 60 seconds. After the initial contact time, a continuous flow of 2 µl/min HBS resulted quick removal of loosely associated LiNA, but thereafter the signal approached a new baseline due to the remaining anchored LiNAs (+200-300 response units after 1000 s). LiNA presence was corroborated by testing their ability to form duplexes with complementary DNA strands (non-modified), which were likewise injected slowly (contact time 1600–2100 s, 2 µl/min) giving rise to approximately a doubling of the response at the end of contact time.
Figure 5.
SPR detection of LiNA-binding to immobilized liposomes. (A) Surface Plasmon Resonance (SPR) assay for detecting spontaneous binding of LiNAs to surface immobilized liposomes. After liposome capture (2 µl/min, 900 seconds), equilibration period (900 s) and buffer wash (20 µl/min) LiNA solution (5 µM in HBS, 600 s, 2 µl/min) or HBS (reference measurement) was injected followed by a dissociation phase under a flow of HBS (600 s, 2 µl/min). Then, complementary DNA (without lipid-modification) is injected in the same manner in both experiments. The complementary oligonucleotides showed no affinity to the liposomes on the sensor surface. The response and time axes of both runs are aligned at the point of LiNA injection (red asterisk) and the reference values subtracted. (B) Average response of LiNA-insertion for LiNA-1 (XE) and LiNA-3 (XEP3) and subsequent hybridization of complementary DNA to LiNA strands that remain bound to the surface after the dissociation phase. Measurements carried out at 25 °C, the solid line shows the mean signal of two injections, the dotted lines show the total error.
This is expected for an efficient hybridization as the surface bound mass approximately doubles when LiNAs hybridize with their complementary strand. After contact of the complementary DNA, a slow signal decrease was observed during HBS flow over the surface, which must either be due to de-hybridization of DNA or “de-anchoring” of LiNA/DNA duplexes (anchoring must become less effective after nearly doubling the molecular mass by a second hybridized DNA).
LiNA-induced liposome fusion and content mixing
For fusion experiments, the fusogens LiNA1-4 were engrafted onto liposomes (DOPC/DOPE/Cholesterol, 2:1:1, ~135 nm avg. diameter (Figure S3), incubated with LiNAs for 15 min at r.t), thereby providing two populations of liposomes each carrying one of the complementary strands of a LiNA pair. The LiNAs hybridize upon mixing the two populations causing the liposomes to cross-link and fuse. Figure 4 schematically illustrates LiNA functionalized liposomes and the recognition sequences used in the present study.
The fusion efficiency was assayed by measuring the content mixing (CM) of the two liposome populations using a well-established fluorescence assay. One population encapsulates Sulforhodamine B (SRB) at a self-quenching concentration (20 mM)21,41 while the other is filled with buffer (unlabeled). In brief, the assay works as follows: upon opening of a fusion pore between docked liposomes, the content volumes rapidly mix and SRB is diluted causing an increase in fluorescence signal intensity (dequenching). SRB efflux and water influx (in the following termed “leakage”) also lead to signal increase. Such leakage could either occur passively or as a result of membrane lesions during fusion. Thus, we refer to this experiment as “apparent CM” (Fig. 6, green curve) and for each experiment a leakage control was performed in parallel. For these controls both populations contained SRB at the same concentration and content mixing thus gives rise to zero dilution. Any observed signal increase must, in this case, be due to SRB-dilution via leakage processes (Fig. 6, grey curve). In another control, the measurement of the apparent CM was repeated with non-complementary LiNAs (LiNA-1 + LiNA-1, Fig. 6, blue curve). All fusion experiments were carried out at 20, 37 and 50 °C and in independent duplicates. The approx. number of LiNA strands on the SRB-labeled and unlabeled liposomes were 195 and 65, respectively, and the populations were mixed in a 1:3 ratio. This allows each SRB-labeled liposome to statistically fuse with up to three unlabeled ones, diluting encapsulated [SRB] from 20 mM to 10 mM, 6.7 mM and 5 mM, after the first, second and third round, respectively.
Figure 6.
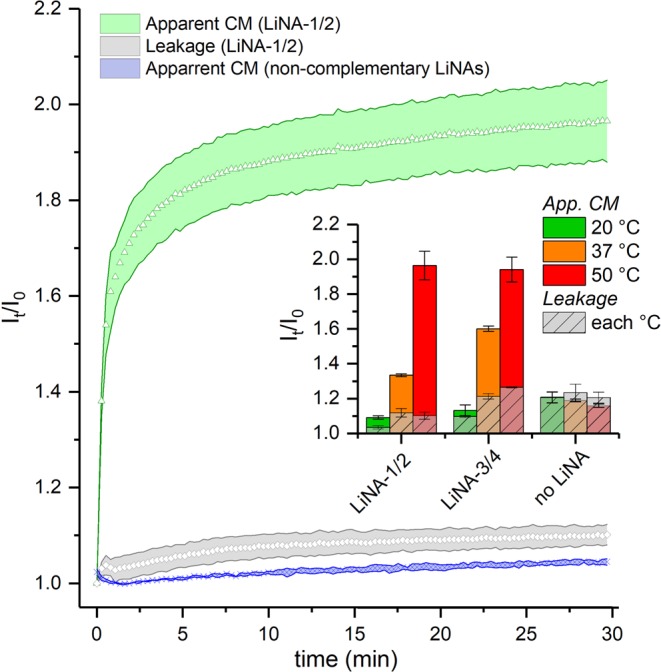
Content mixing during fusion experiments. Increase in relative fluorescence during time course experiments upon mixing populations functionalized with LiNA-1/2 (Apparent CM, green) including controls, i.e. leakage during fusion (grey) and apparent CM when LiNAs are non-complementary (blue). Obtained at 50 °C, ratio labeled/unlabeled liposomes 1:3, ~195 and 65 LiNA/liposome, respectively. The inset summarizes the observed data with complementary LiNAs, or in absence of LiNAs, illustrating contribution of leakage to the observed signal. Error bars: total error between duplicates.
In addition to measuring CM, the particle size distribution was measured at intervals throughout the course of the experiment. This was done by taking aliquots from a fusion experiment using only unlabeled liposomes and diluting these approx. 50-fold in buffer at room temperature. The samples were then analyzed using nanoparticle tracking analysis (NTA, see Figures S4 and S5).
As previously observed, the LiNAs mediated fusion most efficiently at 50 °C as summarized in Fig. 6. The inset illustrates the amount of leakage that contributed to the apparent CM signal, from which it is clear, that in absence of LiNAs the signal increase was due to background processes entirely (see Figure S6 for time course overlay with control experiments). At the same time, the presence of non-complementary LiNAs only gave rise to a minute signal increase (Fig. 6, blue curve) suggesting that engrafting liposomes with polyaza crown ether modified LiNAs had the effect of decreasing passive SRB permeability as observed previously for the anchors with a 3-aminopropane-backbone12,13. As hypothesized earlier, this effect is likely due to the charge repulsion between liposomes by the polyanionic DNA on the liposome surface12,42.
In the current study, elevated leakage was observed for LiNAs with a P3 spacer (LiNA-3/4), where both leakage and content mixing scaled with temperature, in comparison to LiNA-1/2, where leakage remained low. In previous LiNA systems, based on a simpler anchor structure (3-aminopropane-1,2-diol)12, the presence of a P3 spacer did not lead to an increased leakage. Based on this result, we speculate, that the spacer can affect membrane stability.
After correcting for leakage contribution to the apparent CM, the resulting net signal was used to estimate the percentage of full fusion events of SRB-filled liposomes relative to the total number of SRB-filled liposomes, hereafter termed fusion yield. To this end, a calibration curve relating the net I/I0 to the remaining fraction of liposomal SRB-concentration (χSRB) follows:
where is the intensity of a series of samples with different χSRB (χSRB = 0.8, 0.5, 0.33; corresponding to [SRB] 16 mM, 10 mM or 6.7 mM, respectively) divided by the intensity at the initial SRB concentration ([SRB]0 = 20 mM); a and b are linear coefficients and were found to be independent on the temperature at which the samples were recorded (see Methods and Ref.13). The fusion yield was then defined as following:
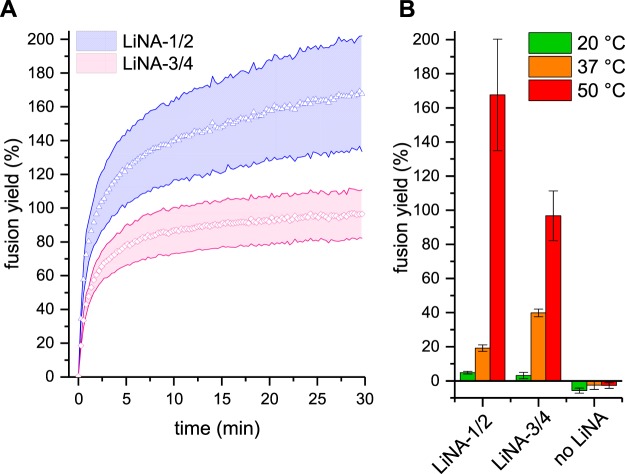
As indicated above, yields >100% can occur, which signifies that labeled liposomes on average fused with more than one unlabeled liposome. The fusion yields at 50 °C are plotted as a function of time in Fig. 7A, and the yields at 30 min are summarized in Fig. 7B.
Figure 7.
Calculated fusion yield. Fusion yield of SRB-labeled population after leakage correction and applying standard curve data of a series of liposome-encapsulated SRB (see SI). (A) Time course experiments depicting fusion yield over time for XE (LiNA-1/2) and XE-P3 (LiNA-3/4) systems at 50 °C; (B) Summary of calculated fusion yields after 30 min at the indicated temperatures. Error bands/bars represent total error between duplicate experiments.
The temperature dependence of LiNA-induced membrane fusion; ~5% fusion yield at 20 °C, 20-40% at 37 °C and 95-170% at 50 °C (Fig. 7B); is congruent with the trend of increasing spontaneous fusion with temperature29. Temperature increase has a two-fold effect: i) increasing the energy of thermal collisions between membranes brought in proximity by the hybridized LiNAs and ii) decreasing the interfacial tension of the membrane due to an increased surface area – each lipid simply has a higher effective surface area43. The consequence of this effect was demonstrated by Parolini et al. using giant liposomes that were tethered to each other via long oligonucleotides: microscopy images showed that the contact area between the liposomes increased with temperature, without changing the number of available tethers44. For LiNA induced fusion, this finding suggests that higher temperatures support membrane deformation, increasing inter-liposome contact area and the number of LiNA duplexes acting cooperatively, prolonging contact time, again amplifying the number thermal collisions between liposomes. Furthermore, decreased surface tension is supportive of extreme membrane curvature, i.e., negative during fusion stalk formation and highly positive during pore opening45,46. A similar trend was observed by Sadek et al. in the membrane fusion mediated by a β-peptide nucleic acid based SNARE-mimic47.
In the present study, the more rigid pair LiNA-1/2 exhibited a higher maximal fusion yield (~170%) when compared to LiNA-3/4 (~95%), a result that might be explained by the shorter intermembrane distance LiNA-1/2 system, than in presence of the two P3 spacers. In contrast, the apparent CM signal was comparable between these systems, which prompts us to speculate whether the triethylene glycol linkers in the LiNA-3/4 play a role in the process from docking to fusion. If the linkers are innocent, the observed increase in leakage could be explained by a prolonged docked state in a configuration that is not quite as inducive to fusion than in the case of LiNA-1/2. If, however, the ethylene glycol interacts strongly with the lipid headgroups of the membrane, it might increase fusogenicity and permeability by disturbing the hydration layer around the membrane. We looked at the initial fusion rates (dYield/dtt=0) of the studied systems and compared them with other LiNA pairs from previous studies (see Table 1). The data suggests that the addition of a spacer slowed down the observed fusion rate both in case of the aza-crown ether and the aminopropane backbone.
Table 1.
Comparison of fusion yield and initial rates at 50 °C.
| Strands | Anchor Design | Fusion yield at 30 min (%)b | Initial fusion rate (%/min)b,c |
|---|---|---|---|
| LiNA-1/2 | T-XE-17nt | 170 ± 25 | 110 ± 10 |
| LiNA-3/4 | T-XE-P3-17nt | 95 ± 10 | 65 ± 5 |
| LiNA-ref1/ref2a | T-XN-17nt | 105 ± 5 | 85 ± 5 |
| LiNA-ref3/ref4a | T-XN-P3-17nt | 280 ± 50 | 70 ± 5 |
The initial fusion rate for LiNA-1/2 was found to be very high (100% fusion yield within ~2 minutes). In the presence of the P3 linker (LiNA-3/4), the initial rate was slower. A similar trend was observed for LiNAs with the aminopropane backbone, that is, for LiNA-ref1/ref2 and LiNA-ref3/ref4, respectively (Table 1). The system with P3 and aminopropane backbone (LiNA-ref3/ref4), however, showed the highest fusion yield, i.e. the SRB-labeled population in average fused with 2.8 of 3 equivalents of unlabeled liposomes, as reported earlier, despite the lower initial rate. For the best aza-crown ether anchored setup (LiNA-1/2), the labeled liposomes fused with 1.7 equivalents of unlabeled liposomes on average. Based on these and the current results, we were intrigued to further understand the parameters controlling fusion kinetics. Different anchor designs may benefit from different optimal linker. On the other hand, linker length is inversely proportional to fusion rate. In an elegant array of coiled-coil based fusogens with different linker lengths, Daudey et al39. were able to suggest optimal linker lengths for phospholipid and cholesterol anchors, respectively. Also, for cholesterol-anchored peptides, a slightly shorter linker on one of the fusogens resulted in increased initial rate. Our previous work tested many combinations of linker positions, lengths and anchor structure based on the aminopropane design, were similar trends in fusion efficiency were observed. On the other hand, it is more challenging to rationalize the influence of backbone structure as well as the spacing between and structure of anchor-moiety on fusion efficiency. Nonetheless, the fact that structurally rather different anchor designs could both lead to very efficient fusion underlines the robustness of using LiNAs as a tool for programmable liposome fusion.
Conclusion
Herein we describe the development of new lipid-nucleic acid conjugates (LiNAs) able to efficiently induce liposomal fusion of inner membrane leaflets and subsequently enable efficient content mixing. An improved synthesis of anchor building blocks based on a polyaza crown ether scaffold with flexible late stage introduction of lipid moieties is reported. LiNAs anchored into the outer liposomes leaflet, these anchors provided strong fusogens. When labeled liposomes were fused with 3:1 excess of unlabeled ones, rapid and efficient fusion was observed. The best system provided complete turnover of the labeled population with low leakage after only 2 min when fusion was carried out at 50 °C. While anchors used in this study are structurally rather different from our previously reported LiNAs, the high fusion efficiency at 37 and 50 °C reported in our earlier study was reproduced. The findings underline LiNA-based fusogens as a robust tool for the fusion of biological membranes.
Methods
Materials and general methods
1,2-Dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl) ammonium salt (DPPE-NBD), 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) ammonium salt (DPPE-LissRhod) were purchased from Avanti Polar Lipids. Cholesterol (Chol) was purchased from Riedel-de Haën in extra pure quality. 9-O-Dimethoxytrityl-triethylene glycol 1-[(2-cyanoethyl)-(N,N-diisopropyl)]-phosphoramidite (Spacer 9, modification P3) was purchased from Glen Research and other phosphoramidites from Proligo. All other reagents were purchased from standard suppliers in reagent grade purity or higher. High purity water (MilliQ, Merck Millipore Advantage A10 system) was used throughout. The liposome fusion experiments, thermal denaturation, circular dichroism (CD) and membrane binding experiments were all carried out in a HEPES buffered saline (HBS, 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 100 mM NaCl in water, adjusted to pH 7.0 with a NaOH solution ([Na+] = 108 to 110 mM). Fluorescence spectroscopy was performed on a Varian Cary Eclipse Fluorescence Spectrophotometer equipped with a 4 position multi-cell holder connected to a Varian Temperature Controller (Peltier device heater; regulated with flow of liquid coolant). Hydrodynamic diameters were determined on a Malvern Nanosight LM14 instrument (details follow below).
Synthesis and characterization of lipid-nucleic acid conjugates
Solid-phase automated DNA synthesis was used to synthesize LiNAs, incl. the DMT-protected phosphoramidite building blocks 5a (0.05 M in DCE/ACN, 2:1, 400 µl, 20 min reaction time) either near the 5′- or 3′-end as previously described3. All LiNAs were purified using RP-HPLC after final DMT-cleavage (Table S1) and verified by MALDI-TOF mass spectrometry. Non-modified sequences (Sigma-Aldrich) were used as received. LiNA hybridization was verified by thermal denaturation experiments (Abs. 260 nm (A260), 20-85 °C and by CD spectroscopy. LiNA stock solutions (50% acetonitrile/water) were kept at 4 °C. Prior to fusion experiments, LiNAs were diluted to 1 µM in HBS (based on A260 and calculated extinction coefficients, stored at RT and used within 48 h (1 µM solutions stored at 4 °C showed surface adsorption issues).
General liposome preparation
To produce unlabeled (DOPC/DOPE/Chol; 2:1:1, molar ratio) or membrane-labeled (additionally 0.25% Lissamine-Rhodamine-DPPE) lipid films, stock solutions in chloroform or methanol were mixed and evaporated under a stream of N2 and further dried under high vacuum. The lipid films were rehydrated in buffer or SRB solution (below). The suspension was vortexed and extruded 21x through a 100 nm polycarbonate membrane (Whatman Nucleopore Track-Etched Membranes) using a hand-extruder (Avanti Polar Lipids) to form unilamellar liposomes (stored at room temperature and used within 36 h. The mean diameter was determined to be 134 ± 3 nm by Nanoparticle Tracking Analysis (NTA).
Encapsulation of Sulforhodamine B (SRB)
Liposomes for content mixing and leakage experiments were obtained by rehydrating lipid films HBS containing 20 mM SRB, placed in a bath sonicator for 10 min at 50 °C, extruded as above and used within 36 h (mean particle diameter 135 ± 4 nm). To remove unentrapped dye, 50 µL batches of the suspensions were purified by spin-column size exclusion chromatography (GE Healthcare Illustra Microspin S-200 HR Columns) pre-equilibrated with HBS. Relative concentrations after spin-column purification was determined by fluorimetry using liposomes labeled with 0.25 mol% DPPE-Lissamine Rhodamine. After size exclusion, the following aliquots were mixed and filled to 100 µl with HBS: blank (10 µL Triton X-100 (1 wt%)), reference (10 µL of untreated liposome dispersion, 10 µL Triton X-100 (1 wt%)) and sample (10 µL of spin-column eluate, 10 µL Triton X-100 (1 wt%)). Average fluorescence was measured from five replicates (excitation wavelength (λex), 560 nm; emission wavelength (λem), 583 nm; emission (em.) filter open, excitation/emission (ex./em). slits 5 nm, PMT 550V) and concentration determined from the average intensity (I) between three independent preparations using the formula: c = (Isample − Iblank)/(Ireference − Iblank) × cstock. Measurement in presence of detergent Triton X-100 (i.e. measuring a non-liposomal dispersion) gave the best reproducibility when using 384-well plates).
Sulforhodamine B (SRB) content mixing assay
Upon content mixing (CM) with unlabeled liposomes – or leakage into the outer medium – the dye is diluted, leading to an increase in fluorescence. The measured fluorescence increase is thus based on both content mixing and leakage (Apparent CM) was corrected by a leakage (L) control, where both liposome populations contain the SRB and fluorescence increase must stem from leakage.
Apparent CM (complementary and non-complementary)
SRB-filled liposomes ({SRB}) and unlabeled liposomes ({buffer}) were prepared by incubation with the respective LiNA for 15 min. at room temperature. Samples were equilibrated to the experiment temperature prior to mixing ({SRB}/{buffer} = 1:3)) in a quartz cuvette (500 µl). SRB fluorescence (λex, 545 nm; λem, 583 nm, em. filter 550-1100 nm, ex./em. slits 5 nm, PMT 550V) was monitored for 30 min after population mixing at controlled temperature (cell block as temperature reference, cuvettes pre-heated for ≥ 5 min). Final concentrations: {SRB}, 46 µM total lipid concentration ([lipid] = [DOPC] + [DOPE] + [Chol]), 0.105 µM DNA (encapsulating 20 mM SRB, lipid/DNA ratio 437:1, ~195 strands per 100 nm liposome); {buffer} = 0.138 mM [lipid], 0.105 µM DNA, lipid/DNA ratio 1310:1, ~65 strands per 100 nm liposome).
Leakage
Two populations of SRB-filled liposomes were incubated with complementary LiNAs, equilibrated, mixed and monitored as above. Final concentrations: {SRB}I and {SRB}II both 46 µM [lipid], 0.105 µM DNA (encapsulating 20 mM SRB, lipid/DNA ratio 437:1, ~200 strands per 100 nm liposome)
Calibration of SRB assay
A calibration curve with standard samples with different fractions of the starting concentration of entrapped SRB (χSRB) were measured. When normalizing the signal to the intensity measured at the starting point for content mixing (i.e., I/I0 = Iχ/Iχ=1) a linear relationship to χSRB was obtained. The normalized signal was largely independent on temperature (20, 37, 50 °C) and current liposome concentration (80, 138 and 275 µM total lipid concentration). The giving the effective χSRB was calculated for each sample based on the SRB concentration after lysis in 0.1% w/v Triton X-100 (obtained against the standard curve of unentrapped SRB). The linear regression in the main text was calculated from a concatenate plot of three samples for each χSRB = 1, 0.8, 0.5, 0.33; corresponding to [SRB] 20 mM,16 mM, 10 mM or 6.7 mM, respectively.
Calculation of effective content mixing
Data from fusion experiments was expressed as I/I0, i.e., the fluorescence intensity at time t (It) divided by the initial intensity (I0). Then, the It/I0 values for leakage (L) signal were subtracted from It/I0 for apparent CM giving the net. fluorescence increase (CM - L). The calibrated content mixing was calculated as χSRB = (2.38 - It/I0)/1.39 using the above linear relation. The fusion yield was calculated as described in the text.
Nanoparticle Tracking Analysis (NTA) Size distributions
The size distributions of freshly extruded liposomes (DOPC, DOPE, Cholesterol (Chol); 2:1:1, molar ratio) and samples from fusion experiments were analyzed using a NanoSight (Wiltshire, UK) instrument, equipped with the NanoSight LM14 flow-cell and laser assembly (405 nm diode laser), a 20 × objective and CCD camera. Data was recorded and analyzed using the NTA v. 2.3 software (Recording: 5 × 30 s with Shutter 387 (25 fps), Gain 200-350, Histogram: 520 lower threshold); Analysis: Detection threshold 10, Blur ‘auto’, Min. Track Length 10, Min. expected size ‘auto’). Liposome preparations were measured at 2 or 5 µM total lipid concentration.
NTA-monitored fusion
Samples were prepared identically as for content mixing. Plain liposomes were used for both populations a 1:1 ratio (I, 138 µM [lipid] with 0.105 µM LiNA-1 and II, 138 µM [lipid] with 0.105 µM LiNA-2, lipid/DNA ratio 1310:1, 65 strands per 100 nm liposome, 15 min incubation at room temperature). The two different liposome populations were pre-heated to 50 °C, then mixed and incubated for to 30 min at 50 °C. At 1, 5, 10, 15 and 30 min, aliquots of the fusion mixtures were transferred and diluted [lipid] = 5 µM (55-fold dilution) in buffer at 20 °C and analyzed as immediate as possible. The strong dilution effectively slowed down further fusion and aggregation processes. The t0 sample corresponds a preparation with unpaired oligonucleotide (LiNA-1) after 15 min incubation. A non-complementary control (LiNA-1/1) was prepared and analyzed after 30 min at 50 °C.
Surface Plasmon Resonance (SPR) Binding Assay
SPR sensorgrams were recorded on a BiacoreTM X100 system using L1 sensor chip (GE Lifesciences) at 25 °C. A method for immobilizing lipidated DNA, introduced by Makishi et al., was used with modified conditioning40. The conditioning of the L1 chip was done by four injections of iPrOH/50 mM NaOH 4:6 (120 s, flow rate: 10 µL/min). After the injection and immobilization of plain liposomes (DOPC, DOPE, Chol, 2:1:1, molar ratio, nominal diameter 100 nm, 1 mM, 1200 s, flow rate: 2 µL/min). Excess liposome material was removed by increasing the flow rate of the running buffer to 10 µL/min for 300 s to achieve a stable baseline (treatment with dilute alkaline was avoided due to the presence of ionizable DOPE). The lipidated DNA was injected (5 µM either LiNA-1 or LiNA-3, or running buffer (negative control); 600 s, flow rate: 2 µL/min), followed by a flow of running buffer to enable dissociation (800 s, flow rate: 2 µL/min). Subsequently, an unmodified complementary or non-complementary DNA strand was injected (A’, 5 µM, 600 s, flow rate: 2 µL/min) After a dissociation time (800 s, flow rate: 2 µL/min), the L1 chip was regenerated by two injections of 50 mM NaOH in 50% v/v isopropanol/water (20 µL, flow rate: 10 µL/min).
Supplementary information
Lipidated Polyaza Crown Ethers as Membrane Anchors for DNA-Controlled Content Mixing between Liposomes
Acknowledgements
The authors gratefully acknowledge funding by the Biomolecular Nanoscale Engineering Center (BioNEC), a Centre of Excellence funded by The VILLUM Foundation, grant no. VKR022710.
Author Contributions
P.M.G.L, A.H.H., O.R. U.J. and S.V. Designed the study. P.M.G.L, A.H.H., O.R. A.R. K. T. S., and K. G. carried out the experiments. P.M.G.L, A.H.H. and S.V analyzed the data and wrote the manuscript. All authors have reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-49862-y.
References
- 1.Gissot A, Camplo M, Grinstaff MW, Barthelemy P. Nucleoside, nucleotide and oligonucleotide based amphiphiles: a successful marriage of nucleic acids with lipids. Organic & Biomolecular Chemistry. 2008;6:1324–1333. doi: 10.1039/B719280K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schade M, Berti D, Huster D, Herrmann A, Arbuzova A. Lipophilic nucleic acids — A flexible construction kit for organization and functionalization of surfaces. Advances in Colloid and Interface Science. 2014;208:235–251. doi: 10.1016/j.cis.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 3.Jakobsen U, Simonsen AC, Vogel S. DNA-Controlled Assembly of Soft Nanoparticles. Journal of the American Chemical Society. 2008;130:10462–10463. doi: 10.1021/ja8030054. [DOI] [PubMed] [Google Scholar]
- 4.Jakobsen U, Vogel S. Assembly of Liposomes Controlled by Triple Helix Formation. Bioconjugate Chemistry. 2013;24:1485–1495. doi: 10.1021/bc300690m. [DOI] [PubMed] [Google Scholar]
- 5.Vogel, S. In DNA in Supramolecular Chemistry and Nanotechnology (eds Stulz, E. & Clever, H.) 397–409 (John Wiley & Sons, Ltd., 2015).
- 6.Jakobsen U, Vogel S. DNA-Controlled Assembly of Liposomes in Diagnostics. Methods in Enzymology. 2009;464:233–248. doi: 10.1016/S0076-6879(09)64012-X. [DOI] [PubMed] [Google Scholar]
- 7.Höök F, et al. Supported lipid bilayers, tethered lipid vesicles, and vesicle fusion investigated using gravimetric, plasmonic, and microscopy techniques. Biointerphases. 2008;3:FA108–FA116. doi: 10.1116/1.2948313. [DOI] [PubMed] [Google Scholar]
- 8.Stengel G, Zahn R, Höök F. DNA-Induced Programmable Fusion of Phospholipid Vesicles. Journal of the American Chemical Society. 2007;129:9584–9585. doi: 10.1021/ja073200k. [DOI] [PubMed] [Google Scholar]
- 9.Stengel G, Simonsson L, Campbell RA, Hook F. Determinants for membrane fusion induced by cholesterol-modified DNA zippers. J Phys Chem B. 2008;112:8264–8274. doi: 10.1021/jp802005b. [DOI] [PubMed] [Google Scholar]
- 10.Chan YHM, van Lengerich B, Boxer SG, Lipid-anchored DNA. mediates vesicle fusion as observed by lipid and content mixing. Biointerphases. 2008;3:Fa17–Fa21. doi: 10.1116/1.2889062. [DOI] [PubMed] [Google Scholar]
- 11.Meng Zhuojun, Yang Jian, Liu Qing, de Vries Jan Willem, Gruszka Agnieszka, Rodríguez-Pulido Alberto, Crielaard Bart J., Kros Alexander, Herrmann Andreas. Efficient Fusion of Liposomes by Nucleobase Quadruple-Anchored DNA. Chemistry - A European Journal. 2017;23(39):9391–9396. doi: 10.1002/chem.201701379. [DOI] [PubMed] [Google Scholar]
- 12.Ries O, Löffler PMG, Rabe A, Malavan JJ, Vogel S. Efficient liposome fusion mediated by lipid-nucleic acid conjugates. Organic & Biomolecular Chemistry. 2017;15:8936–8945. doi: 10.1039/C7OB01939D. [DOI] [PubMed] [Google Scholar]
- 13.Löffler PMG, et al. A DNA-Programmed Liposome Fusion Cascade. Angewandte Chemie International Edition. 2017;56:13228–13231. doi: 10.1002/anie.201703243. [DOI] [PubMed] [Google Scholar]
- 14.Nickel W, et al. Content mixing and membrane integrity during membrane fusion driven by pairing of isolated v-SNAREs and t-SNAREs. Proc Natl Acad Sci U S A. 1999;96:12571–12576. doi: 10.1073/pnas.96.22.12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jahn R, Lang T, Sudhof TC. Membrane fusion. Cell. 2003;112:519–533. doi: 10.1016/S0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- 16.Schuette CG, et al. Determinants of liposome fusion mediated by synaptic SNARE proteins. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:2858–2863. doi: 10.1073/pnas.0400044101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weber T, et al. SNAREpins: Minimal Machinery for Membrane Fusion. Cell. 1998;92:759–772. doi: 10.1016/S0092-8674(00)81404-X. [DOI] [PubMed] [Google Scholar]
- 18.Marsden HR, Elbers NA, Bomans PHH, Sommerdijk NAJM, Kros A. A Reduced SNARE Model for Membrane Fusion. Angewandte Chemie International Edition. 2009;48:2330–2333. doi: 10.1002/anie.200804493. [DOI] [PubMed] [Google Scholar]
- 19.Zheng T, et al. Controlling the rate of coiled coil driven membrane fusion. Chemical Communications. 2013;49:3649–3651. doi: 10.1039/C3CC38926J. [DOI] [PubMed] [Google Scholar]
- 20.Hernandez JM, et al. Membrane fusion intermediates via directional and full assembly of the SNARE complex. Science. 2012;336:1581–1584. doi: 10.1126/science.1221976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lygina AS, Meyenberg K, Jahn R, Diederichsen U. Transmembrane Domain Peptide/Peptide Nucleic Acid Hybrid as a Model of a SNARE Protein in Vesicle Fusion. Angew. Chem. Int. Ed. 2011;50:8597–8601. doi: 10.1002/anie.201101951. [DOI] [PubMed] [Google Scholar]
- 22.Wehland Jan-Dirk, Lygina Antonina S., Kumar Pawan, Guha Samit, Hubrich Barbara E., Jahn Reinhard, Diederichsen Ulf. Role of the transmembrane domain in SNARE protein mediated membrane fusion: peptide nucleic acid/peptide model systems. Molecular BioSystems. 2016;12(9):2770–2776. doi: 10.1039/C6MB00294C. [DOI] [PubMed] [Google Scholar]
- 23.Rabe A, Löffler PMG, Ries O, Vogel S. Programmable fusion of liposomes mediated by lipidated PNA. Chemical Communications. 2017;53:11921–11924. doi: 10.1039/C7CC06058K. [DOI] [PubMed] [Google Scholar]
- 24.Marsden HR, Tomatsu I, Kros A. Model systems for membrane fusion. Chemical Society Reviews. 2011;40:1572–1585. doi: 10.1039/C0CS00115E. [DOI] [PubMed] [Google Scholar]
- 25.Ma M, Bong D. Controlled fusion of synthetic lipid membrane vesicles. Acc. Chem. Res. 2013;46:2988–2997. doi: 10.1021/ar400065m. [DOI] [PubMed] [Google Scholar]
- 26.Hernandez JM, Kreutzberger AJ, Kiessling V, Tamm LK, Jahn R. Variable cooperativity in SNARE-mediated membrane fusion. Proc Natl Acad Sci U S A. 2014;111:12037–12042. doi: 10.1073/pnas.1407435111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuhrmans M, Marelli G, Smirnova YG, Müller M. Mechanics of membrane fusion/pore formation. Chemistry and Physics of Lipids. 2015;185:109–128. doi: 10.1016/j.chemphyslip.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 28.Francois-Martin C, Rothman JE, Pincet F. Experimental Measurement of the Activation Energy of Phospholipid Membrane Fusion. Biophysical Journal. 2016;110:520a–520a. doi: 10.1016/j.bpj.2015.11.2778. [DOI] [Google Scholar]
- 29.Francois-Martin C, Rothman JE, Pincet F. Low energy cost for optimal speed and control of membrane fusion. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:1238–1241. doi: 10.1073/pnas.1621309114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang J, et al. Drug Delivery via Cell Membrane Fusion Using Lipopeptide Modified Liposomes. ACS Central Science. 2016;2:621–630. doi: 10.1021/acscentsci.6b00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oude Blenke EE, van den Dikkenberg J, van Kolck B, Kros A, Mastrobattista E. Coiled coil interactions for the targeting of liposomes for nucleic acid delivery. Nanoscale. 2016;8:8955–8965. doi: 10.1039/C6NR00711B. [DOI] [PubMed] [Google Scholar]
- 32.Yang J, et al. Application of Coiled Coil Peptides in Liposomal Anticancer Drug Delivery Using a Zebrafish Xenograft Model. Acs Nano. 2016;10:7428–7435. doi: 10.1021/acsnano.6b01410. [DOI] [PubMed] [Google Scholar]
- 33.Sun L, et al. Guiding protein delivery into live cells using DNA-programmed membrane fusion. Chemical Science. 2018;9:5967–5975. doi: 10.1039/C8SC00367J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ries O, Löffler PMG, Vogel S. Convenient synthesis and application of versatile nucleic acid lipid membrane anchors in the assembly and fusion of liposomes. Organic & Biomolecular Chemistry. 2015;13:9673–9680. doi: 10.1039/C5OB01207D. [DOI] [PubMed] [Google Scholar]
- 35.Lehn JM, Vierling P. The [18]-N3O3 aza-oxa macrocycle: a selective receptor unit for primary ammonium cations. Tetrahedron Letters. 1980;21:1323–1326. doi: 10.1016/S0040-4039(00)74566-7. [DOI] [Google Scholar]
- 36.Vogel, S., Rohr, K., Dahl, O. & Wengel, J. A substituted triaza crown ether as a binding site in DNA conjugates. Chemical Communications, 1006–1007, 10.1039/B301100C (2003). [DOI] [PubMed]
- 37.Rohr K, Vogel S. Polyaza Crown Ethers as Non-Nucleosidic Building Blocks in DNA Conjugates: Synthesis and Remarkable Stabilization of dsDNA. ChemBioChem. 2006;7:463–470. doi: 10.1002/cbic.200500392. [DOI] [PubMed] [Google Scholar]
- 38.Anaya M, Kwak M, Musser AJ, Müllen K, Herrmann A. Tunable Hydrophobicity in DNA Micelles: Design, Synthesis, and Characterization of a New Family of DNA Amphiphiles. Chemistry – A European Journal. 2010;16:12852–12859. doi: 10.1002/chem.201001816. [DOI] [PubMed] [Google Scholar]
- 39.Daudey GA, Zope HR, Voskuhl J, Kros A, Boyle AL. Membrane-Fusogen Distance Is Critical for Efficient Coiled-Coil-Peptide-Mediated Liposome Fusion. Langmuir. 2017;33:12443–12452. doi: 10.1021/acs.langmuir.7b02931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Makishi S, Shibata T, Okazaki M, Dohno C, Nakatani K. Modulation of binding properties of amphiphilic DNA containing multiple dodecyl phosphotriester linkages to lipid bilayer membrane. Bioorganic & Medicinal Chemistry Letters. 2014;24:3578–3581. doi: 10.1016/j.bmcl.2014.05.042. [DOI] [PubMed] [Google Scholar]
- 41.Versluis F, et al. In Situ Modification of Plain Liposomes with Lipidated Coiled Coil Forming Peptides Induces Membrane Fusion. Journal of the American Chemical Society. 2013;135:8057–8062. doi: 10.1021/ja4031227. [DOI] [PubMed] [Google Scholar]
- 42.Banga RJ, Chernyak N, Narayan SP, Nguyen ST, Mirkin CA. Liposomal Spherical Nucleic Acids. J. Am. Chem. Soc. 2014;136:9866–9869. doi: 10.1021/ja504845f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kučerka N, Nieh M-P, Katsaras J. Fluid phase lipid areas and bilayer thicknesses of commonly used phosphatidylcholines as a function of temperature. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2011;1808:2761–2771. doi: 10.1016/j.bbamem.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 44.Parolini L, et al. Volume and porosity thermal regulation in lipid mesophases by coupling mobile ligands to soft membranes. Nat. Commun. 2015;6:5948. doi: 10.1038/ncomms6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cevc G, Richardsen H. Lipid vesicles and membrane fusion. Advanced Drug Delivery Reviews. 1999;38:207–232. doi: 10.1016/S0169-409x(99)00030-7. [DOI] [PubMed] [Google Scholar]
- 46.Kozlov MM, Chernomordik LV. Membrane tension and membrane fusion. Current Opinion in Structural Biology. 2015;33:61–67. doi: 10.1016/j.sbi.2015.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sadek M, Berndt D, Milovanovic D, Jahn R, Diederichsen U. Distance Regulated Vesicle Fusion and Docking Mediated by β-Peptide Nucleic Acid SNARE Protein Analogues. ChemBioChem. 2016;17:479–485. doi: 10.1002/cbic.201500517. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Lipidated Polyaza Crown Ethers as Membrane Anchors for DNA-Controlled Content Mixing between Liposomes