Abstract
We dissected the genetic basis of total spikelet number (TSN) along with other traits, viz. spike length (SL) and flowering time (FT) in a panel of 518 elite European winter wheat varieties. Genome-wide association studies (GWAS) based on 39,908 SNP markers revealed highly significant quantitative trait loci (QTL) for TSN on chromosomes 2D, 7A, and 7B, for SL on 5A, and FT on 2D, with 2D-QTL being the functional marker for the gene Ppd-D1. The physical region of the 7A-QTL for TSN revealed the presence of a wheat ortholog (TaAPO-A1) to APO1–a rice gene that positively controls the spikelet number on the panicles. Interspecific analyses of the TaAPO-A1 orthologs showed that it is a highly conserved gene important for floral development and present in a wide range of terrestrial plants. Intraspecific studies of the TaAPO-A1 across wheat genotypes revealed a polymorphism in the conserved F-box domain, defining two haplotypes. A KASP marker developed on the polymorphic site showed a highly significant association of TaAPO-A1 with TSN, explaining 23.2% of the total genotypic variance. Also, the TaAPO-A1 alleles showed weak but significant differences for SL and grain yield. Our results demonstrate the importance of wheat sequence resources to identify candidate genes for important traits based on genetic analyses.
Subject terms: Agricultural genetics, Plant genetics
Introduction
The wheat spike and its architecture are key components for improving grain yield. In the recent past, several genes controlling spike morphology have been investigated and described in temperate cereals1,2. Most spike morphological traits in wheat such as spike length and spikelet number behave as quantitative traits, and various QTL and association studies have recently been published3–8. High associations and prediction abilities for total and fertile spikelet number as well as spike length and grain yield were also reported9.
Only a few cloned genes for the trait number of spikelet pairs in wheat are available; among them is the Q-gene which played a major role in wheat domestication and encodes an AP2 transcription factor10. The domesticated allele Q confers a free-threshing character, a sub-compact spike11, and is regulated by microRNA17212. Also, genes related to heading date are involved in spikelet meristem identity determination. For example, the photoperiodism gene Ppd was reported to influence spikelet primordia initiation13. Mutants of the FLOWERING LOCUS T2 (FT2) in wheat showed a significant increase in the number of spikelets per spike with an extended spike development period accompanied by delayed heading time14. Moreover, Ppd-1 and FT were reported as regulators of paired spikelet formation resulting in an increased number of grain-producing spikelets15. Mutants of the MADS-box genes, e.g., VRN1 or FUL2 showed an increased number of spikelets per spike, likely due to a delayed formation of the terminal spikelet16 and a putative ortholog to rice MOC1 regulating axillary meristem initiation and outgrowth was associated with spikelet number per spike in wheat17.
The ABERRANT PANICLE ORGANIZATION 1 (APO1) gene in rice was reported essential for regulating the inflorescence structure by controlling floral organ identity and floral determinacy18,19. APO1 was shown important for maintaining proper inflorescence architecture and spikelet number by preventing precocious conversion of inflorescence meristem to spikelet meristems. On the molecular level, APO1 encodes an F-box protein, an ortholog of UNUSUAL FLORAL ORGAN (UFO) in Arabidopsis, which regulates floral organ identity19–22. Four dominant mutants with elevated expression levels of APO1 produced an increased number of spikelets by a delay in the programmed shift to spikelet formation. Ectopic overexpression of APO1 resulted in increased meristem size caused by different rates of cell proliferation. It was concluded that the level of APO1 activity regulates the inflorescence form through the control of meristematic cell proliferation20.
In the present study, we investigated the inheritance and genetic basis of total spikelet number (TSN) per spike, spike length and flowering time as component traits of grain yield in an elite European winter wheat panel. Our findings show the complex genetic architecture of the investigated traits, and that TaAPO-A1–an ortholog of rice APO1, which is vital for inflorescence development–is associated with the TSN determination in wheat. Intraspecific sequence analyses of TaAPO-A1 revealed that polymorphisms were forming distinct haplotypes while interspecific studies showed the conserved nature of this gene across terrestrial plant species.
Results
Total spikelet number per spike is significantly correlated with spike length, flowering time, and grain yield
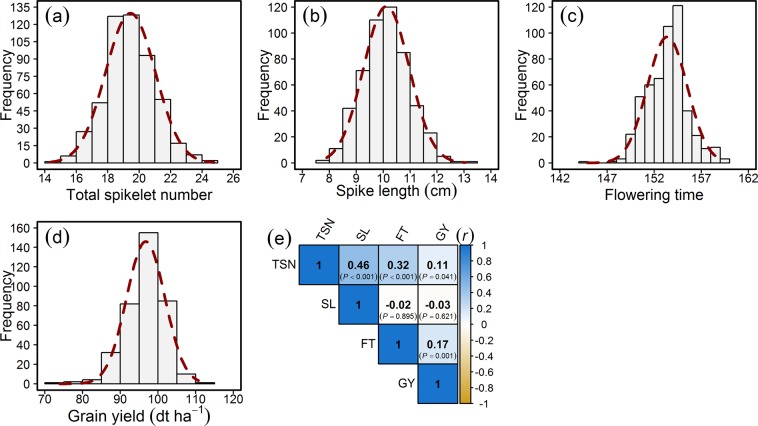
The assessment of total spikelet number (TSN) per spike, spike length (SL), and flowering time (FT) were performed in the field trials on 518 elite European winter wheat varieties (including 15 spring type wheat varieties as an outgroup). The trait grain yield (GY) was assessed in multiple environment field trials on a subset (in total 372) of varieties in a previous study23. The best linear unbiased estimations (BLUEs) of all traits approximated normal distribution and showed wide variation (Fig. 1a–d; Table S1a). The ANOVA showed that genotypic (σG2) and environmental (σE2) variation was significantly (P < 0.001) larger than zero (Table 1). The broad-sense heritability estimates ranging from 0.68 to 0.89 indicated the good quality of the phenotypic data and its potential for use in genome-wide association studies (GWAS) to map the quantitative trait loci (QTL) underlying the traits (Table 1). We analyzed the Pearson’s product-moment correlation (r) among the best linear unbiased estimations (BLUEs) of the investigated traits, which revealed that TSN was positively and significantly correlated with SL, FT, and GY (Fig. 1e). The TSN and SL showed the highest correlation among the investigated traits (r = 0.46; P < 0.001) whereas SL showed almost a null correlation with FT and GY suggesting that FT augments GY mainly by influencing the TSN in wheat.
Figure 1.
Distribution and correlation of the investigated traits in a panel of 518 elite European winter wheat varieties. Distribution of (a) Total spikelet number (TSN) per spike, (b) Spike length (SL), (c) Flowering time (FT), and (d) Grain yield (GY); (e) Pearson’s product moment correlation (r) among the investigated traits. P-value denotes the significance of the respective correlation.
Table 1.
Summary statistics of the investigated traits, namely total spikelet number (TSN) per spike, spike length (SL; cm), flowering time (FT), and grain yield (GY; dt ha−1).
| Parameter | TSN | SL | FT | GY |
|---|---|---|---|---|
| Minimum | 14.38 | 7.90 | 144.96 | 73.94 |
| Mean | 19.45 | 10.12 | 153.38 | 96.74 |
| Maximum | 24.75 | 13.05 | 159.61 | 110.71 |
| σ G 2 | 1.71a | 0.50a | 3.42a | 22.89a |
| σ E 2 | 1.75a | 1.63a | 6.30a | 94.51a |
| σ e 2 | 1.60 | 0.44 | 1.90 | 23.74 |
| H 2 | 0.68 | 0.70 | 0.84 | 0.89 |
| nE | 2 | 2 | 3 | 8 |
σG2 = genotypic variance; σE2 = environmental variance; σe2 = residual variance; H2 = broad-sense heritability; nE = number of environments; a = significant at < 0.001 probability level.
High-density marker arrays reveal the absence of distinct sub-populations and sharp LD decay in European elite winter wheat
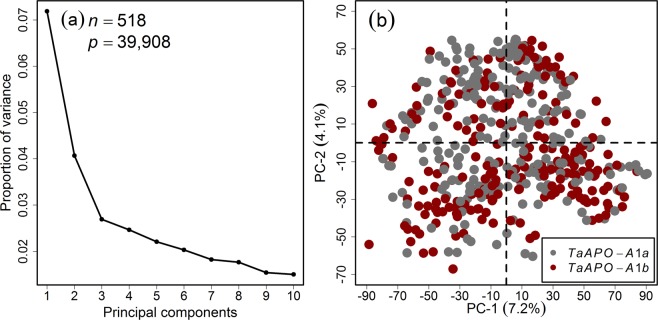
The whole wheat panel was extensively genotyped with high-density SNP arrays and functional markers for the genes Ppd-D1, Rht-B1, Rht-D1, Vrn-A1, Vrn-B1, and Vrn-D1, which resulted in 39,908 high-quality markers. The population structure analyzed with marker genotypes by principal component (PC) analysis resulted in the absence of distinct sub-populations with the first two PCs representing only 11.3% of the variation (Fig. 2). The high familial relatedness and non-existence of distinct sub-populations were further supported by plotting a heat map of the genomic relationships among the wheat varieties (Fig. S1) and by the structure-like inference algorithm LEA, which resulted in the sub-populations being distinguished but with a slight entropy shift. The bar plots indicated admixed and weak sub-populations (Fig. S2).
Figure 2.
Principal component (PC) analysis on the wheat marker loci combined from the 35k and 90k single nucleotide polymorphism arrays. (a) Scree plot showing the first ten PCs and their corresponding proportion of variance, (b) Scatterplot showing the absence of pronounced sub-clustering among the varieties. Different colors represent the TaAPO-A1 alleles. n and p denote the number of varieties and the marker genotypes used in the analysis, respectively.
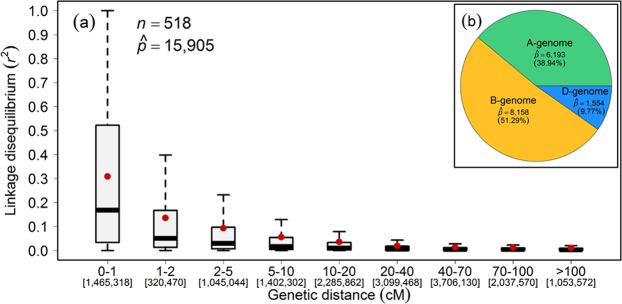
Linkage disequilibrium (LD) between the marker genotypes determines the number of markers needed to perform GWAS. Genome-wide LD analysis was performed with the mapped marker genotypes which resulted in a rapid LD decay by increasing the genetic (cM) distances: first and third quantile dropped to 0.002 and 0.028, respectively, and the mean and median values equaled 0.051 and 0.008, respectively (Fig. 3a). The sub-genome-wise distribution of the markers varied: the highest number of markers mapped on B-genome, followed by A- and D-genomes (Fig. 3b). Although the whole panel was genotyped with state-of-the-art genotyping arrays, the sub-genome-wise distribution of marker genotypes suggests that the marker density could be improved especially for D-genome.
Figure 3.
Genome-wide decay of linkage disequilibrium (LD; r2) as a function of genetic map distance (cM) between the marker loci in the population of European winter wheat varieties. (a) Boxplots represent the LD-decay, (b) Sub-genome-wise distribution of mapped marker loci. Red dots within the boxplots represent the mean. The numbers on second row of x-axis represent the number of marker-pairs present in the corresponding genetic distance. n and denote the number of varieties and mapped marker loci, respectively.
GWAS identifies large-effect QTL for TSN on chromosome 7A in European winter wheat
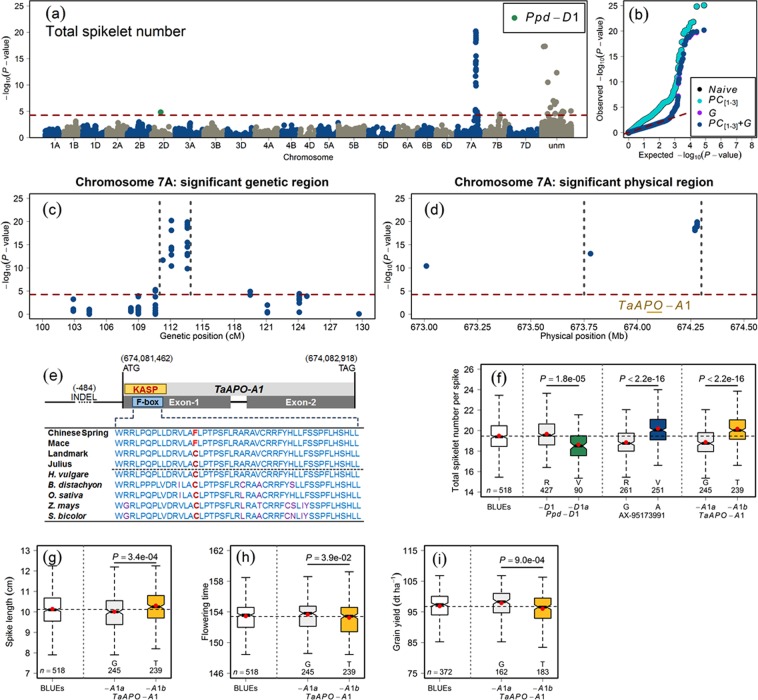
Among the different GWAS models used in our study, we observed that the PC[1–3] + G model could best control the spurious marker-trait associations (MTA). Our GWAS analyses identified QTL on chromosomes 2D, 7A, and 7B for TSN (Fig. 4a,b; Table S2a), for SL on chromosome 5A (Fig. S3, Table S2b), and for FT on chromosome 2D (Fig. S4; Table S2c). The QTL on chromosome 2D identified for TSN and FT was most likely the gene Ppd-D1. The photoperiod insensitive allele Ppd-D1a significantly reduced the TSN in wheat (Fig. 4a,f). The phenotypic data for GY were analyzed to investigate if there exists any significant correlation between the identified marker alleles and GY (Fig. 4i). The total proportion of genotypic variance (pG) imparted by the identified mapped QTL amounted to 65.44% for TSN, 15.15% for SL, and 31.58% for FT. A relatively low pG explained for SL and FT is the result of the identification of only one mapped marker for each trait.
Figure 4.
Summary of the genome-wide association studies (GWAS) of total spikelet number per spike in the population of 518 European winter wheat varieties. (a) Manhattan plot shows the distribution of marker significance −log10(P − value) along the chromosomes. The correction for population stratification and familial relatedness was performed by using the first three principal components (PC[1–3]) and an additive genomic relationship matrix (G) in a linear mixed-effect model. The red dashed line marks the multiple testing criteria of false discovery rate (FDR) <0.05, (b) Quantile-quantile plot showing the distribution of observed versus expected (red dashed line) −log10(P − value). The naïve model represents the GWAS without the correction of population structure, the PC[1–3] model represents the population structure corrected with the first three PCs, the G model represents the familial relatedness corrected with a genomic relationship matrix, and the PC[1–3] + G model represents the population structure and familial relatedness corrected with the first three PCs and the G matrix. The color code for different models is given in the figure legend, (c) Significant genetic region on chromosome 7A for TSN in wheat. The gray vertical dashed lines mark the highly significant genetic region, (d) Significant physical region on chromosome 7A for TSN in wheat. The gray vertical dashed lines mark the highly significant physical region, (e) Gene structure of the TaAPO-A1. The orange box represents the location of the KASP marker developed to exploit the variation in the F-box domain (highlighted in blue color). The horizontal line preceding the first exon depicts the promotor region harboring an INDEL and its corresponding position. The first four rows represent the F-box sequences of wheat varieties (courtesy: The 10+ Wheat Genomes Project) and the second five rows represent the F-box domain of closely related species viz. Hordeum vulgare, Brachypodium distachyon, Oryza sativa, Zea mays, and Sorghum bicolor. The non-synonymous mutation is highlighted in red color. The location of start and stop codons on chromosome 7A are given in the figure, (f) Allele-wise phenotypic distribution of the most significant markers and the KASP marker for TaAPO-A1 associated with (f) TSN, (g) Spike length, (h) Flowering time, and (i) Grain yield. P-value denotes the significance value of the two-sided t-test used to compare the mean value of the marker alleles. In sub-figures (f) to (i), the first boxplots represent the distribution of the best linear unbiased estimations (BLUEs) for the respective trait.
For TSN, we identified 43 MTA: one mapped to chromosome 2D (Ppd-D1), 24 on chromosome 7A, three on chromosome 7B, while 15 MTA were unmapped according to our mapping (ITMI) resources (Table S2a). We used additional consensus maps24,25 to assign the chromosomes to the 15 unmapped markers. By following this approach, we could assign 14 out of 15 unmapped markers (13 to chromosome 7A and one to 7B) whereas one marker was unmapped according to all mapping resources (Table S2a). Moreover, we analyzed the similarity between the chromosomal assignments of our mapped markers with other consensus maps. The chromosomal assignments of our mapped markers corresponded to other maps with a 100% congruence (Table S2a,b). However, although the chromosomes assigned to the SNPs are the same in all mapping resources, the genetic (cM) positions differ–a most likely reason is that the mapping populations used in different resources are dissimilar. Nevertheless, of interest is a large-effect QTL identified for TSN on chromosome 7A–for which the most significant marker AX-95173991 is located at 112.10 cM and explained 25.70% of the total genotypic variance (Fig. 4a,b, Table S2a). This warrants, on the one hand, that the use of 7A-QTL would be beneficial for efficient marker-assisted selection. On the other hand, it made possible the further investigation of 7A-QTL at the physical sequence level to search for candidate genes.
Significant physical region of chromosome 7A-QTL harbors TaAPO-A1–a putative candidate gene for TSN in wheat
The significant 7A-QTL genetic region for TSN spanned initially from 110.6 to 124.1 cM (Table S2a). We narrowed down the genetic region with the highly significant MTA with −log10(P − value) >10 within 2.3 cM starting from 111.3 to 113.6 cM (Fig. 4c). The alignment of marker sequences present within this most significant genetic region onto chromosome 7A revealed a physical region starting from 673.78 to 674.30-Mb (Fig. 4d) that harbored only ten genes (Table S3). The functional annotations of these ten genes revealed an interesting candidate gene TraesCS7A01G481600; (physical map position: 674,081,462–674,082,918-bp) with functional annotation as Aberrant panicle organization 1 (APO1) protein (Table S3). The APO1 in rice regulates inflorescence architecture and positively controls the total spikelet number by suppressing the precocious conversion of inflorescence meristems to spikelet meristems18,19.
A KASP marker developed for the TaAPO-A1 shows significant association with TSN in wheat varieties
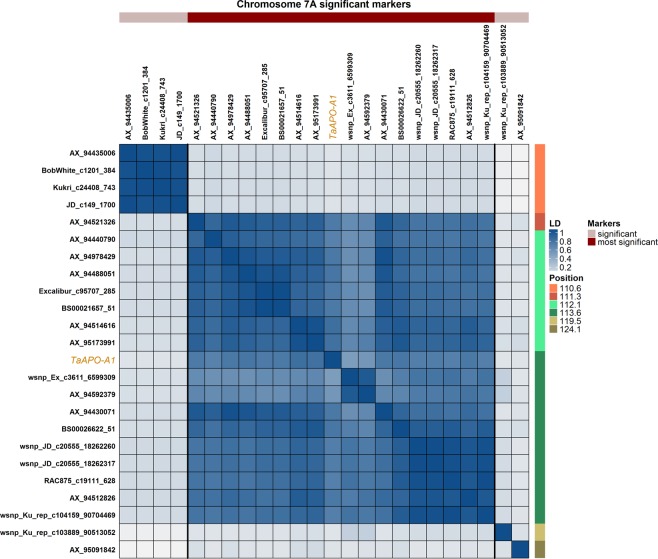
TaAPO-A1 is a 1,457-bp long gene and, like APO1 in rice, it has two exons separated by one intron (Fig. 4e). We investigated the variation of TaAPO-A1 in ten wheat varieties which revealed two haplotypes–the sequences were taken from The 10+ Wheat Genome Project (Figs 4e and S6). The first exon harbors a highly conserved F-box domain of 46 amino acid residues across the wheat varieties and other species (Figs 4e, S6 and S7). Intraspecific sequence analysis of TaAPO-A1 revealed a non-synonymous mutation in the F-box domain: out of ten wheat varieties, four (including Chinese Spring) harbored T while six had G allele. We developed a KASP marker for TaAPO-A1 harboring this non-synonymous mutation in the F-box domain (Table S1b). The KASP marker for TaAPO-A1 was highly significantly associated with TSN (Fig. 4f) and the marker alleles were evenly distributed in the variety panel (Fig. 2b; Table S1a). The second round of GWAS was performed by the TaAPO-A1 KASP marker integrated into the original SNP matrix which further confirmed the significant association of TaAPO-A1 with TSN, explaining 23.21% of the total genotypic variance (Fig. S5; Table S2a). The reference allele in the population (represented by TaAPO-A1a, with nucleotide G translating to cysteine) was present in 50.62% of the investigated varieties and resulted in an average TSN of 18.83, whereas the variant allele (represented by TaAPO-A1b, with nucleotide T translating to phenylalanine) was present in 49.38% of the varieties and revealed an average TSN of 20.13 (Fig. 4f, Table S1a). The analysis of local linkage disequilibrium performed with the markers present in the 7A-QTL genetic region and the KASP marker for TaAPO-A1 showed that TaAPO-A1 was in tight linkage with other markers (Fig. 5). Furthermore, we also observed a rather weak but significant association of the TaAPO-A1 KASP marker alleles with SL, FT, and GY (Fig. 4g–i).
Figure 5.
Pairwise linkage disequilibrium (LD; r2) among the marker loci (including the KASP marker for the gene TaAPO-A1) present in the significant genetic region of TSN on chromosome 7A in wheat. Based on the linkage blocks, markers are divided into two categories viz. significant, and most significant. The color key is given in the figure.
The single nucleotide substitution G (low TSN allele) to T (high TSN allele) in the conserved functional domain of TaAPO-A1 resulted in a non-synonymous amino acid substitution from cysteine (C) to phenylalanine (F). The amino acid cysteine appears to be well conserved across various grass species at this position potentially indicating the conservation of C residue across grasses. However, the SIFT (Sorting Intolerant from Tolerant) score26 analysis showed no potential deleterious effect from C to F substitution at this position (Table S4). We then looked at the promoter region of TaAPO-A1 in ten genotypes from The 10+ Wheat Genome Project and identified a 115-bp INDEL (insertion-deletion) polymorphism at −484-bp upstream of the transcription start site of TaAPO-A1. Interestingly, the low TSN haplotype “G” (coding for cysteine) always had a deletion of 115-bp in the promoter, whereas the high TSN haplotype “T” (coding for phenylalanine) had 115-bp insertion. It, nevertheless, remains to be established via functional studies if this INDEL affects the transcription rate of TaAPO-A1 contributing to the observed phenotypic differences for TSN in two haplogroups.
Phylogenetic analyses show that TaAPO-A1, an ortholog of UFO in Arabidopsis, is conserved across terrestrial plant species
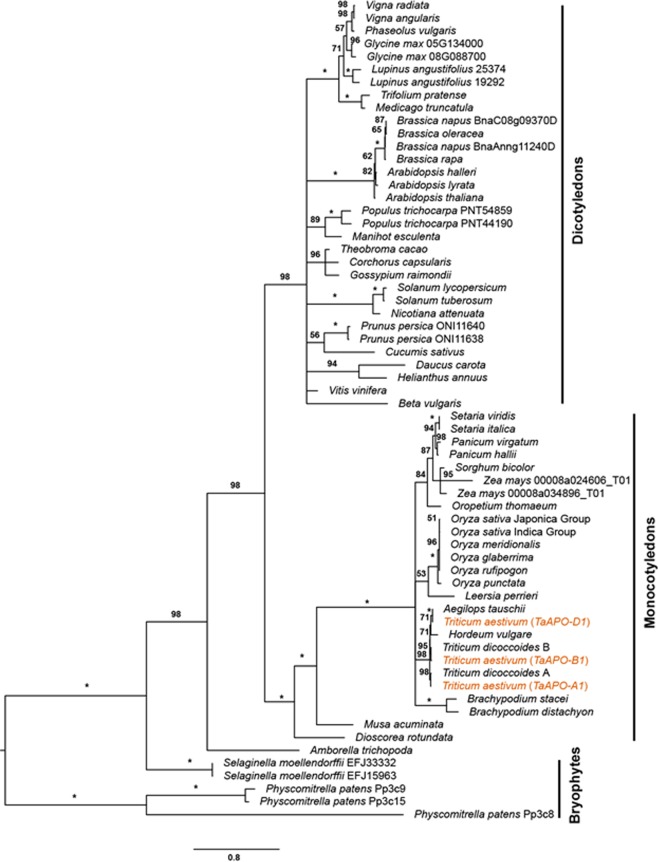
The BLAST search of TaAPO-A1 orthologs across diverse plant species from the EnsemblPlants and the protein databases Phytozome v12.1 retrieved 64 protein sequences from 37 genera (52 species, Table S5) including Bryophytes, monocotyledons, and eudicotyledons. The final alignment consisted of 670 positions. The obtained maximum likelihood (ML) topology reflects the evolution of terrestrial plants with Amborella trichopoda at the base of the two main clades, monocotyledons and eudicotyledons (Fig. 6). The protein is relatively well conserved as seen from the tiny branches especially within the grass tribe Triticeae, including Triticum aestivum and Hordeum vulgare, which diverged about ten million years ago (Ma)27 or even the Poaceae, whose most recent common ancestor probably occurred 50–75 Ma28.
Figure 6.
Maximum likelihood phylogenetic tree of TaAPO-A1 orthologous proteins across terrestrial plant species. Bootstrap values are indicated along the braches. Asterisks indicate >99% bootstrap values. The TaAPO homoeologs are highlighted in orange color. The bars on the right side indicate the major clades. The amino acid substitution scale is indicated at the bottom of the figure.
Discussion
Exploiting significant, heritable genetic variation of TSN as well as a positive correlation with other traits can help to improve the grain yield in wheat
Grain yield (GY) improvement is considered as the top focus of virtually every wheat breeding program. However, an extremely complex genetic nature of GY often hampers its genetic improvement as it is the product of several yield components, e.g., the number of spikes per plant, grains per spike, thousand-grain weight. The number of grains per spike is the product of total spikelet number (TSN) per spike and spikelet fertility. Therefore, an essential consideration in wheat breeding has been to employ a reductionist approach, i.e., to exploit the information about the individual component traits–most of which are negatively associated with each other. In this study, we analyzed a winter wheat panel comprising 518 varieties for GY component traits such as TSN and spike length (SL) along with the flowering time (FT). The GY data based on previous studies were taken for comparison purposes23. In all observed traits, besides significant genetic variation, we observed a significant genotype-by-environment (year) interaction. Nevertheless, the broad-sense heritability estimates ranging from 0.68 to 0.89 suggested that genetic variation is heritable–an essential indicator of high selection response (Table 1). Similar heritability values for the investigated traits have been reported recently in other diverse mapping populations4,6.
In addition to significant genetic variation, TSN showed a positive and significant correlation with SL, FT, and GY (Fig. 1e). A relatively low correlation of TSN with GY in comparison to SL and FT may not be a true reflection of the relationship between TSN and GY since GY data was taken from a study that investigated different number of lines (372) in different sets of environments. In this set of varieties, the Chinese Spring allele coding for “T” was associated with an increase in TSN, but a decrease in GY. However, albeit being weak (which is by virtue of the extreme quantitative genetic nature of GY), TSN’s correlation with GY could help in selection and improve the genetic gains. Moreover, it should be noted that the genetic architecture of yield component traits per se is also important which means that if the component traits possess complex genetic architecture, the problem of grain yield improvement would be further compounded. Nevertheless, a reasonably high heritability value suggests that TSN is strongly genetically inherited and that the mapping of the underlying quantitative trait loci (QTL) would be efficient.
High marker density governs the efficacy of genetic and physical mapping
The efficiency of genome-wide association studies (GWAS) depends on the size of the population and genetic diversity. Therefore, genome-wide marker density with many polymorphic sites is vital, and coupled with a sharp decline in linkage disequilibrium (LD) between the marker loci, it increases the GWAS resolution. In our study, the size of the population, high-density genotyping, and the use of stringent linear mixed-effect models warranted the genetic mapping of true marker-trait-associations (MTA). As noted in another study based on a subset of varieties, the absence of distinct sub-populations in this panel suggests that the European winter wheat varieties have been bred, by and large, from a narrow genetic base and with similar goals29 which is in line with other reports based on studies using similar genetic material but different marker platforms30,31.
To identify the candidate genes, high marker density in the genetic regions of the quantitative trait loci (QTL) is necessary since it helps to narrow down to the physical regions harboring the gene(s) underlying the trait. Moreover, since GWAS hinges on the principle that the markers work as proxies to the genes/QTL underlying the traits, a high density of markers in the QTL genetic region becomes vital for the success of fine mapping. In this study, we exploited this premise to identify a candidate gene physically.
Physical mapping shows that TaAPO-A1 is a likely candidate gene for TSN in wheat
Our GWAS analyses revealed a significant QTL for TSN on chromosome 7A, which explained ~25% of the total genotypic variance. Also, Würschum et al.6, recently reported a QTL for TSN on chromosome 7A in a similar type of elite winter wheat germplasm. Zhang et al.17, reported a putative MOC1 ortholog to be associated with spikelet number, which is also located on chromosome 7A.
The strategy to investigate orthologous genes of rice with the known function was already successfully applied for various genes associated with grain size, grain weight, and yield in wheat32–37. The highly significant region of the detected TSN-QTL in our study corresponded to a physical interval of <1-Mb, containing a block of only ten genes, all in high LD (Fig. 5). Based on the functional annotations, the rice gene ABERRANT PANICLE ORGANIZATION 1 (APO1), an ortholog of Arabidopsis UFO18,19,21,22, was considered as the most likely candidate gene and was named as TaAPO-A1 in wheat. The functional analyses in both rice and Arabidopsis revealed that the F-box containing proteins are involved in the regulation and development of floral organs–more specifically, APO1 in rice controls the number of spikelets per panicle by regulating the cell proliferation in meristems20. Recently, two independent studies based on GWAS and linkage mapping reported APO1 as the best candidate gene affecting the TSN per spike in wheat38,39.
Functional diversity among the orthologs of TaAPO-A1 reveals the conserved F-box domain
The availability of genomic data for several wheat varieties from The 10+ Wheat Genome Project allowed the investigation of the intraspecific diversity of TaAPO-A1 gene. The TaAPO-A1 contains two exons, each containing a SNP which causes an amino acid substitution. In the first exon, a T/G polymorphism at base 140 was related to the exchange of phenylalanine to cysteine, and in the second exon, at base 1,284, a G/A polymorphism mutated aspartic acid to asparagine (Fig. S6). It was possible to develop a functional KASP marker for the SNP in the first exon and screen the whole germplasm panel. Both alleles were present in almost identical frequencies with 49.38% of the varieties carrying the allele of Chinese Spring with nucleotide T (referred to as TaAPO-A1b) and 50.62% of the varieties carrying the G nucleotide (referred to as TaAPO-A1a). The Chinese Spring allele was strongly associated (P < 2.2e-16) with an increase in TSN and moderately associated with an increase in SL (P = 3.4e-04) and a decrease in GY (P = 9.0e-04) (Fig. 4f–i). For the B- and D-genomes, the orthologs of TaAPO-A1 were related to the genes TraesCS7B01G384000 and TraesCS7D01G468700. However, no MTA were discovered on these genomes. The identified TaAPO-A1 variants reflect natural allelic diversity with mild phenotypic effects, which is beneficial for practical breeding.
The presence of TaAPO-A1 orthologs in a wide range of plants including Bryophytes, monocotyledons and eudicotyledons suggests a central role of this gene class in the evolution and development of terrestrial plants (Figs 6 and S7). The Arabidopsis gene UFO and rice APO1 (orthologs of TaAPO-A1) encode for an F-box containing protein. It has been shown that the rice APO1 and Arabidopsis UFO are important for floral development in respective species19,21. Molecularly, the proteins SKP1, cullin like, and F-box containing polypeptides form the SCF protein complexes to function as E3-ubiquitin ligases that target specific proteins for degradation40,41. For example, it was shown that Arabidopsis UFO indirectly regulates the expression of class B floral homeotic gene APETALA 3 by targeting the degradation of proteins which negatively regulate its transcription21. The rice apo1 mutants show a reduction in the number of primary branches and, thereby, the number of spikelets due to the precocious conversion of inflorescence meristem (IM) to spikelet meristem (SM). Such a mutant phenotype offers an indication that APO1 might target proteins that promote the precocious conversion of IM to SM for degradation in a functional state. In line with this idea, the dominant gain of function APO1 alleles with an elevated expression as well as overexpression transgenic lines of APO1 showed prolonged inflorescence development resulting in more branch iterations and consequently more spikelets20.
From our promoter analysis, we found an INDEL where the 115-bp insertion was always associated with high TSN haplotype, whereas the deletion with low TSN haplotype. From this finding, it may be inferred that winter wheat genotypes in the haplogroup with insertion polymorphism have slightly elevated expression of TaAPO-A1 leading to prolonged maturation of inflorescence meristem and eventually producing more spikelets per spike. Conversely, the deletion haplotype has a comparatively reduced expression level of TaAPO-A1, leading to less number of spikelets. Nevertheless, validation of the INDEL haplotype across the whole winter wheat panel as well as expression analysis of TaAPO-A1 in the two haplogroups with high and low TSN may offer further insights into the regulation of TSN in wheat.
Conclusions
Our results demonstrate that with the availability of modern genomic tools such as the wheat reference sequence and the access to The 10+ Wheat Genome Project, the way from phenotype to a candidate gene is shortened considerably. Nevertheless, robust genetic analyses including appropriate mapping populations, accurate and high-density genotyping, and proper phenotypic analyses are prerequisites to detecting significant QTL regions from which the causative genes could be deduced.
Materials and Methods
Phenotypic data analyses
The data for total spikelet number (TSN), spike length (SL), and flowering time (FT) were collected on an elite European winter wheat panel comprising of 518 varieties. The whole panel was grown in the experimental fields of Leibniz Institute of Plant Genetics and Crop Plant Research (IPK) Gatersleben, Germany in plots of 2 m2 as single replication in three cropping seasons (2015/16; 2016/17; and 2017/18), henceforth called environments. The traits TSN and SL were recorded in two environments (2016/17 and 2017/18) from ten spikes per plot as the total number of spikelets and spike length in centimeters (cm) from basal spikelet to the top of a spike by excluding the awns. The arithmetic mean of TSN and SL from ten spikes were calculated to represent the phenotypic value of traits in the individual environments. Flowering time was recorded in all three environments by counting the number of days from the first of January to when approximately half of the spikes in a plot flowered. The phenotypic data for grain yield estimated in eight environments were taken from the previous study for comparison purposes23. A linear mixed-effect model was used for across environment phenotypic data analysis as:
where, yik is the phenotypic record of the ith genotype in the jth environment, μ is the common intercept term, Gi is the effect of the ith genotype, Ej is the effect of the jth environment, and eij denotes the corresponding error term. All effects, except the intercept, were assumed to be random to calculate the individual variance components. The broad-sense heritability (H2) was calculated as:
where, σG2 and σe2 denote the variance components of the genotype and the error, respectively; nE denotes the number of environments. To calculate the best linear unbiased estimations (BLUEs), the intercept and the genotypic effects were assumed to be fixed in the above model.
Genotypic data analyses, population structure, and linkage disequilibrium
All 518 varieties were extensively genotyped with the 35k Affymetrix and 90k iSELECT single nucleotide polymorphism (SNP) arrays24,25 which generated in total 116,730 SNP markers (35k = 35,143; 90k = 81,587). Moreover, we genotyped the whole panel with functional markers for the candidate genes such as photoperiodism (Ppd-D1), reduced height (Rht), and vernalization (Vrn1). The quality of the marker data was improved by removing the markers harboring >10% heterozygous or missing calls and markers with a minor allele frequency of <0.05. The mean of both alleles imputed the remaining missing data. The quality control resulted in a total of 39,908 markers, which were used in subsequent analyses.
Population structure based on marker genotypes was examined by principal component (PC) analysis. The first two PCs were drawn to see the sub-clustering among varieties. Furthermore, the genetic relatedness among varieties was evaluated by an additive variance-covariance genomic relationship matrix. To infer the hidden population sub-structuring, an inference algorithm LEA (Landscape and Ecological Association Studies) was used by assuming ten ancestral populations (K = 1–10). The function snmf, which provides the least-squares estimates of ancestry proportions and estimates an entropy criterion to evaluate the quality fit of the model by cross-validation, was used. The number of ancestral populations best explaining the data can be chosen by using the entropy criterion. We performed ten repetitions for each K, and the optimal repetition demonstrating the minimum cross-entropy value was used to visualize clustering among varieties via bar plots42.
Linkage disequilibrium (LD), the non-random association of alleles at different loci, was measured as the squared correlation (r2) among markers. The genetic mapping positions of the markers for both arrays were adopted from the data generated for the International Triticeae Mapping Initiative (ITMI) DH population, as described in Sorrells et al.43. Although inter and intra-chromosomal LD among the loci varies, genome-wide calculation of LD gives a global estimate about the genetic map distance over which the LD decays in a given population. The genome-wide (global) LD was calculated only from the mapped markers.
Genome-wide association studies
Genome-wide association studies (GWAS) were performed on data taken from the individual environment and SNPs passing the quality criteria plus the functional gene markers. Let n be the number of varieties and p be the predictor marker genotypes. A standard linear mixed-effect model following Yu et al.44, was used to perform GWAS as:
where, y is the n × 1 vector of phenotypic record of each genotype in each environment, μ is the common intercept, τ, β, v, u and e are the vectors of the environment, marker, population (principal components), polygenic background, and the error effects, respectively; E, X, P and Z are the corresponding design matrices. In the model, μ, τ, β and v were assumed to be fixed while u and e as random with u ~ N(0,Gσa2), and e ~ N(0,Iσe2). The n × n variance-covariance additive relationship matrix (G) was calculated from n × p matrix W = (wik) of marker genotypes (being 0, 1 or 2) as where, wik and wjk are the profiles of the kth marker for the ith and jth variety, respectively; pk is the estimated frequency of one allele in kth marker, as described by VanRaden45.
As population stratification and familial relatedness can severely impact the power to detect true marker-trait association (MTA) in GWAS, different statistical models were used to avoid spurious MTA viz., (1) general linear model (naive), (2) population structure correction via principal components (PCs), (3) correction of familial relatedness via genomic relationship matrix (G), and (4) correction of population structure and relatedness via PCs and G. It is expected that using both PCs and G in the model can enhance the accuracy of GWAS. Along with this, environmental fixed effects were assigned in all model scenarios. The models described above were compared by plotting the expected versus the observed in a quantile-quantile plot and the best model was determined by checking how well the observed aligned with the expected.
To declare the presence of MTA, a false discovery rate (FDR) < 0.05 to account for multiple testing was applied46. Following Utz et al.47, the percentage of total genotypic variance (pG) explained by all the QTL passing the FDR threshold was determined as pG = [Radj2/H2] × 100 where, Radj2 was calculated by fitting all the MTA in a multiple linear regression model in the order of ascending P-values and H2 is the broad-sense heritability. The pG values of individual QTL were accordingly derived from the sum of squares of the QTL (SSQTL) in the linear model.
Candidate gene identification, haplotype analysis by exploiting resources from The 10+ Wheat Genome Project, and the KASP marker development
We narrowed-down the QTL region, and BLASTed sequences of all the significant markers present within the genetically defined region onto the physical map of the corresponding chromosome of the reference sequence of the wheat genome which yielded significant physical region48,49. Afterward, the gene identifiers (gene-IDs) present within the physical region and their annotated functional descriptions were retrieved. Among them was a most likely candidate gene TaAPO-A1 for TSN.
The 10+ Wheat Genome Project is an international collaborative effort that aims to assemble the genomes of more than ten wheat varieties bred in different countries to characterize the wheat pan-genome (http://www.10wheatgenomes.com/). We retrieved the genomic sequence of TaAPO-A1 for ten wheat varieties from The 10+ Wheat Genome Project and aligned the sequences to observe the haplotype structures. The SNP that revealed a clear haplotype structure was used to design a Kompetitive Allele Specific PCR (KASP) marker in the candidate gene. The allele-wise phenotypic distribution of the investigated traits with the gene-specific KASP marker was analyzed by plotting the boxplots. The significance (P-values) between the mean values of genotypes harboring different KASP marker alleles was determined by two-sided t-test. Moreover, we performed a second round of GWAS by incorporating the gene-specific KASP marker in the original SNP matrix to determine whether it associates with the phenotypes. The GWAS parameters were kept the same as described above.
Multiple sequence alignment and phylogenetic analyses
The TaAPO-A1 protein sequence (corresponding to TraesCS7A01G481600) was used as a BLAST query to retrieve the monocot, dicot and Bryophyte orthologs from EnsemblPlants (http://plants.ensembl.org/index.html) and Phytozome v12.1 (https://phytozome.jgi.doe.gov/pz/portal.html) databases. The orthologous protein sequences were aligned using ClustalW in Geneious v11.0.550. The protein alignment was used to infer a maximum likelihood (ML) phylogeny. The JTT matrix51 was identified as the best-fitting model of protein evolution with ProtTest 352,53 and the Akaike Information Criterion (AIC). The evolutionary history among TaAPO-A1 orthologs across various plant species was inferred using RAxML v8.2.1254 with PROTGAMMAJTT model, rapid bootstrapping of 100 replicates, and search for best-scoring ML tree (options “-f a -x 1 -# 100”). The consensus tree was further processed to collapse branches with bootstrap support lower than 50%, and the tree was rooted with the Bryophytes Physcomitrella patens and Selaginella moellendorffii as an outgroup.
Supplementary information
Acknowledgements
The genotyping data were produced in the project VALID funded by the German Federal Ministry of Education and Research (BMBF; project number 0315947). We are grateful to Ellen Weiß, Anette Heber, Ute Ostermann, and Sonja Allner for help in phenotypic data collection. We are thankful to The 10+ Wheat Genome Project for making the resources available before publication. We gratefully acknowledge two anonymous reviewers whose comments helped to improve this manuscript.
Author Contributions
Q.H.M. and M.S.R. conceived the idea. Q.H.M. analyzed the data, interpreted the results, and wrote the manuscript. J.B. and R.K. contributed to sequence and phylogenetic analyses. J.P. and M.W.G. contributed the genotypic data. R.K. and M.S.R. contributed to the interpretation of results and writing of the manuscript.
Competing Interests
On behalf of all authors, the corresponding author states that there is no conflict of interest. J.P. and M.W.G. are members of the company TraitGenetics GmbH. This does, however, in no way limit the availability or sharing of data and materials.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-50331-9.
References
- 1.Koppolu R, Schnurbusch T. Developmental pathways for shaping spike inflorescence architecture in barley and wheat. Journal of Integrative Plant Biology. 2019;61(3):278–295. doi: 10.1111/jipb.12771. [DOI] [PubMed] [Google Scholar]
- 2.Gauley A, Boden SA. Genetic pathways controlling inflorescence architecture and development in wheat and barley. Journal of Integrative Plant Biology. 2019;61(3):296–309. doi: 10.1111/jipb.12732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deng Z, et al. Discovery of consistent QTLs of wheat spike-related traits under nitrogen treatment at different development stages. Frontiers in Plant Science. 2017;8:2120. doi: 10.3389/fpls.2017.02120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo Z, et al. Genome‐wide association analyses of 54 traits identified multiple loci for the determination of floret fertility in wheat. New Phytologist. 2017;214(1):257–270. doi: 10.1111/nph.14342. [DOI] [PubMed] [Google Scholar]
- 5.Liu J, et al. A genome-wide association study of wheat spike related traits in China. Frontiers in Plant Science. 2018;9:1584. doi: 10.3389/fpls.2018.01584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Würschum T, et al. Phenotypic and genetic analysis of spike and kernel characteristics in wheat reveals long-term genetic trends of grain yield components. Theoretical and Applied Genetics. 2018;131(10):2071–2084. doi: 10.1007/s00122-018-3133-3. [DOI] [PubMed] [Google Scholar]
- 7.Zhai H, et al. QTL analysis of spike morphological traits and plant height in winter wheat (Triticum aestivum L.) using a high-density SNP and SSR-based linkage map. Frontiers in Plant Science. 2016;7:1617. doi: 10.3389/fpls.2016.01617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakuma S, et al. Unleashing floret fertility in wheat through the mutation of a homeobox gene. Proceedings of the National Academy of Sciences. 2019;116(11):5182–5187. doi: 10.1073/pnas.1815465116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo Z, et al. Manipulation and prediction of spike morphology traits for the improvement of grain yield in wheat. Scientific Reports. 2018;8(1):14435. doi: 10.1038/s41598-018-31977-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faris JD, et al. A bacterial artificial chromosome contig spanning the major domestication locus Q in wheat and identification of a candidate gene. Genetics. 2003;164(1):311–321. doi: 10.1093/genetics/164.1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greenwood JR, et al. New alleles of the wheat domestication gene Q reveal multiple roles in growth and reproductive development. Development. 2017;144(11):1959–1965. doi: 10.1242/dev.146407. [DOI] [PubMed] [Google Scholar]
- 12.Debernardi JM, et al. microRNA172 plays a crucial role in wheat spike morphogenesis and grain threshability. Development. 2017;144(11):1966–1975. doi: 10.1242/dev.146399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ochagavía H, et al. Dynamics of leaf and spikelet primordia initiation in wheat as affected by Ppd-1a alleles under field conditions. Journal of Experimental Botany. 2018;69(10):2621–2631. doi: 10.1093/jxb/ery104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaw LM, et al. FLOWERING LOCUS T2 regulates spike development and fertility in temperate cereals. Journal of Experimental Botany. 2018;70(1):193–204. doi: 10.1093/jxb/ery350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boden SA, et al. Ppd-1 is a key regulator of inflorescence architecture and paired spikelet development in wheat. Nature Plants. 2015;1(2):14016. doi: 10.1038/nplants.2014.16. [DOI] [PubMed] [Google Scholar]
- 16.Li, C. et al. Wheat VRN1 and FUL2 play critical and redundant roles in spikelet meristem identity and spike determinacy. bioRxiv, 510388 (2019). [DOI] [PMC free article] [PubMed]
- 17.Zhang B, et al. Novel function of a putative MOC1 ortholog associated with spikelet number per spike in common wheat. Scientific Reports. 2015;5:12211. doi: 10.1038/srep12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikeda K, Nagasawa N, Nagato Y. ABERRANT PANICLE ORGANIZATION 1 temporally regulates meristem identity in rice. Developmental Biology. 2005;282(2):349–360. doi: 10.1016/j.ydbio.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 19.Ikeda K, et al. Rice ABERRANT PANICLE ORGANIZATION 1, encoding an F‐box protein, regulates meristem fate. The Plant Journal. 2007;51(6):1030–1040. doi: 10.1111/j.1365-313X.2007.03200.x. [DOI] [PubMed] [Google Scholar]
- 20.Ikeda-Kawakatsu K, et al. Expression level of Aberrant Panicle Organization1 determines rice inflorescence form through control of cell proliferation in the meristem. Plant Physiology. 2009;150(2):736–747. doi: 10.1104/pp.109.136739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samach A, et al. The Unusual Floral Organs gene of Arabidopsis thaliana is an F‐box protein required for normal patterning and growth in the floral meristem. The Plant Journal. 1999;20(4):433–445. doi: 10.1046/j.1365-313x.1999.00617.x. [DOI] [PubMed] [Google Scholar]
- 22.Wilkinson MD, Haughn GW. Unusual Floral Organs controls meristem identity and organ primordia fate in Arabidopsis. The Plant Cell. 1995;7(9):1485–1499. doi: 10.2307/3870137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schulthess AW, et al. The roles of pleiotropy and close linkage as revealed by association mapping of yield and correlated traits of wheat (Triticum aestivum L.) Journal of Experimental Botany. 2017;68(15):4089–4101. doi: 10.1093/jxb/erx214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang SC, et al. Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnology Journal. 2014;12(6):787–796. doi: 10.1111/pbi.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allen AM, et al. Characterization of a Wheat Breeders’ Array suitable for high-throughput SNP genotyping of global accessions of hexaploid bread wheat (Triticum aestivum) Plant Biotechnology Journal. 2017;15(3):390–401. doi: 10.1111/pbi.12635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sim N-L, et al. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Research. 2012;40(W1):W452–W457. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernhardt N, et al. Dated tribe-wide whole chloroplast genome phylogeny indicates recurrent hybridizations within Triticeae. BMC Evolutionary Biology. 2017;17(1):141. doi: 10.1186/s12862-017-0989-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bouchenak-Khelladi Y, et al. Biogeography of the grasses (Poaceae): a phylogenetic approach to reveal evolutionary history in geographical space and geological time. Botanical Journal of the Linnean Society. 2010;162(4):543–557. doi: 10.1111/j.1095-8339.2010.01041.x. [DOI] [Google Scholar]
- 29.Muqaddasi QH, et al. Genome-wide association mapping and prediction of adult stage Septoria tritici blotch infection in European winter wheat via high-density marker arrays. Plant. Genome. 2019;12:180029. doi: 10.3835/plantgenome2018.05.0029. [DOI] [PubMed] [Google Scholar]
- 30.Kollers S, et al. Genetic architecture of resistance to Septoria tritici blotch (Mycosphaerella graminicola) in European winter wheat. Molecular Breeding. 2013;32(2):411–423. doi: 10.1007/s11032-013-9880-6. [DOI] [Google Scholar]
- 31.Würschum T, et al. Population structure, genetic diversity and linkage disequilibrium in elite winter wheat assessed with SNP and SSR markers. Theoretical and Applied Genetics. 2013;126(6):1477–1486. doi: 10.1007/s00122-013-2065-1. [DOI] [PubMed] [Google Scholar]
- 32.Su Z, et al. Identification and development of a functional marker of TaGW2 associated with grain weight in bread wheat (Triticum aestivum L.) Theoretical and Applied Genetics. 2011;122(1):211–223. doi: 10.1007/s00122-010-1437-z. [DOI] [PubMed] [Google Scholar]
- 33.Zhang L, et al. TaCKX6‐D1, the ortholog of rice OsCKX2, is associated with grain weight in hexaploid wheat. New Phytologist. 2012;195(3):574–584. doi: 10.1111/j.1469-8137.2012.04194.x. [DOI] [PubMed] [Google Scholar]
- 34.Zheng J, et al. TEF-7A, a transcript elongation factor gene, influences yield-related traits in bread wheat (Triticum aestivum L.) Journal of Experimental Botany. 2014;65(18):5351–5365. doi: 10.1093/jxb/eru306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, et al. TaGS-D1, an ortholog of rice OsGS3, is associated with grain weight and grain length in common wheat. Molecular breeding. 2014;34(3):1097–1107. doi: 10.1007/s11032-014-0102-7. [DOI] [Google Scholar]
- 36.Wang S, et al. A single-nucleotide polymorphism of TaGS5 gene revealed its association with kernel weight in Chinese bread wheat. Frontiers in Plant Science. 2015;6:1166. doi: 10.3389/fpls.2015.01166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma L, et al. TaGS5‐3A, a grain size gene selected during wheat improvement for larger kernel and yield. Plant Biotechnology Journal. 2016;14(5):1269–1280. doi: 10.1111/pbi.12492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voss-Fels, K. P. et al. High-resolution mapping of rachis nodes per rachis, a critical determinant of grain yield components in wheat. Theoretical and Applied Genetics, 1–13 (2019). [DOI] [PubMed]
- 39.Kuzay, S. et al. Identification of a candidate gene for a QTL for spikelet number per spike on wheat chromosome arm 7AL by high-resolution genetic mapping. Theoretical and Applied Genetics, 1–17 (2019). [DOI] [PMC free article] [PubMed]
- 40.Patton EE, Willems AR, Tyers M. Combinatorial control in ubiquitin-dependent proteolysis: don’t Skp the F-box hypothesis. Trends in Genetics. 1998;14(6):236–243. doi: 10.1016/S0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- 41.Kaiser P, et al. Cdc34 and the F-box protein Met30 are required for degradation of the Cdk-inhibitory kinase Swe1. Genes & Development. 1998;12(16):2587–2597. doi: 10.1101/gad.12.16.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frichot E, François O. LEA: An R package for landscape and ecological association studies. Methods in Ecology and Evolution. 2015;6(8):925–929. doi: 10.1111/2041-210X.12382. [DOI] [Google Scholar]
- 43.Sorrells ME, et al. Reconstruction of the Synthetic W7984 × Opata M85 wheat reference population. Genome. 2011;54(11):875–882. doi: 10.1139/g11-054. [DOI] [PubMed] [Google Scholar]
- 44.Yu JM, et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nature Genetics. 2006;38(2):203–208. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- 45.VanRaden PM. Efficient methods to compute genomic predictions. Journal of Dairy Science. 2008;91(11):4414–4423. doi: 10.3168/jds.2007-0980. [DOI] [PubMed] [Google Scholar]
- 46.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57(1):289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 47.Utz HF, Melchinger AE, Schön CC. Bias and sampling error of the estimated proportion of genotypic variance explained by quantitative trait loci determined from experimental data in maize using cross validation and validation with independent samples. Genetics. 2000;154(4):1839–1849. [PMC free article] [PubMed] [Google Scholar]
- 48.Consortium IWGS. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science. 2018;361(6403):eaar7191. doi: 10.1126/science.aar7191. [DOI] [PubMed] [Google Scholar]
- 49.Altschul SF, et al. Basic local alignment search tool. Journal of Molecular Biology. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 50.Kearse M, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Bioinformatics. 1992;8(3):275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 52.Darriba D, et al. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27(8):1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 54.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.