Abstract
The immunological roles of the nuclear factor-kappaB (NF-κB) pathway are mediated via the canonical components in immune responses and via non-canonical components in immune organogenesis and homeostasis, although the two components are capable of crosstalk. Regulatory CD4 T cells (Tregs) are homeostatically functional and represent an interesting potential meeting point of these two NF-κB components. We show that mice deficient in the non-canonical NF-κB component gene Nfkb2 (p100) had normal thymic development and suppressive function of Tregs. However, they had enhanced frequencies of peripheral ‘effector-phenotype’ Tregs (eTregs). In bi-parental chimeras of wild-type (WT) and Nfkb2−/− mice, the Nfkb2−/− genotype was over-represented in Tregs, with a further increase in the relative prominence of eTregs. Consistent with distinct properties of eTregs, the Nfkb2−/− genotype was more prominent in Tregs in extra-lymphoid tissues such as liver in the bi-parental chimeras. The Nfkb2−/− Tregs also displayed greater survival, activation and proliferation in vivo. These Nfkb2−/− Tregs showed higher nuclear NF-κB activity mainly comprising of RelB-containing dimers, in contrast to the prominence of cRel- and RelA-containing dimers in WT Tregs. Since p100 is an inhibitor of RelB activation as well as a participant as cleaved p52 in RelB nuclear activity, we tested bi-parental chimeras of WT and Relb−/− mice, and found normal frequencies of Relb−/− Tregs and eTregs in these chimeric mice. Our findings confirm and extend recent data, and indicate that p100 normally restrains RelB-mediated Treg activation, and in the absence of p100, p50-RelB dimers can contribute to Treg activation.
Subject terms: Regulatory T cells, NF-kappaB
Introduction
Regulatory T cells expressing the characteristic lineage determining transcription factor FOXP3 are actively involved in suppressing overt activation of the immune system and the development of autoimmunity1. Tregs are characterised by constitutive expression of IL-2Rα (CD25)2 and of several other molecules that contribute to their suppressive function such as CTLA-43, ICOS4 and CD735. The peripheral Treg population includes thymic-origin Tregs (‘natural’ Tregs (nTregs)) and Tregs that have been induced in the periphery from conventional CD4 T cells (‘induced’ Tregs (iTregs))6. Tregs perform their functions through a number of mechanisms such as by competing for IL-2 with activated conventional T cells7,
by secreting cytokines such as IL-10 which regulates T cell activation and cytokine production8, through direct killing mediated by perforin or granzyme production9, and by interfering with T cell-dendritic cell interactions10.
Tregs in the periphery exhibit heterogeneity and can be categorized into ‘naive’ and ‘effector’ subsets like conventional T cells11. Most Tregs in secondary lymphoid organs express high levels of CD62L12 and CCR713 like naive conventional T cells, and are termed central Tregs (cTregs)14. These cTregs are activated in the periphery11 and give rise to effector Tregs (eTregs) showing surface phenotypes similar to recently activated conventional T cells, such as lower levels of CCR7 and CD62L14. The eTregs are more prominent in non-lymphoid tissues13, short lived11, express lower levels of Bcl-214 and contain a greater proportion of homeostatically proliferating cells compared to cTregs11. Additionally, eTregs also express higher levels of CD44, GITR, ICOS11,15 and chemokine receptors such as CCR416, CCR917, CCR618, etc which direct their migration to specific nonlymphoid tissues.
The cytokine and signaling requirements for maintenance also differ between cTregs and eTregs; while the former depend on IL-2 mediated survival signals, the latter require signaling through ICOS14. In addition, sustained T cell receptor (TCR)-mediated signals are also essential for eTreg generation and maintenance. The absence of TCR signals leads to loss of expression of target genes of NFAT indicating its possible role in eTreg generation and maintenance19,20. One such NFAT target, IRF4, is indispensable for eTreg generation as well as their trafficking. IRF4 in turn regulates the expression of Blimp1, which is indispensable for the production IL-10 by eTregs and regulates their migration to lymphoid organs as well as non-lymphoid tissues20,21.
In addition to IRF4 and BLIMP1, the transcription factors of the canonical NF-κB family have been shown to play a role in eTreg generation and/or maintenance. The NF-κB family of dimeric transcription factors consists of NF-κB1 (p105/p50), NF-κB2 (p100/p52), cRel, RelA (p65) and RelB22. The canonical NF-κB pathway, activated downstream of TCR, the tumor necrosis factor receptor (TNFR) and toll like receptor (TLR)-interleukin (IL)-1R superfamilies among others, triggers nuclear translocation of primarily RelA:p50 and cRel:p50 heterodimers by inducing the degradation of IkBα, IkBβ and IkBε22,23. Ex vivo Tregs, unlike conventional T cells, show p50:RelA and p50:p50 activity under homeostatic conditions and RelA has been shown to regulate eTreg homeostasis24. In addition to RelA, cRel also plays a role in activated Treg (aTreg) generation and/or maintenance25. Hetero-dimers of p52 and RelB constitute the activity of the non-canonical NF-κB pathway, which is activated through a select set of TNFR superfamily members like CD40, OX-40, GITR and LT-βR26,27.
Previous studies have also suggested a role for the noncanonical NF-κB pathway in Treg development and maintenance. The kinases NIK and IKKα both act upstream of NF-κB2 and RelB and are involved in activation of the non-canonical NF-κB signaling26. Mice bearing a T cell specific deletion of IKKα show compromised Treg development28 whereas NIK deficiency leads to reduced Treg numbers in the periphery29. Mice lacking RelB display inflammatory cell infiltration in several organs, myeloid hyperplasia and splenomegaly due to extramedullary hematopoiesis. While these effects were seen to depend on T cells, the inflammation seen in Relb−/− mice was shown to be due to a role of RelB in non-hematopoietic cells30.
Signals activating the non-canonical NF-κB pathway induce processing of the NF-κB precursor protein p100, encoded by Nfkb2, into the mature NF-κB2 subunit p52. In resting cells, p100 sequesters RelB in the cytoplasm and signal-induced p100 processing allows for nuclear activation of RelB:p52 heterodimers26. However, p100 retains RelA- and cRel-containing heterodimers as well31. Indeed, p100 deficiency triggers sustained RelA activity in TLR4-stimulated embryonic fibroblasts32, and also prolongs cRel responses in maturing B cells33.
Interestingly, a lack of p100 in myeloma cells, attributed to genetic aberrations, provokes an alternate RelB:p50 NF-κB response via the canonical pathway34. RelB is encoded by an NF-κB target gene, and its expression is upregulated by canonical NF-κB signals35. In the absence of p100, de novo synthesized RelB accumulates in the nucleus as RelB:p50 heterodimers and regulates NF-κB-driven gene expression34. These studies underline a complex role of Nfkb2 at the crossroad of canonical and noncanonical NF-κB pathways; p52 constitutes the noncanonical RelB:p52 activity while p100 inhibits the activity of multiple p50-containing NF-κB dimers, including RelB:p50 dimer, in the canonical pathway. Despite the involvement of Nfkb2 in diverse physiological and pathological settings, the role of Nfkb2-dependent pathways in Treg biology has been less explored.
Here, we report that mice deficient in Nfkb2 showed normal thymic development of Tregs, but enhanced frequencies of peripheral eTregs. This effect was cell-autonomous, since it was recapitulated in bi-parental chimeras of wild-type (WT) and Nfkb2−/− mice. These Nfkb2−/− Tregs exhibited normal suppressive function, showed greater survival, activation and proliferation in vivo, and trafficked better to non-lymphoid tissues. However, the frequencies of Tregs and eTregs were normal in bi-parental chimeras of WT and knockout mice lacking RelB, which regulates gene-expressions in association with p52. Our biochemical analyses revealed that Nfkb2 deficiency in Tregs did not alter RelA and cRel activities but triggered instead an additional strong NF-κB activity comprising of RelB:p50 heterodimers. Together, our data indicate, as shown recently as well, that Nfkb2 functions in Treg are dictated primarily by the inhibitory p100, which restrains RelB:p50 mediated Treg activation, and not by p52. Nuclear induction of RelB-p50 in the absence of p100 contributes to a greater prominence of activated Tregs. Our data provide further insights into the complex roles that p100 plays in Treg biology.
Results
Comparable thymic Treg development but higher splenic Treg numbers in Nfkb2−/− mice
Thymic Treg development has been shown to consist of two steps: a TCR- and co-stimulation-dependent step that generates thymic Treg precursors and a cytokine-dependent second step that results in the differentiation of Treg precursors into mature Tregs36. Thymic Treg precursors in WT (B6.SJL: expressing CD45.1) and Nfkb2−/− (expressing CD45.2) mice were identified as CD90+CD4+CD8− (CD4 single positive: CD4SP) thymocytes expressing CD25 but not FOXP3, while mature thymic Tregs were identified as CD25+FOXP3+CD4SP thymocytes (Supplementary Information, Fig. S1)36. While thymi from WT and Nfkb2−/− mice had equivalent frequencies of Treg precursors, the frequency of Tregs was significantly higher in Nfkb2−/− mice than in WT mice (Fig. 1A). The cell numbers of Treg precursors and Tregs in the thymus of WT and Nfkb2−/− mice respectively were equivalent (Fig. 1B) Equivalent Treg numbers despite higher frequency of Tregs in Nfkb2−/− mice might partly be explained by a reduction in total thymic cell numbers seen in Nfkb2−/− mice.
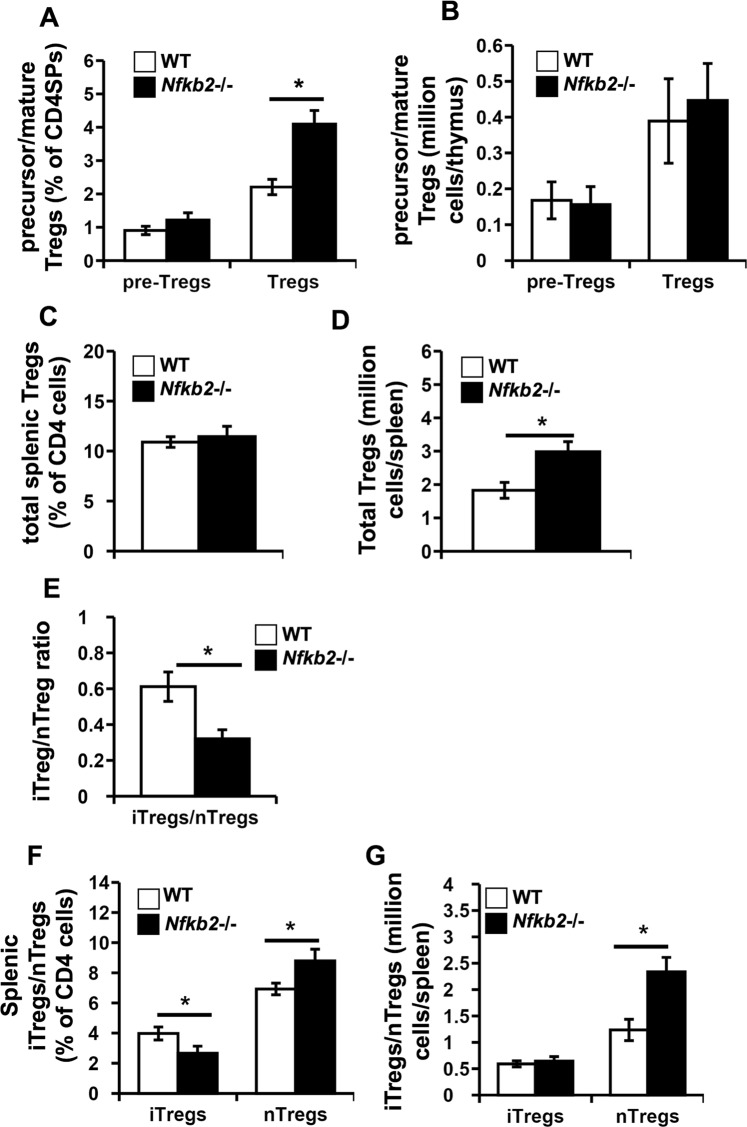
Figure 1.
Unaltered thymic Treg numbers but higher splenic Treg numbers in Nfkb2−/− mice. (A,B) Thymocytes isolated from WT (B6.SJL) and Nfkb2−/− mice were fixed, permeabilised and stained with anti-mouse CD45.1, CD45.2, CD90, CD4, CD8, CD25 and FOXP3 to identify Treg precursors (pre-Tregs) and mature Tregs (Supplementary Fig. S1). The bar graphs represent the (A) frequencies and (B) cell numbers per thymus of pre-Tregs and mature Tregs in CD90+CD4+CD8− thymocytes (CD4SP: CD4 single-positive thymocytes). (C–G) Spleen cells isolated from WT and Nfkb2−/− mice were fixed, permeabilised and stained with anti-mouse CD45.1, CD45.2, CD4, CD25, FOXP3 and HELIOS to identify Tregs as well as the iTreg and nTreg subpopulations (Supplementary Fig. S2). The bar graphs represent (C) the frequencies and (D) numbers per spleen of the Tregs in total CD4 cells. The ratio of iTreg/nTreg within the total Tregs (E), and the frequencies (F) and numbers per spleen (G) of iTregs and nTregs in the spleens of WT and Nfkb2−/− mice are also shown. *p < 0.05; n ≥ 12 mice/group.
In the periphery, the frequency of splenic Tregs (CD4+CD25+FOXP3+; Supplementary Information, Fig. S2)37 was comparable between WT and Nfkb2−/− mice but the latter contained significantly higher Treg numbers (Fig. 1C,D). This is likely to be a consequence of the higher numbers of CD4 T cells in Nfkb2−/− mouse spleens (Supplementary Information, Fig. S2). In addition to nTregs of thymic origin, the splenic Treg population also includes peripherally induced Tregs (iTregs) generated from conventional CD4 T cells. The transcription factor HELIOS appears to be generally expressed in thymic Tregs but not in iTregs6, although this is subject to caveats38. When we analyzed splenic Tregs as either HELIOS-high nTregs or HELIOS-low iTregs (Supplementary Information, Fig. S2), the ratio of iTregs to nTregs among the splenic Tregs in Nfkb2−/− mice was less than in WT mice (Fig. 1E). This was reflected in the reduced frequencies (Fig. 1F) of iTregs in the spleens of Nfkb2−/− mice compared to WT mice. In contrast, the frequencies and numbers of nTregs were increased in the spleens of Nfkb2−/− mice (Fig. 1F,G).
Splenic Tregs in Nfkb2−/− mice have a higher proportion of effector Tregs
In addition to developmental heterogeneity, peripheral Tregs can also be distinguished into central Tregs and effector Tregs, based on the expression of CD62L and CD44 (Supplementary Information, Fig. S2)14. CD62LlowCD44high effector Tregs were relatively more prominent than CD62LhighCD44low central Tregs in both iTreg (Fig. 2A) and nTreg (Fig. 2B) compartments in Nfkb2−/− spleens compared to their WT counterparts. Thus, it was notable that, while total splenic iTreg numbers were comparable in Nfkb2−/− mice and WT mice (Fig. 1G), the numbers of effector iTregs tended to be higher in Nfkb2−/− mice (Fig. 2C). As expected, both the frequencies (Fig. 2D) and numbers of effector nTregs (Fig. 2E) were higher in the spleens of Nfkb2−/− mice compared to WT mice.
Figure 2.
Nfkb2−/− mice have higher proportions of peripheral effector Tregs compared to WT mice. Spleen cells were isolated from WT and Nfkb2−/− mice, fixed, permeabilised and stained with anti-mouse CD4, CD25, FOXP3, HELIOS, CD62L and CD44 to identify Tregs and Treg subsets (Supplementary Fig. S2). (A,B) The bar graphs represent the ratios of effector (eTreg) to central (cTregs) phenotype Tregs among (A) iTregs and (B) nTregs. (C–E) The bar graphs represent the numbers per spleen of central and effector iTregs (C), and the frequencies (D) and numbers per spleen (E) of central and effector nTregs in WT and Nfkb2−/− mice. *p < 0.05; n = 15 mice/group.
NF-κB2 has a cell intrinsic role in regulating the splenic Treg population
The data above regarding splenic Tregs in WT and Nfkb2−/− mice suggested a possible role of NF-κB2 in regulating both splenic iTreg population and the proportions of effector Tregs among both iTregs and nTregs. Defects in both thymic and splenic organization have been reported in Nfkb2−/− mice due to a critical role of the non-canonical NF-κB pathway in the stromal cell compartments involved39,40. Therefore, it was necessary to examine whether the observed effects of the Nfkb2−/− genotype on the Treg compartment were cell-intrinsic or not. For this, we generated bi-parental bone marrow chimeras using bone marrow cells from WT and Nfkb2−/− mice, and the Treg populations in the thymus and spleen of bi-parental bone marrow chimeras were analyzed after reconstitution.
In the thymi of the bi-parental bone marrow chimeras, the frequencies of Treg precursors (pre-Tregs) and mature Treg were equivalent (Fig. 3A) between WT and Nfkb2−/− donors. In the spleen, there was a large increase in Nfkb2−/− Treg frequency compared to the WT Tregs (Fig. 3B). As in the parent strain, the ratio of iTregs to nTregs in splenic Tregs was lower in Nfkb2−/− Tregs (Fig. 3C). The effector and central Treg populations in the spleens of bi-parental bone marrow chimeras showed patterns similar to those observed in the parental strains, namely, an increase in the effector to central Treg ratio among both iTregs (Fig. 3D) and nTregs (Fig. 3E) in the Nfkb2−/− donor compared to WT donor. Thus, the absence of NF-κB2 led to reduced proportion of iTregs and higher effector Tregs in a cell-intrinsic manner.
Figure 3.
In bi-parental mixed bone marrow chimeras, the Nfkb2−/− genotype shows greater prominence of peripheral Tregs and higher proportions of effector Tregs. (A) Thymocytes were isolated from WT and Nfkb2−/− bi-parental bone marrow chimeras, fixed, permeabilized and stained to identify precursor (pre-Tregs) and mature Tregs (Supplementary Fig. S1). The bar graph shows the frequencies of pre-Tregs and mature Tregs in CD4SP thymocytes. (B–D) Spleen cells were isolated from WT and Nfkb2−/− bi-parental bone marrow chimeras and stained to identify Tregs and their subsets (Supplementary Fig. S2). The bar graphs show frequencies of total splenic Tregs (B), ratios of iTregs to nTregs (C), and the ratios of effector (eTregs) to central (cTreg) Tregs among iTregs (D) and nTregs (E) in the WT and Nfkb2−/− donors in bi-parental bone marrow chimeras. *p < 0.05; n ≥ 10 mice/group.
Differential expression of several characteristic Treg markers between WT and Nfkb2−/− Tregs
Since NF-κB2 regulated the relative prominence of specific Treg sub-populations, we tested if the absence of NF-κB2 also led to alterations in the phenotype of these Tregs. We therefore examined the expression of Treg specific transcription factor FOXP31, molecules contributing to Treg suppressive function namely CTLA-4 (CD152)41, Neuropilin-1 (CD304)42,43 and CD2844, markers highly expressed by effector Tregs or representative of Treg activation such as GITR (CD357), IL-2Rβ (CD122)11, IRF4, TIGIT, Egr2, PD-1 (CD279)20, GARP45, Fas (CD95) and IL-15Rα (CD215) which plays a role in Treg proliferation46 on Tregs from the bi-parental bone marrow chimeras (Supplementary Information, Fig. S3). Of the markers analyzed, the expression of FOXP3, CD28, PD-1, Fas and IL-15Rα was found to be unaltered between WT and Nfkb2−/− Tregs (Fig. 4A). The expression of Neuropilin-1 and GITR was found to be higher whereas the expression of CTLA-4, IRF4, IL-2Rβ and GARP was lower on Nfkb2−/− Tregs compared to WT Tregs (Supplementary Information; Fig. S3 and Fig. 4A). The expression of TIGIT and Egr2 on Tregs displayed a bimodal distribution, as reported previously20, and hence the frequencies of TIGIT+ and Egr2+ WT and Nfkb2−/− Tregs respectively in the bi-parental bone marrow chimeras were analyzed (Supplementary Information, Fig. S3). While the frequencies of TIGIT+ cells were found to be higher among Nfkb2−/− Tregs, the frequency of Egr2+ cells were comparable between WT and Nfkb2−/− Tregs (Fig. 4B,C).
Figure 4.
Qualitative differences between WT and Nfkb2−/− Tregs. (A–D) Spleen cells were isolated from the bi-parental bone marrow chimeras, fixed, permeabilised and stained with anti-mouse CD45.1, CD45.2, CD4, CD25, FOXP3 and either CD152 (CTLA-4), CD304 (Neuropilin-1), CD28, CD357 (GITR), CD122 (IL-2Rβ), IRF4, CD279 (PD-1), GARP, Fas(CD95), CD215 (IL-15Rα), TIGIT, Egr2 and CD103 (Supplementary Fig. S3). Mean fluorescence indices (MFI) of the indicated molecules were calculated for WT and Nfkb2−/− Tregs and normalized to WT. The bar graphs indicate the relative levels (MFI) of the indicated surface/intracellular markers expressed by WT versus Nfkb2−/− Tregs (A), frequencies of TIGIT+ total Tregs (B), frequencies of Egr2+ total Tregs (C), and frequencies of CD103+ iTregs and nTregs (D), among WT and Nfkb2−/− donors respectively in the bi-parental bone marrow chimeras. (E,F) Leukocytes were isolated from the liver of the bi-parental bone marrow chimeras, fixed, permeabilised and stained with anti-mouse CD45.1, CD45.2, CD4, CD25, FOXP3 and HELIOS to identify Tregs and their subsets (Supplementary Fig. S2). The bar graphs depict frequencies of total WT and Nfkb2−/− Tregs (E), as well as frequencies of both iTregs and nTregs (F) in the liver of the bi-parental bone marrow chimeras. *p < 0.05; n ≥ 5 mice/group.
Additionally, CD103+ Tregs constitute a subset of eTregs47 and as the overall eTreg compartment was expanded among Nfkb2−/− Tregs, we further analyzed the CD103+ Treg compartment (Supplementary Information, Fig. S3). Interestingly, we observed that both Nfkb2−/− iTregs and nTregs showed higher proportions of CD103+ cells compared to the corresponding WT populations (Fig. 4D). Effector Tregs are reported to make up the majority of Tregs present in non-lymphoid organs12. As the proportion of effector Tregs among the Nfkb2−/− Tregs was higher compared to WT Tregs in a cell-intrinsic manner, we hypothesized that there would be a higher representation of these Nfkb2−/− Tregs in non-lymphoid organs such as liver in the bi-parental bone marrow chimeras. Indeed, we found that there was a higher frequency of Nfkb2−/− Tregs than that of WT Tregs in the liver (Fig. 4E) of the mixed bone marrow chimeras, and this included both nTregs and iTregs (Fig. 4F).
Generation and functions of Nfkb2−/− Tregs
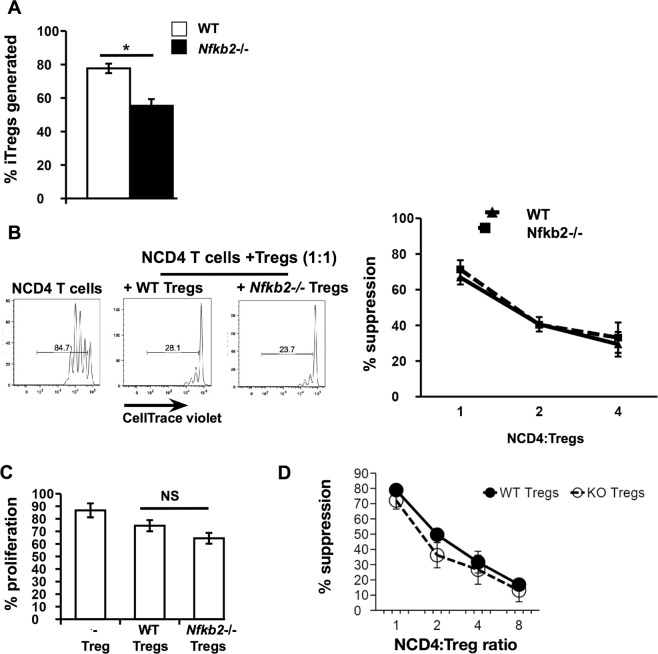
The iTreg compartment was reduced in Nfkb2−/− Tregs compared to WT Tregs, both in the parental strain and in the bi-parental bone marrow chimeras. The reduction indicated a possible role of NF-κB2 in the generation and/or survival of iTregs. To test the former possibility, WT and Nfkb2−/− naïve CD4 (NCD4) T cells were sorted from bi-parental bone marrow chimeras and cultured in vitro under Treg differentiation conditions (Supplementary Information; Fig. S4). At the end of five days, it was observed that the Nfkb2−/− NCD4 T cells gave rise to a lower frequency of Tregs in vitro compared to WT NCD4 T cells (Fig. 5A). FOXP3 staining profiles were comparable between WT and Nfkb2−/− iTregs (Supplementary Information; Fig. S4).
Figure 5.
Nfkb2−/− CD4 T cells show poor Treg generation in vitro, but Nfkb2−/− Tregs show normal suppressive capacity. (A) WT and Nfkb2−/− naïve CD4 (NCD4) T cells were sorted from bi-parental bone marrow chimeras and stimulated in vitro under Treg differentiation conditions (Supplementary Information, Fig. S4). The bar graph indicates frequencies of Tregs generated in vitro from WT and Nfkb2−/− NCD4 T cells. The figure is an average of 3 independent experiments. (B) NCD4 T cells isolated from B6.GFP mice labelled with CellTrace violet dye were cultured with CD90− C57BL/6 irradiated spleen cells and aqueous-phase anti-mouse CD3 (2 μg/ml) in the absence or presence of WT or Nfkb2−/− Tregs isolated from the spleens of bi-parental bone marrow chimeras at varying responder to suppressor ratios for 60 h. NCD4 T cell proliferation was then analysed by CellTrace violet dye dilution. The line graph represents the percentage of suppression at each responder to suppressor ratio calculated as [(%proliferation without Tregs − %proliferation with Tregs)/(%proliferation without Tregs) × 100] with % suppression mediated by WT Tregs represented as solid and Nfkb2−/− Tregs as dashed lines respectively. The figure is an average of 3 independent experiments. (C) NCD4 T cells were isolated from B6.GFP mice, labelled with CellTrace violet dye and transferred into TCRβδ−/− mice either alone or with WT or Nfkb2−/− Tregs respectively for 3 days. The proliferation of B6.GFP NCD4 T cells was assessed in the spleens of recipient mice 3 days after transfer to estimate the in vivo suppressive effects of WT and Nfkb2−/− Tregs. n = 3mice/group. (D) NCD4 T cells isolated from B6.GFP mice labelled with CellTrace violet dye were cultured with CD90− C57BL/6 irradiated spleen cells and aqueous-phase anti-mouse CD3 (2 μg/ml) in the absence or presence of Tregs isolated from the spleens of WT or Nfkb2−/− mice at varying responder to suppressor ratios for 60 h. NCD4 T cell proliferation was then analysed by CellTrace violet dye dilution. The line graph represents the percentage of suppression at each responder to suppressor ratio calculated as [(%proliferation without Tregs − %proliferation with Tregs)/(%proliferation without Tregs) × 100] with % suppression mediated by WT Tregs represented as solid and Nfkb2−/− Tregs as dashed lines respectively. The figure is an average of 2 independent experiments.
Since the Nfkb2−/− Tregs contained a higher proportion of effector Tregs that have been shown to be potent producers of IL-1021, we tested the ability of WT and Nfkb2−/− Tregs to suppress NCD4 T cell proliferation. Interestingly, the Nfkb2−/− Tregs showed equivalent suppression of naïve T cell proliferation in vitro compared to WT Tregs (Fig. 5B). In addition, when we compared the ability of bi-parental bone marrow chimera-derived WT and Nfkb2−/− Tregs to suppress NCD4 T cell proliferation upon transfer in TCRβδ−/− mice, for a duration of 3 days, the in vivo suppressive function of WT and Nfkb2−/− Tregs was also found to be comparable (Fig. 5C). Additionally, parent strain-derived WT and Nfkb2−/− Tregs also showed equivalent capacity to suppress NCD4 T cell proliferation in vitro (Fig. 5D).
The Nfkb2−/− Tregs show altered survival, proliferation and effector differentiation
The higher proportions of Nfkb2−/− donor Tregs in the bi-parental chimeras could be plausibly due to differential survival and/or proliferation and activation of Nfkb2−/− central and effector Tregs compared to WT Tregs. We first compared survival, proliferation and activation of WT (CD45.1+) and Nfkb2−/− (CD45.2+) central Tregs (CD4+CD25+CD62Lhigh) sorted from bi-parental bone marrow chimeras by transferring them into recipient congenic mice in equal numbers (Supplementary Information; Fig. S5). Higher frequencies of Nfkb2−/− Tregs was observed in the spleen 10 days post-transfer (Fig. 6A) Proliferation among transferred Tregs at ten days post-transfer was also estimated using Ki67 detection, which showed that a higher proportion of transferred Nfkb2−/− Tregs were Ki67+ compared to WT Tregs (Fig. 6B). In addition to displaying greater proliferation, it was evident that Nfkb2−/− cTregs showed greater conversion to an effector phenotype (Fig. 6C).
Figure 6.
Nfkb2−/− central and effector Tregs show higher survival, activation and proliferation in vivo. (A–C) WT and Nfkb2−/− central Tregs were sorted from the spleens of bi-parental bone marrow chimeras and parked in vivo for 10 days. Transferred populations were then analysed (Supplementary Fig. S5). Bar graphs represent (A) frequencies of the indicated transferred Treg populations in the spleen of recipients at the end of 10 days, (B) frequencies of proliferating cells in respective transferred Treg population as indicated by Ki67 expression frequencies, and (C) ratios of central (CD62Lhigh) to effector (CD62Llow) Tregs. (D) WT and Nfkb2−/− central Tregs were sorted from the spleens of bi-parental bone marrow chimeras and parked in vivo for 3 days. The bar graph represents the frequencies of the indicated transferred Treg populations in the spleen of recipients at the end of 3 days. (E–G) WT and Nfkb2−/− effector Tregs were sorted from the spleens of bi-parental bone marrow chimeras and parked in vivo for 10 days as above. Bar graphs represent (E) frequencies of the indicated transferred Treg populations in the spleens of recipients at the end of 10 days, (F) frequencies of proliferating cells in respective transferred Treg populations as indicated by Ki67 expression frequencies, and (G) ratios of central (CD62Lhigh) to effector (CD62Llow) Tregs. *p < 0.05; n ≥ 4 mice/group.
In order to distinguish survival versus proliferation differences between WT and Nfkb2−/− cTregs we assessed the survival of WT and Nfkb2−/− cTregs in vivo 3 days after transfer, a time point at which there is neither major detectable proliferation nor transition to an effector phenotype (Supplementary Information, Fig. S6)11, and observed that Nfkb2−/− cTregs displayed enhanced survival compared to WT Tregs (Fig. 6D). Nonetheless, these data do not account for differences potentially contributed by differences in proliferation below detection limits for this assay.
Thus, central-phenotype Nfkb2−/− Tregs showed better survival, proliferation and effector transition compared to WT central Tregs. We next tested if effector-phenotype Tregs also showed similar differences, and if re-conversion from the effector to central phenotype differed between the two genotypes. To examine these issues, WT (CD45.1+) and Nfkb2−/− (CD45.2+) effector Tregs (CD4+CD25+CD62Llow) purified from bi-parental bone marrow chimeras were parked in vivo as above (Supplementary Information; Fig. S5). Ten days later, the transferred populations were analyzed in the spleens of recipient mice. Similar to the observation in central Tregs, Nfkb2−/− effector Tregs showed enhanced survival and proliferation (Fig. 6E,F). Interestingly, when the effector/central phenotype was analyzed in the transferred effector Tregs, it was evident that higher frequencies of WT effector Tregs shifted to a central phenotype, despite proliferating and/or surviving less well (Fig. 6G). Thus, NF-κB2 in Tregs appears to control the efficiency of survival, proliferation and transition between resting and effector phenotypes.
Altered NF-κB activity in Nfkb2−/− Tregs
The observations that Nfkb2−/− Tregs showed enhanced survival, proliferation and activation compared to WT Tregs indicated a negative regulatory role for NF-κB2 in these contexts. Nfkb2 deficiency causes loss of both p52 and p100. Because p52 predominantly functions as RelB:p52 heterodimers during non-canonical signaling26, we first asked if a lack of RelB similarly affected the frequencies of total Tregs and specific Treg subsets in the spleens of bi-parental chimeras of WT and Relb−/− parental strains. We found that the frequency of splenic Relb−/− and WT Tregs were not significantly different (Fig. 7A). Additionally, the frequencies of iTreg and nTreg populations were also comparable (Fig. 7B). Further, the distribution of cTregs and eTregs was comparable between WT and Relb−/− genotypes in both the iTreg and nTreg populations (Fig. 7C). These results indicated that Nfkb2 functions in Tregs were independent of RelB:p52 and that chronic non-canonical signaling only marginally contributed in the observed eTreg phenotype.
Figure 7.
RelB activity is likely responsible for altered Treg homeostasis in the absence of Nfkb2. (A–C) Tregs and Treg subsets in the spleens of bi-parental bone marrow chimeras of WT and Relb−/− mice were identified (Supplementary Fig. S2) and their frequencies estimated. The bar graphs represent frequencies of (A) total Tregs, (B) iTregs and nTregs, and (C) ratios of central to effector Tregs among the iTregs and nTregs present in the spleen of the bi-parental bone marrow chimeras of WT and Relb−/− mice. n = 7 mice/group. (D) Tregs were generated in vitro from WT and Nfkb2−/− NCD4 T cells under Treg-inducing conditions using anti-CD3 + anti-CD28 + TGFβ + IL-2 for 72 h as described, and subjected to an EMSA to assess nuclear NF-κB activity. Supershift assays were done with anti-RelA, anti-RelA + anti-cRel and anti-RelA + anti-RelB antibodies to identify the relative prominence of RelA, RelB and cRel in these assays. Arrow and arrowhead represent RelA-, RelB- and cRel- containing NF-κB activity, respectively. Asterisk represents supershifted complexes. The panels for detection of NF-κB activity and the relative prominence of RelA, RelB and cRel by means of supershifts are a part of a single gel (shown in Supplementary Fig. S7). The panel for loading control, using Oct1, is from a separate gel (also depicted in Supplementary Fig. S7). ssAb:supershift antibody. The data represent two independent experiments.
We then investigated if Nfkb2 deficiency triggered anomalous activation of p50-containing NF-κB heterodimers because of a lack of p100-mediated inhibition.
To this end, we examined nuclear extracts derived from WT and Nfkb2−/− Tregs in the electrophoretic mobility shift assay (EMSA) for NF-κB DNA binding activities and distinguished between RelA-, cRel- and RelB-containing heterodimers using antibody-mediated supershift assays (Supplementary Information, Fig. S7; Fig. 7D). While NF-κB activity was detectable in both WT and Nfkb2−/− Tregs, the latter showed strikingly higher nuclear NF-κB activity. Supershift assays showed that NF-κB activity was mainly composed of RelA- and cRel-containing dimers in WT Tregs. In the Nfkb2−/− Tregs, however, the high NF-κB activity was mainly due to RelB-containing dimers with a minimal contribution of cRel-containing dimers. Our data thus indicate that increased nuclear RelB:p50 activity in Nfkb2−/−Tregs, in the absence of p100, may be responsible for their higher activation, proliferation and survival. In other words, p100-mediated inhibition of RelB activation, rather than p100 processing-dependent activation of RelB:p52, determines Nfkb2 functions in Tregs.
Effects of Nfkb2 deficiency on the Treg transcriptome
We also evaluated the differences in gene expression between WT and Nfkb2−/− Tregs by RNA-seq analysis. It must be noted that these analyses were done ex vivo, with no further activation of the isolated Tregs in vitro as previously reported48, and therefore the outcomes of the two transcriptome analyses are complementary rather than simply confirmatory. Some 854 genes exhibited differential expression between WT and Nfkb2−/− Tregs (Supplementary Information, Table S1). We further investigated the enrichment of pathways annotated in KEGG database49 as well as Reactome database50 among these differentially expressed genes (Fig. 8A). Chemokine and cytokine signaling pathways, the TNF signaling pathway and apoptosis-related pathways were enriched in the set of genes upregulated in Nfkb2−/− Tregs, while primary immunodeficiency related pathways, hematopoietic cell-lineage pathways, and the NF-κB pathway were enriched in the downregulated gene set. Since Nfkb2−/− Tregs showed better survival, we asked if any of the curated gene sets in MolSigDB that are associated with cell proliferation or survival were enriched among differentially expressed genes. When we examined the enrichment of curated gene sets in MSigDB using GSEA software, we noted that four gene sets associated with cell proliferation and two terms related to cell survival/cell death were substantially enriched in Nfkb2−/− Tregs (Fig. 8B). Interestingly, our analyses revealed that gene sets associated with FOXP3 functions were also enriched in genes differentially expressed between WT and Nfkb2−/− Tregs (Fig. 8B). We then analyzed the RNA-seq data for the expression levels of several known genes associated with cell survival and cell proliferation (Fig. 8C). A comparison showed that several pro-survival genes, including those encoding Traf151 and cIAP152 (Birc2) were upregulated in Nfkb2−/− Tregs (Fig. 8C). Conversely, some pro-apoptotic genes, such as Bim, Bid and Bad53, were downregulated in the absence of Nfkb2-p100 (Fig. 8C). However, Nfkb2 deficiency did not substantially alter the expression of cIAP2 (Birc3)52, Bcl2 and Mcl153. Similarly, WT and Nfkb2−/− Tregs expressed almost equivalent levels of some genes such as Bax and Bak153 (Supplementary Information, Fig. S8). Finally, we noted that pro-proliferative genes, such as Cdk654 and Med3155, were also upregulated in Nfkb2−/− Tregs (Fig. 8C). These observations suggested that complex interactions between different pathways involved in cell survival likely dictates the Nfkb2−/− Treg phenotype.
Figure 8.
Transcriptome analysis shows misregulation of multiple pathways in Nfkb2−/− Tregs. (A) KEGG pathways enriched in Nfkb2−/− Tregs, - upregulated genes (left panel) and downregulated genes (right panel), - are plotted. Pathway analysis was done using g:profiler. (B) Gene sets related to cell proliferation, cell survival/death, and Foxp3 targets that are significantly enriched in Nfkb2−/− cells are plotted against their -log10(p-value). (C) Heatmap showing log2-fold changes of genes associated with cell survival/death and cell proliferation in Nfkb2−/− Tregs compared to WT cells. Genes associated with cell survival and proliferation are annotated. Genes were selected from the gene sets enriched in GSEA analysis.
Discussion
The NF-κB signaling pathways occupy an important place in Treg biology. The canonical NF-κB signaling axis promotes Treg development and is also important in maintaining the peripheral Treg population. Absence of IKKβ, cRel or RelA leads to a reduction in the thymic Treg numbers56,57, implicating the importance of canonical NF-κB axis in Treg development. Further, p50−/− mice show a reduction in splenic Treg numbers57, extending the significance of the canonical NF-κB pathway in regulating peripheral Tregs. Both cRel and RelA also regulate peripheral Treg homeostasis and function with some redundancy between them, and a deficiency of either cRel or RelA in Tregs leads to compromised Treg function as well as lymphoproliferative disease58.
Non-canonical NF-κB signaling also plays a role in Treg development and homeostasis. NIK- or IKKα-deficient mice show a reduction in thymic Treg numbers, although the effect of the NIK deficiency appears to be Treg-extrinsic28,29. Nonetheless, the absence of NIK does result in reduced peripheral Treg numbers29. T cell-specific deletion of IKK-alpha also results in reduced Treg proliferation and suppressive function in vivo28. However, the kinases NIK and IKKα can also activate canonical NF-κB signaling59; and hence, Treg alterations in the absence of these kinases might not reflect direct effects on non-canonical NF-κB signaling. We have therefore extended these studies by analyzing mice with a deletion of the Nfkb2 gene, which plays a critical role in non-canonical NF-κB signaling. Our results both confirm and extend recently reported findings48,60.
The thymic development of Tregs was largely comparable between WT and Nfkb2−/− mice. However, the prominence of splenic iTregs was notably lower in the Nfkb2−/− mice. The difference persisted in bi-parental bone marrow chimeric mice, indicating that the effect was likely cell-autonomous. This suggested that iTreg induction from conventional NCD4 T cells would be less efficient in the absence of functional NF-κB2 in those cells. Our data from in vitro generation of iTregs validates this expectation. The crosstalk involving the NF-κB2 protein and TGF-β signaling involved in iTreg induction remains to be elucidated.
The peripheral Treg phenotype of Nfkb2−/− mice was also notable for higher proportions of splenic effector Tregs. Despite the reduction in iTreg prominence, this effector phenotype was observed in both nTregs and iTregs. Again, the difference remained quite evident in bi-parental bone marrow chimeric mice as well, indicating a cell-autonomous effect of NF-kB2 deficit. Moreover, higher Nfkb2−/− Treg frequencies were also found in the liver of bi-parental bone marrow chimeras, consistent with the greater prominence of Nfkb2−/− effector Tregs. In addition, the proportions of CD103-expressing Tregs, which constitute a subpopulation of the effector Tregs that have been reported to be more efficient at suppressing T cell-induced colitis47, were also found to be higher in the Nfkb2−/− splenic Tregs.
In addition to these quantitative differences, Nfkb2−/− Tregs also differed from WT Tregs in the levels of expression of Treg activation-associated molecules as well as of molecules highly expressed on effector Tregs. The levels of GITR on Nfkb2−/− Tregs and the frequencies of TIGIT+ Nfkb2−/− Tregs were strikingly higher than WT Tregs consistent with the higher proportions of effector Tregs seen in the former20. Nonetheless, expression of other surface molecules known to show higher expression on eTregs, such as IL-2Rβ11 and GARP45, were found to be reduced on Nfkb2−/− Tregs. The levels of IRF4 were also reduced in Nfkb2−/− Tregs albeit modestly, though IRF4 is known to be involved in the expansion of eTregs21. The higher Neuropilin-1 expression on Nfkb2−/− Tregs may also in part reflect changes in iTreg and nTreg distribution, since Neuropilin-1 has been proposed to be a marker of nTregs61. The expression of molecules contributing to Treg suppressive functions, namely CTLA-441, CD2844 and Neuropilin-142,43, also showed diverse patterns. CD28 levels were comparable between WT and Nfkb2−/− Tregs, while there was higher expression of Neuropilin-1 and lower CTLA-4 expression in Nfkb2−/− Tregs. Despite lower levels of CTLA-4, Nfkb2−/− Tregs showed unaltered capacities compared to WT Tregs to suppress NCD4 T cell proliferation in vitro and in vivo. These data suggest that the absence of NF-kB2, in addition to increasing eTreg frequencies, leads to qualitative, aberrant changes in Nfkb2−/− Tregs, with consequences for their activation as well as functional status.
Notably, a recent report showed impaired Nfkb2−/− Treg function in older mice, associated with immune inflammation48, while the present data showed no impairment. It is thus possible that Nfkb2−/− Tregs begin to be compromised in their suppressive functions over time, rather than being poorly functional ab initio, and such a role for Nfkb2 in maintaining Treg responsiveness over time would be of interest.
Interestingly, Nfkb2−/− Tregs, both cTregs and eTregs, survived better and underwent greater proliferation in vivo. Further, Nfkb2−/− cTregs converted to eTregs more efficiently, while Nfkb2−/− eTregs displayed lower reversion to the cTreg phenotype in vivo. These data help explain the Treg phenotype in Nfkb2−/− mice, and indicate that Nfkb2 likely contributes a net negative effect on Treg proliferation, activation and survival, although our data do not provide definitive evidence for separate effects on each of these parameters. All these factors possibly contribute to the expansion of Tregs in total and eTregs in particular seen in the bi-parental chimeras within the Nfkb2−/− donor.
Thus, while the absence of NIK and IKKα led to a reduction in Treg numbers in the spleen28,29, the absence of Nfkb2−/− actually led to Treg expansion. Notably, RelB deficiency did not recapitulate this phenotype. This indicated differential modes of action of NIK and IKKα in Tregs and suggested that they act by means of activating the canonical NF-κB pathway rather than through the activity of RelB:p52 dimers. Thus, these results are consistent at first glance with the mechanistic possibility that the non-canonical NF-κB pathway negatively regulates processes involved in promoting Treg activation, proliferation and survival.
In addition to the role of NF-κB2/p100 in mediating non-canonical NF-κB signaling, it is also known to be able to sequester the major mediators of both the canonical and the non-canonical NF-κB pathways, RelA, cRel and RelB, in the cytoplasm, via its C-terminal ankyrin repeats26,31. Thus, NF-κB2/p100 can also function as an inhibitor of both NF-κB pathways. There is evidence that NF-κB2/p100 plays an inhibitory role in TCR-mediated canonical NF-κB activation62,63. The generation of effector Tregs is dependent on signaling through the TCR, with the likely involvement of cRel-dependent canonical NF-κB signaling19,20. Additionally, both cRel25 and RelA24 have been shown to regulate homeostasis of effector Tregs. Thus, it is possible that the absence of NF-κB2/p100 in Tregs can result in enhanced canonical NF-κB activity via RelA and cRel. This could well explain the increased proliferation, activation and survival of effector Tregs.
When we tested this possibility by generating bi-parental chimeras of WT and Relb−/−mice, it was evident that Tregs of the Relb−/− genotype did not show any expansion of peripheral Tregs or any increase in the prominence of effector Tregs. Thus, non-canonical RelB-dependent NF-κB activity does not provide any negative regulatory signals to Tregs. It is thus plausible that NF-κB2/p100 primarily functions non-redundantly as a negative regulator of NF-κB activity. The NF-κB2/p100 protein is typically proteolytically processed to p52, which dimerizes with RelB to complete non-canonical NF-κB signaling26, indicating that NF-κB2/p100 is important for induction of non-canonical NF-κB signaling. However, in the absence of NF-κB2, RelB can also use the p50 protein from the canonical pathway as a dimer partner for transcriptional activity34,64. In this context, it was noteworthy that, while NF-κB activity in WT Tregs consisted primarily of RelA and cRel, Nfkb2−/− Tregs showed higher NF-κB activity mainly involving RelB. Interestingly, multiple myeloma cells with mutations that lead to complete degradation of NF-κB2/p100 have been reported to show a similar enhancement of TNF-alpha-mediated RelB:p50 NF-κB activity, mediating the expression of pro-survival genes34.
A recent report has also shown that a Treg-specific deletion of RelB, did not bring about any alteration in the peripheral Treg numbers or their suppressive function60. This indicates, consistent with our results, that RelB is dispensable for the regulation of peripheral Treg homeostasis. Additionally, our data further indicate that RelB activity also likely participates in the greater prominence of effector Tregs in the absence of Nfkb2, making it possible that RelB regulates the transcription of genes involved in eTreg generation in addition to those involved in Treg proliferation and survival.
NF-κB2/p100 in T cells has been reported to have a negative regulatory effect, through regulation of co-stimulation, on TCR activation-driven events such as proliferation65. Treg cells show substantial TCR-mediated tonic signalling in vivo66. It is therefore possible that Nfkb2−/− Tregs show such heightened TCR-induced signaling, contributing to enhanced activation and proliferation as well as effector conversion in vivo, including the formation of CD103+ Tregs. Similarly, while our study finds altered prominence of iTregs in the absence of Nfkb2, it must be kept in mind that our definition of the iTreg/nTreg distinction is based on the expression of Helios. As already noted38, this is a tentative identification at best. Further, Helios-expressing Tregs have been shown to be enriched in eTregs67,68.Thus, it is in fact possible that Helios expression and iTreg and eTreg prominence in the Treg compartment are also modified by the enhancement of TCR-mediated tonic signalling in vivo. In this context, cRel25 and RelA69 have also been implicated in regulating Helios expression, making it possible that removal of the negative regulatory effect of NF-κB2/p100 on either cRel or RelA activity contributes to the observed differences in iTreg to nTreg ratio among Nfkb2−/− Tregs. Finally, despite these caveats, our finding that deficiency of Nfkb2 results in poor Treg induction in vitro from naïve conventional CD4 T cells confirms previous similar findings48,70, and is consistent with our in vivo observation of a reduction in the iTreg compartment among Nfkb2−/− Tregs.
Another recent study has used a Treg-specific deletion of Nfkb2 and has reported findings similar to ours in terms of increased prominence of Tregs and of the effector phenotype amongst them48. This report also showed that, while Treg-specific Nfkb2-deficient mice did not develop any disease at a young age, from about one year of age they began showing immune inflammatory lesions associated with poor Treg suppressive capacity48. These features required RelB activity, since mice with Tregs lacking both RelB and Nfkb2 showed normal Treg functionality and no immune inflammation48.
However, when we compared the suppressive functions of WT and Nfkb2−/− Tregs sorted from younger mice, either parental mice or bi-parental chimeras, we found no differences. We have not yet examined suppressive effects of Tregs from aged bi-parental chimeras. It remains an intriguing possibility that, as Nfkb2−/− Treg compartments age, with the enhanced activation and proliferation observed in these cells, suppressive activity declines perhaps as a result of induction of Th17 programming as suggested in the previous report48.
Despite being apparently functionally normal in young mice, the Nfkb2−/− Tregs in our study do show some altered characteristics reported previously as well, namely, reduced levels of CTLA-4 and enhanced GITR levels possibly related to heightened noncanonical NF-κB signaling48,71. It has been shown earlier that an absence of Nfkb2-p100 provokes enhanced RelB:p50 activity via the canonical NF-κB pathway in mouse embryonic fibroblasts, and that RelB:p50 induces a distinct set of genes, which are not activated by RelA:p50 and are associated with cell survival and cell division72. Our biochemical analyses revealed heightened nuclear activity of RelB:p50 in functionally normal Nfkb2−/− Tregs, while our global transcriptomic studies established an altered gene expression pattern in them favoring pro-survival and pro-proliferative functions. These analyses indicated that Nfkb2-p100 may also modulate Treg functions by tuning FOXP3 activity. Taken together, we argue that increased nuclear RelB:p50 activity in Nfkb2−/− Tregs, in the absence of Nfkb2-p100, may contribute to their higher proliferation and survival. It is therefore now attractive to hypothesize that, in the absence of Nfkb2, it is the persistence of unchecked RelB-NF-kappaB activity that over time leads to compromised Treg phenotype, and eventually, function. Thus, pathways mediating enhanced Treg activation, proliferation and survival may also in time lead to loss of function, a balance that may be maintained by NF-κB2. Our study highlights interesting interplay between non-canonical and canonical NF-κB pathway intermediates involving Nfkb2.
Materials and Methods
Mice
All mouse strains used in this study: C57BL/6, B6.TCRsß∂−/−, C57BL/6-Tg(UBC-GFP)30Scha/J (B6.GFP), B6.SJL-Ptprca Pepcb/BoyJ (B6.SJL), B6.SJLXC57BL/6 F1(CD45.1+CD45.2+) and Nfkb2(p100)−/−73 were obtained from The Jackson Laboratory (Bar Harbor, ME, USA) and were bred in the small animal facility of the National Institute of Immunology, New Delhi, India. All mice were used at 6–12 weeks of age and bi-parental chimeras were used 2–4 months post reconstitution. All mice were maintained and used in accordance with the guidelines and with the prior approval of the duly constituted ‘Institutional Animal Ethics Committee’ (IAEC) of the National Institute of Immunology. All methods were performed in accordance with relevant guidelines and regulations. All experimental protocols were approved by the Institutional Animal Ethics Committee (IAEC) authorised for this purpose.
Ex-vivo cell isolation
Mice were euthanised by cervical dislocation and various organs were dissected. Erythrocyte lysis using Gey’s solution was used where necessary. Livers were perfused prior to preparation of single-cell suspensions, and a Percoll gradient centrifugation step was used to isolate leukocytes.
Flow cytometry
Cells from various tissues were stained with combinations of the following antibodies to either identify NCD4 T cells, non-T cells (CD90−) and various Treg subsets, and to analyse expression of specific intracellular/extracellular markers on Tregs. The antibodies used were; CD90 (5E10), CD4 (RM4-5), CD8 (53–6.7), CD44 (IM7), CD62L (MEL-14), CD25 (PC61.5), CD45.1 (A20), CD45.2 (104), HELIOS(22-F6), FOXP3(FJK-16s), CD357(GITR) (DTA-1), CD103(2E7), CD152(CTLA-4) (UC10-4B9), CD28(CD28.2), TIGIT(MBSA43), CD279(PD-1) (J43), IRF4(3E4), CD304(Neuropilin-1) (3DS304M), GARP(YGIC86), CD122(IL-2Rβ)(5H4), CD95(Fas)(15A7), CD215(IL-15Rα)(DNT15Ra) and Egr2 (erongr2) (eBioscience, ThermoFisher Scientific, Carlsbad, CA). Surface marker staining was performed in staining buffer (phosphate-buffered saline (PBS) + 1% fetal bovine serum (FBS) + 0.05%NaN3) at 4 °C for 20 min followed by two washes with staining buffer. For intracellular proteins, cells were first fixed and permeabilised using Foxp3/Transcription Factor Staining Buffer Set (eBioscience, ThermoFisher Scientific, Carlsbad, CA) and then stained with appropriate combinations of antibodies. Sytox dead cell stain (MolecularProbes, ThermoFisher Scientific, Carlsbad, CA) was used to stain dead cells in order to assess cell viability. Fixable Violet (Invitrogen, ThermoFisher Scientific, Carlsbad, CA) was used to label dead cells before fixation and permeabilisation. Flow cytometric data collection was done (FACSVerse; BD Biosciences) and data analysed with FlowJo software (Treestar, Ashland, OR).
Bi-parental bone marrow chimera generation
B6.GFP or C57BL/6 X B6.SJL F1s (CD45.1+CD45.2+) were subjected to a 10 Gy dose of gamma-radiation (Source: Co60, BARC, Mumbai). Five million bone marrow cells each from WT (B6.SJL: expressing CD45.1) and Nfkb2−/− (expressing CD45.2) mice were transferred intravenously into the gamma-irradiated mice and were allowed to reconstitute. After 8 weeks, reconstitution of both donors within the recipient was assessed in the peripheral blood of the recipients. Mice with >80% overall donor reconstitution were used for analysis.
Cell sorting
Ex-vivo spleen cells were suspended in complete RPMI1640 medium (Biological Industries, Beit Haemek, Israel) supplemented with 10% FBS, 2 mM L-glutamine, 1.35 g/l sodium bicarbonate, 5 mM HEPES, 50 μM β-mercaptoethanol, 100 μg/ml streptomycin and 100 IU penicillin (Millipore Sigma, Merck KGaA, Germany) and stained with the relevant antibody combinations and incubated at 4 °C for 20 min. The cells were then washed, re-suspended at a density of 30 million cells/ml in complete RPMI and filtered through a 40-micron size sieve. This was followed by analysis and sorting (BD FACSAria III; BD Biosciences, CA). Sorted populations showed >90% purity.
Treg suppression assay
WT and Nfkb2−/− Tregs (CD4+CD25+) were sorted from bi-parental bone marrow chimeras and NCD4 T cells were sorted from B6.GFP mice and labelled with CellTrace violet dye. For estimating suppression in vitro, labelled NCD4 T cells were cultured in the absence or presence of varying Treg numbers with aqueous phase anti-mouse CD3 (2 µg/ml) and gamma-irradiated (10 Gy) CD90− splenocytes from C57BL/6 mice in complete RPMI. After 60 h of culture, the degree of proliferation of the live NCD4 T cells was determined by dye dilution to test the suppressive function of the respective Treg population. For in vivo suppression assays, labelled NCD4 T cells were transferred into B6.TCRβδ−/− mice either without Tregs or with purified WT or Nfkb2−/− Tregs from bi-parental bone marrow chimeras respectively at a ratio of 3:1 (NCD4:Tregs).Three days post-transfer, proliferation of the transferred NCD4 T cells was assessed in the spleens of the recipient mice by dye dilution.
In vitro iTreg generation
Generation of iTreg cells was performed according to a protocol described previously74. WT and Nfkb2−/− NCD4 T cells were sorted from the mixed bone marrow chimeras, mixed in a 1:1 ratio (0.1 million cells of each) and cultured in vitro with plate-bound anti-CD3 (5 µg/ml) and anti-CD28 (5 µg/ml) in the presence of recombinant IL-2 (10 U/ml) and recombinant TGF-β1 (10 ng/ml) in complete RPMI. After 3–5 days of culture, cells were stained with fixable violet dye, fixed and permeabilised and then stained with anti-mouse CD45.1, CD45.2, CD4, CD25 and FOXP3 to identify the frequency of Tregs generated from WT and Nfkb2−/− NCD4 T cells.
In vivo parking experiment
WT and Nfkb2−/− central and effector Tregs respectively were sorted from mixed bone marrow chimeras and 0.3 million cells of each type were mixed together equally and intravenously transferred into CD45.1+CD45.2+ F1 mice. After 3–10 days of parking in vivo, spleen cells of the recipients were harvested and stained with anti-mouse CD45.1, CD45.2, CD4, CD25, FOXP3, CD62L and Ki67. Tregs were identified as CD4+CD25+FOXP3+ cells and among them the transferred populations were identified as WT (CD45.1+) or Nfkb2−/− (CD45.2+) Tregs. The transition from central to effector Treg phenotype (or vice versa) was determined by analysing the expression of CD62L on the transferred Tregs. The frequency of proliferating cells among the transferred populations was determined by assessing frequency of Ki67+ cells.
RNA-seq analysis
WT and Nfkb2−/− Tregs were isolated as described above from spleens of bi-parental bone marrow chimeras. Sorted cells were rested in complete RPMI at 37 °C for 2 h before being re-suspended in 1 ml TRIzol reagent (Thermofisher Scientific, CA) and stored at −80 °C. Three such biological repeats of WT and Nfkb2−/− Tregs were collected. RNA was isolated by treatment with chloroform, isopropanol and ethanol according to the RNA isolation protocol using TRIzol reagent as recommended. The RNA extracted was quantified using NanoDrop1000 (Thermofisher Scientific, CA). Unstranded RNAseq libraries were prepared using TruSeq RNA Sample Prep Kit v2 (Ilumina, San Diego) according to the kit protocol, and barcoded libraries were sequenced in Illumina HiSeq2500 (Macrogen, S. Korea). Fastq files generated by paired-end sequencing reactions were quality controlled using FastQC package75, followed by mapping to the mouse genome using HISAT2 tool76. The BAM file generated was used to count the reads mapped to each gene with htseq-count77. Read quality check, mapping and counting were done in galaxy public server78. The read counts generated were analysed for differential gene expression using DEseq2 package79. g:profiler80 was used for gene ontology analysis of upregulated genes. Genomic sequence and the annotation file was obtained from ENSEMBL81. For gene set enrichment analysis, the GSEA tool (Broad institute) was used with the collections H, C2 and C582. Log2-fold changes for genes belonging to the significantly enriched sets related to proliferation and survival were used for heatmaps.
Statistical analyses
Student’s ‘t’ test was used to calculate statistical significance of experimental data.
Supplementary information
Acknowledgements
We thank Mr. Inder Jit Singh for help with animal maintenance and Mr. Rajesh Kumar for extensive help with electronic cell sorting. The study was supported in part by grants from the Department of Biotechnology (to AG # BT/PR12849/MED/15/35/2009; to VB # BT/PR14420/Med/29/213/2010; and to SR # BT/PR‐14592/BRB/10/858/2010), and from the Department of Science and Technology, Government of India (to VB # SR/SO/HS‐0005/2011 and #EMR/2015/001074; to SR # SB/SO/HS/210/2013). The National Institute of Immunology is supported by the Department of Biotechnology, Government of India.
Author Contributions
A.D. planned and performed critical experiments, analysed data and wrote the manuscript. A.D. also drew the schemas provided in the Supplementary Information (Figs S4, S5, S7). M.C. helped in planning, performing and analysing some key experiments critical for the study. S.C., N.O., D.U. and S.G. helped both in providing some key reagents and in the performance of some crucial experiments. V.B., A.G., S.R., G.A.A. and S.B. conceived the questions and approach, planned experiments, analysed data, wrote the manuscript, and share senior authorship. All authors read and approved the final manuscript.
Data Availability
The RNA-seq data generated and analysed in the manuscript have been made available by submission to the Geo repository (accession number awaited).
Competing Interests
S.R. is a non‐executive director of Ahammune Biosciences Private Limited, Pune, India, and a member of the scientific advisory boards of Curadev Pharma Private Limited, NOIDA, India, and Mynvax Private Limited, Bangalore, India. Other authors have no competing interests to declare.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-50454-z.
References
- 1.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- 2.Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. Journal of immunology. 1995;155:1151–1164. [PubMed] [Google Scholar]
- 3.Read S, Malmstrom V, Powrie F. Cytotoxic T lymphocyte-associated antigen 4 plays an essential role in the function of CD25(+)CD4(+) regulatory cells that control intestinal inflammation. The Journal of experimental medicine. 2000;192:295–302. doi: 10.1084/jem.192.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burmeister Y, et al. ICOS controls the pool size of effector-memory and regulatory T cells. Journal of immunology. 2008;180:774–782. doi: 10.4049/jimmunol.180.2.774. [DOI] [PubMed] [Google Scholar]
- 5.Deaglio S, et al. Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. The Journal of experimental medicine. 2007;204:1257–1265. doi: 10.1084/jem.20062512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curotto de Lafaille MA, Lafaille JJ. Natural and adaptive foxp3+ regulatory T cells: more of the same or a division of labor? Immunity. 2009;30:626–635. doi: 10.1016/j.immuni.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Kastenmuller W, et al. Regulatory T cells selectively control CD8+ T cell effector pool size via IL-2 restriction. Journal of immunology. 2011;187:3186–3197. doi: 10.4049/jimmunol.1101649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joetham A, et al. Naturally occurring lung CD4(+)CD25(+) T cell regulation of airway allergic responses depends on IL-10 induction of TGF-beta. Journal of immunology. 2007;178:1433–1442. doi: 10.4049/jimmunol.178.3.1433. [DOI] [PubMed] [Google Scholar]
- 9.Gondek DC, Lu LF, Quezada SA, Sakaguchi S, Noelle RJ. Cutting edge: contact-mediated suppression by CD4+CD25+ regulatory cells involves a granzyme B-dependent, perforin-independent mechanism. Journal of immunology. 2005;174:1783–1786. doi: 10.4049/jimmunol.174.4.1783. [DOI] [PubMed] [Google Scholar]
- 10.Oderup C, Cederbom L, Makowska A, Cilio CM, Ivars F. Cytotoxic T lymphocyte antigen-4-dependent down-modulation of costimulatory molecules on dendritic cells in CD4+ CD25+ regulatory T-cell-mediated suppression. Immunology. 2006;118:240–249. doi: 10.1111/j.1365-2567.2006.02362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisson S, et al. Continuous activation of autoreactive CD4+ CD25+ regulatory T cells in the steady state. The Journal of experimental medicine. 2003;198:737–746. doi: 10.1084/jem.20030686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee JH, Kang SG, Kim CH. FoxP3+ T cells undergo conventional first switch to lymphoid tissue homing receptors in thymus but accelerated second switch to nonlymphoid tissue homing receptors in secondary lymphoid tissues. Journal of immunology. 2007;178:301–311. doi: 10.4049/jimmunol.178.1.301. [DOI] [PubMed] [Google Scholar]
- 13.Szanya V, Ermann J, Taylor C, Holness C, Fathman CG. The subpopulation of CD4+CD25+ splenocytes that delays adoptive transfer of diabetes expresses L-selectin and high levels of CCR7. Journal of immunology. 2002;169:2461–2465. doi: 10.4049/jimmunol.169.5.2461. [DOI] [PubMed] [Google Scholar]
- 14.Smigiel KS, et al. CCR7 provides localized access to IL-2 and defines homeostatically distinct regulatory T cell subsets. The Journal of experimental medicine. 2014;211:121–136. doi: 10.1084/jem.20131142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liston A, Gray DH. Homeostatic control of regulatory T cell diversity. Nature reviews. Immunology. 2014;14:154–165. doi: 10.1038/nri3605. [DOI] [PubMed] [Google Scholar]
- 16.Sather BD, et al. Altering the distribution of Foxp3(+) regulatory T cells results in tissue-specific inflammatory disease. The Journal of experimental medicine. 2007;204:1335–1347. doi: 10.1084/jem.20070081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo Z, et al. CD4+CD25+ regulatory T cells in the small intestinal lamina propria show an effector/memory phenotype. International immunology. 2008;20:307–315. doi: 10.1093/intimm/dxm143. [DOI] [PubMed] [Google Scholar]
- 18.Kitamura K, Farber JM, Kelsall BL. CCR6 marks regulatory T cells as a colon-tropic, IL-10-producing phenotype. Journal of immunology. 2010;185:3295–3304. doi: 10.4049/jimmunol.1001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levine AG, Arvey A, Jin W, Rudensky AY. Continuous requirement for the TCR in regulatory T cell function. Nature immunology. 2014;15:1070–1078. doi: 10.1038/ni.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vahl JC, et al. Continuous T cell receptor signals maintain a functional regulatory T cell pool. Immunity. 2014;41:722–736. doi: 10.1016/j.immuni.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 21.Cretney E, et al. The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nature immunology. 2011;12:304–311. doi: 10.1038/ni.2006. [DOI] [PubMed] [Google Scholar]
- 22.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annual review of immunology. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 23.Gerondakis S, Fulford TS, Messina NL, Grumont RJ. NF-kappaB control of T cell development. Nature immunology. 2014;15:15–25. doi: 10.1038/ni.2785. [DOI] [PubMed] [Google Scholar]
- 24.Messina N, et al. The NF-kappaB transcription factor RelA is required for the tolerogenic function of Foxp3(+) regulatory T cells. Journal of autoimmunity. 2016;70:52–62. doi: 10.1016/j.jaut.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 25.Grinberg-Bleyer Y, et al. NF-kappaB c-Rel is Crucial for the Regulatory T Cell Immune Checkpoint in Cancer. Cell. 2017;170:1096–1108 e1013. doi: 10.1016/j.cell.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun SC. The non-canonical NF-kappaB pathway in immunity and inflammation. Nature reviews. Immunology. 2017;17:545–558. doi: 10.1038/nri.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schaer DA, et al. GITR pathway activation abrogates tumor immune suppression through loss of regulatory T cell lineage stability. Cancer immunology research. 2013;1:320–331. doi: 10.1158/2326-6066.CIR-13-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen X, et al. IKKalpha is required for the homeostasis of regulatory T cells and for the expansion of both regulatory and effector CD4 T cells. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2015;29:443–454. doi: 10.1096/fj.14-259564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murray SE. Cell-intrinsic role for NF-kappa B-inducing kinase in peripheral maintenance but not thymic development of Foxp3+ regulatory T cells in mice. PloS one. 2013;8:e76216. doi: 10.1371/journal.pone.0076216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weih F, et al. Both multiorgan inflammation and myeloid hyperplasia in RelB-deficient mice are T cell dependent. Journal of immunology. 1996;157:3974–3979. [PubMed] [Google Scholar]
- 31.Tao Z, et al. p100/IkappaBdelta sequesters and inhibits NF-kappaB through kappaBsome formation. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:15946–15951. doi: 10.1073/pnas.1408552111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shih VF, et al. Kinetic control of negative feedback regulators of NF-kappaB/RelA determines their pathogen- and cytokine-receptor signaling specificity. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:9619–9624. doi: 10.1073/pnas.0812367106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Almaden JV, et al. A pathway switch directs BAFF signaling to distinct NFkappaB transcription factors in maturing and proliferating B cells. Cell reports. 2014;9:2098–2111. doi: 10.1016/j.celrep.2014.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roy P, et al. Non-canonical NFkappaB mutations reinforce pro-survival TNF response in multiple myeloma through an autoregulatory RelB:p50 NFkappaB pathway. Oncogene. 2017;36:1417–1429. doi: 10.1038/onc.2016.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bren GD, et al. Transcription of the RelB gene is regulated by NF-kappaB. Oncogene. 2001;20:7722–7733. doi: 10.1038/sj.onc.1204868. [DOI] [PubMed] [Google Scholar]
- 36.Lio CW, Hsieh CS. A two-step process for thymic regulatory T cell development. Immunity. 2008;28:100–111. doi: 10.1016/j.immuni.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez-Perea AL, Arcia ED, Rueda CM, Velilla PA. Phenotypical characterization of regulatory T cells in humans and rodents. Clinical and experimental immunology. 2016;185:281–291. doi: 10.1111/cei.12804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin X, et al. Advances in distinguishing natural from induced Foxp3(+) regulatory T cells. International journal of clinical and experimental pathology. 2013;6:116–123. [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu M, et al. NF-kappaB2 is required for the establishment of central tolerance through an Aire-dependent pathway. The Journal of clinical investigation. 2006;116:2964–2971. doi: 10.1172/JCI28326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caamano JH, et al. Nuclear factor (NF)-kappa B2 (p100/p52) is required for normal splenic microarchitecture and B cell-mediated immune responses. The Journal of experimental medicine. 1998;187:185–196. doi: 10.1084/jem.187.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tai X, et al. Basis of CTLA-4 function in regulatory and conventional CD4(+) T cells. Blood. 2012;119:5155–5163. doi: 10.1182/blood-2011-11-388918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao YL, et al. Neuropilin-1highCD4(+)CD25(+) Regulatory T Cells Exhibit Primary Negative Immunoregulation in Sepsis. Mediators Inflamm. 2016;2016:7132158. doi: 10.1155/2016/7132158. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Sarris M, Andersen KG, Randow F, Mayr L, Betz AG. Neuropilin-1 expression on regulatory T cells enhances their interactions with dendritic cells during antigen recognition. Immunity. 2008;28:402–413. doi: 10.1016/j.immuni.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang R, et al. An obligate cell-intrinsic function for CD28 in Tregs. The Journal of clinical investigation. 2013;123:580–593. doi: 10.1172/JCI65013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou AX, Kozhaya L, Fujii H, Unutmaz D. GARP-TGF-beta complexes negatively regulate regulatory T cell development and maintenance of peripheral CD4+ T cells in vivo. Journal of immunology. 2013;190:5057–5064. doi: 10.4049/jimmunol.1300065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clark RA, Kupper TS. IL-15 and dermal fibroblasts induce proliferation of natural regulatory T cells isolated from human skin. Blood. 2007;109:194–202. doi: 10.1182/blood-2006-02-002873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lehmann J, et al. Expression of the integrin alpha Ebeta 7 identifies unique subsets of CD25+ as well as CD25- regulatory T cells. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:13031–13036. doi: 10.1073/pnas.192162899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grinberg-Bleyer Y, et al. The Alternative NF-kappaB Pathway in Regulatory T Cell Homeostasis and Suppressive Function. Journal of immunology. 2018;200:2362–2371. doi: 10.4049/jimmunol.1800042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fabregat A, et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018;46:D649–D655. doi: 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edilova MI, Abdul-Sater AA, Watts TH. TRAF1 Signaling in Human Health and Disease. Front Immunol. 2018;9:2969. doi: 10.3389/fimmu.2018.02969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smolewski P, Robak T. Inhibitors of apoptosis proteins (IAPs) as potential molecular targets for therapy of hematological malignancies. Curr Mol Med. 2011;11:633–649. doi: 10.2174/156652411797536723. [DOI] [PubMed] [Google Scholar]
- 53.Kale J, Osterlund EJ, Andrews DW. BCL-2 family proteins: changing partners in the dance towards death. Cell Death Differ. 2018;25:65–80. doi: 10.1038/cdd.2017.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tigan AS, Bellutti F, Kollmann K, Tebb G, Sexl V. CDK6-a review of the past and a glimpse into the future: from cell-cycle control to transcriptional regulation. Oncogene. 2016;35:3083–3091. doi: 10.1038/onc.2015.407. [DOI] [PubMed] [Google Scholar]
- 55.Risley MD, Clowes C, Yu M, Mitchell K, Hentges KE. The Mediator complex protein Med31 is required for embryonic growth and cell proliferation during mammalian development. Dev Biol. 2010;342:146–156. doi: 10.1016/j.ydbio.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 56.Schmidt-Supprian M, et al. Mature T cells depend on signaling through the IKK complex. Immunity. 2003;19:377–389. doi: 10.1016/S1074-7613(03)00237-1. [DOI] [PubMed] [Google Scholar]
- 57.Isomura I, et al. c-Rel is required for the development of thymic Foxp3+ CD4 regulatory T cells. The Journal of experimental medicine. 2009;206:3001–3014. doi: 10.1084/jem.20091411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oh H, et al. An NF-kappaB Transcription-Factor-Dependent Lineage-Specific Transcriptional Program Promotes Regulatory T Cell Identity and Function. Immunity. 2017;47:450–465 e455. doi: 10.1016/j.immuni.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zarnegar B, Yamazaki S, He JQ, Cheng G. Control of canonical NF-kappaB activation through the NIK-IKK complex pathway. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3503–3508. doi: 10.1073/pnas.0707959105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li J, et al. Role of the NF-kappaB Family Member RelB in Regulation of Foxp3(+) Regulatory T Cells In Vivo. Journal of immunology. 2018;200:1325–1334. doi: 10.4049/jimmunol.1701310. [DOI] [PubMed] [Google Scholar]
- 61.Weiss Jonathan M., Bilate Angelina M., Gobert Michael, Ding Yi, Curotto de Lafaille Maria A., Parkhurst Christopher N., Xiong Huizhong, Dolpady Jayashree, Frey Alan B., Ruocco Maria Grazia, Yang Yi, Floess Stefan, Huehn Jochen, Oh Soyoung, Li Ming O., Niec Rachel E., Rudensky Alexander Y., Dustin Michael L., Littman Dan R., Lafaille Juan J. Neuropilin 1 is expressed on thymus-derived natural regulatory T cells, but not mucosa-generated induced Foxp3+T reg cells. The Journal of Experimental Medicine. 2012;209(10):1723–1742. doi: 10.1084/jem.20120914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ishimaru N, Kishimoto H, Hayashi Y, Sprent J. Regulation of naive T cell function by the NF-kappaB2 pathway. Nature immunology. 2006;7:763–772. doi: 10.1038/ni1351. [DOI] [PubMed] [Google Scholar]
- 63.Legarda-Addison D, Ting AT. Negative regulation of TCR signaling by NF-kappaB2/p100. Journal of immunology. 2007;178:7767–7778. doi: 10.4049/jimmunol.178.12.7767. [DOI] [PubMed] [Google Scholar]
- 64.Derudder E, et al. RelB/p50 dimers are differentially regulated by tumor necrosis factor-alpha and lymphotoxin-beta receptor activation: critical roles for p100. The Journal of biological chemistry. 2003;278:23278–23284. doi: 10.1074/jbc.M300106200. [DOI] [PubMed] [Google Scholar]
- 65.Giardino Torchia ML, Conze DB, Jankovic D, Ashwell JD. Balance between NF-kappaB p100 and p52 regulates T cell costimulation dependence. Journal of immunology. 2013;190:549–555. doi: 10.4049/jimmunol.1201697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moran AE, et al. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. The Journal of experimental medicine. 2011;208:1279–1289. doi: 10.1084/jem.20110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thornton AM, et al. Helios(+) and Helios(−) Treg subpopulations are phenotypically and functionally distinct and express dissimilar TCR repertoires. Eur J Immunol. 2019;49:398–412. doi: 10.1002/eji.201847935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zabransky DJ, et al. Phenotypic and functional properties of Helios+ regulatory T cells. PloS one. 2012;7:e34547. doi: 10.1371/journal.pone.0034547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vasanthakumar A, et al. The TNF Receptor Superfamily-NF-kappaB Axis Is Critical to Maintain Effector Regulatory T Cells in Lymphoid and Non-lymphoid Tissues. Cell reports. 2017;20:2906–2920. doi: 10.1016/j.celrep.2017.08.068. [DOI] [PubMed] [Google Scholar]
- 70.Xiao X, et al. GITR subverts Foxp3(+) Tregs to boost Th9 immunity through regulation of histone acetylation. Nat Commun. 2015;6:8266. doi: 10.1038/ncomms9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shimizu J, Yamazaki S, Takahashi T, Ishida Y, Sakaguchi S. Stimulation of CD25(+)CD4(+) regulatory T cells through GITR breaks immunological self-tolerance. Nature immunology. 2002;3:135–142. doi: 10.1038/ni759. [DOI] [PubMed] [Google Scholar]
- 72.Chatterjee B, et al. Immune Differentiation Regulator p100 Tunes NF-kappaB Responses to TNF. Front Immunol. 2019;10:997. doi: 10.3389/fimmu.2019.00997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Banoth, B. et al. Stimulus-selective crosstalk via the NF-kappaB signaling system reinforces innate immune response to alleviate gut infection. eLife4, 10.7554/eLife.05648 (2015). [DOI] [PMC free article] [PubMed]
- 74.Fantini MC, Dominitzki S, Rizzo A, Neurath MF, Becker C. In vitro generation of CD4+ CD25+ regulatory cells from murine naive T cells. Nature protocols. 2007;2:1789–1794. doi: 10.1038/nprot.2007.258. [DOI] [PubMed] [Google Scholar]
- 75.Simon, A. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc (2012).
- 76.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Afgan E, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46:W537–W544. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. 2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reimand J, Kull M, Peterson H, Hansen J, Vilo J. g:Profiler–a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35:W193–200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zerbino DR, et al. Ensembl 2018. Nucleic Acids Res. 2018;46:D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data generated and analysed in the manuscript have been made available by submission to the Geo repository (accession number awaited).