Abstract
BACKGROUND
Highly upregulated in liver cancer (HULC) is a long non-coding RNA (lncRNA) which has recently been identified as a key regulator in hepatocellular carcinoma (HCC) progression. However, its role in the secretion of exosomes from HCC cells remains unknown.
AIM
To explore the mechanism by which HULC promotes the secretion of exosomes from HCC cells.
METHODS
Serum and liver tissue samples were collected from 30 patients with HCC who had not received chemotherapy, radiotherapy, or immunotherapy before surgery. HULC expression in serum exosomes and liver cancer tissues of patients was measured, and compared with the data obtained from healthy controls and tumor adjacent tissues. The effect of HULC upregulation in HCC cell lines and the relationship between HULC and other RNAs were studied using qPCR and dual-luciferase reporter assays. Nanoparticle tracking analysis was performed to detect the quantity of exosomes.
RESULTS
HULC expression in serum exosomes of patients with HCC was higher than that in serum exosomes of healthy controls, and HULC levels were higher in liver cancer tissues than in tumor adjacent tissues. The expression of HULC in serum exosomes and liver cancer tissues correlated with the tumor-node-metastasis (TNM) classification, and HULC expression in tissues correlated with that in serum exosomes. Upregulation of HULC promoted HCC cell growth and invasion and repressed apoptosis. Notably, it also facilitated the secretion of exosomes from HCC cells. Moreover, qPCR assays showed that HULC repressed microRNA-372-3p (miR-372-3p) expression. We also identified Rab11a as a downstream target of miR-372-3p. Dual-luciferase reporter assays suggested that miR-372-3p could directly bind both HULC and Rab11a.
CONCLUSION
Our findings illustrate the importance of the HULC/miR-372-3p/Rab11a axis in HCC and provide new insights into the molecular mechanism regulating the secretion of exosomes from HCC cells.
Keywords: Long non-coding RNA, Exosomes, Hepatocellular carcinoma, miR-372-3p, Rab11a
Core tip: We found that (1) HULC expression was higher in serum exosomes of patients with HCC than in those of healthy controls, and higher in liver cancer tissues than in tumor adjacent tissues; (2) HULC expression in serum exosomes and liver cancer tissues were correlated with the TNM classification, and HULC expression in tissues was correlated with that in serum exosomes; (3) Increased HULC expression was associated with increased proliferation and invasion and reduced apoptosis of HCC cells; and (4) HULC repressed miR-372-3p expression, and miR-372-3p could directly bind both HULC and Rab11a. Furthermore, the HULC/ miR-372-3p /Rab11a axis promoted the secretion of exosomes from HCC cells.
INTRODUCTION
Hepatocellular carcinoma (HCC) is a malignant tumor with high mortality, and more than 700000 patients with HCC die every year worldwide. Tumor invasion, metastasis, and recurrence are the main causes of death in these patients[1]. Despite advances in the understanding of the molecular mechanisms underlying HCC and improved therapeutic methods to treat this disease, the 5-year overall survival (OS) for patients with HCC is still unsatisfactory[2].
Long noncoding RNAs (lncRNAs) are transcribed RNAs that regulate gene expression through various mechanisms. LncRNAs exert vital roles in many processes such as transcriptional control, posttranscriptional regulation, and epigenetic regulation[3]. In recent years, it has been demonstrated that several lncRNAs play an important role in the progression of HCC[4]. One of these lncRNAs is HULC, located on chromosome 6p24.3, which was found to be highly expressed in HCC tissue by Panzitt in 2007[5]. HULC transcription produces an RNA of approximately 500 nt that is located in the cytoplasm and participates in the development of HCC[6]; it acts as a competitive endogenous RNA (ceRNA), which regulates mRNA through competitive miRNA sharing. For example, HULC, in combination with miR-15a, inhibits PTEN expression and participates in autophagy to promote the progression of HCC[7]. Via sequestrating miR-124, HULC participates in endothelial cell angiogenesis[8]. However, the mechanism of HULC-mediated invasion and metastasis of HCC remains unclear.
In recent years, exosomes have become a research hotspot, and they have been found to be closely related to tumor progression[9]. Exosomes are membranous structures derived from cells that play various roles in normal physiology[10]. In cancer, exosomes have been shown to contribute to essential cancer-related processes such as sustaining the proliferative signalling, evading growth suppression, resisting cell death, enabling replicative immortality, inducing angiogenesis, promoting genome instability and mutations, increasing tumor-promoting inflammation, and especially activating invasion and metastasis. In the tumor, exosomes participate in basic cancer-related processes such as promoting cancer cell proliferation, reducing apoptosis, achieving replication immortality, inducing angiogenesis, promoting genomic instability and mutations, increasing tumors and promoting inflammation, and especially activating tumors. For example, exosomal miR-335 could be a novel therapeutic strategy for HCC[11]. Tumor-derived exosomes could elicit tumor suppression in murine HCC models and humans in vivo[12]. Additionally, exosomes are frequently released by tumor cells and may facilitate the communication between the primary tumor and its local microenvironment[13]. The link between HULC and exosomes in tumor genesis and development has not been clarified to date.
In this study, we investigated the expression of HULC in serum-derived exosomes and hepatic tissues, analysed the correlation between HULC expression and other RNAs, and demonstrated the mechanism of action of HULC in HCC. Increased HULC expression was associated with increased proliferation and invasion, and reduced apoptosis of HCC cells. HULC was found to function as a ceRNA of miR-372-3p, which, in turn, inhibits the expression of Rab11a. Rab proteins are key regulators of intracellular membrane trafficking, and Rab11a has been reported to promote exosome secretion[14,15]. Thus, HULC induces exosome secretion and contributes to tumor growth and metastasis.
MATERIALS AND METHODS
Subjects
Serum samples and tissues (n = 30) and paired adjacent liver tissues (n = 30) were obtained from patients with HCC who underwent surgery at the Tianjin First Central Hospital (China) from January to August 2017. Normal human serum was taken from patients undergoing a physical examination at the Tianjin First Central Hospital (China). All our patients had hepatitis B virus associated cancer with cirrhosis. The average age of the patients was 53.9 ± 16.2 years. Patient clinical data are shown in Table 1. Tumor differentiation and TNM stage were assessed based on the WHO grading system. Each HCC case was confirmed by histopathological diagnosis. Blood samples from the patients were taken one week before surgery. Tissue samples were taken from the resected HCC tissues and tumor-adjacent liver tissues (histopathological diagnosis confirmed that the tumor had not invaded the adjacent tissues). The serum and tissue samples were immediately stored at -80 °C before use. This study was approved by the Tianjin First Central Hospital Medical Ethics Committee, and all patients provided written informed consent for the use of their samples for clinical research.
Table 1.
Relationship between expression of long non-coding RNA highly up-regulated in liver cancer and clinicopathological factors in patients with hepatocellular carcinoma
| Clinicopathological factor | Number of patients (n = 30) |
HULC in serum exosomes |
χ2 | P-value |
HULC in HCC tissue |
χ2 | P-value | ||
| High | Low | High | Low | ||||||
| Age (yr) | 0.391 | 0.532 | 0.382 | 0.497 | |||||
| ≤ 55 | 18 | 11 | 7 | 12 | 8 | ||||
| > 55 | 12 | 7 | 5 | 5 | 5 | ||||
| sex | 0.370 | 0.492 | 0.365 | 0.592 | |||||
| Female | 21 | 11 | 7 | 13 | 8 | ||||
| Male | 9 | 6 | 6 | 4 | 5 | ||||
| Tumor size (cm) | 2.323 | 0.124 | 2.225 | 0.126 | |||||
| ≤ 5 | 10 | 4 | 6 | 5 | 5 | ||||
| > 5 | 20 | 11 | 9 | 10 | 10 | ||||
| Tumor differentiation | 2.224 | 0.115 | 2.356 | 0.098 | |||||
| Well/moderate | 21 | 12 | 9 | 11 | 8 | ||||
| Poor | 9 | 5 | 4 | 6 | 5 | ||||
| Clinical stage (TNM) | 4.812 | 0.032 | 4.924 | 0.026 | |||||
| I-II | 10 | 5 | 5 | 4 | 6 | ||||
| III-IV | 20 | 11 | 9 | 12 | 8 | ||||
HULC: Highly upregulated in liver cancer; HCC: Hepatocellular carcinoma; TNM: Tumor-node-metastasis.
The inclusion criteria were: (1) Patients with complete clinical data; and (2) Patients had not received chemotherapy, radiotherapy, or immunotherapy before surgery. The exclusion criteria were: (1) Lack of clinical data; (2) Area of necrosis in the tumor tissue > 60%; and (3) Presence of additional malignant tumors.
Cell culture
Human HCC cell lines (HepG2 and SMMC7721) and a normal hepatic cell line (LO2) were purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in DMEM or RPMI-1640 (GIBCO, Grand Island, NY, United States) containing 10% foetal bovine serum (FBS; BioWest, Nuaille, France), and 1% bivalent antibiotics (Hyclone, South Logan, UT, United States). Cells were incubated at 37 °C with 5% CO2.
Exosome extract
Exosomes were isolated from HCC cells as follows. Exosome-depleted FBS was obtained by ultracentrifugation at 110000 g (CP80MX; HITACHI, Japan) for 16 h and used for culturing HCC cells. HCC cell culture medium supernatants were collected after incubation for 48 h, and then centrifuged at 10000 g for 40 min to remove HCC cell debris. Supernatants were filtrated through a 0.22 μm-pore filter (Millipore). The ExoQuick solution (EXOTC10A-1, System Biosciences, Mountain View, United States) was added to the filtered solution at a 1:5 ratio. The mixed solution was stored at 4 °C for 12 h, and then centrifuged at 1500 g for 30 min. The supernatants were discarded, and the precipitate (containing the exosomes) was dissolved in phosphate buffered saline (PBS).
Exosomes were isolated from patients’ serum as follows. The serum was filtrated through a 0.22 μm-pore filter (Millipore). The ExoQuick solution (EXOQ5A-1, System Biosciences, Mountain View, United States) was added to the filtered solution at a 1:4 ratio. The mixed solution was stored at 4 °C for 12 h, and then centrifuged at 1500 g for 30 min. The supernatants were discarded, and the precipitate (containing the exosomes) was dissolved in PBS.
Transfection
HULC (HULC over-expression plasmid), HULC-NC (HULC over-expression normal control plasmid), HULC-siRNA (HULC low-expression plasmid), HULC-siRNA-NC (HULC low-expression control plasmid), miRNA-372-3p (miRNA-372-3p over-expression plasmid), miRNA-372-3p inhibitor, WT-HULC (HULC wild type luciferase reporter plasmid), MUT-HULC (HULC mutant luciferase reporter plasmid), WT-Rab11a (Rab11a wild type luciferase reporter plasmid), and MUT-Rab11a (Rab11a mutant luciferase reporter plasmids) (Genechem, Shanghai, China) were used for transfection experiments. HepG2 and SMMC7721 cells were seeded in 6-well plates and transfected with HULC inhibitors or plasmids causing HULC overexpression, using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The transfected cells were incubated for 48 h before analysis.
Western blot analysis
Protein samples were collected from cells or exosomes in RIPA lysis buffer (Solarbio, Beijing, China). The processed protein samples were separated by 10% SDS–polyacrylamide gel electrophoresis (SDS-PAGE), transferred to 0.22 μm PVDF membranes (Millipore), and incubated with primary antibodies against CD63, TSG101, Rab11a, and GAPDH (Abcam, Cambridge, MA, United States) at 4 °C. The secondary antibody was goat anti-rabbit antibody (Abcam, Cambridge, MA, United States) and incubation was performed at 24 °C. Chemiluminescence reagent (Millipore) was used to detect the proteins.
Nanoparticle tracking analysis (NTA)
NTA with a ZetaView PMX (Particle Metrix, Dusseldorf, Germany) was used to measure concentration, mean particle size, and distribution of exosomes. Exosomes were resuspended in 0.05 × PBS (filtered through a 0.22 μm-pore filter) and added to the instrument. The same parameters were used for size and concentration measurements. Data were analysed using the software ZetaView 8.02.28 and Microsoft Excel 2016 (Microsoft, Seattle, WA, United States).
Transmission electron microscopy
The purified exosomes were fixed with 1% glutaraldehyde in PBS. A drop of the suspension (approximately 20 μL) was pipetted onto formvar/carbon-coated grids and stained with aqueous phosphotungstic acid for 1 min. The morphology of isolated exosomes was identified using a transmission electron microscope (HT7800, Hitachi, Japan).
Quantitative PCR
Total RNA was extracted from HCC tissues and cells using Trizol reagent (Invitrogen, Carlsbad, CA, United States) and total RNA was extracted from exosomes using the SeraMir Exosome RNA Purification Kit (System Biosciences, Mountain View, United States) following the manufacturer’s instructions. After determining the concentration, degree, and purity of RNA, cDNA was synthesised using a reverse transcription kit (Takara Biotechnology, Japan) and PCR was performed with a commercial kit (Takara Biotechnology). With GAPDH and U6 as references, the expression levels of HULC, miRNA-372-3p, and Rab11a were calculated using relative quantitative analysis. The relative standard curve method (2-△△CT) was used to determine the relative RNA expression in cells and -△CT was used to determine the relative RNA expression in HCC tissues and serum exosomes. The primers used for quantitative PCR (qPCR) are shown in Table 2.
Table 2.
Primers used for quantitative PCR
| Gene | Primer sequence |
| HULC | Forward: 5’-GCAAGCCAGGAAGAGTCGTC-3' |
| Reverse: 5’-GCTGTGCTTAGTTTATTGCCAGG-3' | |
| GAPDH | Forward: 5'-GTCAACGGATTTGGTCTGTATT-3' |
| Reverse: 5'-AGTCTTCTGGGTGGCAGTGAT-3' | |
| miRNA-372-3p | Forward: 5'-CAACAGAAGGCTCGAGCAACCTGCGGAGAAGATAC-3' |
| Reverse: 5'-TTCTGATCAGGATCCCATTACAGCCAGACGCTGTAAG-3' | |
| U6 | Forward: 5'-GCGCGTCGTGAAGCGTTC-3' |
| Reverse: 5'-GTGCAGGGTCCGAGGT-3' | |
| Rab11a | Forward: 5'-ACATCAGCATATTATCGTGGAGC-3' |
| Reverse: 5'-GGAACTGCCCTGAGATGACG-3' |
HULC: Highly upregulated in liver cancer.
Dual-luciferase reporter assay
HULC could specifically bind to miR-372-3p, acting as a "microRNA sponge"[16,17]. At the same time, miR-372-3p was predicted to regulate Rab11a expression using TargetScan (http://www.targetscan.org/) and starBase (http://starbase.sysu.edu.cn/). HULC and Rab11a 3′-UTR double luciferase recombinant plasmids (Genechem, Shanghai, China) were constructed and transfected into HCC cells. Luciferase assays were performed using a luciferase reporter assay kit (Genecopoeia) according to the manufacturer’s recommendations. Before detection, HCC cells were washed 2-3 times with PBS, and 150 μL of fresh passive cell lysate was added into each well (24-well plates) to lyse the cells. The supernatant was centrifuged at 13000 g for 1 min, and 20 μL of the supernatant was transferred to 96-well plates. Then, 100 μL of LAR II was added to detect the firefly luciferase activity. The Stop&Glo reagent (100 mL) was used to detect the fluorescence of the Renilla luciferase. Luciferase was measured using a microplate reader, with 1-2 s delay and 5-10 s readings for measurement. The final luciferase activity is expressed as the ratio of luciferase activity of firefly/luciferase activity of Renilla.
Cell counting kit-8 (CCK8)
Cells (50000 cells per well) were cultured overnight in a 96-well plate. At 0, 24, 36, 48, and 72 h, respectively, 100 μL of CCK8 solution (BOSTER, Wuhan, China) was added. The assay was performed according to the manufacturer’s recommendations. A microplate reader (Bio Tek Synergy2, United States) was used to measure the absorbance at 450 nm.
Transwell assay
Cells were seeded in the upper chamber of Matrigel-coated Transwells (BD Bioscience, CA, United States). Serum-free medium was added to the upper chamber, and medium with 10% FBS was added into the lower chamber. Cells were incubated for 24 h; the non-migrating or non-invading cells were removed, and the remaining cells (on the bottom of the filters) were fixed using methanol and stained with crystal violet. Five random fields were chosen and counted per well using a microscope (×100), (Olympus, Tokyo, Japan).
TUNEL assay
Apoptosis was detected using TUNEL assay. Cells were fixed using 4% paraformaldehyde for 30 min, and the assay was carried out using a TUNEL kit (Roche, Basel, Switzerland) according to the manufacturers’ recommendations. Nuclei were stained with DAPI. Apoptotic cells with green nuclear staining were observed. Five random fields were chosen and counted per well by using a microscope (×100). The number of apoptotic cells was counted, and the apoptotic rate (%) was calculated as (apoptotic cell count/total cell number) × 100%.
Statistical analysis
The SPSS statistical software version 20.0 was used for statistical analyses. The data, expressed as the mean ± SD, were analysed using one-way ANOVA. Count data, expressed as percentages (%), were analysed using the χ2 tests. P-values < 0.05 were considered statistically significant.
RESULTS
Exosome extraction from HCC cells and serum of patients with HCC
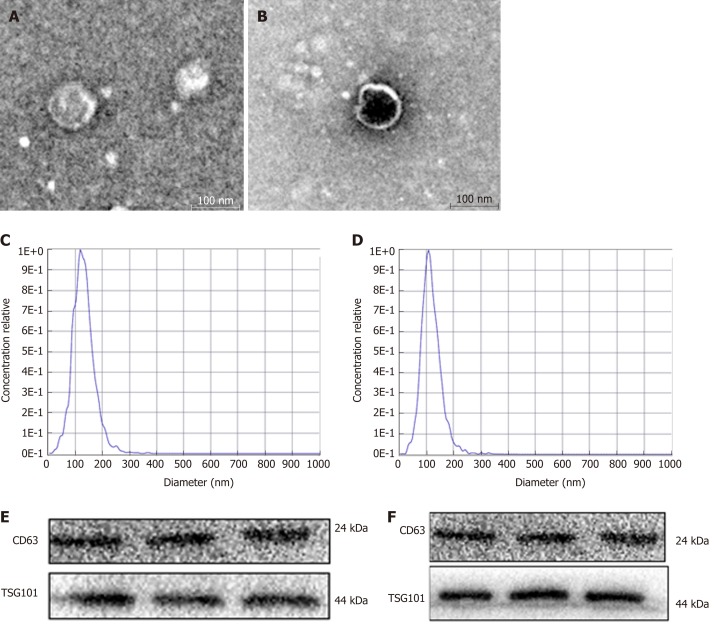
Exosomes were successfully extracted from the serum of patients with HCC and HCC cell medium (Figure 1). First, we examined the shape and size of the exosomes isolated from HCC serum (Figure 1A) and the culture medium of HepG2 or SMMC7721 cells (Figure 1B) by transmission electron microscopy. The exosomes were homogeneous in shape and size, and the vesicles had the characteristic appearance of exosomes with a membrane-capsulated spherical shape. The median size of the vesicles, calculated by NTA, was 110.2 nm and 106.9 nm in HCC serum (Figure 1C) and the culture medium of HepG2 or SMMC7721 cells (Figure 1D), respectively, similar to the reported size of exosomes (30–120 nm)[18]. Additionally, Western blot assays showed that the TSG101 and CD63 exosome markers were part of the exosome cargo (Figure 1E, 1F), confirming that the isolated vesicles were indeed exosomes.
Figure 1.
Identification of the exosomes. A: Transmission electronc microscopy (TEM) of exosomes extracted from the serum of patients with hepatocellular carcinoma (HCC); B: TEM of exosomes isolated from HCC cell medium; C: Western blot analysis of exosomal markers (CD63 and TSG101) in HCC serum; D: Western blot analysis of exosomal markers (CD63 and TSG101) in HCC cell medium; E: Nanoparticle tracking analysis (NTA) for detection of exosomes extracted from the HCC serum; F: NTA detection of exosomes extracted from HCC cell medium.
HULC is upregulated in liver cancer serum-derived exosomes, hepatic tissues, and HCC cells
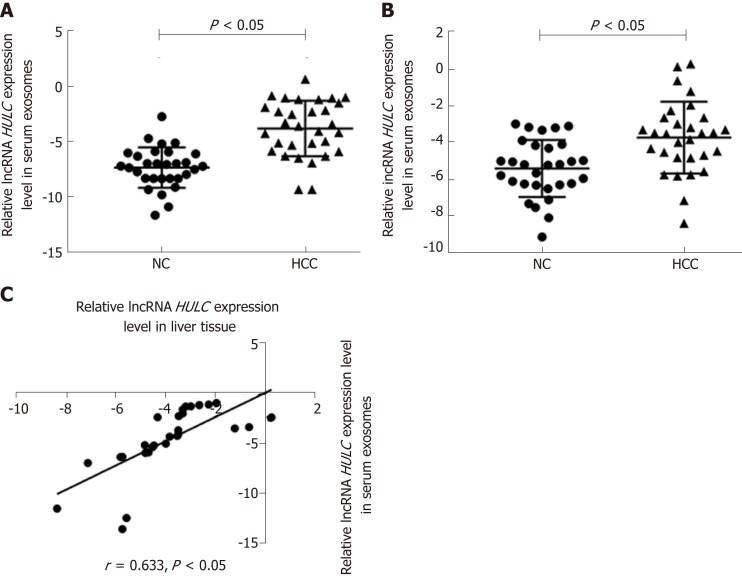
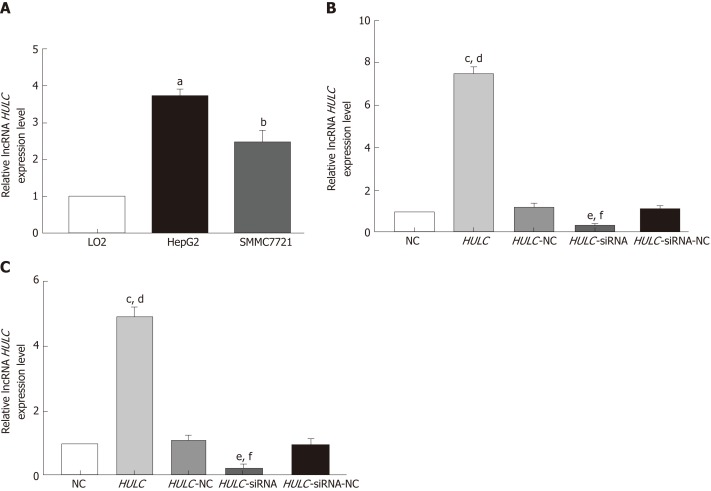
The baseline characteristics of the 30 patients (9 men and 21 women) enrolled in this study are summarised in Table 1. The average age was 53.9 ± 16.2 years. The expression of HULC in the serum exosomes of patients with HCC was higher than that in exosomes from healthy subjects (P < 0.05; Figure 2A). HULC levels in liver cancer tissues were higher than those in adjacent tissues (P < 0.05; Figure 2B). The expression of HULC in tissues correlated with that in exosomes (r = 0.633, P < 0.05; Figure 2C). Additionally, the expression of HULC in serum exosomes correlated with the TNM stage of the patients (P < 0.05; Table 1). Finally, HULC was upregulated in HepG2 and SMMC7721 cells (Figure 3A).
Figure 2.
Detection of long non-coding RNA highly up-regulated in liver cancer in hepatocellular carcinoma. A: Expression of HULC in the serum exosomes of patients with HCC; B: Expression of HULC in liver cancer tissues; C: Correlation between HULC expression in HCC serum exosomes and tissue. Error bars stand for the mean ± SD of at least triplicate experiments. HULC: Highly upregulated in liver cancer; HCC: Hepatocellular carcinoma.
Figure 3.
Detection of long non-coding RNA highly up-regulated in liver cancer in hepatocellular carcinoma cells. A:Expression of HULC in HepG2, SMMC7721, and LO2 cells; B: Expression of HULC in HepG2 transfected with HULC inhibitors or plasmids causing HULC overexpression; C: Expression of HULC in SMMC7721 transfected with HULC inhibitors or plasmids causing HULC overexpression. Error bars stand for the mean ± SD of at least triplicate experiments. aP < 0.05 vs LO2 group; bP < 0.05 vs LO2 group; cP < 0.01 vs NC group; dP < 0.01 vs HULC-NC group; eP < 0.05 vs NC group; fP < 0.05 vs HULC-siRNA-NC group. HULC: Highly upregulated in liver cancer; NC: Normal control group; HULC: HULC over-expression plasmid group; HULC-NC: HULC over-expression normal control plasmid group; HULC-siRNA: HULC low-expression plasmid group; HULC-siRNA-NC: HULC low-expression control plasmid group.
HULC induces HCC cell proliferation and invasion and inhibits apoptosis
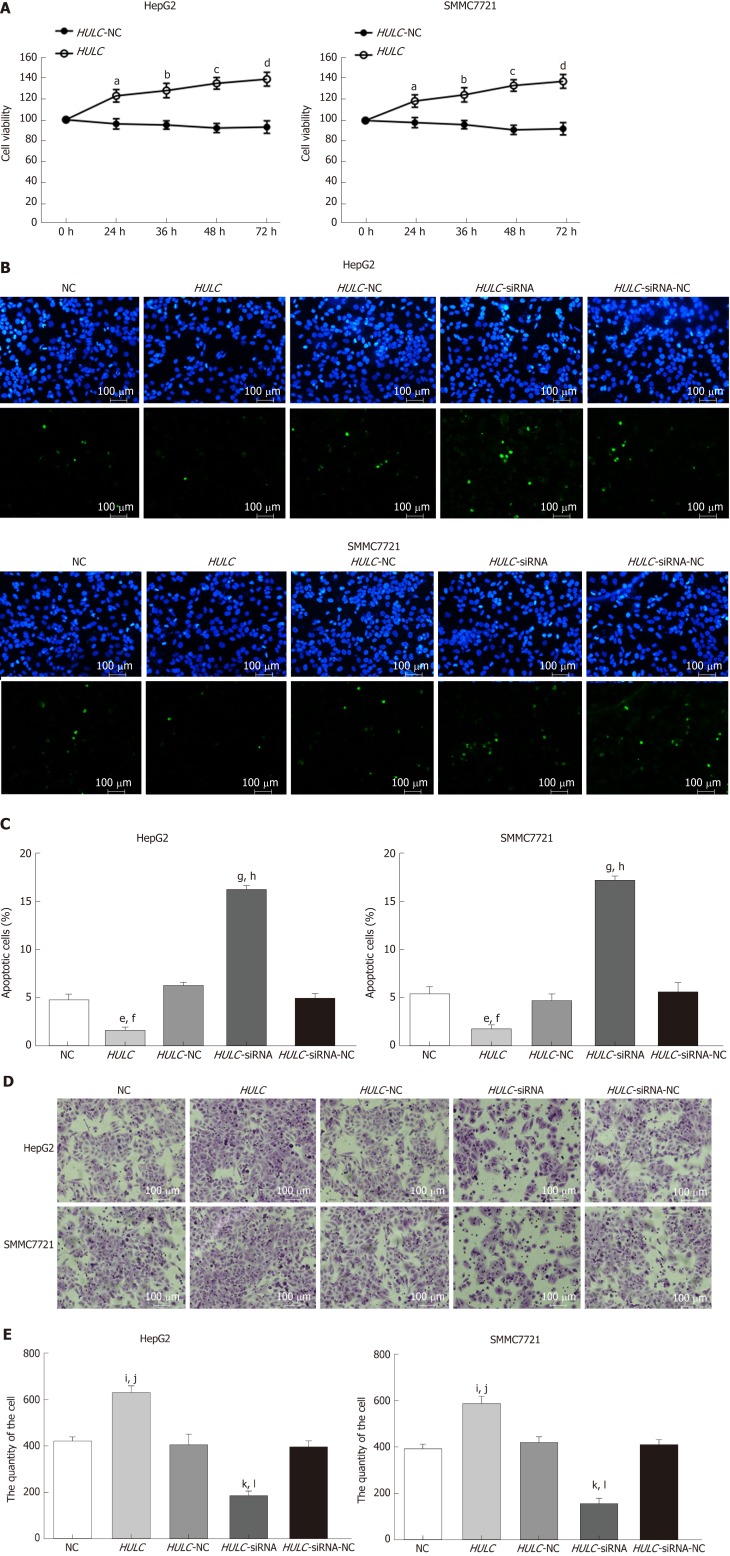
After transfection of HULC over-expression and inhibiting plasmids into HCC cells, the expression of HULC changed significantly (Figure 3B, 3C). To explore the role of HULC in HCC cells, we performed CCK8 assays. We observed that HULC upregulation was associated with the growth of HepG2 and SMMC7721 cells (Figure 4A). Next, TUNEL assays showed that apoptosis was significantly inhibited by HULC (Figure 4B, 4C). Finally, we tested the effect of HULC on the invasion capabilities of HCC cells using Transwell assays, which showed that HULC induced the invasion of HCC cells (Figure 4D, 4E). These results suggest that HULC remarkably promoted HCC development.
Figure 4.
Long non-coding RNA highly up-regulated in liver cancer induces hepatocellular carcinoma cell proliferation and invasion and inhibits apoptosis in vitro. A: Proliferation of hepatocellular carcinoma (HCC) cells transfected with HULC inhibitors or plasmids causing HULC overexpression; B: Apoptotic pictures of HCC cells (×100). The blue fluorescence is DAPI nuclear staining and green fluorescence is the apoptotic cell. The index of apoptosis is the number of apoptotic cells/total number of visual field × 100%; C: Column chart of index of apoptosis in different groups; D: Effect of HULC on invasion of HCC cells evaluated bTranswell assay (×100); E: Column chart of number of invasive HCC cells in different groups. Error bars stand for the mean ± SD of at least triplicate experiments. aP < 0.05 vs HULC-NC 24 h group; bP < 0.05 vs HULC-NC 36 h group; cP < 0.05 vs HULC-NC 48 h group; dP < 0.05 vs HULC-NC 72 h group; eP < 0.05 vs NC group; fP < 0.05 vs HULC-NC group; gP < 0.01 vs NC group; hP < 0.01 vs HULC-siRNA-NC group; iP < 0.05 vs NC group; jP < 0.05 vs HULC-NC group; kP < 0.05 vs NC group; lP < 0.05 vs HULC-siRNA-NC group. HULC: Highly upregulated in liver cancer; NC: Normal control group; HULC: HULC over-expression plasmid group; HULC-NC: HULC over-expression normal control plasmid group; HULC-siRNA: HULC low-expression plasmid group; HULC-siRNA-NC: HULC low-expression control plasmid group.
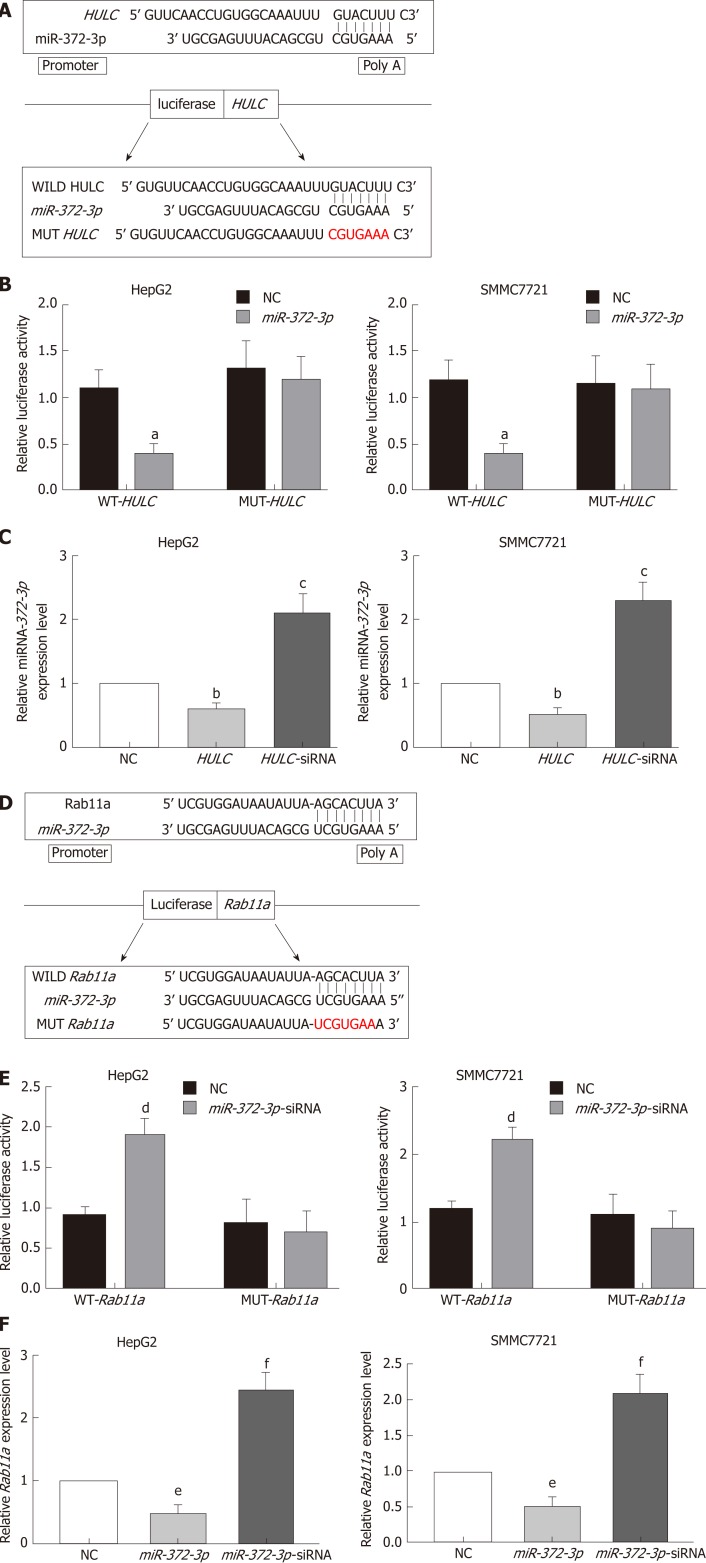
HULC reduces the expression of miR-372-3p by sponging it
The relationship between lncRNA HULC and miR-372-3p has been reported in many studies[16,17]. Therefore, we identified the binding sites between miR-372-3p and HULC (Figure 5A). To confirm the direct interaction between miR-372-3p and HULC, we generated luciferase reporter plasmids with the wild type or mutated HULC sequence (Figure 5A). A decreased reporter activity was observed when wild type HULC and miR-372-3p mimics were co-transfected into HCC cells (Figure 5B). To investigate whether miR-372-3p is regulated by HULC in vitro, we overexpressed HULC in HepG2 and SMMC7721 cells and measured miR-372-3p expression (Figure 5C). We found that miR-372-3p levels decreased after HULC upregulation (Figure 5C), indicating a negative correlation between miR-372-3p and HULC. These results showed that miR-372-3p is a direct target of HULC in HCC cells.
Figure 5.
Long non-coding RNA highly up-regulated in liver cancer mediates secretion of exosomes from hepatocellular carcinoma cells via miR-372-3p /Rab11a. A: Sequence of binding sites between HULC and miR-372-3p; B: Dual-luciferase reporter assay for miR-372-3p and HULC; C: Relative expression of miR-372-3p after HCC cells were transfected with HULC inhibitors or plasmids causing HULC overexpression; D: Sequence of binding sites with Rab11a and miR-372-3p; E: Dual-luciferase reporter assay for miR-372-3p and Rab11a; F: Relative expression of Rab11a after HCC cells were transfected with miR-372-3p-siRNA or plasmids causing miR-372-3p overexpression. Error bars stand for the mean ± SD of at least triplicate experiments. aP < 0.05 vs WT-HULC-NC group; bP < 0.05 vs NC group; cP < 0.01 vs NC group; dP < 0.05 vs WT-Rab11a -NC group; eP < 0.05 vs NC group; fP < 0.01 vs NC group. HULC: Highly upregulated in liver cancer; NC: Normal control group; HULC: HULC over-expression plasmid group; HULC-siRNA: HULC low-expression plasmid group; miRNA-372-3p: miRNA-372-3p over-expression plasmid group; miR-372-3p-siRNA: miR-372-3p low-expression plasmid group; WT-HULC: HULC wild type luciferase reporter plasmid group; MUT-HULC: HULC mutant luciferase reporter plasmids group; WT-Rab11a: Rab11a wild type luciferase reporter plasmid group; MUT-Rab11a: Rab11a mutant luciferase reporter plasmids group.
Rab11a is a direct target of miR-372-3p in HCC cells
Next, we used bioinformatic analyses to predict the downstream target of miR-372-3p. We identified Rab11a as a putative target of miR-372-3p. To validate the interaction between miR-372-3p and Rab11a, we performed luciferase reporter assays using plasmids with wild type Rab11a and mutated Rab11a (Figure 5D, 5E). Co-transfection of the luciferase reporter plasmid containing wild type Rab11a and miR-372-3p inhibitors in HCC cells led to increased luciferase activity (Figure 5E). Additionally, miR-372-3p inhibitors increased Rab11a expression (Figure 5F).
HULC acts via the miR-372-3p /Rab11a axis
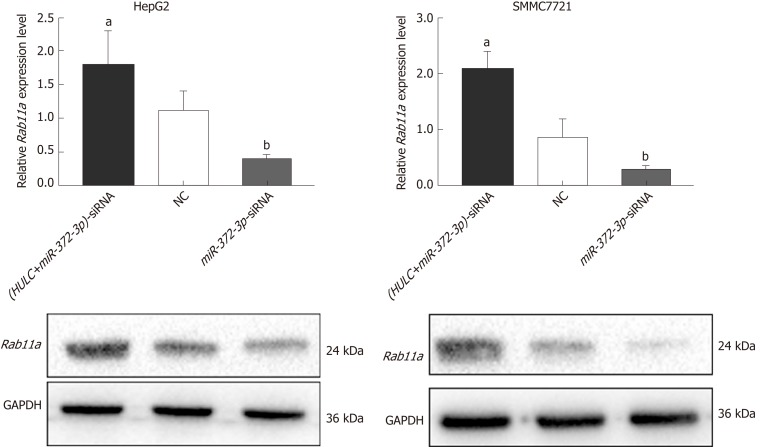
To further confirm whether HULC can regulate the secretion of exosomes via the miR-372-3p/Rab11a axis, HULC-siRNA and miR-372-3p-siRNA were co-transfected into HCC cells. We found that the expression of Rab11a was inhibited by HULC silencing, whereas miR-372-3p downregulation rescued Rab11a expression (Figure 6). Western blot assays showed similar results (Figure 6). These findings indicate that HULC can promote the secretion of exosomes by decreasing miR-372-3p and increasing Rab11a expression.
Figure 6.
Expression of Rab11a in hepatocellular carcinoma cells transfected with highly up-regulated in liver cancer-siRNA and miR-372-3p-siRNA. Silencing HULC inhibited Rab11a expression, while silencing HULC and miR-372-3p increased Rab11a expression in vitro. Error bars stand for the mean ± SD of at least triplicate experiments. aP < 0.05 vs NC group; bP < 0.05 vs NC group. HULC: Highly upregulated in liver cancer; NC: Normal control group; HULC-siRNA: HULC low-expression plasmid group; miR-372-3p-siRNA: miR-372-3p low-expression plasmid group.
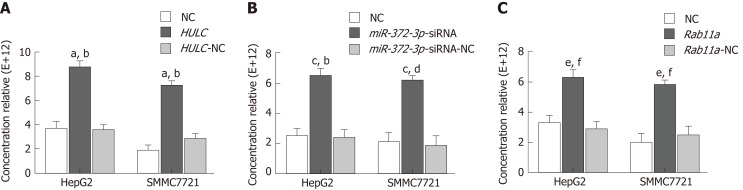
Increased HULC and Rab11a and decreased miR-372-3p promote secretion of exosomes from HCC cells
NTA was used to detect the expression of exosomes in the following groups. The number of exosomes from HepG2 and SMMC7721 cells in the HULC group was higher than that in the HULC-NC or NC group. However, there was no significant difference between the HULC-NC and NC groups (Figure 7A).
Figure 7.
Increased long non-coding RNA highly up-regulated in liver cancer and Rab11a and decreased miR-372-3p promote the secretion of exosomes from hepatocellular carcinoma cells. A: Quantification of exosomes after transfecting HULC over-expression plasmid into HCC cells; B: Quantification of exosomes after transfecting miR-372-3p low-expression plasmid into HCC cells; C: Quantification of exosomes after transfecting Rab11a over-expression plasmid into HCC cells. Error bars stand for the mean ± SD of at least triplicate experiments. aP < 0.01 vs NC group; bP < 0.01 vs HULC-NC group; cP < 0.01 vs NC group; dP < 0.01 vs miRNA-372-3p-siRNA-NC group; eP < 0.01 vs NC group; fP < 0.01 vs Rab11a-NC group. HULC: Highly upregulated in liver cancer; HULC: HULC over-expression group; HULC-NC: HULC (over-expression plasmid normal control group); NC: Normal control; miR-372-3p-siRNA: miR-372-3p low-expression group; miR-372-3p-siRNA-NC: miR-372-3p low-expression plasmid normal control group; Rab11a: Rab11a over-expression group; Rab11a-NC: Rab11a over-expression plasmid normal control group.
The number of exosomes from HepG2 and SMMC7721 cells in the miR-372-3p-siRNA group was higher than that in the miR-372-3p-siRNA-NC or NC groups. However, there was no significant difference between the miR-372-3p-NC and NC groups (Figure 7B).
The number of exosomes from HepG2 and SMMC7721 cells in the Rab11a group was higher than that in the Rab11a-NC or NC groups. However, there was no significant difference between the Rab11a-NC and NC groups (Figure 7C).
DISCUSSION
Many studies have reported that lncRNAs participate in various cellular processes and have key functions in cancer epigenetics. Specifically, some lncRNAs are associated with recurrence, metastasis, and prognosis of various cancers, such as HCC, gastric cancer, and breast cancer[19-22]. LncRNA HULC is one of them, playing an important role in the development of liver tumors. HULC is highly expressed in HCC and is involved in the regulation of tumorigenesis and development[5]. Phosphorylation of the cAMP response element binding protein (CREB) activates the transcription of HULC[16]. Du et al[23] found that the mechanism for hepatitis B virus X protein (HBx)-activated HULC to promote the proliferation of HCC cells is closely related to downregulation of p18. HULC mediates abnormal lipid metabolism in HCC cells by sequestering miR-9[24]. HULC is also expressed in gastric cancer[25], pancreatic cancer[26], and other cancers.
Our clinical data showed that HULC expression is increased in HCC tissues compared with adjacent liver tissues. Moreover, we found that HULC expression was higher in the exosomes derived from the serum of patients with HCC than in those derived from the serum of healthy controls. Notably, HULC expression in HCC tissues and serum exosomes was significantly related to HCC clinical stage. Therefore, HULC can be used as a potential biomarker candidate in HCC. Although HULC has been shown to participate in neoplasm invasion and metastasis, the underlying mechanism remains unclear. In recent years, the relation between exosomes and tumor progression has become evident[27]. However, the role of HULC in this process has not been demonstrated.
Exosomes play a key role in HCC progression[13]. Exosome-mediated miRNAs regulate TAK1 expression and lead to deterioration of HCC[28]. In addition, lncRNA H19 in exosomes derived from CD90+ HCC cells influence tumor microenvironment balance by accelerating angiogenesis[29]. Exosome-mediated lysyl oxidase-like 4 (LOXL4) activation promotes tumor metastasis by modulating the FAK/SRC pathway in HCC[30]. Several studies have confirmed that lncRNAs in blood-circulating exosomes can be used as diagnostic markers for cancers: LINC00152[31] and ZFAS1[32] in gastric cancer, MALAT-1[33] in lung cancer, BCAR4[34] and CRNDE-h[35] in colon cancer, and HOTAIR[36] in laryngeal squamous cell carcinoma.
In this study, we showed that the expression of HULC in exosomes from HCC serum and in HCC tissues were higher than that in the control group, and that there was a linear correlation between them. Therefore, HULC in exosomes from the blood of patients with HCC can be used as a diagnostic marker. We showed that the expression of HULC in exosomes correlated with TNM stage and therefore, with the progression of HCC. These findings are in agreement with previous studies[5,37].
We also investigated the relationship between HULC and exosome secretion. Bioinformatic analyses suggest that HULC mediated the secretion of exosomes from HCC cells via the miR-372-3p/Rab11a axis. Rab11a is a key regulatory protein that induces exosome secretion. In addition, we found that the number of exosomes from HCC cell lines overexpressing HULC was higher than that of control groups, indicating that HULC promotes exosome secretion.
Many studies have reported that lncRNAs act as sponges of miRNAs and inhibit their function[38]. For example, HULC mediates abnormal lipid metabolism in HCC cells by sequestering miR-9[24]. Here, we observed a negative correlation between HULC and miR-372-3p. A previous report showed that low miR-372-3p expression correlates with a poor prognosis and tumor metastasis in HCC[39]. Our results showed that when HULC was upregulated in HCC cells, miR-372-3p was repressed. Dual-luciferase reporter assays confirmed that HULC directly interacted with miR-372-3p. In addition, decreased levels of miR-372-3p promoted the secretion of exosomes from HCC cells.
Rab11a is a member of the subfamily of the Rab small molecule GTPases and is a key regulatory protein that induces exosome secretion[14,15]. Rab11a plays an important role in the regulation of endosome recycling and is highly expressed in many tumors, such as esophageal adenocarcinoma[40] and skin cancer[41]. In addition, the Rab11 family-interacting protein 4 is involved in the metastasis of HCC[42]. In our study, we found that Rab11a is a downstream target of miR-372-3p and that it promotes the secretion of exosomes from HCC cells. Collectively, our results suggest that HULC negatively interferes with miR-372-3p-mediated inhibition of Rab11a, leading to secretion of exosomes in HCC.
In conclusion, we demonstrated that HULC enhances the secretion of exosomes by sponging miR-372-3p which in turn targets Rab11a. Our findings provide novel insights into the mechanism of action of HULC in HCC. We plan to continue investigating the precise mechanisms through which Rab11a and exosomes regulate HCC development. Whether HULC targets other miRNAs to promote exosome secretion also warrants additional research. In addition, we need to further explore the relationship between silencing of HULC and angiogenesis, epidermal mesenchymal transition, and lipid metabolism in HCC. Furthermore, the relationship between silencing of HULC and survival and prognosis of patients with HCC needs to be elucidated.
ARTICLE HIGHLIGHTS
Research background
Long non-coding RNA highly up-regulated in liver cancer (lncRNA HULC) is highly expressed in hepatocellular carcinoma (HCC), and involved in the development and regulation of tumorigenesis. The relationship between HULC and exosomes has not yet been reported. Studying the mechanism of exosome secretion is helpful to reveal the pathogenesis of HCC.
Research motivation
Although HULC has long been known to play important roles in HCC, its function and regulatory mechanism are unclear. Since exosomes participate in the progression of HCC, they have been actively studied. We are interested in exploring whether HULC regulates exosome secretion and is involved in the occurrence and development of hepatocellular carcinoma. This study will provide novel insights into the mechanism of action of HULC in HCC.
Research objectives
The main objective of the study was to explore the molecular mechanism of HULC to regulate exosome secretion and and provide novel insights into the mechanism of action of HULC in HCC.
Research methods
We collected samples from serum and tissues of 30 patients with HCC. We measured HULC expression in the serum exosomes and liver cancer tissues of patients, and compared the data to those obtained from controls. We further explored the effect of HULC upregulation in HCC cell lines and studied the relationship between HULC and other RNAs using qPCR and dual-luciferase reporter assays. NTA was used to detect the quantity of exosomes.
Research results
HULC expression in serum exosomes of patients with HCC was higher than that in serum exosomes of healthy controls, and HULC levels were higher in liver cancer tissues than in adjacent tissues. The expression of HULC in serum exosomes and liver cancer tissues correlated with the tumor-node-metastasis (TNM) classification and HULC expression in tissues correlated with that in serum exosomes. HULC promoted HCC cell growth and invasion and repressed apoptosis. Additionly, it also facilitated the secretion of exosomes from HCC cells. Moreover, qPCR assays show that HULC repressed microRNA-372-3p (miR-372-3p) expression. We also identified Rab11a as a downstream target of miR-372-3p. Dual-luciferase reporter assays suggest that miR-372-3p could directly bind both HULC and Rab11a.
Research conclusions
HULC regulates Rab11a to promote secretion of exosomes by competitive miR-372-3p sharing, and this finding provides new insights into the molecular mechanism to regulate the secretion of exosomes from HCC cells.
Research perspectives
We aim to further explore the potential of serum exosome HULC as a sensitive preoperative marker for HCC and the role of HULC in the staging and prognosis evaluation of HCC.
ACKNOWLEDGEMENTS
We thank NHC Key Laboratory of Critical Care Medicine for allowing this work to be performed there.
Footnotes
Institutional review board statement: The study was reviewed and approved by the Tianjin First Central Hospital Medical Ethics Committee.
Conflict-of-interest statement: The authors declare no potential conflict of interest in this paper.
Peer-review started: April 26, 2019
First decision: May 24, 2019
Article in press: May 24, 2019
Specialty type: Medicine, research and experimental
Country of origin: China
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Bramhall C, Hoyos S, Ocker M, Yu J S-Editor: Ma YJ L-Editor: Wang TQ E-Editor: Ma YJ
Contributor Information
Shun-Qi Cao, Tianjin First Central Hospital Clinic Institute, Tianjin Medical University, Tianjin 300070, China.
Hong Zheng, Department of Organ Transplantation, Tianjin First Central Hospital, Tianjin 300192, China.
Bao-Cun Sun, Department of Pathology, Tianjin Medical University, Tianjin 300070, China.
Zheng-Lu Wang, Department of Pathology, Tianjin First Central Hospital, Tianjin 300192, China.
Tao Liu, NHC Key Laboratory of Critical Care Medicine, Tianjin First Central Hospital, Tianjin 300192, China.
Dong-Hui Guo, Tianjin First Central Hospital Clinic Institute, Tianjin Medical University, Tianjin 300070, China.
Zhong-Yang Shen, Department of Organ Transplantation, Tianjin First Central Hospital, Tianjin 300192, China. shen_zhongyang@126.com.
References
- 1.Global Burden of Disease Liver Cancer Collaboration. Akinyemiju T, Abera S, Ahmed M, Alam N, Alemayohu MA, Allen C, Al-Raddadi R, Alvis-Guzman N, Amoako Y, Artaman A, Ayele TA, Barac A, Bensenor I, Berhane A, Bhutta Z, Castillo-Rivas J, Chitheer A, Choi JY, Cowie B, Dandona L, Dandona R, Dey S, Dicker D, Phuc H, Ekwueme DU, Zaki MS, Fischer F, Fürst T, Hancock J, Hay SI, Hotez P, Jee SH, Kasaeian A, Khader Y, Khang YH, Kumar A, Kutz M, Larson H, Lopez A, Lunevicius R, Malekzadeh R, McAlinden C, Meier T, Mendoza W, Mokdad A, Moradi-Lakeh M, Nagel G, Nguyen Q, Nguyen G, Ogbo F, Patton G, Pereira DM, Pourmalek F, Qorbani M, Radfar A, Roshandel G, Salomon JA, Sanabria J, Sartorius B, Satpathy M, Sawhney M, Sepanlou S, Shackelford K, Shore H, Sun J, Mengistu DT, Topór-Mądry R, Tran B, Ukwaja KN, Vlassov V, Vollset SE, Vos T, Wakayo T, Weiderpass E, Werdecker A, Yonemoto N, Younis M, Yu C, Zaidi Z, Zhu L, Murray CJL, Naghavi M, Fitzmaurice C. The Burden of Primary Liver Cancer and Underlying Etiologies From 1990 to 2015 at the Global, Regional, and National Level: Results From the Global Burden of Disease Study 2015. JAMA Oncol. 2017;3:1683–1691. doi: 10.1001/jamaoncol.2017.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hung AK, Guy J. Hepatocellular carcinoma in the elderly: Meta-analysis and systematic literature review. World J Gastroenterol. 2015;21:12197–12210. doi: 10.3748/wjg.v21.i42.12197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Furuno M, Pang KC, Ninomiya N, Fukuda S, Frith MC, Bult C, Kai C, Kawai J, Carninci P, Hayashizaki Y, Mattick JS, Suzuki H. Clusters of internally primed transcripts reveal novel long noncoding RNAs. PLoS Genet. 2006;2:e37. doi: 10.1371/journal.pgen.0020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balas MM, Johnson AM. Exploring the mechanisms behind long noncoding RNAs and cancer. Noncoding RNA Res. 2018;3:108–117. doi: 10.1016/j.ncrna.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder R, Trauner M, Zatloukal K. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330–342. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 6.Hämmerle M, Gutschner T, Uckelmann H, Ozgur S, Fiskin E, Gross M, Skawran B, Geffers R, Longerich T, Breuhahn K, Schirmacher P, Stoecklin G, Diederichs S. Posttranscriptional destabilization of the liver-specific long noncoding RNA HULC by the IGF2 mRNA-binding protein 1 (IGF2BP1) Hepatology. 2013;58:1703–1712. doi: 10.1002/hep.26537. [DOI] [PubMed] [Google Scholar]
- 7.Xin X, Wu M, Meng Q, Wang C, Lu Y, Yang Y, Li X, Zheng Q, Pu H, Gui X, Li T, Li J, Jia S, Lu D. Long noncoding RNA HULC accelerates liver cancer by inhibiting PTEN via autophagy cooperation to miR15a. Mol Cancer. 2018;17:94. doi: 10.1186/s12943-018-0843-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin D, Li Y, Fu C, Feng Y. Pro-Angiogenic Role of LncRNA HULC in Microvascular Endothelial Cells via Sequestrating miR-124. Cell Physiol Biochem. 2018;50:2188–2202. doi: 10.1159/000495060. [DOI] [PubMed] [Google Scholar]
- 9.Hu C, Chen M, Jiang R, Guo Y, Wu M, Zhang X. Exosome-related tumor microenvironment. J Cancer. 2018;9:3084–3092. doi: 10.7150/jca.26422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colombo M, Moita C, van Niel G, Kowal J, Vigneron J, Benaroch P, Manel N, Moita LF, Théry C, Raposo G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci. 2013;126:5553–5565. doi: 10.1242/jcs.128868. [DOI] [PubMed] [Google Scholar]
- 11.Wang F, Li L, Piontek K, Sakaguchi M, Selaru FM. Exosome miR-335 as a novel therapeutic strategy in hepatocellular carcinoma. Hepatology. 2018;67:940–954. doi: 10.1002/hep.29586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rao Q, Zuo B, Lu Z, Gao X, You A, Wu C, Du Z, Yin H. Tumor-derived exosomes elicit tumor suppression in murine hepatocellular carcinoma models and humans in vitro. Hepatology. 2016;64:456–472. doi: 10.1002/hep.28549. [DOI] [PubMed] [Google Scholar]
- 13.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koles K, Nunnari J, Korkut C, Barria R, Brewer C, Li Y, Leszyk J, Zhang B, Budnik V. Mechanism of evenness interrupted (Evi)-exosome release at synaptic boutons. J Biol Chem. 2012;287:16820–16834. doi: 10.1074/jbc.M112.342667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Savina A, Vidal M, Colombo MI. The exosome pathway in K562 cells is regulated by Rab11. J Cell Sci. 2002;115:2505–2515. doi: 10.1242/jcs.115.12.2505. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, Chen N, Sun F, Fan Q. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang WT, Ye H, Wei PP, Han BW, He B, Chen ZH, Chen YQ. LncRNAs H19 and HULC, activated by oxidative stress, promote cell migration and invasion in cholangiocarcinoma through a ceRNA manner. J Hematol Oncol. 2016;9:117. doi: 10.1186/s13045-016-0348-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 19.Huo X, Han S, Wu G, Latchoumanin O, Zhou G, Hebbard L, George J, Qiao L. Dysregulated long noncoding RNAs (lncRNAs) in hepatocellular carcinoma: implications for tumorigenesis, disease progression, and liver cancer stem cells. Mol Cancer. 2017;16:165. doi: 10.1186/s12943-017-0734-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun M, Nie F, Wang Y, Zhang Z, Hou J, He D, Xie M, Xu L, De W, Wang Z, Wang J. LncRNA HOXA11-AS Promotes Proliferation and Invasion of Gastric Cancer by Scaffolding the Chromatin Modification Factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76:6299–6310. doi: 10.1158/0008-5472.CAN-16-0356. [DOI] [PubMed] [Google Scholar]
- 21.Li Z, Xu L, Liu Y, Fu S, Tu J, Hu Y, Xiong Q. LncRNA MALAT1 promotes relapse of breast cancer patients with postoperative fever. Am J Transl Res. 2018;10:3186–3197. [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuoka T, Yashiro M. Biomarkers of gastric cancer: Current topics and future perspective. World J Gastroenterol. 2018;24:2818–2832. doi: 10.3748/wjg.v24.i26.2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du Y, Kong G, You X, Zhang S, Zhang T, Gao Y, Ye L, Zhang X. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J Biol Chem. 2012;287:26302–26311. doi: 10.1074/jbc.M112.342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cui M, Xiao Z, Wang Y, Zheng M, Song T, Cai X, Sun B, Ye L, Zhang X. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res. 2015;75:846–857. doi: 10.1158/0008-5472.CAN-14-1192. [DOI] [PubMed] [Google Scholar]
- 25.Zhao Y, Guo Q, Chen J, Hu J, Wang S, Sun Y. Role of long non-coding RNA HULC in cell proliferation, apoptosis and tumor metastasis of gastric cancer: a clinical and in vitro investigation. Oncol Rep. 2014;31:358–364. doi: 10.3892/or.2013.2850. [DOI] [PubMed] [Google Scholar]
- 26.Peng W, Gao W, Feng J. Long noncoding RNA HULC is a novel biomarker of poor prognosis in patients with pancreatic cancer. Med Oncol. 2014;31:346. doi: 10.1007/s12032-014-0346-4. [DOI] [PubMed] [Google Scholar]
- 27.Ruivo CF, Adem B, Silva M, Melo SA. The Biology of Cancer Exosomes: Insights and New Perspectives. Cancer Res. 2017;77:6480–6488. doi: 10.1158/0008-5472.CAN-17-0994. [DOI] [PubMed] [Google Scholar]
- 28.Kogure T, Lin WL, Yan IK, Braconi C, Patel T. Intercellular nanovesicle-mediated microRNA transfer: a mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology. 2011;54:1237–1248. doi: 10.1002/hep.24504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conigliaro A, Costa V, Lo Dico A, Saieva L, Buccheri S, Dieli F, Manno M, Raccosta S, Mancone C, Tripodi M, De Leo G, Alessandro R. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol Cancer. 2015;14:155. doi: 10.1186/s12943-015-0426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li R, Wang Y, Zhang X, Feng M, Ma J, Li J, Yang X, Fang F, Xia Q, Zhang Z, Shang M, Jiang S. Exosome-mediated secretion of LOXL4 promotes hepatocellular carcinoma cell invasion and metastasis. Mol Cancer. 2019;18:18. doi: 10.1186/s12943-019-0948-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Q, Shao Y, Zhang X, Zheng T, Miao M, Qin L, Wang B, Ye G, Xiao B, Guo J. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumour Biol. 2015;36:2007–2012. doi: 10.1007/s13277-014-2807-y. [DOI] [PubMed] [Google Scholar]
- 32.Pan L, Liang W, Fu M, Huang ZH, Li X, Zhang W, Zhang P, Qian H, Jiang PC, Xu WR, Zhang X. Exosomes-mediated transfer of long noncoding RNA ZFAS1 promotes gastric cancer progression. J Cancer Res Clin Oncol. 2017;143:991–1004. doi: 10.1007/s00432-017-2361-2. [DOI] [PubMed] [Google Scholar]
- 33.Zhang R, Xia Y, Wang Z, Zheng J, Chen Y, Li X, Wang Y, Ming H. Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem Biophys Res Commun. 2017;490:406–414. doi: 10.1016/j.bbrc.2017.06.055. [DOI] [PubMed] [Google Scholar]
- 34.Dong L, Lin W, Qi P, Xu MD, Wu X, Ni S, Huang D, Weng WW, Tan C, Sheng W, Zhou X, Du X. Circulating Long RNAs in Serum Extracellular Vesicles: Their Characterization and Potential Application as Biomarkers for Diagnosis of Colorectal Cancer. Cancer Epidemiol Biomarkers Prev. 2016;25:1158–1166. doi: 10.1158/1055-9965.EPI-16-0006. [DOI] [PubMed] [Google Scholar]
- 35.Liu T, Zhang X, Gao S, Jing F, Yang Y, Du L, Zheng G, Li P, Li C, Wang C. Exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget. 2016;7:85551–85563. doi: 10.18632/oncotarget.13465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J, Zhou Y, Lu J, Sun Y, Xiao H, Liu M, Tian L. Combined detection of serum exosomal miR-21 and HOTAIR as diagnostic and prognostic biomarkers for laryngeal squamous cell carcinoma. Med Oncol. 2014;31:148. doi: 10.1007/s12032-014-0148-8. [DOI] [PubMed] [Google Scholar]
- 37.Xie H, Ma H, Zhou D. Plasma HULC as a promising novel biomarker for the detection of hepatocellular carcinoma. Biomed Res Int. 2013;2013:136106. doi: 10.1155/2013/136106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu G, Wang Y, Lu X, He H, Liu H, Meng X, Xia S, Zheng K, Liu B. Low mir-372 expression correlates with poor prognosis and tumor metastasis in hepatocellular carcinoma. BMC Cancer. 2015;15:182. doi: 10.1186/s12885-015-1214-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldenring JR, Ray GS, Lee JR. Rab11 in dysplasia of Barrett's epithelia. Yale J Biol Med. 1999;72:113–120. [PMC free article] [PubMed] [Google Scholar]
- 41.Gebhardt C, Breitenbach U, Richter KH, Fürstenberger G, Mauch C, Angel P, Hess J. c-Fos-dependent induction of the small ras-related GTPase Rab11a in skin carcinogenesis. Am J Pathol. 2005;167:243–253. doi: 10.1016/S0002-9440(10)62969-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu F, Deng X, Yang X, Jin H, Gu D, Lv X, Wang C, Zhang Y, Huo X, Shen Q, Luo Q, Zhao F, Ge T, Zhao F, Chu W, Shu H, Yao M, Fan J, Qin W. Hypoxia upregulates Rab11-family interacting protein 4 through HIF-1α to promote the metastasis of hepatocellular carcinoma. Oncogene. 2015;34:6007–6017. doi: 10.1038/onc.2015.49. [DOI] [PubMed] [Google Scholar]