Abstract
Ovarian cancer (OC) causes significant morbidity and mortality as neither detection nor screening of OC is currently feasible at an early stage. Difficulty to promptly diagnose OC in its early stage remains challenging due to non-specific symptoms in the early-stage of the disease, their presentation at an advanced stage and poor survival. Therefore, improved detection methods are urgently needed. In this article, we summarize the potential clinical utility of epigenetic signatures like DNA methylation, histone modifications, and microRNA dysregulation, which play important role in ovarian carcinogenesis and discuss its application in development of diagnostic, prognostic, and predictive biomarkers. Molecular characterization of epigenetic modification (methylation) in circulating cell free tumor DNA in body fluids offers novel, non-invasive approach for identification of potential promising cancer biomarkers, which can be performed at multiple time points and probably better reflects the prevailing molecular profile of cancer. Current status of epigenetic research in diagnosis of early OC and its management are discussed here with main focus on potential diagnostic biomarkers in tissue and body fluids. Rapid and point of care diagnostic applications of DNA methylation in liquid biopsy has been precluded as a result of cumbersome sample preparation with complicated conventional methods of isolation. New technologies which allow rapid identification of methylation signatures directly from blood will facilitate sample-to answer solutions thereby enabling next-generation point of care molecular diagnostics. To date, not a single epigenetic biomarker which could accurately detect ovarian cancer at an early stage in either tissue or body fluid has been reported. Taken together, the methodological drawbacks, heterogeneity associated with ovarian cancer and non-validation of the clinical utility of reported potential biomarkers in larger ovarian cancer populations has impeded the transition of epigenetic biomarkers from lab to clinical settings. Until addressed, clinical implementation as a diagnostic measure is a far way to go.
Keywords: biomarker, cell free DNA, diagnosis, DNA methylation, epigenetics, epithelial ovarian cancer
KeyPoints
Prompt diagnosis remains challenging due to non-specific symptoms in the early-stage of the disease, their presentation at an advanced stage and poor survival.
DNA methylation occurs very early in malignant transformation and their utility as biomarker holds great promise to overcome the false positive detection of ovarian cancer using current standard serum marker CA125.
Not even a single report has suggested or demonstrated a good epigenetic marker for early and accurate detection of OC in either tissue or fluid. Thus, early detection still remains a huge unmet need. However, analysis of a panel of aberrant methylation based epigenetic markers in blood-based non-invasive assay could pave its way into clinical implementation.
Introduction
Ovarian cancer, a molecularly heterogeneous disease, remains the most lethal disease among gynecological malignancies. Representing as the third most frequent cancer among female gynecological system carcinoma, ovarian cancer is associated with the highest mortality rates. Despite constituting only 3% of all female cancer, the annual incidence of ovarian cancer worldwide is 220,000 with 21,290 estimated numbers of new cases and 14,600 estimated deaths annually (Siegel et al., 2015). Typical diagnosis of more than 70% of OC cases, at an advanced disease stage is one of the potent reasons for high fatality rate and carries poor prognosis with current therapies. The median age of disease presentation in ovarian cancer is 60 years and its lifetime risk is one in seventy with an overall lifetime mortality of one in ninety five (Cannistra, 2004; Howe et al., 2006).
Epithelial ovarian cancer (E0C) comprises 90% of all forms of OC cases and is characterized by heterogeneity at histopathological, clinical and molecular level. The exact cause for the ovarian malignancy still remains unknown. A strong familiar history either of ovarian or breast cancer has been described as important risk factors associated with OC. More than one-fifth of ovarian carcinomas (about 23%) have hereditary susceptibility and germline mutations of BRCA1 and BRCA2 tumor suppressor genes; in particular contribute to 65–85% of these cases (Ramus et al., 2007). An association of hormonal risk in postmenopausal women is suggested by over 50% of deaths. In addition, parity, pregnancy, lactation, tubal ligation, and oral contraceptive use are associated with reduced risk and have been found to be protective factors against disease development.
Rapid growth, non-specific clinical symptoms at early stage of the disease and lack of early diagnostic methods make prompt diagnosis challenging. As a result, EOC is typically diagnosed at an advanced stage (FIGO III/IV), when the tumor has spread beyond the pelvis and even unlikely to be completely removed by surgery. The long term survival rates for women with disseminated malignancies are low (10–30%). However, diagnosis of ovarian cancer at the localized stage (confinement of lesion still to the ovaries) is highly curable (over 95% 5 year survival rate; Siegel et al., 2011). To improve the overall survival of women diagnosed with EOC and to overcome the non-specific clinical manifestation of EOC, identification of molecular biomarkers of preclinical or early stage EOC tumors is critically needed. A better understanding of EOC genome portrait will help in the identification of promising biomarkers of clinical utility for early diagnosis of OC.
Molecular Classification
The primary OC were classified into epithelial (60%), germ cell (30%), and sex-cord stromal tumors (8%), by the World Health Organization (WHO) classification and tumor morphology system (2014). A large majority of OC, almost 80–85%, are of epithelial origin. However, a small proportion accounting approximately 10% of all OC falls into germ cell and sex-cord stromal tumor categories (Devouassoux-Shisheboran and Genestie, 2015). Further on the basis of disease dissemination, the American Joint committee on Cancer/Tumor Node Metastasis (AJCC/TNM) and International Federation of Gynecology and Obstetrics (FIGO) staging systems, classified ovarian cancer into various stages. The confinement of tumors to the ovaries is represented by stage I and II whereas stage III is associated with local metastasis (usually lymph) and stage IV with distal organ metastases (Yarbro et al., 1999).
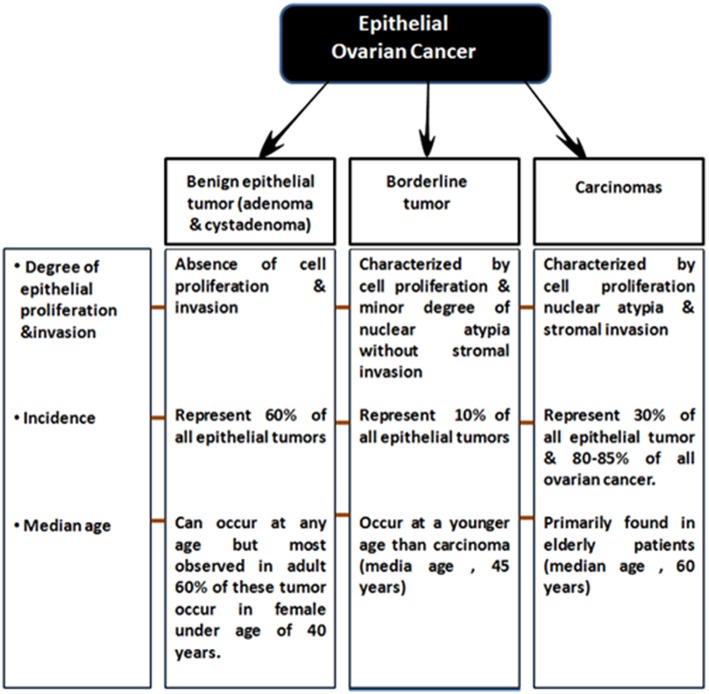
EOCs have been further sub-categorized based on following two criteria: (a) firstly, on the degree of proliferation, grade and extent of invasion into Benign (adenoma and cystadenoma), low malignant potential (LMP) and malignant (b) and secondly based on tumor histopathological grade and molecular characteristics, EOC malignant tumors are classified into serous (70%, most common), endometrioid (10–20%), clear cell (12%), mucinous (3%) and less commonly, transitional (6%), squamous, mixed, and undifferentiated (<1%) subtypes (Bowtell, 2010; Devouassoux-Shisheboran and Genestie, 2015; Earp and Cunningham, 2015; Figure 1) On the basis of histological type and grade, these tumors exhibit different genetic and epidemiological risk factors, pattern of spread, molecular abnormalities, response to targeted therapies and disease prognosis (Devouassoux-Shisheboran and Genestie, 2015; Earp and Cunningham, 2015).
Figure 1.
Sub-classification of epithelial ovarian tumors.
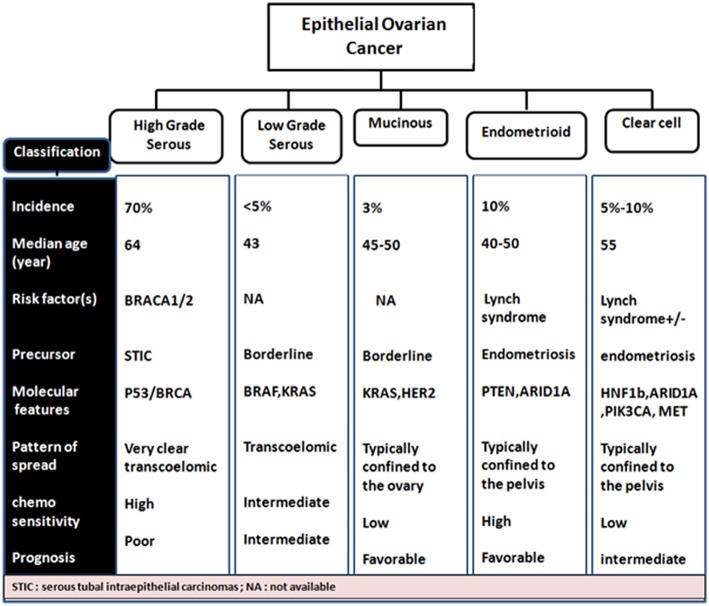
Almost a decade ago, a dualistic classification system recognized Type I and Type II EOC tumors (Shih and Kurman, 2004; Vang et al., 2009). Type I EOCs are generally low grade serous carcinomas but also include mucinous, endometrioid, and clear cell subtype tumors. They are thought to arise from a low malignant potential precursor, are characterized as slow growing with low levels of chromosomal instability, intact DNA repair machinery and harbor mutations in KRAS, BRAF, and ERBB2 at a high frequency. Type II EOCs arise de novo and are comprised of high-grade serous carcinoma. These aggressive tumors also include malignant mixed mesodermal and undifferentiated carcinomas, are characterized by rapid growth with no identified precursor lesions, high levels of chromosomal aberrations along with high frequency of TP53, BRCA1/2 mutations. They constitute 70% of EOC cases (Jayson et al., 2014; Figure 2).
Figure 2.
Phenotypic- genotypic classification of epithelial ovarian cancer subtypes.
The cells of origin of ovarian cancer are still debated. Two models with respect to the origin of ovarian cancer have been proposed: (1) origin from ovarian surface epithelium (OSE), (2) from the fallopian tube. Taken together, the pro-inflammatory environment due to ovulation events, expression pattern of ovarian inclusion cysts and biomarkers which are shared by OSE and malignant growth, form the basis of first model. On contrary, tubal precursor lesions, genetic evidence of BRCA1/2 mutation carriers and recent studies strongly implicate a non-ovarian origin and form the basis of the later model. To date, neither model has evidently revealed superiority over the other. Thus, it is speculated that the HGSOC could have arisen from two different sites which undergo similar changes and could be a possible reason for tumor heterogeneity (Klotz and Wimberger, 2017). It has also been postulated that aberrantly methylated Mullerian duct cells migrate into ovarian stroma where they are supported by the epigenetically/ genetically altered stromal environment, facilitating a cascade of events which culminate in ovarian carcinogenesis. Epigenetic profiling of endocervical glandular cells would facilitate in prediction of risk or early detection of ovarian cancer (Jones et al., 2010).
Screening and Early Detection
OC is generally characterized by few non-specific early symptoms, presentation of the disease at a late stage and poor survival. Difficulty to diagnose it in its early stages still remains challenging. Early diagnosis, screening and personalized treatment is still the biggest unmet need to combat this devastating disease. Unavailability of early cancer-specific diagnostic markers and ubiquitous acquisition of drug resistance to targeted therapies are the most striking obstacles for the effective OC treatment.
Clinically, serum antigen-125 (CA125) is the most extensively studied, established and utilized diagnostic marker of EOC, despite its elevation marked by only 47% of early-stage EOC (Woolas et al., 1993). Additionally, aberrantly elevated serum CA125 have been reported in several benign conditions of endometriosis, pregnancy, peritonitis, pelvic inflammatory disease, uterine fibroids, menstrual cycle, liver cirrhosis. Its elevation is also associated with several malignancies such as lung and colorectal cancer (Jacobs and Bast, 1989). Moreover, poor specificity, high false positive rate, and low positive prediction value make CA125 alone unsuitable as an EOC diagnostic marker. However, CA125 is more suitable markers for tumor recurrence (Clarke-Pearson, 2009).
For clinical needs to diagnose OC at an early stage, the conventional screening methods such as serum cancer antigen 125 (CA125) concentrations, transvaginal ultrasound probe and magnetic resonance imaging have not shown reliability in reducing population mortality or morbidity due to high false-negatives rates and lower sensitivity and specificity (Menon and Jacobs, 2000; Jacobs and Menon, 2004; Munkarah et al., 2007). Therefore, methods for early detection are critically required. Owing to the low incidence rate of OC amongst postmenopausal women, a logistic diagnostic screening test warrants the need of high sensitivity (>75%) and specificity (>99.6%) to attain a positive prediction value (PPV) of 10%. Novel biomarkers for early-stage diagnosis are being explored and it is more likely that a combination of biomarkers could achieve these required diagnostic criteria (Moore et al., 2010).
To determine the effect of screening on OC mortality, several randomized controlled trial in general population had been undertaken. Recently, both CA125 and transvaginal sonography (TVS) was evaluated in the Prostate, Lung, Colorectal, and Ovarian (PLCO) cancer screening trial, however no significant difference was observed in OC mortality between screening and conventional care arms (Buys et al., 2011). The United Kingdom Collaborative Trial of Ovarian Cancer Screening (UKCTOCS), being considered as the largest prospective randomized trial, comprised of over 200,000 asymptomatic postmenopausal women who were screened with TVS alone and combined TVS and CA125. Although improvement in specificity of detection was achieved on combining CA125 with TVS, however these trials failed to attain the requisite diagnostic accuracy of 99.6% specificity (Menon et al., 2009). CA125 together with HE4 has somewhat improved sensitivity and specificity of detection which correctly identified 76.4% of cancer samples and 95% of cancer negative samples. This accuracy was notably higher than either marker alone. However further validation is still required (Moore et al., 2010). According to the Guide to Clinical Preventive Services 2010–2011, it has been mentioned that neither of any screening test [serum antigen-125 (CA-125), ultrasound imaging, pelvic examination or any earlier diagnosis methods] was able to improve OC survival rates U. S. Preventive Services Task Force (2010).
The Risk of Malignancy Index, widely used at present, particularly UK, is a score based on ultrasound variables, menopausal status and CA125 (Jacobs et al., 1990). Its sensitivity is the determining criteria for a patient to be sent to experts by referring gynecologist provided objective assessment score is lower (78%) (Geomini et al., 2009). Transvaginal sonography (TVS) is based on a formal scoring model system. Though highly sensitive and being considered as an ideal method for second stage diagnosis, the major limitation associated with this method is its high dependency on individual expertise (Yazbek et al., 2008). Therefore, in clinical practice to discriminate benign and malignant ovarian tumors is still a significant challenge. The availability of biomarker or their combination which can potentially detect ovarian cancer at its earliest stage with required sensitivity and specificity would help in improving clinical outcomes.
Markers for Ovarian Cancer Diagnosis and Management
Protein Markers
As discussed before, a suitable screening test for OC early stage diagnosis will require high sensitivity and high specificity. Current practices for screening of OC include transvaginal ultrasonography, biomarker analysis, or a combination of both. To date, a number of potential biomarkers for early diagnosis of OC have been identified through intense research in proteomic and genomic. Here, we summarize a comprehensive account of recent researches on explored novel and robust serum based biomarkers for the non-invasive early stage screening of ovarian cancer (Table 1).
Table 1.
Novel tumor biochemical markers for early detection of ovarian cancer.
| Biochemical marker | Nature | Origin/function | Status | Source | Clinical utility | References |
|---|---|---|---|---|---|---|
| CA125 | Glycoprotein antigen | Expressed by fetal amniotic epithelium and coelomic epithelium | Elevated Early stage: 47% Advanced stage: >90% |
Serum | Valuable marker for tumor recurrence Limitations: Unsuitable for early detection due to insufficient sensitivity and being elevated in other conditions |
Clarke-Pearson, 2009; Moore et al., 2010 |
| HE4 | Protease | Serum maturation | Elevated Serous: 93% Endometrioid: 100% |
Serum | Histotype differentiation and screening | Hough et al., 2000; Lu et al., 2004; Moore et al., 2008 |
| Mesothelin | Glycosylphosphatidylinositol- linked cell surface molecule | *Expressed by mesothelial cells *Involved in metastasis |
Elevated Early stage: 60% |
Urine | Early screening | McIntosh et al., 2004; Tang et al., 2013 |
| Transthyretin | An acute phase reactant and major carrier of serum thyroxine | Tumor development | Downregulated in EOC patients | Serum | Early stage screening | Mählck and Grankvist, 1994; Schweigert and Sehouli, 2005; Nosov et al., 2009 |
| ApoA1 | Constituent of high density lipoproteins | Prevents tumor development | Downregulated in ovarian cancer patients | Serum | Early stage screening | Gadomska et al., 2005; Kim et al., 2012 |
| Kallikrein | Family of Serine proteases *Human KLK family: 15 members *Chromosome position: 19q13.3–4 |
Regulates proteolytic cascades | Elevated: 12KLK/15 | Serum | Elevated KLK-6 and−10 in OC cases with low level of CA125. Useful marker for OC detection |
Borgoño and Diamandis, 2004; Rosen et al., 2005 |
| Osteopontin | An adhesive glycoprotein | *Synthesized by osteoblasts and vascular endothelial cells *Associated with bone remodeling and immune function |
Elevated in invasive and borderline ovarian cancer tumors | Plasma | Early stage screening | Kim et al., 2002 |
Although being considered as the “gold standard” biomarker for detection of OC, its clinical relevance mainly falls in evaluating disease recurrence. Other biochemical markers such as lysophosphatidic acid, human epididymis protein 4 (HE4), inhibins (which are members of TGF-β subfamily), Mesothelin (associated with migration and metastasis) (Huang et al., 2006), Osteopontin, and YKL-40 have been reported to be elevated in sera of patients with OC amongst various studies, which could be of diagnostic significance for improved cancer detection, most likely in various combination with one another and /or with CA125 (Rosenthal et al., 2006; Moore et al., 2010). The most promising molecular biomarker of all these, to date are HE4 and Mesothelin. So far, US FDA has only approved CA125 and HE4 for monitoring disease progression/recurrence, but not for screening purpose (Rosenthal et al., 2006).
For the triage of pelvic mass, the multivariate index assay OVA1, constituting measurements of 5-proteins: CA125-II, apolipoprotein A1, transthyretin, beta 2 microglobulin, and transferrin, has been approved by FDA since 2009. Although, the test had improved sensitivity but compromised in revealing diagnostic potential with its low specificity upon replacement of CA125 with the multivariate index assay (Nguyen et al., 2013). Elevated levels of Kallikrein 6 and 7 (KLK6 and KLK7) was reported in sera of ovarian carcinoma subtypes, depicting their potential to improve early detection of OC. Other biomarkers with potential clinical significance for early diagnosis in women with EOC include GSTT1, Prostasin (PRSS8), KLK6, KLK7, FOLR1, and ALDH1, which are currently under research and clinical trials (Sarojini et al., 2012).
Evaluation of several prediagnostic multimarker panels along with PLCO screening trial has identified promising biomarkers which are able to distinguish ovarian cancer cases from normal control groups; for instance, a four biomarker panel consisting of CA-125, HE4, CEA, and VCAM-1 effectively discriminated early stage OC from healthy controls with sensitivity of 86% at 98% specificity (Lin et al., 2009). Another panel constituting of CA-125, ApoA1, TTR, and H418, was able to differentiate OC patients at early stage of disease from cancer-free healthy control samples with 74% sensitivity at 97% specificity (Zhang et al., 2004). Still to date, no panel of biomarkers that has been examined amongst numerous studies could outperform CA125 alone, in distinguishing between the two groups. The sensitivity and specificity of serum based non-invasive biomarkers for improved ovarian cancer detection from various studies as well as the currently active/completed clinical trials evaluating potent biochemical markers of clinical significance for early diagnosis of EOC are summarized in Tables 2, 3 respectively.
Table 2.
Specificity and sensitivity of early detection biomarkers for ovarian cancer from various studies.
| Biochemical biomarker | Source | Population tested | Clinical prediction | References | |
|---|---|---|---|---|---|
| Sensitivity | Specificity | ||||
| HE4 | Serum | 147 Cancer (111 ovarian cancer cases), 285 Benign, 66 controls | 79.3% | 98.9% | Molina et al., 2011 |
| HE4 + CA125 | Serum | 383 Benign and 89 Cancer | 100% 92.30% |
74.20% 76.0 % |
Moore et al., 2011 |
| Osteopontin | Plasma | 46 Benign, 47 cancer, 51 ovarian cancer, 107 controls | - | - | Kim et al., 2002 |
| Prostasin + CA125 | Serum | 64 cancer, 137 control | 92% | 94% | Mok et al., 2001 |
| KLK6 | Serum | 141 Benign, 146 ovarian cancer, 97 controls | 21–26% | 95% | Diamandis et al., 2003 |
| KLK6+ CA125 | Serum | 141 Benign, 146 ovarian cancer, 97 controls | 42% | 90% | Diamandis et al., 2003 |
| B7-H4 | Serum | 1,023 cancer, 997 Benign (236 ovarian cancer cases, 260 controls) | 45% | 97% | Simon et al., 2006 |
| B7-H4 + CA125 | Serum | 1,023 cancer, 997 Benign (236 ovarian cancer cases, 260 controls) | 65% | 97% | Simon et al., 2007 |
| CA125/IL-6/IL-8/VEGF/EGF | Serum | 44 Early-stage cancers 37 Benign, 45 controls |
84% | 95% | Gorelik, 2005 |
| CA125/IL-6/G-CSF/VEGF/EGF | Serum | 44 Early-stage cancers 37 Benign, 45 controls |
86.5% | 93% | Gorelik, 2005 |
| CA125/HE4/Glycodelin/ PLAUR/MUC1/PAI-1 | Serum | 200 Cancers (133 stage I/II), 396 Healthy controls | 80.5% | 96.5% | Havrilesky et al., 2008 |
| Leptin/Prolactin/ Osteopontin/IGF2 | Serum | 100 Cancers, 106 controls | 95% | 94% | Mor et al., 2005 |
| CA125 HE4 Mesothelin |
Serum | 143 Cancers, 124 benign, 344 controls | 78% 68–82% 31–44% |
98% 98% 98% |
Shah et al., 2009 |
| Leptin/Prolactin/ Osteopontin/IGF2/MIF/ CA125 | Serum | Training: 113 cancers, 181 controls Test: 43 cancers, 181 Controls | 95.3% | 99.4% | Visintin et al., 2008 |
| CA125/ CA19- 9 /EGFR /CRP/ Myoglobin/APOA1/ APOC3/MIP1A/ IL-6/IL-18/ Tenascin C |
Serum | 115 Cancers, 93 benign 24 Controls, 13 non-ovarian cancers | 91.3% | 88.5% | Amonkar et al., 2009 |
| CA-125, HE4, SI | Serum | 74 cancer, 137 controls | 84% | 98.5% | Andersen et al., 2010 |
| RIM, ROMA, CA-125, HE4, | Serum | 445 Benign, 31 borderline, and 162 malignant tumors | Postmenopausal 89, 91, 92, and 72% Premenopausal 87, 87, 96, and 83% |
Postmenopausal 80, 77, 80, and 92% Premenopausal 90, 81, 60, 91% |
Lycke et al., 2018 |
| CA-125 HE4 TTR |
Serum | 130 Benign, 126 ovarian cancer, 400 controls | 64.29% 46.4% 78.6% |
53.57% 43.3% 68.8% |
Zheng et al., 2018 |
| CA-125, HE4, TK1 | Serum | 75 ovarian cancer, 40 Benign, 35 controls | 94.18% | 79.53% | Xi et al., 2017 |
| CA-125+HE4, HE4+FOLR1 |
Serum | 150 benign controls, 216 ovarian cancer, 20 controls | 67% 65% |
95% 95% |
Leung et al., 2016 |
| CA-125, ApoA1, TTR | Serum | 200 cancer, 142 controls | 74% | 97% | Zhang et al., 2004 |
| CA125, HE4, MMP-7, CA72-4 | Serum | 142 stage I cancer, 217 controls | 83.2% | 98% | Simmons et al., 2016 |
| CA-125, CA 72–4, MCSF | Plasma | 123 cancer, 224 controls | 70% | 98% | Edgell et al., 2010 |
| CA-125, TTR, ApoA1 | Serum | 20 cancer, 82 controls | 89% | 92% | Su et al., 2007 |
| CA-125, HE4, CEA, VCAM-1 | Serum | 456 cancer, 2,000 controls | 86–93% | 98% | Lin et al., 2009 |
| CA-125, CRP, SAA, IL-6, IL-8 | Plasma | 150 cancer, 212 controls | 94.1% | 91.3% | Edgell et al., 2010 |
| S100A4 | Serum | 160 cancer, 52 Benign, 52 controls | 78% | 92% | Lv et al., 2018 |
| KPNA2 | Serum | 162 cancer, 48 controls | 71.4% | 81.2% | Huang et al., 2017 |
| Septin-9 Clusterin |
Plasma | 137 EOC, 12 borderline,51 benign, 58 controls | 82.5% 71.5% |
50.0% 41.4% |
Lyu et al., 2018 |
Table 3.
Clinical trials (currently active or completed) for evaluating novel biomarkers of ovarian cancer.
| Biochemical marker | Setting | Phase | Samples (n) | Status | Clinical trial no. | Primary clinical outcome | References |
|---|---|---|---|---|---|---|---|
| All biomarkers | Adnexal mass | 1 | 500 (E) | Completed | NCT01466049 | Screening | NA |
| HE4 + CA125 | Pelvic mass | 0 | 566 | Completed | NCT00315692 | Cancer vs. benign disease | Moore et al., 2009 |
| CA125 | Low risk women | 1 | 9500 (E) | Recruiting | NCT00539162 | Rate of increase in CA125 over time | NA |
| HE4 + CA125 | Adnexal mass | 1 | 512 | Completed | NCT00987649 | Initial cancer risk assessment | NA |
| CA125+ HE4 | High risk women | 1 | 1208 (E) | Active, not recruiting | NCT01121640 | PPV of screening protocol | NA |
| CA125 | High risk women | 2 | 2400 (E) | Withdrawn | NCT00080639 | Screening | NA |
| Mesothelin | Low risk women | 0 | 250 (E) | Unknown | NCT000155740 | Screening | NA |
| FOLR1 | Stage I ovarian cancer | 2 | 50 (E) | Terminated | NCT01511055 | Sensitivity and specificity of Intraoperative imaging (IOI) with folate | NA |
| CA125 + TVU | Ovarian disease | 0 | 750 (E) | Terminated | NCT01292733 | CA125 measurement in blood over time | NA |
| CA125± TVU | Postmenopausal | 0 | 48230 | Completed | NCT00058032 | Screening post menopausal women | Menon et al., 2009; Jacobs et al., 2011 |
| CA125 | High risk women | 0 | 2430 | Unknown | NCT00039559 | Sensitivity and specificity of early detection for ovarian cancer | NA |
| CA125+ TVU | High genetic risk women | 0 | 5000 (E) | Completed | NCT00033488 | Screening women at high genetic risk for ovarian cancer | NA |
| CA125 | High risk women | 0 | 6000 (E) | Recruiting | NCT00005095 | Screening | NA |
| Combined methods | Ovarian neoplasms | 0 | 36000 | Unknown | NCT01178736 | Low- cost screening | NA |
| Interventional | High risk women | 0 | 1500 | Recruiting | NCT00849199 | Genetic testing, screening | NA |
| All biomarkers | High risk women | 0 | 250 (E) | Unknown | NCT00854399 | Overall survival | NA |
| Tumor markers | High risk women | 0 | 5000 | Completed | NCT00267072 | Early stage detection | NA |
| DNA markers | Ovarian cancer | 0 | 118 (E) | Active, not recruiting | NCT00879840 | Assessment of screening modalities | NA |
| BRCA1/2 Mutation | Ovarian neoplasms | 0 | 1500 | Completed | NCT00001468 | Identifying BRCA1/2 mutation | NA |
| BRCA | Epithelial ovarian cancer | 0 | 600 (E) | Completed | NCT03229122 | Genetic Testing of BRCA | NA |
| All biomarkers | High risk women | 0 | 500(E) | Recruiting | NCT03150121 | Identification of uterine lavage-based biomarkers for early detection | NA |
| All biomarkers | High risk women | 0 | 6000 | Recruiting | NCT00005095 | Early stage detection and prevention | NA |
| CA125 | High genetic risk women | 0 | 40 | Completed | NCT00043472 | Screening | NA |
| DNA markers | Women with serous epithelial ovarian cancer | 0 | 250 | Not yet recruiting | NCT03622385 | Early detection of high grade serous epithelial ovarian cancer | NA |
TVU, transvaginal ultrasonography; (w), women; (E), estimated enrollment; IOI, intraoperative imaging. Source: http://clinicaltrials.gov/.
Genetic Marker
About 23% of ovarian tumors have been associated with hereditary conditions and the genetic abnormalities in about 65–85% of hereditary ovarian carcinomas is the germline mutation in BRCA (breast cancer early onset genes BRCA1 and BRCA2) genes which are essential for DNA repair as well as in maintaining genomic stability and integrity. The cumulative lifetime risk of EOC for a woman with BRCA1 and BRCA2 mutation is 39–46% and 12–20%, respectively (Ramus et al., 2007). Lifetime risk to develop breast cancer and ovarian cancer is enhanced up to 85% and up to 54% respectively in the carriers of BRCA1 and BRCA2 mutations. Association of several tumor suppressor genes and oncogenes (tumor suppressor gene TP53 in Li- Fraumeni syndrome, mismatch repair genes (MMR) in Lynch syndrome, genes involved with double strand break repair system: BARD1, CHEK2, RAD51, and PALB2) with hereditary ovarian cancer has been reported. Till date, around 16 genes have been reported to be associated with hereditary ovarian carcinogenesis while several other mutations are yet unknown and need to be further explored (Toss et al., 2015).
Epigenetic Marker
Epigenetics is the mechanism for the regulation of gene expression without any alternation in the primary DNA sequence (Jones and Laird, 1999; Jones and Baylin, 2002; Feinberg and Tycko, 2004). DNA methylation, modification of histone proteins and miRNAs are the key modulator in regulating several cellular processes such as cell differentiation, embryogenesis, inactivation of X chromosome, genome imprinting, and many others (Jones, 2001; Reik and Lewis, 2005; Kacem and Feil, 2009; Portela and Esteller, 2010). The epigenetic alternations involve interplay between DNA methylation, histone modification and micro RNA expression to modulate gene expression during development and cancer progression. (1) The global hypomethylation, largely of repetitive DNA which results in demethylation of several oncogenes and (2) localized hypermethylation at promoters of various tumor suppressor genes leading to their transcriptional silencing, are two opposite epigenetic phenomenon involved in tumorigenesis (Sharma et al., 2010). DNA methyltransferase (DNMT) mediated methylation of deoxycytosine located within the CpG dinucleotides is the best known and widely studied epigenetic mechanism leading to transcription repression in cancer (Bird and Wolffe, 1999; Hendrich and Bird, 2000; Bird, 2002). DNA methylation is known to be the earliest event during carcinogenesis and plays a crucial role in silencing of tumor suppressor genes (Sharma et al., 2010; Teschendorff and Widschwendter, 2012; Teschendorff et al., 2012, 2016; Bartlett et al., 2016). Promoter methylation mediated epigenetic silencing of gene is regulated by the recruitment of MBD (methyl CpG binding proteins such as MeCP2, MBD1, MBD2, and MBD4) which in turn regulates chromatin state by recruiting histone modifying and chromatin-remodeling complexes (repressors) at the site of methylation, which subsequently generates condensed chromatin structure and results in transcriptional repression (Esteller, 2007; Lopez-Serra and Esteller, 2008). On contrary, epigenetic activation of gene is regulated by recruitment of Cfp1 and histone methyltransferase Setd1 which aids in generating an open chromatin structure by creating domains which are enriched with active histone marks (acetylation and H3K4 trimethylation) (Thomson et al., 2010, p. 1; Jones and Baylin, 2007; Supplementary Figure 1). Increasing evidences has revealed the significant role of DNA methylation in cancer development and progression, right from transcriptional silencing of tumor suppressor genes to the activation of oncogenes and consequently promoting metastasis (Costello and Plass, 2001; Herman and Baylin, 2003; Wilting and Dannenberg, 2012). Apparently, it is quite evident now that DNA methylation plays an equal or possibly even greater role than the genetic lesion such as mutations, deletion and translocations which have been associated for long, with malignant transformations and carcinogenesis (Chan T. A. et al., 2008). For instance, though the familial breast cancer susceptibility gene 1 (BRCA1) mutations contributes to 5–10% of EOC, promoter hypermethylation of non-mutated BRCA1 allele is the second disruptive event to the development of this cancer (Barton et al., 2008).
Tissue Biomarkers
Diagnosis
So far, several methylation based signatures have been reported in EOC. Here, we summarize an overview of some of the extensively studied potential biomarkers of diagnostic utility in ovarian cancer (Table 6). In ovarian cancer, a large number of tumor suppressor genes have been identified to be silenced by promoter hypermethylation and downregulated includes DAPK, LOT1, TMS1/ASC, and PAR4 (pro-apoptotic function and cell cycle regulation), p16, SPARC, ANGPTL2, and CTGF (tumor suppressor activity), ICAM-1 and CDH1 (cell adhesion), PEG31 (role in imprinting) and many others (Tables 4, 5). In ovarian cancer, some of the most frequently methylated genes include OPCML (tumor suppressor activity), TES (involved in regulation of cell motility) and RASSF1A (tumor suppressor activity as well as an inhibitor of the anaphase-promoting complex) (Barton et al., 2008). Promoter methylation of HOXA10 and HOXA11, which are involved in very early ovarian tumor initiation effectively distinguished normal and malignant ovaries (Fiegl et al., 2008; Widschwendter et al., 2009). Methylation induced silencing of PTEN has also been frequently observed in primary epithelial ovarian carcinomas (Kurose et al., 2001). CTGF (encodes the connective tissue growth factor) (Kikuchi et al., 2007; Barbolina et al., 2009), CCBE1 (hypothesized to be involved in regulation of cell motility) (Barton et al., 2010), HIC1 (a p53 target gene) (Rathi et al., 2002), CDH13 (Makarla et al., 2005), and CDH1 (the loss of which correlates with the upregulation of matrix metalloproteinases and metastasis- promoting protein a 5- integrin) (Sawada et al., 2008) act as metastasis suppressors. Methylation induced repression of these suppressors correlates with invasive EOC.
Table 6.
Epigenetic biomarkers for ovarian cancer detection.
| Epigenetic marker | Source | Sensitivity /specificity | Technique | References |
|---|---|---|---|---|
| GTF2A1 and HAAO | Tumor tissue | Presence of malignancy 95/88%; 89/82% |
qMSP | Huang et al., 2009 |
| HOXA9 and SCGB3A1 | Tumor tissue | Early stage carcinoma (18/24, p = 0.002) (5/25, p = 0.020) |
MSP | Wu et al., 2007 |
| RASSF1A and HIC1 | Tumor tissue | Early stage ovarian carcinomas 34/100, 2/68, OR = 0.3 34/100, 10/68, OR = 0.4 |
MSP | Feng Q. et al., 2008 |
| RASSF1A, GSTP1, MGMT, APC | Tumor tissue | Presence of invasive tumors RASSF1A: 30% vs. 0%; GSTP1: 9% vs. 0%; MGMT: 9% vs. 0%; APC:22% vs. 0%; |
MSP | Barton et al., 2008 |
| SPARC | Tumor tissue | Association with tumor gradeMethylation frequency: 68% | MSP | Socha et al., 2009 |
| CDH13, CRABP1, HOXA9, and SCGB3A1 | Tumor tissue | Histological subtype differentiation CDH13, CRABP1, HOXA9 and SCGB3A1 (P = 0.041, P <0.001, P = 0.007, P <0.001) |
MSP | Wu et al., 2007 |
| CTGF | Tumor tissue | Inversely correlated with invasive disease | cDNA microarray analysis | Barbolina et al., 2009 |
| CCBE1 | Tumor tissue | Inversely correlated with metastasis 6/11 (55%) in OC cell lines 38/81 (41%) OC tumors. |
Small-interfering RNA (siRNA)-mediated knockdown | Barton et al., 2010 |
| HIC1 | Tumor tissue | Presence of malignancy Methylation frequency: 35% |
MSP | Rathi et al., 2002 |
| CDH1 | Tumor tissue | Inversely correlated with metastasis | Small-interfering RNA (siRNA)-mediated knockdown | Sawada et al., 2008 |
| SFN, TMS1, and WTI | Tumor tissue | Methylation exclusive for Clear cell subtype | MSP | Barton et al., 2008 |
| hMLH1, CDKN2A, and MGMT | Matched tumors | Associated with development of Synchronous endometrial and ovarian cancer Methylation frequency: 39, 41, and 48% |
MSP | Furlan, 2006 |
| 14–3-3s | Tumor tissue | Advanced stage ovarian carcinomas | MSP, quantitative reverse transcription-PCR | Akahira et al., 2004b |
| HOXA11 | Tumor tissue | Presence of malignancy, Suboptimal tumor debulking and poor outcome (relative risk for death = 3.4) | MethyLight assay | Fiegl et al., 2008 |
| 10 gene panel | Tumor tissue | Presence of serous adenocarcinoma (69.4% sensitivity and 70.2% specificity) | Microarray Mediated Methylation Assay (MethDet test) | Melnikov et al., 2009 |
| Polycomb group target genes in particular HOXA9 | Normal endometrium | Hoxa9 hypermethylation association with increased risk (12.3 fold) of ovarian cancer | MethyLight assay | Widschwendter et al., 2009 |
| SNCG, MASPIN, and CLDN4 | Tumor tissue | Advanced stage ovarian carcinomas | Small-interfering RNA (siRNA)-mediated knockdown, qRT-PCR | Gupta et al., 2003; Rose et al., 2006; Choi et al., 2007; Honda et al., 2007 |
| PCDH17 | Tumor tissue | Presence of malignancy | Next-generation sequencing, Methylation-sensitive high-resolution melting Analysis |
Baranova et al., 2018 |
| EGFL7 and RASSF1 | Tumor tissue | Early stage disease detection and progression | DNA methylation microarray assay, Bisulfite pyrosequencing | Rattanapan et al., 2018 |
| LDOC1 | Ovarian cancer cell line | Early stage disease detection | RT-PCR and real-time PCR | Buchholtz et al., 2014 |
| GPR150, ITGA8, and HOXD11 | Ovarian cancer cell line | Tumor marker | Methylation-sensitive-representational difference analysis (MS-RDA) and MSP | Cai et al., 2007 |
| TGFBI | Tumor tissue and ovarian cancer cell line | Presence of malignancy | Real-time RT-PCR, MS-PCR, and bisulfite sequencing | Kang et al., 2010 |
| DAPK1 and SOX1 | Tumor tissue | Early stage disease | MethyLight | Kaur et al., 2016 |
| BORIS | Tumor tissue | Presence of malignancy | qRT-PCR | Woloszynska-Read et al., 2007 |
| long-intergenic non-coding RNA (lincRNA) gene (LOC134466) | Tumor tissue and ovarian cancer cell line | Potential novel diagnostic biomarker for high grade (Type II) serous ovarian carcinoma (HGSOC) | MeDIP-Chip and Sequenom massARRAY methylation analysis | Gloss et al., 2012 |
Table 4.
List of most frequently epigenetically dysregulated genes in ovarian cancer.
| S. No | Gene | Function | Epigenetic event | References |
|---|---|---|---|---|
| 1. | Survivin | Apoptotic inhibitor | Downregulated | Mirza et al., 2002 |
| 2. | GATA 4 | Transcription factor | Downregulated | Caslini et al., 2006; Cai et al., 2009 |
| 3. | APC | Regulation of cell adhesion | Downregulated | Tam et al., 2007 |
| 4. | ARHI | Ras homolog, maternally imprinted putative tumor suppressor. Negative regulator of cancer growth and progression | Downregulated | Yu et al., 2003; Fu et al., 2015 |
| 5. | BRCA 1 | DNA damage response | Downregulated | Baldwin et al., 2000; Wilcox et al., 2005; Press et al., 2008 |
| 6. | DAPK | Apoptosis | Downregulated | Bai et al., 2004 |
| 7. | Estrogen receptor β | Transcriptional activator | Downregulated | Suzuki et al., 2008 |
| 8. | hMLH 1 | DNA mismatch repair | Downregulated | Meng et al., 2008; Zhang H. et al., 2008 |
| 9. | hMSH 2 | DNA mismatch repair | Downregulated | Zhang H. et al., 2008 |
| 10. | ICAM 1 | Cell/matrix adhesion | Downregulated | Arnold et al., 2001 |
| 11. | LOT1 | Apoptosis | Downregulated | Abdollahi et al., 2003; Kamikihara et al., 2005 |
| 12. | OPCML | Cell adhesion | Downregulated | Sellar et al., 2003; Teodoridis et al., 2005; Zhang J. et al., 2006 |
| 13. | PACE-4 | Serine protease | Downregulated | Fu et al., 2003 |
| 14. | RASSFIA | Microtubule stability | Downregulated | Agathanggelou et al., 2001; Yoon et al., 2001 |
| 15. | PEG 3 | Apoptosis | Downregulated | Feng W. et al., 2008 |
| 16. | DLEC 1 | Unknown | Downregulated | Kwong et al., 2006 |
| 17. | ARLTS 1 | Premature termination of translation | Downregulated | Petrocca et al., 2006 |
| 18. | TCEAL 7 | Cell death regulation | Downregulated | Chien et al., 2005 |
| 19. | P16 | Cell cycle control | Downregulated | Milde-Langosch et al., 1998; Katsaros et al., 2004 |
| 20. | TMS1 | Apoptosis | Downregulated | Akahira et al., 2004a |
| 21. | WT1 | Transcription factor | Downregulated | Kaneuchi et al., 2005 |
| 22. | 14-3-3 SIGMA | Regulation of cell growth and differentiation | Downregulated | Kaneuchi et al., 2004 |
| 23. | DR 4 | Apoptosis | Downregulated | Horak et al., 2005 |
| 24. | FBXO 32 | Apoptosis (?) | Downregulated | Chou et al., 2010 |
| 25. | IGFBP-3 | Antiproliferative, pro-apoptosis, and invasion suppressor | Downregulated | Torng et al., 2009 |
| 26. | SFRP5 | Modulator of Wnt signaling | Downregulated | Su et al., 2010 |
| 27. | CCBE 1 | Migration and cell survival | Downregulated | Barton et al., 2010 |
| 28. | RUNX3 | Transcription factor | Downregulated | Zhang et al., 2009 |
| 29. | CHFR | Cell cycle control | Downregulated | Gao et al., 2009 |
| 30. | Maspin(SERPINBS) | Protease inhibitor | Expressed | Rose et al., 2006 |
| 31. | FANCF | DNA repair(?) | Up regulated | Taniguchi et al., 2003 |
| 32. | Synuclein gamma | Unknown | Up regulated | Gupta et al., 2003 |
| 33. | TUBB3 | Formation of microtubules | Up regulated | Izutsu et al., 2008 |
| 34. | CLDN3 | Tight junction protein | Up regulated | Honda et al., 2007 |
| 35. | HOXA10 | Transcription factor | Up regulated | Cheng et al., 2010 |
| 36. | FBXW7 | Ubiquitin ligase mediates ubiquitylation of oncoproteins | Downregulated | Kitade et al., 2016 |
| 37. | SPARC | Membrane-associated glycoprotein, normal development, anti-proliferative, and de-adhesive properties angiogenesis, ECM production | Downregulated | Socha et al., 2009 |
| 38. | HIC1 | Transcriptional repressor | Downregulated | Pieretti et al., 1995; Rathi et al., 2002; Teodoridis et al., 2005; Tam et al., 2007 |
| 39. | Rarβ | Transcriptional regulator of cell growth | Downregulated | Makarla et al., 2005; Tam et al., 2007 |
| 40. | GSTP1 | Intra cellular detoxification | Downregulated | Makarla et al., 2005; Bol et al., 2010 |
| 41. | TBX15 | Development of mesodermal derivative | Downregulated | Gozzi et al., 2016 |
| 42. | HUSIF 1 | Modulate heparin binding growth factor signaling promote Wnt signaling pathway | Downregulated | Staub et al., 2007 |
| 43. | SFRP1 | Inhibitor of Wnt/β-catenin signaling pathway | Downregulated | Su et al., 2009; Kardum et al., 2017 |
| 44. | RunX1T1 | Putative SMAD4 target/TGFβ/SMAD4 signaling | Downregulated | Yeh et al., 2011 |
| 45. | ANGPTL2 | Secreted glycoprotein involved in angiogenesis | Downregulated | Kikuchi et al., 2008 |
| 46. | CTGF | Adhesion molecule, motility modulator | Downregulated | Kikuchi et al., 2007 |
| 47. | FOXD3 | Transcriptional regulator of development, cell maintenance, and lineage specification | Downregulated | Luo et al., 2019 |
| 48. | NISCH | Encodes imidazoline receptor Nischarin, regulates cellular migration, and invasion upon interacting with PAK1, LIMK, Rac1, and LKB1 | Downregulated | Li J. et al., 2015 |
| 49. | ABCA1 | A TGF-β target, regulator of cholesterol efflux and metabolism | Downregulated | Chou et al., 2015, p. 1 |
| 50. | TIMP2 | A EZH2 target, endogenous regulator of matrix metalloproteinases, repressor of metastasis | Downregulated | Yi et al., 2017 |
| 51. | PCDH17 | Transmembrane protein belonging to cadherin superfamily, potential calcium-dependent cell-adhesion protein | Downregulated | Baranova et al., 2018 |
| 52. | LDOC1 | A nuclear transcription factor, regulator of NFκB Signaling | Downregulated | Buchholtz et al., 2014 |
| 53. | RGS2 | Regulator of GTPase activity of G protein subunits. Negative regulator of angiotensin-activated signaling pathway | Downregulated | Cacan, 2017 |
| 54. | PRTFDC1 | Unknown | Downregulated | Cai et al., 2007 |
| 55. | DDR | Subclass of RTKs, associated with cell differentiation, proliferation, adhesion, migration, and invasion | Downregulated | Chung et al., 2017 |
| 56. | ARNTL | Circadian gene | Downregulated | Yeh et al., 2014 |
| 57. | GULP1 | Apoptosis, lipid homeostasis, regulator of Arf6-mediated signaling | Downregulated | Maldonado et al., 2018, p. 1 |
| 58. | TGFB1 | Adhesion, essential for function of microfibrils and interacts with fibronectin and integrins | Downregulated | Kang et al., 2010 |
| 59. | SALL2 | Cellular quiescence factor, neural development | Downregulated | Sung et al., 2013 |
| 60. | PDZ-LIM | Ubiquitination of nuclear p65, inflammation | Downregulated | Zhao et al., 2016 |
| 61. | KLF11 | Inhibitory regulator of TGFβ signaling, promotes apoptosis | Downregulated | Wang et al., 2015 |
| 62. | GBGT1 | Encodes glycosyltransferase which plays role in synthesis of Forssman glycolipid | Downregulated | Jacob et al., 2014 |
Table 5.
List of hypermethylated genes in ovarian cancer.
| Gene | Frequency of hypermethylation in ovarian cancer | Ovarian cancer subtype | Frequency of hypermethylation in normal tissue | Method | References |
|---|---|---|---|---|---|
| RASSF1A | 15.6–50% | S, M, E, CC | 0–13% | MSP | Yoon et al., 2001; Makarla et al., 2005; Choi et al., 2006; Bol et al., 2010 |
| DAPK | 50–67% (full) | S, CC, E, M, CS, PDA | 50% (Partial) | MSP | Collins et al., 2006; Häfner et al., 2011 |
| p16(CDKN2A) | 16.9–42% | S, M, E | 0–25% | MSP | Strathdee et al., 2001; Rathi et al., 2002; Tam et al., 2007 |
| HIC1 | 15.9–51.7% | Not specific | 12.5–19%; 11.1% | MSP | Strathdee et al., 2001; Ongenaert et al., 2008 |
| OPCML | 46.5–83.3% | Not specific | 0% | Restriction enzyme cut analysis, MSP | Czekierdowski et al., 2006a; Zhang J. et al., 2006 |
| MLH1 | 10% | S, E, M, CC, MIX | 0% ADJ NLS | MSP | Strathdee et al., 2001 |
| TERT | 29.8% | S, M, E, CC | 30% | qMSP | Widschwendter A. et al., 2004 |
| PTEN | 16.9% | E, S, M, CC | 0% | MSP | Ongenaert et al., 2008 |
| ING1 | 24% | S, M, E, CC, PDA | 0% | MSP | Shen et al., 2005 |
| ITGA8 | 13.3% | S, E, M, CC, SCC | 0% END cyst | MSP | Cai et al., 2007 |
| MGMT | 9% | S, M, CC, E, UN | 16% | MSP | Makarla et al., 2005 |
| MINT25 | 16% | S, E, M, CC, MIX | 0% ADJ NLS | MSP | Strathdee et al., 2001 |
| APC | 18–47.2% | S, M, CC, E, UN | 0–25% | MSP | Rathi et al., 2002; Makarla et al., 2005; Ongenaert et al., 2008 |
| BRCA1 | 10–24% | S, E, M, CC, MIX | 0–5.5% ADJ NLS | MSP | Strathdee et al., 2001; Rathi et al., 2002; Ibanez de Caceres et al., 2004 |
| CASP8 | 3% | S, E, M, CC, MIX | 0% ADJ NLS | MSP | Strathdee et al., 2001 |
| CDH1 (E-cadherin) | 26–29% | S, M, CC, E, UN | 6% | MSP | Rathi et al., 2002; Makarla et al., 2005 |
| CDH13 (H-cadherin) | 18–22% | S,M, CC, E, UN | 8–13% | MSP | Rathi et al., 2002; Makarla et al., 2005 |
| DCR1 | 43% | NS | 0% | MSP | Shivapurkar et al., 2004 |
| GPR150 | 26.6% | S, E, M, CC, SCC | 0% END cyst | MSP | Cai et al., 2007 |
| Htr (TERC) | 24% | S, E, M, CC, MIX | 0% ADJ NLS | MSP | Strathdee et al., 2001 |
ADJ NLS, Adjacent normals; CC, Clear cell; CS, Carcinosarcoma; E, Endometroid; END, Endometrial; M, Mucinous; MIX, Mixed; MSP, Methylation-specific PCR; NS, Not specified; PDA, Poorly differentiated adenocarcinoma; QMSP, Quantitative methylation-specific PCR; S, Serous; SCC, Squamous cell carcinoma; UN, Undifferentiated.
Several studies have identified the association of tumor-specific gene methylation with molecular, clinical, and pathological characteristics of epithelial ovarian carcinomas. For instance, highest degree of promoter methylation of SFN (an inhibitor of cell cycle progression), TMS1 and WT1 has been demonstrated in clear-cell ovarian tumors than in other histological types (Kaneuchi et al., 2004, p. 14; Terasawa et al., 2004; Kaneuchi et al., 2005; Teodoridis et al., 2005). Another finding suggests that promoter methylation of RASSF1A, APC, GSTP1, and MGMT correlates with the presence of invasive ovarian carcinomas (Makarla et al., 2005). Hypermethylation of FOXD3 correlated with tumor suppressive role (inhibition of proliferation, migration and promotion of apoptosis) in ovarian cancer cells and thus could serve as a potential therapeutic target for diagnosis of ovarian cancer (Luo et al., 2019).
Using a high–throughput approach to screen genes that showed highest differential methylation between ovarian cancer and normal tissue, Melnikov et al. identified 10 genes to be informative in tissue samples which include: BRCA1, EP300, NR3C1, MLH1, DNAJC15, CDKN1C, TP73, PGR, THBS1, and TMS1. A maximum sensitivity of 69% with 70% specificity was attained on testing the potential of several combinations of these genes to discriminate normal from cancer tissue. Since, all tumors analyzed were of advanced stage (either stage IIIA or higher), therefore, the potential of this panel to diagnose EOC at an early stage is unknown (Melnikov et al., 2009). Ibanez de Caceres et al. demonstrated that hypermethylation of atleast one of the six genes in panel (BRCA1, RASSF1A, APC, p14arf, p16ink4a, and DAPK) could be detected in 70/ 71 (99%) of EOCs using methylation specific PCR. Furthermore, none of the normal non-neoplastic tissue showed methylation, revealing a specificity of 100%. Additionally, across all histological subtypes, grades, stages as well as age, hypermethylation of TSGs was observed (Ibanez de Caceres et al., 2004). Taken together, these results support hypermethylation of these tumor suppressor genes as a relatively early event in ovarian carcinogenesis and could serve as a potential biomarker for detection and accurate discrimination of EOC at early stage.
Using 7- genes panel [secreted frizzled receptor proteins 1, 2 4, 5 (SFRP1, 2, 4, 5), SRY box1 (SOX1), paired box gene 1(PAX1), and LIM homeobox transcription factor 1, alpha (LMX1A)], Sui et al. investigated methylation in 126 primary ovarian tumors, 75 benign ovarian tumors and 14 borderline ovarian tumors and in 26 OC serum samples. Their findings indicated that promoter methylation of any one of SOX1, PAX1, and SFRP1 could distinguish EOC patients from normal control with a sensitivity of 73.08% and a specificity of 75%. Though these test scores are higher than those of CA125 alone, however it is probably not high enough to warrant its implementation as a diagnostic test for individual patients. Moreover, as no specification of tumor stage within the studied group was provided, the performance of this panel in detection of EOC at an early stage therefore remains unclear (Su et al., 2009).
Hypomethylation induced abnormal expression of several oncogenes such as CLDN4 (encodes an integral component of tight junctions) (Honda et al., 2006; Litkouhi et al., 2007), MAL (mal, T-cell differentiation protein) (Lee et al., 2010), BORIS (a cancer testis antigen family candidate oncogenes) (Woloszynska-Read et al., 2007), and IGF2 (an imprinted gene involved in other malignancies) (Murphy et al., 2006) has been demonstrated in ovarian carcinomas. Promoter hypomethylation induced upregulation of other cancer-associated genes in ovarian cancer includes maspin (SERPINB5) (Rose et al., 2006), MCJ (Strathdee et al., 2004, 2005), and SNCG (synucelin-γ) (Gupta et al., 2003; Czekierdowski et al., 2006b), which encodes an activator of the MAPK and Elk-1 signaling cascades. Hypomethylation of SNCG, MASPIN, and CLDN4 correlates with advanced-stage and metastasis while that of BORIS is linked with disease presence.
Hypomethylation of Sat2 (satellite 2) DNA in the juxtacentromeric region of chromosome 1 and 16 has been reported in ovarian cancer (Qu et al., 1999). A significant increase in hypomethylation of chromosome 1 Sat2 and chromosome 1 satellite α from non-neoplastic tissue toward ovarian cancer tissue was observed. Higher hypomethylation levels were observed in serous and endometrioid tumors in comparison to mucinous. Moreover, extensive hypomethylation was prevalent in high grade or advanced stage tumors (Widschwendter M. et al., 2004). Taken together, consistent higher expression levels along with hypomethylation of L1 and human endogenous retrovirus- W retrotransposons (repetitive sequences widely distributed throughout the genome) has been reported in malignant ovarian tumors against normal control samples (Menendez et al., 2004). It has been hypothesized that promotion in homologous recombination as a result of increased hypomethylation, leads to chromosomal aberrations associated with carcinogenesis (Kolomietz et al., 2002; Symer et al., 2002).
Prognosis
Potential prognostic biomarker includes FBXO32, which correlates with advanced stage and shorter disease free survival (Chou et al., 2010), Ribosomal DNA (18S and 28S) linked with prolonged disease free survival (Chan, 2005), IGFBP-3, correlates with disease progression and death in early stage EOC (Wiley et al., 2006b) and HOXA11, association with postsurgical residual tumor and poor outcome (Fiegl et al., 2008). Methylation of ≥1 gene of SFRP1, SFRP2, and SOX1 correlated with short disease free survival while SOX1, LMX1A, and SFRP1 methylation was associated with recurrence and short overall survival (Su et al., 2009). A progression-free survival prediction accuracy of 95% is reported by Wei et al. with hMLH1, IGFP3, and NEUROD1 among a panel of 112 highly discriminatory loci (Wei, 2006). Furthermore, detection of prognostic epigenetic biomarker has also been described in plasma as well as peritoneal fluid. Methylation of hMLH1, analyzed in 138 plasma samples predicted poor survival (hazard ratio: 1.99) (Gifford, 2004) while CDH1, CDH13, and APC (out of a 15 gene panel) analyzed in peritoneal fluid from 57 ovarian cancer patients could predict overall survival (Suehiro et al., 2008). Huang et al. recently reported that the epigenetic loss of heparin sulfate 3-O-sulfation makes ovarian cancer cells sensitive to oncogenic signals and could predict prognosis, thereby reflecting the utility of HS3ST2 for targeted therapy (Huang et al., 2018).
Recently using genome-wide methylation data analysis, five-methylation signature (SLC39A14, PREX2, KCNIP2, CORO6, and EFNB1) were reported as novel independent prognostic biomarker for patients with ovarian serous cystadenocarcinoma, which significantly associated with OS of patients. Moreover, these signatures exhibited higher sensitivity and specificity to predict OSC prognosis (AUC = 0.715), which reflects their clinical significance in improving outcome prediction. Furthermore, these 5- methylation signatures were more accurate over known biomarkers in predicting prognostic survival of OSC patients (Guo T. Y. et al., 2018). Promoter methylation of BRCA1 has been reported to be associated significantly with increased PFS of patients with OC undergoing adjuvant platinum–taxane-based chemotherapy (P = 0.008) as well for the patients with disease recurrence (PFS = 18.5 months over 12.8 months for patients without BRCA1 promoter methylation), thereby reflecting that promoter methylation of BRCA1 could be a better predictive marker of response to platinum–taxane-based chemotherapy in sporadic Epithelial ovarian carcinoma (Ignatov et al., 2014).
Another study highlights the potential of CDH1, DLEC1, and SFRP5 gene methylation panel as a prognostic biomarker in advanced stage OC patients. Presence of two or more methylated genes in patients significantly correlated with disease recurrence (hazard ratio: 1.91; p = 0.002) and shorter overall survival and disease free survival (hazard ratio: 1.96; p = 0.006) (Lin et al., 2018). Liu et al. reported the prognostic potential of C/EBPβ (a transcription factor) which augments chemoresistance of ovarian cancer cells by maintaining an open chromatin state via reprogramming H3K79 methylation of multiple drug-resistance genes upon direct interaction with DOT1L (DNA methyltransferase), thus provides a new insight for more precise therapeutics options in OC by identifying and targeting the key regulators of epigenetics (Liu et al., 2018).
Several recent researches have suggested the hypermethylation and reduced expression is prognostic for shorter progression free survival. For instance, using genome wide array analyses, Hafner et al. reported 220 differentially methylated region with short and long PFS. Validation experiments on a large cohort of type II EOC revealed the association of RUNX3/CAMK2N1 with poor clinical outcome (Lower PFS), indicating the prognostic potential of these genes (Häfner et al., 2016). Few studies have highlighted the tight link between promoter methylation and metastasis. For instance, stimulation of ovarian cancer cell lines by TGFβ, which is a key player in metastasis, extensively change promoter methylation of genes that are associated with EMT (Epithelial-mesenchymal transition) and progression of cancer (Cardenas et al., 2014). Deng et al. reported the tumor suppressive role of IQGAP2 which suppresses the ovarian cancer progression via suppressing Epithelial-mesenchymal transition by regulating Wnt/β signaling, thereby providing a potential biomarker and therapeutic strategy to combat ovarian cancer diagnosis (Deng et al., 2016).
Brachova et al. studied the association of oncomorphic TP53 mutation on patient outcome diagnosed with advanced EOC. Oncomorphic TP53 mutation correlated with worse progression free survival, higher risk of recurrence and higher rate of platinum resistance (Brachova et al., 2015). Dai et al. explored the association of methylation-based prognostic biomarkers within key ovarian cancer-related pathways with progression free survival to platinum based chemotherapy in HGSOC. NKD1, VEGFB, and PRDX2 were identified as the best predictors of progression free survival (PFS: HR = 2.3 p = 3.3 × 10–5; Overall Survival: HR = 1.9, p = 0.007). Further validation using independent TCGA data set revealed the significant association of VEGFA, VEGFB, and VEGFC promoter methylation with progression free survival (Dai et al., 2013).
Promoter hypomethylation and expression of PRAME correlates with increased survival in high grade serous ovarian carcinoma (Zhang et al., 2016). Promoter hypomethylation and increased expression of proto-oncogenes is predictive for more aggressiveness and metastasis of disease and thereby lower survival, which is evident from recent studies on GABRP, SLC6A12, MGAT3, CT45, CA9, MUC13, and AGR2 (Sung et al., 2014a,b,c, 2017a,b; Zhang et al., 2015; Kohler et al., 2016). Hypomethylation of Sat2 DNA (Chr 1) was associated relapse and poor prognosis (Widschwendter M. et al., 2004), and LINE1 was linked with poorer overall survival and lower progression free survival (Pattamadilok et al., 2008; Table 7).
Table 7.
Epigenetic biomarkers for ovarian cancer prognostication.
| Epigenetic marker | Source | Clinical prediction | References |
|---|---|---|---|
| 182-gene panel | Tumor tissue | HR 2.5 for PFS | Wei et al., 2002 |
| 112-gene panel | Tumor tissue | Prediction accuracy of 95% for shorter Disease free survival | Wei, 2006 |
| SFRP1, SFRP2, and SOX1 | Tumor tissue | Correlates with shorter Disease free survival | Su et al., 2009 |
| SOX1, LMX1A, and SFRP1 | Tumor tissue | Correlates with shorter overall survival | Su et al., 2009 |
| hMLH1 | Plasma | If hypermethylated HR:1.99 for OS | Gifford, 2004 |
| HOXA10, HOXA11 | Tumor tissue | RR for death:3.4, if HOXA11 methylated | Fiegl et al., 2008, p. 11 |
| 18S and 28S rDNA | Tumor tissue | Prolonged DFS | Chan, 2005 |
| EN2 | Tumor tissue | Short progression free survival | McGrath et al., 2018 |
| MYLK3 | Tumor tissue | Higher methylation level significantly predicted better overall survival with least residual disease | Phelps et al., 2017 |
| FBXO32 | Tumor tissue | Advanced stage and short DFS | Chou et al., 2010 |
| Panel of IGFFBP3, p16, BRCA1, GSTP1, ER-α, hMLH1 | Tumor tissue | Seven fold increased risk of short DFS HR: 6.53 for disease progression |
Wiley et al., 2006a |
| RUNX3, CAMK2N1 | Tumor tissue and ovarian cancer cell line | Short overall survival | Häfner et al., 2016 |
| ABCA1 | Tumor tissue and ovarian cancer cell line | Shorter overall survival | Chou et al., 2015 |
| GULP1 | ovarian cancer cell line | Residual disease, worse overall survival, and disease specific survival | Maldonado et al., 2018, p. 1 |
| FZD4, DVL1, and ROCK1 | Tumor tissue | Correlated with early disease relapse | Dai et al., 2011 |
| DNA hypomethylated genes | |||
| 15 gene panel | Peritoneal fluid | Short overall survival | Muller, 2004 |
| Sat 2 DNA (Chr1) | Tumor tissue | RR for relapse:4.1, RR for death:9.4 if region methylated | Widschwendter M. et al., 2004 |
| LINE1 | Tumor tissue | Lower methylation level significantly predicted poor OS and PFS | Pattamadilok et al., 2008 |
| ATG4A, HIST1H2BN | Tumor tissue | Poor progression free survival | Liao et al., 2014 |
Another important study by Wei et al. reported 112 methylated loci which were prognostic for reduced PFS and could predict PFS with an accuracy of 95% using Significance Analysis of Microarray and Prediction Analysis of Microarray algorithm (Wei, 2006). Twenty-two hypermethylated loci were identified by global methylation profiling of 485 tumor samples of clear-cell ovarian cancer in a recent study. These hypermethylated loci were associated with 9 genes (VWA1, FOXP1, FGFRL1, LINC00340, KCNH2, ANK1, ATXN2, NDRG21, and SLC16A11). Further, methylation induced silencing of KCNH2 (HERG, a potassium channel) could be a better prognostic factor for poor survival provided increased proliferation was mediated by overexpression of Eag family members. However, further validation on larger cohort is still warranted (Cicek et al., 2013). Huang et al. identified 63 differentially methylated regions of prognostic relevance which significantly correlated with poor PFS. Further, epigenetic silencing of regulators of hedgehog signaling pathway ZIC1 and ZIC4 was associated with increased proliferation, migration, and invasion. Additionally, promoter hypermethylation of ZIC1 significantly correlated with poor survival and thus could serve as prognostic determinant for patient outcome (Huang et al., 2013).
Another study describes that the global methylome status of HGSOC PDX (patient-derived xenografts) resembled with global methylation in corresponding patient tumor over several generations and could be efficiently modulated by demethylating agents. C-terminal Src kinase (CSK), a novel epigenetically regulated gene and associated pathways were also identified. Low CSK methylation significantly correlated with improved PFS and OS in HGSOC patients (Tomar et al., 2016). Koestler et al. using integrative global methylation and single nucleotide polymorphisms analysis identified DNA methylation marks (13 unique CpGs and 17 unique SNPs) which could mediate EOC genetic risk (Koestler et al., 2014).
Recently, Sharma et al. investigated epigenetic regulation of POTE gene family, which is localized to autosomal pericentromeric region. POTE gene family is over-expressed in HGSOC. Epigenetic silencing of POTE gene was functionally verified by experiments involving treatment with Decitabine and DNMT knockout cell lines. In addition expression of individual gene in POTE gene family correlated with chemoresistance and poor clinical outcome in HGSOC patients. Furthermore, several epigenetic alternations (pericentromeric activation, global and locus-specific L1 hypomethylation, and locus-specific 5' CpG hypomethylation) served as a determinant for regulation of epigenetic activation of POTE gene (Sharma et al., 2019).
In conclusion, these studies provides insight to the association of several potential methylation based prognostic biomarkers with clinical outcome in ovarian carcinoma and further suggest that these reports on epigenome wide interrogation of DNA methylation warrants detailed functional analysis of loci sufficiently discriminating OC with normal state. New targets identified through comprehensive methylome analysis in OC have significant translational potential to pave the design of future clinical investigations and therapeutics.
Predictive
Methylation mediated transcriptional repression of specific drug-response genes results in acquisition of drug resistance and significantly extends its impact on different facets of chemotherapeutic actions: membrane entry/exit, drug metabolism, response to cellular injury, DNA repair, apoptosis etc., in cancer cells. Hypermethylated genes such as hMLH1, ASS1 (arginine biosynthesis-related gene), ESR2 (encoding ER-β), and SFRP5 (encodes an inhibitor of oncogenic WNT signaling pathway) have been implicated in platinum resistance. Three studies well defined in ovarian cancer includes: Methylation of either BRCA1, GSTP1, or MGMT significantly correlates with improved response to chemotherapy (p = 0.013) (Teodoridis et al., 2005). Hypermethylation of RASSF1A and CABIN1 have been reported to correlate with response to adjuvant therapy. Patients who responded to therapy had moderately higher frequencies of RASSF1A hypermethylation (OR = 0.4) and significantly higher frequencies of CABIN1 hypermethylation (OR = 0.1) (Feng Q. et al., 2008). Strathdee et al. demonstrated that high levels of MCJ methylation significantly correlated with poor response to therapy (p = 0.027) and poor overall survival (p = 0.023; HR = 2.9) (Strathdee et al., 2005). Hypomethylation induced upregulation of ABCG2 (multidrug transporter) MAL (determinant of platinum resistance) and TUBB3 (determinant of taxane resistance) genes have been described in advanced ovarian carcinoma cases with drug-acquired chemoresistance (Izutsu et al., 2008; Balch et al., 2010; Lee et al., 2010; Table 8).
Table 8.
Epigenetic biomarkers for ovarian cancer prediction.
| Epigenetic marker | Source | Clinical prediction | References |
|---|---|---|---|
| Methylation of >1 of BRCA1, GSTP1, and MGMT | Tumor tissue | Association with improved response to chemotherapy | Teodoridis et al., 2005 |
| RASSF1A, CABIN1 | Tumor tissue | Association with response to chemotherapy | Feng Q. et al., 2008 |
| ASS1 | Tumor tissue | Determinant of Platinum resistance | Nicholson et al., 2009 |
| HSulf1 | Tumor tissue | Determinant of Platinum resistance | Staub et al., 2007 |
| SFRP5 | Tumor tissue | Determinant of Platinum resistance | Su et al., 2010 |
| hMLH1 | Plasma | Relapse of Chemoresistant tumor | Gifford, 2004 |
| ESR2 | Tumor tissue | Determinant of Platinum resistance | Yap et al., 2009 |
| MCJ | Tumor tissue | Association with response to chemotherapy and overall survival | Strathdee et al., 2005 |
| TUBB3 | Tumor tissue | Taxane resistance | Izutsu et al., 2008 |
| MSX1 | Ovarian cancer cell line | Sensitivity to platinum drug | Bonito et al., 2016 |
| TBX2 | Tumor tissue | Sensitivity to platinum drug | Tasaka et al., 2017 |
| MAL | Tumor tissue | Platinum resistance | Lee et al., 2010 |
Recently Pulliam et al. demonstrated the combinatorial effect of DNA methyltransferase inhibitor (DNMTi) guadecitabine and the Poly (ADP-ribose) polymerase (PARP) inhibitors (PARPi) talazoparib in resensitizing PARPi resistant breast and ovarian cancer irrespective of BRCA status. Synergistic effect of guadecitabine and talazoparib increased ROS accumulation, and further sensitized the breast and ovarian cancer cells toward PARPi sensitivity by subsequent activation of cAMP/PKA signaling which in turn promoted PARP activation. Furthermore, DNMTi augmented PARP “trapping” by talazoparib. The finding of this complementary model supports further clinical exploration of this combination therapy in PARPi-resistant cancers (Pulliam et al., 2018). Another study using integrated global methylation analysis on extreme chemoresponsive HGSOC patients identified four genes of clinical relevance (FZD10, FAM83A, MYO18B, and MKX) as epigenetic marker of platinum based chemoresponse, of which, FZD10 was reported as functionally validated marker of platinum sensitivity (Tomar et al., 2017). Promoter methylation of OPCML was significantly associated with poor overall survival of OC patients and thus could be of use in predicting disease prognosis (Zhou et al., 2014).
A recent study has described induction of hypomethylation in resistant ovarian cancer patients upon treatment with cisplatin, though, in the intergenic regions, the loss of methylation was primarily observed (Lund et al., 2017). Hypomethylation of developmental genes MSX1 and TMEM88 correlated with platinum resistance in patients with ovarian cancer (Bonito et al., 2016; de Leon et al., 2016). Stimulation of EMT by non-coding RNA HOTAIR has been reported to be regulated by DNA methylation and is indicative of resistance to carboplatin (Teschendorff et al., 2015). Likewise, another study highlights promotion of platinum resistance by TET. Induction of EMT by TET is mediated by demethylation of Vimentin promoter in ovarian carcinoma (Han et al., 2017).
A recent study has described how methylome-targeting strategies could bring forth anti-tumor effect. Guadecitabine-mediated induction of global hypomethylation not only affects metabolic and immune responses but also activates tumor suppressor genes which eventually contribute to platinum drug re-sensitization in ovarian cancer. This might offer utility in improving survival outcomes of patients with ovarian cancer (Fang et al., 2018). Another recent study has highlighted the tumor suppressor role of ZNF671 and its methylation could act as a predictor for early recurrence of serous ovarian carcinoma (Mase et al., 2019). Another important study by M. Keita et al. has for the first time reported the exclusive association of massive DNA hypomethylation with poorly differentiated tumors, which correlates with disease aggressiveness and progression. This report also raises concern over the adverse effect of use of demethylating agents which probably aid the activation of oncogenes and prometastatic genes (Keita et al., 2013).
In conclusion, it is speculated that the combinatorial therapies utilizing epigenetic inhibitors holds promise and would be most effective for chemo-resensitization of resistant tumors, possibly by restoration of pathways associated with drug response, and thus would subsequently implicate improved survival outcomes as well as personalized treatment for this devastating disease.
Histone Modifications in Ovarian Cancer
Compared with DNA methylation, the evidence on chromatin modification in development of ovarian cancer is limited. Histone modification mediated regulation of cell cycle regulatory proteins such as cyclin B1 (Valls et al., 2005), p21 (Richon et al., 2000), and ADAM19 (Chan M. W. et al., 2008) have been described in various reports. Association of histone modifications with aberrant class III β tubulin protein expression (Izutsu et al., 2008), reduction of PACE3 expression (Fu et al., 2003) and silencing of survivin (Mirza et al., 2002) has been reported in ovarian tumorigenesis. Upregulation of tumor suppressor Rb and CDKN1 (cyclin-dependent kinase inhibitor) by histone acetylation was described by Strait et al. (2002). Moreover, the overexpression of HDACs 1–3 in ovarian cancer has been reported to be associated with high grade tumors and resulting poor prognosis (Weichert et al., 2008). On the other note, the derepression of claudin-3 and claudin-4 was found to be associated with loss of trimethylated histone 3 lysine 27 (H3K27me3) (Kwon et al., 2010). The transcriptional repression of osteoprotegerin (OPG) has been reported to be mediated by reduced histone 3 lysine 4 trimethylation (H3K4me3) and increased H3K27me3 (Lu et al., 2009). Similarly, the association of transcriptional silencing of GATA4 and GATA6 with hypoacetylation of histones H3 and H4 and loss of trimethylated histone 3l ysine 4 (H3K4me3) has been described by Caslini et al. (2006).
A very recent report has provided insight into the mechanism associated with development and progression of OC. Early Loss of E3 ubiquitin ligase RNF20 and histone H2B monoubiquitylation (H2Bub1) has been reported to drive ovarian tumorigenesis by altering chromatin accessibility and thereby activating immune signaling pathways (IL6), and this loss has been defined by majority of high grade serous ovarian carcinomas tumors (Hooda et al., 2019). Cacan et al. reported that the loss of FAS expression which contributes to drug resistance is mediated by histone deacetylase 1 (HDAC1) in chemoresistant OC cells (Cacan, 2016). Recently Tang et al. highlighted the repression of histone H3 lysine 27 trimethylation (H3K27me3) which was mitigated by AMP-activated protein kinase (AMPK) phosphorylation upon treatment with metformin thus implicated the antitumor effect of metformin and suggested its utility in the treatment of EOC patients who are not diabetic (Tang et al., 2018).
In another study, the mechanism associated with upregulation of ABCB1 was conferred to chromatin remodeling (via p300 mediated H3K9ac and AR complex binding to ARE4) which in turn leads to the development of taxol resistant phenotype. It was shown that the upregulation of p300 and GCN5 (HATs) was associated with overexpression of ABCB1 and resistance to taxol and PI3K/AKT pathway which is activated by taxol, mediates the regulation of the expression of p300 and AR. These results further reveal the significance of AKT/p300/AR axis as a novel treatment strategy in combating taxol resistance (Sun et al., 2019). Using ChIP-seq approach, Curry et al. identified genome-wide bivalent domains (H3K27me3 and H3K4me3) at gene promoter in tumor samples which were collected pre and post platinum resistance acquisition, and showed that these representative poised gene sets are pre-disposed to hypermethylation induced epigenetic silencing during acquisition of drug resistance, thus provides novel insights to prevent emergence of drug resistance (Curry et al., 2018).
Yi et al. reported that Enhancer of zeste homolog 2 (EZH2) mediates repression of tissue inhibitor of metalloproteinases 2(TIMP2) by H3K27me3 and DNA methylation thereby facilitating ovarian cancer metastasis (Yi et al., 2017). In similar context, another study highlighted silencing of ARHI in ovarian cancer which was synergistically mediated by Enhancer of zeste homolog 2 (EZH2) induced H3K27me3 and DNA methylation. Furthermore increased EZH2 expression correlated with worse overall survival rates, implicating prognostic potential of EZH2 in EOC (Fu et al., 2015). Repression of Regulator of G-protein signaling 2 (RGS2) via histone deacetylases (HDACs) and DNA methyltransferase I in chemoresistant OC cells has been reported recently by Cacan et al., and utility of their inhibitors might serve as a novel approach to overcome chemoresistance in ovarian cancer (Cacan, 2017).
Clinical Application of Epigenetic Biomarker in Liquid Biopsies for Ovarian Cancer Management
Cell Free DNA Biology
Advancement in the understanding underlying molecular pathogenesis of cancer, along with advancements in molecular techniques has facilitated the study of molecular alternations associated with cancer development at an early stage in body fluids. Circulating cell free DNA which are believed to have derived from tumor cells, reflect specific genetic and epigenetic alternations, and thus may offer potential non-invasive viable biomarkers for several cancer, capable of providing valuable information regarding disease progression and response to therapy in real time.
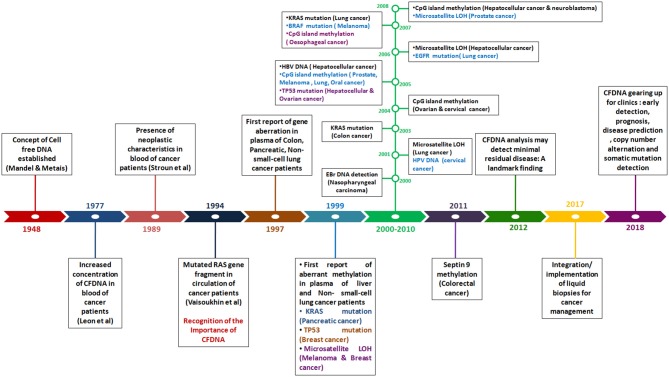
In 1948, the existence of cell free DNA was first described by Mandel and Métais. Cell free DNA are derived from necrotic and apoptotic cells, commonly released by all cell types. Further, numerous subsequent studies confirmed that the tumor-specific pattern of alterations, such as chromosomal abnormality, somatic mutations, resistance mutation, aberrant methylation and copy number variations could be found in cfDNA, which can serve as potential target for diagnosis of cancer through non-invasive approach (Leon et al., 1977; Polivka et al., 2015; Figure 3).
Figure 3.
Timeline reflecting the detection of genetic and epigenetic alternations in Cell-free DNA in blood of patients with different cancer type.
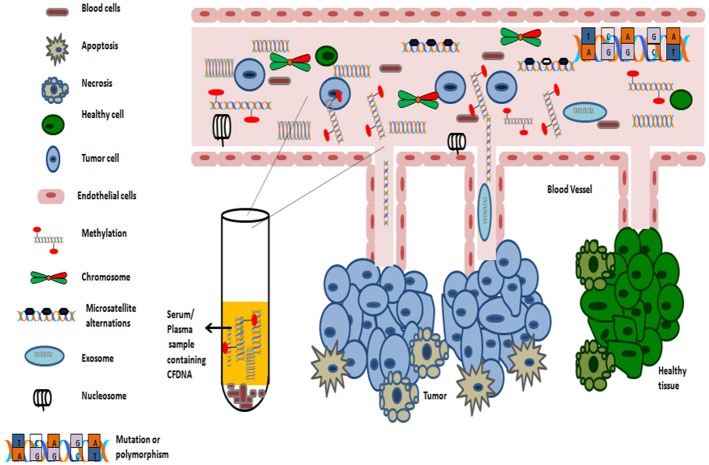
Numerous studies support the detection of methylation signature in almost any body fluid (such as serum, plasma, smears, nipple fluid aspirate, and vaginal fluid etc.). As sampling of blood can be considered as minimal invasive process, thus serves as an ideal substrate for methylation analysis. The average concentrations of circulating cell free DNA in healthy subjects is 30 ng/ml. However, in cancer patients, the average concentration of cell free serum DNA is higher, approximately 180 ng/ml as dying cancer cells release tumor DNA into the blood (Gormally et al., 2007). The average length of circulating cfDNA, which are usually fragmented, is 140 to 170 bp and of which, only a fraction of few thousand amplifiable copies of cfDNA /ml of blood, might be of diagnostic relevance (Gormally et al., 2007; Polivka et al., 2015). The levels of circulating cell free DNA in serum is abnormally high in early as well as advanced-stage tumors (Perlin and Moquin, 1972; Leon et al., 1977). For this phenomenon, the proposed two primary mechanisms includes: either cells in cancer tissue undergoes in situ apoptosis and/or necrosis or cells might detach from tumors and extravasate into bloodstream where they undergo lysis (Figure 4).
Figure 4.
Origin of Cell-free DNA in blood.
Since its first validation, the potential application of circulating DNA in research settings and for non-invasive management of cancer as “liquid biopsy” is expanding with improvement in molecular and genomic techniques. Numerous studies have demonstrated that tumor specific aberrant methylation can also be detected in cfDNA of patients with different tumor types such as lung, prostate, breast and colorectal cancer and further confirmed altered methylation as an independent diagnostic/ prognostic marker (Board et al., 2008; Brock et al., 2008; Lofton-Day et al., 2008; Vlassov et al., 2010). Warren et al. developed a highly sensitive non-invasive test for screening of colorectal cancer based on methylation of SEPT9 in plasma which could specifically detect all stages and locations of colorectal cancers (Warren et al., 2011). Hypermethylation of Vim gene is strongly correlated with the occurrence of colorectal cancer. Similarly hypermethylation of SHOX2 in sputum has been used as biomarker for distinguishing malignant and benign lung diseases (Kneip et al., 2011). Gstp1 methylation status in urine is strongly correlated with early onset of prostate cancer (Belinsky, 2004).
Numerous reports have highlighted the potential of DNA methylation based biomarkers for non-invasive detection of cancer utilizing cell free DNA. Recently, using integrated methylome analysis Wei et al. reported hypermethylation of SPG20, a putative STAT3 target, for non-invasive detection of gastric cancer at an early stage (Wei et al., 2019). Yang et al. explored the potential of eight gene panel for non-invasive detection of lung cancer using qMSP and revealed that the promoter methylation of any of the eight gene could detect the disease with a sensitivity of 72% with 91% specificity, reflecting the utility of plasma DNA methylation as a novel approach for detection of lung cancer at early stage (Yang et al., 2018).
Similarly, promoter methylation of OPCML and HOXD9 assessed in serum cell free DNA using methylation-sensitive high-resolution melting, was detected with a sensitivity of 62.50% with specificity of 100%, thus could serve as a non-invasive differential biomarker to prevent misdiagnosis of cholangiocarcinoma (CCA) and other biliary diseases (Wasenang et al., 2019). Further, for the management of pancreatic cancer and its early detection Eissa et al. analyzed the methylation of ADAMTS1 and BNC1 in cfDNA using qMSP, which exhibited a sensitivity of 94.8% and specificity of 91.6% with a AUC of 0.95 reflecting diagnostic potential of this blood based two-gene panel in detection of pancreatic cancer at an early stage (Eissa et al., 2019). Methylation of APC, FOXA1, and RASSF1A in cell free DNA served as a best performing cassette in terms of diagnostic and prognostic value, revealing a sensitivity, specificity and accuracy over 70% suggesting its putative utility in management of breast cancer (Salta et al., 2018).
Other studies using genome-wide methylation profiling of serum/plasma cell-free DNA have identified potential biomarkers for clinical utility. For instance, Xu et al. using MeDIP-seq approach reported 10 significant differentially methylated genes as potent biomarker for lung cancer clinical application (Xu et al., 2019). Similarly, using genome-wide methylome profiling and Sequenom MassARRAY approach, it was reported that promoter methylation of CASZ1, CDH13, and ING2 could serve as a potent noninvasive biomarker for detection of esophageal cancer at early stage (Wang H.Q. et al., 2018).
Challenges
The analysis of blood borne cell-free DNA has tremendous potential to enable rapid, non-invasive molecular diagnosis of cancer. They are of great clinical relevance as they provide specific targets for initial diagnosis, permit monitoring of treatment efficacy as well as information about tumor profile and its dynamics which are critical for treatment decisions (De Mattos-Arruda et al., 2014; Lewis et al., 2015).
The advantages of analyzing tumor specific DNA methylation in cell free serum DNA includes, improved sensitivity as cfDNA can be easily amplified by PCR, fewer false positive rate as methylation pattern is generally conserved throughout the progression of disease, stability during sample collection as abnormal DNA methylation is chemically as well as biologically stable and remains relatively unaffected by physiological condition at the time of sample collection, increased technical sensitivity and specificity for gene specific assays as well as offers assay design advantages over genetic alternation that might be interspersed throughout a given gene. Furthermore, DNA methylation is a positively detectable signal, unlike a loss of signal as in chromosomal deletions (Wittenberger et al., 2014).
Several limitations in the methylation detection of cell free serum DNA includes extremely low amount of available cfDNA, missing bisulfite conversions as they are usually fragmented, low sensitivity demonstrated by a single marker and time-consuming, complicated and expensive conventional techniques for cfDNA isolation. The most commonly used technique for methylation detection is MSP PCR (methylation specific PCR) which is a bisulfite-conversion- based method. The limitation of bisulfite conversion of cfDNA is the missing DNA. Because of the technical difficulties of DNA methylation analysis, only few DNA methylation based markers has been identified to date, which apply only to a fraction of gynecological cancers including breast, ovarian and endometrial cancers (Wittenberger et al., 2014; Lewis et al., 2015).
The two technological challenges to be addressed include (1) the detection of low abundant tumor-specific DNA methylation patterns through methylation specific PCR priming or probing with high signal-to-noise ratio (2) the determination of methylation status of consecutive sites in individual DNA molecules with single base-pair resolution. This requires methylation-independent priming and sequence analysis of combined PCR product. Clinically the major problem associated with DNA methylation assays is to detect scarcely abundant alleles within high background levels of non-target molecules. However, with the advent of digital MethyLight assay together with rapid advances in next generation sequencing based technologies, these issues can be overcome. One example of this novel approach is the development of the PraenaTest™ (LifeCodexx, Germany) (Weisenberger et al., 2008).
Serum Based Epigenetic Biomarker
Tumor-specific methylation-based biomarkers might possibly prove valuable for monitoring disease prognosis and different pathological determinants; however, non-invasive analysis and characterization of biomarkers in body fluids offers more feasibility in early screening and detection of the disease as well in monitoring the response to therapy. Numerous studies have reported aberrant methylation in ovarian cancer as discussed earlier; there are relatively few reports of serum/plasma methylation biomarkers for earlier detection of OC. Various studies that demonstrated striking detection sensitivities and specificities in non-invasive assays, thereby supporting the promising utility of these biomarkers for early screening and detection of OC has been summarized in Table 9.
Table 9.
Non-invasive epigenetic DNA methylation biomarkers for ovarian cancer.
| Epigenetic marker(s) | Source | Patient/sample | Clinical prediction | Technology | References |
|---|---|---|---|---|---|
| Methylation of ≥1 gene of BRCA1, RASSF1A, APC, p14ARF, p16INK4A, or DAPK | Serum and cytologically negative peritoneal fluid | 50 serum, 40 peritoneal fluid from EOC patients along with 40 control serum and peritoneal fluid samples | Presence of malignancy Sensitivity: 41/50 (82% for serum) 28/30 (93% for peritoneal fluid), Specificity: 100% (all tumor stages). |
Methylation-specific-PCR (MSP) | Ibanez de Caceres et al., 2004 |
| DAPK | Whole peripheral blood DNA | 26 peripheral blood samples | Sensitivity: 14/16 (54%) for DAPK-methylation-positive samples: Specificity: 10/10 (100%) for DAPK methylation negative |
Methylation-specific PCR (MSP) | Collins et al., 2006 |
| Methylation of ≥1 gene of SOX1, PAX1 or SFRP1 | Serum | 46 (26 ovarian cancer cases and 20 patients with a benign condition | Sensitivity: 73.08% Specificity: 75% | Methylation-specific PCR (MSP) | Su et al., 2009 |
| Methylation of 7-gene panel (APC, RASSF1A, CDH1, RUNX3, TFPI2, SFRP5, and OPCML | Serum | 202 patients (87 EOC cases, 53 benign cases and 62 controls) | Sensitivity of 7-gene panel: 85.3% Specificity of 7-gene panel: 90.5% in stage I EOC Sensitivity of CA125: 56.1% Specificity of CA125: 64.15% |
Multiplex methylation-specific PCR (MSP) | Zhang et al., 2013 |
| 10-gene panel (Combination of BRCA1, HIC1,PAX5,PGR, THBS1) | Plasma | 66 (33 cancer cases and 33 control) | Presence of malignancy Sensitivity: 61% Specificity: 85% |
Microarray based multiplex assay(MethDet56 technique) | Melnikov et al., 2009 |
| Several gene panel (RASSF1A and PGR-PROX) (RASSF1A, CALCA and EP300) |
Serum | 90 (30 EOC cases, 30 cases with Benign disease along with 30 controls non neoplastic samples) | Methylation of RASSF1A and PGR-PROX Sensitivity: 80.0% Specificity:73.3% Methylation of RASSF1A, CALCA and EP300 Sensitivity: 90.0% Specificity: 86.7%. |
Microarray based Assay(MethDet 56) | Liggett et al., 2011 |
| OPCML | Serum | 194 (71 EOC, 43 benign and 80 controls non neoplastic samples) | Sensitivity: 87.18% Specificity: 93.75% Accuracy: 90.14% |
Nested Methylation -specific PCR (MSP) | Wang et al., 2017 |
| RASSF1A | Plasma | 53 samples including OC tumors, adjacent tumor cell free tissues and paired plasma circulating tumor DNA | Sensitivity: 33/53 (62.3%), RASSF1A methylation of paired plasma CtDNA showed slight concordant with primary tumor samples (P = 0.227, 2-sided Pearson χ2 test, k = 0.156). Significantly correlates with overall survival |
Real-time methylation specific-PCR (real-time MSP) and methylation-sensitive high-resolution melting analysis (MS-HRMA) | Giannopoulou et al., 2017 |
| ESR1 | Plasma | Group A: 66 OC cases Group B: 53 OC case along with 50 paired plasma samples |
Sensitivity of detection: 38%. ESR1 methylation predicted better clinical outcomes: overall survival (P = 0.027), progression-free survival (P = 0.041) |
Real-time methylation-specific PCR (real-time MSP) assay | Giannopoulou et al., 2018 |
| 3-gene panel | Serum | For assay development: 151 cases and for validation study 250 cases with different conditions in 3 sets | Pre-chemotherapy Sensitivity: 41.4% Specificity: 90.7% Post chemotherapy Responders: 78% non-responders: 86% |
Targeted ultra-high coverage bisulfite sequencing | Widschwendter et al., 2017 |
MicroRNAs in Ovarian Cancer
Aberrant expression of microRNAs has been confirmed in ovarian carcinogenesis. A decrease in mRNA levels of the miR- processing enzymes in OC malignant cases against normal controls, strongly implicates an overall tumor suppressive role of miRs in ovarian tumorigenesis (Merritt et al., 2008; Pampalakis et al., 2010). Overexpression of Drosha and Dicer was significantly associated with better survival, while low expression of Drosha was associated with suboptimal surgical cytoreduction and low expression of Dicer with advanced tumor stage, thereby further implicating the tumor suppressive role of microRNAs in OC (Merritt et al., 2008; Faggad et al., 2010). With respect to ovarian cancer, the potential targets for several upregulated miRs includes pro-apoptotic, metastasis-suppressing or antiproliferation gene products while those for the downregulated miRs includes growth signaling, prometastatic- or anti-apoptosis-associated proteins. A list of upregulated/downregulated miRs involved in ovarian cancer development is shown in Table 10. A list of aberrantly expressed miRs which could serve as a promising biomarker for detection of ovarian cancer has been summarized in Table 11. Chao et al. reported that in advanced stage cancer, miR-187 regulates carcinogenesis through Dab2 dependent EMT (epithelial-to-mesenchymal transition) (Chao et al., 2012, p. 2). Furthermore, other studies have described miR-199a, miR-200a, miR-200b, miR-200c, and miR-214 as significantly overexpressed and miR-100 and miRNAlet-7i as significantly downregulated in ovarian tumors (Iorio et al., 2007; Yang H. et al., 2008; Yang N. et al., 2008). Several miRNA signatures that could distinguish ovarian tumors based on histological subtypes has been studied such as miR-200b and miR-141 was observed to be overexpressed in serous and endometrioid subtypes; upregulated of miR-21, miR-203, and miR-205 correlated with endometrioid histotype; downregulated miR-145 correlated with serous and clear cell subtype, while downregulated miR 222 was associated with endometrioid and clear cell subtype (Iorio et al., 2007).
Table 10.
List of dysregulated miRNAs in ovarian cancer.
| Mechanism(s) | miR(s) | Targets | Consequence(s) leading to tumorigenesis | References | |
|---|---|---|---|---|---|
| Upregulated | c-myc activation | 130a | MCSF, GAX and HOXA5 | Chemotherapy resistance, angiogenesis, and dedifferentiation | Taylor and Gercel-Taylor, 2008; Eitan et al., 2009 |
| Copy gain | 27a and 451 | ZBTB10, Myt-1, HMGB2, HOXA2, and CYP1B1 | Multidrug export, oncogenic signaling and reduced apoptotic potential | Shibata et al., 2006; Zhang L. et al., 2006 | |
| 213 | APP and SATB2 | Chemoresistance | Boren et al., 2009 | ||
| 199a, 200a, b, c and 335 | TGFβ, ZEB1, ZEB2, BAP1, GATA4, GATA6, TNC, FN1, EXOC5, and TUBB3 | Mesenchymal–epithelial transition | Weisenberger et al., 2008; Su et al., 2009; Liggett et al., 2011; Giannopoulou et al., 2017; Wang et al., 2017 | ||
| Hypomethylation | 203 | p63, SOCS-3, ABL1, MCEF, and ADAMTS6 | Unknown | Iorio et al., 2007; Lee et al., 2009 | |
| 205 | ZEB1, ZEB2, E2F1, ERBB3, PKCe, and SHIP2 | Mesenchymal–epithelial transition, oncogenic signal transduction | Iorio et al., 2007; Yu et al., 2008; Lee et al., 2009 | ||
| 21 | PDCD4†, RPS7†, NCAPG†, TPM1, and PTEN | Reduced apoptotic potential and anchorage independence | Iorio et al., 2007; Laios et al., 2008; Sorrentino et al., 2008; Eitan et al., 2009; Lee et al., 2009; Resnick et al., 2009; Yang et al., 2009 | ||
| Unknown | 340 | PAM, RTN3, PPL, RNF34, and ZNF513 | Chemoresistance | Boren et al., 2009 | |
| 221/222 | KIT, AIP1, p21, p57, TCF12, RIMS3, and ARNT | Cell cycle progression and angiogenesis | Merritt et al., 2008; Vlassov et al., 2010; Kneip et al., 2011 | ||
| Downregulated | C/EBPα Downregulation | 1 | FOXP1, HDAC4 c-Met, Pim1 and HAND2 | Tumor growth, cell motility and proliferation | Iorio et al., 2007 |
| DNA methylation and copy loss | 137 | CDK6, MITF, KLF12, and PDLIM3 | Cell cycle progression and dedifferentiation | Iorio et al., 2007; Zhang L. et al., 2008 | |
| 140 | c-SRK, MMP13 and FGF2 | Oncogenic signaling | Iorio et al., 2007; Zhang L. et al., 2008 | ||
| 150 | c-Myb, MAK9, Akt3, and MAP2K4 | Oncogenic signaling | Zhang L. et al., 2008 | ||
| 551a | LPHN1, ERBB4, and ZFP36 | Oncogenic signaling | Dahiya et al., 2008 | ||
| 9 | NF-kB†, Bcl2†, Bcl6, FGF†, and b-Raf | Oncogenic signaling | Leon et al., 1977; Weisenberger et al., 2008; Faggad et al., 2010; Kneip et al., 2011 | ||
| 184 | TTK69, K10, and Sax(A) | Dedifferentiation | Zhang L. et al., 2008 | ||
| Unknown | 30b and d | CTGF | Invasion/metastasis | Laios et al., 2008 | |
| 98 | HMGA2, LIN28B, and HIC2 | Oncogenic signaling and cancer stemness | Dahiya et al., 2008 | ||
| 517a and b | CREAP-1, MAPKAPK5, NFKBIE, and PTK2B | Chemoresistance and oncogenic signaling | Lee et al., 2009 | ||
| Let-7i | HMGA2, LIN28Bm TRIM71, and IGF2BP1 | Chemoresistance | Yang N. et al., 2008 | ||
| 662 | NEGR1, MKX, and CSF3 | Unknown | Dahiya et al., 2008 |
Table 11.
List of misexpressed miRNAs in ovarian cancer.
| Epigenetic marker | Alternations in miRNAs | Source | Methodology | Clinical prediction | References |
|---|---|---|---|---|---|
| Dicer, Drosha mRNA | Downregulated | Tumor tissue and ovarian cancer cell lines | Quantitative RT-PCR, gene expression array | Advanced tumor stage and Suboptimal tumor debulking | Merritt et al., 2008; Faggad et al., 2010; Pampalakis et al., 2010 |
| let-7i, miR-221,−30c,−152 and−193 miR-185,−106a,−181a, −210,−423,−103,−107 and let-7c | Downregulated Upregulated | Tumor tissue from Endometrial cancer, normal endometrial and atypical hyperplasia | Quantitative RT-PCR, microarray analysis | Association with Endometrial cancer development | Boren et al., 2008 |
| miR-124-1,−124-2,−124-3,−127,−132,−137,−193A, 375 and−339 | Downregulated | Tumor tissue | Quantitative RT-PCR, MSP, direct Sanger sequencing | EOC metastasis (including peritoneal macro-metastases) | Loginov et al., 2018 |
| let-7i | Downregulated | Tumor tissue and ovarian cancer cell lines | miRNA Microarray, Stem-loop real-time RT-PCR (TaqMan miRNA assay) | Associated with increased resistance to chemotherapy drugs, cis-platinum, and short progression-free survival | Yang N. et al., 2008 |
| miR-30c,−130a, and−335 | Downregulated | Paclitaxel and cisplatin resistant cancer cell lines | miRNA Microarray, qPCR, Northern blots | Association with development of chemoresistance | Sorrentino et al., 2008 |
| miR- 199b-5p | Downregulated | Cisplatin-sensitive and -resistant ovarian cancer cell lines | miRCURY LNATM microRNA array and Q-PCR | Development of acquired chemoresistance through the activation of JAG1-Notch1 signaling cascade | Liu et al., 2014 |
| miR-34a, miR-34b*/c | Downregulated | Tumor tissue | Quantitative RT-PCR, in- situ hybridization | Associated with motility and invasion by regulation of MET, progression of disease to advanced stages | Corney et al., 2010 |
| miR−34a,−200a,−200b,−449b,−509-3p,−509-3-5p,−513a-5p and −574-5p | Upregulated | Tumor tissue | MicroRNA microarray | Differentially expressed in Stage I disease | Eitan et al., 2009 |
| miR-302b,−22, and−373 miR-148b and−211 | Upregulated Downregulated | Tumor tissue and ovarian cancer cell lines | MicroRNA microarray, Quantitative Real time PCR (Taqman based) | Discriminates serous vs. non-serous disease | Iorio et al., 2007 |
| miR-7, 34c-5p, 146b-5p and 449a | Upregulated | Tumor tissue | Massively parallel pyrosequencing, TaqMan® qRT-PCR assays | Serous adenocarcinoma | Wyman et al., 2009 |
| miR-23p,−125a-3p,−125a-5p,−130a,−146b-5p,−193a-3p,−193a-5p,−423-5p,−451 and−491-5p | Upregulated | Tumor tissue | MicroRNA microarray | Differentially expressed in Stage III disease | Eitan et al., 2009 |
| miRs 100, 199a, 200a, and 214 | Upregulated | Tumor tissue and ovarian cancer cell line | miRNA array and Northern blot analysis, quantitative reverse transcription-PCR. | Late clinical stage and high-grade tumors, negative regulation of PTEN by miR-214 thereby inducing cell survival and cisplatin resistance | Yang H. et al., 2008 |
| miR-302b,−325,−299-5p,−222, and−324-3p miR-212 and−150 | Upregulated Downregulated | Tumor tissue and ovarian cancer cell lines | MicroRNA microarray, Quantitative Real time PCR (Taqman based) | Differentiates Serous vs. endometrioid disease | Iorio et al., 2007 |
| miR-325,−22,−302c,−299–5p,−373, and−196b miR-9 and−18 | Upregulated Downregulated | Tumor tissue and ovarian cancer cell lines | MicroRNA microarray, Quantitative Real time PCR (Taqman based) | Associated with Poor differentiation | Iorio et al., 2007 |
| miR-30a,−30a*, and−486-5p | Upregulated | Tumor tissue and ovarian cancer cell lines | MicroRNA microarray, Quantitative Real time PCR (Taqman based) | Clear cell disease | Iorio et al., 2007 |
| Methylation of let-7a-3 | Downregulated | Tumor tissue | Real-time methylation-specific PCR and real-time reverse Transcription-PCR, direct Sanger sequencing | Favorable prognosis | Lu et al., 2007, p. 3 |
| miR-449b | Upregulated | Tumor tissue | MicroRNA microarray | Good prognosis | Eitan et al., 2009 |
| let-7e | Upregulated | Paclitaxel and cisplatin resistant cancer cell lines | miRNA Microarray, qPCR, Northern blots | Associated with resistance to Paclitaxel | Sorrentino et al., 2008 |
| miR-200a,−200b, and−429 | Upregulated | Primary tumor and ovarian cancer cell line | Real-time reverse transcription-PCR | Long disease free survival and delayed recurrence, prognostic marker in advanced ovarian cancer | Cochrane et al., 2009; Hu et al., 2009 |
| miRs 100, 199a, 200a, and 214 | Upregulated | Tumor tissue and ovarian cancer cell line | miRNA array and Northern blot analysis, quantitative reverse transcription-PCR. | Late clinical stage and high-grade tumors | Yang H. et al., 2008 |
| Methylation of miR-34a | Downregulated | Tumor tissue | Quantitative reverse transcription-PCR, MethyLight assay | Inversely associated with grade, p53 mutation, and dualistic tumor type. Reduced progression free survival and worsen overall survival. | Schmid et al., 2016 |
| Methylation of miR-199a-3p | Downregulated | Tumor tissue and ovarian cancer cell line | Methylation-specific PCR and bisulphite sequencing | Tumor aggressiveness and enhanced cisplatin resistance through promoting DDR1 expression | Deng et al., 2017 |
| Methylation of 10 miRNA genes (miR-124-2,−124-3,−125B-1,−127,−129-2,−137, −193A,−203A,−339,−375) | Downregulated | Tumors tissue and matched peritoneal metastases | Methylation-specific PCR | Involved in metastasis | Loginov et al., 2018 |
| (miR-34b/c, miR-9-1, miR-124-3, miR-129-2, and miR-107) | Downregulated | Tumor tissue | Methylation-specific PCR | Associated with clinical grade and metastasis. High sensitivity and specificity reveals its diagnostic potential (87–94%, AUC = 0.92). | Braga et al., 2018b |
| miR-150 | Downregulated | Tumor tissue | Real time PCR | Correlated with shorter progression free survival | Wilczynski et al., 2018 |
| miR-4443 and miR-5195-3p | Downregulated | Tumor tissue | Real time PCR | Correlates with metastasis and tumor progression | Ebrahimi and Reiisi, 2019 |
| miR-148a | Downregulated | Plasma samples and ovarian cancer cell line | Real time PCR | Associated with poor prognosis, tumor growth and metastasis | Gong et al., 2016 |
| (hsa-miR-135, 150,−340, 625, 1908, 3187,−96,−196b,−449c, and−1275) | Downregulated | Tumor tissue | Small RNA sequencing, quantitative RT-PCR | Associated with survival | Chen et al., 2018 |
| miR-595 | Downregulated | Tumor tissue | qRT-PCR | Correlated with shorter overall survival | Zhou et al., 2017 |
| miR-498 | Downregulated | Tumor tissue | qPCR | Correlated with shorter overall survival and progression free survival | Cong et al., 2015 |
| miR-9 | Downregulated | Tumor tissue and ovarian cancer cell line | Real time PCR, luciferase reporter assay, Western Blot, Methylation study, RNAi technique, and cytotoxicity Assay | Resistance to paclitaxel by targeting CCNG1. | Li X. et al., 2015 |
| miR-508-3p | Downregulated | Tumor tissue and ovarian cancer cell line | System biology approach, qRT-PCR, methylation PCR, RNA sequencing, immuno blot | Predictor for mesenchymal subtype and metastasis | Zhao et al., 2019 |
| hsa-miR-1273g-3p | Downregulated | Serum samples of recurrent epithelial ovarian cancer patients | Microarray and qRT-PCR | Prognostic biomarker for recurrence | Günel et al., 2018 |
Recently, Braga et al. described methylation of miR-9-1, miR-9-3, and miR-130b which strongly correlated with progression of OC (Braga et al., 2018a). Different histotype of ovarian carcinomas reflect differential expression of specific miRNAs which might serve as a valid biomarker. Agostini et al. reported significant overexpression of miR-192, miR-194, and miR-215 in mucinous subtype of ovarian carcinomas. However their expression was downregulated in other subtypes and sex cord-stromal tumors (Agostini et al., 2018).
A list of promising aberrantly expressed miRs which could be of prognostic and predictive relevance in ovarian cancer has been summarized in Table 11. A lower ratio of miR-221 to miR-222 significantly correlated with worse overall survival in predominantly high grade, advanced stage sporadic ovarian carcinomas (Wurz et al., 2010). Downregulation of miR-141, miR−200a, miR-200b, miR-200c, and miR-429 correlated with poor progression free survival. Moreover, multivariate analysis of relevant clinicopathological variables such as debulking status, stage and grade of tumor revealed the correlation of miR-429 expression with recurrence-free survival (Leskela et al., 2010). Downregulated miR-422b and miR-34c correlated with decreased disease-specific survival in HGSOC patients with BRCA1/2 abnormalities (Lee et al., 2009).
In ovarian cancer, overexpression of miR-214 has been specifically associated with the degradation of PTEN mRNA which further leads to the activation of Akt pathway and has been correlated with platinum resistance (Yang H. et al., 2008). Downregulation of miR-Let7i has been reported in platinum-resistance ovarian tumors; however its gain of function resulted in restoration of drug sensitivity of chemoresistance OC cells (Yang N. et al., 2008).
Several studies have recently highlighted the diagnostic and prognostic relevance of several miRNAs, their association with overall survival of patients and have shown that they could serve as putative biomarker as well as therapeutic target for ovarian cancer management. For instance, Li et al. have reported tumor suppressive role of miR-542-3p, which directly targets CDK14 and was observed significantly downregulated in EOC tissue and OC cell lines (Li et al., 2019).
Si et al. highlighting the therapeutic significance of miR-27a in OC, reported miR-27a mediated regulation of proliferation, chemosensitivity and invasion of OC by targeting Cullin 5 (CUL5) (Si et al., 2019, p. 5). Another study by Jia et al. reported the tumor suppressive role of miR-34 in regulation of tumor proliferation via inducting autophagy and apoptosis and suppression of cell invasion by targeting Notch 1 (Jia et al., 2019, p. 1). Wang et al. utilizing integrated meta-analysis approach have shown the oncogenic role of miRNA-27a by mediating FOXO1 and its inhibition could serve as a new strategy in combating ovarian cancer (Wang Z. et al., 2018, p. 1). Hu et al. identified miR-934 as an oncogene in OC by directly targeting BRMS1L, and thus could serve as a therapeutic marker (Hu et al., 2019). It has been reported that miR-1294 was identified to be downregulated in EOC and correlated with tumor progression and shorter overall survival, thereby could serve as an independent prognostic indicator (Guo W. et al., 2018).
Liu et al. provided insights into the oncogenic role of microRNA-96 (miR-96-5p) in ovarian cancer. Its significant overexpression was found in tissue as well as serum samples. Overexpression of miR-96-5p was correlated with increased proliferation and migration by suppressing Caveolae1 (CAV1) and inhibiting AKT signaling pathway and its downstream proteins (Cyclin D1 and P70), thus implying that miR-96-5p could serve as a promising therapeutic target for ovarian cancer (Liu et al., 2019, p. 1). Similarly, Chaluvally-Raghavan et al. reported that miR551b-3p which is an oncogenic microRNA, directly upregulates STAT3 expression and further deregulates proliferation and metastasis in vivo and in vitro. Reduced expression of STAT3 in OC cells in vitro and in vivo via anti-miR551b-3p leads to reduction in growth of ovarian tumor in vivo, thereby implying that it could serve as promising therapeutic target in future for ovarian cancer (Chaluvally-Raghavan et al., 2016).
In another study, miR-152 mediated suppression of tumor proliferation along with promotion of apoptosis via repression of ERBB3 was reported, thus demonstrating miR-152 as a potential therapeutic target (Li et al., 2017, p. 3). Liu et al. reported association of miR-506 with better response to therapy as well as long PFS and overall survival in OC patients. Further, it sensitized cancer cells to chemotherapy by directly targeting RAD51 and thus could be of therapeutic importance (Liu et al., 2015).
10 miRs which were identified using genome wide MicroRNA expression profiling were capable to discriminate malignant tissue samples from normal with a sensitivity of 97% and specificity of 92% (Wang et al., 2014). Biamonte et al. have reported tumor suppressive role of miR-let-7g and significant association of its reduced expression in both tissue as well as serum with chemoresistance in advanced stage EOC patient which reflects its potential as a predictive biomarker to monitor response to chemotherapy (Biamonte et al., 2019). Kobayashi et al. have shown significant overexpression of serum miR-1290 in advance stage HGSOC in comparison to early stage. Moreover, it was capable to discriminate patients with HGSOC from patients with malignancies of other histological subtypes with a sensitivity of 47% and specificity of 85% (AUC = 0.76), thus reflecting diagnostic potential of miR-1290 for HGSOC (Kobayashi et al., 2018).
Mahmoud et al. examined the diagnostic significance of serum miR-21 and reported that its upregulation was significantly negatively correlated with Programmed Cell Death-4 (PDCD4) expression in EOC patients (Mahmoud et al., 2018). Another study highlighted significantly elevated expression of serum exosomal miR-93, miR-145, and miR-200c in OC. Moreover, the sensitivity for miR-145 and miR-200c was 91.6 and 90.0% which was far superior in comparison with CA125, thus these serum exosomal microRNAs could be of diagnostic relevance for preoperative diagnosis of OC (Kim et al., 2019). miR-21 was observed significantly overexpressed in the sera of EOC patients and its elevated expression correlated with shorter overall survival (Xu et al., 2013). Further, downregulation of serum miR-25 and miR-93 and upregulation of miR-7 and miR-429 have been reported in OC patients. In addition, the sensitivity and specificity achieved by these four serum miRs were 93 and 92% to discriminate cancer patients from non-neoplastic control samples, deciphering their diagnostic significance in EOC. Moreover serum miR-429 correlated with overall survival and could serve as an independent prognostic indicator (Meng et al., 2015). Findings from another study reveal the relevance of serum miR-141 and miR-200c in OC diagnosis and prognosis. Both of these miRs were identified to be overexpressed in serum of EOC patients; however miR-200c displayed a descending expression trend across tumor stage (early to advance) while an escalating expression trend was observed in case of miR-141. Moreover, the sensitivity for miR-141 and miR-200c were 0.69 and 0.72 with a specificity of 0.72 and 0.70, respectively, to discriminate cancerous samples from normal control [AUC = 0.75 and 0.79, respectively]. Furthermore, high serum miR-200c correlated with higher survival rate. On contrary, low serum miR-141 correlated with higher survival rate (Gao and Wu, 2015).
Langhe et al. using Exiqon platform explored a 4-miR panel in serum of EOC patients for their diagnostic utility and found that these miRs were significantly downregulated in EOC patients. Furthermore these miRs target WNT signaling, AKT/mTOR signaling and TLR-4/MyD88 to regulate ovarian cancer progression and resistance (Langhe et al., 2015). Overexpression of serum miR-200a, miR-200b, and miR-200c which have been observed in EOC patients, correlates with aggressive disease progression and could be indicative of disease prognosis and patient survival (Zuberi et al., 2015). Higher serum concentration of exosomal miR-200b and miR-200c correlated with shorter overall survival, which suggests its prognostic relevance. (Meng et al., 2016b) Serum miR-200a, miR-200b, and miR-200c differentiated cancerous and benign tumors with 83% sensitivity and 100% specificity, which reflect that these miRs could be of diagnostic utility (Meng et al., 2016a).
These miRs though hold great potential for their utility in ovarian cancer management; however its therapeutical implementation still remains a challenge. To address this, well-designed clinical study as well as validated methodologies is essentially warranted.
Expert Commentary
It is now well-established that DNA methylation occurs very early in malignant transformation and their utility as biomarker holds great promise to overcome the false positive detection of ovarian cancer using current standard serum marker CA125. In this review, we highlight the recent epigenetic biomarkers analyzed in tissue and body fluids for early detection of OC. Strikingly; to date no single epigenetic biomarker facilitating early diagnosis of OC has made transition to the clinics. The probable reasons for this could be: the heterogeneous nature of EOC, difference in sample processing, assay design, technique used and approach could explain the variations observed in methylation frequencies amongst various studies for individual genes. Most of the studies for methylation analysis of genes were conducted on small sample size and in particular the normal control samples were insufficient to conclude the specificity of the assay. Therefore, further studies on larger sample size are necessary to be conducted to determine the potential of methylation if it could serve as biomarker for early EOC screening or not. Another limitation is the absence of standardized reference value for methylation analysis when trying to analyze if a particular locus is hyper or hypomethylated. To overcome this, currently, methylation cut off points which are based on already published reports or consensus are used.
The majority of the reports highlight the methylation status of gene or genes in a panel. No epigenetic biomarker screening study has been performed till date. However, for the detection figures approaching current screening modalities (89.5% sensitivity and 99.8% specificity) has been achieved by Ibanez de Caceres et al. (2004) with 82% sensitivity and 100% specificity (Ibanez de Caceres et al., 2004). All 30 control cases showed 0 false positive rate and further replication of the study on the basis of this sample size would give a false positive rate between 0 and 11.4% (95% confidence interval), thereby indicating that perfect specificity would unlikely hold up in the follow-up studies. In view of these considerations regarding the study of Ibanez de Caceres et al. are left to follow-up studies to shed light on. However, none of the report has been further validated undertaking follow up studies on a larger cohort and prospective study design thereby limiting the utility of the reported findings.
Molecular analysis of epigenetic modification (methylation) in circulating cell free tumor DNA in fluids serves as a novel, non-invasive approach for identification of potential promising cancer biomarkers, which can be performed at multiple time points and probably better reflects the prevailing molecular profile of cancer. Very few studies analyzing the methylation status of genes in blood-based assay for ovarian cancer diagnosis has been reported. Careful precision handling and processing of liquid biopsy for cell free DNA extraction is critically needed.
Future Prospects
Over the last decade, an exponential progress in DNA methylation based biomarker development has been witnessed. Owing to the stability of DNA and methylation pattern, a number of cfDNA as well as tissue based screening assay has paved its way into clinics. The commercial success of several tests based on DNA methylation biomarkers for early detection of colon, lung and prostate cancer and prediction of bladder cancer along with various markers under validation study shows that the time for transition into clinics can be relatively rapid. New technologies which allow rapid identification of methylation signatures directly from blood will facilitate sample-to answer solutions thereby enabling next-generation point of care molecular diagnostics. Moreover, ongoing work on liquid biopsies together with the recent advanced technologies such as digital PCR, bisulfite sequencing, methyl immune-precipitation coupled with next-generation sequencing, and methylation arrays along with advanced statistical data analysis may mitigate the problematic issues for the development of non-invasive method thereby overcoming the existing challenges to personalized medicine.
Author Contributions
AS wrote the manuscript. SG and MS edited the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2019.00182/full#supplementary-material
References
- Abdollahi A., Pisarcik D., Roberts D., Weinstein J., Cairns P., Hamilton T. C. (2003). LOT1 (PLAGL1/ZAC1), the candidate tumor suppressor gene at chromosome 6q24-25, is epigenetically regulated in cancer. J. Biol. Chem. 278, 6041–6049. 10.1074/jbc.M210361200 [DOI] [PubMed] [Google Scholar]
- Agathanggelou A., Honorio S., Macartney D. P., Martinez A., Dallol A., Rader J., et al. (2001). Methylation associated inactivation of RASSF1A from region 3p21.3 in lung, breast and ovarian tumours. Oncogene 20, 1509–1518. 10.1038/sj.onc.1204175 [DOI] [PubMed] [Google Scholar]
- Agostini A., Brunetti M., Davidson B., Trop,é C. G., Eriksson A. G. Z., Heim S., et al. (2018). The microRNA miR-192/215 family is upregulated in mucinous ovarian carcinomas. Sci. Rep. 8:11069. 10.1038/s41598-018-29332-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akahira J., Sugihashi Y., Ito K., Niikura H., Okamura K., Yaegashi N. (2004a). Promoter methylation status and expression of TMS1 gene in human epithelial ovarian cancer. Cancer Sci. 95, 40–43. 10.1111/j.1349-7006.2004.tb03168.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akahira J., Sugihashi Y., Suzuki T., Ito K., Niikura H., Moriya T., et al. (2004b). Decreased expression of 14-3-3 sigma is associated with advanced disease in human epithelial ovarian cancer: its correlation with aberrant DNA methylation. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 10, 2687–2693. 10.1158/1078-0432.CCR-03-0510 [DOI] [PubMed] [Google Scholar]
- Amonkar S. D., Bertenshaw G. P., Chen T.-H., Bergstrom K. J., Zhao J., Seshaiah P., et al. (2009). Development and preliminary evaluation of a multivariate index assay for ovarian cancer. PLoS ONE 4:e4599. 10.1371/journal.pone.0004599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen M. R., Goff B. A., Lowe K. A., Scholler N., Bergan L., Drescher C. W., et al. (2010). Use of a symptom index, CA125, and HE4 to predict ovarian cancer. Gynecol. Oncol. 116, 378–383. 10.1016/j.ygyno.2009.10.087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold J. M., Cummings M., Purdie D., Chenevix-Trench G. (2001). Reduced expression of intercellular adhesion molecule-1 in ovarian adenocarcinomas. Br. J. Cancer 85, 1351–1358. 10.1054/bjoc.2001.2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai T., Tanaka T., Yukawa K., Maeda M., Umesaki N. (2004). Reduced expression of death-associated protein kinase in human uterine and ovarian carcinoma cells. Oncol. Rep. 11, 661–665. 10.3892/or.11.3.661 [DOI] [PubMed] [Google Scholar]
- Balch C., Matei D. E., Huang T. H.-M., Nephew K. P. (2010). Role of epigenomics in ovarian and endometrial cancers. Epigenomics 2, 419–447. 10.2217/epi.10.19 [DOI] [PubMed] [Google Scholar]
- Baldwin R. L., Nemeth E., Tran H., Shvartsman H., Cass I., Narod S., et al. (2000). BRCA1 promoter region hypermethylation in ovarian carcinoma: a population-based study. Cancer Res. 60, 5329–5333. [PubMed] [Google Scholar]
- Baranova I., Kovarikova H., Laco J., Dvorak O., Sedlakova I., Palicka V., et al. (2018). Aberrant methylation of PCDH17 gene in high-grade serous ovarian carcinoma. Cancer Biomark. 23, 125–133. 10.3233/CBM-181493 [DOI] [PubMed] [Google Scholar]
- Barbolina M. V., Adley B. P., Kelly D. L., Shepard J., Fought A. J., Scholtens D., et al. (2009). Downregulation of connective tissue growth factor by three-dimensional matrix enhances ovarian carcinoma cell invasion. Int. J. Cancer 125, 816–825. 10.1002/ijc.24347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett T. E., Chindera K., McDermott J., Breeze C. E., Cooke W. R., Jones A., et al. (2016). Epigenetic reprogramming of fallopian tube fimbriae in BRCA mutation carriers defines early ovarian cancer evolution. Nat. Commun. 7:11620. 10.1038/ncomms11620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton C. A., Gloss B. S., Qu W., Statham A. L., Hacker N. F., Sutherland R. L., et al. (2010). Collagen and calcium-binding EGF domains 1 is frequently inactivated in ovarian cancer by aberrant promoter hypermethylation and modulates cell migration and survival. Br. J. Cancer 102, 87–96. 10.1038/sj.bjc.6605429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton C. A., Hacker N. F., Clark S. J., O'Brien P. M. (2008). DNA methylation changes in ovarian cancer: implications for early diagnosis, prognosis and treatment. Gynecol. Oncol. 109, 129–139. 10.1016/j.ygyno.2007.12.017 [DOI] [PubMed] [Google Scholar]
- Belinsky S. A. (2004). Gene-promoter hypermethylation as a biomarker in lung cancer. Nat. Rev. Cancer 4, 707–717. 10.1038/nrc1432 [DOI] [PubMed] [Google Scholar]
- Biamonte F., Santamaria G., Sacco A., Perrone F. M., Di Cello A., Battaglia A. M., et al. (2019). MicroRNA let-7g acts as tumor suppressor and predictive biomarker for chemoresistance in human epithelial ovarian cancer. Sci. Rep. 9:5668. 10.1038/s41598-019-42221-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. (2002). DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21. 10.1101/gad.947102 [DOI] [PubMed] [Google Scholar]
- Bird A. P., Wolffe A. P. (1999). Methylation-induced repression–belts, braces, and chromatin. Cell 99, 451–454. 10.1016/S0092-8674(00)81532-9 [DOI] [PubMed] [Google Scholar]
- Board R. E., Knight L., Greystoke A., Blackhall F. H., Hughes A., Dive C., et al. (2008). DNA methylation in circulating tumour DNA as a biomarker for cancer. Biomark. Insights 2, 307–319. 10.1177/117727190700200003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bol G. M., Suijkerbuijk K. P. M., Bart J., Vooijs M., van der Wall E., van Diest P. J. (2010). Methylation profiles of hereditary and sporadic ovarian cancer. Histopathology 57, 363–370. 10.1111/j.1365-2559.2010.03642.x [DOI] [PubMed] [Google Scholar]
- Bonito N. A., Borley J., Wilhelm-Benartzi C. S., Ghaem-Maghami S., Brown R. (2016). Epigenetic regulation of the homeobox gene MSX1 associates with platinum-resistant disease in high-grade serous epithelial ovarian cancer. Clin. Cancer Res. 22, 3097–3104. 10.1158/1078-0432.CCR-15-1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boren T., Xiong Y., Hakam A., Wenham R., Apte S., Chan G., et al. (2009). MicroRNAs and their target messenger RNAs associated with ovarian cancer response to chemotherapy. Gynecol. Oncol. 113, 249–255. 10.1016/j.ygyno.2009.01.014 [DOI] [PubMed] [Google Scholar]
- Boren T., Xiong Y., Hakam A., Wenham R., Apte S., Wei Z., et al. (2008). MicroRNAs and their target messenger RNAs associated with endometrial carcinogenesis. Gynecol. Oncol. 110, 206–215. 10.1016/j.ygyno.2008.03.023 [DOI] [PubMed] [Google Scholar]
- Borgoño C. A., Diamandis E. P. (2004). The emerging roles of human tissue kallikreins in cancer. Nat. Rev. Cancer 4, 876–890. 10.1038/nrc1474 [DOI] [PubMed] [Google Scholar]
- Bowtell D. D. L. (2010). The genesis and evolution of high-grade serous ovarian cancer. Nat. Rev. Cancer 10, 803–808. 10.1038/nrc2946 [DOI] [PubMed] [Google Scholar]
- Brachova P., Mueting S. R., Carlson M. J., Goodheart M. J., Button A. M., Mott S. L., et al. (2015). TP53 oncomorphic mutations predict resistance to platinum- and taxane-based standard chemotherapy in patients diagnosed with advanced serous ovarian carcinoma. Int. J. Oncol. 46, 607–618. 10.3892/ijo.2014.2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga E. A., Loginov V. I., Burdennyi A. M., Filippova E. A., Pronina I. V., Kurevlev S. V., et al. (2018a). Five Hypermethylated MicroRNA genes as potential markers of ovarian cancer. Bull. Exp. Biol. Med. 164, 351–355. 10.1007/s10517-018-3988-y [DOI] [PubMed] [Google Scholar]
- Braga E. A., Loginov V. I., Filippova E. A., Burdennyi A. M., Pronina I. V., Kazubskaya T. P., et al. (2018b). Diagnostic value of a group of MicroRNA genes hypermethylated in ovarian carcinoma. Bull. Exp. Biol. Med. 166, 253–256. 10.1007/s10517-018-4326-0 [DOI] [PubMed] [Google Scholar]
- Brock M. V., Hooker C. M., Ota-Machida E., Han Y., Guo M., Ames S., et al. (2008). DNA methylation markers and early recurrence in stage I lung cancer. N. Engl. J. Med. 358, 1118–1128. 10.1056/NEJMoa0706550 [DOI] [PubMed] [Google Scholar]
- Buchholtz M.-L., Brüning A., Mylonas I., Jückstock J. (2014). Epigenetic silencing of the LDOC1 tumor suppressor gene in ovarian cancer cells. Arch. Gynecol. Obstet. 290, 149–154. 10.1007/s00404-014-3177-9 [DOI] [PubMed] [Google Scholar]
- Buys S. S., Partridge E., Black A., Johnson C. C., Lamerato L., Isaacs C., et al. (2011). Effect of screening on ovarian cancer mortalitys (PLCO) cancer screening randomized controlled trial. JAMA 305, 2295–2303. 10.1001/jama.2011.766 [DOI] [PubMed] [Google Scholar]
- Cacan E. (2016). Histone deacetylase-1-mediated suppression of FAS in chemoresistant ovarian cancer cells. Anticancer Res. 36, 2819–2826. [PubMed] [Google Scholar]
- Cacan E. (2017). Epigenetic regulation of RGS2 (Regulator of G-protein signaling 2) in chemoresistant ovarian cancer cells. J. Chemother. 29, 173–178. 10.1080/1120009X.2016.1277007 [DOI] [PubMed] [Google Scholar]
- Cai K. Q., Caslini C., Capo-chichi C. D., Slater C., Smith E. R., Wu H., et al. (2009). Loss of GATA4 and GATA6 expression specifies ovarian cancer histological subtypes and precedes neoplastic transformation of ovarian surface epithelia. PLoS ONE 4:e6454. 10.1371/journal.pone.0006454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L., Abe M., Izumi S., Imura M., Yasugi T., Ushijima T. (2007). Identification of PRTFDC1 silencing and aberrant promoter methylation of GPR150, ITGA8 and HOXD11 in ovarian cancers. Life Sci. 80, 1458–1465. 10.1016/j.lfs.2007.01.015 [DOI] [PubMed] [Google Scholar]
- Cannistra S. A. (2004). Cancer of the ovary. N. Engl. J. Med. 351, 2519–2529. 10.1056/NEJMra041842 [DOI] [PubMed] [Google Scholar]
- Cardenas H., Vieth E., Lee J., Segar M., Liu Y., Nephew K. P., et al. (2014). TGF-β induces global changes in DNA methylation during the epithelial-to-mesenchymal transition in ovarian cancer cells. Epigenetics 9, 1461–1472. 10.4161/15592294.2014.971608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caslini C., Capo-Chichi C. D., Roland I. H., Nicolas E., Yeung A. T., Xu X. X. (2006). Histone modifications silence the GATA transcription factor genes in ovarian cancer. Oncogene 25:5446. 10.1038/sj.onc.1209533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaluvally-Raghavan P., Jeong K. J., Pradeep S., Silva A. M., Yu S., Liu W., et al. (2016). Direct upregulation of STAT3 by MicroRNA-551b-3p deregulates growth and metastasis of ovarian cancer. Cell Rep. 15, 1493–1504. 10.1016/j.celrep.2016.04.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan M. W., Huang Y.-W., Hartman-Frey C., Kuo C.-T., Deatherage D., Qin H., et al. (2008). Aberrant transforming growth factor beta1 signaling and SMAD4 nuclear translocation confer epigenetic repression of ADAM19 in ovarian cancer. Neoplasia 10, 908–919. 10.1593/neo.08540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan M. W. Y. (2005). Hypermethylation of 18S and 28S Ribosomal DNAs predicts progression-free survival in patients with ovarian cancer. Clin. Cancer Res. 11, 7376–7383. 10.1158/1078-0432.CCR-05-1100 [DOI] [PubMed] [Google Scholar]
- Chan T. A., Glockner S., Yi J. M., Chen W., Van Neste L., Cope L., et al. (2008). Convergence of mutation and epigenetic alterations identifies common genes in cancer that predict for poor prognosis. PLoS Med. 5:e114. 10.1371/journal.pmed.0050114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao A., Lin C.-Y., Lee Y.-S., Tsai C.-L., Wei P.-C., Hsueh S., et al. (2012). Regulation of ovarian cancer progression by microRNA-187 through targeting disabled homolog-2. Oncogene 31, 764–775. 10.1038/onc.2011.269 [DOI] [PubMed] [Google Scholar]
- Chen S. F., Liu Z., Chaurasiya S., Dellinger T. H., Lu J., Wu X., et al. (2018). Identification of core aberrantly expressed microRNAs in serous ovarian carcinoma. Oncotarget 9, 20451–20466. 10.18632/oncotarget.24942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W., Jiang Y., Liu C., Shen O., Tang W., Wang X. (2010). Identification of aberrant promoter hypomethylation of HOXA10 in ovarian cancer. J. Cancer Res. Clin. Oncol. 136, 1221–1227. 10.1007/s00432-010-0772-4 [DOI] [PubMed] [Google Scholar]
- Chien J., Staub J., Avula R., Zhang H., Liu W., Hartmann L. C., et al. (2005). Epigenetic silencing of TCEAL7 (Bex4) in ovarian cancer. Oncogene 24, 5089–5100. 10.1038/sj.onc.1208700 [DOI] [PubMed] [Google Scholar]
- Choi Y.-L., Kang S. Y., Shin Y. K., Choi J. S., Kim S. H., Lee S.-J., et al. (2006). Aberrant hypermethylation of RASSF1A promoter in ovarian borderline tumors and carcinomas. Virchows Arch. Int. J. Pathol. 448, 331–336. 10.1007/s00428-005-0091-3 [DOI] [PubMed] [Google Scholar]
- Choi Y.-L., Kim J., Kwon M. J., Choi J. S., Kim T.-J., Bae D.-S., et al. (2007). Expression profile of tight junction protein claudin 3 and claudin 4 in ovarian serous adenocarcinoma with prognostic correlation. Histol. Histopathol. 22, 1185–1195. 10.14670/HH-22.1185 [DOI] [PubMed] [Google Scholar]
- Chou J.-L., Huang R.-L., Shay J., Chen L.-Y., Lin S.-J., Yan P. S., et al. (2015). Hypermethylation of the TGF-β target, ABCA1 is associated with poor prognosis in ovarian cancer patients. Clin. Epigenetics 7:1. 10.1186/s13148-014-0036-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J.-L., Su H.-Y., Chen L.-Y., Liao Y.-P., Hartman-Frey C., Lai Y.-H., et al. (2010). Promoter hypermethylation of FBXO32, a novel TGF-beta/SMAD4 target gene and tumor suppressor, is associated with poor prognosis in human ovarian cancer. Lab. Investig. J. Tech. Methods Pathol. 90, 414–425. 10.1038/labinvest.2009.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung V. Y., Tan T. Z., Huang R.-L., Lai H.-C., Huang R. Y.-J. (2017). Loss of discoidin domain receptor 1 (DDR1) via CpG methylation during EMT in epithelial ovarian cancer. Gene 635, 9–15. 10.1016/j.gene.2017.09.001 [DOI] [PubMed] [Google Scholar]
- Cicek M. S., Koestler D. C., Fridley B. L., Kalli K. R., Armasu S. M., Larson M. C., et al. (2013). Epigenome-wide ovarian cancer analysis identifies a methylation profile differentiating clear-cell histology with epigenetic silencing of the HERG K+ channel. Hum. Mol. Genet. 22, 3038–3047. 10.1093/hmg/ddt160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke-Pearson D. L. (2009). Clinical practice. Screening for ovarian cancer. N. Engl. J. Med. 361, 170–177. 10.1056/NEJMcp0901926 [DOI] [PubMed] [Google Scholar]
- Cochrane D. R., Spoelstra N. S., Howe E. N., Nordeen S. K., Richer J. K. (2009). MicroRNA-200c mitigates invasiveness and restores sensitivity to microtubule-targeting chemotherapeutic agents. Mol. Cancer Ther. 8, 1055–1066. 10.1158/1535-7163.MCT-08-1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins Y., Dicioccio R., Keitz B., Lele S., Odunsi K. (2006). Methylation of death-associated protein kinase in ovarian carcinomas. Int. J. Gynecol. Cancer Off. J. Int. Gynecol. Cancer Soc. 16 (Suppl. 1), 195–199. 10.1136/ijgc-00009577-200602001-00031 [DOI] [PubMed] [Google Scholar]
- Cong J., Liu R., Wang X., Wang J., Wang H., Hou J. (2015). Low miR-498 expression levels are associated with poor prognosis in ovarian cancer. Eur. Rev. Med. Pharmacol. Sci. 19, 4762–4765. [PubMed] [Google Scholar]
- Corney D. C., Hwang C.-I., Matoso A., Vogt M., Flesken-Nikitin A., Godwin A. K., et al. (2010). Frequent downregulation of miR-34 family in human ovarian cancers. Clin. Cancer Res. 16, 1119–1128. 10.1158/1078-0432.CCR-09-2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello J. F., Plass C. (2001). Methylation matters. J. Med. Genet. 38, 285–303. 10.1136/jmg.38.5.285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curry E., Zeller C., Masrour N., Patten D. K., Gallon J., Wilhelm-Benartzi C. S., et al. (2018). Genes predisposed to DNA hypermethylation during acquired resistance to chemotherapy are identified in ovarian tumors by bivalent chromatin domains at initial diagnosis. Cancer Res. 78, 1383–1391. 10.1158/0008-5472.CAN-17-1650 [DOI] [PubMed] [Google Scholar]
- Czekierdowski A., Czekierdowska S., Szymanski M., Wielgos M., Kaminski P., Kotarski J. (2006a). Opioid-binding protein/cell adhesion molecule-like (OPCML) gene and promoter methylation status in women with ovarian cancer. Neuro Endocrinol. Lett. 27, 609–613. [PubMed] [Google Scholar]
- Czekierdowski A., Czekierdowska S., Wielgos M., Smolen A., Kaminski P., Kotarski J. (2006b). The role of CpG islands hypomethylation and abnormal expression of neuronal protein synuclein-gamma (SNCG) in ovarian cancer. Neuro Endocrinol. Lett. 27, 381–386. [PubMed] [Google Scholar]
- Dahiya N., Sherman-Baust C. A., Wang T.-L., Davidson B., Shih I.-M., Zhang Y., et al. (2008). MicroRNA expression and identification of putative miRNA targets in ovarian cancer. PLoS ONE 3:e2436. 10.1371/journal.pone.0002436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W., Teodoridis J. M., Zeller C., Graham J., Hersey J., Flanagan J. M., et al. (2011). Systematic CpG Islands methylation profiling of genes in the Wnt pathway in epithelial ovarian cancer identifies biomarkers of progression-free survival. Clin. Cancer Res. 17, 4052–4062. 10.1158/1078-0432.CCR-10-3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W., Zeller C., Masrour N., Siddiqui N., Paul J., Brown R. (2013). Promoter CpG Island methylation of genes in key cancer pathways associates with clinical outcome in high-grade serous ovarian cancer. Clin. Cancer Res. 19, 5788–5797. 10.1158/1078-0432.CCR-13-1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leon M., Cardenas H., Vieth E., Emerson R., Segar M., Liu Y., et al. (2016). Transmembrane protein 88 (TMEM88) promoter hypomethylation is associated with platinum resistance in ovarian cancer. Gynecol. Oncol. 142, 539–547. 10.1016/j.ygyno.2016.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mattos-Arruda L., Weigelt B., Cortes J., Won H. H., Ng C. K. Y., Nuciforo P., et al. (2014). Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 25, 1729–1735. 10.1093/annonc/mdu239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Zhao F., Hui L., Li X., Zhang D., Lin W., et al. (2017). Suppressing miR-199a-3p by promoter methylation contributes to tumor aggressiveness and cisplatin resistance of ovarian cancer through promoting DDR1 expression. J. Ovarian Res. 10. 10.1186/s13048-017-0333-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z., Wang L., Hou H., Zhou J., Li X. (2016). Epigenetic regulation of IQGAP2 promotes ovarian cancer progression via activating Wnt/β-catenin signaling. Int. J. Oncol. 48, 153–160. 10.3892/ijo.2015.3228 [DOI] [PubMed] [Google Scholar]
- Devouassoux-Shisheboran M., Genestie C. (2015). Pathobiology of ovarian carcinomas. Chin. J. Cancer 34, 50–55. 10.5732/cjc.014.10273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamandis E. P., Scorilas A., Fracchioli S., Van Gramberen M., De Bruijn H., Henrik A., et al. (2003). Human kallikrein 6 (hK6): a new potential serum biomarker for diagnosis and prognosis of ovarian carcinoma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 21, 1035–1043. 10.1200/JCO.2003.02.022 [DOI] [PubMed] [Google Scholar]
- Earp M. A., Cunningham J. M. (2015). DNA methylation changes in epithelial ovarian cancer histotypes. Genomics 106, 311–321. 10.1016/j.ygeno.2015.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi S. O., Reiisi S. (2019). Downregulation of miR-4443 and miR-5195-3p in ovarian cancer tissue contributes to metastasis and tumorigenesis. Arch. Gynecol. Obstet. 299, 1453–1458. 10.1007/s00404-019-05107-x [DOI] [PubMed] [Google Scholar]
- Edgell T., Martin-Roussety G., Barker G., Autelitano D. J., Allen D., Grant P., et al. (2010). Phase II biomarker trial of a multimarker diagnostic for ovarian cancer. J. Cancer Res. Clin. Oncol. 136, 1079–1088. 10.1007/s00432-009-0755-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissa M. A. L., Lerner L., Abdelfatah E., Shankar N., Canner J. K., Hasan N. M., et al. (2019). Promoter methylation of ADAMTS1 and BNC1 as potential biomarkers for early detection of pancreatic cancer in blood. Clin. Epigenetics 11:59. 10.1186/s13148-019-0650-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitan R., Kushnir M., Lithwick-Yanai G., David M. B., Hoshen M., Glezerman M., et al. (2009). Tumor microRNA expression patterns associated with resistance to platinum based chemotherapy and survival in ovarian cancer patients. Gynecol. Oncol. 114, 253–259. 10.1016/j.ygyno.2009.04.024 [DOI] [PubMed] [Google Scholar]
- Esteller M. (2007). Epigenetic gene silencing in cancer: the DNA hypermethylome. Hum. Mol. Genet. 16, R50–R59. 10.1093/hmg/ddm018 [DOI] [PubMed] [Google Scholar]
- Faggad A., Budczies J., Tchernitsa O., Darb-Esfahani S., Sehouli J., Müller B. M., et al. (2010). Prognostic significance of Dicer expression in ovarian cancer-link to global microRNA changes and oestrogen receptor expression. J. Pathol. 220, 382–391. 10.1002/path.2658 [DOI] [PubMed] [Google Scholar]
- Fang F., Cardenas H., Huang H., Jiang G., Perkins S. M., Zhang C., et al. (2018). Genomic and epigenomic signatures in ovarian cancer associated with resensitization to platinum drugs. Cancer Res. 78, 631–644. 10.1158/0008-5472.CAN-17-1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Tycko B. (2004). The history of cancer epigenetics. Nat. Rev. Cancer 4, 143–153. 10.1038/nrc1279 [DOI] [PubMed] [Google Scholar]
- Feng Q., Deftereos G., Hawes S. E., Stern J. E., Willner J. B., Swisher E. M., et al. (2008). DNA hypermethylation, Her-2/neu overexpression and p53 mutations in ovarian carcinoma. Gynecol. Oncol. 111, 320–329. 10.1016/j.ygyno.2008.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W., Marquez R. T., Lu Z., Liu J., Lu K. H., Issa J.-P. J., et al. (2008). Imprinted tumor suppressor genes ARHI and PEG3 are the most frequently down-regulated in human ovarian cancers by loss of heterozygosity and promoter methylation. Cancer 112, 1489–1502. 10.1002/cncr.23323 [DOI] [PubMed] [Google Scholar]
- Fiegl H., Windbichler G., Mueller-Holzner E., Goebel G., Lechner M., Jacobs I. J., et al. (2008). HOXA11 DNA methylation–a novel prognostic biomarker in ovarian cancer. Int. J. Cancer 123, 725–729. 10.1002/ijc.23563 [DOI] [PubMed] [Google Scholar]
- Fu Y., Campbell E. J., Shepherd T. G., Nachtigal M. W. (2003). Epigenetic regulation of proprotein convertase PACE4 gene expression in human ovarian cancer cells. Mol. Cancer Res. 1, 569–576. [PubMed] [Google Scholar]
- Fu Y., Chen J., Pang B., Li C., Zhao J., Shen K. (2015). EZH2-Induced H3K27me3 is Associated with epigenetic repression of the ARHI tumor-suppressor gene in ovarian cancer. Cell Biochem. Biophys. 71, 105–112. 10.1007/s12013-014-0168-1 [DOI] [PubMed] [Google Scholar]
- Furlan D. (2006). The high frequency of de novo promoter methylation in synchronous primary endometrial and ovarian carcinomas. Clin. Cancer Res. 12, 3329–3336. 10.1158/1078-0432.CCR-05-2679 [DOI] [PubMed] [Google Scholar]
- Gadomska H., Grzechocinska B., Janecki J., Nowicka G., Powolny M., Marianowski L. (2005). Serum lipids concentration in women with benign and malignant ovarian tumours. Eur. J. Obstet. Gynecol. Reprod. Biol. 120, 87–90. 10.1016/j.ejogrb.2004.02.045 [DOI] [PubMed] [Google Scholar]
- Gao Y., Lou G., Zhang G.-M., Sun X.-W., Ma Y.-Y., Yang Y.-M., et al. (2009). CHFR promoter hypermethylation and reduced CHFR mRNA expression in ovarian cancer. Int. J. Biol. Markers 24, 83–89. 10.1177/172460080902400204 [DOI] [PubMed] [Google Scholar]
- Gao Y., Wu J. (2015). MicroRNA-200c and microRNA-141 as potential diagnostic and prognostic biomarkers for ovarian cancer. Tumor Biol. 36, 4843–4850. 10.1007/s13277-015-3138-3 [DOI] [PubMed] [Google Scholar]
- Geomini P., Kruitwagen R., Bremer G. L., Cnossen J., Mol B. W. J. (2009). The accuracy of risk scores in predicting ovarian malignancy: a systematic review. Obstet. Gynecol. 113, 384–394. 10.1097/AOG.0b013e318195ad17 [DOI] [PubMed] [Google Scholar]
- Giannopoulou L., Chebouti I., Pavlakis K., Kasimir-Bauer S., Lianidou E. S. (2017). RASSF1A promoter methylation in high-grade serous ovarian cancer: a direct comparison study in primary tumors, adjacent morphologically tumor cell-free tissues and paired circulating tumor DNA. Oncotarget 8:21429. 10.18632/oncotarget.15249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannopoulou L., Mastoraki S., Buderath P., Strati A., Pavlakis K., Kasimir-Bauer S., et al. (2018). ESR1 methylation in primary tumors and paired circulating tumor DNA of patients with high-grade serous ovarian cancer. Gynecol. Oncol. 150, 355–360. 10.1016/j.ygyno.2018.05.026 [DOI] [PubMed] [Google Scholar]
- Gifford G. (2004). The Acquisition of hMLH1 Methylation in Plasma DNA after chemotherapy predicts poor survival for ovarian cancer patients. Clin. Cancer Res. 10, 4420–4426. 10.1158/1078-0432.CCR-03-0732 [DOI] [PubMed] [Google Scholar]
- Gloss B. S., Patterson K. I., Barton C. A., Gonzalez M., Scurry J. P., Hacker N. F., et al. (2012). Integrative genome-wide expression and promoter DNA methylation profiling identifies a potential novel panel of ovarian cancer epigenetic biomarkers. Cancer Lett. 318, 76–85. 10.1016/j.canlet.2011.12.003 [DOI] [PubMed] [Google Scholar]
- Gong L., Wang C., Gao Y., Wang J. (2016). Decreased expression of microRNA-148a predicts poor prognosis in ovarian cancer and associates with tumor growth and metastasis. Biomed. Pharmacother. 83, 58–63. 10.1016/j.biopha.2016.05.049 [DOI] [PubMed] [Google Scholar]
- Gorelik E. (2005). Multiplexed immunobead-based cytokine profiling for early detection of ovarian cancer. Cancer Epidemiol. Biomarkers Prev. 14, 981–987. 10.1158/1055-9965.EPI-04-0404 [DOI] [PubMed] [Google Scholar]
- Gormally E., Caboux E., Vineis P., Hainaut P. (2007). Circulating free DNA in plasma or serum as biomarker of carcinogenesis: practical aspects and biological significance. Mutat. Res. Mutat. Res. 635, 105–117. 10.1016/j.mrrev.2006.11.002 [DOI] [PubMed] [Google Scholar]
- Gozzi G., Chelbi S. T., Manni P., Alberti L., Fonda S., Saponaro S., et al. (2016). Promoter methylation and downregulated expression of the TBX15 gene in ovarian carcinoma. Oncol. Lett. 12, 2811–2819. 10.3892/ol.2016.5019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günel T., Gumusoglu E., Dogan B., Ertem F. B., Hosseini M. K., Cevik N., et al. (2018). Potential biomarker of circulating hsa-miR-1273g-3p level for detection of recurrent epithelial ovarian cancer. Arch. Gynecol. Obstet. 298, 1173–1180. 10.1007/s00404-018-4913-3 [DOI] [PubMed] [Google Scholar]
- Guo T. Y., Xu H.-Y., Chen W. J., Wu M.-X., Dai X. (2018). Downregulation of miR-1294 associates with prognosis and tumor progression in epithelial ovarian cancer. Eur. Rev. Med. Pharmacol. Sci. 22, 7646–7652. [DOI] [PubMed] [Google Scholar]
- Guo W., Zhu L., Yu M., Zhu R., Chen Q., Wang Q. (2018). A five-DNA methylation signature act as a novel prognostic biomarker in patients with ovarian serous cystadenocarcinoma. Clin. Epigenetics 10:142. 10.1186/s13148-018-0574-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Godwin A. K., Vanderveer L., Lu A., Liu J. (2003). Hypomethylation of the synuclein gamma gene CpG island promotes its aberrant expression in breast carcinoma and ovarian carcinoma. Cancer Res. 63, 664–673. [PubMed] [Google Scholar]
- Häfner N., Diebolder H., Jansen L., Hoppe I., Dürst M., Runnebaum I. B. (2011). Hypermethylated DAPK in serum DNA of women with uterine leiomyoma is a biomarker not restricted to cancer. Gynecol. Oncol. 121, 224–229. 10.1016/j.ygyno.2010.11.018 [DOI] [PubMed] [Google Scholar]
- Häfner N., Steinbach D., Jansen L., Diebolder H., Dürst M., Runnebaum I. B. (2016). RUNX3 and CAMK2N1 hypermethylation as prognostic marker for epithelial ovarian cancer: prognostic methylation markers for EOC. Int. J. Cancer 138, 217–228. 10.1002/ijc.29690 [DOI] [PubMed] [Google Scholar]
- Han X., Zhou Y., You Y., Lu J., Wang L., Hou H., et al. (2017). TET1 promotes cisplatin-resistance via demethylating the vimentin promoter in ovarian cancer: TET1 in ovarian cancer. Cell Biol. Int. 41, 405–414. 10.1002/cbin.10734 [DOI] [PubMed] [Google Scholar]
- Havrilesky L. J., Whitehead C. M., Rubatt J. M., Cheek R. L., Groelke J., He Q., et al. (2008). Evaluation of biomarker panels for early stage ovarian cancer detection and monitoring for disease recurrence. Gynecol. Oncol. 110, 374–382. 10.1016/j.ygyno.2008.04.041 [DOI] [PubMed] [Google Scholar]
- Hendrich B., Bird A. (2000). Mammalian methyltransferases and methyl-CpG-binding domains: proteins involved in DNA methylation. Curr. Top. Microbiol. Immunol. 249, 55–74. 10.1007/978-3-642-59696-4_4 [DOI] [PubMed] [Google Scholar]
- Herman J. G., Baylin S. B. (2003). Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 349, 2042–2054. 10.1056/NEJMra023075 [DOI] [PubMed] [Google Scholar]
- Honda H., Pazin M. J., D'Souza T., Ji H., Morin P. J. (2007). Regulation of the CLDN3 gene in ovarian cancer cells. Cancer Biol. Ther. 6, 1733–1742. 10.4161/cbt.6.11.4832 [DOI] [PubMed] [Google Scholar]
- Honda H., Pazin M. J., Ji H., Wernyj R. P., Morin P. J. (2006). Crucial roles of Sp1 and epigenetic modifications in the regulation of the CLDN4 promoter in ovarian cancer cells. J. Biol. Chem. 281, 21433–21444. 10.1074/jbc.M603767200 [DOI] [PubMed] [Google Scholar]
- Hooda J., Novak M., Salomon M. P., Matsuba C., Ramos R. I., MacDuffie E., et al. (2019). Early loss of histone H2B monoubiquitylation alters chromatin accessibility and activates key immune pathways that facilitate progression of ovarian cancer. Cancer Res. 79, 760–772. 10.1158/0008-5472.CAN-18-2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horak P., Pils D., Haller G., Pribill I., Roessler M., Tomek S., et al. (2005). Contribution of epigenetic silencing of tumor necrosis factor-related apoptosis inducing ligand receptor 1 (DR4) to TRAIL resistance and ovarian cancer. Mol. Cancer Res. 3, 335–343. 10.1158/1541-7786.MCR-04-0136 [DOI] [PubMed] [Google Scholar]
- Hough C. D., Sherman-Baust C. A., Pizer E. S., Montz F. J., Im D. D., Rosenshein N. B., et al. (2000). Large-scale serial analysis of gene expression reveals genes differentially expressed in ovarian cancer. Cancer Res. 60, 6281–6287. [PubMed] [Google Scholar]
- Howe H. L., Wu X., Ries L. A. G., Cokkinides V., Ahmed F., Jemal A., et al. (2006). Annual report to the nation on the status of cancer, 1975–2003, featuring cancer among U.S. Hispanic/Latino populations. Cancer 107, 1711–1742. 10.1002/cncr.22193 [DOI] [PubMed] [Google Scholar]
- Hu X., Macdonald D. M., Huettner P. C., Feng Z., El Naqa I. M., Schwarz J. K., et al. (2009). A miR-200 microRNA cluster as prognostic marker in advanced ovarian cancer. Gynecol. Oncol. 114, 457–464. 10.1016/j.ygyno.2009.05.022 [DOI] [PubMed] [Google Scholar]
- Hu Y., Zhang Q., Cui J., Liao Z.-J., Jiao M., Zhang Y.-B., et al. (2019). Oncogene miR-934 promotes ovarian cancer cell proliferation and inhibits cell apoptosis through targeting BRMS1L. Eur. Rev. Med. Pharmacol. Sci. 23, 5595–5602. 10.26355/eurrev_201907_18293 [DOI] [PubMed] [Google Scholar]
- Huang C.-Y., Cheng W.-F., Lee C.-N., Su Y.-N., Chien S.-C., Tzeng Y.-L., et al. (2006). Serum mesothelin in epithelial ovarian carcinoma: a new screening marker and prognostic factor. Anticancer Res. 26, 4721–4728. [PubMed] [Google Scholar]
- Huang L., Zhou Y., Cao X.-P., Lin J.-X., Zhang L., Huang S.-T., et al. (2017). KPNA2 is a potential diagnostic serum biomarker for epithelial ovarian cancer and correlates with poor prognosis. Tumor Biol. 39:101042831770628. 10.1177/1010428317706289 [DOI] [PubMed] [Google Scholar]
- Huang R.-L., Chen H.-J., Chen L.-Y., Chao T.-K., Lin W.-Y., Liew P.-L., et al. (2018). Epigenetic loss of heparan sulfate 3-O-sulfation sensitizes ovarian carcinoma to oncogenic signals and predicts prognosis: less 3-O-sulfated HS sensitizes prognostic signals. Int. J. Cancer 143, 1943–1953. 10.1002/ijc.31580 [DOI] [PubMed] [Google Scholar]
- Huang R.-L., Gu F., Kirma N. B., Ruan J., Chen C.-L., Wang H.-C., et al. (2013). Comprehensive methylome analysis of ovarian tumors reveals hedgehog signaling pathway regulators as prognostic DNA methylation biomarkers. Epigenetics 8, 624–634. 10.4161/epi.24816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.-W., Jansen R. A., Fabbri E., Potter D., Liyanarachchi S., Chan M. W. Y., et al. (2009). Identification of candidate epigenetic biomarkers for ovarian cancer detection. Oncol. Rep. 22, 853–861. 10.3892/or_00000509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanez de Caceres I., Battagli C., Esteller M., Herman J. G., Dulaimi E., Edelson M. I., et al. (2004). Tumor cell-specific BRCA1 and RASSF1A hypermethylation in serum, plasma, and peritoneal fluid from ovarian cancer patients. Cancer Res. 64, 6476–6481. 10.1158/0008-5472.CAN-04-1529 [DOI] [PubMed] [Google Scholar]
- Ignatov T., Eggemann H., Costa S. D., Roessner A., Kalinski T., Ignatov A. (2014). BRCA1 promoter methylation is a marker of better response to platinum–taxane-based therapy in sporadic epithelial ovarian cancer. J. Cancer Res. Clin. Oncol. 140, 1457–1463. 10.1007/s00432-014-1704-5 [DOI] [PubMed] [Google Scholar]
- Iorio M. V., Visone R., Di Leva G., Donati V., Petrocca F., Casalini P., et al. (2007). MicroRNA signatures in human ovarian cancer. Cancer Res. 67, 8699–8707. 10.1158/0008-5472.CAN-07-1936 [DOI] [PubMed] [Google Scholar]
- Izutsu N., Maesawa C., Shibazaki M., Oikawa H., Shoji T., Sugiyama T., et al. (2008). Epigenetic modification is involved in aberrant expression of class III beta-tubulin, TUBB3, in ovarian cancer cells. Int. J. Oncol. 32, 1227–1235. 10.3892/ijo_32_6_1227 [DOI] [PubMed] [Google Scholar]
- Jacob F., Hitchins M. P., Fedier A., Brennan K., Nixdorf S., Hacker N. F., et al. (2014). Expression of GBGT1 is epigenetically regulated by DNA methylation in ovarian cancer cells. BMC Mol. Biol. 15:24. 10.1186/1471-2199-15-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs I., Bast R. C. (1989). The CA 125 tumour-associated antigen: a review of the literature. Hum. Reprod. Oxf. Engl. 4, 1–12. 10.1093/oxfordjournals.humrep.a136832 [DOI] [PubMed] [Google Scholar]
- Jacobs I., Gentry-Maharaj A., Burnell M., Manchanda R., Singh N., Sharma A., et al. (2011). Sensitivity of transvaginal ultrasound screening for endometrial cancer in postmenopausal women: a case-control study within the UKCTOCS cohort. Lancet Oncol. 12, 38–48. 10.1016/S1470-2045(10)70268-0 [DOI] [PubMed] [Google Scholar]
- Jacobs I., Oram D., Fairbanks J., Turner J., Frost C., Grudzinskas J. G. (1990). A risk of malignancy index incorporating CA 125, ultrasound and menopausal status for the accurate preoperative diagnosis of ovarian cancer. Br. J. Obstet. Gynaecol. 97, 922–929. 10.1111/j.1471-0528.1990.tb02448.x [DOI] [PubMed] [Google Scholar]
- Jacobs I. J., Menon U. (2004). Progress and challenges in screening for early detection of ovarian cancer. Mol. Cell. Proteomics 3, 355–366. 10.1074/mcp.R400006-MCP200 [DOI] [PubMed] [Google Scholar]
- Jayson G. C., Kohn E. C., Kitchener H. C., Ledermann J. A. (2014). Ovarian cancer. Lancet 384, 1376–1388. 10.1016/S0140-6736(13)62146-7 [DOI] [PubMed] [Google Scholar]
- Jia Y., Lin R., Jin H., Si L., Jian W., Yu Q., et al. (2019). MicroRNA-34 suppresses proliferation of human ovarian cancer cells by triggering autophagy and apoptosis and inhibits cell invasion by targeting Notch 1. Biochimie 160, 193–199. 10.1016/j.biochi.2019.03.011 [DOI] [PubMed] [Google Scholar]
- Jones A., Lechner M., Fourkala E.-O., Kristeleit R., Widschwendter M. (2010). Emerging promise of epigenetics and DNA methylation for the diagnosis and management of women's cancers. Epigenomics 2, 9–38. 10.2217/epi.09.47 [DOI] [PubMed] [Google Scholar]
- Jones P. A. (2001). The role of DNA methylation in mammalian epigenetics. Science 293, 1068–1070. 10.1126/science.1063852 [DOI] [PubMed] [Google Scholar]
- Jones P. A., Baylin S. B. (2002). The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 3:415. 10.1038/nrg816 [DOI] [PubMed] [Google Scholar]
- Jones P. A., Baylin S. B. (2007). The epigenomics of cancer. Cell 128, 683–692. 10.1016/j.cell.2007.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. A., Laird P. W. (1999). Cancer-epigenetics comes of age. Nat. Genet. 21, 163–167. 10.1038/5947 [DOI] [PubMed] [Google Scholar]
- Kacem S., Feil R. (2009). Chromatin mechanisms in genomic imprinting. Mamm. Genome 20, 544–556. 10.1007/s00335-009-9223-4 [DOI] [PubMed] [Google Scholar]
- Kamikihara T., Arima T., Kato K., Matsuda T., Kato H., Douchi T., et al. (2005). Epigenetic silencing of the imprinted geneZAC by DNA methylation is an early event in the progression of human ovarian cancer. Int. J. Cancer 115, 690–700. 10.1002/ijc.20971 [DOI] [PubMed] [Google Scholar]
- Kaneuchi M., Sasaki M., Tanaka Y., Shiina H., Verma M., Ebina Y., et al. (2004). Expression and methylation status of 14-3-3 sigma gene can characterize the different histological features of ovarian cancer. Biochem. Biophys. Res. Commun. 316, 1156–1162. 10.1016/j.bbrc.2004.02.171 [DOI] [PubMed] [Google Scholar]
- Kaneuchi M., Sasaki M., Tanaka Y., Shiina H., Yamada H., Yamamoto R., et al. (2005). WT1 and WT1-AS genes are inactivated by promoter methylation in ovarian clear cell adenocarcinoma. Cancer 104, 1924–1930. 10.1002/cncr.21397 [DOI] [PubMed] [Google Scholar]
- Kang S., Dong S. M., Park N.-H. (2010). Frequent promoter hypermethylation of TGFBI in epithelial ovarian cancer. Gynecol. Oncol. 118, 58–63. 10.1016/j.ygyno.2010.03.025 [DOI] [PubMed] [Google Scholar]
- Kardum V., Karin V., Glibo M., Skrtic A., Martic T. N., Ibisevic N., et al. (2017). Methylation-associated silencing of SFRP1 gene in high-grade serous ovarian carcinomas. Ann. Diagn. Pathol. 31, 45–49. 10.1016/j.anndiagpath.2017.07.002 [DOI] [PubMed] [Google Scholar]
- Katsaros D., Cho W., Singal R., Fracchioli S., Rigault De La Longrais I. A., Arisio R., et al. (2004). Methylation of tumor suppressor gene p16 and prognosis of epithelial ovarian cancer. Gynecol. Oncol. 94, 685–692. 10.1016/j.ygyno.2004.06.018 [DOI] [PubMed] [Google Scholar]
- Kaur M., Singh A., Singh K., Gupta S., Sachan M. (2016). Development of a multiplex MethyLight assay for the detection of DAPK1 and SOX1 methylation in epithelial ovarian cancer in a north Indian population. Genes Genet. Syst. 91, 175–181. 10.1266/ggs.15-00051 [DOI] [PubMed] [Google Scholar]
- Keita M., Wang Z.-Q., Pelletier J.-F., Bachvarova M., Plante M., Gregoire J., et al. (2013). Global methylation profiling in serous ovarian cancer is indicative for distinct aberrant DNA methylation signatures associated with tumor aggressiveness and disease progression. Gynecol. Oncol. 128, 356–363. 10.1016/j.ygyno.2012.11.036 [DOI] [PubMed] [Google Scholar]
- Kikuchi R., Tsuda H., Kanai Y., Kasamatsu T., Sengoku K., Hirohashi S., et al. (2007). Promoter hypermethylation contributes to frequent inactivation of a putative conditional tumor suppressor gene connective tissue growth factor in ovarian cancer. Cancer Res. 67, 7095–7105. 10.1158/0008-5472.CAN-06-4567 [DOI] [PubMed] [Google Scholar]
- Kikuchi R., Tsuda H., Kozaki K., Kanai Y., Kasamatsu T., Sengoku K., et al. (2008). Frequent inactivation of a putative tumor suppressor, angiopoietin-like protein 2, in ovarian cancer. Cancer Res. 68, 5067–5075. 10.1158/0008-5472.CAN-08-0062 [DOI] [PubMed] [Google Scholar]
- Kim J.-H., Skates S. J., Uede T., Wong K., Schorge J. O., Feltmate C. M., et al. (2002). Osteopontin as a potential diagnostic biomarker for ovarian cancer. JAMA 287, 1671–1679. 10.1001/jama.287.13.1671 [DOI] [PubMed] [Google Scholar]
- Kim S., Choi M. C., Jeong J.-Y., Hwang S., Jung S. G., Joo W. D., et al. (2019). Serum exosomal miRNA-145 and miRNA-200c as promising biomarkers for preoperative diagnosis of ovarian carcinomas. J. Cancer 10, 1958–1967. 10.7150/jca.30231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.-W., Bae S. M., Lim H., Kim Y. J., Ahn W. S. (2012). Development of multiplexed bead-based immunoassays for the detection of early stage ovarian cancer using a combination of serum biomarkers. PLoS ONE 7:e44960. 10.1371/journal.pone.0044960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitade S., Onoyama I., Kobayashi H., Yagi H., Yoshida S., Kato M., et al. (2016). FBXW7 is involved in the acquisition of the malignant phenotype in epithelial ovarian tumors. Cancer Sci. 107, 1399–1405. 10.1111/cas.13026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz D. M., Wimberger P. (2017). Cells of origin of ovarian cancer: ovarian surface epithelium or fallopian tube? Arch. Gynecol. Obstet. 296, 1055–1062. 10.1007/s00404-017-4529-z [DOI] [PubMed] [Google Scholar]
- Kneip C., Schmidt B., Seegebarth A., Weickmann S., Fleischhacker M., Liebenberg V., et al. (2011). SHOX2 DNA methylation is a biomarker for the diagnosis of lung cancer in plasma. J. Thorac. Oncol. 6, 1632–1638. 10.1097/JTO.0b013e318220ef9a [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Sawada K., Nakamura K., Yoshimura A., Miyamoto M., Shimizu A., et al. (2018). Exosomal miR-1290 is a potential biomarker of high-grade serous ovarian carcinoma and can discriminate patients from those with malignancies of other histological types. J. Ovarian Res. 11:81. 10.1186/s13048-018-0458-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koestler D. C., Chalise P., Cicek M. S., Cunningham J. M., Armasu S., Larson M. C., et al. (2014). Integrative genomic analysis identifies epigenetic marks that mediate genetic risk for epithelial ovarian cancer. BMC Med. Genomics 7:260. 10.1158/1538-7445.AM2014-260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler R. S., Anugraham M., López M. N., Xiao C., Schoetzau A., Hettich T., et al. (2016). Epigenetic activation of MGAT3 and corresponding bisecting GlcNAc shortens the survival of cancer patients. Oncotarget 7:51674–51686. 10.18632/oncotarget.10543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolomietz E., Meyn M. S., Pandita A., Squire J. A. (2002). The role of Alu repeat clusters as mediators of recurrent chromosomal aberrations in tumors. Genes. Chromosomes Cancer 35, 97–112. 10.1002/gcc.10111 [DOI] [PubMed] [Google Scholar]
- Kurose K., Zhou X.-P., Araki T., Cannistra S. A., Maher E. R., Eng C. (2001). Frequent loss of PTEN Expression Is Linked to Elevated Phosphorylated Akt levels, but not associated with p27 and Cyclin D1 expression, in primary epithelial ovarian carcinomas. Am. J. Pathol. 158, 2097–2106. 10.1016/S0002-9440(10)64681-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M. J., Kim S.-S., Choi Y.-L., Jung H. S., Balch C., Kim S.-H., et al. (2010). Derepression of CLDN3 and CLDN4 during ovarian tumorigenesis is associated with loss of repressive histone modifications. Carcinogenesis 31, 974–983. 10.1093/carcin/bgp336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong J., Lee J.-Y., Wong K.-K., Zhou X., Wong D. T. W., Lo K.-W., et al. (2006). Candidate tumor-suppressor gene DLEC1 is frequently downregulated by promoter hypermethylation and histone hypoacetylation in human epithelial ovarian cancer. Neoplasia 8, 268–278. 10.1593/neo.05502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laios A., O'Toole S., Flavin R., Martin C., Kelly L., Ring M., et al. (2008). Potential role of miR-9 and miR-223 in recurrent ovarian cancer. Mol. Cancer 7:35. 10.1186/1476-4598-7-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langhe R., Norris L., Saadeh F. A., Blackshields G., Varley R., Harrison A., et al. (2015). A novel serum microRNA panel to discriminate benign from malignant ovarian disease. Cancer Lett. 356, 628–636. 10.1016/j.canlet.2014.10.010 [DOI] [PubMed] [Google Scholar]
- Lee C.-H., Subramanian S., Beck A. H., Espinosa I., Senz J., Zhu S. X., et al. (2009). MicroRNA profiling of BRCA1/2 mutation-carrying and non-mutation-carrying high-grade serous carcinomas of ovary. PLoS ONE 4:e7314. 10.1371/journal.pone.0007314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P. S., Teaberry V. S., Bland A. E., Huang Z., Whitaker R. S., Baba T., et al. (2010). Elevated MAL expression is accompanied by promoter hypomethylation and platinum resistance in epithelial ovarian cancer. Int. J. Cancer 126, 1378–1389. 10.1002/ijc.24797 [DOI] [PubMed] [Google Scholar]
- Leon S. A., Shapiro B., Sklaroff D. M., Yaros M. J. (1977). Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 37, 646–650. [PubMed] [Google Scholar]
- Leskela S., Leandro-Garcia L. J., Mendiola M., Barriuso J., Inglada-Perez L., Munoz I., et al. (2010). The miR-200 family controls -tubulin III expression and is associated with paclitaxel-based treatment response and progression-free survival in ovarian cancer patients. Endocr. Relat. Cancer 18, 85–95. 10.1677/ERC-10-0148 [DOI] [PubMed] [Google Scholar]
- Leung F., Bernardini M. Q., Brown M. D., Zheng Y., Molina R., Bast R. C., et al. (2016). Validation of a novel biomarker panel for the detection of ovarian cancer. Cancer Epidemiol. Biomarkers Prev. 25, 1333–1340. 10.1158/1055-9965.EPI-15-1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. M., Heineck D. P., Heller M. J. (2015). Detecting cancer biomarkers in blood: challenges for new molecular diagnostic and point-of-care tests using cell-free nucleic acids. Expert Rev. Mol. Diagn. 15, 1187–1200. 10.1586/14737159.2015.1069709 [DOI] [PubMed] [Google Scholar]
- Li J., He X., Dong R., Wang Y., Yu J., Qiu H. (2015). Frequent loss of NISCH promotes tumor proliferation and invasion in ovarian cancer via inhibiting the FAK signal pathway. Mol. Cancer Ther. 14, 1202–1212. 10.1158/1535-7163.MCT-14-0911 [DOI] [PubMed] [Google Scholar]
- Li J., Shao W., Feng H. (2019). MiR-542-3p, a microRNA targeting CDK14, suppresses cell proliferation, invasiveness, and tumorigenesis of epithelial ovarian cancer. Biomed. Pharmacother. 110, 850–856. 10.1016/j.biopha.2018.11.104 [DOI] [PubMed] [Google Scholar]
- Li L.-W., Xiao H.-Q., Ma R., Yang M., Li W., Lou G. (2017). miR-152 is involved in the proliferation and metastasis of ovarian cancer through repression of ERBB3. Int. J. Mol. Med. 41, 1529–1535. 10.3892/ijmm.2017.3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Pan Q., Wan X., Mao Y., Lu W., Xie X., et al. (2015). Methylation-associated Has-miR-9 deregulation in paclitaxel- resistant epithelial ovarian carcinoma. BMC Cancer 15:1. 10.1186/s12885-015-1509-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y.-P., Chen L.-Y., Huang R.-L., Su P.-H., Chan M. W. Y., Chang C.-C., et al. (2014). Hypomethylation signature of tumor-initiating cells predicts poor prognosis of ovarian cancer patients. Hum. Mol. Genet. 23, 1894–1906. 10.1093/hmg/ddt583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liggett T. E., Melnikov A., Yi Q., Replogle C., Hu W., Rotmensch J., et al. (2011). Distinctive DNA methylation patterns of cell-free plasma DNA in women with malignant ovarian tumors. Gynecol. Oncol. 120, 113–120. 10.1016/j.ygyno.2010.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B., White J. T., Wu J., Lele S., Old L. J., Hood L., et al. (2009). Deep depletion of abundant serum proteins reveals low-abundant proteins as potential biomarkers for human ovarian cancer. Proteomics Clin. Appl. 3, 853–861. 10.1002/prca.200800141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H.-W., Fu C.-F., Chang M.-C., Lu T.-P., Lin H.-P., Chiang Y.-C., et al. (2018). CDH1, DLEC1 and SFRP5 methylation panel as a prognostic marker for advanced epithelial ovarian cancer. Epigenomics 10, 1397–1413. 10.2217/epi-2018-0035 [DOI] [PubMed] [Google Scholar]
- Litkouhi B., Kwong J., Lo C.-M., Smedley J. G., McClane B. A., Aponte M., et al. (2007). Claudin-4 overexpression in epithelial ovarian cancer is associated with hypomethylation and is a potential target for modulation of tight junction barrier function using a C-terminal fragment of Clostridium perfringens enterotoxin. Neoplasia 9, 304–314. 10.1593/neo.07118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Zhang J., Yang D. (2019). miR-96-5p promotes the proliferation and migration of ovarian cancer cells by suppressing Caveolae1. J. Ovarian Res. 12:57. 10.1186/s13048-019-0533-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Zhang X.-X., Li M.-C., Cao C.-H., Wan D.-Y., Xi B.-X., et al. (2018). C/EBPβ enhances platinum resistance of ovarian cancer cells by reprogramming H3K79 methylation. Nat. Commun. 9:1739. 10.1038/s41467-018-03590-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Yang D., Rupaimoole R., Pecot C. V., Sun Y., Mangala L. S., et al. (2015). Augmentation of response to chemotherapy by microRNA-506 through regulation of RAD51 in serous ovarian cancers. J. Natl. Cancer Inst. 107:djv108. 10.1093/jnci/djv108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M. X., Siu M. K. Y., Liu S. S., Yam J. W. P., Ngan H. Y. S., Chan D. W. (2014). Epigenetic silencing of microRNA-199b-5p is associated with acquired chemoresistance via activation of JAG1-Notch1 signaling in ovarian cancer. Oncotarget 5, 944–958. 10.18632/oncotarget.1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lofton-Day C., Model F., Devos T., Tetzner R., Distler J., Schuster M., et al. (2008). DNA methylation biomarkers for blood-based colorectal cancer screening. Clin. Chem. 54, 414–423. 10.1373/clinchem.2007.095992 [DOI] [PubMed] [Google Scholar]
- Loginov V. I., Pronina I. V., Burdennyy A. M., Filippova E. A., Kazubskaya T. P., Kushlinsky D. N., et al. (2018). Novel miRNA genes deregulated by aberrant methylation in ovarian carcinoma are involved in metastasis. Gene 662, 28–36. 10.1016/j.gene.2018.04.005 [DOI] [PubMed] [Google Scholar]
- Lopez-Serra L., Esteller M. (2008). Proteins that bind methylated DNA and human cancer: reading the wrong words. Br. J. Cancer 98, 1881–1885. 10.1038/sj.bjc.6604374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K. H., Patterson A. P., Wang L., Marquez R. T., Atkinson E. N., Baggerly K. A., et al. (2004). Selection of potential markers for epithelial ovarian cancer with gene expression arrays and recursive descent partition analysis. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 10, 3291–3300. 10.1158/1078-0432.CCR-03-0409 [DOI] [PubMed] [Google Scholar]
- Lu L., Katsaros D., Rigault de la Longrais I. A., Sochirca O., Yu H. (2007). Hypermethylation of let-7a-3 in Epithelial Ovarian Cancer Is Associated with Low Insulin-like Growth Factor-II Expression and Favorable Prognosis. Cancer Res. 67, 10117–10122. 10.1158/0008-5472.CAN-07-2544 [DOI] [PubMed] [Google Scholar]
- Lu T.-Y., Kao C.-F., Lin C.-T., Huang D.-Y., Chiu C.-Y., Huang Y.-S., et al. (2009). DNA methylation and histone modification regulate silencing of OPG during tumor progression. J. Cell. Biochem. 108, 315–325. 10.1002/jcb.22256 [DOI] [PubMed] [Google Scholar]
- Lund R. J., Huhtinen K., Salmi J., Rantala J., Nguyen E. V., Moulder R., et al. (2017). DNA methylation and transcriptome changes associated with cisplatin resistance in ovarian cancer. Sci. Rep. 7:1469. 10.1038/s41598-017-01624-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G., Chen C., Wang J., Yue H., Tian Y., Yang P., et al. (2019). FOXD3 may be a new cellular target biomarker as a hypermethylation gene in human ovarian cancer. Cancer Cell Int. 19:44. 10.1186/s12935-019-0755-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Y., Niu Z., Guo X., Yuan F., Liu Y. (2018). Serum S100 calcium binding protein A4 (S100A4, metatasin) as a diagnostic and prognostic biomarker in epithelial ovarian cancer. Br. J. Biomed. Sci. 75, 88–91. 10.1080/09674845.2017.1394052 [DOI] [PubMed] [Google Scholar]
- Lycke M., Kristjansdottir B., Sundfeldt K. (2018). A multicenter clinical trial validating the performance of HE4, CA125, risk of ovarian malignancy algorithm and risk of malignancy index. Gynecol. Oncol. 151, 159–165. 10.1016/j.ygyno.2018.08.025 [DOI] [PubMed] [Google Scholar]
- Lyu N., Wang Y., Wang J., Zhang Z., Kong W. (2018). Study on early diagnosis of epithelial ovarian cancer by analysis of plasma septin-9 and clusterin level. J. Cancer Res. Ther. 14:444. 10.4103/0973-1482.181178 [DOI] [PubMed] [Google Scholar]
- Mählck C. G., Grankvist K. (1994). Plasma prealbumin in women with epithelial ovarian carcinoma. Gynecol. Obstet. Invest. 37, 135–140. 10.1159/000292542 [DOI] [PubMed] [Google Scholar]
- Mahmoud E. H., Fawzy A. A., Elshimy R. A. (2018). Serum MicroRNA-21 negatively relates to expression of programmed cell death-4 in patients with epithelial ovarian cancer. Asian Pac. J. Cancer Prev. 19, 33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarla P. B., Saboorian M. H., Ashfaq R., Toyooka K. O., Toyooka S., Minna J. D., et al. (2005). Promoter hypermethylation profile of ovarian epithelial neoplasms. Clin. Cancer Res. 11, 5365–5369. 10.1158/1078-0432.CCR-04-2455 [DOI] [PubMed] [Google Scholar]
- Maldonado L., Brait M., Izumchenko E., Begum S., Chatterjee A., Sen T., et al. (2018). Integrated transcriptomic and epigenomic analysis of ovarian cancer reveals epigenetically silenced GULP1. Cancer Lett. 433, 242–251. 10.1016/j.canlet.2018.06.030 [DOI] [PubMed] [Google Scholar]
- Mase S., Shinjo K., Totani H., Katsushima K., Arakawa A., Takahashi S., et al. (2019). ZNF671 DNA methylation as a molecular predictor for the early recurrence of serous ovarian cancer. Cancer Sci. 110, 1105–1116. 10.1111/cas.13936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath S. E., Annels N., Madhuri T. K., Tailor A., Butler-Manuel S. A., Morgan R., et al. (2018). Engrailed-2 (EN2) – a novel biomarker in epithelial ovarian cancer. BMC Cancer 18:943. 10.1186/s12885-018-4816-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh M. W., Drescher C., Karlan B., Scholler N., Urban N., Hellstrom K. E., et al. (2004). Combining CA 125 and SMR serum markers for diagnosis and early detection of ovarian carcinoma. Gynecol. Oncol. 95, 9–15. 10.1016/j.ygyno.2004.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov A., Scholtens D., Godwin A., Levenson V. (2009). Differential methylation profile of ovarian cancer in tissues and plasma. J. Mol. Diagn. 11, 60–65. 10.2353/jmoldx.2009.080072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez L., Benigno B. B., McDonald J. F. (2004). L1 and HERV-W retrotransposons are hypomethylated in human ovarian carcinomas. Mol. Cancer 3:12. 10.1186/1476-4598-3-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng C.-F., Dai D.-Q., Guo K.-J. (2008). [Effects of 5-Aza-2′-deoxycytidine and trichostatin A on DNA methylation and expression of hMLH1 in ovariancancer cell line COC1/DDP]. Ai Zheng 27, 1251–1255. [PubMed] [Google Scholar]
- Meng X., Joosse S. A., Müller V., Trillsch F., Milde-Langosch K., Mahner S., et al. (2015). Diagnostic and prognostic potential of serum miR-7, miR-16, miR-25, miR-93, miR-182, miR-376a and miR-429 in ovarian cancer patients. Br. J. Cancer 113, 1358–1366. 10.1038/bjc.2015.340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Müller V., Milde-Langosch K., Trillsch F., Pantel K., Schwarzenbach H. (2016a). “Circulating Cell-Free miR-373, miR-200a, miR-200b and miR-200c in Patients with Epithelial Ovarian Cancer,” in Circulating Nucleic Acids in Serum and Plasma – CNAPS IX, eds Gahan P. B., Fleischhacker M., Schmidt B. (Cham: Springer International Publishing; ), 3–8. 10.1007/978-3-319-42044-8_1 [DOI] [PubMed] [Google Scholar]
- Meng X., Müller V., Milde-Langosch K., Trillsch F., Pantel K., Schwarzenbach H. (2016b). Diagnostic and prognostic relevance of circulating exosomal miR-373, miR-200a, miR-200b and miR-200c in patients with epithelial ovarian cancer. Oncotarget 7. 10.18632/oncotarget.7850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon U., Gentry-Maharaj A., Hallett R., Ryan A., Burnell M., Sharma A., et al. (2009). Sensitivity and specificity of multimodal and ultrasound screening for ovarian cancer, and stage distribution of detected cancers: results of the prevalence screen of the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS). Lancet Oncol. 10, 327–340. 10.1016/S1470-2045(09)70026-9 [DOI] [PubMed] [Google Scholar]
- Menon U., Jacobs I. J. (2000). Recent developments in ovarian cancer screening. Curr. Opin. Obstet. Gynecol. 12, 39–42. 10.1097/00001703-200002000-00007 [DOI] [PubMed] [Google Scholar]
- Merritt W. M., Lin Y. G., Han L. Y., Kamat A. A., Spannuth W. A., Schmandt R., et al. (2008). Dicer, Drosha, and outcomes in patients with ovarian cancer. N. Engl. J. Med. 359, 2641–2650. 10.1056/NEJMoa0803785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milde-Langosch K., Ocon E., Becker G., Löning T. (1998). p16/MTS1 inactivation in ovarian carcinomas: high frequency of reduced protein expression associated with hyper-methylation or mutation in endometrioid and mucinous tumors. Int. J. Cancer 79, 61–65. [DOI] [PubMed] [Google Scholar]
- Mirza A., McGuirk M., Hockenberry T. N., Wu Q., Ashar H., Black S., et al. (2002). Human survivin is negatively regulated by wild-type p53 and participates in p53-dependent apoptotic pathway. Oncogene 21, 2613–2622. 10.1038/sj.onc.1205353 [DOI] [PubMed] [Google Scholar]
- Mok S. C., Chao J., Skates S., Wong K., Yiu G. K., Muto M. G., et al. (2001). Prostasin, a potential serum marker for ovarian cancer: identification through microarray technology. J. Natl. Cancer Inst. 93, 1458–1464. 10.1093/jnci/93.19.1458 [DOI] [PubMed] [Google Scholar]
- Molina R., Escudero J. M., Aug,é J. M., Filella X., Foj L., Torn,é A., et al. (2011). HE4 a novel tumour marker for ovarian cancer: comparison with CA 125 and ROMA algorithm in patients with gynaecological diseases. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 32, 1087–1095. 10.1007/s13277-011-0204-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R. G., Brown A. K., Miller M. C., Skates S., Allard W. J., Verch T., et al. (2008). The use of multiple novel tumor biomarkers for the detection of ovarian carcinoma in patients with a pelvic mass. Gynecol. Oncol. 108, 402–408. 10.1016/j.ygyno.2007.10.017 [DOI] [PubMed] [Google Scholar]
- Moore R. G., MacLaughlan S., Bast R. C. (2010). Current state of biomarker development for clinical application in epithelial ovarian cancer. Gynecol. Oncol. 116, 240–245. 10.1016/j.ygyno.2009.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R. G., McMeekin D. S., Brown A. K., DiSilvestro P., Miller M. C., Allard W. J., et al. (2009). A novel multiple marker bioassay utilizing HE4 and CA125 for the prediction of ovarian cancer in patients with a pelvic mass. Gynecol. Oncol. 112, 40–46. 10.1016/j.ygyno.2008.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R. G., Miller M. C., Disilvestro P., Landrum L. M., Gajewski W., Ball J. J., et al. (2011). Evaluation of the diagnostic accuracy of the risk of ovarian malignancy algorithm in women with a pelvic mass. Obstet. Gynecol. 118, 280–288. 10.1097/AOG.0b013e318224fce2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor G., Visintin I., Lai Y., Zhao H., Schwartz P., Rutherford T., et al. (2005). Serum protein markers for early detection of ovarian cancer. Proc. Natl. Acad. Sci.U.S.A. 102, 7677–7682. 10.1073/pnas.0502178102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. M. (2004). Analysis of methylated genes in peritoneal fluids of ovarian cancer patients: a new prognostic tool. Clin. Chem. 50, 2171–2173. 10.1373/clinchem.2004.034090 [DOI] [PubMed] [Google Scholar]
- Munkarah A., Chatterjee M., Tainsky M. A. (2007). Update on ovarian cancer screening: Curr. Opin. Obstet. Gynecol. 19, 22–26. 10.1097/GCO.0b013e328011ec99 [DOI] [PubMed] [Google Scholar]
- Murphy S. K., Huang Z., Wen Y., Spillman M. A., Whitaker R. S., Simel L. R., et al. (2006). Frequent IGF2/H19 domain epigenetic alterations and elevated IGF2 expression in epithelial ovarian cancer. Mol. Cancer Res. 4, 283–292. 10.1158/1541-7786.MCR-05-0138 [DOI] [PubMed] [Google Scholar]
- Nguyen L., Cardenas-Goicoechea S. J., Gordon P., Curtin C., Momeni M., Chuang L., et al. (2013). Biomarkers for early detection of ovarian cancer. Womens Health 9, 171–187. 10.2217/WHE.13.2 [DOI] [PubMed] [Google Scholar]
- Nicholson L. J., Smith P. R., Hiller L., Szlosarek P. W., Kimberley C., Sehouli J., et al. (2009). Epigenetic silencing of argininosuccinate synthetase confers resistance to platinum-induced cell death but collateral sensitivity to arginine auxotrophy in ovarian cancer. Int. J. Cancer 125, 1454–1463. 10.1002/ijc.24546 [DOI] [PubMed] [Google Scholar]
- Nosov V., Su F., Amneus M., Birrer M., Robins T., Kotlerman J., et al. (2009). Validation of serum biomarkers for detection of early-stage ovarian cancer. Am. J. Obstet. Gynecol. 200, 639.e1–5. 10.1016/j.ajog.2008.12.042 [DOI] [PubMed] [Google Scholar]
- Ongenaert M., Van Neste L., De Meyer T., Menschaert G., Bekaert S., Van Criekinge W. (2008). PubMeth: a cancer methylation database combining text-mining and expert annotation. Nucl. Acids Res. 36, D842–D846. 10.1093/nar/gkm788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pampalakis G., Diamandis E. P., Katsaros D., Sotiropoulou G. (2010). Down-regulation of dicer expression in ovarian cancer tissues. Clin. Biochem. 43, 324–327. 10.1016/j.clinbiochem.2009.09.014 [DOI] [PubMed] [Google Scholar]
- Pattamadilok J., Huapai N., Rattanatanyong P., Vasurattana A., Triratanachat S., Tresukosol D., et al. (2008). LINE-1 hypomethylation level as a potential prognostic factor for epithelial ovarian cancer. Int. J. Gynecol. Cancer 18, 711–717. 10.1111/j.1525-1438.2007.01117.x [DOI] [PubMed] [Google Scholar]
- Perlin E., Moquin R. B. (1972). Serum DNA levels in patients with malignant disease. Am. J. Clin. Pathol. 58, 601–602. 10.1093/ajcp/58.5.601 [DOI] [PubMed] [Google Scholar]
- Petrocca F., Iliopoulos D., Qin H. R., Nicoloso M. S., Yendamuri S., Wojcik S. E., et al. (2006). Alterations of the tumor suppressor gene ARLTS1 in ovarian cancer. Cancer Res. 66, 10287–10291. 10.1158/0008-5472.CAN-06-2289 [DOI] [PubMed] [Google Scholar]
- Phelps D. L., Borley J. V., Flower K. J., Dina R., Darb-Esfahani S., Braicu I., et al. (2017). Methylation of MYLK3 gene promoter region: a biomarker to stratify surgical care in ovarian cancer in a multicentre study. Br. J. Cancer 116, 1287–1293. 10.1038/bjc.2017.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieretti M., Cavalieri C., Conway P. S., Gallion H. H., Powell D. E., Turker M. S. (1995). Genetic alterations distinguish different types of ovarian tumors. Int. J. Cancer 64, 434–440. 10.1002/ijc.2910640614 [DOI] [PubMed] [Google Scholar]
- Polivka J., Pesta M., Janku F. (2015). Testing for oncogenic molecular aberrations in cell-free DNA-based liquid biopsies in the clinic: are we there yet? Expert Rev. Mol. Diagn. 15, 1631–1644. 10.1586/14737159.2015.1110021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portela A., Esteller M. (2010). Epigenetic modifications and human disease. Nat. Biotechnol. 28, 1057–1068. 10.1038/nbt.1685 [DOI] [PubMed] [Google Scholar]
- Press J. Z., De Luca A., Boyd N., Young S., Troussard A., Ridge Y., et al. (2008). Ovarian carcinomas with genetic and epigenetic BRCA1 loss have distinct molecular abnormalities. BMC Cancer 8:17. 10.1186/1471-2407-8-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulliam N., Fang F., Ozes A. R., Tang J., Adewuyi A., Keer H., et al. (2018). An effective epigenetic-PARP inhibitor combination therapy for breast and ovarian cancers independent of BRCA mutations. Clin. Cancer Res. 24, 3163–3175. 10.1158/1078-0432.CCR-18-0204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu G., Dubeau L., Narayan A., Yu M. C., Ehrlich M. (1999). Satellite DNA hypomethylation vs. overall genomic hypomethylation in ovarian epithelial tumors of different malignant potential. Mutat. Res. 423, 91–101. 10.1016/S0027-5107(98)00229-2 [DOI] [PubMed] [Google Scholar]
- Ramus S. J., Harrington P. A., Pye C., DiCioccio R. A., Cox M. J., Garlinghouse-Jones K., et al. (2007). Contribution of BRCA1 and BRCA2 mutations to inherited ovarian cancer. Hum. Mutat. 28, 1207–1215. 10.1002/humu.20599 [DOI] [PubMed] [Google Scholar]
- Rathi A., Virmani A. K., Schorge J. O., Elias K. J., Maruyama R., Minna J. D., et al. (2002). Methylation profiles of sporadic ovarian tumors and nonmalignant ovaries from high-risk women. Clin. Cancer Res. 8, 3324–3331. [PubMed] [Google Scholar]
- Rattanapan Y., Korkiatsakul V., Kongruang A., Chareonsirisuthigul T., Rerkamnuaychoke B., Wongkularb A., et al. (2018). EGFL7 and RASSF1 promoter hypermethylation in epithelial ovarian cancer. Cancer Genet. 224–225, 37–40. 10.1016/j.cancergen.2018.04.117 [DOI] [PubMed] [Google Scholar]
- Reik W., Lewis A. (2005). Co-evolution of X-chromosome inactivation and imprinting in mammals. Nat. Rev. Genet. 6, 403–410. 10.1038/nrg1602 [DOI] [PubMed] [Google Scholar]
- Resnick K. E., Alder H., Hagan J. P., Richardson D. L., Croce C. M., Cohn D. E. (2009). The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol. Oncol. 112, 55–59. 10.1016/j.ygyno.2008.08.036 [DOI] [PubMed] [Google Scholar]
- Richon V. M., Sandhoff T. W., Rifkind R. A., Marks P. A. (2000). Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc. Natl. Acad. Sci. U.S.A. 97, 10014–10019. 10.1073/pnas.180316197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose S. L., Fitzgerald M. P., White N. O., Hitchler M. J., Futscher B. W., De Geest K., et al. (2006). Epigenetic regulation of maspin expression in human ovarian carcinoma cells. Gynecol. Oncol. 102, 319–324. 10.1016/j.ygyno.2005.12.025 [DOI] [PubMed] [Google Scholar]
- Rosen D. G., Wang L., Atkinson J. N., Yu Y., Lu K. H., Diamandis E. P., et al. (2005). Potential markers that complement expression of CA125 in epithelial ovarian cancer. Gynecol. Oncol. 99, 267–277. 10.1016/j.ygyno.2005.06.040 [DOI] [PubMed] [Google Scholar]
- Rosenthal A. N., Menon U., Jacobs I. J. (2006). Screening for ovarian cancer. Clin. Obstet. Gynecol. 49, 433–447. 10.1097/00003081-200609000-00004 [DOI] [PubMed] [Google Scholar]
- Salta S. P, Nunes S., Fontes-Sousa M., Lopes P., Freitas M., et al. (2018). A DNA methylation-based test for breast cancer detection in circulating cell-free DNA. J. Clin. Med. 7:420. 10.3390/jcm7110420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarojini S., Tamir A., Lim H., Li S., Zhang S., Goy A., et al. (2012). Early detection biomarkers for ovarian cancer. J. Oncol. 2012, 1–15. 10.1155/2012/709049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada K., Mitra A. K., Radjabi A. R., Bhaskar V., Kistner E. O., Tretiakova M., et al. (2008). Loss of E-cadherin promotes ovarian cancer metastasis via alpha 5-integrin, which is a therapeutic target. Cancer Res. 68, 2329–2339. 10.1158/0008-5472.CAN-07-5167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid G., Notaro S., Reimer D., Abdel-Azim S., Duggan-Peer M., Holly J., et al. (2016). Expression and promotor hypermethylation of miR-34a in the various histological subtypes of ovarian cancer. BMC Cancer 16:102. 10.1186/s12885-016-2135-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweigert F. J., Sehouli J. (2005). Transthyretin, a biomarker for nutritional status and ovarian cancer. Cancer Res. 65:1114; author reply 1114. [PubMed] [Google Scholar]
- Sellar G. C., Watt K. P., Rabiasz G. J., Stronach E. A., Li L., Miller E. P., et al. (2003). OPCML at 11q25 is epigenetically inactivated and has tumor-suppressor function in epithelial ovarian cancer. Nat. Genet. 34, 337–343. 10.1038/ng1183 [DOI] [PubMed] [Google Scholar]
- Shah C. A., Lowe K. A., Paley P., Wallace E., Anderson G. L., McIntosh M. W., et al. (2009). Influence of ovarian cancer risk status on the diagnostic performance of the serum biomarkers mesothelin, HE4, and CA125. Cancer Epidemiol. Biomarkers Prev. 18, 1365–1372. 10.1158/1055-9965.EPI-08-1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Albahrani M., Zhang W., Kufel C. N., James S. R., Odunsi K., et al. (2019). Epigenetic activation of POTE genes in ovarian cancer. Epigenetics 14, 185–197. 10.1080/15592294.2019.1581590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Kelly T. K., Jones P. A. (2010). Epigenetics in cancer. Carcinogenesis 31, 27–36. 10.1093/carcin/bgp220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D.-H., Chan K. Y.-K., Khoo U.-S., Ngan H. Y.-S., Xue W.-C., Chiu P.-M., et al. (2005). Epigenetic and genetic alterations of p33ING1b in ovarian cancer. Carcinogenesis 26, 855–863. 10.1093/carcin/bgi011 [DOI] [PubMed] [Google Scholar]
- Shibata D., Mori Y., Cai K., Zhang L., Yin J., Elahi A., et al. (2006). RAB32 hypermethylation and microsatellite instability in gastric and endometrial adenocarcinomas. Int. J. Cancer 119, 801–806. 10.1002/ijc.21912 [DOI] [PubMed] [Google Scholar]
- Shih I.-M., Kurman R. J. (2004). Ovarian tumorigenesis: a proposed model based on morphological and molecular genetic analysis. Am. J. Pathol. 164, 1511–1518. 10.1016/S0002-9440(10)63708-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivapurkar N., Toyooka S., Toyooka K. O., Reddy J., Miyajima K., Suzuki M., et al. (2004). Aberrant methylation of trail decoy receptor genes is frequent in multiple tumor types. Int. J. Cancer 109, 786–792. 10.1002/ijc.20041 [DOI] [PubMed] [Google Scholar]
- Si L., Jia Y., Lin R., Jian W., Yu Q., Yang S. (2019). 10.1016/j.biochi.2019.03.011. Arch. Biochem. Biophys. 668, 9–15. 10.1016/j.abb.2019.04.009 [DOI] [PubMed] [Google Scholar]
- Siegel R., Ward E., Brawley O., Jemal A. (2011). Cancer statistics, 2011: The impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J. Clin. 61, 212–236. 10.3322/caac.20121 [DOI] [PubMed] [Google Scholar]
- Siegel R. L., Miller K. D., Jemal A. (2015). Cancer Statistics, 2015. CA Cancer J. Clin. 65, 5–29. 10.3322/caac.21254 [DOI] [PubMed] [Google Scholar]
- Simmons A. R., Clarke C. H., Badgwell D. B., Lu Z., Sokoll L. J., Lu K. H., et al. (2016). Validation of a biomarker panel and longitudinal biomarker performance for early detection of ovarian cancer. Int. J. Gynecol. Cancer 26, 1070–1077. 10.1097/IGC.0000000000000737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon I., Katsaros D., Rigault de la Longrais I., Massobrio M., Scorilas A., Kim N. W., et al. (2007). B7-H4 is over-expressed in early-stage ovarian cancer and is independent of CA125 expression. Gynecol. Oncol. 106, 334–341. 10.1016/j.ygyno.2007.03.035 [DOI] [PubMed] [Google Scholar]
- Simon I., Zhuo S., Corral L., Diamandis E. P., Sarno M. J., Wolfert R. L., et al. (2006). B7-h4 is a novel membrane-bound protein and a candidate serum and tissue biomarker for ovarian cancer. Cancer Res. 66, 1570–1575. 10.1158/0008-5472.CAN-04-3550 [DOI] [PubMed] [Google Scholar]
- Socha M. J., Said N., Dai Y., Kwong J., Ramalingam P., Trieu V., et al. (2009). Aberrant Promoter Methylation of Sparc in Ovarian Cancer. Neoplasia 11, 126–135. 10.1593/neo.81146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino A., Liu C.-G., Addario A., Peschle C., Scambia G., Ferlini C. (2008). Role of microRNAs in drug-resistant ovarian cancer cells. Gynecol. Oncol. 111, 478–486. 10.1016/j.ygyno.2008.08.017 [DOI] [PubMed] [Google Scholar]
- Staub J., Chien J., Pan Y., Qian X., Narita K., Aletti G., et al. (2007). Epigenetic silencing of HSulf-1 in ovarian cancer:implications in chemoresistance. Oncogene 26, 4969–4978. 10.1038/sj.onc.1210300 [DOI] [PubMed] [Google Scholar]
- Strait K. A., Dabbas B., Hammond E. H., Warnick C. T., Iistrup S. J., Ford C. D. (2002). Cell cycle blockade and differentiation of ovarian cancer cells by the histone deacetylase inhibitor trichostatin A are associated with changes in p21, Rb, and Id proteins. Mol. Cancer Ther. 1, 1181–1190. [PubMed] [Google Scholar]
- Strathdee G., Appleton K., Illand M., Millan D. W., Sargent J., Paul J., et al. (2001). Primary ovarian carcinomas display multiple methylator phenotypes involving known tumor suppressor genes. Am. J. Pathol. 158, 1121–1127. 10.1016/S0002-9440(10)64059-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathdee G., Davies B. R., Vass J. K., Siddiqui N., Brown R. (2004). Cell type-specific methylation of an intronic CpG island controls expression of the MCJ gene. Carcinogenesis 25, 693–701. 10.1093/carcin/bgh066 [DOI] [PubMed] [Google Scholar]
- Strathdee G., Vass J. K., Oien K. A., Siddiqui N., Curto-Garcia J., Brown R. (2005). Demethylation of the MCJ gene in stage III/IV epithelial ovarian cancer and response to chemotherapy. Gynecol. Oncol. 97, 898–903. 10.1016/j.ygyno.2005.03.023 [DOI] [PubMed] [Google Scholar]
- Su F., Lang J., Kumar A., Ng C., Hsieh B., Suchard M. A., et al. (2007). Validation of candidate serum ovarian cancer biomarkers for early detection. Biomark. Insights 2, 369–375. 10.1177/117727190700200011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H.-Y., Lai H.-C., Lin Y.-W., Chou Y.-C., Liu C.-Y., Yu M.-H. (2009). An epigenetic marker panel for screening and prognostic prediction of ovarian cancer. Int. J. Cancer 124, 387–393. 10.1002/ijc.23957 [DOI] [PubMed] [Google Scholar]
- Su H.-Y., Lai H.-C., Lin Y.-W., Liu C.-Y., Chen C.-K., Chou Y.-C., et al. (2010). Epigenetic silencing of SFRP5 is related to malignant phenotype and chemoresistance of ovarian cancer through Wnt signaling pathway. Int. J. Cancer 127, 555–567. 10.1002/ijc.25083 [DOI] [PubMed] [Google Scholar]
- Suehiro Y., Okada T., Okada T., Anno K., Okayama N., Ueno K., et al. (2008). Aneuploidy predicts outcome in patients with endometrial carcinoma and is related to lack of CDH13 hypermethylation. Clin. Cancer Res. 14, 3354–3361. 10.1158/1078-0432.CCR-07-4609 [DOI] [PubMed] [Google Scholar]
- Sun N., Kohli A., Huang S., Chang T., Chao C. C. -K. (2019). Androgen receptor transcriptional activity and chromatin modifications on the ABCB1/MDR gene are critical for taxol resistance in ovarian cancer cells. J. Cell. Physiol. 234, 8760–8775. 10.1002/jcp.27535 [DOI] [PubMed] [Google Scholar]
- Sung C. K., Li D., Andrews E., Drapkin R., Benjamin T. (2013). Promoter methylation of the SALL2 tumor suppressor gene in ovarian cancers. Mol. Oncol. 7, 419–427. 10.1016/j.molonc.2012.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H. Y., Choi E. N., Lyu D., Park A. K., Ju W., Ahn J.-H. (2014a). Aberrant hypomethylation-mediated AGR2 overexpression induces an aggressive phenotype in ovarian cancer cells. Oncol. Rep. 32, 815–820. 10.3892/or.2014.3243 [DOI] [PubMed] [Google Scholar]
- Sung H. Y., Ju W., Ahn J.-H. (2014b). DNA hypomethylation-mediated overexpression of carbonic anhydrase 9 induces an aggressive phenotype in ovarian cancer cells. Yonsei Med. J. 55:1656. 10.3349/ymj.2014.55.6.1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H. Y., Park A. K., Ju W., Ahn J.-H. (2014c). overexpression of mucin 13 due to promoter methylation promotes aggressive behavior in ovarian cancer cells. Yonsei Med. J. 55:1206. 10.3349/ymj.2014.55.5.1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H. Y., Yang S.-D., Ju W., Ahn J.-H. (2017a). Aberrant epigenetic regulation of GABRP associates with aggressive phenotype of ovarian cancer. Exp. Mol. Med. 49:e335. 10.1038/emm.2017.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H. Y., Yang S.-D., Park A. K., Ju W., Ahn J.-H. (2017b). Aberrant hypomethylation of solute carrier family 6 member 12 promoter induces metastasis of ovarian cancer. Yonsei Med. J. 58:27. 10.3349/ymj.2017.58.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki F., Akahira J.-I., Miura I., Suzuki T., Ito K., Hayashi S.-I., et al. (2008). Loss of estrogen receptor beta isoform expression and its correlation with aberrant DNA methylation of the 5'-untranslated region in human epithelial ovarian carcinoma. Cancer Sci. 99, 2365–2372. 10.1111/j.1349-7006.2008.00988.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symer D. E., Connelly C., Szak S. T., Caputo E. M., Cost G. J., Parmigiani G., et al. (2002). Human l1 retrotransposition is associated with genetic instability in vivo. Cell 110, 327–338. 10.1016/S0092-8674(02)00839-5 [DOI] [PubMed] [Google Scholar]
- Tam K. F., Liu V. W. S., Liu S. S., Tsang P. C. K., Cheung A. N. Y., Yip A. M. W., et al. (2007). Methylation profile in benign, borderline and malignant ovarian tumors. J. Cancer Res. Clin. Oncol. 133, 331–341. 10.1007/s00432-006-0178-5 [DOI] [PubMed] [Google Scholar]
- Tang G., Guo J., Zhu Y., Huang Z., Liu T., Cai J., et al. (2018). Metformin inhibits ovarian cancer via decreasing H3K27 trimethylation. Int. J. Oncol. 52, 1899–1911. 10.3892/ijo.2018.4343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z., Qian M., Ho M. (2013). The role of mesothelin in tumor progression and targeted therapy. Anticancer Agents Med. Chem. 13, 276–280. 10.2174/1871520611313020014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T., Tischkowitz M., Ameziane N., Hodgson S. V., Mathew C. G., Joenje H., et al. (2003). Disruption of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian tumors. Nat. Med. 9, 568–574. 10.1038/nm852 [DOI] [PubMed] [Google Scholar]
- Tasaka R., Fukuda T., Shimomura M., Inoue Y., Wada T., Kawanishi M., et al. (2017). TBX2 expression is associated with platinum-sensitivity of ovarian serous carcinoma. Oncol. Lett. 15, 3085–3090. 10.3892/ol.2017.7719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D. D., Gercel-Taylor C. (2008). MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 110, 13–21. 10.1016/j.ygyno.2008.04.033 [DOI] [PubMed] [Google Scholar]
- Teodoridis J. M., Hall J., Marsh S., Kannall H. D., Smyth C., Curto J., et al. (2005). CpG island methylation of DNA damage response genes in advanced ovarian cancer. Cancer Res. 65, 8961–8967. 10.1158/0008-5472.CAN-05-1187 [DOI] [PubMed] [Google Scholar]
- Terasawa K., Sagae S., Toyota M., Tsukada K., Ogi K., Satoh A., et al. (2004). Epigenetic inactivation of TMS1/ASC in ovarian cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 10, 2000–2006. 10.1158/1078-0432.CCR-0932-03 [DOI] [PubMed] [Google Scholar]
- Teschendorff A. E., Gao Y., Jones A., Ruebner M., Beckmann M. W., Wachter D. L., et al. (2016). DNA methylation outliers in normal breast tissue identify field defects that are enriched in cancer. Nat. Commun. 7:10478. 10.1038/ncomms10478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff A. E., Jones A., Fiegl H., Sargent A., Zhuang J. J., Kitchener H. C., et al. (2012). Epigenetic variability in cells of normal cytology is associated with the risk of future morphological transformation. Genome Med. 4:24. 10.1186/gm323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff A. E., Lee S.-H., Jones A., Fiegl H., Kalwa M., Wagner W., et al. (2015). HOTAIR and its surrogate DNA methylation signature indicate carboplatin resistance in ovarian cancer. Genome Med. 7:108. 10.1186/s13073-015-0233-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff A. E., Widschwendter M. (2012). Differential variability improves the identification of cancer risk markers in DNA methylation studies profiling precursor cancer lesions. Bioinformatics 28, 1487–1494. 10.1093/bioinformatics/bts170 [DOI] [PubMed] [Google Scholar]
- Thomson J. P., Skene P. J., Selfridge J., Clouaire T., Guy J., Webb S., et al. (2010). CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature 464, 1082–1086. 10.1038/nature08924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomar T., Alkema N. G., Schreuder L., Meersma G. J., de Meyer T., van Criekinge W., et al. (2017). Methylome analysis of extreme chemoresponsive patients identifies novel markers of platinum sensitivity in high-grade serous ovarian cancer. BMC Med. 15:116. 10.1186/s12916-017-0870-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomar T., de Jong S., Alkema N. G., Hoekman R. L., Meersma G. J., Klip H. G., et al. (2016). Genome-wide methylation profiling of ovarian cancer patient-derived xenografts treated with the demethylating agent decitabine identifies novel epigenetically regulated genes and pathways. Genome Med. 8:107. 10.1186/s13073-016-0361-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torng P.-L., Lin C.-W., Chan M. W., Yang H.-W., Huang S.-C., Lin C.-T. (2009). Promoter methylation of IGFBP-3 and p53 expression in ovarian endometrioid carcinoma. Mol. Cancer 8:120. 10.1186/1476-4598-8-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toss A., Tomasello C., Razzaboni E., Contu G., Grandi G., Cagnacci A., et al. (2015). Hereditary ovarian cancer: not only BRCA 1 and 2 genes. BioMed Res. Int. 2015, 1–11. 10.1155/2015/341723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- U. S. Preventive Services Task Force (2010). The Guide to Clinical Preventive Services 2010 - 2011: Recommendations of the U.S. Preventive Services Task Force. Rockville, MD: Agency for Healthcare Research and Quality (US) Available online at: http://www.ncbi.nlm.nih.gov/books/NBK56707/ (accessed December 19, 2017). [PubMed] [Google Scholar]
- Valls E., Sánchez-Molina S., Martínez-Balbás M. A. (2005). Role of histone modifications in marking and activating genes through mitosis. J. Biol. Chem. 280, 42592–42600. 10.1074/jbc.M507407200 [DOI] [PubMed] [Google Scholar]
- Vang R., Shih I.-M., Kurman R. J. (2009). Ovarian low-grade and high-grade serous carcinoma: pathogenesis, clinicopathologic and molecular biologic features, and diagnostic problems. Adv. Anat. Pathol. 16, 267–282. 10.1097/PAP.0b013e3181b4fffa [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin I., Feng Z., Longton G., Ward D. C., Alvero A. B., Lai Y., et al. (2008). Diagnostic markers for early detection of ovarian cancer. Clin. Cancer Res. 14, 1065–1072. 10.1158/1078-0432.CCR-07-1569 [DOI] [PubMed] [Google Scholar]
- Vlassov V. V., Laktionov P. P., Rykova E. Y. (2010). Circulating nucleic acids as a potential source for cancer biomarkers. Curr. Mol. Med. 10, 142–165. 10.2174/156652410790963295 [DOI] [PubMed] [Google Scholar]
- Wang B., Yu L., Luo X., Huang L., Li Q., Shao X., et al. (2017). Detection of OPCML methylation, a possible epigenetic marker, from free serum circulating DNA to improve the diagnosis of early-stage ovarian epithelial cancer. Oncol. Lett. 14, 217–223. 10.3892/ol.2017.6111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Li X., Tian W., Wang Y., Wu D., Sun Z., et al. (2015). Promoter DNA methylation is associated with KLF11 expression in epithelial ovarian cancer: promoter Dna Methylation Is Associated With KLF11 Expression. Genes. Chromosomes Cancer 54, 453–462. 10.1002/gcc.22257 [DOI] [PubMed] [Google Scholar]
- Wang H.-Q., Yang C.-Y., Wang S.-Y., Wang T., Han J.-L., Wei K., et al. (2018). Cell-free plasma hypermethylated CASZ1, CDH13 and ING2 are promising biomarkers of esophageal cancer. J. Biomed. Res. 32, 424–433. 10.7555/JBR.32.20170065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Zhu M.-J., Ren A.-M., Wu H.-F., Han W.-M., Tan R.-Y., et al. (2014). A Ten-MicroRNA signature identified from a genome-wide MicroRNA expression profiling in human epithelial ovarian cancer. PLoS ONE 9:e96472. 10.1371/journal.pone.0096472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Ji G., Wu Q., Feng S., Zhao Y., Cao Z., et al. (2018). Integrated microarray meta-analysis identifies miRNA-27a as an oncogene in ovarian cancer by inhibiting FOXO1. Life Sci. 210, 263–270. 10.1016/j.lfs.2018.08.043 [DOI] [PubMed] [Google Scholar]
- Warren J. D., Xiong W., Bunker A. M., Vaughn C. P., Furtado L. V., Roberts W. L., et al. (2011). Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC Med. 9:133. 10.1186/1741-7015-9-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasenang W., Chaiyarit P., Proungvitaya S., Limpaiboon T. (2019). Serum cell-free DNA methylation of OPCML and HOXD9 as a biomarker that may aid in differential diagnosis between cholangiocarcinoma and other biliary diseases. Clin. Epigenetics 11:39. 10.1186/s13148-019-0634-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei K.-L., Chou J.-L., Chen Y.-C., Jin H., Chuang Y.-M., Wu C.-S., et al. (2019). Methylomics analysis identifies a putative STAT3 target, SPG20, as a noninvasive epigenetic biomarker for early detection of gastric cancer. PLoS ONE 14:e0218338. 10.1371/journal.pone.0218338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S. H. (2006). Prognostic DNA methylation biomarkers in ovarian cancer. Clin. Cancer Res. 12, 2788–2794. 10.1158/1078-0432.CCR-05-1551 [DOI] [PubMed] [Google Scholar]
- Wei S. H., Chen C.-M., Strathdee G., Harnsomburana J., Shyu C.-R., Rahmatpanah F., et al. (2002). Methylation microarray analysis of late-stage ovarian carcinomas distinguishes progression-free survival in patients and identifies candidate epigenetic markers. Clin. Cancer Res. 8, 2246–2252. [PubMed] [Google Scholar]
- Weichert W., Denkert C., Noske A., Darb-Esfahani S., Dietel M., Kalloger S. E., et al. (2008). Expression of class I histone deacetylases indicates poor prognosis in endometrioid subtypes of ovarian and endometrial carcinomas. Neoplasia 10, 1021–1027. 10.1593/neo.08474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenberger D. J., Trinh B. N., Campan M., Sharma S., Long T. I., Ananthnarayan S., et al. (2008). DNA methylation analysis by digital bisulfite genomic sequencing and digital MethyLight. Nucleic Acids Res. 36, 4689–4698. 10.1093/nar/gkn455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widschwendter A., Müller H. M., Hubalek M. M., Wiedemair A., Fiegl H., Goebel G., et al. (2004). Methylation status and expression of human telomerase reverse transcriptase in ovarian and cervical cancer. Gynecol. Oncol. 93, 407–416. 10.1016/j.ygyno.2004.01.036 [DOI] [PubMed] [Google Scholar]
- Widschwendter M., Apostolidou S., Jones A. A., Fourkala E. O., Arora R., Pearce C. L., et al. (2009). HOXA methylation in normal endometrium from premenopausal women is associated with the presence of ovarian cancer: a proof of principle study. Int. J. Cancer 125, 2214–2218. 10.1002/ijc.24599 [DOI] [PubMed] [Google Scholar]
- Widschwendter M., Jiang G., Woods C., Müller H. M., Fiegl H., Goebel G., et al. (2004). DNA hypomethylation and ovarian cancer biology. Cancer Res. 64, 4472–4480. 10.1158/0008-5472.CAN-04-0238 [DOI] [PubMed] [Google Scholar]
- Widschwendter M., Zikan M., Wahl B., Lempiäinen H., Paprotka T., Evans I., et al. (2017). The potential of circulating tumor DNA methylation analysis for the early detection and management of ovarian cancer. Genome Med. 9:116. 10.1186/s13073-017-0500-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox C. B., Baysal B. E., Gallion H. H., Strange M. A., DeLoia J. A. (2005). High-resolution methylation analysis of the BRCA1 promoter in ovarian tumors. Cancer Genet. Cytogenet. 159, 114–122. 10.1016/j.cancergencyto.2004.12.017 [DOI] [PubMed] [Google Scholar]
- Wilczynski M., Zytko E., Danielska J., Szymanska B., Dzieniecka M., Nowak M., et al. (2018). Clinical significance of miRNA-21,−103,−129,−150 in serous ovarian cancer. Arch. Gynecol. Obstet. 297, 741–748. 10.1007/s00404-018-4660-5 [DOI] [PubMed] [Google Scholar]
- Wiley A., Katsaros D., Chen H., Rigault de la Longrais I. A., Beeghly A., Puopolo M., et al. (2006a). Aberrant promoter methylation of multiple genes in malignant ovarian tumors and in ovarian tumors with low malignant potential. Cancer 107, 299–308. 10.1002/cncr.21992 [DOI] [PubMed] [Google Scholar]
- Wiley A., Katsaros D., Fracchioli S., Yu H. (2006b). Methylation of the insulin-like growth factor binding protein-3 gene and prognosis of epithelial ovarian cancer. Int. J. Gynecol. Cancer 16, 210–218. 10.1111/j.1525-1438.2006.00299.x [DOI] [PubMed] [Google Scholar]
- Wilting R. H., Dannenberg J.-H. (2012). Epigenetic mechanisms in tumorigenesis, tumor cell heterogeneity and drug resistance. Drug Resist. Updat. 15, 21–38. 10.1016/j.drup.2012.01.008 [DOI] [PubMed] [Google Scholar]
- Wittenberger T., Sleigh S., Reisel D., Zikan M., Wahl B., Alunni-Fabbroni M., et al. (2014). DNA methylation markers for early detection of women's cancer: promise and challenges. Epigenomics 6, 311–327. 10.2217/epi.14.20 [DOI] [PubMed] [Google Scholar]
- Woloszynska-Read A., James S. R., Link P. A., Yu J., Odunsi K., Karpf A. R. (2007). DNA methylation-dependent regulation of BORIS/CTCFL expression in ovarian cancer. Cancer Immun. 7, 21. [PMC free article] [PubMed] [Google Scholar]
- Woolas R. P., Xu F. J., Jacobs I. J., Yu Y. H., Daly L., Berchuck A., et al. (1993). Elevation of multiple serum markers in patients with stage I ovarian cancer. J. Natl. Cancer Inst. 85, 1748–1751. 10.1093/jnci/85.21.1748 [DOI] [PubMed] [Google Scholar]
- Wu Q., Lothe R. A., Ahlquist T., Silins I., Tropé C. G., Micci F., et al. (2007). DNA methylation profiling of ovarian carcinomas and their in vitro models identifies HOXA9, HOXB5, SCGB3A1, and CRABP1 as novel targets. Mol. Cancer 6:45. 10.1186/1476-4598-6-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurz K., Garcia R. L., Goff B. A., Mitchell P. S., Lee J. H., Tewari M., et al. (2010). MiR-221 and MiR-222 alterations in sporadic ovarian carcinoma: relationship to CDKN1B, CDKNIC and overall survival. Genes. Chromosomes Cancer. 49, 577–584. 10.1002/gcc.20768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman S. K., Parkin R. K., Mitchell P. S., Fritz B. R., O'Briant K., Godwin A. K., et al. (2009). Repertoire of microRNAs in epithelial ovarian cancer as determined by next generation sequencing of small RNA cDNA Libraries. PLoS ONE 4:e5311. 10.1371/journal.pone.0005311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Q.-P., Pu D.-H., Lu W.-N. (2017). Research on application value of combined detection of serum CA125, HE4 and TK1 in the diagnosis of ovarian cancer. Eur. Rev. Med. Pharmacol. Sci. 21, 4536–4541. [PubMed] [Google Scholar]
- Xu W., Lu J., Zhao Q., Wu J., Sun J., Han B., et al. (2019). Genome-wide plasma cell-free DNA methylation profiling identifies potential biomarkers for lung cancer. Dis. Markers 2019, 1–7. 10.1155/2019/4108474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y.-Z., Xi Q.-H., Ge W.-L., Zhang X.-Q. (2013). Identification of serum MicroRNA-21 as a biomarker for early detection and prognosis in human epithelial ovarian cancer. Asian Pac. J. Cancer Prev. 14, 1057–1060. 10.7314/APJCP.2013.14.2.1057 [DOI] [PubMed] [Google Scholar]
- Yang H., Kong W., He L., Zhao J.-J., O'Donnell J. D., Wang J., et al. (2008). MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 68, 425–433. 10.1158/0008-5472.CAN-07-2488 [DOI] [PubMed] [Google Scholar]
- Yang N., Kaur S., Volinia S., Greshock J., Lassus H., Hasegawa K., et al. (2008). MicroRNA microarray identifies Let-7i as a novel biomarker and therapeutic target in human epithelial ovarian cancer. Cancer Res. 68, 10307–10314. 10.1158/0008-5472.CAN-08-1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Chaerkady R., Beer M. A., Mendell J. T., Pandey A. (2009). Identification of miR-21 targets in breast cancer cells using a quantitative proteomic approach. Proteomics 9, 1374–1384. 10.1002/pmic.200800551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Qi W., Sun L., Zhou H., Zhou B., Hu Y. (2018). 10.1186/s13148-019-0634-0. Adv. Clin. Exp. Med. 28, 361–366. 10.17219/acem/84935 [DOI] [Google Scholar]
- Yap O. W. S., Bhat G., Liu L., Tollefsbol T. O. (2009). Epigenetic modifications of the Estrogen receptor beta gene in epithelial ovarian cancer cells. Anticancer Res. 29, 139–144. [PMC free article] [PubMed] [Google Scholar]
- Yarbro J. W., Page D. L., Fielding L. P., Partridge E. E., Murphy G. P. (1999). American Joint Committee on Cancer prognostic factors consensus conference. Cancer 86, 2436–2446. [DOI] [PubMed] [Google Scholar]
- Yazbek J., Raju S. K., Ben-Nagi J., Holland T. K., Hillaby K., Jurkovic D. (2008). Effect of quality of gynaecological ultrasonography on management of patients with suspected ovarian cancer: a randomised controlled trial. Lancet Oncol. 9, 124–131. 10.1016/S1470-2045(08)70005-6 [DOI] [PubMed] [Google Scholar]
- Yeh C.-M., Shay J., Zeng T.-C., Chou J.-L., Huang T. H.-M., Lai H.-C., et al. (2014). Epigenetic silencing of ARNTL, a circadian gene and potential tumor suppressor in ovarian cancer. Int. J. Oncol. 45, 2101–2107. 10.3892/ijo.2014.2627 [DOI] [PubMed] [Google Scholar]
- Yeh K.-T., Chen T.-H., Yang H.-W., Chou J.-L., Chen L.-Y., Yeh C.-M., et al. (2011). Aberrant TGFβ/SMAD4 signaling contributes to epigenetic silencing of a putative tumor suppressor, RunX1T1 in ovarian cancer. Epigenetics 6, 727–739. 10.4161/epi.6.6.15856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X., Guo J., Guo J., Sun S., Yang P., Wang J., et al. (2017). EZH2-mediated epigenetic silencing of TIMP2 promotes ovarian cancer migration and invasion. Sci. Rep. 7:3568. 10.1038/s41598-017-03362-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J. H., Dammann R., Pfeifer G. P. (2001). Hypermethylation of the CpG island of the RASSF1A gene in ovarian and renal cell carcinomas. Int. J. Cancer 94, 212–217. 10.1002/ijc.1466 [DOI] [PubMed] [Google Scholar]
- Yu J., Ryan D. G., Getsios S., Oliveira-Fernandes M., Fatima A., Lavker R. M. (2008). MicroRNA-184 antagonizes microRNA-205 to maintain SHIP2 levels in epithelia. Proc. Natl. Acad. Sci. U.S.A. 105, 19300–19305. 10.1073/pnas.0803992105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Fujii S., Yuan J., Luo R. Z., Wang L., Bao J., et al. (2003). Epigenetic regulation of ARHI in breast and ovarian cancer cells. Ann. N. Y. Acad. Sci. 983, 268–277. 10.1111/j.1749-6632.2003.tb05981.x [DOI] [PubMed] [Google Scholar]
- Zhang H., Zhang S., Cui J., Zhang A., Shen L., Yu H. (2008). Expression and promoter methylation status of mismatch repair gene hMLH1 and hMSH2 in epithelial ovarian cancer. Aust. N. Z. J. Obstet. Gynaecol. 48, 505–509. 10.1111/j.1479-828X.2008.00892.x [DOI] [PubMed] [Google Scholar]
- Zhang J., Ye F., Chen H., Ye D., Lu W., Xie X. (2006). [Deletion of OPCML gene and promoter methylation in ovarian epithelial carcinoma]. Zhongguo Yi Xue Ke Xue Yuan Xue Bao 28, 173–177. [PubMed] [Google Scholar]
- Zhang L., Huang J., Yang N., Greshock J., Megraw M. S., Giannakakis A., et al. (2006). microRNAs exhibit high frequency genomic alterations in human cancer. Proc. Natl. Acad. Sci. U.S.A. 103, 9136–9141. 10.1073/pnas.0508889103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Volinia S., Bonome T., Calin G. A., Greshock J., Yang N., et al. (2008). Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc. Natl. Acad. Sci. U.S.A. 105, 7004–7009. 10.1073/pnas.0801615105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Hu G., Yang Q., Dong R., Xie X., Ma D., et al. (2013). A multiplex methylation-specific PCR assay for the detection of early-stage ovarian cancer using cell-free serum DNA. Gynecol. Oncol. 130, 132–139. 10.1016/j.ygyno.2013.04.048 [DOI] [PubMed] [Google Scholar]
- Zhang S., Wei L., Zhang A., Zhang L., Yu H. (2009). RUNX3 gene methylation in epithelial ovarian cancer tissues and ovarian cancer cell lines. Omics J. Integr. Biol. 13, 307–311. 10.1089/omi.2009.0030 [DOI] [PubMed] [Google Scholar]
- Zhang W., Barger C. J., Eng K. H., Klinkebiel D., Link P. A., Omilian A., et al. (2016). PRAME expression and promoter hypomethylation in epithelial ovarian cancer. Oncotarget 7, 45352–45369. 10.18632/oncotarget.9977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Barger C. J., Link P. A., Mhawech-Fauceglia P., Miller A., Akers S. N., et al. (2015). DNA hypomethylation-mediated activation of Cancer/Testis Antigen 45 (CT45) genes is associated with disease progression and reduced survival in epithelial ovarian cancer. Epigenetics 10, 736–748. 10.1080/15592294.2015.1062206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Bast R. C., Yu Y., Li J., Sokoll L. J., Rai A. J., et al. (2004). Three biomarkers identified from serum proteomic analysis for the detection of early stage ovarian cancer. Cancer Res. 64, 5882–5890. 10.1158/0008-5472.CAN-04-0746 [DOI] [PubMed] [Google Scholar]
- Zhao L., Wang W., Xu L., Yi T., Zhao X., Wei Y., et al. (2019). Integrative network biology analysis identifies miR-508-3p as the determinant for the mesenchymal identity and a strong prognostic biomarker of ovarian cancer. Oncogene 38, 2305–2319. 10.1038/s41388-018-0577-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Yu C., Zhou S., Lau W. B., Lau B., Luo Z., et al. (2016). Epigenetic repression of PDZ-LIM domain-containing protein 2 promotes ovarian cancer via NOS2-derived nitric oxide signaling. Oncotarget 7, 1408–1420. 10.18632/oncotarget.6368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Chen S., Li L., Liu X., Liu X., Dai S., et al. (2018). Evaluation of HE4 and TTR for diagnosis of ovarian cancer: comparison with CA-125. J. Gynecol. Obstet. Hum. Reprod. 47, 227–230. 10.1016/j.jogoh.2018.03.010 [DOI] [PubMed] [Google Scholar]
- Zhou F., Tao G., Chen X., Xie W., Liu M., Cao X. (2014). Methylation of OPCML promoter in ovarian cancer tissues predicts poor patient survival. Clin. Chem. Lab. Med. 52, 735–742. 10.1515/cclm-2013-0736 [DOI] [PubMed] [Google Scholar]
- Zhou Q.-H., Zhao Y.-M., Jia L.-L., Zhang Y. (2017). Mir-595 is a significant indicator of poor patient prognosis in epithelial ovarian cancer. Eur. Rev. Med. Pharmacol. Sci. 21, 4278–4282. [PubMed] [Google Scholar]
- Zuberi M., Mir R., Das J., Ahmad I., Javid J., Yadav P., et al. (2015). Expression of serum miR-200a, miR-200b, and miR-200c as candidate biomarkers in epithelial ovarian cancer and their association with clinicopathological features. Clin. Transl. Oncol. 17, 779–787. 10.1007/s12094-015-1303-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.