Abstract
Many studies correlate changes in human gut microbiome with the onset of various diseases, mostly by 16S rRNA gene sequencing. Setting up the optimal sampling and DNA isolation procedures is crucial for robustness and reproducibility of the results. We performed a systematic comparison of several sampling and DNA isolation kits, quantified their effect on bacterial gDNA quality and the bacterial composition estimates at all taxonomic levels. Sixteen volunteers tested three sampling kits. All samples were consequently processed by two DNA isolation kits. We found that the choice of both stool sampling and DNA isolation kits have an effect on bacterial composition with respect to Gram-positivity, however the isolation kit had a stronger effect than the sampling kit. The proportion of bacteria affected by isolation and sampling kits was larger at higher taxa levels compared to lower taxa levels. The PowerLyzer PowerSoil DNA Isolation Kit outperformed the QIAamp DNA Stool Mini Kit mainly due to better lysis of Gram-positive bacteria while keeping the values of all the other assessed parameters within a reasonable range. The presented effects need to be taken into account when comparing results across multiple studies or computing ratios between Gram-positive and Gram-negative bacteria.
Subject terms: Next-generation sequencing, Bacterial genes, Metagenomics
Introduction
The gut microbiome plays a key role in shaping human health and has been the subject of an increasing number of studies in the context of disease development, diagnostics and treatment. Important progress has been made especially in investigating uncultured bacteria, which constitute the main part of the gut microbiome and were previously difficult to characterize with standard techniques such as cloning, Sanger sequencing or Denaturing Gradient Gel Electrophoresis (DGGE)1,2. Next generation sequencing (NGS) provides new and more detailed means to study the human microbiome and helps uncovering its impact on the human immune system development3–5, or on the development of chronic diseases6,7. However, human microbiome is very dynamic and can change rapidly in response to many factors such as diet, antibiotic use, lifestyle or environment8–16. Many diseases were associated with a phenomenon called dysbiosis – microbial imbalance. Unfortunately, due to the huge microbiome variability it is very difficult to define a normality baseline for an individual. To extract disease-relevant information and generate new or confirm existing biological hypotheses, large cohort microbiome studies are needed. These studies face multiple challenges with the microbiome sampling. First, successful compliance of participants with the established protocol demands both motivation and an easy sampling procedure. Especially, sampling of the stool at home can induce a “yuck effect” and positive education and uncomplicated sampling workflow can significantly decrease the number of study drop-outs17,18.
Another major problem is the large variability of methodological approaches employed by different microbiome studies. The final composition of bacteria as assessed by sequencing the 16S rRNA gene is influenced by many factors: sampling method19–22, sample storage conditions20,22–29, DNA extraction8,21,22,26,30–39, primers targeting different parts of the 16S rRNA gene40,41 and data analysis42. All of these factors may lead to the misinterpretation of changes in the microbiome and thus hamper direct comparisons of results between individual studies43–45. These technical problems, along with an as yet unknown gut microbiome diversity in the healthy population, lead to challenges in the implementation of metagenomics into cohort studies and, in consequence, delay the translation of the knowledge to clinical practice.
Most studies focused on the technical factors influencing the assessment of bacterial composition often provide only a description of the observed differences on a limited number of samples, while the comparison of the effect sizes of these factors, or combination thereof remains unexplored. The effect of sampling was previously described with respect to storage conditions (such as temperatures20,23,26,28,29, periods at room temperature20,24 or a presence and type of stabilizer19,21,22,27,28). None of these studies reported on the volunteers’ compliance or the differences in preprocessing steps specific to different sampling kits. Multiple studies describe the effect of stool homogenization prior DNA extraction25,46, but they only report its overall effect on the interindividual variation, without quantifying this effect at different bacterial taxon levels.
The DNA extraction method was highlighted as a critical factor influencing the observed bacterial composition39,47. Commercially available extraction kits use different lysis procedures such as enzymatic, chemical or mechanical bacterial cell disruption methods. Generally, the combination of enzymatic and mechanical disruption is recommended as more effective in the lysis of Gram-positive bacteria8,22,26,34,35,37,39. However, these DNA extraction comparison studies are limited to a rather small number of individuals (from 2 to 9) and none of them compared the kits in terms of DNA yield and quality, presence of PCR inhibitors, the human to bacterial DNA ratio, the efficiency of Gram-positive bacteria cell wall lysis and the observed bacterial composition at different taxa levels all at once.
The aim of our study was therefore to perform systematic assessment of effect of sampling and DNA isolation kits and their combinations on a full range of parameters of bacterial DNA quality, bacterial diversity and composition, with respect to user acceptance.
Results
We analyzed stool samples from sixteen volunteers. Each volunteer collected the samples from the same stool sample using three different sampling kits (SK): a stool container (SK1); a flocked swab (SK2) and a cotton swab (SK3). The DNA was extracted using two isolation kits PowerLyzer PowerSoil DNA Isolation Kit (PS) and QIAamp DNA Stool Mini Kit (QS) (see Methods), totaling 96 samples for the analysis.
Evaluation of user acceptance of the sampling kits
The participants were asked to select the best and the worst kit based on their ease of manipulation including the time spent using it. All 16 volunteers selected the stool container as the easiest to use and 13 out of 16 (81.25%) volunteers indicated the flocked swab as the worst sampling kit. We believe that the manipulation with cotton and flocked swabs is uncomfortable due to the small size and the necessity to insert the swab stick back into the tube without touching the tube wall. On the contrary, the stool container is easy to manipulate even for people with reduced motoric skills. In addition, the flocked swab is designed for sampling of liquid samples and the solid stool samples do not adhere on its synthetic fibers.
The effect of sampling and DNA isolation kits on the bacterial gDNA quality
DNA yield, purity and integrity
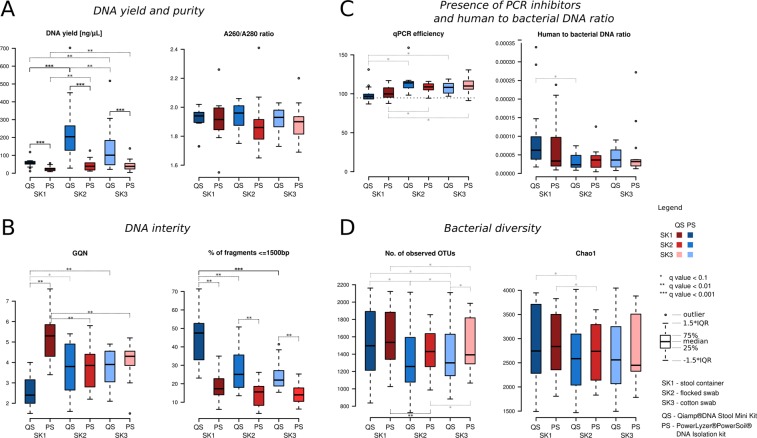
Significantly higher DNA yields were obtained with the QS isolation kit, regardless of the sampling kit used (q < 0.01) (Fig. 1, Supplementary Table S1). The median values of the A260/A280 ratio (the measure of purity of DNA) were well within the expected range (1.8–2) and did not differ significantly between the DNA isolation kits or between the sampling kits (Fig. 1, Supplementary Table S1).
Figure 1.
Comparison of sample DNA quality and diversity using different sampling and isolation kits. (A) DNA yield and purity comparison. dthe samples were five times diluted prior the DNA extraction (see Methods); (B) DNA integrity comparison; (C) Presence of PCR inhibitors and human to bacterial DNA ratio comparison. Horizontal dotted line represents median efficiency value of the positive control; (D) Bacterial diversity comparison.
The DNA integrity was determined using the GQN measure (on a scale from 1 to 10; low GQN indicates strongly degraded gDNA sample) and the proportion of short fragments (≤1500 bp; the larger the proportion the more degraded gDNA). We observed interaction effects of isolation and sampling kit for both DNA integrity measures. We found significantly lower proportion of short fragments when using the PS isolation kit (Fig. 1, Supplementary Table S1) and this difference was much larger when the stool container was used for sampling. There was no difference in GQN measure between the isolation kits when cotton or flocked swabs were used. However, for stool container samples, the QS kit provided much lower GQN values compared to the PS kit. These results point to worse DNA integrity for the QS isolation kit compared to the PS isolation kit mostly when stool container is used for sampling.
Presence of PCR inhibitors
The presence of PCR inhibitors in the samples decreases the sensitivity of the PCR reaction and even can lead to the impossibility of amplification of the selected region of 16S rRNA. It is usually measured by median efficiency values estimated from inhibition plots. Ideally, the efficiency should be 100%, meaning the template doubles in each cycle. Usually, the efficiency within 90–110% range is considered acceptable, where lower efficiency is caused by non-optimal reagent concentration or lower enzyme quality, while higher efficiency values are caused by the presence of PCR inhibitors. In our data, the efficiency values ranged from 96.7% to 114.0% (Fig. 1, Supplementary Table S2). In each of the isolation/sampling kit combinations, there were minimum two samples which exceeded the efficiency of 110%. The efficiency values of all isolation/sampling kit combinations, except for stool container samples after DNA isolation with the QS kit, were significantly increased compared to control samples without PCR inhibitors (efficiencymed = 94.7%). No difference in efficiency values was observed between the isolation kits. The samples from stool containers (regardless of the isolation kit used) contained less PCR inhibitors in comparison to all other sampling/DNA isolation kit combinations (significantly lower efficiency, Supplementary Table S2). We hypothesize that this sampling kit effect is due to the sample dilution step prior to the DNA isolation step.
Human to bacterial DNA ratio
In all samples, the quantity of human DNA was lower than that of the bacterial DNA (ranging from 2947x to 221239x, median 29369x, see Fig. 1, Supplementary Table S2). No difference was found between sampling/isolation kit combinations in terms of human to bacterial DNA ratio, except for the increased ratio in the stool container compared to flocked swab samples after isolation with the QS kit (q = 0.03).
The effect of sampling and DNA isolation kits on bacterial diversity and composition
Bacterial diversity
In total, 96 stool samples were sequenced. The number of reads after quality filtering and removal of chimeras ranged from 27680 to 67809, with median of 46192. We assessed the bacterial diversity using the number of observed OTUs and the Chao 1 diversity metric (Fig. 1, Supplementary Table S1). Overall, both diversity measures were independent of the DNA yield in all sampling/DNA isolation kit combinations.
While there was no difference in Chao 1 measure between the isolation kits, the number of observed OTUs was significantly increased after isolation with the PS kit, but only for cotton swab samples (q-value = 0.029). When comparing diversity measures between the sampling kits within each isolation kit separately, the stool container resulted in significantly higher number of observed OTUs in both DNA isolation kits (Fig. 1, Supplementary Table S1). In addition, we observed significantly higher number of OTUs in flocked swab samples compared to cotton swab samples after DNA isolation with the PS kit (q-value = 0.04) and significantly lower number of OTUs in flocked swab samples compared to cotton swab samples after DNA isolation with the QS kit (q-value = 0.09). For the Chao 1 diversity metric, significant differences were found in stool container samples compared to flocked swab samples in both PS and QS isolation kits (q = 0.04 and q = 0.09, respectively).
Bacterial composition
We identified 12,948 OTUs belonging to 13 bacterial phyla.
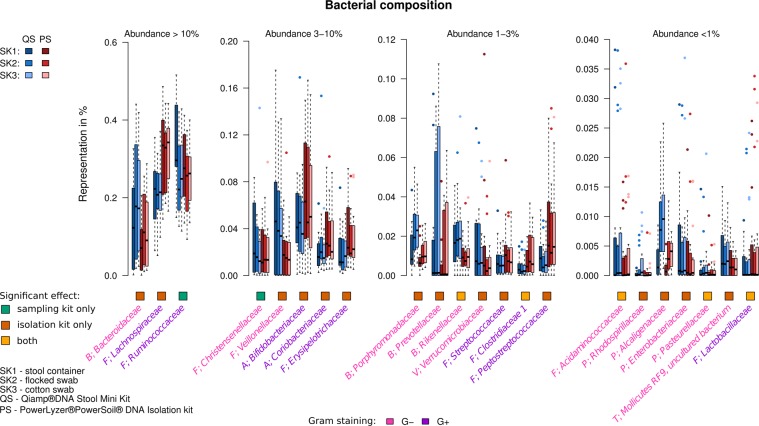
In order to quantify the effect of the sampling and isolation kits on bacterial composition, we performed mixed linear regression on each taxon that passed the filtering criteria (maximum abundance across all samples ≥1%) at all the seven taxonomical levels (phylum, class, order, family, genus, species, OTUs) separately. Interestingly, the proportion of taxa significantly affected by isolation or sampling kit differed between taxonomical levels (Fig. 2). The choice of sampling or DNA isolation kit affected 100% of taxa at phylum, class and order levels, and had decreasing trend from family to OTU level. The effects of sampling and isolation kits on the ten most abundant taxa at different taxa levels are summarized in Table 1 (see Supplementary Tables S3–S8 for complete results), the composition of significantly affected families is shown in Fig. 3. Overall, the choice of the isolation kit affected the abundance of more taxa than the choice of the sampling kit. In most of the cases where the taxa was affected by both factors, the p-values associated with the effect of the isolation kit were smaller than those of the sampling kit, indicating a more significant contribution of isolation kit to the overall model.
Figure 2.
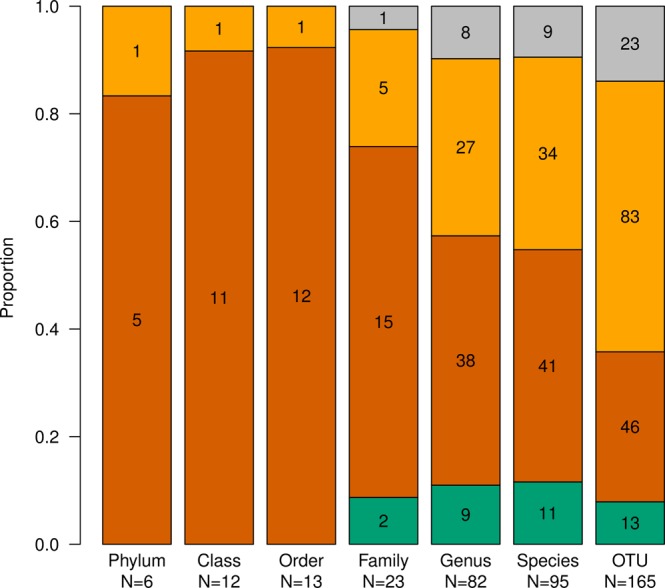
The proportion of taxa significantly affected by sampling or isolation kit at different taxonomical levels. Proportion of the tested taxa significantly affected by the sampling kit only (green), by the isolation kit only (dark yellow) and by both sampling and isolation kit (brick red). Grey indicates taxa not affected by sampling or isolation kit. The significance level was chosen at FDR < 10%, only taxa that met the selection criteria (maximum abundance >1%) were tested.
Table 1.
Summary of taxa at all levels and detailed results for top 10 taxa significantly affected by sampling or DNA isolation kit.
| Taxonomic level (# of all and significantly affected taxa) | Taxa (show ten most abundant) | q-value | Sign of the estimated effect size of the isolation or sampling kit | Relative abundance: total sum % | Gram stain | ||||
|---|---|---|---|---|---|---|---|---|---|
| isolation kit effect | sampling kit effect | PS to QS | SK2 to SK1 | SK3 to SK1 | SK3 to SK2 | ||||
|
Phylum All taxa: 14 Max >1% taxa: 6 Significantly affected taxa: 6 Isolation only: 1 Sampling only: 0 Both: 5 |
Firmicutes | 1.27E-15 | 4.43E-11 | + | − | − | + | 68.3 | G+ |
| Bacteroidetes | 3.42E-02 | 1.81E-02 | − | + | + | + | 18.5 | G− | |
| Actinobacteria | 3.67E-17 | 5.13E-04 | + | − | − | − | 7.1 | G+ | |
| Proteobacteria | 5.05E-08 | 3.47E-04 | − | + | + | + | 1.1 | G− | |
| Verrucomicrobia | 1.69E-03 | 2.39E-03 | − | − | − | + | 0.5 | G− | |
| Tenericutes | 2.08E-03 | 4.05E-01 | − | − | − | − | 0.1 | G− | |
|
Class All taxa: 30 Max >1% taxa: 12 Significantly affected taxa: 12 Isolation only: 1 Sampling only: 0 Both: 11 |
F; Clostridia | 2.66E-15 | 9.45E-12 | + | − | − | + | 61.2 | G+ |
| B; Bacteroidia | 1.02E-02 | 2.15E-02 | − | + | + | + | 18.5 | G− | |
| A; Actinobacteria | 5.80E-14 | 3.04E-04 | + | − | − | − | 4.6 | G+ | |
| F; Negativicutes | 2.58E-15 | 3.52E-04 | − | − | − | − | 2.9 | G−/var | |
| A; Coriobacteria | 5.80E-14 | 4.09E-03 | + | − | − | − | 2 | G+ | |
| F; Erysipelotrichia | 2.66E-15 | 8.57E-05 | + | − | − | + | 1.8 | G+ | |
| F; Bacilli | 2.58E-15 | 3.61E-05 | + | − | − | + | 1 | G+ | |
| V; Verrucomicrobiae | 1.94E-03 | 1.33E-06 | − | − | − | + | 0.5 | G− | |
| P; Betaproteobacteria | 2.14E-05 | 1.49E-09 | − | + | + | + | 0.4 | G− | |
| P; Gammaproteobacteria | 3.67E-07 | 7.22E-03 | − | − | − | + | 0.2 | G− | |
|
Order All taxa: 49 Max >1% taxa: 13 Significantly affected taxa: 13 Isolation only: 1 Sampling only: 0 Both: 12 |
F; Clostridiales | 9.46E-13 | 1.37E-11 | + | − | − | + | 61.2 | G+ |
| B; Bacteroidales | 4.58E-03 | 1.69E-02 | − | + | + | + | 18.5 | G− | |
| A; Bifidobacteriales | 5.63E-12 | 2.30E-03 | + | − | − | − | 4.5 | G+ | |
| F; Selenomonadales | 9.08E-17 | 2.42E-03 | − | − | − | + | 2.9 | G−/var | |
| A; Coriobacteriales | 5.73E-12 | 2.30E-02 | + | − | − | − | 2 | G+ | |
| F; Erysipelotrichales | 3.05E-13 | 2.16E-04 | + | − | − | + | 1.8 | G+ | |
| F; Lactobacillales | 3.05E-13 | 1.98E-04 | + | − | − | + | 1 | G+ | |
| V; Verrucomicrobiales | 3.29E-04 | 1.22E-05 | − | − | − | + | 0.5 | G− | |
| P; Burkholderiales | 1.33E-05 | 1.70E-09 | − | + | + | + | 0.4 | G− | |
| T; Mollicutes | 1.04E-05 | 6.43E-05 | − | − | − | + | 0.1 | G− | |
|
Family All taxa: 85 Max >1% taxa: 23 Significantly affected taxa: 22 Isolation only: 5 Sampling only: 2 Both: 15 |
F; Ruminococcaceae | 9.20E-01 | 6.81E-13 | − | + | + | + | 27.1 | G+ |
| F; Lachnospiraceae | 5.68E-20 | 1.60E-03 | + | + | + | + | 25.3 | G+ | |
| B; Bacteroidaceae | 7.55E-03 | 1.35E-02 | − | + | + | + | 10.2 | G− | |
| A; Bifidobacteriaceae | 5.90E-11 | 6.87E-03 | + | + | + | + | 4.5 | G+ | |
| F; Veillonellaceae | 1.90E-12 | 1.55E-04 | − | + | + | + | 2.4 | G+ | |
| A; Coriobacteriaceae | 7.68E-11 | 4.62E-02 | − | + | + | + | 2 | G+ | |
| F; Erysipelotrichaceae | 1.74E-12 | 8.08E-04 | − | + | + | + | 1.8 | G+ | |
| F; Christensenellaceae | 8.43E-01 | 1.24E-08 | − | + | + | + | 1.4 | G− | |
| B; Rikenellaceae | 5.90E-11 | 7.24E-01 | − | + | + | + | 1.3 | G− | |
| B; Porphyromonadaceae | 5.57E-04 | 4.03E-03 | − | + | + | + | 1.1 | G− | |
|
Genus All taxa: 277 Max >1% taxa: 82 Significantly affected taxa: 74 Isolation only: 27 Sampling only: 9 Both: 38 |
B; Bacteroides | 6.18E-03 | 1.54E-02 | − | + | + | + | 10.2 | G− |
| F; Faecalibacterium | 1.37E-02 | 9.70E-05 | + | − | − | + | 7.2 | G+ | |
| F; Blautia | 1.24E-24 | 1.05E-01 | + | − | − | + | 5 | G+ | |
| A; Bifidobacterium | 1.48E-10 | 2.48E-02 | + | − | − | − | 4.5 | G+ | |
| F; Subdoligranulum | 4.64E-03 | 3.18E-01 | − | − | − | + | 3.7 | G− | |
| F; Pseudobutyrivibrio | 9.63E-10 | 6.32E-01 | + | − | − | + | 2.8 | G− | |
| F; Dialister | 3.17E-09 | 5.86E-03 | − | − | − | + | 2.2 | G− | |
| F; Roseburia | 1.85E-02 | 4.95E-01 | + | − | − | + | 1.5 | G+ | |
| A; Collinsella | 8.59E-05 | 3.91E-01 | + | + | − | − | 1.4 | G+ | |
| F,Christensenellaceae R-7 group | 6.06E-01 | 1.50E-07 | − | − | − | − | 1.4 | G− | |
The significant q - values are shown in bold. SK1- stool container; SK2 – flocked swabs; SK3 - cotton swabs; PS – PowerLyzer PowerSoil DNA Isolation Kit; QS - QIAamp DNA Stool Mini Kit. All taxa – number of taxa found at the respective taxa level; Max >1% taxa – number of taxa that fulfilled the selection criteria for the analysis; Significantly affected taxa – the overall number of taxa at the respective taxa level affected by the isolation or sampling kit; Isolation only – number of taxa at the respective taxa level affected by the isolation kit only; Sampling only – number of taxa at the respective taxa level affected by the sampling kit only; Both – number of taxa at the respective taxa level affected by both sampling and isolation kit.
Figure 3.
Distributions of relative abundances of significantly affected taxa at family level. Four graphs represent families divided according to third quartile of their abundance. Only taxa that passed the filtering criteria (maximum abundance >1%), significantly affected by isolation or sampling kit are shown. The colored squares below the graph indicate whether the family was affected significantly by the sampling kit only, the isolation kit only or both.
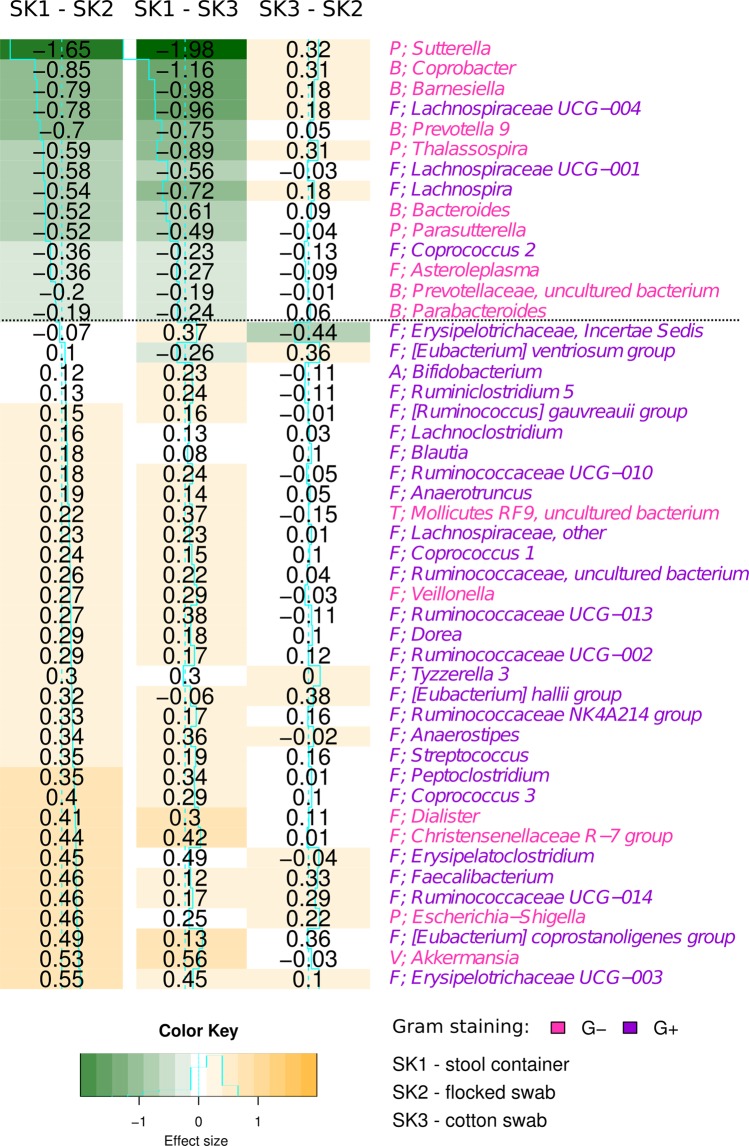
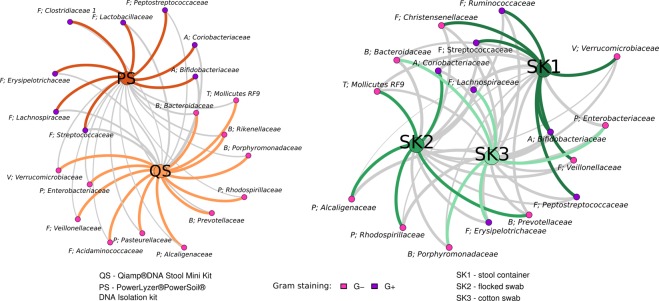
We hypothesized that the observed effect of the isolation kit was a result of different efficiency of the kit-specific bacterial cell walls lysis procedure. In this case, one of the kits would be more successful in isolating Gram-positive (G+) bacterial species. The Table 2 shows the numbers of significantly affected G+ taxa in all taxonomic levels and statistical pairwise comparison of their proportion after both isolation methods and all sampling methods. We found significantly higher proportions of G+ bacteria after the isolation using the PS kit at all the taxon levels. (96.4% to 100%, Table 2), compared to the QS isolation kit (G+ proportion varying from 0 to 44%). Similar observations were made for the effect of the sampling kit (Table 2), but this trend was not significant on any of the taxa levels except for the comparison of cotton swab (SK2) and stool container (SK1) on the genus level. We hypothesize that these differences are attributed to the dilution of the samples during the preprocessing steps specific to the stool container (see Methods for more details), resulting in lower sample density thus increasing the efficiency of the bead beating procedure. No difference in proportion of Gram-positive bacteria was found between flocked and cotton swabs. Figure 4 shows estimated effect sizes pairwise between the sampling kits on the genus level. Figure 5 visualizes bacteria with significant changes in abundance between isolation or sampling kits, with nodes colored according to Gram-positivity, where we can observe association of Gram-positive bacteria with the PS isolation kit.
Table 2.
Results of statistical testing of the proportion of G+ bacteria between significantly more abundant taxa within the selected isolation or sampling kit (pairwise).
| Phylum | Class | Order | Family | Genus | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample groups | Signif. more abundant | % of G+ phyla | q-val | % of G+ classes | q-val | % of G+ orders | q-val | % of G+ families | q-val | % of G+ geni | q-val |
| PS to QS | in PS | 100% (2/2) | 6.67E-02 | 100% (5/5) | 2.71E-03 | 100% (5/5) | 2.10E-03 | 100% (8/8) | 1.98E-05 | 96.4% (27/28) | 1.98E-05 |
| in QS | 0% (0/4) | 0% (0/6) | 0% (0/7) | 0% (0/12) | 44.1% (15/34) | ||||||
| SK2 to SK1 | in S1 | 66.7% (2/3) | 4.00E-01 | 71.4% (5/7) | 2.27E-01 | 62.5% (5/8) | 2.27E-01 | 58.3% (7/12) | 1.10E-01 | 80.8% (21/26) | 4.29E-02 |
| in S2 | 0% (0/2) | 0% (0/3) | 0% (0/3) | 0% (0/5) | 44.4% (4/12) | ||||||
| SK3 to SK1 | in S1 | 66.7% (2/3) | 4.00E-01 | 71.4% (5/7) | 2.27E-01 | 62.5% (5/8) | 2.27E-01 | 58.3% (7/12) | 1.10E-01 | 80.0% (20/25) | 1.10E-01 |
| in S3 | 0% (0/2) | 0% (0/3) | 0% (0/3) | 0% (0/5) | 38.5% (5/13) | ||||||
| SK3 to SK2 | in S3 | 25% (1/4) | 5.37E-01 | 37.5% (3/8) | 5.37E-01 | 33.3% (3/9) | 5.37E-01 | 35.7% (5/14) | 5.37E-01 | 60.7% (17/28) | 5.37E-01 |
| in S2 | 50% (1/1) | 100% (2/2) | 100% (2/2) | 66.7% (2/3) | 80.0% (8/10) | ||||||
The significant q – values are shown in bold. SK1- stool container; SK2 – flocked swabs; SK3 – cotton swabs; PS – PowerLyzer PowerSoil DNA Isolation Kit; QS – QIAamp DNA Stool Mini Kit. Sample groups – which pairwise comparison was performed; Signif. more abundant – in which group the taxa were significantly more abundant; % of G+ taxa – proportion of G+ in the significantly more abundant taxa within the respective group and level.
Figure 4.
Comparison of sampling kits effects at genus level. Each column corresponds to a pair of sampling kits and each row corresponds to a specific bacteria genus. The values represent log fold changes of bacterial abundances (effect size) between the sampling kits, color coded from green (less abundant) to orange (more abundant). Only significantly affected taxa are shown.
Figure 5.
Association of bacterial families significantly differentially abundant between different sampling and isolation kits. The strength of the edges is weighted by relative abundance of taxa between the different kits (the stronger the edge, the larger the difference). Color-coding of the edges highlights taxa belonging to the same community, as detected by network modularity (see Methods for details). Grey edges represent connections between different communities.
Discussion
The gut microbiome seems to be crucial factor influencing human health and to date, a number of different diseases were correlated with microbiome dysbiosis. Understanding the true role of microbiome and fully comprehending its variability will require many cohort studies and, most probably, comparison of their results in large-scale meta-analyses. As with any other scientific domain, the incoherent methodological approaches constitute an important obstacle for such comparisons44. In an attempt to elucidate some of the factors determining the success of such studies, we focused on the effects of sampling and DNA extraction methods on a number of relevant variables from DNA integrity to final bacterial composition at different taxa levels. For this purpose, we selected sampling and DNA isolation kits that are the most common and accessible and hence are probably the most relevant for majority of cohort studies.
Our group of sixteen healthy volunteers used three different sampling kits – stool container, flocked swabs and cotton swabs. Without exception, the stool container was indicated as the most acceptable by the volunteers. Moreover, stool in the container can be easily diluted, homogenized and aliquoted for different analyses. Unfortunately, the stool container is inconvenient for sampling diarrhea or baby stool. Importantly, as we discuss below, the pre-processing specific to stool container samples influences both DNA quality and bacterial composition and these effects seem to interact with the DNA isolation kit.
For measuring the effect of different DNA extraction procedures, we used PowerLyzer PowerSoil DNA Isolation Kit (PS) and QIAamp DNA Stool Mini Kit (QS).
While the PS kit cell-wall lysis procedure is based on combination of bead-beating step and enzymatic lysis, the standard protocol of the QS kit comprises only enzymatic lysis. Considering the fact that the beat-beating step leads to higher DNA yield and higher number of observed OTUs from difficult-to-lyse bacteria, we added the bead-beating step also into the QS protocol, as commonly recommended8,30,34,35,39.
DNA isolation by the QS kit resulted in significantly higher DNA yields compared to the PS kit (regardless of the sampling kit). Similar results were observed in other studies30,32. In agreement with previous studies30,35,37, we found no significant correlation between DNA yield and alpha diversity.
On the other hand, the PS kit produced DNA of better integrity, even though in the PS protocol we applied more rigorous mechanical lysis (or higher speed of bead beating), which, according to the literature, should result in more degraded DNA48. We hypothesize that the observed differences might be caused by another factor, such as the type of the beads (0.1 mm glass in PS vs 0.1 mm zirconia in QS), the buffer composition, or the incubation temperature. Overall, for preparation of the shotgun libraries or sequencing using third generation of sequencers, we consider DNA integrity to be more important factor than the DNA yield, which favors PS kit over the QS kit.
To properly homogenize the samples from the stool container, we included a preprocessing procedure comprising five times dilution. This naturally resulted in lower yields of isolated DNA, but after adjustment for this dilution we obtained higher final DNA concentrations compared to undiluted stool samples from flocked and cotton swabs. It seems that the dilution step also affected the DNA integrity. Compared to the undiluted samples from flocked and cotton swabs, stool container samples resulted in less degraded DNA after isolation using the PS kit and, in contrast, in more degraded DNA after isolation using the QS kit. Interestingly, two other independent studies, where different isolation kits were used, showed either a negative34 or a positive48 effect of sample dilution on the DNA integrity. This, together with our results leads us to conclude, that the effect of dilution step on DNA integrity is dependent on the isolation kit.
PCR inhibitors persisted in the DNA of the samples after isolation with both kits. Presence of PCR inhibitors could complicate the use of conventional molecular methods for the detection of low abundance or rare pathogenic microorganisms49,50. The dilution of stool container samples prior to processing has led to significantly lower proportion of PCR inhibitors, hence for some applications, this approach might be preferred.
Both DNA extraction kits isolated preferentially bacterial DNA, independently on the sampling kit used and the amount of human DNA was negligible. From practical point of view, there is no superiority of any of the DNA isolation vs sampling kit combinations with respect to amount of residual human DNA. Some of the studies, however, use these kits to estimate the concentration of human DNA in stool samples as an indicator of inflammation that might predict onset of certain bowel diseases51–55. From this perspective, based on our results, we do not consider these kits eligible for human DNA quantification.
As for the alpha diversity, we observed increased number of OTUs after DNA isolation with the PS kit in all sampling kits, but the difference was significant only for cotton swab samples. We observed significant differences in number of OTUs between all sampling kits combinations, with the stool container resulting in the highest number of OTUs. We attribute the observed differences to higher effectivity of bead beating process in the less dense samples (the dilution preprocessing step used for the stool container). This is in contrast with the results of Santiago et al.34, who report no changes in alpha diversity after sample dilution. In that study, however, a different isolation kit was used, so the results are not directly comparable.
The final bacterial composition was more affected by the choice of the DNA isolation kit than by the choice of the sampling kit. The preference of the PS isolation kit for Gram-positive bacteria was confirmed by statistical testing on all taxa levels and we believe that it is a result of more effective lysis of the Gram-positive cell wall bacteria when using the PS kit, despite the additional bead-beating step we introduced into the QS protocol. This is in agreement with previously published results8,26. It has to be taken into account, that Gram staining not always corresponds with the cell wall structure (e.g. Pseudobutyrivibrio56 or Deinococcus57, which is for many bacteria unknown. The efficiency of the lysis procedure can be as well influenced by atypical composition of the cell wall, presence of S-layer or capsules. The bacterial cell wall type also plays a role in the sampling effect: in our study it was associated with the dilution preprocessing step of the stool container, although less significantly.
There is a common belief that the effect of the individual is the most influential on the final bacterial composition8,32. Indeed, many metagenomic studies are reporting differences between groups of interest at the OTU level, where the effect of isolation and sampling is less important, as we showed in this study. However, some hypotheses are connecting particular disease with higher or lower bacterial abundance at the phylum or family level. An example is the commonly used Bacteroidetes/Firmicutes ratio58–64. Our results show, that this ratio is very dependent on both the selected DNA isolation method and sampling kit (dilution step). In our study, the PS kit and the dilution step (stool container) led to significantly higher proportion of e.g. Firmicutes (G+) and Actinobacteria (G+) and significantly lower proportion of Proteobacteria (G−) and Bacteroidetes (G−).
Another example of the cell wall structure effect is the Gram-positive genus Blautia. Blautia is a common and highly prevalent bacteria in the gastrointestinal tract, which is connected with healthy gut, since it is an effective short-chain fatty acid producer65,66. Lower abundance of Blautia in the gut is associated with many diseases66–73. In our study, Blautia was bacteria the most significantly affected by DNA isolation (across all the taxonomic levels). Similar observations were also described as the effect of isolation in other studies26,34.
The sampling kit (dilution effect) influenced most significantly the abundance of genus Sutterella, bacteria correlated with many diseases such as celiac diseases67, Down syndrome74, autism75 or irritable bowel syndrome76. Clearly, the dilution step represents an important batch effect, which raises a question, whether it is related only to the artificial dilution, or this effect could also be observed in diarrheic samples. The effect of stool consistency was described previously as an important factor12,77,78 influencing the bacterial composition, but this effect was not connected with effect of higher water content (dilution), rather with the transit time. As previously recommended77, we also suggest to control for the stool consistency as a potential confounding factor to avoid the effect of sample water content in this kind of studies, especially if one of the illness symptoms is diarrhea.
Despite the fact that the significance of the sampling and isolation dependent batch effects is repeatedly reported, no systematic study of these effects was performed yet on samples from larger numbers of individuals. Efforts for standardization of laboratory practices in metagenomics have been made in large international projects such as Metagenomic Research Group (MGRG), Genomic Standard Consortium (GSC), The Microbiome Quality Control Project (MBQC) and International Human Microbiome Standards (IHMS). IHMS recommends a procedure for fecal sample DNA extraction, based on study of Costea et al., where 21 extraction protocols were compared, including protocols similar to ours – protocol 3 (with PowerLyzer PowerSoil DNA Isolation Kit) and 11 (with QIAamp DNA Stool Mini Kit and bead beating step)39. They selected the protocol with QIAamp DNA Stool Mini Kit as the best choice for its accuracy and reproducibility. In contrast to our results, both protocol 3 and 11, provide good lysis of Gram-positive bacteria, but protocol 3 was excluded for insufficient DNA quality. The main difference between the studies is that the Costea study was based on the results of whole metagenomics sequencing and only compared bacterial composition annotated at the species level.
All these above mentioned studies and our results confirm that meta-analytical studies are extremely challenging due to the many sources of batch effects that need to be accounted for. Incorporation of a standardized mock community to the sequencing workflow, followed by normalization of the results to these reference values could be solution in future. The increased cost per run and slightly more complex library preparation is a small price to pay for robustness, consistency and comparability of results.
Conclusions
We performed systematic study of effects of DNA isolation and sampling kit on DNA quality and bacterial composition based on sequencing of gene for 16S rRNA on a the largest number of individuals to day (96 samples from 16 individuals).
We found significant effect of both DNA isolation and sampling kits on DNA purity, DNA integrity, alpha diversity and bacterial composition. Overall, the DNA isolation effect was stronger than that of the sampling kit. Interestingly the proportion of taxa affected by isolation or sampling was decreasing with decreasing taxonomical level.
We confirmed previously reported effect of DNA isolation kit on bacterial composition due to bacterial cell wall structure, namely the better efficacy of The PowerLyzer PowerSoil DNA Isolation Kit in lysis of Gram-positive bacteria. In addition, we report that the dilution pre-processing step of the stool container samples favored Gram-positive bacteria, although mostly at the genus level.
Both the choice of isolation and sampling kits significantly affected the Firmicutes to Bacteroidetes ratio. We conclude that the choice of DNA isolation and sampling kit (dilution step, and by extension the stool consistency) is an important batch effect that has to be taken into account mainly when comparing results between studies.
Methods
Sample collection
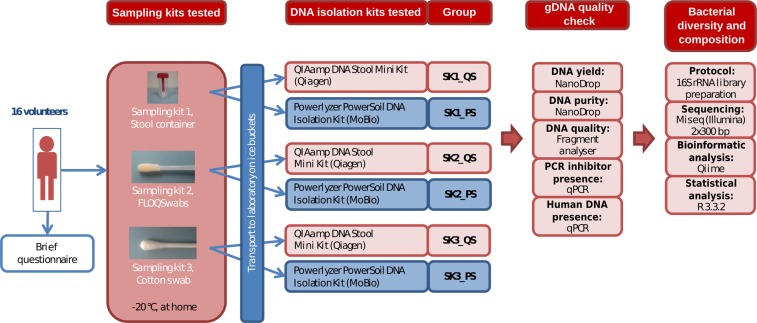
Stool samples were collected from a group of 16 volunteers. The subjects were 23–65 years old with an average age of 40.9 and none of them suffered from diarrhea during sample collection. Stool samples were collected at home. Volunteers received three stool sampling kits: sampling kit 1 (SK1) comprising 1x stool container (FL Medical, Italy); sampling kit 2 (SK2) comprising 2x flocked swabs (Copan, Italy) and sampling kit 3 (SK3) comprising 2x cotton swabs (SceneSafe, Great Britain). Sampling kits also contained disposable gloves and hand and surface disinfectant wipes for more convenient sampling. Each volunteer was instructed to collect all the samples from the same stool and from the same spot. Stool samples were then stored in a freezer at −20 °C overnight to freeze completely and the next day were transported on ice buckets to the laboratory, where they were stored at −20 °C prior to processing. Each group of samples was processed at the same time and by the same person. Participants filled out a brief questionnaire about satisfaction with individual sampling kits after stool sample collection. The study design is summarized in Fig. 6.
Figure 6.
Study design. Flowchart summarizing the study design and methods used.
This study was carried out in accordance with the recommendations of the ELSPAC Steering Committee of Masaryk University with written informed consent from all subjects. All subjects gave written informed consent in accordance with the Declaration of Helsinki. The protocols were approved by the ELSPAC Steering Committee of Masaryk University.
DNA extraction
Stool in the stool container (SK1) was diluted 5x with molecular grade water and homogenized by vortexing with Zirconia beads 2.3 mm (BioSpec, USA) to receive identical aliquots. This step is not necessary for the swabs, since each swab serves as an aliquot itself. Stool suspension (250 µl) was used for DNA extractions. Flocked swabs (SK2) and cotton swabs (SK3) were transferred into 2 ml tubes to be prepared for subsequent DNA extraction. DNA extractions were performed using a PowerLyzer PowerSoil DNA Isolation Kit (Mo Bio, USA) (PS) and QIAamp DNA Stool Mini Kit (Qiagen, USA) (QS) according to the manufacturer’s instructions.
Deviations from PS protocol:
750 µl of Bead Solution and 60 µl of C1 Solution were added to swab samples (SK2 and SK3) after defrosting. Samples were thoroughly vortexed and centrifuged for 4 min at 36,220 RCF. The swabs were then removed. Next, the samples were homogenized using the FastPrep-24 (MP Biomedicals, USA) 45 s 6.5 m/s.
Deviations from QS protocol:
A homogenization step with 0.1 mm zirconia beads (BioSpec, USA) was added to the protocol after the third step (i.e. after the suspension was heated for 5 min at 95 °C).
1.4 µl Buffer ASL was added to swab samples (SK2 and SK3) after defrosting. Samples were vortexed continuously for 1 min and the suspension was heated for 5 min at 70 °C. Next, the samples were homogenized using the FastPrep-24 (MP Biomedicals, USA) 45 s 5.5 m/s.
Evaluation of DNA yield, purity and quality
The final yield of extracted DNA was determined spectrophotometrically using theNanoDropND-1000 (Thermo Fisher SCIENTIFIC, USA). The purity of extracted DNA was indicated by an A260/A280 nm ratio. The quality of extracted DNA was assessed using the Fragment Analyzer (Advanced Analytical Technologies, USA) and High Sensitivity Genomic DNA Analysis Kit (Advanced Analytical Technologies, USA). The percentage of short fragments (≤1,500 bp) and Genomic Quality Number (GQN threshold of 10,000 bp) were calculated by PROSize 2.0 (Advanced Analytical Technologies, USA). Extracted DNA from each sample was diluted approximately to 5 ng/µl, aliquoted and stored at −20 °C. Aliquots were subsequently used in all further methods as starting material.
Presence of PCR inhibitors after different DNA extractions
The presence of inhibitors was tested by qPCR. A primer pair specific for the conservative regions of 16S rRNA gene (Table 3) was used. qPCR was performed on the TOptical Thermocycler (Analytik Jena - Biometra, Ireland) using a KAPA SYBR FAST qPCR Kit (Kapa Biosystems, USA). Cycling conditions are described in Table 2. Melting temperature was determined after PCR to verify the correctness of each PCR product. Extracted DNA from four different isolates of Escherichia coli DH10B served as a positive control without PCR inhibitors. Each extracted DNA from sample and positive control (concentration approximately 5 ng/µl) was diluted three times (10x, 100x, 1,000x). The subsequent qPCR reactions were performed using both diluted and undiluted samples. Inhibition plots were created from Ct values and efficiency (=10(−1/slope)−1) was calculated for each sample and positive control.
Table 3.
Primers and cycling conditions used in this study.
| Target region/gene | Amplicon size | Primer name | Primer Sequences (5′ → 3′) | Cycling conditions | Reference | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 16S rRNA gene (bacterial DNA) | 146 bp | q16S-univF | GTGSTGCAYGGYTGTCGTCA | 95 °C | 45x | 95 °C | 53 °C | 72 °C | Maeda et al.90 | |
| q16S-univR | ACGTCRTCCMCACCTTCCTC | 10 min | 20 s | 30 s | 20 s | — | ||||
| GAPDH (human DNA) | 74 bp | TGCACCACCAACTGCTTAGC | 95 °C | 40x | 95 °C | 65 °C | Vandesompele et al.91 | |||
| GGCATGGACTGTGGTCATGAG | 10 min | 10 s | 60 s | — | — | |||||
| V3/V4 16S rRNA gene (library preparation) | ~460 bp | s16S_F | TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG-InnerTag-CCTACGGGNGGCWGCAG | 95 °C | 25x | 95 °C | 55 °C | 72 °C | 72 °C | Klindworth et al.79 |
| s16S_R | GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG- InnerTag-GACTACHVGGGTATCTAATCC | 3 min | 30 s | 30 s | 30 s | 5 min | ||||
Proportion of human DNA to bacterial DNA after different DNA extractions
The ratio of human and bacterial DNA in samples was tested by qPCR. Bacterial DNA was assessed using a primer pair specific for the conservative regions of 16S rRNA gene and human DNA using a primer pair specific for protein kinase (Table 3). qPCRwas performed on the TOptical Thermocycler (Analytik Jena - Biometra, Ireland) with KAPA SYBR FAST qPCR Kit (Kapa Biosystems, USA). Cycling conditions are described in Table 3. Melting temperature was determined after PCR to verify the correctness of each PCR product. The amount of human DNA to bacterial DNA was calculated as 2ΔCt. Ct value of 40 was used for all samples under the limit of detection.
PCR amplification and Illumina library preparation
Extracted DNA was used as a template in amplicon PCR to target the hypervariable V3 and V4 regions of the bacterial 16S rRNA gene. The 16S metagenomics library was prepared according to the Illumina 16S Metagenomic sequencing Library Preparation protocol with some deviations described below (for workflow diagram see Supplementary Fig. S1). Each PCR was performed in triplicate, with the primer pair consisting of Illumina overhang nucleotide sequences, an inner tag and gene-specific sequences79. The Illumina overhang served to ligate the Illumina index and adapter. Each inner tag, i.e. a unique sequence of 7–9 bp, was designed to differentiate samples into groups. Primer sequences and PCR cycling conditions are summarized in Table 3. After PCR amplification, triplicates were pooled and the amplified PCR products were determined by gel electrophoresis. PCR clean-up was performed with Agencourt AMPure XP beads (Beckman Coulter Genomics, USA). Samples with different inner tags were equimolarly pooled based on fluorometrically measured concentration using Qubit dsDNA HS Assay Kit (Invitrogen, USA) and microplate reader Synergy Mx (BioTek, USA). Pools were used as a template for a second PCR with Nextera XT indexes (Illumina, USA). Differently indexed samples were quantified using the KAPA Library Quantification Complete Kit (Kapa Biosystems, USA) and equimolarly pooled according to the measured concentration. The prepared library was checked with a 2100 Bioanalyzer Instrument (Agilent Technologies, USA) and concentration was measured with qPCR shortly before sequencing. The library was diluted to a final concentration of 8 pM and 20% of PhiX DNA (Illumina, USA) was added. Sequencing was performed with the Miseq reagent kit V3 using a MiSeq. 2000 instrument according to the manufacturer’s instructions (Illumina, USA).
Data analysis
Forward and reverse pair-end reads, that fulfilled the condition of both quality and length filtering, were merged using the fastq-join method within the join_pair_ends.py command in QIIME 1.9.180. Data were demultiplexed and barcodes and primers were trimmed using package Biostrings81 in R 3.3.282. Operational taxonomic units (OTUs) were constructed by binding sequences into clusters of greater than 97% sequence similarity using QIIME. In the next step, chimeras were detected on the set of representative sequences of each OTU with UCHIME in USEARCH v6.1.54483. These chimera OTUs were subsequently excluded from the analysis. Taxonomy was assigned to each OTU based on SILVA 123 reference database84. The observed species metric and the Chao1 index were used to estimate alpha diversity for each sample in QIIME. Beta diversity was computed in QIIME using both weighted and unweighted UniFrac metrics85. All statistical analysis was performed in R 3.3.282.
The data were treated as compositional (proportions of total read count in each sample, non-rarefied) and prior to all statistical analyses were transformed using centered log-ratio transformation86. The analyses were performed on each of the seven taxonomy levels (Phylum, Class, Order, Family, Genus, Species and OTUs) separately and the resulting p-values were adjusted for multiple hypothesis testing using Benjamini-Hochberg procedure. Results were considered significant at FDR = 10%. The adjusted p-values are referred to as q-values.
To estimate the effects of isolation and sampling kits on bacterial composition while accounting for repeated measurements (effect of individual), we applied linear mixed model with sampling and izolation kits as fixed effects and individual as random effect (intercept). Log-likelihood test was performed to detect significance of each of the fixed effects – each time we compared the full model to the model without the fixed effect of interest.
A non-parametric Wilcoxon paired test, was used for comparison of effect of isolation kits on DNA quality. We used Spearman’s rank order correlation coefficient to discover the strength of the link between the number of observed species and DNA concentration.
Bipartite networks were used to visualize the influence of different kits on detection of Gram-positive and Gram-negative bacteria. These networks were reconstructed according to Sedlar et al.87 using R 3.3.2 and visualized in Gephi 0.9.288,89. Communities within networks were extracted using modularity optimization criterion88.
Supplementary information
Acknowledgements
We acknowledge the help of the volunteers enrolled in this study. This study was funded by the Ministry of Education, Youth and Sports of the Czech Republic and European Structural and Investment Funds (CETOCOEN PLUS project: CZ.02.1.01/0.0/0.0/15_003/0000469 and the RECETOX research infrastructure: LM2015051 and CZ.02.1.01/0.0/0.0/16_013/0001761) and by the Ministry of Health, the Czech Republic (FNBr, 65269705).
Author Contributions
P.V. and E.B. designed the study, drafted the manuscript and interpreted the results; Kr.S. and L.M. processed the samples in the laboratory; B.Z. and Ka.S. performed bioinformatic analysis; E.B. and V.P. performed statistical data analysis. All authors read and approved the final manuscript.
Data Availability
Sequencing data were uploaded to the European Nucleotide Archive under accession number PRJEB24411. Read counts per sample at different taxa levels and sample information table are available in Supplementary Files S9–S11.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-49520-3.
References
- 1.Suau A, et al. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 1999;65:4799–807. doi: 10.1128/aem.65.11.4799-4807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zoetendal EG, et al. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl. Environ. Microbiol. 2002;68:3401–7. doi: 10.1128/AEM.68.7.3401-3407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Russell SL, et al. Perinatal antibiotic treatment affects murine microbiota, immune responses and allergic asthma. Gut Microbes. 2013;4:158–64. doi: 10.4161/gmic.23567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jandhyala SM, et al. Role of the normal gut microbiota. World J. Gastroenterol. 2015;21:8787–803. doi: 10.3748/wjg.v21.i29.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matamoros S, Gras-Leguen C, Le Vacon F, Potel G, De La Cochetiere MF. Development of intestinal microbiota in infants and its impact on health. Trends in Microbiology. 2013;21:167–173. doi: 10.1016/j.tim.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 6.Underwood MA. Intestinal dysbiosis: Novel mechanisms by which gut microbes trigger and prevent disease. Prev. Med. (Baltim). 2014;65:133–137. doi: 10.1016/j.ypmed.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y-J, et al. Impacts of gut bacteria on human health and diseases. Int. J. Mol. Sci. 2015;16:7493–519. doi: 10.3390/ijms16047493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mackenzie BW, Waite DW, Taylor MW. Evaluating variation in human gut microbiota profiles due to DNA extraction method and inter-subject differences. Front. Microbiol. 2015;6:130. doi: 10.3389/fmicb.2015.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abeles SR, et al. Microbial diversity in individuals and their household contacts following typical antibiotic courses. Microbiome. 2016;4:39. doi: 10.1186/s40168-016-0187-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korpela K, de Vos W. Antibiotic use in childhood alters the gut microbiota and predisposes to overweight. Microb. Cell. 2016;3:296–298. doi: 10.15698/mic2016.07.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhernakova A, et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352:565–9. doi: 10.1126/science.aad3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graf D, et al. Contribution of diet to the composition of the human gut microbiota. Microb. Ecol. Health Dis. 2015;26:26164. doi: 10.3402/mehd.v26.26164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorvitovskaia A, Holmes SP, Huse SM. Interpreting Prevotella and Bacteroides as biomarkers of diet and lifestyle. Microbiome. 2016;4:15. doi: 10.1186/s40168-016-0160-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Claus SP, Guillou H, Ellero-Simatos S. The gut microbiota: a major player in the toxicity of environmental pollutants? npj Biofilms Microbiomes. 2016;2:16003. doi: 10.1038/npjbiofilms.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madan JC, Farzan SF, Hibberd PL, Karagas MR. Normal neonatal microbiome variation in relation to environmental factors, infection and allergy. Curr. Opin. Pediatr. 2012;24:753–9. doi: 10.1097/MOP.0b013e32835a1ac8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schultze A, et al. Comparison of stool collection on site versus at home in a population-based study. Bundesgesundheitsblatt - Gesundheitsforsch. - Gesundheitsschutz. 2014;57:1264–1269. doi: 10.1007/s00103-014-2051-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feigelson HS, et al. Feasibility of self-collection of fecal specimens by randomly sampled women for health-related studies of the gut microbiome. BMC Res. Notes. 2014;7:204. doi: 10.1186/1756-0500-7-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loftfield E, et al. Comparison of collection methods for fecal samples for discovery metabolomics in epidemiologic studies. Cancer Epidemiol. Biomarkers Prev. 2016;25:1483–1490. doi: 10.1158/1055-9965.EPI-16-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tedjo DI, et al. The effect of sampling and storage on the fecal microbiota composition in healthy and diseased subjects. PLoS One. 2015;10:e0126685. doi: 10.1371/journal.pone.0126685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathay C, et al. Method Optimization for Fecal Sample Collection and Fecal DNA Extraction. Biopreserv. Biobank. 2015;13:79–93. doi: 10.1089/bio.2014.0031. [DOI] [PubMed] [Google Scholar]
- 22.Panek M, et al. Methodology challenges in studying human gut microbiota – effects of collection, storage, DNA extraction and next generation sequencing technologies. Sci. Rep. 2018;8:5143. doi: 10.1038/s41598-018-23296-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lauber CL, Zhou N, Gordon JI, Knight R, Fierer N. Effect of storage conditions on the assessment of bacterial community structure in soil and human-associated samples. FEMS Microbiol. Lett. 2010;307:80–86. doi: 10.1111/j.1574-6968.2010.01965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cardona. Storage conditions of intestinal microbiota matter in metagenomic analysis. BMC Microbiol. 12, 158 (2012). [DOI] [PMC free article] [PubMed]
- 25.Gorzelak MA, et al. Methods for improving human gut microbiome data by reducing variability through sample processing and storage of stool. PLoS One. 2015;10:e0134802. doi: 10.1371/journal.pone.0134802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maukonen J, Simões C, Saarela M. The currently used commercial DNA-extraction methods give different results of clostridial and actinobacterial populations derived from human fecal samples. FEMS Microbiol. Ecol. 2012;79:697–708. doi: 10.1111/j.1574-6941.2011.01257.x. [DOI] [PubMed] [Google Scholar]
- 27.Hill CJ, et al. Effect of room temperature transport vials on DNA quality and phylogenetic composition of faecal microbiota of elderly adults and infants. Microbiome. 2016;4:19. doi: 10.1186/s40168-016-0164-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choo JM, Leong LEX, Rogers GB. Sample storage conditions significantly influence faecal microbiome profiles. Sci. Rep. 2015;5:16350. doi: 10.1038/srep16350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim D, et al. Optimizing methods and dodging pitfalls in microbiome research. Microbiome. 2017;5:52. doi: 10.1186/s40168-017-0267-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan S, Cohen DB, Ravel J, Abdo Z, Forney LJ. Evaluation of Methods for the Extraction and Purification of DNA from the Human Microbiome. PLoS One. 2012;7:e33865. doi: 10.1371/journal.pone.0033865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Janabi AHD, Kerkhof LJ, McGuinness LR, Biddle AS, McKeever KH. Comparison of a modified phenol/chloroform and commercial-kit methods for extracting DNA from horse fecal material. J. Microbiol. Methods. 2016;129:14–19. doi: 10.1016/j.mimet.2016.07.019. [DOI] [PubMed] [Google Scholar]
- 32.Kennedy NA, et al. The impact of different DNA extraction kits and laboratories upon the assessment of human gut microbiota composition by 16S rRNA gene sequencing. PLoS One. 2014;9:e88982. doi: 10.1371/journal.pone.0088982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith B, Li N, Andersen AS, Slotved HC, Krogfelt KA. Optimising bacterial DNA extraction from faecal samples: comparison of three methods. Open Microbiol. J. 2011;5:14–7. doi: 10.2174/1874285801105010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santiago A, et al. Processing faecal samples: a step forward for standards in microbial community analysis. BMC Microbiol. 2014;14:112. doi: 10.1186/1471-2180-14-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerasimidis K, et al. The effect of DNA extraction methodology on gut microbiota research applications. BMC Res. Notes. 2016;9:365. doi: 10.1186/s13104-016-2171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Claassen S, et al. A comparison of the efficiency of five different commercial DNA extraction kits for extraction of DNA from faecal samples. J. Microbiol. Methods. 2013;94:103–110. doi: 10.1016/j.mimet.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salonen A, et al. Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: Effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J. Microbiol. Methods. 2010;81:127–134. doi: 10.1016/j.mimet.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 38.Lim MY, Song E-J, Kim SH, Lee J, Nam Y-D. Comparison of DNA extraction methods for human gut microbial community profiling. Syst. Appl. Microbiol. 2018;41:151–157. doi: 10.1016/j.syapm.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 39.Costea PI, et al. Towards standards for human fecal sample processing in metagenomic studies. Nat. Biotechnol. 2017;35:1069–1076. doi: 10.1038/nbt.3960. [DOI] [PubMed] [Google Scholar]
- 40.Walker AW, et al. 16S rRNA gene-based profiling of the human infant gut microbiota is strongly influenced by sample processing and PCR primer choice. Microbiome. 2015;3:26. doi: 10.1186/s40168-015-0087-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cruaud P, et al. Influence of DNA Extraction Method, 16S rRNA Targeted Hypervariable Regions, and Sample Origin on Microbial Diversity Detected by 454 Pyrosequencing in Marine Chemosynthetic Ecosystems. Appl. Environ. Microbiol. 2014;80:4626–4639. doi: 10.1128/AEM.00592-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schloss PD, Gevers D, Westcott SL. Reducing the Effects of PCR Amplification and Sequencing Artifacts on 16S rRNA-Based Studies. PLoS One. 2011;6:e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clooney AG, et al. Comparing Apples and Oranges?: Next Generation Sequencing and Its Impact on Microbiome Analysis. PLoS One. 2016;11:e0148028. doi: 10.1371/journal.pone.0148028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lozupone CA, et al. Meta-analyses of studies of the human microbiota. Genome Res. 2013;23:1704–1714. doi: 10.1101/gr.151803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu BC, et al. Characterization of the gut microbiome in epidemiologic studies: the multiethnic cohort experience. Ann. Epidemiol. 2016;26:373–379. doi: 10.1016/j.annepidem.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsieh Y-H, et al. Impact of Different Fecal Processing Methods on Assessments of Bacterial Diversity in the Human Intestine. Front. Microbiol. 2016;7:1643. doi: 10.3389/fmicb.2016.01643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–30. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bürgmann H, Pesaro M, Widmer F, Zeyer J. A strategy for optimizing quality and quantity of DNA extracted from soil. J. Microbiol. Methods. 2001;45:7–20. doi: 10.1016/S0167-7012(01)00213-5. [DOI] [PubMed] [Google Scholar]
- 49.Schrader C, Schielke A, Ellerbroek L, Johne R. PCR inhibitors - occurrence, properties and removal. J. Appl. Microbiol. 2012;113:1014–1026. doi: 10.1111/j.1365-2672.2012.05384.x. [DOI] [PubMed] [Google Scholar]
- 50.Oikarinen S, et al. PCR inhibition in stool samples in relation to age of infants. J. Clin. Virol. 2009;44:211–214. doi: 10.1016/j.jcv.2008.12.017. [DOI] [PubMed] [Google Scholar]
- 51.Lewis JD, et al. Inflammation, Antibiotics, and Diet as Environmental Stressors of the Gut Microbiome in Pediatric Crohn’s Disease. Cell Host Microbe. 2015;18:489–500. doi: 10.1016/j.chom.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.TEIXEIRA Y, et al. Human dna quantification in the stools of patients with colorectal cancer. Arq. Gastroenterol. 2015;52:293–298. doi: 10.1590/S0004-28032015000400008. [DOI] [PubMed] [Google Scholar]
- 53.Varela E, et al. Faecal DNA and calprotectin as biomarkers of acute intestinal toxicity in patients undergoing pelvic radiotherapy. Aliment. Pharmacol. Ther. 2009;30:175–85. doi: 10.1111/j.1365-2036.2009.04019.x. [DOI] [PubMed] [Google Scholar]
- 54.Zou H, Harrington JJ, Klatt KK, Ahlquist DA. A sensitive method to quantify human long DNA in stool: relevance to colorectal cancer screening. Cancer Epidemiol. Biomarkers Prev. 2006;15:1115–9. doi: 10.1158/1055-9965.EPI-05-0992. [DOI] [PubMed] [Google Scholar]
- 55.Klaassen CHW, et al. Quantification of human DNA in feces as a diagnostic test for the presence of colorectal cancer. Clin. Chem. 2003;49:1185–7. doi: 10.1373/49.7.1185. [DOI] [PubMed] [Google Scholar]
- 56.Hespell RB, Kato K, Costerton JW. Characterization of the cell wall of Butyrivibrio species. Can. J. Microbiol. 1993;39:912–921. doi: 10.1139/m93-139. [DOI] [PubMed] [Google Scholar]
- 57.Thompson BG, Murray RG. Isolation and characterization of the plasma membrane and the outer membrane of Deinococcus radiodurans strain Sark. Can. J. Microbiol. 1981;27:729–34. doi: 10.1139/m81-111. [DOI] [PubMed] [Google Scholar]
- 58.Karlsson FH, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 59.Li E, et al. Inflammatory Bowel Diseases Phenotype, C. difficile and NOD2 Genotype Are Associated with Shifts in Human Ileum Associated Microbial Composition. PLoS One. 2012;7:e26284. doi: 10.1371/journal.pone.0026284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gevers D, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gao Z, Guo B, Gao R, Zhu Q, Qin H. Microbiota disbiosis is associated with colorectal cancer. Front. Microbiol. 2015;6:20. doi: 10.3389/fmicb.2015.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–31. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 63.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 64.Larsen N, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One. 2010;5:e9085. doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tanaka S, Yamamoto K, Yamada K, Furuya K, Uyeno Y. Relationship of Enhanced Butyrate Production by Colonic Butyrate-Producing Bacteria to Immunomodulatory Effects in Normal Mice Fed an Insoluble Fraction of Brassica rapa L. Appl. Environ. Microbiol. 2016;82:2693–9. doi: 10.1128/AEM.03343-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Murri M, et al. Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case-control study. BMC Med. 2013;11:46. doi: 10.1186/1741-7015-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cheng J, et al. Duodenal microbiota composition and mucosal homeostasis in pediatric celiac disease. BMC Gastroenterol. 2013;13:113. doi: 10.1186/1471-230X-13-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One. 2012;7:e39743. doi: 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hong P-Y, Croix JA, Greenberg E, Gaskins HR, Mackie RI. Pyrosequencing-based analysis of the mucosal microbiota in healthy individuals reveals ubiquitous bacterial groups and micro-heterogeneity. PLoS One. 2011;6:e25042. doi: 10.1371/journal.pone.0025042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bajaj JS, et al. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 2012;303:G675–85. doi: 10.1152/ajpgi.00152.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schnabl B, Brenner DA. Interactions between the intestinal microbiome and liver diseases. Gastroenterology. 2014;146:1513–1524. doi: 10.1053/j.gastro.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Org E, et al. Relationships between gut microbiota, plasma metabolites, and metabolic syndrome traits in the METSIM cohort. Genome Biol. 2017;18:70. doi: 10.1186/s13059-017-1194-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lippert K., Kedenko L., Antonielli L., Kedenko I., Gemeier C., Leitner M., Kautzky-Willer A., Paulweber B., Hackl E. Gut microbiota dysbiosis associated with glucose metabolism disorders and the metabolic syndrome in older adults. Beneficial Microbes. 2017;8(4):545–556. doi: 10.3920/BM2016.0184. [DOI] [PubMed] [Google Scholar]
- 74.Biagi E, et al. Gut microbiome in Down syndrome. PLoS One. 2014;9:e112023. doi: 10.1371/journal.pone.0112023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang L, et al. Increased abundance of Sutterella spp. and Ruminococcus torques in feces of children with autism spectrum disorder. Mol. Autism. 2013;4:42. doi: 10.1186/2040-2392-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mukhopadhya I, et al. A Comprehensive Evaluation of Colonic Mucosal Isolates of Sutterella wadsworthensis from Inflammatory Bowel Disease. PLoS One. 2011;6:e27076. doi: 10.1371/journal.pone.0027076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vandeputte D, et al. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut. 2016;65:57–62. doi: 10.1136/gutjnl-2015-309618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Falony G, et al. Population-level analysis of gut microbiome variation. Science. 2016;352:560–4. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 79.Klindworth A, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen HM, Lifschitz CH. Preparation of fecal samples for assay of volatile fatty acids by gas-liquid chromatography and high-performance liquid chromatography. Clin. Chem. 1989;35:74–76. [PubMed] [Google Scholar]
- 81.Pagés, H., Aboyout, P., Gentleman, R. & Biostrings, D. S. String objects representing biological sequences, and matching algorithms (2016).
- 82.R Core Team (2016). R: A language and environment for statistical computing. (R Foundation for Statistical Computing, Vienna, Austria, 2016).
- 83.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pruesse E, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lozupone C, Knight R. UniFrac: a New Phylogenetic Method for Comparing Microbial Communities. Appl. Environ. Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Aitchison, J. (John). The statistical analysis of compositional data. (Chapman and Hall, 1986).
- 87.Sedlar, K., Videnska, P., Skutkova, H., Rychlik, I. & Provaznik, I. Bipartite graphs for visualization analysis of microbiome data. Evol. Bioinforma. 12 (2016). [DOI] [PMC free article] [PubMed]
- 88.Blondel VD, Guillaume J-L, Lambiotte R, Lefebvre E. Fast unfolding of communities in large networks. J. Stat. Mech. Theory Exp. 2008;2008:P10008. doi: 10.1088/1742-5468/2008/10/P10008. [DOI] [Google Scholar]
- 89.Bastian, M., Heymann, S. & Jacomy, M. Gephi: An Open Source Software for Exploring and Manipulating Networks Visualization and Exploration of Large Graphs.
- 90.Maeda Hiroshi, Fujimoto Chiyo, Haruki Yasuhiro, Maeda Takemasa, Kokeguchi Susumu, Petelin Millan, Arai Hideo, Tanimoto Ichiro, Nishimura Fusanori, Takashiba Shogo. Quantitative real-time PCR using TaqMan and SYBR Green forActinobacillus actinomycetemcomitans,Porphyromonas gingivalis,Prevotella intermedia,tetQgene and total bacteria. FEMS Immunology & Medical Microbiology. 2003;39(1):81–86. doi: 10.1016/S0928-8244(03)00224-4. [DOI] [PubMed] [Google Scholar]
- 91.Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, RESEARCH0034 (2002). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data were uploaded to the European Nucleotide Archive under accession number PRJEB24411. Read counts per sample at different taxa levels and sample information table are available in Supplementary Files S9–S11.