Abstract
The fungal cell wall is a structure in constant contact with the external environment. It confers shape to the cell and protects it from external threats. During host adaptation, the cell wall structure of fungal pathogens is continuously reshaped by the orchestrated action of numerous genes. These genes respond to environmental stresses and challenging growth conditions, influencing the infective potential of the fungus. Here, we aimed to identify cell wall biosynthesis-related genes that putatively encode virulence factors in Trichophyton rubrum. We used RNA-seq to examine the impact of two drugs, namely undecanoic acid, and acriflavine as well as the effects of the carbon source switching from glucose to keratin on T. rubrum cell wall metabolism. By using functional annotation based on Gene Ontology terms, we identified significantly differentially expressed cell wall-related genes in all stress conditions. We also exposed T. rubrum to osmotic and other cell wall stressors and evaluated the susceptibility and gene modulation in response to stress. The changes in the ambient environment caused continuous cell wall remodeling, forcing the fungus to undergo modulatory restructuring. The influence of the external challenges indicated a highly complex response pattern. The genes that were modulated simultaneously in the three stress conditions highlight potential targets for antifungal development.
Keywords: ambient stress, dermatophyte, drug targets, host-pathogen interactions, RNA-seq
Introduction
The fungal cell wall directly interacts with the external environment and is vital to the survival of the organism. Its structural integrity is actively modulated in response to external and internal stresses. The cell wall supports fungal growth and development and enables the organism to endure hostile ambient conditions (Perlin, 2015; Valiante et al., 2015).
Pathogenic fungi use their cell walls to sense the host milieu and detect nutrients there. By modulating their cell wall components, fungal pathogens escape host immunity and invade the organism (Muszewska et al., 2017; Beauvais and Latge, 2018). Dermatophytes are fungal pathogens that infect keratinized tissues and dead epidermis in humans and animals. They produce several cell wall components that prevent them from being recognized by the host and function as virulence factors. Genome sequencing revealed that LysM binding domains are abundant in dermatophytes and probably mask pathogen cell wall components to confound the host immune response (Martinez et al., 2012; Kar et al., 2019). Also, the enrichment of several chitinase-encoding gene domains produces proteins supporting pathogen growth on a wide variety of substrates including soil and human skin. Thus, they enhance fungal infectiveness (Martinez et al., 2012). Differential protease secretion intensifies the inflammatory response induced by dermatophyte infection (Vermout et al., 2008). Moreover, through the production of extracellular vesicles the host innate immunity can be modulated (Bitencourt et al., 2018).
The constituents of the cell wall of the dermatophyte Trichophyton rubrum determine pathogen virulence. Mannans are associated with lymphocyte inhibition (Blake et al., 1991), a subtilisin homolog induces both immediate and delayed host immune responses (Woodfolk et al., 2000), and LysM proteins hide chitin and glucans from the host immune system (Kar et al., 2019). However, little is known about the relationships among dermatophyte pathogenesis, cell wall biosynthesis, and cell wall morphology.
Fungal virulence is determined by the coordinated expression of various genes mediating host-fungus interactions. Identification of the specific gene products or pathways crucial for fungal survival and infectiveness may potentially direct the development of new antifungal therapeutics (Bok et al., 2005; Martinez-Rossi et al., 2017). Changes in the ambient environment and external stressors continuously remodel the cell wall. Therefore, elucidation of the cell wall-related genes modulated in response to external challenges may disclose potential targets for antifungal drug development.
The cell wall is highly pertinent to antifungal drug discovery as it has a unique polysaccharide composition. For example, echinocandins inhibit β-(1,3)-D-glucan synthase biosynthesis in fungal cell walls. Thus, it is important to broaden our knowledge of fungal cell wall structure and the pathways regulating its formation (Grover, 2010; Perlin, 2015; Beauvais and Latge, 2018). Cell wall modulation in response to stressors may reveal putative targets for antifungal drug development (Martinez-Rossi et al., 2018).
The present study aimed to establish the modulation profile of cell wall genes in response to ambient challenges in the dermatophyte T. rubrum. We evaluated RNA-seq results under stress conditions wherein the pathogen was exposed to two drugs, or forced to switch its carbon source from glucose to keratin. Keratin mimics the host environment, and the drugs chosen, acriflavine and undecanoic acid, present a non-specific antifungal activity against T. rubrum. We identified a wide range of genes putatively encoding the virulence factors of T. rubrum. Those genes whose expression levels were altered under all stress conditions were considered as possible targets for antifungal drug development. We also challenged T. rubrum with various osmotic and cell wall stressor agents and evaluated their effects on the modulation of different genes controlling cell wall morphology.
Materials and Methods
T. rubrum Strain and Culture Conditions for RNA-seq
T. rubrum strain CBS118892 (Centraalbureau voor Schimmelcultures, Netherlands) was maintained on malt extract agar (MEA; 2% (w/v) glucose, 2% (w/v) malt extract, 0.1% (w/v) peptone, pH 5.7) for 17 d at 28°C. Approximately 106 conidia mL–1, obtained as previously described (Fachin et al., 1996; Jacob et al., 2012), were inoculated into 100 mL Sabouraud dextrose broth (SDB; 2% (w/v) glucose, 1% (w/v) peptone) and incubated at 28°C for 96 h under agitation; one flask was used for each time point of each tested condition (keratin or glucose). The mycelia in each flask were aseptically filtered and transferred into a new flask containing 100 mL minimal medium (MM) at pH 5.0 (Cove, 1966) supplemented with 70 mM nitrate (Sigma Aldrich Corp., St. Louis, MO, United States) and either 50 mM glucose (Sigma Aldrich Corp., St. Louis, MO, United States) (control) or 0.5% (w/v) bovine keratin (treatment) as the carbon source. All experiments were performed in three biological replicates. The cultures were incubated under agitation at 28°C for 24 h, 48 h, or 96 h. Mycelia were collected and the RNA was extracted from them. Drug-related library data are available in the GEO database under accession nos. GSE102872 and GSE40425; these results were obtained through the exposure of 96 h-grown SDB mycelia to 1.75 μg mL–1 acriflavine (ACF; Sigma Aldrich Corp., St. Louis, MO, United States) (Persinoti et al., 2014) or 17.5 μg mL–1 undecanoic acid (UDA; Sigma Aldrich Corp., St. Louis, MO, United States) (Mendes et al., 2018) in RPMI 1640 medium (Thermo Fisher Scientific, Waltham, MA, United States). T. rubrum susceptibility, as previously described, was determined by assessing MIC using the microdilution approach (M38-A) proposed by the Clinical and Laboratory Standards Institute (CLSI) (Persinoti et al., 2014; Mendes et al., 2018).
Stress Conditions
Susceptibility of T. rubrum to stressors was evaluated using plates containing SDB medium (1.5% (w/v) agar) supplemented with KCl (0.5 M), NaCl (0.5 M), SDS (0.01% (w/v)), sorbitol (1.2 M), Congo Red (CR; 200 μg mL–1), or Calcofluor White (CFW; 200 μg mL–1). Radial growth was measured using the diameter of propagated mycelia in centimeters on the 6th day of development. Data correspond to the means of three measurements. For qRT-PCR, mycelia grown for 96 h in flasks containing SDB were filtered, transferred, and incubated for 1 or 3 h in SDB containing the afore mentioned stressor agents. After incubation, mycelia from each experiment were frozen in liquid nitrogen, stored at −80°C, and used for RNA extraction.
RNA Isolation, Sequencing, and Data Analysis
Total RNA was isolated from ∼100 mg mycelia using an Illustra RNAspin mini isolation kit (GE Healthcare, Chicago, IL, United States). The RNA concentrations were determined using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States). RNA quality was validated with an Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA, United States). Equal amounts of RNA from three independent biological replicates of T. rubrum keratin/glucose cultures at 24, 48, or 96 h were used for the synthesis of cDNA with the TruSeq RNA library Kit (Illumina, San Diego, CA, United States), and sequenced with a Hiseq 2000 sequencer (Illumina, San Diego, CA, United States) according to the manufacturer’s instructions. Paired-end reads 150 bp in size were generated. Raw read data obtained via RNA-seq were filtered for quality control by FastQC tool and trimmed with Trimmomatic (Bolger et al., 2014) to remove adapters and Illumina-specific sequences. Trimmed paired-end reads from each sample were aligned to the T. rubrum reference genome1 with STAR aligner (Dobin et al., 2013). Gene-level read counts were quantified with STAR’s ‘-quantModeGeneCounts’ parameter. Differential expression was analyzed with the DESeq2 Bioconductor package (Love et al., 2014). A Benjamini-Hochberg correction (Benjamini and Hochberg, 1995) adjusted the P threshold and was applied to reveal statistically significant changes in gene expression levels. It was set to 0.05, with a log2 fold change ± 1.5 and postulated as a significantly modulated transcript abundance level. Genes surpassing these thresholds are hereinafter referred to as differentially expressed genes (DEG). They were functionally categorized with the Gene Ontology (GO) terms assigned by the Blast2GO algorithm (Robinson et al., 2011; Thorvaldsdottir et al., 2012). Highly represented categories were determined by enrichment analysis with the BayGO algorithm (Vencio et al., 2006). After functional annotation analysis, the cell wall-related genes in T. rubrum RNA-seq were identified with the R script mapping modulated Genes and Gene Ontology terms. A customized R script detected the descendant terms of the GO cellular component ‘cell wall’ with the Bioconductor GO.db package (Carlson, 2018). The genes are listed in Supplementary Table S1. Nine of these genes were arbitrarily selected to validate the results obtained through RNA-seq.
cDNA Synthesis, and qRT-PCR Analysis
Total RNA was treated with DNase I (Sigma Aldrich Corp., St. Louis, MO, United States) to remove residual genomic DNA. The complementary DNA (cDNA) was generated with a high-capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA, United States). The qRT-PCR was performed with a StepOnePlus Real-Time PCR system (Applied Biosystems, Foster City, CA, United States). Gene expression was analyzed to confirm keratin RNA-seq and evaluate transcriptional responses to osmotic- and cell wall stressors. Reactions were run in a total volume of 12.5 μL with Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, United States) and 50 ng template cDNA. Primer sequences, concentrations, and reaction efficiencies are listed in Supplementary Table S2. Glyceraldehyde-3-phosphate dehydrogenase (gapdh) and DNA-dependent RNA polymerase II (rpb 2) were used as internal controls (Jacob et al., 2012). Data were derived from three independent replicates. The 2–ΔΔct relative expression quantification method was used to calculate gene responsiveness (Livak and Schmittgen, 2001). Data were statistically analyzed with Student’s t-test (RNA-seq validation) or one-way ANOVA followed by Tukey’s post hoc test. The statistical software was GraphPad Prism v. 5.1 (GraphPad Software, La Jolla, CA, United States).
Results
Global DEG Identification in Response to Keratin
For comprehensive analysis of the T. rubrum genes expressed in response to keratin, we performed high-throughput sequencing (RNA-seq) using glucose as the control. Approximately 192 million high-quality reads generated ∼167 million mapped paired-end sequences (Supplementary Table S3). The dataset comprised 2,797 genes that were modulated in response to keratin relative to glucose. The upregulated and downregulated transcripts were defined using a 1.5-fold change cutoff (≥ 2.8-fold difference and a stringent statistical significance threshold of P < 0.05). We used the Blast2Go tool (Gotz et al., 2008) to depict the functional distribution of the modulated genes. This information elucidated the functionality of the DEGs in response to changing growth conditions.
Identification of the Genes Associated With Cell Wall Synthesis in the RNA-seq Libraries
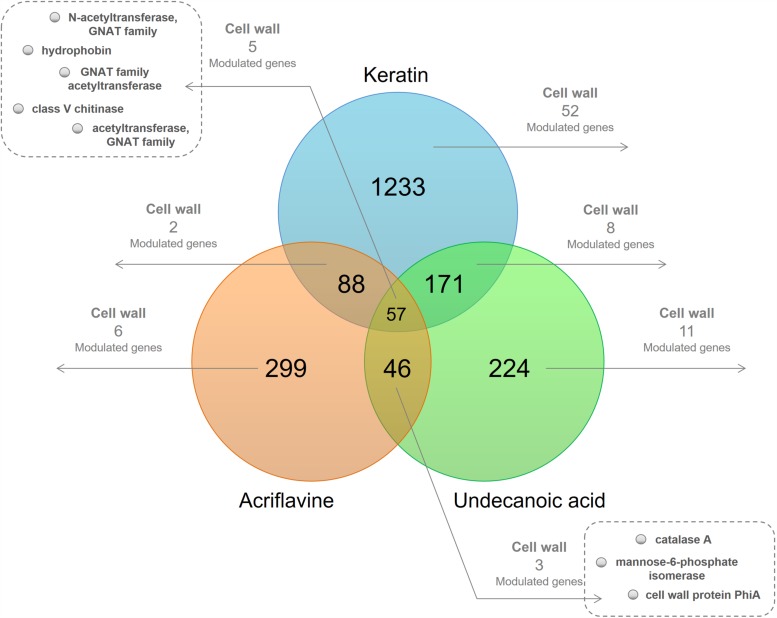
We used functional annotation based on GO terms to identify cell wall-related genes among the DEGs for the keratin, acriflavine, and undecanoic acid RNA-seq (Figure 1 and Supplementary Table S1). The R script found cell wall-related genes by detecting direct child terms of the cellular component ‘cell wall’ in GO.
FIGURE 1.
Venn diagram of the differentially expressed genes (DEGs) in the libraries. At the intersections, number of genes modulated in at least one of the time points of each of the three libraries. The number of modulated cell wall-related genes present in at least one of the time points are highlighted.
We identified genes common to all three libraries and those that were common to two of them. We also determined those unique to each library. We obtained 20 downregulated and 47 upregulated genes in response to keratin culture. Acriflavine treatment induced five genes and repressed 12 others. There were 19 downregulated and nine upregulated genes in response after undecanoic acid exposure. Five of the genes modulated in response to acriflavine were common to all three libraries. Four were altered after acriflavine undecanoic acid treatment. Two others changed after exposure to acriflavine keratin (Supplementary Table S1).
Genes Modulated in Response to Acriflavine
Acriflavine treatment repressed acetyltransferases of the GNAT (Gcn5-related N-acetyltransferase) family (TERG_02517, TERG_05545, and TERG_07408) and induced genes essential for cell wall resistance such as catalases (TERG_01252, TERG_06053) and the hydrophobin (TERG_04234) (Supplementary Table S1 and results extracted from Persinoti et al., 2014).
Genes Modulated in Response to Undecanoic Acid
Undecanoic acid treatment upregulated genes encoding glycosyl hydrolases (TERG_12281, TERG_12282, TERG_06016), a cytosolic Cu/Zn superoxide dismutase (TERG_08969), and catalase A (TERG_01252). The latter two were also induced by acriflavine exposure.
Undecanoic acid also downregulated the cell wall glucanase scw11 (TERG_05576), genes encoding chitinases (TERG_05626, TERG_05625, and TERG_02350), and other cell wall-associated genes (TERG_03624, TERG_06144, and TERG_07456). Both undecanoic acid and acriflavine downregulated genes encoding three GNAT family N-acetyltransferases (TERG_02517, TERG_05545, and TERG_07408). Whereas undecanoic acid downregulated the gene encoding hydrophobin (TERG_04234), acriflavine upregulated it (Supplementary Table S1 and results extracted from Mendes et al., 2018).
Genes Modulated in Response to Keratin
The shift from culture media containing glucose to those with keratin induced significantly more genes than it repressed. Media with keratin downregulated certain genes encoding GNAT family acetyltransferases (TERG_05545, TERG_07987, TERG_08211, and TERG_07408), and the Wiskott-Aldrich syndrome protein family member 2 (TERG_00693).
Genes encoding glycosyl hydrolases (TERG_05530, TERG_01837, TERG_02742, and others), glucanases (TERG_04887 and TERG_07817), chitinases (TERG_05626 and TERG_06925), and an alpha-1,2-mannosyltransferase (TERG_06397) were upregulated in response to keratin. Hydrophobin (TERG_04234) overexpression was observed as it was for the acriflavine treatment.
Osmotic and Cell Wall Stressors Influence T. rubrum Development
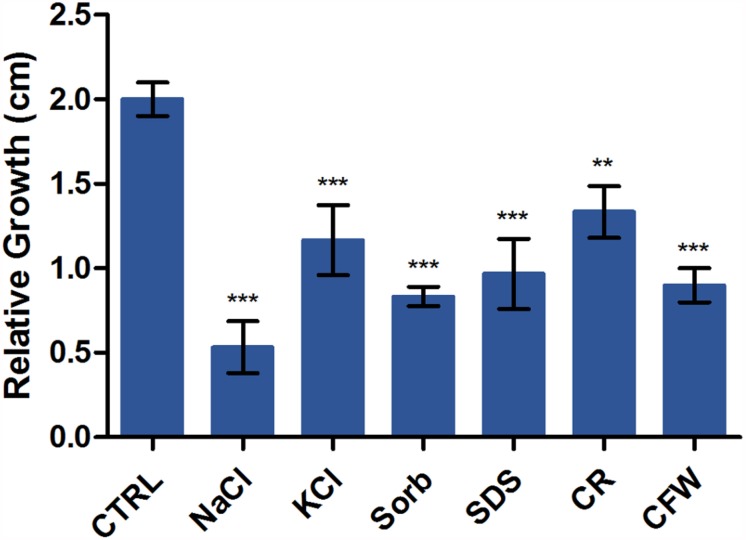
We evaluated the relative sensitivities of T. rubrum grown on SDB medium with or without KCl, NaCl, SDS, sorbitol, CR, and CFW. Figure 2 shows radial colony growth inhibition induced by each stressor. A qRT-PCR analysis evaluated the effects of these stressors on the transcription profiles of STE/STE20/YSK protein kinase kic1 (TERG_01721), cell morphogenesis protein sog2 (TERG_07599), conidiophore development protein hym1 (TERG_00759), AGC/NDR/NDR protein kinase cbk1 (TERG_03379), kinase activator protein mob2 (TERG_02863), cell morphogenesis protein tao3 (TERG_01788), 1,3-β-glucan synthase component fks (TERG_01127), chitin synthase 2 chs (TERG_12319), class III chitinase cts (TERG_02705), and cell wall glucanase scw11 (TERG_05576).
FIGURE 2.
Susceptibility of T. rubrum to various stressors. Strains were inoculated on Sabouraud dextrose broth with or without the stressors NaCl, KCl, sorbitol (Sorb), SDS, Congo Red (CR), and Calcofluor White (CFW) at 28°C for 6 d. The graph represents the relative colony diameters (in centimeters) of each strain in the presence of the stressors relative to the control (CTRL), without the stressors. Data are means of three biological replicates. The error bar indicates the standard deviation (SD). Asterisks indicate statistical significance determined by ANOVA followed by Tukey’s ad hoc test (∗∗P < 0.01; ∗∗∗P < 0.001).
RNA-Seq Validation by qRT-PCR
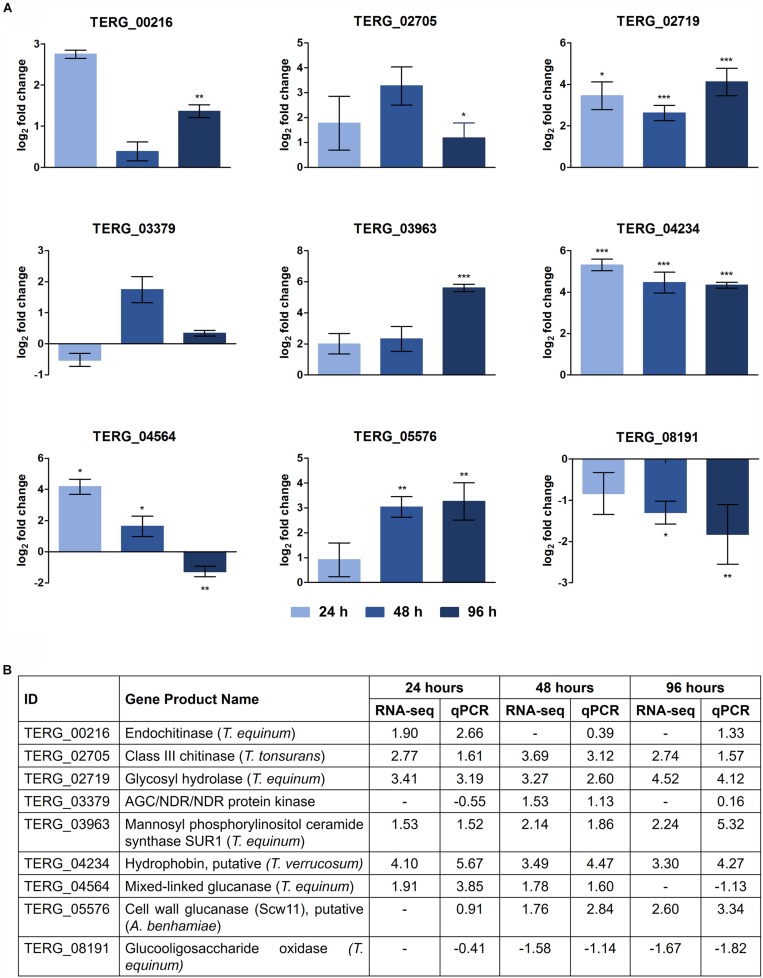
The DEGs selected and assayed through qRT-PCR validated the RNA-seq results. The expression patterns were observed in the samples, examined in triplicate, and confirmed the reliability of the RNA-seq results (Pearson’s correlation, r > 0.82; P < 0.001) (Figure 3).
FIGURE 3.
Validation of DEGs under keratin cultivation by qRT-PCR. (A) Gene expression levels are represented as log2-fold changes relative to each control condition (24 h keratin × 24 h glucose; 48 h keratin × 48 h glucose; 96 h keratin × 96 h glucose). Asterisks indicate statistical significance determined by Student’s t-tests comparing treatment and control conditions at each time point (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001). (B) Comparison of gene expression levels determined by RNA-seq with those evaluated by qRT-PCR (Pearson’s correlation, r > 0.82; P < 0.001).
Discussion
The Cell Wall Is a Virulence Factor Modulated in Response to Environmental Conditions
The fungal cell wall is the most promising virulence factor target for drug discovery because of its location, mediation of fungal-host interactions, uniqueness of composition, and absolute necessity for the survival of the pathogen. The roles of the cell wall in adhesion, colonization, signaling, and immune recognition make it vital for pathogen infection (Arana et al., 2009; Hasim and Coleman, 2019). As cell wall of fungi enables them to interact dynamically with the ambient environment, evaluation of the genes responsive to environmental stresses may lead to a better comprehension of the role of the cell wall in T. rubrum.
Only a Few Cell Wall-Related Genes Were Modulated in Response to Acriflavine
T. rubrum was subjected to sublethal doses of acriflavine which has antiseptic and anticancer properties but whose use is limited by its toxicity (Persinoti et al., 2014). RNA sequencing analysis revealed modulation of only a few cell wall-related genes. Thus, acriflavine had minimal influence on T. rubrum cell growth, shape, or protection.
Two catalase genes were upregulated in response to acriflavine. Acriflavine resistance was observed in Dictyostelium, as a result of catalase A gene (catA) disruption. This gene has peroxidase activity that modifies acriflavine into its cytotoxic form (Garcia et al., 2002). Catalases protect cells against peroxide-induced damage. They are also essential for pathogen virulence and cell wall integrity during host invasion (Skamnioti et al., 2007). The observed upregulation of two catalases in T. rubrum suggests that the pathogen metabolized the drug and induced catalase during cell wall formation and/or maintenance in the attempt to increases its virulence.
Hydrophobin is a small cysteine-rich protein secreted only by filamentous fungi. It regulates cell wall integrity (Wosten, 2001; Mankel et al., 2002) and is upregulated in response to acriflavine exposure. Acriflavine increases hydrophobicity, decreases hydrophilicity, induces impermeability, and alters cell wall thickness (Glynn and Priest, 1970; Kawai et al., 2009). Hydrophobins are amphiphilic and lower the surface tension of water (Linder et al., 2005). The hydrophobin induction detected in T. rubrum in response to acriflavine exposure suggests that the fungus was attempting to counterbalance the increase in cell wall hydrophobicity caused by the drug.
Only a few genes are repressed in response to acriflavine including three members of the GNAT family of acetyltransferases. They are involved in post-translational modification by transferring acetyl groups from acetyl-CoA to their cognate substrates (Favrot et al., 2016). GNATs regulate transcriptional responses to various environmental stressors such as heat, cold, oxidative stress, and low nutrient availability (Huisinga and Pugh, 2004; O’Meara et al., 2010). The GNAT repression observed in our results suggests increasing stress-mediated activation and drug susceptibility in T. rubrum.
Undecanoic Acid Mainly Represses T. rubrum Cell Wall-Associated Genes
Undecanoic acid exposure, especially for 3 h, downregulated several genes associated with cell wall morphogenesis, including scw11, which encodes the cell wall-based endo-1,3-β-glucanase (Millet et al., 2018), the SUN domain protein Uth1-like, encoding a β-(1,3)-glucan-hydrolyzing enzyme (Gastebois et al., 2013), a cell wall serine-threonine-rich galactomannoprotein Mp1 (Gautam et al., 2011), and a cell wall structural protein PhiA. Repression of genes related to cell wall maintenance was reported for Aspergillus fumigatus exposed to the antimalarial drug artemisinin, which is known to have antifungal activity (Gautam et al., 2011). This same effect was also reported in T. rubrum subjected to ketoconazole and amphotericin B (Yu et al., 2007). It was shown that the antifungal agent undecanoic acid effectively impairs T. rubrum cell wall formation (Mendes et al., 2018).
Undecanoic acid represses GNAT family N-acetyltransferase genes, similarly to what was observed for acriflavine. The gene encoding a hydrophobin, upregulated by acriflavine was downregulated in response to undecanoic acid exposure. Undecanoic acid damages the cell wall of T. rubrum by reducing its ergosterol content and altering its fatty acid metabolism (Mendes et al., 2018). Ergosterol controls water penetration and regulates enzymes involved in protein transport and chitin synthesis. Excessive intracellular water uptake may occur when ergosterol metabolism is deregulated. The hydrophobin gene appears to be involved in the water flux across the fungal cell wall (Abe et al., 2009; Zuza-Alves et al., 2017) and is associated with fungal pathogenesis, mediating the attachment of fungal infective structures to their targets (Wosten, 2001). Undecanoic acid exposure in T. rubrum may have negatively impacted its pathogenicity by downregulating hydrophobins.
The gene encoding a class V chitinase (glycoside hydrolase family 18 protein-GH18) and another encoding an endochitinase were repressed by undecanoic acid. As chitin is absent in mammalian cells and essential for fungal cell wall integrity, it is a promising target for antifungal drug development (Ruiz-Herrera and San-Blas, 2003). The observed repressive effect against chitinases emphasize the promising use of undecanoic acid in fungal treatment. Acriflavine also repressed the GH18; therefore, both drugs may disrupt important biochemical events involved in the establishment and maintenance of fungal infection in the host (Persinoti et al., 2014). In T. rubrum, this chitinase presents LysM domains and may be involved in keratin degradation (Lopes et al., 2019).
Three glycosyl hydrolase genes were upregulated in response to undecanoic acid. The TERG_06016, identified as a PHO system negative regulator, responsible for the hydrolysis of O-glycosyl compounds, and two cell wall acid trehalases, that shelter fungi against several physiological and environmental stressors (Arguelles, 2000). These inductions were detected 3 h after undecanoic acid treatment. Thus, T. rubrum immediately responded to this exposure and attempted to offset the damage caused by the stress in the cell walls. We also observed that the genes improving fungal stress tolerance were upregulated after 12 h incubation. These included the cytosolic Cu/Zn superoxide dismutase and catalase A. Moreover, cell wall formation was repressed.
The Cell Wall Is Actively Synthesized When T. rubrum Is Cultured With Keratin
We supplied T. rubrum with keratin to mimic the initial host infection stages. Mycelia harvested after 96 h incubation in glucose-rich media were shifted to keratin or glucose (test and control, respectively) and forced to adapt to a stress-inducing environment.
After 96 h culture in keratin, T. rubrum had upregulated more cell wall-related genes than it had downregulated. The highest number of DEGs was observed at 96 h. Thus, T. rubrum endeavored to grow, develop, establish, and maintain cell wall integrity in the keratin-containing medium.
Among the genes repressed in keratin only two were also repressed in both the acriflavine and undecanoic acid treatments. They belonged to the stress-responsive GNAT acetyltransferase family. A gene encoding Wiskott-Aldrich syndrome protein family member 2 was also downregulated. It encodes a β-1,6-glucan putatively belonging to the KRE family. β-1,6-glucan integrates into the fungal cell wall, acts as the central core of the protein-carbohydrate network, interconnecting chitin and β-1,3-glucan, and associating mannoproteins there (Shahinian and Bussey, 2000). As this gene was repressed only after 96 h of fungal adaptation to keratin, the pathogen avoided excessive remodeling by restraining cell wall formation.
We observed the up-modulation of the hydrophobin gene, also modulated in response to acriflavine and undecanoic acid, attesting its role in the stress response of T. rubrum. Thus, the hydrophobin plays a role in the stress response of T. rubrum. The upregulation of hydrophobins in T. rubrum at all measurement time points of keratin incubation indicates that the pathogen tries to increase its cell wall hydrophobicity and, by extension, its virulence, during the early stages of host infection.
Genes encoding glycosyl hydrolase, glucanase, and chitinase were induced in response to keratin. These are the most commonly occurring hydrolases in the fungal cell wall. They are associated with cell wall polymer branching and cross-linking and the maintenance of cell wall plasticity during morphogenesis (Adams, 2004). They also influence fungal virulence as they participate in non-self cell wall degradation and enable the pathogen to penetrate and invade the host (Gruber and Seidl-Seiboth, 2012).
Keratin incubation also upregulated α-1,2-mannosyltransferase. In the human pathogenic mold A. fumigatus, deletion of an orthologous gene resulted in attenuated virulence. This gene is a potential target for novel antifungal therapies as it is unique in fungi (Wagener et al., 2008). The observed induction of α-1,2-mannosyltransferase in T. rubrum grown with keratin indicates that the fungus was trying to maintain its virulence and sustain its development under these conditions.
Genes Involved in Hyphal Extension and Cell Growth Are Induced in T. rubrum Challenged With Cell Wall Stressors
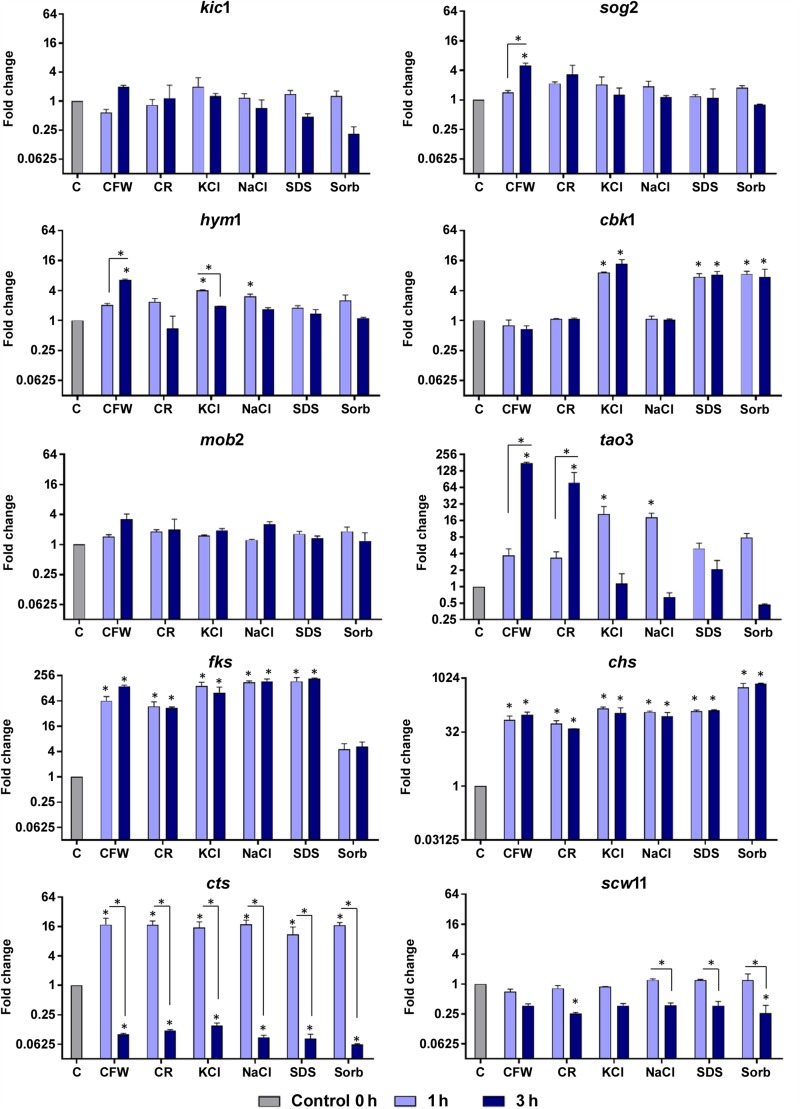
Various stressors (CFW, CR, SDS, NaCl, KCl, and sorbitol) were added to the culture medium to determine their effects on radial colony growth (Figure 2). The fungal cell walls were sensitive to osmotic and cell wall stresses, and mycelial growth and development were inhibited. We then used qRT-PCR to evaluate the expression of genes involved in polarized growth, cell wall remodeling, and hyphal growth after exposure to the abovementioned cell wall stressors. The genes selected for evaluation are either regulated by the Ace2 transcription factor or belong to the RAM (regulation of Ace2 activity and cellular morphogenesis) signaling pathway. The RAM network is a protein kinase-signaling pathway associated to the maintenance of the cell wall integrity. RAM regulates the zinc finger transcriptional factor Ace2 which governs fks, chs, cts, and scw11 genes, encoding 1,3-β-glucan synthase, chitin synthase, chitinase, and β-glucanase, respectively. These play vital roles in cell wall morphogenesis (Sbia et al., 2008; Saputo et al., 2012).
Figure 4 shows that osmotic stressors did not affect the expression levels of the mob2 and kic1 genes. The leucine-rich repeat-containing sog2 was induced only in response to CFW and scw11 was repressed upon challenge with CR and sorbitol; both genes responded at the latest time of exposure. Saline stress induced an immediate response by the hym1 gene and CFW exposure resulted in its induction after 3 h of. At both time points, cbk1 responded to KCl, SDS, and sorbitol. The protein kinase tao3 is strongly induced upon challenge with the cell wall stressors CFW and CR and is upmodulated in response to saline stress after 3h. The unique transcriptional pattern observed suggests that, although all these genes are directly related to cell wall integrity, as part of the RAM signaling network, their activation is time- and stress-responsive.
FIGURE 4.
Relative expression levels of genes encoding components of the RAM network or putatively regulated by Ace2 after exposure of T. rubrum to Calcofluor White (CFW), Congo Red (CR), KCl, NaCl, SDS, and sorbitol (Sorb), for 1 or 3 h. Control (C) is the growth of T. rubrum for 96 h before addition of the stressors. Data are means and SD of three independent experiments. Asterisks indicate statistical significance determined by ANOVA followed by Tukey’s ad hoc test (P < 0.05).
Fungal cell walls increase their chitin content in reaction to stress conditions (Walker et al., 2008; Fortwendel et al., 2009). This response is mediated by the upregulation of chitin synthases and/or the downregulation of chitinases (Heilmann et al., 2013). In the present study, we observed that T. rubrum induced chs and fks1 and repressed cts following exposure to osmotic stressors (Figure 4). Cell wall restoration and selective modulatory activation of the RAM pathway genes are indicative of the efforts of T. rubrum to restore and maintain homeostasis in response to external stressors.
Cell Wall, Virulence, and Antifungal Drug Development
Increasing global transcriptome data favors more robust studies on fungal gene modulation that may elucidate fungal metabolism, pathogenesis, and drug resistance. The results obtained herein may also afford strategies for the identification of novel drug development targets.
Here, five genes were modulated in T. rubrum in response to the various stressors. Three belonged to the GNAT family, one was a class V chitinase, and the fifth was a hydrophobin. The observed stress-mediated response of the hydrophobin gene suggests its possible use as a therapeutic antifungal drug target. Antifungal targets must be essential proteins, have a high degree of sequential similarity across pathogenic fungal species, and be absent in the human genome (Liu et al., 2006). Hydrophobins present with low overall sequence conservation. Nevertheless, they have a broad range of functions including the regulation of cell wall integrity. They can also change the hydrophilic/hydrophobic properties of cell surfaces and are unique to filamentous fungi (Wosten, 2001; Linder et al., 2005; Bayry et al., 2012).
The GNAT superfamily is ubiquitous across many taxa, including humans and fungi. However, a few of its members are potential antifungal drug targets. Identification of the structural differences between human and fungal GNAT members may facilitate selective antifungal drug design (Masubuchi et al., 2003; Hurtado-Guerrero et al., 2008).
A class V chitinase was repressed in response to acriflavine and undecanoic acid exposure but induced in the presence of keratin. Thus, it plays a vital role in substrate adaptation. It was reported that in T. rubrum this gene participates in keratin degradation (Lopes et al., 2019). Fungal cell walls contain high levels of chitin and they are cleaved by chitinases during cell wall remodeling. Thus, disruption of this process is expected to affect fungal virulence and survival and points to chitinases as antifungal drug targets (Rush et al., 2010).
Conclusion
Environmental changes and external stresses induce continuous remodeling of fungal cell walls, and the transcriptional responses of T. rubrum to those observed in this study are essential for its resistance and pathogenicity. We have generated a robust dataset, which underscores the relevance of the cell wall in pathogen-host interactions, advancing our knowledge on the cell wall regulatory mechanisms of dermatophyte fungi. Our results indicate that stress conditions forced T. rubrum to restructure its cell wall by modifying the cell wall composition; thus, affecting fungal virulence. The modulation of genes controlling the fungal cell-wall structure has great potential in the identification of putative targets for the development of novel antifungal drug therapies.
Data Availability Statement
The RNA-seq data is available at the GEO database under the accession number GSE134406.
Author Contributions
MM and LGS drafted the manuscript. MM, LGS, and LDS performed the experiments. PS performed the computational analyses. AR and NM-R supervised the study and prepared the manuscript. NM-R designed the project. All authors participated in the data analysis and critical revision of the manuscript, and approved the final version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank V. M. Oliveira and M. Mazucato for technical assistance and M. D. Martins for laboratory services.
Funding. This study was supported by grants from the following Brazilian funding agencies: Research Support Foundation of São Paulo State - FAPESP (Grant No. 2014/03847-7, Postdoctoral Fellowship No. 2018/11319-1 to MM, and Doctoral Fellowship No. 2010/15017-8 to LGS), National Council for Scientific and Technological Development - CNPq (Grant Nos. 305252/2013-5 and 304222/2013-5), Coordination for the Improvement of Higher Education Personnel - CAPES (Finance Code 001), and Foundation for Support to Teaching, Research and Assistance - FAEPA of the HCFMRP-USP.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02168/full#supplementary-material
References
- Abe F., Usui K., Hiraki T. (2009). Fluconazole modulates membrane rigidity, heterogeneity, and water penetration into the plasma membrane in Saccharomyces cerevisiae. Biochemistry 48 8494–8504. 10.1021/bi900578y [DOI] [PubMed] [Google Scholar]
- Adams D. J. (2004). Fungal cell wall chitinases and glucanases. Microbiology 150 2029–2035. 10.1099/mic.0.26980-0 [DOI] [PubMed] [Google Scholar]
- Arana D. M., Prieto D., Roman E., Nombela C., Alonso-Monge R., Pla J. (2009). The role of the cell wall in fungal pathogenesis. Microb. Biotechnol. 2 308–320. 10.1111/j.1751-7915.2008.00070.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguelles J. C. (2000). Physiological roles of trehalose in bacteria and yeasts: a comparative analysis. Arch. Microbiol. 174 217–224. 10.1007/s002030000192 [DOI] [PubMed] [Google Scholar]
- Bayry J., Aimanianda V., Guijarro J. I., Sunde M., Latge J. P. (2012). Hydrophobins-unique fungal proteins. PLoS Pathog. 8:e1002700. 10.1371/journal.ppat.1002700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauvais A., Latge J. P. (2018). Special issue: fungal cell wall. J. Fungi 4:91. 10.3390/jof4030091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57 289–300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- Bitencourt T. A., Rezende C. P., Quaresemin N. R., Moreno P., Hatanaka O., Rossi A., et al. (2018). Extracellular vesicles from the dermatophyte Trichophyton interdigitale modulate macrophage and keratinocyte functions. Front. Immunol. 9:2343. 10.3389/fimmu.2018.02343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake J. S., Dahl M. V., Herron M. J., Nelson R. D. (1991). An immunoinhibitory cell wall glycoprotein (mannan) from Trichophyton rubrum. J. Invest. Dermatol. 96 657–661. [DOI] [PubMed] [Google Scholar]
- Bok J. W., Balajee S. A., Marr K. A., Andes D., Nielsen K. F., Frisvad J. C., et al. (2005). LaeA, a regulator of morphogenetic fungal virulence factors. Eukaryot. Cell 4 1574–1582. 10.1128/ec.4.9.1574-1582.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M. (2018). GO.db: A Set of Annotation Maps Describing the Entire Gene Ontology, in: R Package Version 3.7.0. [Google Scholar]
- Cove D. J. (1966). The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim. Biophys. Acta 113 51–56. 10.1016/s0926-6593(66)80120-0 [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fachin A. L., Maffei C. M., Martinez-Rossi N. M. (1996). In vitro susceptibility of Trichophyton rubrum isolates to griseofulvin and tioconazole. Induction and isolation of a resistant mutant to both antimycotic drugs. Mutant of Trichophyton rubrum resistant to griseofulvin and tioconazole. Mycopathologia 135 141–143. 10.1007/bf00632334 [DOI] [PubMed] [Google Scholar]
- Favrot L., Blanchard J. S., Vergnolle O. (2016). Bacterial GCN5-Related N-Acetyltransferases: from resistance to regulation. Biochemistry 55 989–1002. 10.1021/acs.biochem.5b01269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortwendel J. R., Juvvadi P. R., Pinchai N., Perfect B. Z., Alspaugh J. A., Perfect J. R., et al. (2009). Differential effects of inhibiting chitin and 1,3-β D-glucan synthesis in ras and calcineurin mutants of Aspergillus fumigatus. Antimicrob. Agents Chemother. 53 476–482. 10.1128/aac.01154-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M. X., Roberts C., Alexander H., Stewart A. M., Harwood A., Alexander S., et al. (2002). Methanol and acriflavine resistance in Dictyostelium are caused by loss of catalase. Microbiology 148 333–340. 10.1099/00221287-148-1-333 [DOI] [PubMed] [Google Scholar]
- Gastebois A., Aimanianda V., Bachellier-Bassi S., Nesseir A., Firon A., Beauvais A., et al. (2013). SUN proteins belong to a novel family of beta-(1,3)-glucan-modifying enzymes involved in fungal morphogenesis. J. Biol. Chem. 288 13387–13396. 10.1074/jbc.M112.440172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam P., Upadhyay S. K., Hassan W., Madan T., Sirdeshmukh R., Sundaram C. S., et al. (2011). Transcriptomic and proteomic profile of Aspergillus fumigatus on exposure to artemisinin. Mycopathologia 172 331–346. 10.1007/s11046-011-9445-3 [DOI] [PubMed] [Google Scholar]
- Glynn A. A., Priest C. M. (1970). The effect of acriflavine on complement sensitive and resistant strains of Escherichia coli and on complement resistant mutants. Immunology 18 19–22. [PMC free article] [PubMed] [Google Scholar]
- Gotz S., Garcia-Gomez J. M., Terol J., Williams T. D., Nagaraj S. H., Nueda M. J., et al. (2008). High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36 3420–3435. 10.1093/nar/gkn176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover N. D. (2010). Echinocandins: a ray of hope in antifungal drug therapy. Indian J. Pharmacol. 42 9–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber S., Seidl-Seiboth V. (2012). Self versus non-self: fungal cell wall degradation in Trichoderma. Microbiology 158 26–34. 10.1099/mic.0.052613-0 [DOI] [PubMed] [Google Scholar]
- Hasim S., Coleman J. J. (2019). Targeting the fungal cell wall: current therapies and implications for development of alternative antifungal agents. Future Med. Chem. 11 869–883. 10.4155/fmc-2018-0465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann C. J., Sorgo A. G., Mohammadi S., Sosinska G. J., De Koster C. G., Brul S., et al. (2013). Surface stress induces a conserved cell wall stress response in the pathogenic fungus Candida albicans. Eukaryot. Cell 12 254–264. 10.1128/EC.00278-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisinga K. L., Pugh B. F. (2004). A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell. 13 573–585. 10.1016/s1097-2765(04)00087-5 [DOI] [PubMed] [Google Scholar]
- Hurtado-Guerrero R., Raimi O. G., Min J., Zeng H., Vallius L., Shepherd S., et al. (2008). Structural and kinetic differences between human and Aspergillus fumigatus D-glucosamine-6-phosphate N-acetyltransferase. Biochem. J. 415 217–223. 10.1042/BJ20081000 [DOI] [PubMed] [Google Scholar]
- Jacob T. R., Peres N. T., Persinoti G. F., Silva L. G., Mazucato M., Rossi A., et al. (2012). rpb2 is a reliable reference gene for quantitative gene expression analysis in the dermatophyte Trichophyton rubrum. Med. Mycol. 50 368–377. 10.3109/13693786.2011.616230 [DOI] [PubMed] [Google Scholar]
- Kar B., Patel P., Free S. J. (2019). Trichophyton rubrum LysM proteins bind to fungal cell wall chitin and to the N-linked oligosaccharides present on human skin glycoproteins. PLoS One 14:e0215034. 10.1371/journal.pone.0215034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai M., Yamada S., Ishidoshiro A., Oyamada Y., Ito H., Yamagishi J. (2009). Cell-wall thickness: possible mechanism of acriflavine resistance in meticillin-resistant Staphylococcus aureus. J. Med. Microbiol. 58 331–336. 10.1099/jmm.0.004184-0 [DOI] [PubMed] [Google Scholar]
- Linder M. B., Szilvay G. R., Nakari-Setala T., Penttila M. E. (2005). Hydrophobins: the protein-amphiphiles of filamentous fungi. FEMS Microbiol. Rev. 29 877–896. 10.1016/j.femsre.2005.01.004 [DOI] [PubMed] [Google Scholar]
- Liu M., Healy M. D., Dougherty B. A., Esposito K. M., Maurice T. C., Mazzucco C. E., et al. (2006). Conserved fungal genes as potential targets for broad-spectrum antifungal drug discovery. Eukaryot. Cell 5 638–649. 10.1128/ec.5.4.638-649.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lopes L., Bitencourt T. A., Lang E. A. S., Sanches P. R., Peres N. T. A., Rossi A., et al. (2019). Genes coding for LysM domains in the dermatophyte Trichophyton rubrum: a transcription analysis. Med. Mycol 10.1093/mmy/myz068 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankel A., Krause K., Kothe E. (2002). Identification of a hydrophobin gene that is developmentally regulated in the ectomycorrhizal fungus Tricholoma terreum. Appl. Environ. Microbiol. 68 1408–1413. 10.1128/aem.68.3.1408-1413.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez D. A., Oliver B. G., Graser Y., Goldberg J. M., Li W., Martinez-Rossi N. M., et al. (2012). Comparative genome analysis of Trichophyton rubrum and related dermatophytes reveals candidate genes involved in infection. mBio 3 e259–12. 10.1128/mBio.00259-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Rossi N. M., Bitencourt T. A., Peres N. T. A., Lang E. A. S., Gomes E. V., Quaresemin N. R., et al. (2018). Dermatophyte resistance to antifungal drugs: mechanisms and prospectus. Front. Microbiol. 9:1108. 10.3389/fmicb.2018.01108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Rossi N. M., Peres N. T., Rossi A. (2017). Pathogenesis of dermatophytosis: sensing the host tissue. Mycopathologia 182 215–227. 10.1007/s11046-016-0057-9 [DOI] [PubMed] [Google Scholar]
- Masubuchi M., Ebiike H., Kawasaki K., Sogabe S., Morikami K., Shiratori Y., et al. (2003). Synthesis and biological activities of benzofuran antifungal agents targeting fungal N-myristoyltransferase. Bioorg. Med. Chem. 11 4463–4478. 10.1016/s0968-0896(03)00429-2 [DOI] [PubMed] [Google Scholar]
- Mendes N. S., Bitencourt T. A., Sanches P. R., Silva-Rocha R., Martinez-Rossi N. M., Rossi A. (2018). Transcriptome-wide survey of gene expression changes and alternative splicing in Trichophyton rubrum in response to undecanoic acid. Sci. Rep. 8:2520. 10.1038/s41598-018-20738-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet N., Latge J. P., Mouyna I. (2018). Members of glycosyl-hydrolase family 17 of A. fumigatus differentially affect morphogenesis. J. Fungi 4:E18. 10.3390/jof4010018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muszewska A., Pilsyk S., Perlinska-Lenart U., Kruszewska J. S. (2017). Diversity of cell wall related proteins in human pathogenic fungi. J. Fungi 4:E6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Meara T. R., Hay C., Price M. S., Giles S., Alspaugh J. A. (2010). Cryptococcus neoformans histone acetyltransferase Gcn5 regulates fungal adaptation to the host. Eukaryot. Cell 9 1193–1202. 10.1128/EC.00098-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlin D. S. (2015). Mechanisms of echinocandin antifungal drug resistance. Ann. N. Y. Acad. Sci. 1354 1–11. 10.1111/nyas.12831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persinoti G., De Aguiar Peres N., Jacob T., Rossi A., Vencio R., Martinez-Rossi N. (2014). RNA-sequencing analysis of Trichophyton rubrum transcriptome in response to sublethal doses of acriflavine. BMC Genomics 15:S1. 10.1186/1471-2164-15-S7-S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E. S., Getz G., et al. (2011). Integrative genomics viewer. Nat. Biotechnol. 29 24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Herrera J., San-Blas G. (2003). Chitin synthesis as target for antifungal drugs. Curr. Drug Targets. Infect. Disord. 3 77–91. 10.2174/1568005033342064 [DOI] [PubMed] [Google Scholar]
- Rush C. L., Schuttelkopf A. W., Hurtado-Guerrero R., Blair D. E., Ibrahim A. F., Desvergnes S., et al. (2010). Natural product-guided discovery of a fungal chitinase inhibitor. Chem. Biol. 17 1275–1281. 10.1016/j.chembiol.2010.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saputo S., Chabrier-Rosello Y., Luca F. C., Kumar A., Krysan D. J. (2012). The RAM network in pathogenic fungi. Eukaryot. Cell 11 708–717. 10.1128/EC.00044-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbia M., Parnell E. J., Yu Y., Olsen A. E., Kretschmann K. L., Voth W. P., et al. (2008). Regulation of the yeast Ace2 transcription factor during the cell cycle. J. Biol. Chem. 283 11135–11145. 10.1074/jbc.M800196200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahinian S., Bussey H. (2000). β-1,6-Glucan synthesis in Saccharomyces cerevisiae. Mol. Microbiol. 35 477–489. 10.1046/j.1365-2958.2000.01713.x [DOI] [PubMed] [Google Scholar]
- Skamnioti P., Henderson C., Zhang Z., Robinson Z., Gurr S. J. (2007). A novel role for catalase B in the maintenance of fungal cell-wall integrity during host invasion in the rice blast fungus Magnaporthe grisea. Mol. Plant Microbe Interact. 20 568–580. 10.1094/mpmi-20-5-0568 [DOI] [PubMed] [Google Scholar]
- Thorvaldsdottir H., Robinson J. T., Mesirov J. P. (2012). Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14 178–192. 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiante V., Macheleidt J., Foge M., Brakhage A. A. (2015). The Aspergillus fumigatus cell wall integrity signaling pathway: drug target, compensatory pathways, and virulence. Front. Microbiol. 6:325. 10.3389/fmicb.2015.00325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vencio R. Z., Koide T., Gomes S. L., Pereira C. A. (2006). BayGO: bayesian analysis of ontology term enrichment in microarray data. BMC Bioinformatics 7:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermout S., Tabart J., Baldo A., Mathy A., Losson B., Mignon B. (2008). Pathogenesis of dermatophytosis. Mycopathologia 166 267–275. 10.1007/s11046-008-9104-5 [DOI] [PubMed] [Google Scholar]
- Wagener J., Echtenacher B., Rohde M., Kotz A., Krappmann S., Heesemann J., et al. (2008). The putative alpha-1,2-mannosyltransferase AfMnt1 of the opportunistic fungal pathogen Aspergillus fumigatus is required for cell wall stability and full virulence. Eukaryot. Cell 7 1661–1673. 10.1128/EC.00221-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L. A., Munro C. A., De Bruijn I., Lenardon M. D., Mckinnon A., Gow N. A. (2008). Stimulation of chitin synthesis rescues Candida albicans from echinocandins. PLoS Pathog. 4:e1000040. 10.1371/journal.ppat.1000040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodfolk J. A., Sung S. S., Benjamin D. C., Lee J. K., Platts-Mills T. A. (2000). Distinct human T cell repertoires mediate immediate and delayed-type hypersensitivity to the Trichophyton antigen, Tri r 2. J. Immunol. 165 4379–4387. 10.4049/jimmunol.165.8.4379 [DOI] [PubMed] [Google Scholar]
- Wosten H. A. (2001). Hydrophobins: multipurpose proteins. Annu. Rev. Microbiol. 55 625–646. 10.1146/annurev.micro.55.1.625 [DOI] [PubMed] [Google Scholar]
- Yu L., Zhang W., Wang L., Yang J., Liu T., Peng J., et al. (2007). Transcriptional profiles of the response to ketoconazole and amphotericin B in Trichophyton rubrum. Antimicrob. Agents Chemother. 51 144–53. 10.1128/AAC.00755-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuza-Alves D. L., Silva-Rocha W. P., Chaves G. M. (2017). An update on Candida tropicalis based on basic and clinical approaches. Front. Microbiol. 8:1927. 10.3389/fmicb.2017.01927 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data is available at the GEO database under the accession number GSE134406.