Abstract
We propose using a modified amplification refractory mutation system real-time polymerase chain reaction (ARMS RTPCR) technique to exclude the invasive prenatal diagnosis for a non-paternally inherited beta thalassemia mutation in couples atrisk for having a baby with CHBT. The ARMS RT-PCR method was performed for 36 at-risk couples by using isolated fetal cell-free DNA from maternal plasma. The modified ARMS RT-PCR primers targeted one of the following paternally inherited beta thalassemia mutation: -28 A→G, CD17 A→T, CD 26 G→A, IVS1-1 G→T and CD 41-42 -CTTT. The method could be successfully employed for NIPST starting with the 7th week of gestation. The results showed that 19 pregnant women were negative for PIBTM (53%). After an on-track and on-time of one year, including postnatal thalassemia blood tests, none of the babies showed symptoms or signs of beta thalassemia disease. We concluded that the modified ARMS RT-PCR method was an accurate, cost-effective and feasible method for use as a NIPST for at-risk couples with the potential of having a baby with CHBT.
Key words: NIPST, ARMS RT-PCR, Maternal cell-free fetal DNA, Paternally inherited beta thalassemia mutation
Introduction
Compound heterozygous beta thalassemia (CHBT) occurs in live births and is the most common form of severe beta thalassemia disease in Southeast Asian countries. It is a single autosomal genetic disorder with a prevalence rate of 1-9%.1 CHBT disease originates from inheriting combination of two different mutated beta globin genes, either β0β0 or β0β+ thalassemia. Both types cause serious hereditary hematological diseases that manifest through progressive deterioration of chronic anemia and iron overload. The optimal treatments are an economic burden. Therefore, prenatal diagnosis is necessary to prevent the birth of babies with these conditions. Current prenatal diagnostics use invasive methods, such as chorionic villus sampling, amniocentesis and cordocenthesis, which have a risk of fetal loss. Fortunately, the discovery of cell-free fetal DNA in maternal plasma has provided options for noninvasive prenatal testing.2 To date, there are many molecular methods available for the noninvasive prenatal testing of paternally inherited beta thalassemia mutation (PIBTM) in maternal plasma.3 However, some of these methods require sophisticated biochemical techniques to obtain highly accurate results and are not readily available for use as routine screening tests. Our approach focuses on the development of a noninvasive prenatal screening test (NIPST) to reduce the use of invasive prenatal testing in the non-paternally inherited at-risk fetus.
Materials and Methods
From 2016 to 2018, 36 couples at-risk for having a baby with CHBT volunteered to participate in the NIPST program. The following paternal alleles were identified, which for both partners were one or two of the following mutations: -28 A→G, CD17 A→T, CD 41-42 CTTT, IVS1-1 G→T and CD26 G→A. The at-risk couples were from the Antenatal Care Unit, Provincial Phayao Hospital, Phayao, Thailand. The fetal age at the time of the study was 7-24 weeks. Ethical approval was obtained from the Medical Ethics Committee of Phayao University (3/011/59). Informed and written consent were collected from the pregnant women and their spouses before blood collection.
Workflow
The plasma cell-free DNA from 20 mL of fresh whole blood of each individual was isolated. The modified amplification refractory mutation system (ARMS) RT-PCR methods were validated using paternal plasma cell-free DNA for the proper fragment size and melt temperature specificity. The isolated maternal plasma cell-free DNA was tested for the presence or absence of the PIBTM using the modified ARMS RT- PCR method. All pregnant women were counselled before undergoing prenatal diagnosis.
The PIBTM negative pregnant women did not undergo invasive PND. After birth, the babies had a blood test for thalassemia. All attended a one year of clinical thalassemia follow up at the Well Baby Clinic of the Provincial Phayao Hospital.
Design of specific ARMS RT- PCR primers
All primers were designed using the Primer-Blast program (https://www.ncbi. nlm.nih.gov/tools/ primer-blast/). The U01317.1 (HBB gene; nucleotide number 62137-63472) DNA sequence of the HBB gene was derived from NCBI’s GenBank. Five ARMS real-time PCR primer pairs were designed for targeting the CD41/42 – CTTT, CD26 G→A (HbE trait), CD17 A→T, -28 A→G and IVS1-1 G→T and they were modified according to the allelespecific priming principle for ARMS RTPCR. 4,5 This allowed for the detection of short DNA segments ranging between 60 and 120 base pairs.6 All primers were purchased from the Invitrogen Corporation (Carlsbad, CA, USA). Each primer pair was validated by real-time PCR against the paternal beta thalassemia mutation in plasma cell-free DNA (Table 1).
Isolation of cell-free DNA in plasma
Twenty ml of an EDTA blood sample was separated into plasma fraction using standard procedures. Cell-free DNA was extracted using the QIAamp Circulating Nucleic Acid kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions.
Assessing the PIBTM in maternal plasma
The ARMS RT- PCR master-mix (25 μL total reaction) contained 2.5 μL of 10x PCR buffer, 0.75 μL of 50 mM MgCl2, 0.5 μL of 10 mM dNTPs, 1.0 μL of 5 mM primer, 1.0 μL of 50 mM SYTO9, 0.2 μL of 5 units/μL Platinum Taq, 9.05 μL of dH2O and 10 μL of DNA solution. The ARMS real-time PCR was performed to identify the PIBTM in maternal plasma cell-free DNA. The paternal and maternal cell-free DNA from the at-risk couples was simultaneously assessed. Thermal cycling was performed using a Bio-Rad CFX96 real-time system (Bio-Rad Laboratories, Hercules, CA, USA) starting with an initial Taq DNA polymerase activation step at 94°C for 2 minutes. The complete PCR underwent 44 cycles. Each cycle consisted of DNA denaturation at 94°C for 10 seconds, annealing at 64°C for 10 seconds and extension at 72°C for 10 seconds. The fluorescence activity was measured on a SYBR Green channel (533 nm) at the end of each cycle. The amplification cycle and melt peak evaluation was performed using the Bio-Rad Precision Melt Analysis Software (Bio-Rad Laboratories, Hercules, CA, USA). The melting program started at 95°C for 10 seconds, followed by a melting cycle from 75°C to 90°C at a transitional rate of 0.2°C every 10 seconds.
Postnatal thalassemia diagnosis
The babies of at-risk couple who participated in the NIPST program were requested for blood test to evaluate their thalassemia status and clinical physical examination at the time of the first-year compulsory immunization. Blood test for beta thalassemia diagnosis was assessed by hemoglobin separation and quantification of hemoglobin types were, according to the manufacturer’s instructions using the capillary electrophoresis system (SEBIA, France).7 The beta thalassemia mutation was carried out using high resolution DNA melting analysis and by direct DNA sequencing technique.8
Results
The HBB gene, DNA sequences and nucleotides from the five developed series of ARMS RT-PCR primers were shown in Table 1. The CD41/42 -CTTT ARMS RTPCR primers were used to detect the paternally inherited beta thalassemia mutation from CD41/42 -CTTT carriers. The detected DNA fragment was 73 base pairs, and was identified by a specific melt peak at 82.00±0.25°C (Figure 1). The CD26 G→A ARMS real-time PCR primers were for the hemoglobin E trait, which had a 111 base pair DNA fragment and a melt peak at 84.7±0.30°C. The CD17 A→T ARMS realtime PCR primers were for the CD17 A→T carrier. The DNA fragment size was 69 base pairs and the melt peak was at 80.93±0.23°C. The -28 A→G and IVS1-1 G→T ARMS real-time PCR primers were for the -28 A→G and IVS1-1 G→T traits, respectively; these primer pairs detected 84 and 61 base pair DNA fragments, with melted peaks at 83.6±0.00 and 78.87±0.12°C, respectively. The paternal cycle threshold (Ct) for the beta thalassemia mutations in paternal plasma cell-free DNA was 30±2.80 cycles (N=10), while the cycle threshold for paternally inherited cell-free fetal DNA in maternal plasma was 35±2.10 cycles (N=17). Nineteen of the at-risk pregnancies using specific ARMS real-time PCR primers were negative for PIBTM. After birth, based on the postnatal thalassemia blood tests, the babies either inherited beta thalassemia maternally and carriers or homozygous wild type alleles (Table 2). While, there were 17 out of 36 pregnancies that were positive for PIBTM. After amniocentesis 5 fetuses were shown to have compound heterozygous beta thalassemia (Table 3). In the pregnant women, the paternal allele was detected at a fetal age of seven weeks.
Discussion
The discovery of fetal DNA in maternal plasma has allowed for many approaches for noninvasive prenatal diagnosis. There are examples showing concentrations of total plasma DNA, ranging from 3.4 to 6.2% during early and late pregnancy. Cellfree fetal DNA can be readily detected as early as the 7th week of gestation 2. The size of cell-free fetal DNA fragments in maternal plasma had been reported as ranging from 193-313 base pairs.6,9 All of these discoveries are very important factors to consider for the successful development of ARMS RT-PCR primers. Fortunately, the 36 at-risk couples recruited were known to be carriers of the 5 common beta thalassemia mutations, specifically CD17 A-T, CD41-42-CTTT, IVS1-1 G-T, -28 A-G and the hemoglobin E mutation (CD26 G-A) (Tables 2 and 3). In this study, 19 of 36 atrisk pregnancies showed negative results for PIBTM. When notified about the results of the mother’s blood test, most of the mothers chose to not undergo additional checks, such as invasive prenatal diagnosis.10-12 Currently, there are many techniques for detecting paternally inherited beta thalassemia mutations, including nextgeneration sequencing,13 COLD-PCR and microarrays,14 next-generation sequencing of SNPs,15 TaqMan genotyping assay,16 droplet digital PCR17 and simple fetal DNA enrichment with allele-specific real-time PCRs.18-22 The ARMS RT- PCR technique was simple, accurate, cost-effective, and able to detect paternally inherited beta thalassemia mutations directly from cell-free DNA in maternal plasma. The method was successfully employed for noninvasive prenatal screening tests in at-risk couples starting from the 7th gestational week. The remaining 17 pregnancies whose PIBTM was positive required further investigation of the fetus for a maternal inherited beta thalassemia mutation (Table 3). In the meantime, methods for the detection of maternally inherited beta thalassemia mutation of the at-risk fetus remained in the experimental stage. Some of these methods, namely, droplet digital PCR, may be available in the near future for use in noninvasive prenatal testing.23,24
Table 1.
The DNA sequence primers, DNA fragment size and melt peak temperature of the modified ARMS RT- PCR using for the detection of the paternally inherited beta thalassemia mutations.
| Primers | Nucleotide sequence (5'->3') | PCR size (base pairs) | Melt peak (°C) | NCBI GenBank (U01317.1) |
|---|---|---|---|---|
| F- -28 | GGTTGGCCAATCTACTCCCA | 62055-62074 | ||
| R- -28 | GTAAGCAATAGATGGCTCTGCCCTGACGTC | 84 | 83.6±0.00 | 62109-62138 |
| F-CD17 | AGAAGTCTGCCGTTACTGCC | 62209-62228 | ||
| R-CD17 | CTCACCACCAACTTCATCCACGTTCAGCTA | 69 | 80.93±0.23 | 62238-62267 |
| F-CD26 | ACCATGGTGCACCTGACTC | 62184-62202 | ||
| R-CD26 | TAACCTTGATACCAACCTGCCCAGGGCATT | 111 | 84.70±0.30 | 62265-62294 |
| F-IVSI-I | TGGATGAAGTTGGTGGTGAGG | 62248-62268 | ||
| R-IVSI-I | TTAAACCTGTCTTGTAACCTTGATACCGAA | 61 | 78.87±0.12 | 62278-62308 |
| F-CD41-42 | TTTTCCCACCCTTAGGCTGCT | 62394-62414 | ||
| R-CD41-42 | GAGTGGACAGATCCCCAAAGGACTCAACCT | 73 | 82.00±0.25 | 62437-62470 |
Figure 1.
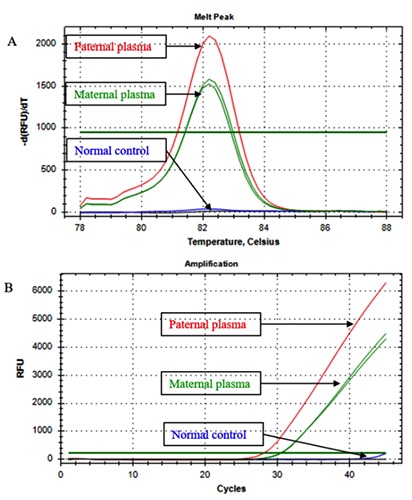
Image of the PIBTM melt peak and cycle threshold of the CD41/42-CTTT using specific ARMS RT- PCR primers. The paternal and the paternally inherited CD41/42-CTTT melt appearance were similar and were at the same temperature at 82.00±0.25°C (A). The paternal cycle threshold for the beta thalassemia mutations in paternal plasma and the cycle threshold of the paternally inherited cell-free fetal DNA in maternal plasma (B).
Table 2.
Negative cases of the NIPST using modified ARMS RT- PCR for the detection of PIBTM in maternal plasma cell-free DNA in the at-risk couples.
| Couple No. | Maternal mutation | Paternal mutation | Gestation age (week) | Paternal ARMS test | ARMS results | Postnatal diagnosis |
|---|---|---|---|---|---|---|
| 1 | CD17 A→T | CD26 G→A | 16 | CD26 | NEG | CD17 trait |
| 2 | IVS1-1 G→T | CD26 G→A | 16 | CD26 | NEG | Normal |
| 3 | -28 A→G | CD26 G→A | 10 | IVS1-1 | NEG | -28 trait |
| 4 | CD17 A→T | CD26 G→A | 10 | CD41/42 | NEG | Unaffected |
| 5 | CD26 G→A | CD41/42 –CTTT | 18 | CD41/42 | NEG | Normal |
| 6 | CD26 G→A | CD41/42 –CTTT | 11 | CD41/42 | NEG | Unaffected |
| 7 | CD41/42 –CTTT | -28 A→G | 18 | CD17 | NEG | Unaffected |
| 8 | CD17 A→T | CD26 G→A | 16 | CD41/42 | NEG | CD17 trait |
| 9 | CD17 A→T | CD26 G→A | 19 | CD17 | NEG | Normal |
| 10 | CD41/42 –CTTT | CD17 A→T | 13 | -28 | NEG | CD41/42 trait |
| 11 | CD26 G→A | CD41/42 –CTTT | 15 | CD26 | NEG | CD26 trait |
| 12 | IVS1-1 G→T | CD26 G→A | 16 | CD26 | NEG | Normal |
| 13 | CD41/42 –CTTT | CD26 G→A | 16 | CD26 | NEG | CD41/42 trait |
| 14 | CD41/42 –CTTT | CD26 G→A | 14 | CD26 | NEG | Normal |
| 15 | CD17 A→T | CD41/42 –CTTT | 14 | CD26 | NEG | Normal |
| 16 | CD41/42 –CTTT | CD26 G→A | 10 | CD41/42 | NEG | Normal |
| 17 | CD17 A→T | CD26 G→A | 7 | CD26 | NEG | CD17 trait |
| 18 | CD26 G→A | IVS1-1 G→T | 8 | CD26 | NEG | CD26 trait |
| 19 | CD26 G→A | CD41/42 –CTTT | 10 | CD26 | NEG | CD26 trait |
NEG, Negative result; POS, Positive results; Unaffected baby did not determine thalassemia genotype.
Table 3.
The results of the cycle threshold value of positive cases of NIPST using modified ARMS RT-PCR for the detection of PIBTM in the at-risk pregnancies.
| Couple No. | Maternal mutation | Paternal mutation | Gestation age (week) | Paternal ARMS test | ARMS results | Postnatal diagnosis | Invasive prenatal diagnosis results |
|---|---|---|---|---|---|---|---|
| 1 | CD26 G→A | CD17 A→T | 21 | CD17 | POS | 37 | CD17 trait |
| 2 | CD41/42 –CTTT | CD26 G→A | 17 | CD26 | POS | 34 | CD26/CD41/42 |
| 3 | CD26 G→A | -28 A→G | 10 | -28 | POS | 35 | CD26/-28 |
| 4 | CD17 A→T | CD26 G→A | 9 | CD26 | POS | 36 | CD26 trait |
| 5 | CD26 G→A | CD41/42 –CTTT | 24 | CD41/42 | POS | 30 | CD41/42 trait |
| 6 | CD26 G→A | IVS1-1 G→T | 19 | IVS1-1 | POS | 33 | IVS1-1 trait |
| 7 | CD26 G→A | CD41/42 –CTTT | 21 | CD41/42 | POS | 33 | CD41/42 trait |
| 8 | CD26 G→A | CD41/42 –CTTT | 11 | CD41/42 | POS | 34 | CD41/42 trait |
| 9 | CD26 G→A | CD41/42 –CTTT | 12 | CD41/42 | POS | 37 | CD41/42 trait |
| 10 | CD17 A→T | -28 A→G | 11 | -28 | POS | 36 | CD17/-28 |
| 11 | CD41/42 –CTTT | Homozygous CD26 G→A | 12 | CD26 | POS | 36 | CD26 trait |
| 12 | CD26 G→A | CD41/42 –CTTT | 12 | CD41/42 | POS | 37 | CD26/CD41/42 |
| 13 | CD17 A→T | CD26 G→A | 15 | CD26 | POS | 38 | CD26 trait |
| 14 | CD26 G→A | CD41/42 –CTTT | 9 | CD41/42 | POS | 35 | CD26/CD41/42 |
| 15 | CD26 G→A | CD41/42 –CTTT | 8 | CD41/42 | POS | 36 | CD41/42 trait |
| 16 | CD26 G→A | CD17 A→T | 7 | CD17 | POS | 35 | CD17 A→T trait |
| 17 | CD26 G→A | IVS1-1 G→T | 16 | IVS1-1 | POS | 31 | IVS1-1 trait |
NEG, Negative result; POS, Positive results.
Conclusions
We concluded that the modified ARMS RT- PCR technique could be used as a NIPST that can allow greater than 50% of couples at risk having a baby with CMBT to omit invasive prenatal testing.
Acknowledgments
We would like to thank all the staff members from the Obstetric Department of the Phayao Provincial Hospital for kindly referring the at-risk couples of CHBT for the NIPST investigation. We would like to acknowledge Rector, Professor Dr. Monthon Sanguansermsri and his administrative group for the permission to use the Phayao University endowment fund to support this project.
Funding Statement
Funding: none.
References
- 1.Fucharoen S, Winichagoon P. Haemoglobinopathies in Southeast Asia. Indian J Med Res 2011;134:498-506. [PMC free article] [PubMed] [Google Scholar]
- 2.Lo YM, Tein MS, Lau TK, et al. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am J Hum Genet 1998;62:768-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zafari M, Kosaryan M, Gill P, et al. Non-invasive prenatal diagnosis of β- thalassemia by detection of the cell-free fetal DNA in maternal circulation: a systematic review and meta-analysis. Ann Hematol 2016;95:1341-50. [DOI] [PubMed] [Google Scholar]
- 4.Newton CP, Graham A, Heptinstall LE, et al. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res 1989;17:2503-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Old JM, Varawalla NY, Weatherall DJ. Rapid detection and prenatal diagnosis of β-thalassaemia: studies in Indian and Cypriot populations in the UK. Lancet 1990;336:834-7. [DOI] [PubMed] [Google Scholar]
- 6.Fan HC, Blumenfeld YJ, Chitkara U, et al. Analysis of the size distributions of fetal and maternal cell-free DNA by paired-end sequencing. Clin Chem 2010;56:1279-86. [DOI] [PubMed] [Google Scholar]
- 7.Srivorakun H, Fucharoen G, Sae-Ung N, et al. Analysis of fetal blood using capillary electrophoresis system: a simple method for prenatal diagnosis of severe thalassemia diseases. Eur J Haematol 2009;83:57-65. [DOI] [PubMed] [Google Scholar]
- 8.Charoenkwan P, Sirichotiyakul S, Phusua A, et al. High-resolution melting analysis for prenatal diagnosis of betathalassemia in northern Thailand. Int J Hematol 2017;106:757-64. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, Holzgreve W, Hahn S. Size fractionation of cell-free DNA in maternal plasma improves the detection of a paternally inherited beta-thalassemia point mutation by MALDI-TOF mass spectrometry. Fetal Diagn Ther 2009;25:246-9. [DOI] [PubMed] [Google Scholar]
- 10.Xiong L, Barrett AN, Hua R, et al. Noninvasive prenatal testing for fetal inheritance of maternal β-thalassaemia mutations using targeted sequencing and relative mutation dosage: a feasibility study. BJOG 2018;125:461-8. [DOI] [PubMed] [Google Scholar]
- 11.Wang W, Yuan Y, Zheng H, et al. A pilot study of noninvasive prenatal diagnosis of alpha - and beta thalassemia with target capture sequencing of cell-free fetal DNA in maternal blood. Genet Test Mol Biomarkers 2017;21:433-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carlberg K, Bose N, Deng J, et al. Towards the development of a noninvasive prenatal test for beta-thalassemia: Utilization of probe capture enrichment and next generation sequencing. Blood 2016;128:3622. [Google Scholar]
- 13.Hudecova I, Chiu RW. Non-invasive prenatal diagnosis of thalassemias using maternal plasma cell free DNA. Best Pract Res Clin Obstet Gynaecol 2017;39:63-73. [DOI] [PubMed] [Google Scholar]
- 14.Galbiati S, Monguzzi A, Damin F, et al. COLD-PCR and microarray: two independent highly sensitive approaches allowing the identification of fetal paternally inherited mutations in maternal plasma. J Med Genet 2016;53:481-7. [DOI] [PubMed] [Google Scholar]
- 15.Chen JJ, Tan JAMA, Chua KH, et al. Non-invasive prenatal diagnosis using fetal DNA in maternal plasma: a preliminary study for identification of paternally inherited alleles using single nucleotide polymorphisms. BMJ Open 2015;5:e007648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breveglieri G, Travan A, D'Aversa E, et al. Postnatal and non-invasive prenatal detection of β-thalassemia mutations based on Taqman genotyping assays. PLoS One 2017;24:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Debrand E, Lykoudi A, Bradshaw E, Allen SK. A Non- Invasive Droplet Digital PCR (ddPCR) Assay to Detect Paternal CFTR Mutations in the Cell- Free Fetal DNA (cffDNA) of Three Pregnancies at Risk of Cystic Fibrosis via Compound Heterozygosity. PLoS One 2015;10:e0142729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramezanzadeh M, Salehi M, Farajzadegan Z, et al. Detection of paternally inherited fetal point mutations for β-thalassemia in maternal plasma using simple fetal DNA enrichment protocol with or without whole genome amplification: an accuracy assessment. J Matern Fetal Neonatal Med 2016;29:2645-9. [DOI] [PubMed] [Google Scholar]
- 19.Saba L, Masala M, Capponi V, et al. Non-invasive prenatal diagnosis of beta-thalassemia by semiconductor sequencing: a feasibility study in the Sardinian population. Eur J Hum Genet 2017;25:600-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu S, Chen L, Zhang X, et al. Primerintroduced restriction analysis polymerase chain reaction method for noninvasive prenatal testing of β-thalassemia. Hemoglobin 2015;39:18-23. [DOI] [PubMed] [Google Scholar]
- 21.Khordadpoor-Deilamani F, Akbari MT. The use of cell-free fetal DNA in maternal plasma for noninvasive prenatal linkage analysis in beta globin gene cluster. Brattish Lek Listy 2015;116: 662-5. [DOI] [PubMed] [Google Scholar]
- 22.Yenilmez ED, Tuli A, Evrüke IC. Noninvasive prenatal diagnosis experience in the Çukurova Region of Southern Turkey: detecting paternal mutations of sickle cell anemia and β- thalassemia in cell-free fetal DNA using high-resolution melting analysis. Prenat Diagn 2013;33:1054-62. [DOI] [PubMed] [Google Scholar]
- 23.Chang MY, Ahn S, Kim MY, et al. Onestep noninvasive prenatal testing (NIPT) for autosomal recessive homozygous point mutations using digital PCR. Sci Rep 2018;8:2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perlado S, Bustamante-Aragonés A, Donas M, et al. Fetal Genotyping in Maternal Blood by Digital PCR: Towards NIPD of Monogenic Disorders Independently of Parental Origin. PLoS One 2016:11:e015325. [DOI] [PMC free article] [PubMed] [Google Scholar]


