The Mycobacterium tuberculosis mycolate flippase MmpL3 has been the proposed target for multiple inhibitors with diverse chemical scaffolds. This diversity in chemical scaffolds has made it difficult to predict compounds that inhibit MmpL3 without whole-genome sequencing of isolated resistant mutants.
KEYWORDS: Mycobacterium tuberculosis, antimicrobials, cell envelope, mechanisms of resistance, phenotypic screening
ABSTRACT
The Mycobacterium tuberculosis mycolate flippase MmpL3 has been the proposed target for multiple inhibitors with diverse chemical scaffolds. This diversity in chemical scaffolds has made it difficult to predict compounds that inhibit MmpL3 without whole-genome sequencing of isolated resistant mutants. Here, we describe the identification of four new inhibitors that select for resistance mutations in mmpL3. Using these resistant mutants, we conducted a targeted whole-cell phenotypic screen of 163 novel M. tuberculosis growth inhibitors for differential growth inhibition of wild-type M. tuberculosis compared to the growth of a pool of 24 unique mmpL3 mutants. The screen successfully identified six additional putative MmpL3 inhibitors. The compounds were bactericidal both in vitro and against intracellular M. tuberculosis. M. tuberculosis cells treated with these compounds were shown to accumulate trehalose monomycolates, have reduced levels of trehalose dimycolate, and displace an MmpL3-specific probe, supporting MmpL3 as the target. The inhibitors were mycobacterium specific, with several also showing activity against the nontuberculous mycobacterial species M. abscessus. Cluster analysis of cross-resistance profiles generated by dose-response experiments for each combination of 13 MmpL3 inhibitors against each of the 24 mmpL3 mutants defined two clades of inhibitors and two clades of mmpL3 mutants. Pairwise combination studies of the inhibitors revealed interactions that were specific to the clades identified in the cross-resistance profiling. Additionally, modeling of resistance-conferring substitutions to the MmpL3 crystal structure revealed clade-specific localization of the residues to specific domains of MmpL3, with the clades showing differential resistance. Several compounds exhibited high solubility and stability in microsomes and low cytotoxicity in macrophages, supporting their further development. The combined study of multiple mutants and novel compounds provides new insights into structure-function interactions of MmpL3 and small-molecule inhibitors.
INTRODUCTION
In efforts to identify new tuberculosis (TB) antibiotics, whole-cell-based phenotypic screens have been conducted against the pathogen Mycobacterium tuberculosis. Over the last decade, several of these screens have identified MmpL3 as the proposed target for diverse small-molecule inhibitors, including AU1235, BM212, C215, DA-5, E11, indole carboxamides, HC2091, NITD-349, PIPD1, rimonabant, Spiro {N-benzyl-6′,7′-dihydrospiro(piperidine-4,4′-thieno[3,2-c]pyran)}, TBL-140, THPP (tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide), and SQ109 {N-adamantan-2-yl-N′-[(E)-3,7-dimethyl-octa-2,6-dienyl]-ethane-1,2-diamine} (1–12). MmpL3 is an essential flippase responsible for transporting trehalose monomycolate (TMM) or acylated-trehalose monomycolate synthesized in the cytoplasm to the pseudoperiplasmic space (13–17). These TMMs are then converted into trehalose dimycolate (TDM) by the Ag85 complex in the cell envelope (18). Additionally, MmpL3 has recently been proposed to transport phosphatidylethanolamine (19), expanding the function of MmpL3 lipid transport. MmpL3 is essential, as evidenced by a preexisting rescue allele being required to generate an mmpL3 knockout (1, 14, 17, 20–22), the lack of mutants in high-throughput transposon mutagenesis screens (23, 24), and studies that show rapid killing in vitro and in vivo in acute infection models when mmpL3 expression is conditionally inhibited (14, 20). This makes MmpL3 an attractive target for drug development, with one of its inhibitors, SQ109, currently in clinical trials (25).
MmpL3 inhibitors fall into diverse classes of chemical scaffolds (26–28), making it hard to computationally predict potential MmpL3 inhibitors based on chemical scaffolds. However, given the frequent finding of MmpL3 as a target, it is reasonable to expect that many new hits in a high-throughput screen may be acting against MmpL3. MmpL3 inhibitors have been identified by the isolation and sequencing of resistant mutants with single nucleotide variations (SNVs) mapping to the coding region of mmpL3, which is time consuming and costly. Efforts to discover MmpL3 inhibitors using targeted approaches include generating hypomorphs, where an mmpL3 knockdown strain showed enhanced sensitivity to MmpL3 inhibitors, including AU1235 (14). However, this strain was also shown to be sensitive to isoniazid (INH), an inhibitor of InhA of the fatty acid synthase II (FAS-II) pathway involved in mycolic acid synthesis, suggesting that while an mmpL3 knockdown strain has robust screening potential for inhibitors of mycolic acid synthesis, maturation, and transport, such strains are not specific enough to identify inhibitors that selectively target MmpL3.
An alternative approach, employed in this study, is to use a pool of inhibitor-resistant mmpL3 mutants to discover potential MmpL3 inhibitors. MmpL3 is a member of the resistance nodulation and division (RND) family of proteins, normally associated with efflux pumps in Gram-negative bacteria (1, 13, 17). However, evidence suggests MmpL3 does not act as a general efflux pump in resistant backgrounds, as resistant mutants do not differ in the amounts of inhibitor isolated from cell fractions compared to the results for wild-type (WT) M. tuberculosis (1). Further supporting the idea that MmpL3 does not act as an efflux pump, the low levels of cross-resistance to compounds not associated with MmpL3 inhibition, including INH, suggest that MmpL3 does not act as a general efflux pump (22). This suggests that MmpL3 inhibitor-resistant mutants could be used to screen for other potential MmpL3 inhibitors. The goal of this study was to discover MmpL3 inhibitors from a collection of 163 newly discovered, uncharacterized inhibitors of M. tuberculosis growth (29). Here, we describe the identification of four novel MmpL3 inhibitors by isolation of resistant M. tuberculosis mutants with mutations mapping to mmpL3. These 24 unique M. tuberculosis mmpL3 mutant strains were then pooled into a single batch culture to conduct a targeted whole-cell phenotypic screen, identifying six new scaffolds with reduced activity in the mixed mutant population compared to the results for the wild type. Cross-resistance and compound interaction studies demonstrated specific structure function interactions between the molecules and MmpL3 and defined domains of MmpL3 associated with differential resistance to MmpL3 inhibitors.
RESULTS
Identification of four new MmpL3 inhibitors by isolation of resistant mutants.
Previously, two high-throughput screens were conducted, targeting the two-component regulatory systems DosRST and PhoPR (29–31). In addition to inhibitors targeting these pathways, a series of compounds was identified that inhibited M. tuberculosis growth independently of the targeted pathways (11, 29, 31). A series of high-throughput assays were then conducted to prioritize these compounds (Fig. S1 in the supplemental material), including confirming hits, testing for eukaryotic cytotoxicity in primary murine bone marrow-derived macrophages (BMMϕ) (≤10% cytotoxicity), and testing for the ability of the compounds to inhibit M. tuberculosis growth inside BMMϕ (≥25% growth inhibition). The results of these screens identified 216 compounds, of which 163 commercially available compounds were purchased as fresh powders. In order to identify the mechanisms of action of these M. tuberculosis growth inhibitors, our laboratory selected several compounds with potent M. tuberculosis growth inhibition, both in vitro and in macrophages, as well as low murine macrophage cytotoxicity.
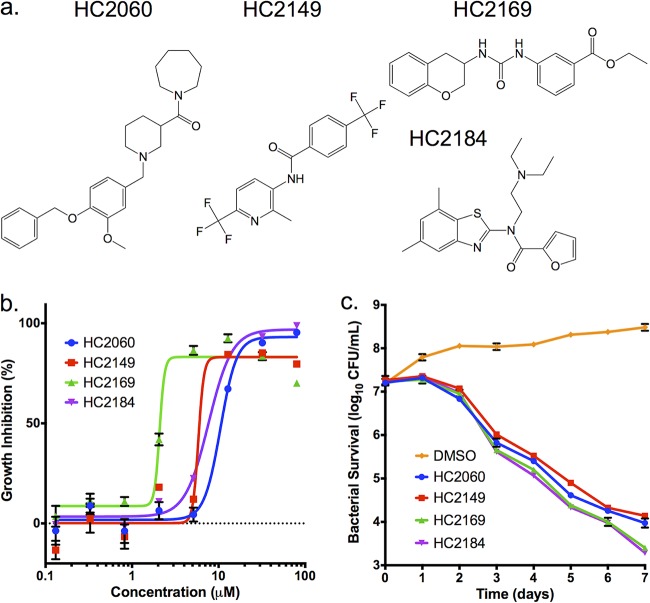
Four compounds of interest, HC2060, HC2149, HC2169, and HC2184 (1-{(1-[4-(benzyloxy)-3-methoxybenzyl]piperidin-3-yl}carbonyl)azepane, N-[2-methyl-6-(trifluoromethyl)pyridin-3-yl]-4-(trifluoromethyl)benzamide, ethyl 3-{[(3,4-dihydro-2H-chromen-3-ylamino)carbonyl]amino}benzoate, and N-(2-diethylaminoethyl)-N-(5,7-dimethyl-1,3-benzothiazol-2-yl)furan-2-carboxamide, respectively) (Fig. 1a) had half-maximal effective concentrations (EC50s) ranging from 1.8 μM to 16.9 μM in vitro (Fig. 1b, Table 1). All four compounds had bactericidal activity when measured at 20 μM (2× the initial screening concentration) (Fig. 1c). To our knowledge, the structures of these compounds are distinct from those of previously described inhibitors of M. tuberculosis growth.
FIG 1.
Four compounds inhibit M. tuberculosis growth in a dose- and time-dependent manner. (a) Structures of HC2060, HC2149, HC2169, and HC2184. (b) Inhibition of M. tuberculosis growth in a dose-dependent manner. (c) Killing of M. tuberculosis in a time-dependent manner when treated with the inhibitors at 20 μM. Error bars indicate the standard deviations from the mean values. Experiments were conducted in biological triplicates.
TABLE 1.
Characterization of MmpL3 inhibitors
| Compound | Clade | EC50 (μM) |
Δψ disruption | CC50 (μM) | Solubility (μM) | Microsome stability (% remaining after 30 min) | |||
|---|---|---|---|---|---|---|---|---|---|
| WT | mmpL3 mutant pool | Differentiala | Mϕ | ||||||
| HC2032 | B | 2.2 | >80 | >36 | 0.8 | Yes | >100 | 18 | 102 |
| HC2060 | B | 16.9 | >80 | >5 | 4.1 | No | >100 | >300 | 44 |
| HC2091 | B | 6.2 | >80 | >13 | 2.2 | No | >100 | >300 | 45 |
| HC2099 | B | 1.7 | 38.9 | 23 | <0.3 | Yes | >100 | 178 | 71 |
| HC2134 | A | 1.4 | >80 | >57 | 7.3 | Yes | >100 | 116 | NDb |
| HC2138 | A | 4.0 | >80 | >20 | <0.3 | Yes | >100 | 66 | 122 |
| HC2149 | A | 6.6 | >80 | >12 | 3.6 | Yes | >100 | 131 | 138 |
| HC2169 | A | 1.8 | >80 | >44 | <0.3 | No | >100 | 17 | 168 |
| HC2178 | B | 3.8 | >80 | >24 | 2.0 | Yes | >100 | >200 | 4 |
| HC2183 | B | 3.2 | 59.9 | 19 | 3.0 | No | >100 | >200 | 25 |
| HC2184 | B | 7.6 | >80 | >11 | 0.7 | Yes | >100 | >300 | 30 |
| C215 | B | 11.2 | 57.5 | 5 | 4.0 | Yes | 14.3 | 87 | 62 |
| SQ109 | B | 2.4 | 6.9 | 2 | <0.3 | Yes | ND | ND | ND |
Differential EC50 is the fold difference between the EC50s of the WT and the mmpL3 mutant pool.
ND, not determined.
To understand the mechanism of action of these four compounds, resistant mutants were isolated using solid agar plates (7H10 oleic acid-albumin-dextrose-catalase [OADC]) amended with 20 or 40 μM of each compound and inoculated with 109 CFU of M. tuberculosis (strain Erdman). Isolated mutants were tested for resistance via dose-response curves. Confirmed resistant clones were isolated as single colonies and retested to confirm resistance (Fig. S2a to d). Genomic DNA was extracted from confirmed resistant mutant strains, and the genomes were sequenced. Analysis of the genome sequences identified SNVs in the coding region of mmpL3 (Rv0206c) in all of the genomes (Table S1). These SNVs encoded nonsynonymous mutations located throughout the gene (Table S1, Fig. S2e). These findings suggest that these compounds may be functioning as MmpL3 inhibitors.
Modulation of TMM and TDM accumulation.
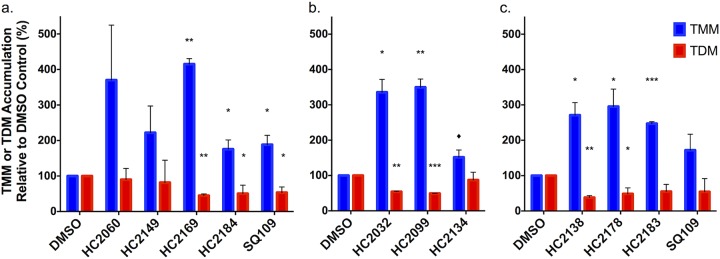
MmpL3 is responsible for the transport of TMM across the inner membrane (14–16, 18). To determine if these compounds inhibited the activity of MmpL3, cultures of M. tuberculosis were grown in the presence of [14C]acetate and treated for 24 h with 20 μM HC2060, HC2149, HC2169, HC2184, or SQ109 or equal volumes of dimethyl sulfoxide (DMSO). Radiolabeled lipids were isolated and analyzed by thin-layer chromatography (TLC) (Fig. 2a; Fig. S3a). The results of the lipid assay show that TMM accumulates in M. tuberculosis samples treated with the proposed MmpL3 inhibitors, as well as in the SQ109-treated samples. Additionally, TDM decreased significantly in cultures treated with HC2169 and HC2184, as well as in those treated with the positive control, SQ109 (Fig. 2a; Fig. S3a). These results are consistent with previously described results for MmpL3 inhibitors (1–5, 8, 10, 11, 27) and support the idea that these four compounds inhibit MmpL3 activity.
FIG 2.
Modulation of TMM and TDM accumulation. (a) Whole-cell 14C-lipids from M. tuberculosis cells treated with 20 μM HC2060, HC2149, HC2169, and HC2184 show increased levels of TMM and decreased levels of TDM. (b and c) Whole-cell 14C-lipids from M. tuberculosis cells treated with a concentration of 20 μM of the six inhibitors identified by the targeted phenotypic screen show increased levels of TMM and decreased levels of TDM. Experiments were conducted in biological duplicates. In both experiments, M. tuberculosis samples were treated with DMSO or 20 μM SQ109 as controls. Error bars indicate the standard deviations. *, P < 0.05; **, <0.005; ***, <0.001; ♦, value that just missed the cut off (P = 0.07 compared to TMM level after HC2134 treatment). The results for HC2060 and HC2149 missed the significance cutoff, but this may be due to the high variability in replicates, as there was a >2-fold difference for HC2060 and HC2149.
Targeted whole-cell phenotypic screen for MmpL3 inhibitors.
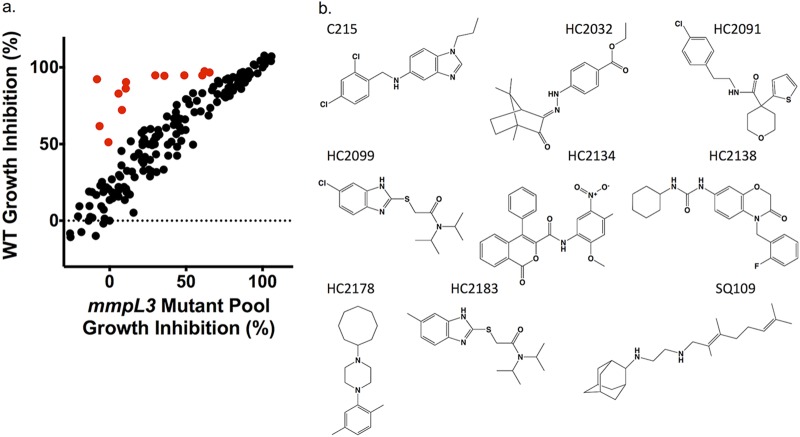
The identification of four new MmpL3 inhibitors, as well as the previously published inhibitor HC2091 (6), suggested that additional MmpL3 inhibitors may exist in the prioritized 163-compound library of M. tuberculosis growth inhibitors (Fig. S1). Review of the known MmpL3 inhibitor scaffolds and those in our compound library identified HC2172 as the previously described MmpL3 inhibitor C215 (3). A recent study by McNeil et al. showed that mmpL3 mutant strains had low levels of cross-resistance against non-MmpL3 inhibitors (22), suggesting that mmpL3 mutants could be used to screen for MmpL3 inhibitors. Additionally, this study also showed that different mutations conferred various levels of cross-resistance between MmpL3 inhibitors. We therefore hypothesized that by pooling unique mmpL3 mutant strains into a single mixed culture, we could overcome limitations of cross-resistance variability. For the targeted phenotypic screen, we directly compared percentages of growth inhibition (% GI) of the WT and a mixed mmpL3 mutant pool consisting of 24 unique mmpL3 mutant strains, including 3 strains previously described as resistant to HC2091 (Table S2) (11). We tested for differences in growth between the WT and the 24 unique mmpL3 strains and did not observe major growth differences following 9 days of incubation. Given that mutant abundances were normalized at the beginning of the assay and rates of growth were similar by day 6 when the assay was completed, the relative abundance of each strain is not anticipated to bias the screen (Fig. S4). The cultures were treated with each of the 163 M. tuberculosis growth inhibitors at 20 μM, as well as DMSO (negative control), rifampin (RIF [positive control]), bedaquiline (BDQ), clofazimine (CLO), INH, para-aminosalicylic acid (PAS), H2O2, or SQ109, for a total of 171 different treatments (Fig. S5a and b). The results of this screen identified 32 compounds with 15% GI in the WT background and 1.5× reduced activity in the mixed mmpL3 mutant background relative to that in the WT background (examples of positive hits are illustrated in red in Fig. 3a). These hits were tested by dose-response experiments conducted in both the WT and mixed mmpL3 mutant background. In total, we identified 13 compounds with reduced activity in the mixed mmpL3 mutant background (Table 1, Fig. S6). Included in our confirmed hits were each of the five inhibitors used to generate the mmpL3 mutant strains (HC2060, HC2091, HC2149, HC2169, and HC2184) and the two control compounds C215 and SQ109. The targeted screen also identified six novel inhibitors, HC2032, HC2099, HC2134, HC2138, HC2178, and HC2183 {ethyl 4-[(2E)-2-(4,7,7-trimethyl-3-oxo-2-bicyclo[2.2.1]heptanylidene)hydrazinyl]benzoate, 2-[(6-chloro-1H-benzimidazol-2-yl)sulfanyl]-N,N-di(propan-2-yl)acetamide, N-(2-methoxy-5-nitrophenyl)-1-oxo-4-phenylisochromene-3-carboxamide, 1-cyclohexyl-3-[4-[(2-fluorophenyl)methyl]-3-oxo-1,4-benzoxazin-7-yl]urea, 1-cyclooctyl-4-(2,5-dimethylphenyl)piperazine, and 2-[(5-methyl-1H-benzimidazol-2-yl)sulfanyl]-N,N-di(propan-2-yl)acetamide, respectively} (Fig. 3b), which have not been previously described as MmpL3 inhibitors. The amount of resistance conferred by the mixed mmpL3 mutant strains against each compound varied, with some compounds, like HC2032, HC2138, and HC2169, losing nearly all activity in the mutant background (Fig. S6), as indicated by the highly differential EC50s (fold difference between the EC50s of the mmpL3 mutant pool and WT) of >36, >20, and >44 (Table 1). Despite the high activity of SQ109 in the WT background, the differential EC50 was only 2 (Table 1); however, this observation is consistent with previous studies which only report marginal increases in MICs in mmpL3 mutant backgrounds (4, 11, 22, 27). Included in our hits were two urea-based compounds, HC2138 and HC2169 (Fig. 1 and Fig. 3b). These urea-based compounds have structures reminiscent of that of the adamantyl-urea MmpL3 inhibitor AU1235 (1). Additionally, two of the compounds identified in the screen, HC2099 and HC2183, had high structure similarity. We also tested our mixed mutant population against rimonabant, an analogue of BM212, previously shown to bind to MmpL3 (12); however, we did not identify any difference between the WT or the mixed mutant population (Fig. S7). This finding is consistent with the mutant pool not containing mutations known to provide resistance to BM212 or rimonabant (2) and demonstrates that the current mutant pool does not comprehensively identify all known MmpL3 inhibitors.
FIG 3.
A targeted whole-cell phenotypic screen identifies six new MmpL3 inhibitors. (a) Results of a direct head-to-head comparison of percentages of growth inhibition of WT M. tuberculosis and a pooled mmpL3 mutant population treated with 163 compounds at 20 μM. Additional treatments included 0.5 μM BDQ, CLO, INH, PAS, or SQ109 or 0.03% H2O2. Examples of hit compounds with reduced activity in the MmpL3 mutant pool are shown in red. (b) Structures of the confirmed hit compounds from the screen, including six new compounds, HC2032, HC2099, HC2134, HC2138, HC2178, and HC2183. Previously described compounds include C215, HC2091, and SQ109.
The compounds were also tested for murine macrophage cytotoxicity, solubility, and stability in mouse microsomes, and the structures were confirmed by mass spectrometry (Table 1). The compounds exhibited low cytotoxicity (50% cytotoxic concentration [CC50] of >100 μM), consistent with our secondary assay screening (Fig. S1). The compounds exhibited various levels of solubility, with HC2169 and HC2138 showing lower solubility (66 μM and 17 μM, respectively) but high microsome stability (122% and 168%, respectively) and compounds like HC2183 showing high solubility (>200 μM) but low microsome stability (25%). Interestingly HC2099, which has high structure similarity to HC2183, showed higher solubility (178 μM) and higher microsome stability (71%). Several of the compounds (e.g., HC2091, HC2099, HC2138, and HC2149) exhibited favorable solubility and microsome stability with no observed macrophage cytotoxicity, supporting their potential for further development.
The phenotypic screen was selective, as it did not identify any of the control treatments known not to target MmpL3, including BDQ, CLO, INH, PAS, H2O2, or HC2051, a proposed Pks13 inhibitor (given its similarity to TAM16 [32]). To confirm the specificity of our screen, we conducted dose-response studies in both the WT and mixed mmpL3 mutant background for each of the aforementioned inhibitors, as well as RIF. The results of the dose-response studies did not identify any significant levels of resistance to these compounds in the mixed mmpL3 mutant background (Table S3, Fig. S8). This was true for inhibitors both of mycolic acid synthesis and maturation (INH and HC2051), suggesting that our screen was specific for inhibitors of MmpL3. Consistent with previous results, we identified increased susceptibility to RIF treatment in the mixed mmpL3 mutant background (Table S3, Fig. S8) (22). The dose-response profiles for BDQ, CLO, and PAS did not show any differences in susceptibility, further supporting the idea that mmpL3 mutations do not confer resistance through general efflux.
Modulation of TDM, membrane potential, and viability.
To determine whether the six compounds identified in the screen can inhibit MmpL3 activity, we examined the accumulation of TMM and TDM as described above. The inhibitors modulated mycolic acid accumulation in whole-cell extracts, with lipids for all treatments showing significant accumulation of TMM (except for HC2134) and treatment with HC2032, HC2099, HC2138, and HC2178 showing significant decreases in TDM relative to the amounts in the DMSO control samples (Fig. 2b and c, Fig. S3b). A recent report has shown that because MmpL3 activity is dependent on the proton motive force (PMF), disruptors of PMF, such as the protonophore carbonyl cyanide m-chlorophenyl hydrazine (CCCP), can also modulate MmpL3 activity (27). Studies have suggested that some proposed MmpL3 inhibitors, such as SQ109 and E11, may indirectly target MmpL3 through disruption of the membrane potential (10, 15, 27), although direct interactions of MmpL3 and SQ109 have recently been reported (12, 33). To determine whether the newly identified inhibitors disrupt membrane potential (Δψ), we conducted dose-response studies using a DiOC2(3)-based assay. Some compounds, including HC2060, HC2169, and HC2183, did not disrupt membrane potential (Table 1, Fig. S9), while others, such as HC2032, HC2099, HC2134, HC2138, HC2149, HC2178, HC2184, and C215, did disrupt membrane potential (Table 1, Fig. S9). Consistent with previous observations, HC2091 did not disrupt membrane potential, while SQ109 did disrupt membrane potential (Table 1, Fig. S9) (11, 15, 27). Surprisingly, there were differences in outcome for the two urea-containing compounds HC2169 and HC2138, as well as between HC2099 and HC2183, which only differ by a chloro and a methyl substitution, respectively. The results for HC2138 and HC2169 are also interesting because the previously described urea-based MmpL3 inhibitor AU1235 does not disrupt the membrane potential (15, 27). These results suggest that the ability to disrupt membrane potential is highly structure specific.
Inhibition of MmpL3 has been shown to have bactericidal effects (20, 27); therefore, we tested these compounds for bactericidal activity using a firefly luciferase reporter strain of M. tuberculosis in conjunction with a luciferase assay (34, 35). This assay relies on active luciferase, generated by the reporter M. tuberculosis strain, and the presence of ATP, which is generated in living cells but rapidly hydrolyzed in lysed cells. All of these compounds showed bactericidal activity (Fig. S10). These results suggest that the growth inhibition is due to compounds killing M. tuberculosis in a dose-dependent manner. The bactericidal activity of these inhibitors is consistent with these compounds targeting MmpL3, which is essential for cell viability (14, 20), although other potential mechanisms, such as modulation of PMF or membrane integrity, are also consistent with bacterial killing.
Direct binding of inhibitors to MmpL3.
To determine if the inhibitors interact directly with MmpL3, we used a recently described fluorescence-based competitive binding assay in a whole-cell Mycobacterium smegmatis mmpL3 mutant expressing M. tuberculosis mmpL3 (mmpL3tb) (M. smegmatis MsmgΔmmpL3/pMVGH1-mmpL3tb) (33). This assay utilizes a fluorophore probe (North 114) consisting of an analogue of the NITD series of MmpL3 inhibitors covalently linked to the fluorophore TAMRA (6-carboxytetramethylrhodamine). North 114 has previously been shown to bind directly to MmpL3, and the addition of MmpL3 inhibitors displaces North 114, thus allowing a competitive binding assay (33). We tested all of the new MmpL3 inhibitors in this assay, along with NITD-349 as a positive control and INH and RIF as negative controls. The results of the assay showed that the inhibitors led to displacement of North 114 similarly to the positive control NITD-349 (Fig. 4) (6). Each of the inhibitors led to displacement of North 114 at concentrations starting at 2 μM, with the exception of HC2178 and C215, which had modest activity at the highest tested concentration of 8 μM (Fig. 4). SQ109 has previously been shown to displace North 114 (33) and was not tested for this study. The results of the competitive binding assays, coupled with those of the lipid assay and the decreased activity in the mixed mmpL3 mutant background, support MmpL3 as a direct target for the compounds identified in our screening approach. However, these findings do not rule out the possibility that secondary activity independent of MmpL3 may contribute to whole-cell activity, particularly for the compounds shown to modulate membrane potential.
FIG 4.
Flow cytometry-based competition binding assay using intact M. smegmatis cells expressing M. tuberculosis MmpL3 (MmpL3tb). The assay was performed in an M. smegmatis mmpL3 deletion mutant expressing the wild-type mmpL3tb gene (M. smegmatis MsmgΔmmpL3/pMVGH1-mmpL3tb). Cells were labeled with 4 mM North 114 and subsequently treated with increasing concentrations of the inhibitors. Shown on the y axis are the mean fluorescence intensities (MFI) of the bacilli from each treatment group expressed relative to the MFI of bacilli not treated with any inhibitor (relative fluorescence intensity [RFI] arbitrarily set to 1). MFIs were determined by analyzing 10,000 bacilli under each condition. The data reported are mean values ± standard deviations of technical duplicates. Replicate samples were analyzed by t test. *, P ≤ 0.05; ♦, P ≤ 0.1.
Spectrum of activity.
While MmpL3 is conserved in mycobacteria, functional orthologs are not found in other bacteria and fungi. Despite this, several proposed MmpL3 inhibitors, including BM212, THPP, and SQ109, have been shown to inhibit multiple bacterial and eukaryotic species (4, 36–38), while other MmpL3 inhibitors, including HC2091, AU1235, and indole carboxamides, are specific to mycobacteria. To define the spectrum of activity, the compounds were tested against several diverse species, including Staphylococcus aureus, Escherichia coli, Pseudomonas aeruginosa, Proteus vulgaris, and Enterococcus faecalis (Table 2). For HC2032, HC2060, HC2099, HC2149, HC2169, HC2178, and HC2184, even at high concentrations (200 μM), no inhibition was observed against nonmycobacteria. However, these inhibitors were positive for activity against other mycobacteria, including the pathogenic nontuberculous mycobacterial species M. abscessus and the saprophytic species M. smegmatis (Table 2). For example, HC2091, HC2099, and HC2134 exhibited MIC50s of 6.25 μM, 25 μM, and 12.5 μM, respectively, against M. abscessus. Additionally, all of the MmpL3 inhibitors tested, except HC2149 and C215, were active against M. smegmatis. This suggests that most of the inhibitors are specific for mycobacteria and may be effective against diverse mycobacterial species. The specificity may be driven by the conservation of MmpL3 in mycobacteria or, should the killing be dependent on a secondary activity independent of MmpL3, by the Mycobacterium specificity of the secondary target(s), differences in cell envelope composition that limit compound permeability, or differences in metabolism of the compounds. The observation that HC2134 and C215 are active against nonmycobacterial species has been seen with other MmpL3 inhibitors (25, 37, 38), and this result may be due to nonspecific activities, such as PMF disruption.
TABLE 2.
Spectrum of activity of MmpL3 inhibitors
| Organism | EC50 (% GI for M. abscessus) of: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HC2032 | HC2060 | HC2091 | HC2099 | HC2134 | HC2138 | HC2149 | HC2169 | HC2178 | HC2184 | C215 | |
| M. tuberculosis Erdman | 3.0 | 14.8 | 7.0 | 5.3 | 2.1 | 2.3 | 11 | 2.4 | 3.7 | 8.9 | 16.2 |
| M. tuberculosis CDC1551 | 2.4 | 12.8 | 6.3 | 4.8 | 1.5 | 2.0 | 10.5 | 2.2 | 2.3 | 7.6 | 14.3 |
| M. abscessus (% GI)a | 34.5 | 13.6 | 96.5 | 81.8 | 81.9 | 8.2 | −28 | −36 | 13.5 | 7.0 | 2.3 |
| M. smegmatis | 2.2 | 80 | 20b | 0.9 | 1.8 | NDc | >200 | 13.2 | 4.5 | 85.7 | >200 |
| S. aureus ATCC 29213 | >200 | >200 | >200b | >200 | 51.0 | ND | >200 | >200 | >200 | >200 | 15.1 |
| S. aureus ATCC 25923 | >200 | >200 | >200b | >200 | 51.8 | ND | >200 | >200 | >200 | >200 | 22.8 |
| E. coli | >200 | >200 | >200b | >200 | >200 | ND | >200 | >200 | >200 | >200 | >200 |
| P. vulgaris | >200 | >200 | >200b | >200 | 35.1 | ND | >200 | >200 | >100 | >200 | >200 |
| E. faecalis | >200 | >200 | >200b | >200 | >200 | ND | >200 | >200 | >200 | >200 | 34 |
| P. aeruginosa | >200 | >200 | >200b | >200 | >200 | ND | >200 | >200 | >200 | >200 | >200 |
All values for M. abscessus are percentages of growth inhibition (% GI) at the single concentration of 20 μM.
Previously published data from reference 9.
ND, not determined.
Activity against intracellular M. tuberculosis.
The compounds were tested against M. tuberculosis growing in BMMϕ using the luciferase-expressing M. tuberculosis strain described above. BMMϕ were infected with M. tuberculosis and treated with the inhibitors for 6 days across a range of concentrations (200 to 0.3 μM). The BMMϕ EC50s are summarized in Table 1 and Fig. S11. The results of the assay show that many of the inhibitors have bactericidal activity in Mϕ that is several magnitudes lower than the eukaryotic CC50, supporting a high selectivity index. The identification of bactericidal effects against M. tuberculosis in BMMϕ is consistent with genetic knockdown studies that show mmpL3 is essential for actively replicating bacteria (4, 5).
Cross-resistance profiles indicate specific MmpL3 protein-inhibitor interactions.
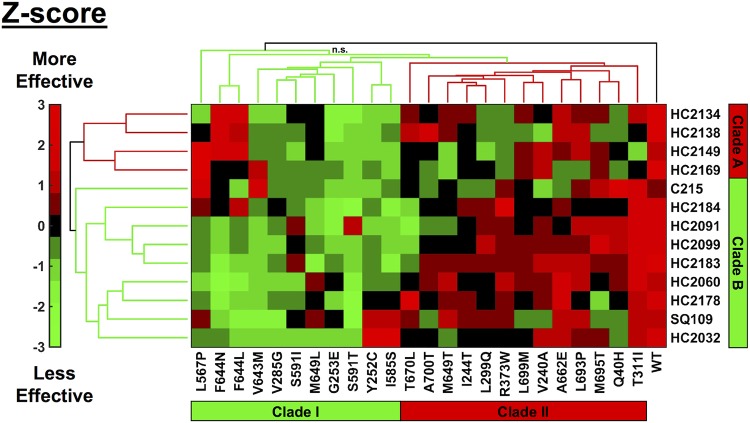
While the results of the screen showed potential for rapid identification of MmpL3 inhibitors, the screen relied on the use of a mixed mutant population. To resolve this issue, we conducted dose-response studies for each combination of the 24 unique mmpL3 mutants against each MmpL3 inhibitor identified from the screen (with WT M. tuberculosis Erdman or CDC1551 as a control). Because there was a complete lack of activity for compounds like HC2169 against HC2169-specific resistant mutants (Fig. S2c), units of measure such as EC50 and MIC cannot be calculated or are not a good measure for comparing responses. Instead, we used the area under the curve (AUC) for each dose response in the mmpL3 mutant backgrounds relative to the AUC for the WT response for a given treatment (Table S4). Because the compounds have differences in potency, the AUC for the WT for each treatment differs, and to account for this issue, we normalized our values by Z-score for each treatment (39). Cluster analysis grouped the data based on both treatment effectiveness and resistance conferred by each mmpL3 mutation. The resulting clustergram (Fig. 5) shows that both compounds and mmpL3 mutant strains, denoted by the amino acid substitutions, fall into distinct clades. The compounds fall into two distinct clades, clade A (Fig. 5, red), which contains HC2134, HC2138, HC2149, and HC2169, and clade B (Fig. 5, green), which contains HC2032, HC2060, HC2091, HC2099, HC2178, HC2183, HC2184, C215, and SQ109. The identification of two distinct clades of compounds suggested that the compounds may be interacting with the MmpL3 protein in distinct ways.
FIG 5.
Cross-resistance profiling identifies clustering of compounds and mutations. Cluster analysis of cross-resistance profiling of 24 mmpL3 strains treated with each of the 13 MmpL3 inhibitors normalized by Z-scoring by treatment. Compounds clustered into two clades: clade A and clade B. Mutant strains, denoted by amino acid substitution, clustered into two clades: clade I and clade II. Colors are based on Z-score normalization of treatment; green indicates when treatments were less effective than the average, and red indicates when treatments were more effective than the average. Black (n.s., not significant) indicates a branch where the approximate unbiased (AU) value was <75. All other branches were significant based on bootstrap AU values of >75.
The resistance mutations also showed specific clustering. Cluster analysis of the strains showed that the WT clustered on its own and the mutants formed a large complex cluster (Fig. 5). Within this large cluster, the mmpL3 mutant strains formed into two subclades, clade I (Fig. 5, green), whose mutations conferred relatively high resistance (lower inhibitor effectiveness), and clade II (Fig. 5, red), whose mutations conferred relatively low resistance (higher inhibitor effectiveness). Clade I contained 11 mmpL3 mutant strains, with mutations Y252C (encoding a change of Y to C at position 252), V285G, G253E, L567P, I585S, S591I, S591T, V643M, F644N, F644L, and M649L. Clade II consisted of the remaining 13 mmpL3 mutant strains, with mutations Q40H, V240A, I244T, L299Q, T311I, R373W, M649T, A662E, T670L, L693P, M695T, L699M, and A700T. Surprisingly, M649T fell into the clade II mutations; this was striking because the mmpL3 mutant with an M649L mutation was clustered with the clade I mmpL3 mutant strains.
Pairwise combination studies using DiaMOND.
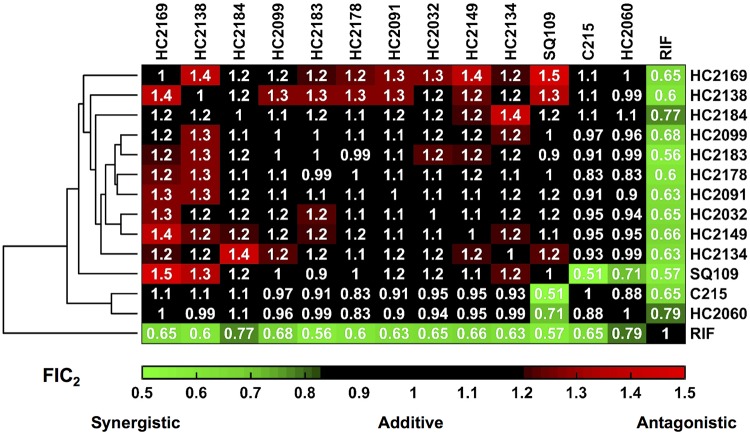
We hypothesized that the clustering of compounds into two clades was due to their having distinct interactions with MmpL3; therefore, combination treatments may reveal antagonistic, additive, or synergistic interactions. In order to test this hypothesis in whole-cell M. tuberculosis, we used the recently described diagonal measurement of n-way drug interactions (DiaMOND) approach (40). RIF was included as a control for these assays, as this drug has been shown to be synergistic when tested with other MmpL3 inhibitors, such as AU1235 and SQ109 (41, 42). The results of the DiaMOND analysis, shown in Fig. 6, identified synergistic interactions (fractional inhibitory concentration of a pairwise combination [FIC2] of <0.82) between all combinations of MmpL3 inhibitors and RIF. Additionally, the results identified mostly additive interactions (FIC2s of 0.82 to 1.18), consistent with the compounds sharing a single target. Interestingly, most combinations between MmpL3 inhibitors and the compounds HC2134, HC2138, HC2149, and HC2169 showed antagonistic interactions (FIC2 of >1.18). These four compounds were clustered together in clade A in the cross-resistance profiles described above (Fig. 5). This antagonistic relationship further supports that the clade A compounds are distinct from the clade B compounds. Another observation from the DiaMOND assay is that pairwise combinations of compounds HC2060, C215, and SQ109 all had synergistic interactions (Fig. 5). The reason for this observation is not clear, as the compounds did not have differential cross-resistance profiles (Fig. 4). Interestingly, combinations of HC2060 and C215, but not SQ109, with the clade A compounds HC2138 and HC2169 did not reveal antagonistic interactions but, instead, additive interactions (Fig. 6). This finding supports that HC2060 and C215 compounds interact with MmpL3 in a manner distinct from the other compounds.
FIG 6.
DiaMOND analysis identifies additive, synergistic, and antagonistic inhibitor interactions. Hierarchical cluster analysis of DiaMOND-based pairwise inhibitor interactions of all combinations of MmpL3 inhibitors and RIF identifies additive (FIC2 of 0.82 to 1.18), antagonistic (FIC2 > 1.18), and synergistic (FIC2 < 0.82) interactions.
3-D modeling of MmpL3 clade I and clade II substitutions.
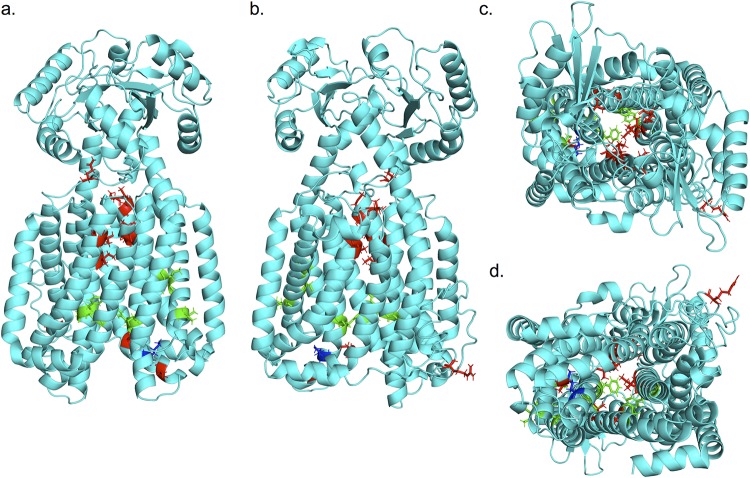
The cross-resistance profiles showed that the mmpL3 mutant strains clustered separately into two clades, clade I and clade II, with the WT clustering on its own as an outgroup (Fig. 5). In order to understand this observation, we generated a three-dimensional (3-D) model of the M. tuberculosis MmpL3 protein aligned to the recently described crystal structure of MmpL3 from M. smegmatis (C score of 0.17; root mean square deviation [RMSD], 8.4 ± 4.5 Å [mean ± standard deviation]) (12) (see https://zhanglab.ccmb.med.umich.edu/I-TASSER/example/cscore.txt for a definition of C score). Substitutions from the mmpL3 mutant strains used in the cross-resistance profiles are highlighted in the model (Fig. 7, red, green, and blue). Consistent with previously described resistant strains of M. tuberculosis, the majority of the substitutions localized along the central vestibule, with the exception of T670, R373, and A662, which did not align along the central vestibule of the model (Fig. 7) (17). This vestibule is conserved among the RND family of proteins and is responsible for the proton translocation that drives protein activity (17, 27). To understand the clustering pattern of the cross-resistance profiling, we highlighted the mutations based on their clades, revealing that the two distinct clades separated spatially in the model. The substitutions of clade I (Fig. 7, green), which conferred higher resistance, localized toward the cytoplasmic face of the protein, while the substitutions of clade II (Fig. 7, red), which generated lower resistance, localized into two separate locations: (i) toward the pseudoperiplasmic face of the protein and (ii) in another region, which does not line the central vestibule (Fig. 7). Interestingly, substitutions for M649 (Fig. 7, blue) separated into either clade I or clade II depending on the substitution. Structure-function profiling by Belardinelli and colleagues (17) had previously described seven essential residues for MmpL3 function (D251, S288, G543, D640, Y641, D710, and R715) that clustered in a single domain (17). This study also modeled substitutions commonly identified from mutants resistant to multiple inhibitors to this same region. To determine whether the two clades separated based on their approximation to this essential region, we highlighted these seven residues in the model (Fig. S12). Notably, the two clades separate based on their proximity to these residues, with clade I substitutions localizing in the same region as the essential residues and clade II substitutions localizing distally from the essential residues. This finding suggests that the strength of resistance conferred by a mutation may be dependent on the proximity of the substitution to residues essential for protein function.
FIG 7.
Mutation substitutions cluster according to cross-resistance clades. (a to d) Front, back, top, and bottom views, respectively, of an I-TASSER-predicted structure of M. tuberculosis MmpL3 based on M. smegmatis MmpL3 structure (PDB code 6AJH). Substitutions conferred by mutations in mmpL3 are indicated. Substitutions are colored based on clade from cross-resistance profiling as follows: green, clade I substitutions; red, clade II substitutions; blue, M649, which fell into both clades depending on the substitution. The model shows a truncated version (732/944 aa) of the MmpL3 protein lacking the C-terminal tail.
DISCUSSION
Over the last decade, MmpL3 has been identified as the proposed target of nearly a dozen small-molecule inhibitors of M. tuberculosis by the whole-genome sequencing of spontaneous resistant mutants. This process, while successful for identifying inhibitors of MmpL3, is time consuming due to the slow growth of M. tuberculosis and costly due to the expense of whole-genome sequencing. Here, we describe a simple growth-based assay to rapidly identify small-molecule inhibitors of M. tuberculosis growth that target MmpL3 using a pool of mmpL3 mutants. This process is specific to small molecules that inhibit MmpL3 activity, with no observable cross-resistance to non-MmpL3 inhibitors. Using a competitive binding assay and lipid accumulation assays, the compounds were found to directly inhibit MmpL3 activity. Cross-resistance profiling of MmpL3 inhibitors against each mmpL3 mutant indicated that both the inhibitors and mutants fell into separate clades. These clades indicated that the compounds interacted differently with MmpL3 and that the mutations conferred different levels of resistance based on the proximity of the substitutions to residues essential for MmpL3 function.
The screening approach described here is fast and accurate in identifying MmpL3 inhibitors; however, this screening approach relies on the potential for cross-resistance between inhibitors and mmpL3 mutant strains present in the mixed mutant population. While our approach was successful at identifying a total of 13 MmpL3 inhibitors, including SQ109, this screening platform could not identify rimonabant, an analogue of BM212 (12, 43). Rimonabant has been shown to have low cross-resistance with mmpL3 mutants isolated from other MmpL3 inhibitors, including SQ109 and AU1235 (12). Additionally, rimonabant has been shown to bind with MmpL3 differently than other MmpL3 inhibitors, including SQ109, AU125, and an indole carboxamide (ICA38), when cocrystalized with MmpL3 (12). While the current version of our screen could not identify rimonabant, future versions of our mixed mutant screening approach could include BM212- or rimonabant-specific mmpL3 mutants in the mixed mutant population, allowing a more robust screening platform. A total of 21 unique mutations in mmpL3 were identified in the genome sequences of the resistant mutants isolated. These mutations translated to substitutions that were a mixture of ones previously described and novel to this study. Included in this list were the previously described substitutions G253E, Y252C, T311I, L567P, S591I, V643M, F644L, and L699M (1, 4, 6–8, 10, 11, 22, 32). Mutations that were unique to this study included ones in positions V240, I244, V285, L299, R373, I585, A662, and L693. We also isolated mutants with mutations in positions Q40, Y252, G253, L567, S591, F644, M649, and L699, where mutations had previously been described to occur (1, 4, 7, 8, 10, 11, 22, 32); however, the exact substitutions in several of these strains’ positions, including Q40H, S591T, F644N, and M649T, were unique to this study. Our cross-resistance profiling found that G253E, V285G, S591I, S591T, L699M, and A700T conferred panresistance, including against SQ109. The number of compounds proposed to target MmpL3 and the large number of substitutions that confer resistance highlight the importance of identifying combinations of drugs that would reduce the frequency of resistance.
Cross-resistance profiling for each combination of mmpL3 mutant and MmpL3 inhibitor identified two clades of mutants, clade I with higher resistance and clade II with lower resistance. Our findings suggest that the strength of the resistance conferred by a substitution is related to the proximity of the substitution to residues essential for MmpL3 function. While we found that the mutants with residues substituted at the same location fell into the same clade for F644 and S591, both falling into clade I, mutants with substitutions at M649 fell into two separate clades (M649L mutant in clade I, and M649T mutant in clade II). Similar findings have recently been described by Li et al. when comparing resistance from different substitutions for the same residue (33). These studies showed that resistance is dependent on the chemical properties of the amino acid substitutions and conformational changes to the protein. However, this does not fully explain our findings, as both the F644 and S591 substitutions fall into the same clades despite the substitutions having different chemical properties. Another possible explanation is the location of the substituted residues. Both the F644 and the S591 residue are located more centrally within the folded protein, while M649 is located near the cytoplasmic face of the protein, potentially allowing more flexibility in terms of the orientation of the specific residue. It therefore may be possible that the orientation of the substituted residue may change, conferring different amounts of resistance to each compound tested. Recent efforts have successfully crystalized MmpL3 from M. smegmatis (12, 19), and efforts to cocrystallize MmpL3 and MmpL3 inhibitors have indicated how different MmpL3 inhibitors disrupt the conformation of MmpL3 (12). Because substitutions in MmpL3 have combined effects of conferring resistance to MmpL3 inhibitors and costs to fitness that are highly dependent on the specific substitution, future efforts to crystallize MmpL3 variants containing substitutions may shed light on how these resistance effects are conferred.
The favorable properties of many of these compounds, including low cytotoxicity, high solubility and microsome stability, and activity in macrophages, suggest that these compounds warrant further development as new therapeutics. It is also possible that combinations of these scaffolds may be developed in a single molecule that can function to reduce the frequency of resistance. Three of the compounds used to isolate resistant mutants in this study, HC2149, HC2169, and HC2184, had a frequency of resistance (FoR) of 3 × 10−7, which is similar to the FoRs of other MmpL3 inhibitors, which range from 10−7 to 10−8 (1, 2, 5, 7, 10, 22). That the FoR for HC2184 was the same as the FoRs for HC2149 and HC2169 is interesting, as the cross-resistance profiles suggested that these compounds interact differently with MmpL3. While the antagonistic interactions identified by DiaMOND analysis suggest that scaffold combinations may lower the activity of a single inhibitor, antagonistic drug combinations have been proposed to decrease the rate of resistance (44, 45). Therefore, it may be possible to design a single inhibitor that fuses more than one scaffold to decrease the rate of resistance. This hypothesis could initially be tested by conducting pairwise combination studies examining compounds for synergistic reductions in the FoR. Given the relative ease with which resistance to MmpL3 inhibitors occurs, a reduced FoR could be a valuable new property for this class of inhibitors.
Many MmpL3 inhibitors with distinct chemical scaffolds have been described. The proposed target of these inhibitors has been driven by the mapping of resistance mutations to mmpL3. The screening platform we describe here greatly accelerates the target identification of such inhibitors. The use of a diverse pool of unique mmpL3 mutants rapidly identified inhibitors of MmpL3 activity, as demonstrated by their ability to modulate TDM and TMM accumulation and direct interaction with MmpL3 by displacement of a probe. A subset of these inhibitors was shown to disrupt membrane potential and, potentially, the PMF, which energizes MmpL3 activity. Two recent studies have suggested that two MmpL3 inhibitors, SQ109 and E11, indirectly inhibit MmpL3 by targeting the PMF, despite the isolation of resistant mmpL3 mutants (10, 15) and cocrystallization of SQ109 to MmpL3 (12). Notably, genetic inhibition of mmpL3 results in an increase in membrane potential, in contrast to inhibitors like BM212, AU1235, and SQ109, which dissipate membrane potential (33). Given that not all MmpL3 inhibitors modulate PMF and that genetic and chemical inhibition of MmpL3 have different outcomes, this suggests that dissipation of membrane potential is not due to direct activity on MmpL3 but, rather, a secondary mechanism. It is therefore possible that some of these new compounds inhibit MmpL3 and also independently dissipate PMF. It is also possible that these inhibitors are killing M. tuberculosis by a secondary mechanism that is independent of MmpL3. Indeed, the inhibitor THPP selects for resistance mutations in mmpL3, consistent with its ability to bind directly to the transporter (33), but has been proposed to also function by an alternative target, EchA6 (46). Continued studies of these inhibitors will further define whether the mechanism of killing is due to direct inhibition of MmpL3, a secondary mechanism, or a combination of multiple mechanisms.
MATERIALS AND METHODS
Media and growth conditions.
Unless otherwise specified, streptomycin-resistant strains of M. tuberculosis Erdman or CDC1551 were cultured in 7H9 medium supplemented with 10% OADC (vol/vol) with 0.05% Tween 80 (vol/vol) in standing T25, T75, or T150 flasks at 37°C with 5% CO2. Spectrum-of-activity studies in different bacterial species (Table 2) were conducted as described by Coulson et al. (31), with the exception of the M. abscessus studies, which are described in Methods in the supplemental material.
Dose-response curves.
M. tuberculosis was grown in rich medium to an optical density at 600 nm (OD600) of 0.5 to 1.0. Cultures were diluted to an OD600 of 0.1 in 7H9 medium and aliquoted into black-walled, clear-bottom, 96-well assay plates. Compounds were tested at concentrations between 80 and 0.13 μM with 2.5-fold dilutions, and the controls included DMSO and 3 μM RIF. Plates were placed in zip lock bags with moistened paper towels and incubated at 37°C for 6 days. Plates were read on a PerkinElmer Enspire plate reader. The percentage of growth inhibition (% GI) was calculated using DMSO and RIF as 0% and 100% inhibition, respectively, and EC50s were calculated using Prism 6 software. Dose-response assays were conducted in biological triplicates and repeated at least once. Significant differences in EC50s were compared using 95% confidence intervals. EC90s were calculated based on the EC50s and the Hillslope.
To examine the spectra of activity of the MmpL3 inhibitors, the EC50 of each compound was also determined for M. smegmatis and other nonmycobacteria, including S. aureus, E. coli, P. aeruginosa, P. vulgaris, and E. faecalis. Tests were performed in 96-well plates in LB broth with shaking at 37°C, with the exception of E. faecalis, which was grown in brain heart infusion medium in standing flasks at 37°C, and M. smegmatis, which was also grown standing at 37°C but in LB broth with 0.05% Tween 80. The cultures were diluted to a starting OD600 of 0.05. Bacteria were incubated in the presence of an 8-point (2-fold) dilution series of each inhibitor, ranging from 200 μM to 1.5 μM, for 6 h, except for M. smegmatis, which was incubated for 72 h. Growth was monitored by measuring optical density and normalized based on kanamycin (100% growth inhibition) and DMSO (0% growth inhibition) controls, with the exception of P. aeruginosa, for which 10 μg/ml tobramycin was used as the control for 100% growth inhibition. The experiments were performed with three technical replicates per plate. EC50s were calculated based on a variable-slope, four-parameter, nonlinear, least-squares regression model in the GraphPad Prism software package (version 8).
Kinetic kill curves.
M. tuberculosis was cultured in 7H9 medium to an OD600 of 0.5 to 1.0 and diluted to an OD600 of 0.1. In triplicate, diluted samples were aliquoted into 96-well plates and inoculated with 20 μM each compound, with DMSO as a negative control. Plates were placed in zip lock bags with moistened paper towels and incubated at 37°C. Daily samples were taken and serially diluted in 96-well plates using 1× phosphate-buffered saline (PBS)–0.05% Tween 80 (vol/vol) and plated on 7H10 quadrant plates supplemented with OADC (10% vol/vol). Plates were incubated at 37°C, and colonies were counted to calculate CFU/ml. Experiments were conducted in biological triplicates and repeated at least twice.
Isolation of resistant mutants.
M. tuberculosis was grown to an OD600 of 0.6 to 1.0, and samples were resuspended in fresh medium for a final cell count of 2 × 109 cells/ml. Cell pellets were resuspended in 1 ml of 7H9 medium and 0.5 ml was plated on 7H10 OADC plates supplemented with 20 μM or 40 μM HC2060, HC2149, HC2169, and HC2184. Plates were incubated at 37°C until isolated colonies appeared. Colonies were picked and inoculated into 5 ml of 7H9 medium in T25 standing flasks and grown to an OD600 of 0.5 to 1.0. Samples were taken and tested for resistance using dose-response curves as described above, along with WT bacteria grown to an OD600 of 0.6 to 1.0, and 3 μM RIF and DMSO were used as controls. Samples were also serially diluted as described above and plated to obtain colony-purified single-colony isolates on quadrant plates containing 7H10 OADC. Single-colony isolates were picked and inoculated into 5 ml of 7H9 OADC in T25 flasks. Resistance was reconfirmed using the same methods described above. Differences in EC50s were deemed significant based on the 95% confidence intervals.
Whole-genome sequencing and analysis.
Whole-genome sequencing was performed as previously described (47). Briefly, cultures of single-colony isolates were grown to an OD600 of ∼1.0 and pelleted. Genomic DNA was extracted and sequenced by Illumina-based whole-genome sequencing using 150-bp reads. Sequencing results were analyzed using the GATK workflow for the identification of single-nucleotide variations (48).
TMM and TDM accumulation assay.
The lipid assay was carried out as previously described (11). Briefly, 30-ml cultures of M. tuberculosis were cultured to an OD600 of 0.6. Samples were diluted to an OD600 of 0.1 in 8-ml cultures in T25 flasks. Cultures were inoculated with 8 μCi of [14C]acetate. Cultures were coinoculated with 20 μM MmpL3 inhibitors and then incubated for 24 h before performing lipid extraction as previously described (11). Total extractable lipid 14C incorporation was determined by scintillation counting, and 5,000 cpm of lipids of each sample were spotted and separated on thin-layer chromatography (TLC) plates with a 24:1:0.5 chloroform/methanol/H2O solvent system. TLC plates were imaged using a Typhoon FLA 7000, and images were quantified using IQ image-quantifying software. Experiments were conducted in biological duplicates. Comparison to the results for the DMSO controls was conducted using the t test.
Growth curves.
Each M. tuberculosis mmpL3 mutant or WT M. tuberculosis (Erdman or CDC1551) was cultured independently in 8 ml of 7H9 medium in T25 standing flasks to an OD600 of ∼0.6. Cultures were then resuspended in 8 ml of 7H9 medium in T25 standing flasks at a starting OD600 of 0.1 in biological triplicates. Cultures were incubated at 37°C in 5% CO2, and 500-μl samples were taken for optical density reading every 3 days.
Targeted whole-cell phenotypic screening.
Each mmpL3 mutant was cultured independently in 8 ml of 7H9 medium in T25 standing flasks to an OD600 of 0.6 to 1.0. Mutant cultures were separately diluted to an OD600 of 0.6 in 1.5 ml of 7H9 medium. The contents of each tube were mixed into a single batch culture in a T75 culture flask. The mixed mutant culture was allowed to recover overnight (∼8 h) at 37°C. Samples of M. tuberculosis Erdman (WT) (OD600 of 0.6) and the mixed mutant population were diluted to an OD600 of 0.1 in 7H9 medium. WT and mutant pools were aliquoted, in technical duplicates, into separate clear-bottom, black-walled, 96-well plates. Samples of WT and mixed mutant cultures were inoculated with each of the 163 compounds from the small-molecule library at 20 μM. Additional treatments included 0.5 μM para-amino salicylic acid (PAS), SQ109, bedaquiline (BDQ), isoniazid (INH), and clofazimine (CLO), as well as DMSO and 0.3 μM RIF. Percentages of growth inhibition (% GI) of the WT and mixed mutant population were calculated for each treatment, and hits were defined as (i) compounds with at least 15% GI in the WT background and (ii) 1.5-fold-decreased inhibition in the mutant pool relative to the inhibition in the WT background. The hit compounds were confirmed by conducting dose-response curves of screen hits as described above against the WT and mmpL3 mutant pools. Dose-response curves were conducted in technical duplicates, and differences between the WT and mmpL3 mutant pool were deemed significant based on the 95% confidence interval. Confirmed hits were reassessed with similar results.
Cross-resistance studies were conducted by generating dose-response curves for every combination of MmpL3 inhibitor and each mmpL3 mutant or a WT M. tuberculosis strain (CDC1551 or Erdman depending on the background of the mmpL3 mutant strain), for a total of 338 dose-response curves. Cross-resistance dose-response assays were conducted singly, unless the dose response identified increased sensitivity in the mmpL3 mutant background, in which case the responses were reexamined using dose-response assays carried out in biological duplicates. The dose responses were then used to calculate the area under the curve (AUC) using Prism 8 software with the default settings. AUCs were compare to the AUC of the respective WT strain by dividing the AUC of the mmpL3 strain by that of the respective WT parent strain. AUC fractions were then standardized by treatment by Z-scoring (20). Z-score-standardized data were then clustered in MatLab by hierarchical agglomerative clustering using the clustergram function with default settings (Euclidean distance model, average linkage clustering). Hierarchical agglomerative clustering using bootstrapped data was conducted in R using pvclust (number of bootstrap replications [nboot] = 1,000) with the Euclidean distance model and average linkage clustering (49).
Membrane potential assays.
The DiOC2 membrane potential assay was carried out as previously described (8, 11). Briefly, M. tuberculosis Erdman cells were labeled with 30 μM DiOC2 (Thermo Scientific) in 1 ml of 1× PBS (pH 7.4) supplemented with 50 mM KCl and were incubated at 37°C for 15 min. Cells were washed twice and suspended in 1× PBS at a final concentration measured by an OD600 of 0.2, and 200-μl amounts of labeled cells were aliquoted to 96-well plates and treated with each of the MmpL3 inhibitors at 80 μM, 20 μM, or 5 μM. Samples were also treated with DMSO (negative control) or 25 μM CCCP (Sigma-Aldrich) (positive control). Each treatment included three technical replicates per plate. The kinetics of fluorescence (excitation, 485 nm; emission, 610 nm/515 nm) was measured every 2 min for 60 min. The red/green (610 nm/515 nm) fluorescence intensity ratio was calculated and used to quantify membrane potential. The experiment was repeated at least twice with similar results. Error bars represent the standard deviations of the geometric mean values (Fig. S9).
Competition binding assays using intact M. smegmatis cells.
Competition binding assays in intact M. smegmatis MsmgΔmmpL3/pMVGH1-mmpL3tb bacilli were conducted by treating the cells with 4 mM probe North 114 for 1 h at 37°C prior to washing the cells twice with 7H9 ADC–0.05% Tween 80 and resuspending them with different concentrations of the test compounds for another hour at 37°C (33). Treated cells washed with 7H9 ADC–0.05% Tween 80 and fixed with 2% paraformaldehyde were finally resuspended in PBS–0.05% Tween 80 and subjected to flow cytometry analysis on a Cytek Aurora Spectral cytometer. Flow cytometry standard (FCS) file format data were analyzed using FlowJo software (Tree Star, Inc., Ashland, OR).
Bactericidal activity in vitro and in macrophages.
An M. tuberculosis CDC1551 strain with a chromosomally encoded firefly luciferase expressed from the hsp60 promoter (CDC1551 pMV306:hsp60:FFLuc) (34) was grown to an OD600 of 0.6 to 1.0 in rich medium. For in vitro experiments, cultures were diluted to an OD600 of 0.1, aliquoted at 100 μl in white-walled 96-well plates, and inoculated with each of the inhibitor compounds, along with DMSO or RIF controls. The luciferase assay was carried out as previously described (34), and plates were read on a PerkinElmer Enspire plate reader.
For studies in macrophages, primary bone marrow-derived macrophages were harvested and infected as previously described (35). Briefly, BMMϕ from C57BL/6 mice were distributed into 96-well white plates and infected for 1 h with M. tuberculosis luciferase reporter strain CDC1551 (34). Following 1 h of infection, cells were treated with MmpL3 inhibitors at concentrations ranging from 200 to 0.2 μM. PAS (20 μM), RIF (3 μM), and DMSO were used as controls. Samples were incubated in the 96-well plates at 37°C and 5% CO2 for 6 days before bacterial survival was monitored by measuring luciferase activity. Experiments were conducted in biological triplicates and repeated at least once with similar results.
Protein modeling.
The 3-D structure for MmpL3 was generated using the I-TASSER server (50). The MmpL3 protein sequence of M. tuberculosis strain H37Rv from Mycobrowser (Rv0206c) (51) was aligned to the MmpL3 crystal structure of M. smegmatis (PDB code 6AJF), with a resulting C score of 0.17 (template modeling [TM] score, 0.74 ± 0.11; RMSD, 8.4 ± 4.5 Å). The resulting structure was modified to remove the C-terminal tail (truncated to 732/944 aa) in PyMol 2.2.3 (52).
DiaMOND.
DiaMOND analysis was carried as described by Cokol et al., with modifications as described previously (40). Briefly, concentration ranges were linearized using the equations ΔD = (M − m)/(N − 1) and ΔD = DN − DN−1, where D is the dose concentration used at dose N (DN), ΔD is the difference between concentrations of each dose, M is the lowest concentration to result in 100% inhibition of M. tuberculosis growth, m is the highest concentration estimated to confer 0% growth inhibition based on the EC50 dose-response curves, and N is the number of doses to be used in DiaMOND. M. tuberculosis strains were then treated with the range of concentrations of each compound by itself (null treatment) at [XN] or in combination with another inhibitor at [1/2(XN)]. Dose responses were used to generate a dose-response curve for each treatment, which was used to interpolate the 50% inhibitory concentration (IC50), which was set for the observed value (o) to calculate the FIC2 (FIC2 = o/e, where e is the expected value), as previously described (40). Dose-response assays were conducted in biological duplicates, and the FIC2 values reported are representative of the geometric mean values of two replicates. Experiments were repeated with similar results.
Eukaryotic cytotoxicity.
Primary BMMϕ were isolated and distributed into white-walled 96-well plates as described above. Cells were treated with inhibitors ranging in concentration from 200 to 0.26 μM. Cells were incubated at 37°C for 3 or 6 days with 5% CO2. Cytotoxicity was tested using the CellTiter-Glo assay kit following the provider’s recommendations. For a negative control, cells were treated with 4% Triton X-100, and DMSO was used as a positive control (29).
Kinetic solubility and microsomal stability assay.
The kinetic solubility assay was conducted as described by Bevan and Lloyd (53). Briefly, the assay was performed with 7-point (2-fold) dilutions of the compounds, from 200 μM to 3.125 μM. Mebendazole, bexarotene, and aspirin were also included as controls. The drug dilutions were added to PBS, pH 7.4, with a final DMSO concentration of 1%, and incubated at 37°C for 2 h. The absorbance at 620 nm was measured for each drug dilution to estimate the compound’s solubility. Three replicates were examined for each dilution. A mouse microsomal stability assay was conducted as described by Obach (54), and the results are presented as the percentages remaining after 30 min. Values greater than 100% are likely due to changes in the solubility of the compounds over the course of the assay and represent high stability in microsomes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Bree Aldridge for assistance in designing the DiaMOND studies. Christopher Colvin and Benjamin Johnson provided technical assistance in the initial prioritization of compounds. Additional technical assistance was provided by Bilal Aleiwi and Tom Dexheimer, in the MSU Medicinal Chemistry Core, to conduct the solubility and microsomal stability studies.
Research in the Abramovitch laboratory was supported by start-up funding from Michigan State University and AgBioResearch and by NIH-NIAID (grants number U54AI057153, R21AI1170181, and R01AI116605), the Bill and Melinda Gates Foundation (grant number OPP1059227), a strategic partnership grant from the MSU Foundation (grant number 14-SPG-Full-2966), and the Jean P. Schultz Endowed Biomedical Research Fund at the MSU College of Human Medicine. This study was also supported by NIH grants to Thomas Dick (grant number R01AI132374) and Mary Jackson (grant number R01AI116525).
J.T.W. and R.B.A. designed the experiments and wrote the manuscript; G.B.C. conducted prioritization assays; J.T.W., E.R.H., and K.N.C. conducted the M. tuberculosis and spectrum of activity experiments; C.C., N.A.-C., and T.D. designed and conducted the M. abscessus experiments; W.L. and M.J. designed and conducted the MmpL3 competitive binding experiments; and E.E. directed the solubility and microsomal stability studies. All authors reviewed the manuscript.
R.B.A. is the founder and owner of Tarn Biosciences, Inc., a company that is working to develop new TB drugs.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00547-19.
REFERENCES
- 1.Grzegorzewicz AE, Pham H, Gundi VA, Scherman MS, North EJ, Hess T, Jones V, Gruppo V, Born SE, Kordulakova J, Chavadi SS, Morisseau C, Lenaerts AJ, Lee RE, McNeil MR, Jackson M. 2012. Inhibition of mycolic acid transport across the Mycobacterium tuberculosis plasma membrane. Nat Chem Biol 8:334–341. doi: 10.1038/nchembio.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.La Rosa V, Poce G, Canseco JO, Buroni S, Pasca MR, Biava M, Raju RM, Porretta GC, Alfonso S, Battilocchio C, Javid B, Sorrentino F, Ioerger TR, Sacchettini JC, Manetti F, Botta M, De Logu A, Rubin EJ, De Rossi E. 2012. MmpL3 is the cellular target of the antitubercular pyrrole derivative BM212. Antimicrob Agents Chemother 56:324–331. doi: 10.1128/AAC.05270-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stanley SA, Grant SS, Kawate T, Iwase N, Shimizu M, Wivagg C, Silvis M, Kazyanskaya E, Aquadro J, Golas A, Fitzgerald M, Dai H, Zhang L, Hung DT. 2012. Identification of novel inhibitors of M. tuberculosis growth using whole cell based high-throughput screening. ACS Chem Biol 7:1377–1384. doi: 10.1021/cb300151m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tahlan K, Wilson R, Kastrinsky DB, Arora K, Nair V, Fischer E, Barnes SW, Walker JR, Alland D, Barry CE III, Boshoff HI. 2012. SQ109 targets MmpL3, a membrane transporter of trehalose monomycolate involved in mycolic acid donation to the cell wall core of Mycobacterium tuberculosis. Antimicrob Agents Chemother 56:1797–1809. doi: 10.1128/AAC.05708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lun S, Guo H, Onajole OK, Pieroni M, Gunosewoyo H, Chen G, Tipparaju SK, Ammerman NC, Kozikowski AP, Bishai WR. 2013. Indoleamides are active against drug-resistant Mycobacterium tuberculosis. Nat Commun 4:2907. doi: 10.1038/ncomms3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rao SP, Lakshminarayana SB, Kondreddi RR, Herve M, Camacho LR, Bifani P, Kalapala SK, Jiricek J, Ma NL, Tan BH, Ng SH, Nanjundappa M, Ravindran S, Seah PG, Thayalan P, Lim SH, Lee BH, Goh A, Barnes WS, Chen Z, Gagaring K, Chatterjee AK, Pethe K, Kuhen K, Walker J, Feng G, Babu S, Zhang L, Blasco F, Beer D, Weaver M, Dartois V, Glynne R, Dick T, Smith PW, Diagana TT, Manjunatha UH. 2013. Indolcarboxamide is a preclinical candidate for treating multidrug-resistant tuberculosis. Sci Transl Med 5:214ra168. doi: 10.1126/scitranslmed.3007355. [DOI] [PubMed] [Google Scholar]
- 7.Remuinan MJ, Perez-Herran E, Rullas J, Alemparte C, Martinez-Hoyos M, Dow DJ, Afari J, Mehta N, Esquivias J, Jimenez E, Ortega-Muro F, Fraile-Gabaldon MT, Spivey VL, Loman NJ, Pallen MJ, Constantinidou C, Minick DJ, Cacho M, Rebollo-Lopez MJ, Gonzalez C, Sousa V, Angulo-Barturen I, Mendoza-Losana A, Barros D, Besra GS, Ballell L, Cammack N. 2013. Tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide and N-benzyl-6′,7′-dihydrospiro[piperidine-4,4′-thieno[3,2-c]pyran] analogues with bactericidal efficacy against Mycobacterium tuberculosis targeting MmpL3. PLoS One 8:e60933. doi: 10.1371/journal.pone.0060933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foss MH, Pou S, Davidson PM, Dunaj JL, Winter RW, Pou S, Licon MH, Doh JK, Li Y, Kelly JX, Dodean RA, Koop DR, Riscoe MK, Purdy GE. 2016. Diphenylether-modified 1,2-diamines with improved drug properties for development against Mycobacterium tuberculosis. ACS Infect Dis 2:500–508. doi: 10.1021/acsinfecdis.6b00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dupont C, Viljoen A, Dubar F, Blaise M, Bernut A, Pawlik A, Bouchier C, Brosch R, Guerardel Y, Lelievre J, Ballell L, Herrmann JL, Biot C, Kremer L. 2016. A new piperidinol derivative targeting mycolic acid transport in Mycobacterium abscessus. Mol Microbiol 101:515–529. doi: 10.1111/mmi.13406. [DOI] [PubMed] [Google Scholar]
- 10.Shetty A, Xu Z, Lakshmanan U, Hill J, Choong ML, Chng SS, Yamada Y, Poulsen A, Dick T, Gengenbacher M. 2018. Novel acetamide indirectly targets mycobacterial transporter MmpL3 by proton motive force disruption. Front Microbiol 9:2960. doi: 10.3389/fmicb.2018.02960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng H, Williams JT, Coulson GB, Haiderer ER, Abramovitch RB. 2018. HC2091 kills Mycobacterium tuberculosis by targeting the MmpL3 mycolic acid transporter. Antimicrob Agents Chemother 62:e02459-17. doi: 10.1128/AAC.02459-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang B, Li J, Yang X, Wu L, Zhang J, Yang Y, Zhao Y, Zhang L, Yang X, Yang X, Cheng X, Liu Z, Jiang B, Jiang H, Guddat LW, Yang H, Rao Z. 2019. Crystal structures of membrane transporter MmpL3, an anti-TB drug target. Cell 176:636–648.e613. doi: 10.1016/j.cell.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 13.Chim N, Torres R, Liu Y, Capri J, Batot G, Whitelegge JP, Goulding CW. 2015. The structure and interactions of periplasmic domains of crucial MmpL membrane proteins from Mycobacterium tuberculosis. Chem Biol 22:1098–1107. doi: 10.1016/j.chembiol.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li W, Obregon-Henao A, Wallach JB, North EJ, Lee RE, Gonzalez-Juarrero M, Schnappinger D, Jackson M. 2016. Therapeutic potential of the Mycobacterium tuberculosis mycolic acid transporter, MmpL3. Antimicrob Agents Chemother 60:5198–5207. doi: 10.1128/AAC.00826-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu ZJ, Meshcheryakov VA, Poce G, Chng SS. 2017. MmpL3 is the flippase for mycolic acids in mycobacteria. Proc Natl Acad Sci U S A 114:7993–7998. doi: 10.1073/pnas.1700062114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamaryo-Botte Y, Rainczuk AK, Lea-Smith DJ, Brammananth R, van der Peet PL, Meikle P, Ralton JE, Rupasinghe TW, Williams SJ, Coppel RL, Crellin PK, McConville MJ. 2015. Acetylation of trehalose mycolates is required for efficient MmpL-mediated membrane transport in Corynebacterineae. ACS Chem Biol 10:734–746. doi: 10.1021/cb5007689. [DOI] [PubMed] [Google Scholar]
- 17.Belardinelli JM, Yazidi A, Yang L, Fabre L, Li W, Jacques B, Angala SK, Rouiller I, Zgurskaya HI, Sygusch J, Jackson M. 2016. Structure-function profile of MmpL3, the essential mycolic acid transporter from Mycobacterium tuberculosis. ACS Infect Dis 2:702–713. doi: 10.1021/acsinfecdis.6b00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varela C, Rittmann D, Singh A, Krumbach K, Bhatt K, Eggeling L, Besra GS, Bhatt A. 2012. MmpL genes are associated with mycolic acid metabolism in mycobacteria and corynebacteria. Chem Biol 19:498–506. doi: 10.1016/j.chembiol.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su CC, Klenotic PA, Bolla JR, Purdy GE, Robinson CV, Yu EW. 2019. MmpL3 is a lipid transporter that binds trehalose monomycolate and phosphatidylethanolamine. Proc Natl Acad Sci U S A 116:11241–11246. doi: 10.1073/pnas.1901346116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Degiacomi G, Benjak A, Madacki J, Boldrin F, Provvedi R, Palu G, Kordulakova J, Cole ST, Manganelli R. 2017. Essentiality of mmpL3 and impact of its silencing on Mycobacterium tuberculosis gene expression. Sci Rep 7:43495. doi: 10.1038/srep43495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li W, Yazidi A, Pandya AN, Hegde P, Tong W, Calado Nogueira de Moura V, North EJ, Sygusch J, Jackson M. 2018. MmpL3 as a target for the treatment of drug-resistant nontuberculous mycobacterial infections. Front Microbiol 9:1547. doi: 10.3389/fmicb.2018.01547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McNeil MB, Dennison D, Parish T. 2017. Mutations in MmpL3 alter membrane potential, hydrophobicity and antibiotic susceptibility in Mycobacterium smegmatis. Microbiology 163:1065–1070. doi: 10.1099/mic.0.000498. [DOI] [PubMed] [Google Scholar]
- 23.Lamichhane G, Tyagi S, Bishai WR. 2005. Designer arrays for defined mutant analysis to detect genes essential for survival of Mycobacterium tuberculosis in mouse lungs. Infect Immun 73:2533–2540. doi: 10.1128/IAI.73.4.2533-2540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffin JE, Gawronski JD, Dejesus MA, Ioerger TR, Akerley BJ, Sassetti CM. 2011. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog 7:e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sacksteder KA, Protopopova M, Barry CE III, Andries K, Nacy CA. 2012. Discovery and development of SQ109: a new antitubercular drug with a novel mechanism of action. Future Microbiol 7:823–837. doi: 10.2217/fmb.12.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee BS, Pethe K. 2018. Therapeutic potential of promiscuous targets in Mycobacterium tuberculosis. Curr Opin Pharmacol 42:22–26. doi: 10.1016/j.coph.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 27.Li W, Upadhyay A, Fontes FL, North EJ, Wang Y, Crans DC, Grzegorzewicz AE, Jones V, Franzblau SG, Lee RE, Crick DC, Jackson M. 2014. Novel insights into the mechanism of inhibition of MmpL3, a target of multiple pharmacophores in Mycobacterium tuberculosis. Antimicrob Agents Chemother 58:6413–6423. doi: 10.1128/AAC.03229-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poce G, Consalvi S, Biava M. 2016. MmpL3 inhibitors: diverse chemical scaffolds inhibit the same target. Mini Rev Med Chem 16:1274–1283. doi: 10.2174/1389557516666160118105319. [DOI] [PubMed] [Google Scholar]
- 29.Zheng H, Colvin CJ, Johnson BK, Kirchhoff PD, Wilson M, Jorgensen-Muga K, Larsen SD, Abramovitch RB. 2017. Inhibitors of Mycobacterium tuberculosis DosRST signaling and persistence. Nat Chem Biol 13:218–225. doi: 10.1038/nchembio.2259. [DOI] [PubMed] [Google Scholar]
- 30.Johnson BK, Colvin CJ, Needle DB, Mba Medie F, Champion PA, Abramovitch RB. 2015. The carbonic anhydrase inhibitor ethoxzolamide inhibits the Mycobacterium tuberculosis PhoPR regulon and Esx-1 secretion and attenuates virulence. Antimicrob Agents Chemother 59:4436–4445. doi: 10.1128/AAC.00719-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coulson GB, Johnson BK, Zheng H, Colvin CJ, Fillinger RJ, Haiderer ER, Hammer ND, Abramovitch RB. 2017. Targeting Mycobacterium tuberculosis sensitivity to thiol stress at acidic pH kills the bacterium and potentiates antibiotics. Cell Chem Biol 24:993–1004.e1004. doi: 10.1016/j.chembiol.2017.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ioerger TR, O’Malley T, Liao R, Guinn KM, Hickey MJ, Mohaideen N, Murphy KC, Boshoff HI, Mizrahi V, Rubin EJ, Sassetti CM, Barry CE III, Sherman DR, Parish T, Sacchettini JC. 2013. Identification of new drug targets and resistance mechanisms in Mycobacterium tuberculosis. PLoS One 8:e75245. doi: 10.1371/journal.pone.0075245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li W, Stevens CM, Pandya AN, Darzynkiewicz Z, Bhattarai P, Tong W, Gonzalez-Juarrero M, North EJ, Zgurskaya HI, Jackson M. 2019. Direct inhibition of MmpL3 by novel antitubercular compounds. ACS Infect Dis 5:1001–1012. doi: 10.1021/acsinfecdis.9b00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andreu N, Zelmer A, Fletcher T, Elkington PT, Ward TH, Ripoll J, Parish T, Bancroft GJ, Schaible U, Robertson BD, Wiles S. 2010. Optimisation of bioluminescent reporters for use with mycobacteria. PLoS One 5:e10777. doi: 10.1371/journal.pone.0010777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson BK, Abramovitch RB. 2015. Macrophage infection models for Mycobacterium tuberculosis. Methods Mol Biol 1285:329–341. doi: 10.1007/978-1-4939-2450-9_20. [DOI] [PubMed] [Google Scholar]
- 36.Garcia-Garcia V, Oldfield E, Benaim G. 2016. Inhibition of Leishmania mexicana growth by the tuberculosis drug SQ109. Antimicrob Agents Chemother 60:6386–6389. doi: 10.1128/AAC.00945-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biava M, Porretta GC, Manetti F. 2007. New derivatives of BM212: a class of antimycobacterial compounds based on the pyrrole ring as a scaffold. Mini Rev Med Chem 7:65–78. doi: 10.2174/138955707779317786. [DOI] [PubMed] [Google Scholar]
- 38.Ballell L, Bates RH, Young RJ, Alvarez-Gomez D, Alvarez-Ruiz E, Barroso V, Blanco D, Crespo B, Escribano J, González R, Lozano S, Huss S, Santos-Villarejo A, Martín-Plaza JJ, Mendoza A, Rebollo-Lopez MJ, Remuiñan-Blanco M, Lavandera JL, Pérez-Herran E, Gamo-Benito FJ, García-Bustos JF, Barros D, Castro JP, Cammack N. 2013. Fueling open-source drug discovery: 177 small-molecule leads against tuberculosis. ChemMedChem 8:313–321. doi: 10.1002/cmdc.201200428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colan SD. 2013. The why and how of Z scores. J Am Soc Echocardiogr 26:38–40. doi: 10.1016/j.echo.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 40.Cokol M, Kuru N, Bicak E, Larkins-Ford J, Aldridge BB. 2017. Efficient measurement and factorization of high-order drug interactions in Mycobacterium tuberculosis. Sci Adv 3:e1701881. doi: 10.1126/sciadv.1701881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li W, Sanchez-Hidalgo A, Jones V, de Moura VC, North EJ, Jackson M. 2017. Synergistic interactions of MmpL3 inhibitors with antitubercular compounds in vitro. Antimicrob Agents Chemother 61:e02399-16. doi: 10.1128/AAC.02399-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen P, Gearhart J, Protopopova M, Einck L, Nacy CA. 2006. Synergistic interactions of SQ109, a new ethylene diamine, with front-line antitubercular drugs in vitro. J Antimicrob Chemother 58:332–337. doi: 10.1093/jac/dkl227. [DOI] [PubMed] [Google Scholar]
- 43.Ramesh R, Shingare RD, Kumar V, Anand A, B S, Veeraraghavan S, Viswanadha S, Ummanni R, Gokhale R, Srinivasa Reddy D. 2016. Repurposing of a drug scaffold: identification of novel sila analogues of rimonabant as potent antitubercular agents. Eur J Med Chem 122:723–730. doi: 10.1016/j.ejmech.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 44.Bollenbach T. 2015. Antimicrobial interactions: mechanisms and implications for drug discovery and resistance evolution. Curr Opin Microbiol 27:1–9. doi: 10.1016/j.mib.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 45.Singh N, Yeh PJ. 2017. Suppressive drug combinations and their potential to combat antibiotic resistance. J Antibiot (Tokyo) 70:1033–1042. doi: 10.1038/ja.2017.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cox JA, Abrahams KA, Alemparte C, Ghidelli-Disse S, Rullas J, Angulo-Barturen I, Singh A, Gurcha SS, Nataraj V, Bethell S, Remuinan MJ, Encinas L, Jervis PJ, Cammack NC, Bhatt A, Kruse U, Bantscheff M, Futterer K, Barros D, Ballell L, Drewes G, Besra GS. 2016. THPP target assignment reveals EchA6 as an essential fatty acid shuttle in mycobacteria. Nat Microbiol 1:15006. doi: 10.1038/nmicrobiol.2015.6. [DOI] [PubMed] [Google Scholar]
- 47.Baker JJ, Abramovitch RB. 2018. Genetic and metabolic regulation of Mycobacterium tuberculosis acid growth arrest. Sci Rep 8:4168. doi: 10.1038/s41598-018-22343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suzuki R, Shimodaira H. 2006. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22:1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- 50.Yang J, Zhang Y. 2015. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res 43:W174–W181. doi: 10.1093/nar/gkv342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kapopoulou A, Lew JM, Cole ST. 2011. The MycoBrowser portal: a comprehensive and manually annotated resource for mycobacterial genomes. Tuberculosis (Edinb) 91:8–13. doi: 10.1016/j.tube.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 52.Schrodinger LLC. 2015. The PyMOL Molecular Graphics System, version 2.2.
- 53.Bevan CD, Lloyd RS. 2000. A high-throughput screening method for the determination of aqueous drug solubility using laser nephelometry in microtiter plates. Anal Chem 72:1781–1787. doi: 10.1021/ac9912247. [DOI] [PubMed] [Google Scholar]
- 54.Obach RS. 1999. Prediction of human clearance of twenty-nine drugs from hepatic microsomal intrinsic clearance data: an examination of in vitro half-life approach and nonspecific binding to microsomes. Drug Metab Dispos 27:1350–1359. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.