Salmonella enterica serovar Goldcoast infection was rare in Taiwan; it was not detected in routine surveillance from 2004 to 2013. This serovar was first identified in 2014, but the frequency of infection remained low until 2017. From 2014 to 2016, all but one isolate was pan-susceptible. S. Goldcoast infections abruptly increased in 2018, and all isolates were multidrug-resistant (MDR).
KEYWORDS: Salmonella enterica, antimicrobial drug resistance, molecular epidemiology, next-generation sequencing, zoonosis
ABSTRACT
Salmonella enterica serovar Goldcoast infection was rare in Taiwan; it was not detected in routine surveillance from 2004 to 2013. This serovar was first identified in 2014, but the frequency of infection remained low until 2017. From 2014 to 2016, all but one isolate was pan-susceptible. S. Goldcoast infections abruptly increased in 2018, and all isolates were multidrug-resistant (MDR). All MDR isolates harbored an IncHI2 plasmid, and the majority carried 14 antimicrobial resistance genes, aac(3)-IId, aadA22, aph(3′)-Ia, aph(6)-Id, blaTEM-1B, blaCTX-M-55, lnu(F), floR, qnrS13, arr-2, sul2, sul3, tet(A), and dfrA14. S. Goldcoast strains recovered in Taiwan and 96 of 99 strains from Germany, the Netherlands, the United Kingdom, and the United States belonged to sequence type 358 (ST358). Whole-genome single-nucleotide polymorphism and core genome multilocus sequence type analyses revealed that all strains of the ST358 clone shared a high degree of genetic relatedness. The present study highlighted that a dramatic increase in S. Goldcoast infections followed the emergence of MDR strains and indicated that a genetically closely related S. Goldcoast ST358 clone may have widespread significance internationally.
INTRODUCTION
Salmonella, a common zoonotic pathogen in humans, comprises two species, Salmonella enterica and Salmonella bongori, and is subdivided into more than 2,600 serotypes (1). Nontyphoidal Salmonella is a leading cause of zoonotic foodborne diseases; it is estimated to cause 93.8 million cases of gastroenteritis and 155,000 deaths globally each year (2). Salmonellosis was the second most commonly reported zoonosis in Europe in 2017 and the second leading foodborne illness in the United States (3, 4). In Taiwan, nontyphoidal Salmonella infections are quite common. As shown in a study, from 2014 to 2016, nontyphoidal Salmonella accounted for 21.8% of acute gastroenteritis in hospitalized children aged less than 5 years (5).
The increase in antimicrobial resistance in Salmonella is a global public health concern. Salmonella strains resistant to the traditional first-line drugs ampicillin, chloramphenicol, and trimethoprim-sulfamethoxazole have emerged worldwide. In the past decade, the emergence of resistance to the second-line drugs extended-spectrum cephalosporins, fluoroquinolones, azithromycin, and even carbapenems has posed a significant threat to public health (6). In Taiwan, a high rate of multidrug resistance, primarily to the first-line drugs, has been found in many nontyphoidal Salmonella serovars among the isolates collected from humans from 1998 to 2002 (7). During that period, only 1 of the 798 tested isolates displayed resistance to the third-generation cephalosporin cefotaxime. However, resistance has increased over time. A comparison study revealed that human isolates of S. enterica serovar Typhimurium from Taiwan collected from 2009 to 2010 displayed significantly higher levels of antimicrobial resistance than those from Denmark, and 10.3% of the isolates were resistant to cefotaxime (8). In another study, 96% of 110 Salmonella isolates recovered from sick pigs were multidrug-resistant (MDR), 44% were resistant to cefotaxime, and 44% had pulsed-field gel electrophoresis (PFGE) patterns which were also found in human isolates (9). The isolates from humans and pigs sharing a common PFGE pattern have identical or very similar resistance patterns, indicating that pigs are one of the major reservoirs of human salmonellosis in Taiwan.
S. enterica serovar Goldcoast was an infrequently isolated serovar from humans in the United States; this serovar was identified in only 38 of the 477,861 Salmonella isolates recovered from 2006 to 2016 (10). In contrast, S. Goldcoast was more common in Europe; it ranked as the 26th most abundant serovar and accounted for 0.26% of the isolates in the Enter-net database recovered from 1994 to 2004 (11). S. Goldcoast was reported to cause foodborne disease outbreaks in Germany and England (12, 13), an international outbreak in tourists returning from Majorca in 2005 (14), and a multistate epidemic outbreak that occurred primarily in Hungary and Italy in 2009 (15, 16). A baseline survey conducted in 24 European Union member states and two nonmember states in 2008 revealed that S. Goldcoast was a common serovar in pigs. It ranked as the 11th most abundant serovar and accounted for 2.1% of Salmonella isolates (17). In Taiwan, no S. Goldcoast was identified among 20,527 isolates collected from hospitals during 2004 to 2013 by the Taiwan Center for Disease Control (Taiwan CDC). S. Goldcoast was first identified in 2014, and only a few isolates were detected from 2014 to 2017. However, S. Goldcoast infections abruptly increased in 2018, and all isolates were multidrug resistant. In the present study, we investigated the epidemiological trend of S. Goldcoast, the mechanism of resistance, and the genetic relatedness among strains collected in Taiwan and other countries with genomic sequences deposited in the database of National Center for Biotechnology and Information (NCBI).
RESULTS AND DISCUSSION
Epidemiological trend of S. Goldcoast.
From 2004 to 2013, no S. Goldcoast was identified among 20,527 Salmonella isolates characterized. Three S. Goldcoast isolates were first identified in 2014. The isolation frequency of this serovar remained low from 2014 to 2017 (Table 1). However, the number of S. Goldcoast infections dramatically increased in 2018, accounting for 3.92% of 1,709 Salmonella isolates characterized that year. It was the 5th most common serovar that year, ranked after S. enterica serovar Enteritidis (31.01%), S. Typhimurium (19.31%), S. enterica serovar Anatum (10.88%), and S. enterica serovar Newport (6.14%).
TABLE 1.
Distribution of S. Goldcoast in Taiwan from 2014 to 2018
| Year | Number of isolates of: |
Ratio (%) | |
|---|---|---|---|
| All serovars | S. Goldcoast | ||
| 2014 | 1,820 | 3 | 0.16 |
| 2015 | 3,037 | 4 | 0.13 |
| 2016 | 3,756 | 6 | 0.16 |
| 2017 | 5,154 | 1 | 0.02 |
| 2018 | 1,709 | 67 | 3.92 |
| Total | 15,476 | 81 | 0.52 |
Antimicrobial susceptibility and resistance genes.
By excluding the testing results of azithromycin, cefoxitin, and nalidixic acid, the 82 isolates were grouped into 8 resistance patterns (see Table S1 in the supplemental material). Of the 13 isolates recovered from 2014 to 2016, 12 were susceptible to all 15 antimicrobials tested; only 1 was resistant to 3 antimicrobials. The first MDR isolate resistant to 10 antimicrobials emerged in 2017. Thereafter, all isolates recovered in 2018 were MDR, among which 2 gained additional resistance to colistin and 10 lost resistance to 1 to 4 antimicrobials. Compared to the pan-susceptible isolates, most MDR isolates were accompanied by elevated MICs for azithromycin, cefoxitin, and nalidixic acid. Among the 68 MDR isolates recovered from humans in 2017 and 2018, 62, 57, and 67 had elevated MIC levels for azithromycin (up from 4 to 8 mg/liter to 16 to 32 mg/liter), cefoxitin (up from 4 to 8 mg/liter to 16 to 64 mg/liter), and nalidixic acid (up from 4 mg/liter to 16 to 32 mg/liter), respectively. The MDR isolate recovered from ground pork in 2019 was also resistant to 10 antimicrobials and displayed elevated MIC levels for these three antimicrobials.
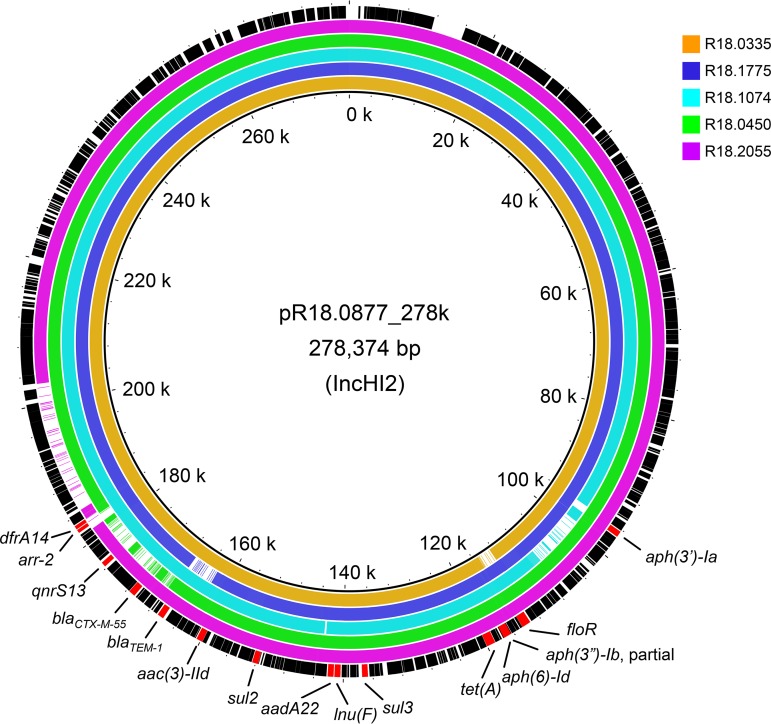
Among the 82 isolates, 29, comprising 1 or a few isolates for each resistance pattern and 1 from food (ground pork), were selected for whole-genome sequencing using the Illumina MiSeq platform to identify resistance genes. R18.0877 was further subjected to Nanopore sequencing to obtain long reads for the assembly of the complete chromosomal and plasmid sequences. A total of 17 acquired antimicrobial resistance genes, aac(6′)-Iaa, aadA, aac(3)-IId, aadA22, aph(3′)-Ia, aph(6)-Id, blaTEM-1B, blaCTX-M-55, lnu(F), floR, qnrS13, arr-2, sul2, sul3, tet(A), dfrA14, and mcr-1, were identified from the 29 isolates. All isolates carried cryptic aac(6′)-Iaa and cryptic aadA genes in their chromosomes. The two genes should not confer aminoglycoside resistance in these isolates, though they could be activated (18, 19). R18.0877 had a chromosome of 4,645,553 bp in length and harbored two plasmids, pR18.0877_278k (incompatibility type IncHI2; 278,374 bp) and pR18.0877_3.3k (unknown incompatibility type; 3,373 bp). The 278-kb plasmid carried 14 antimicrobial resistance genes, aac(3)-IId, aadA22, aph(3′)-Ia, aph(6)-Id, blaTEM-1B, blaCTX-M-55, lnu(F), floR, qnrS13, arr-2, sul2, sul3, tet(A), and dfrA14 (Fig. 1). These genes can fully confer resistance to the 10 antimicrobials for resistance pattern 3 (see Table S1). Of the two isolates which acquired an additional colistin resistance (resistance pattern 7), one carried an additional IncI2 plasmid and an mcr-1 gene. The IncI2 plasmid is one of the plasmid types frequently associated with carriage of the mcr-1 gene in Salmonella (20). The isolates with resistance patterns 4, 6, and 8 lost resistance to 4, 1, and 1 antimicrobial, respectively. Comparison of the plasmid sequences for the isolates of resistance patterns 4, 6, and 8 with the pR18.0877_278k plasmid from strain R18.0877 (resistance pattern 3) indicated that the loss of resistance resulted from deletions over the corresponding resistance genes dfrA14, arr-2, qnrS13, and blaCTX-M-55 in strain R18.0450 (resistance pattern 4), floR in strain R18.0335 (resistance pattern 6), and dfrA14 and arr-2 in strain R18.2055 (resistance pattern 8) (Fig. 1). Deletion over blaTEM-1B occurred in strain R18.1775 (resistance pattern 3) and aph(3′)-Ia in strain R18.1074 (resistance pattern 5), indicating that resistance genes in large plasmids seem to be easy to lose via deletion, resulting in the emergence of a number of diverse resistance patterns over a short period of time (21, 22).
FIG 1.
Genetic maps of plasmids pR18.0877_238k in S. Goldcoast strain R18.0877 and the variants in strains R18.0335, R18.1775, R18.1074, R18.0450, and R18.2055.
No corresponding horizontally transferable genes responsible for the elevated MICs to azithromycin, cefoxitin, and nalidixic acid were identified in the MDR isolates. Among the 82 isolates, 5 with the PFGE pattern SMX.1665 displayed low MIC levels as the pan-susceptible isolates to azithromycin and cefoxitin. This observation suggests that the elevation of MICs to azithromycin, cefoxitin, and nalidixic acid may be mediated by genes. The five isolates can serve as an excellent material for elucidating the mechanism of the elevated levels of resistance to the antimicrobials in the MDR isolates.
Genetic relationships among S. Goldcoast strains.
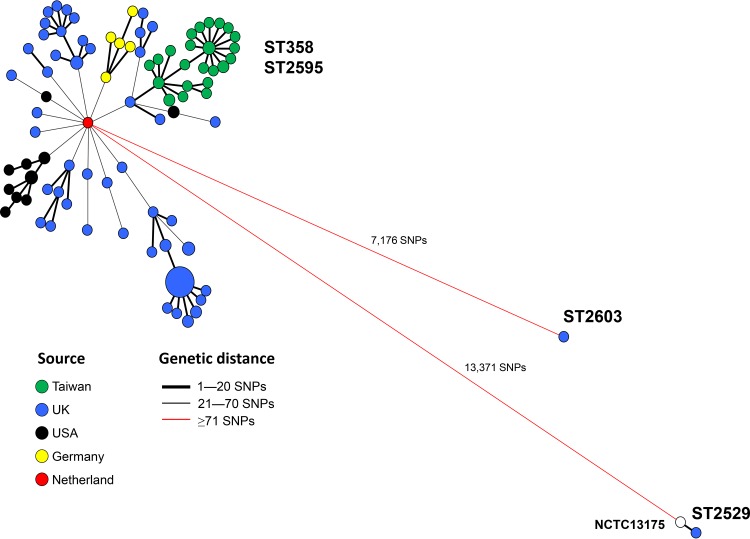
Four sequence types, sequence type 358 (ST358), ST2595, ST2529, and ST2603, were identified in 124, 1, 2, and 1 strain, respectively. ST2595 is a single-locus variant of ST358, but ST2529 and ST2603 differ from ST358 by 5 and 3 loci, respectively. The genetic relationships among the strains were further established using the whole-genome single-nucleotide polymorphism (wgSNP) and core gene multilocus sequence typing (cgMLST) approaches. We identified 17,792 SNPs from the 128 genomic sequences and constructed a phylogenetic tree with the wgSNP profiles using the minimum spanning tree algorithm. The tree revealed a tight cluster for the strains of ST358 and ST2595, even though the strains were from different countries (Fig. 2). These ST358 and ST2595 strains differed by a maximum distance of only 70 SNPs. The ST2529 strains (reference strain NCTC13175 and a U.K. strain) and the ST2603 strain had a distance of 13,371 SNPs and 7,176 SNPs, respectively, from the ST358 clone. All strains from Taiwan belonged to ST358 and were more closely related than those from other countries. The Taiwanese strains had a closer distance to a UK strain, with a distance of only 13 SNPs.
FIG 2.
Genetic relationships among 128 S. Goldcoast genomes from 5 different countries. The tree was constructed with wgSNP profiles using the minimum spanning tree algorithm. The genomic sequence of S. Goldcoast NCTC13175 (GenBank accession number NZ_LR134158) was used as the reference for SNP calling and is marked by a white circle in the diagram.
The genetic relationships among the strains constructed with cgMLST profiles and wgSNP profiles were quite concordant; the ST358 and ST2595 strains were closely related, but the ST2529 and ST2603 strains had a large genetic distance from the ST358 clone (see Fig. S1). Of the 29 strains from Taiwan, 28 were tightly linked with two U.K. strains, but the MDR strain that emerged in 2014 was a little separate from other Taiwanese strains. IncHI2 plasmids were found in all MDR strains from Taiwan and two MDR strains from Germany (strains N2-2 and N2-6). IncHI2 plasmids in most MDR strains recovered in Taiwan carried 14 resistance genes, of which only 3 to 4 were found in the two German strains. Until now, this 14-resistance gene-carrying IncHI2 plasmid was found only in MDR strains from Taiwan, and the resistance plasmid was most likely acquired locally by an indigenous pan-susceptible strain.
This study reports a noted increase in S. Goldcoast infections in Taiwan in 2018, and the increase was associated with the emergence of an MDR strain in 2017. This story is similar to the emergence of MDR S. Anatum in Taiwan (23). S. Anatum infections were not common in Taiwan from 2004 to 2014; however, a dramatic increase in S. Anatum infections was observed in 2016 and 2017. The increase in infections was also associated with the emergence of an MDR S. Anatum strain in 2015. The relationship between the increase of infections and the emergence of MDR S. Goldcoast and MDR Anatum strains is worth further investigation.
In conclusion, S. Goldcoast was rare in Taiwan; it was first identified in 2014 and caused an epidemic in 2018. All but one isolate that appeared between 2014 and 2016 were pan-susceptible; however, all isolates that appeared in 2017 to 2018 were MDR. All isolates recovered in Taiwan belonged to the ST358 clone and, surprisingly, all ST358 strains recovered from different countries, including Taiwan, Germany, the Netherlands, the United Kingdom, and the United States, were genetically closely related. The dramatic increase in S. Goldcoast infections in 2018 was strongly associated with the emergence of an MDR strain in 2017. The greatest cause for concern regarding the emergence of the S. Goldcoast MDR strains is the carriage of blaCTX-M-55, an extended-spectrum β-lactamase gene that can confer resistance to many β-lactam drugs, including third-generation cephalosporins. In addition, the majority of MDR strains were also accompanied by elevated MICs to azithromycin, cefoxitin, and nalidixic acid. It is worth investigating the mechanisms responsible for the elevated resistance to the 3 antimicrobials in the MDR S. Goldcoast strains and the causes leading to the increase in infections following the emergence of the MDR strains.
MATERIALS AND METHODS
Epidemiological trend.
The rate of S. Goldcoast infections from 2004 to 2018 was calculated using the data collected by a national Salmonella surveillance program conducted by the Taiwan CDC since 2004. The program collected Salmonella isolates from participating hospitals around the country and conducted serotyping, genotyping, and antimicrobial susceptibility testing. The laboratory testing results and demographic data are managed using BioNumerics software (Applied Maths, Inc.).
Antimicrobial susceptibility testing.
Salmonella isolates were subjected to antimicrobial susceptibility testing using the broth microdilution method with custom-made 96-well Sensititre MIC panels (Trek Diagnostic Systems, Ltd., West Sussex, UK). The MIC panels contained 15 antimicrobials, azithromycin, colistin, ampicillin, cefoxitin, cefotaxime, ceftazidime, ertapenem, nalidixic acid, ciprofloxacin, gentamicin, chloramphenicol, streptomycin, sulfamethoxazole, tetracycline, and trimethoprim-sulfamethoxazole. The test procedure was performed according to the manufacturer’s instructions, and the interpretation of MIC results followed the guidelines of the Clinical and Laboratory Standards Institute (24) or the European Committee on Antimicrobial Susceptibility Testing breakpoint tables version 9.0 for colistin (25). The interpretive criterion for streptomycin resistance was ≥32 mg/liter (26, 27).
Whole-genome sequencing and data analysis.
Twenty-eight S. Goldcoast isolates recovered from humans and one from ground pork were subjected to whole-genome sequencing using the Illumina MiSeq platform to generate reads of 300 bp in length. Genomic sequences of 99 S. Goldcoast strains originating from Germany, the Netherlands, the United Kingdom (UK), and the United States were obtained from the NCBI database. The reads were assembled using the SPAdes assembler (28). The resistance genes, plasmid incompatibility types, and sequence types were determined from the genomic sequence data using the pipelines ResFinder, PlamidFinder, and MLST provided in the Center for Genomic Epidemiology (http://www.genomicepidemiology.org/; accessed 22 May 2019). Among the 29 Taiwanese isolates, R18.0877 was further subjected to whole-genome sequencing (WGS) using an Oxford Nanopore MinION sequencer to obtain long reads. Two sets of WGS data generated using Illumina and Oxford Nanopore sequencers were together assembled using the Unicycler assembler (29) to complete the circular chromosomal and plasmid sequences. The complete sequence of pR18.0877_278k was employed as the reference for comparison with the plasmid sequences of other isolates using the BLAST Ring Image Generator (BRIG) pipeline (http://brig.sourceforge.net/) (30). The WGS raw reads of 29 isolates were deposited in the SRA database of the NCBI under the BioProject accession number PRJNA478278, and the BioSample accession numbers for the 29 isolates are listed in Table S1. The complete genomic sequences of isolate R18.0877 were submitted to the GenBank database under the accession numbers CP037960 for the chromosomal sequence, CP037959 for plasmid pR18.0877_278k, and CP037958 for plasmid pR18.0877_3.3k.
Whole-genome single-nucleotide polymorphism analysis.
The tools provided in BioNumerics version 7.6.3 were used for SNP calling and cluster analysis of wgSNP profiles. The sequences of raw reads were mapped to the genomic reference sequence of S. Goldcoast strain NCTC13175 (GenBank assembly accession number GCA_900635695), and the mapped sequences and the reference were aligned for SNP calling using the option of strict SNP filtering (closed SNP set). By using this SNP calling criterion, SNPs were called by removing positions with at least one ambiguous base (non-ATGC base), one unreliable base (N), one gap, and noninformative SNPs. Each retained SNP position had a minimum 5× coverage and was covered at least once in both the forward and reverse directions. The minimum distance between the retained SNP positions was 12 bp. The genetic relationships among strains were constructed with wgSNP profiles using the minimal spanning tree algorithm for clustering.
Core gene multilocus sequence typing analysis.
To generate cgMLST profiles, raw reads were assembled using the SPAdes assembler (28). The assembled sequences (contigs) were subjected to generating cgMLST profiles using the in-house-developed BENGA cgMLST profiling tool (Y.-H. Tu, Y.-S. Chen, B.-H. Chen, Y.-Y. Liu, Y.-P. Hong, R.-H. Teng, Y.-W. Wang, and C.-S. Chiou, submitted for review). Profiling was run using a Salmonella allele database constructed in our laboratory, which contains alleles of 3,285 Salmonella core genes. Genetic relationships among the strains were constructed with cgMLST profiles using the single linkage algorithm for clustering.
Supplementary Material
ACKNOWLEDGMENT
This study was funded by the Ministry of Health and Welfare, Taiwan (grant number MOHW108-CDC-C-315-134516).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01122-19.
REFERENCES
- 1.Guibourdenche M, Roggentin P, Mikoleit M, Fields PI, Bockemuhl J, Grimont PA, Weill FX. 2010. Supplement 2003-2007 (no. 47) to the White-Kauffmann-Le Minor scheme. Res Microbiol 161:26–29. doi: 10.1016/j.resmic.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O’Brien SJ, Jones TF, Fazil A, Hoekstra RM, International Collaboration on Enteric Disease “Burden of Illness” Studies. 2010. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis 50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 3.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis 17:7–15. doi: 10.3201/eid1701.09-1101p1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.European Food Safety Authority. 2018. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J 16:5500. doi: 10.2903/j.efsa.2018.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung N, Wang SM, Shen CF, Kuo FC, Ho TS, Hsiung CA, Mu JJ, Wu FT, Huang LM, Huang YC, Huang YC, Chi H, Lin HC, Liu CC, Taiwan Pediatric Infectious Disease A. 2017. Clinical and epidemiological characteristics in hospitalized young children with acute gastroenteritis in southern Taiwan: according to major pathogens. J Microbiol Immunol Infect 50:915–922. doi: 10.1016/j.jmii.2017.07.015. [DOI] [PubMed] [Google Scholar]
- 6.Crump JA, Sjolund-Karlsson M, Gordon MA, Parry CM. 2015. Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive Salmonella infections. Clin Microbiol Rev 28:901–937. doi: 10.1128/CMR.00002-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lauderdale TL, Aarestrup FM, Chen PC, Lai JF, Wang HY, Shiau YR, Huang IW, Hung CL, Hospitals T. 2006. Multidrug resistance among different serotypes of clinical Salmonella isolates in Taiwan. Diagn Microbiol Infect Dis 55:149–155. doi: 10.1016/j.diagmicrobio.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Torpdahl M, Lauderdale TL, Liang SY, Li I, Wei SH, Chiou CS. 2013. Human isolates of Salmonella enterica serovar Typhimurium from Taiwan displayed significantly higher levels of antimicrobial resistance than those from Denmark. Int J Food Microbiol 161:69–75. doi: 10.1016/j.ijfoodmicro.2012.11.022. [DOI] [PubMed] [Google Scholar]
- 9.Kuo HC, Lauderdale TL, Lo DY, Chen CL, Chen PC, Liang SY, Kuo JC, Liao YS, Liao CH, Tsao CS, Chiou CS. 2014. An association of genotypes and antimicrobial resistance patterns among Salmonella isolates from pigs and humans in Taiwan. PLoS One 9:e95772. doi: 10.1371/journal.pone.0095772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Centers for Disease Control. 2018. National enteric disease surveillance: Salmonella annual report, 2016. https://www.cdc.gov/nationalsurveillance/pdfs/2016-Salmonella-report-508.pdf.
- 11.Wollin R. 2007. A study of invasiveness of different Salmonella serovars based on analysis of the Enter-net database. Euro Surveill 12:E0709273. doi: 10.2807/esw.12.39.03275-en. [DOI] [PubMed] [Google Scholar]
- 12.Inns T, Beasley G, Lane C, Hopps V, Peters T, Pathak K, Perez-Moreno R, Adak G, Shankar A, Outbreak Control Team. 2013. Outbreak of Salmonella enterica Goldcoast infection associated with whelk consumption, England, June to October 2013. Euro Surveill 18:20654. doi: 10.2807/1560-7917.es2013.18.49.20654. [DOI] [PubMed] [Google Scholar]
- 13.Bremer V, Leitmeyer K, Jensen E, Metzel U, Meczulat H, Weise E, Werber D, Tschaepe H, Kreienbrock L, Glaser S, Ammon A. 2004. Outbreak of Salmonella Goldcoast infections linked to consumption of fermented sausage, Germany 2001. Epidemiol Infect 132:881–887. doi: 10.1017/S0950268804002699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis H, Outbreak Control Team. 2005. International outbreak of Salmonella Goldcoast infection in tourists returning from Majorca, September-October 2005: final summary. Euro Surveill 10:E051208.3. doi: 10.2807/esw.10.49.02853-en. [DOI] [PubMed] [Google Scholar]
- 15.Scavia G, Ciaravino G, Luzzi I, Lenglet A, Ricci A, Barco L, Pavan A, Zaffanella F, Dionisi AM. 2013. A multistate epidemic outbreak of Salmonella Goldcoast infection in humans, June 2009 to March 2010: the investigation in Italy. Euro Surveill 18:20424. doi: 10.2807/ese.18.11.20424-en. [DOI] [PubMed] [Google Scholar]
- 16.Horvath JK, Mengel M, Krisztalovics K, Nogrady N, Paszti J, Lenglet A, Takkinen J. 2013. Investigation into an unusual increase of human cases of Salmonella Goldcoast infection in Hungary in 2009. Euro Surveill 18:20422. doi: 10.2807/ese.18.11.20422-en. [DOI] [PubMed] [Google Scholar]
- 17.European Food Safety Agency (EFSA). 2009. Analysis of the baseline survey on the prevalence of Salmonella in holdings with breeding pigs in the EU; 2008. EFSA J 7:1377. doi: 10.2903/j.efsa.2009.1377. [DOI] [Google Scholar]
- 18.Koskiniemi S, Pranting M, Gullberg E, Nasvall J, Andersson DI. 2011. Activation of cryptic aminoglycoside resistance in Salmonella enterica. Mol Microbiol 80:1464–1478. doi: 10.1111/j.1365-2958.2011.07657.x. [DOI] [PubMed] [Google Scholar]
- 19.Magnet S, Courvalin P, Lambert T. 1999. Activation of the cryptic aac(6′)-Iy aminoglycoside resistance gene of Salmonella by a chromosomal deletion generating a transcriptional fusion. J Bacteriol 181:6650–6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiou CS, Chen YT, Wang YW, Liu YY, Kuo HC, Tu YH, Lin AC, Liao YS, Hong YP. 2017. Dissemination of mcr-1-carrying plasmids among colistin-resistant Salmonella strains from humans and food-producing animals in Taiwan. Antimicrob Agents Chemother 61:e00338-17. doi: 10.1128/AAC.00338-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiou CS, Lin JM, Chiu CH, Chu CH, Chen SW, Chang YF, Weng BC, Tsay JG, Chen CL, Liu CH, Chu C. 2009. Clonal dissemination of the multi-drug resistant Salmonella enterica serovar Braenderup, but not the serovar Bareilly, of prevalent serogroup C1 Salmonella from Taiwan. BMC Microbiol 9:264. doi: 10.1186/1471-2180-9-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong YP, Wang YW, Huang IH, Liao YC, Kuo HC, Liu YY, Tu YH, Chen BH, Liao YS, Chiou CS. 2018. Genetic relationships among multidrug-resistant Salmonella enterica serovar Typhimurium strains from humans and animals. Antimicrob Agents Chemother 62:e00213-18. doi: 10.1128/AAC.00213-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiou C-S, Hong Y-P, Liao Y-S, Wang Y-W, Tu Y-H, Chen B-H, Chen Y-S. 2019. New multidrug-resistant Salmonella enterica serovar Anatum clone, Taiwan, 2015–2017. Emerg Infect Dis 25:144–147. doi: 10.3201/eid2501.181103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clinical and Laboratory Standards Institute (CLSI). 2018. Performance standards for antimicrobial susceptibility testing, 28th ed Clinical and Laboratory Standards Institute, Wayne (PA). [Google Scholar]
- 25.European Committee on Antimicrobial Susceptibility Testing. 2019. Breakpoint tables for interpretation of MICs and zone diameters. Version 9.0 http://www.eucast.org/clinical_breakpoints.
- 26.Tyson GH, Li C, Ayers S, McDermott PF, Zhao S. 2016. Using whole-genome sequencing to determine appropriate streptomycin epidemiological cutoffs for Salmonella and Escherichia coli. FEMS Microbiol Lett 363:fnw009. doi: 10.1093/femsle/fnw009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia-Migura L, Sunde M, Karlsmose S, Veldman K, Schroeter A, Guerra B, Granier SA, Perrin-Guyomard A, Gicquel-Bruneau M, Franco A, Englund S, Teale C, Heiska H, Clemente L, Boerlin P, Moreno MA, Daignault D, Mevius D, Hendriksen RS, Aarestrup FM. 2012. Establishing streptomycin epidemiological cut-off values for Salmonella and Escherichia coli. Microb Drug Resist 18:88–93. doi: 10.1089/mdr.2011.0064. [DOI] [PubMed] [Google Scholar]
- 28.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.