Selection of extended-spectrum mutations in narrow-spectrum oxacillinases (e.g., OXA-2 and OXA-10) is an emerging mechanism for development of in vivo resistance to ceftolozane-tazobactam and ceftazidime-avibactam in Pseudomonas aeruginosa. Detection of these challenging enzymes therefore seems essential to prevent clinical failure, but the complex phenotypic plasticity exhibited by this species may often lead to their underestimation.
KEYWORDS: OXA, Pseudomonas aeruginosa, antimicrobial resistance, ceftazidime-avibactam, ceftolozane-tazobactam, class D beta-lactamase
ABSTRACT
Selection of extended-spectrum mutations in narrow-spectrum oxacillinases (e.g., OXA-2 and OXA-10) is an emerging mechanism for development of in vivo resistance to ceftolozane-tazobactam and ceftazidime-avibactam in Pseudomonas aeruginosa. Detection of these challenging enzymes therefore seems essential to prevent clinical failure, but the complex phenotypic plasticity exhibited by this species may often lead to their underestimation. The underlying resistance mechanisms of two sequence type 175 (ST175) P. aeruginosa isolates showing multidrug-resistant phenotypes and recovered at early and late stages of a long-term nosocomial infection were evaluated. Whole-genome sequencing (WGS) was used to investigate resistance genomics, whereas molecular and biochemical methods were used for characterization of a novel extended-spectrum OXA-2 variant selected during therapy. The metallo-β-lactamase blaVIM-20 and the narrow-spectrum oxacillinase blaOXA-2 were present in both isolates, although they differed by an inactivating mutation in the mexB subunit, present only in the early isolate, and in a mutation in the blaOXA-2 β-lactamase, present only in the final isolate. The new OXA-2 variant, designated OXA-681, conferred elevated MICs of the novel cephalosporin–β-lactamase inhibitor combinations in a PAO1 background. Compared to OXA-2, kinetic parameters of the OXA-681 enzyme revealed a substantial increase in the hydrolysis of cephalosporins, including ceftolozane. We describe the emergence of the novel variant OXA-681 during treatment of a nosocomial infection caused by a Pseudomonas aeruginosa ST175 high-risk clone. The ability of OXA-681 to confer cross-resistance to ceftolozane-tazobactam and ceftazidime-avibactam together with the complex antimicrobial resistance profiles exhibited by the clinical strains harboring this new enzyme argue for maintaining active surveillance on emerging broad-spectrum resistance in P. aeruginosa.
INTRODUCTION
The recent introduction into the clinical setting of the novel β-lactam–β-lactamase inhibitor combinations ceftolozane-tazobactam and ceftazidime-avibactam represents a step forward in the fight against β-lactam-resistant Pseudomonas aeruginosa infections because they show increased stability against the major mutation-driven β-lactam resistance mechanisms in this species, such as the overexpression of the chromosomal cephalosporinase AmpC and efflux pumps and the inactivation of the porin OprD (1). Nevertheless, P. aeruginosa is a constantly moving target, and the catalogue of reports documenting development of resistance to these new currently available drugs during therapy is growing. Overproduction and structural modification of AmpC have been identified as the main strategies developed by P. aeruginosa to compromise the activity of ceftolozane-tazobactam and ceftazidime-avibactam during therapy (2–4). Compared to AmpC, the contribution of class D OXA enzymes to the resistance profile of P. aeruginosa has been considered of less clinical importance, since narrow-spectrum oxacillinases (e.g., OXA-2 and OXA-10, which are relatively common in P. aeruginosa strains worldwide) exhibit only a very limited substrate profile, and extended-spectrum variants (ES-OXAs) are rarely reported (5, 6). However, recent in vivo studies have evidenced that, like AmpC enzymes, they are prone to developing extended-spectrum mutations during the course of therapy, resulting in derivatives able to confer cross-resistance to ceftolozane-tazobactam and ceftazidime-avibactam (7). It is also worth mentioning that the vast majority of these enzymes are frequently mobilized in transposable elements within class 1 integrons and cotransferred with aminoglycoside-modifying enzymes or other remarkable resistance determinants, such as VIM-type metallo-β-lactamases (MBLs), the acquisition of which restricts the choice of therapy to an alarming extent (8). The overwhelming association of these horizontally acquired enzymes with a few epidemic multidrug-resistant (MDR)/extensively drug-resistant (XDR) P. aeruginosa clones disseminated across hospitals worldwide, the so-called high-risk clones, further adds to the concern (9).
Given this background, whereas studies are being conducted to identify potential strategies to prevent AmpC-mediated β-lactam resistance, such as inactivation of the key AmpG permease involved in AmpC overexpression (10), active surveillance is required to detect strains harboring these challenging horizontally acquired class D OXA β-lactamases and thus contribute to extending the life of these new currently available drugs. However, this is not always straightforward, since P. aeruginosa is equipped with a considerable repertoire of chromosomal genes able to modulate its levels of antimicrobial resistance. As a result, P. aeruginosa exhibits an outstanding phenotypic plasticity that severely hampers the inference of the underlying resistance mechanisms, which can often therefore be underestimated and lead to inadequate treatment, the development of resistance, and clinical failure (11). Here, we dissect the underlying resistance mechanisms of two sequence type 175 (ST175) P. aeruginosa isolates recovered at the early and late stages of a long-term nosocomial infection. A whole-genome sequencing (WGS) approach was used to depict the mutational and transferable resistance mechanisms involved, whereas molecular and biochemical studies were performed to characterize a novel OXA-2 variant that emerged during the course of treatment.
RESULTS AND DISCUSSION
Molecular typing, antimicrobial susceptibility, and genomic features of the clinical isolates.
Molecular typing procedures revealed that PA34 and PA145 were derived from the same progenitor strain that colonized the patient, who received treatment with several antipseudomonal β-lactams. Both isolates yielded identical SpeI pulsed-field gel electrophoresis (PFGE) genotype patterns and were assigned by multilocus sequence typing (MLST) to the epidemic ST175 high-risk clone, which is highly prevalent in multiple European hospitals, particularly in Spain and France. The antimicrobial susceptibility results and main mutational and horizontally acquired resistance determinants for strains PA34 and PA145 are summarized in Table 1. The two isolates demonstrated similar MICs of aminoglycosides, quinolones, colistin, and carbapenems, although major differences were observed for the rest of the β-lactams. Indeed, PA34 showed wide susceptibility to piperacillin-tazobactam, aztreonam, ceftazidime, ceftazidime-avibactam, and cefepime but yielded an unexpected ceftolozane-tazobactam MIC of 16 μg/ml. In contrast, PA145, which was isolated 70 days later after several courses of antimicrobial therapy with piperacillin-tazobactam, cefepime, and ceftazidime, exhibited an XDR phenotype and remained susceptible to only aztreonam and colistin. WGS analysis identified that both isolates shared the classical MexZ mutation (G195E) involved in MexXY overexpression and the triple quinolone resistance-determining region (QRDR) mutations affecting GyrA (T83I and D87N) and ParC (S87W), hallmarks of the mutational resistome of this high-risk clone (12), but lacked other characteristic mutations described in Spanish and French ST175 isolates, such as those affecting AmpR (G154R), the LysR-type transcriptional regulator of AmpC, responsible for AmpC overexpression, and the OprD Q142X inactivating mutation, which was replaced by a 5-bp insertion that also resulted in a truncated OprD. Mutations previously associated with resistance were not identified in the genes coding for the penicillin-binding proteins (PBP1a, PBP1b, PBP2, PBP3, or PBP4), suggesting that β-lactam-binding affinities were not altered in the two isolates.
TABLE 1.
Strain, sequence type, antimicrobial susceptibility, and main mutational and transferable resistance mechanisms exhibited by isolates PA34 and PA145a
| Strain | ST | MIC (μg/ml)b |
Resistance mechanisms [gene(s) (mutations)] |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chromosomal |
Transferable |
||||||||||||||||||||
| TIC (S ≤ 16) | P/T (S ≤ 16) | AZT (S ≤ 16) | CAZ (S ≤ 8) | CTZ (S ≤ 4) | CZA (S ≤ 8) | FEP (S ≤ 8) | TOB (S ≤ 4) | AK (S ≤ 8) | IPM (S ≤ 4) | MRP (S ≤ 2) | CIP (S ≤ 0.5) | COL (S ≤ 2) | MexAB-OprM | MexXY-OprM | AmpC | OprD | QRDR | β-Lactamases | AMEs | ||
| PA34 | 175 | 256 | <4/4 | 0,19 | 4 | 16/4 | 4/4 | 4 | >32 | 32 | 64 | 16 | 16 | 1 | mexB (nt80Δ10) | mexX (W358R), mexZ (G195D) | nt49InsGCACT | gyrA (T83I, D87N), parC (S87W) | blaOXA-2, blaVIM-20 | aadB, aac(6′)-Ib3, aphA-6, aadA13 | |
| PA145 | 175 | >512 | 32/4 | 4 | 64 | >32/4 | 32/4 | 16 | >32 | 32 | 32 | 32 | >16 | 1 | mexX (W358R), MexZ (G195D) | nt49InsGCACT | gyrA (T83I, D87N), parC (S87W) | blaOXA-681, blaVIM-20 | aadB, aac(6′)-Ib3, aphA-6, aadA13 | ||
ST, sequence type; S, sensitivity breakpoint; TIC, ticarcillin; P/T, piperacillin-tazobactam; AZT, aztreonam; CAZ, ceftazidime; CTZ, ceftolozane-tazobactam; CZA, ceftazidime-avibactam; FEP, cefepime; TOB, tobramycin; AK, amikacin; IPM, imipenem; MRP, meropenem; CIP, ciprofloxacin; COL, colistin; QRDR, quinolone resistance-determining region; AMEs, aminoglycoside-modifying enzymes.
2019 EUCAST breakpoints.
Analysis of horizontally acquired resistance determinants revealed the presence of several genes encoding aminoglycoside-modifying enzymes, including aadB, which is frequently detected in this lineage, aadA13, aac(6′)-Ib3, and aphA-6, which explains the high MICs observed for aminoglycosides. Likewise, analysis of horizontally acquired β-lactamases identified the presence of the genes coding for the blaVIM-20 carbapenemase (VIM-2 cluster) and the narrow-spectrum oxacillinase blaOXA-2 in both isolates. These results were partially unexpected, since although the presence of these β-lactamases correlated well with the phenotype exhibited by the final XDR isolate PA145 (susceptible to only colistin and aztreonam), it failed to explain the paradoxical resistance to ceftolozane-tazobactam with respect to susceptibility to piperacillin-tazobactam (MIC = 4/4 μg/ml), ceftazidime (MIC = 4 μg/ml), and cefepime (MIC = 4 μg/ml) of the early isolate PA34.
However, an analysis of the genes coding for the MexAB-OprM transporter, which strongly contributes to P. aeruginosa’s natural resistance to multiple β-lactams (13), revealed the presence of a 10-bp mutational insertion in the mexB gene, leading to a functional loss of MexAB-OprM pumping activity. Abolition of MexAB-OprM efflux explains the particularly low MIC of aztreonam (0.19 μg/ml), which is not hydrolyzed by either VIM-20 or OXA-2 enzymes (14, 15), but also seems to compensate for the effect of carbapenemase production on the MICs of ceftazidime and cefepime, since both are also substrates for this pump and exhibit MICs below the clinical breakpoints (4 μg/ml each). Indeed, a similar effect of the latter has been described in ceftazidime-susceptible cystic fibrosis isolates with deficient MexAB-OprM efflux and stable AmpC overproduction (16). On the other hand, it remains unclear why ceftolozane-tazobactam MICs remained above the clinical breakpoints (16 μg/ml) in this early strain, since this antibiotic bypasses the main intrinsic and mutational resistance mechanisms exhibited by P. aeruginosa and tends to show increased activity compared to other antipseudomonal cephalosporins against both wild-type and resistant strains. A possible explanation is a higher hydrolytic activity of VIM-20 against ceftolozane than against ceftazidime and cefepime, although the precise mechanisms that sustain this complicated resistance phenotype cannot be totally solved using the data presented here and deserve further investigation in future specific studies.
Moreover, it also deserves mention that the deficient MexAB-OprM efflux found in the isolate PA34 indicates that the late isolate PA145 is not the result of the direct evolution of the former, since it is difficult to conceive that the MexAB-OprM system could recover its functionality despite a 10-bp insertion in the mexB gene. Certainly, this was not unexpected, since in patients with chronic respiratory infections, as in this case, it is frequently found that many populations of P. aeruginosa coexist in the lung, and the timing and order of their recovery do not guarantee the evolutionary trajectory of the isolates. However, taking into account the genomic features described above and the results obtained by molecular typing procedures, it is evident that both isolates were derived from the same VIM-20/OXA-2-coproducing ST175 progenitor strain.
Characterization of β-lactamases.
Analysis of the sequences of acquired β-lactamases showed that isolate PA145 harbored a previously undescribed 3-bp (TAG) deletion affecting two codons in the blaOXA-2 gene, leading to the deletion of isoleucine at position 159 and the replacement of glutamic acid by lysine at position 160. This novel OXA-2 variant was designated OXA-681. To our knowledge, this is the first report in which an extended-spectrum OXA β-lactamase is generated from a narrow-spectrum OXA β-lactamase through the concomitant deletion and substitution of two conserved residues. Remarkably, both changes were located within the Trp-X-Glu-X-X-Leu-X-Ile-Ser stretch, a conserved motif in class D β-lactamases, also called the omega loop, which plays a major role in the accommodation of the β-lactam molecule in the active site (17–19). Based on the crystal structure of the OXA-2 enzyme available (PDB accession number 1K38), it is expected that the change of a negatively charged residue (E160) to a positively charged residue (K160), as well as the deletion of a neutral group (I159), would change significantly the charge of this region of the enzyme. As a consequence, this change might alter the binding mode of cephalosporins, as a result of trying to accommodate the side-chain groups of the ligand in a more positive environment.
In order to evaluate the impact of these substitutions on β-lactam resistance, blaOXA-2 and blaOXA-681 genes were cloned in parallel and expressed in a PAO1 background. Comparative MIC data for PAO1 expressing OXA-2 and OXA-681 are detailed in Table 2. Compared to OXA-2, the production of OXA-681 caused a 16-fold increase in the MIC of ceftazidime, a 32-fold increase in the MIC of ceftazidime-avibactam, and a 64-fold increase in the MIC of ceftolozane and ceftolozane-tazobactam, providing good evidence of the increased spectrum of resistance to antipseudomonal cephalosporins conferred by this new OXA-2 variant. In contrast, the production of OXA-681 caused a 4-fold decrease in the MIC of meropenem. Interestingly, the antimicrobial susceptibility profile conferred by OXA-681 on a PAO1 background is almost identical to the one recently reported for OXA-539, an OXA-2-derived extended-spectrum cephalosporinase selected during ceftazidime therapy (7). Therefore, the emergence of OXA-681 adds further evidence of the worrying effect of the classic antipseudomonal cephalosporins on the selection of class D β-lactamase variants with increased activity against the novel β-lactam–β-lactamase inhibitor combinations ceftolozane-tazobactam and ceftazidime-avibactam.
TABLE 2.
MICs for the PAO1 transformants producing OXA-2 and OXA-681 β-lactamasesa
| Strain | MIC (μg/ml) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TIC | TZP | AZT | CAZ | CZA | CTO | CTZ | FEP | IPM | MRP | |
| PAO1(pUCP24) | 16 | ≤4/4 | 4 | ≤1 | ≤1 | ≤0.5 | ≤0.5/4 | ≤1 | 1 | ≤0.5 |
| PAO1(pUCP-OXA-2) | 256 | ≤4/4 | 4 | 4 | 1/4 | 0.5 | 0.5/4 | 2 | 2 | 8 |
| PAO1(pUCP-OXA-681) | 256 | ≤4/4 | 4 | 64 | 32/4 | 32 | 32/4 | 4 | 2 | 2 |
TIC, ticarcillin; TZP, piperacillin-tazobactam; AZT, aztreonam; CAZ, ceftazidime; CZA, ceftazidime-avibactam; CTO, ceftolozane; CTZ, ceftolozane-tazobactam; FEP, cefepime; IPM, imipenem; MRP, meropenem.
Genetic context of β-lactamases.
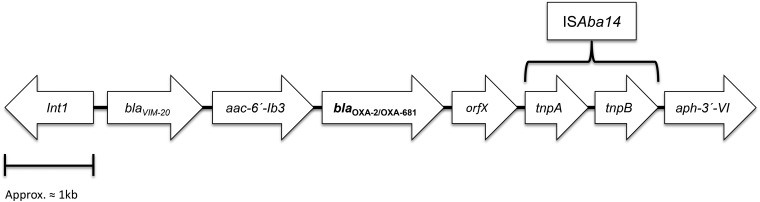
Both the blaOXA-2 and blaOXA-681 genes were documented to be located in a 6.625-kb class 1 integron harboring the blaVIM-20, aac(6′)-Ib3, blaOXA-2/681, and orfX (putative exonuclease) gene cassettes and truncated at its right-hand extremity by the conserved linear organization of ISAba14–aphA(3′)-VI (20) (Fig. 1). All attempts to transfer OXA-2/OXA-681 through conjugation and electroporation yielded persistently negative results. However, an analysis of the whole-genome sequencing data using plasmidSPAdes software identified the integron containing the blaOXA-2 and blaOXA-681 genes in a plasmid contig, therefore suggesting the plasmid location of the β-lactamases.
FIG 1.
Schematic structure of the truncated integron harboring OXA-2 and OXA-681 β-lactamases.
Relative kinetic properties of OXA-681 and OXA-2.
Kinetic measurements were performed on OXA-2 and OXA-681. Consistent with the MIC data, kinetic assays with ceftazidime, ceftolozane, and meropenem revealed major differences between the two proteins (Table 3). As shown, OXA-681 exhibited an enhanced ability to hydrolyze cephalosporins compared to OXA-2. The key amino acid changes I159del and E160K on the OXA-2 protein caused a substantial enhancement of the enzyme-substrate binding affinity (Km) without significantly affecting turnover rates (kcat). This effect is noticeable for both ceftazidime and ceftolozane, yielding kcat/Km (catalytic efficiency) ratios 1,514- and 17-fold higher than those for OXA-2. It is tempting to suggest, therefore, that the cephalosporin sequestration conferred by OXA-681 in the P. aeruginosa background (characterized by high impermeability) may play a greater role in resistance than turnover rates (21). In contrast, OXA-681 exhibited lower turnover rates and catalytic efficiency toward meropenem, illustrating that its newly evolved activity against cephalosporins is detrimental to the natural carbapenemase activity of the parental OXA-2 protein. On the other hand, the 50% inhibitory concentration (IC50) values (Table 4) show that the I159del and E160K mutations present in the OXA-681 enzyme confer not only increased cephalosporin hydrolysis but also a slight, ∼10-fold decrease in the affinity for tazobactam and the novel diazabicyclooctane (DBO) inhibitor avibactam compared to OXA-2.
TABLE 3.
Kinetic parameters of the β-lactamases OXA-2 and OXA-681 for representative substratesa
| Drug | OXA-2 |
OXA-681 |
kcat/Km ratio for OXA-681/OXA-2 | ||||
|---|---|---|---|---|---|---|---|
| Km (μM) ± SD | kcat (s−1) ± SD | kcat/Km (mM−1 s−1) | Km (μM) ± SD | kcat (s−1) ± SD | kcat/Km (mM−1 s−1) | ||
| Nitrocefin | 56.9415 ± 14.5405 | 128.7833 ± 12.4233 | 2,261 | 78.0759 ± 19.0628 | 11.0857 ± 2.0566 | 141.9862 | 0.06 |
| Ceftazidime | 991.2681 ± 175.1077 | 0.0025 ± 0.0002 | 0.0025 | 0.6706 ± 0.2037 | 0.0025 ± 0.0007 | 3.7280 | 1,514 |
| Ceftolozane | 305.2738 ± 35.4224 | 0.0003 ± 0.0001 | 0.0009 | 31.9304 ± 7.9682 | 0.0005 ± 0.0001 | 0.0157 | 17 |
| Meropenem | 0.0384 ± 0.0052 | 0.0097 ± 0.0006 | 252.6042 | 0.0036 ± 0.0006 | 0.0004 ± 0.0001 | 111.1111 | 0.40 |
Data represent the means from three independent experiments.
TABLE 4.
IC50 parameters of the β-lactamase inhibitors tazobactam and avibactam against OXA-2 and OXA-681a
| Drug | IC50 (nM) ± SD |
Ratio of IC50 of OXA-681/IC50 of OXA-2 | |
|---|---|---|---|
| OXA-2 | OXA-681 | ||
| Tazobactam | 1.48 ± 0.4761 | 13 ± 1.8302 | 9 |
| Avibactam | 367 ± 24.1150 | 3,050 ± 902.4786 | 8 |
Data represent the means from three independent experiments.
Concluding remarks.
The new antipseudomonal agents ceftolozane-tazobactam and ceftazidime-avibactam, which are now available, are increasingly recognized as the most suitable options for the treatment of MDR P. aeruginosa infections, and preserving their clinical utility represents a major challenge for the medical community. Although resistance to these new antibiotics in P. aeruginosa emerges mainly through specific mutations leading to the overexpression and structural modification of AmpC (22, 23), the selection of extended-spectrum mutations from narrow-spectrum OXA-type β-lactamases, such as OXA-2, represents another important emerging resistance mechanism. Here, we add evidence of this importance in the characterization of the tight-binding OXA-681 enzyme, which exhibits specific amino acid substitutions and abrogates the utility of the novel cephalosporin–β-lactamase inhibitor combinations. The relevance of these results is emphasized first by the fact that this enzyme emerged in a strain belonging to the epidemic ST175 high-risk clone, which may further disseminate among patients, and second by the fact that OXA-681 was located in a plasmid-encoded class 1 integron coharboring the MBL VIM-20, resulting in a challenging MBL/ES-OXA MDR element that could be disseminated further among strains. Finally, the impaired MexAB-OprM efflux identified in the early isolate of this work, which is a hallmark of strains recovered from long-term infections such as cystic fibrosis or bronchiectasis and is expected to play a minor role in strains recovered from acute infections, appears as a mechanism that could contribute to the emergence of complex resistance phenotypes that make it difficult to infer the relevant underlying resistance mechanisms in P. aeruginosa and therefore deserves further investigation. Altogether, these findings argue for the need for maintaining active surveillance on emerging broad-spectrum resistance in MDR P. aeruginosa strains.
MATERIALS AND METHODS
Case summary and clinical strains.
An 82-year-old patient with bronchiectasis and chronic respiratory failure, with a history of multiple hospital admissions during the preceding months, presented to the emergency department of a tertiary-level academic hospital with severe dyspnea, cough with sputum production, and oxygen desaturation on room air requiring noninvasive ventilatory support. The patient was receiving chronic respiratory treatment with tobramycin and azithromycin. Previous microbiological studies revealed positive respiratory cultures for several nonfermenting Gram-negative bacilli, including P. aeruginosa, Achromobacter xylosoxidans, and Stenotrophomonas maltophilia. After obtaining sputum samples and starting empirical therapy with piperacillin-tazobactam and trimethoprim-sulfamethoxazole (T/S), the patient was admitted to the pneumology department for follow-up. Forty-eight hours later, the sputum sample obtained upon admission grew MDR P. aeruginosa exhibiting an unusual susceptibility profile, being resistant to ceftolozane-tazobactam but susceptible to piperacillin-tazobactam, ceftazidime, ceftazidime-avibactam, cefepime, aztreonam, and colistin (PA34). The patient remained on therapy with piperacillin-tazobactam, and the cough and sputum disappeared. On hospital day 14, piperacillin-tazobactam was discontinued. Three days later, the patient experienced a new episode of severe cough and sputum production. Sputum samples yielded MDR P. aeruginosa with the same susceptibility pattern as the one obtained at admission (PA34), which was treated with cefepime for 29 days. After this time, the patient’s condition alternated between brief periods (3 to 4 days) of clinical improvement and severe acute exacerbations, yielding sputum cultures positive for A. xylosoxidans and P. aeruginosa that were treated for 10 days with T/S and for 15 days with ceftazidime, respectively. At the start of a novel acute exacerbation on hospital day 77, respiratory cultures grew XDR P. aeruginosa susceptible to only aztreonam and colistin (PA145). Therapy with aztreonam was continued and colistin inhalation was maintained, but the respiratory condition of the patient deteriorated, and he died a few days later.
Antimicrobial susceptibility testing.
MICs of ticarcillin, piperacillin-tazobactam, ceftazidime, ceftazidime plus 4 μg/ml avibactam, cefepime, aztreonam, ceftolozane, ceftolozane plus 4 μg/ml tazobactam, imipenem, meropenem, tobramycin, amikacin, ciprofloxacin, and colistin were determined by broth microdilution according to European Committee on Antimicrobial Susceptibility Testing (EUCAST) v.9.0 clinical breakpoints and guidelines (http://www.eucast.org/clinical_breakpoints/).
Molecular typing.
Clonal relatedness was assessed by using SpeI pulsed-field gel electrophoresis (PFGE) according to protocols described previously (24). Multilocus sequence typing was performed according to the scheme proposed by Curran et al. (25). The isolates were assigned a sequence type (ST) according to the allelic profiles available in the MLST Database (http://pubmlst.org/paeruginosa).
Determination of resistance mechanisms by WGS.
The presence of mutations within 146 chromosomal genes involved in the modulation of antimicrobial resistance was determined as described previously (26). Indexed paired-end libraries were generated from genomic DNA using a commercial library preparation kit (Nextera XT DNA library preparation kit; Illumina) and sequenced on an Illumina MiSeq benchtop sequencer with a MiSeq reagent kit (version 3; Illumina Inc., USA). The resulting paired-ended reads were compared to the P. aeruginosa PAO1 reference genome, and sequence variation was further analyzed for the 146 chromosomal genes related to antimicrobial resistance. The presence of horizontally acquired resistance determinants was also evaluated using online databases (https://cge.cbs.dtu.dk//services/ResFinder/).
Characterization of β-lactamases.
blaOXA-2 and blaOXA-681 genes were amplified in parallel using the primer pair OXA-2-F-Xba1 (5′-TCCTCTAGAGTTGGGCATTAGGAAAAG-3′) and OXA-2-R-HindIII (5′-TCCAAGGCTTTTATCGCGCAGCGTCCGAG-3′). The purified PCR products were digested with XbaI and HindIII, ligated into pUCP24, and transformed into Escherichia coli XL1-Blue. The resulting transformants were selected in LB agar plates containing 5 μg/ml gentamicin–X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)-IPTG (isopropyl-β-d-thiogalactopyranoside). Recombinant plasmids were electroporated into reference strain PAO1 and plated in LB agar plates containing 30 μg/ml gentamicin. MICs of ticarcillin, piperacillin-tazobactam, ceftazidime, ceftazidime-avibactam, cefepime, aztreonam, ceftolozane, ceftolozane-tazobactam, imipenem, and meropenem for the resulting PAO1 transformants were determined according to the methodology described above.
Characterization of genetic elements harboring acquired β-lactamases.
The possible plasmid location of acquired β-lactamase genes was preliminarily analyzed through transformation and conjugation experiments according to previously described protocols (27). Moreover, the plasmid location of acquired β-lactamase genes was further evaluated from whole-genome sequencing data containing short Illumina reads using the plasmidSPAdes software tool (28). Sequence analysis of the integron regions harboring blaVIM-20 and blaOXA-2/OXA-681 genes scattered in different contigs was performed by PCR followed by sequencing.
Protein purification.
blaOXA-2 and blaOXA-681 genes were directionally cloned into the p-GEX-6P-1 plasmid (GE Healthcare, Little Chalfont, UK) for protein purification. Genes were amplified using the primer pair OXA-2-F-BamHI (5′-CGCGGATCCGCGCAAGAAGGCACG-3′) and OXA-2-R-SmaI (5′-TCCCCCGGGGGATTATCGCGCAGC-3′), digested with BamHI and SmaI, and ligated into BamHI/SmaI-digested p-GEX-6P-1. The recombinant plasmids were electroporated into the protease-deficient E. coli BL21 strain to generate two fusion proteins: glutathione S-transferase (GST)/OXA-2 and GST/OXA-681. The GST tag was then cleaved off, and the resulting proteins were then purified to homogeneity using the GST gene fusion system (Amersham Pharmacia Biotech, Europe) according to the manufacturer’s instructions. Finally, SDS-PAGE was performed to ascertain the absence of impurities in the final extract (≥95% purity).
Kinetic assays.
The kinetic parameters of purified OXA-2 and OXA-681 β-lactamases were determined at room temperature using a Nicolet Evolution 300 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) or an Epoch 2 microplate spectrophotometer (BioTek, VT, USA). Each experiment was performed in triplicate using 50 mM sodium phosphate with 20 mM sodium bicarbonate in 0.2-cm-path-length cuvettes. Kinetic parameters were determined by measuring the initial hydrolysis rates and using a Hanes-Woolf linearization of the Henri-Michaelis-Menten equation. For each antibiotic, the Km values were measured as the Ki by measuring the residual β-lactamase activity, using nitrocefin (Oxoid, Hampshire, United Kingdom) as the reporter substrate, whereas kcat values were determined by monitoring the direct hydrolysis of the antibiotic at a substrate concentration much higher than the Km. For inhibition kinetics, the IC50s for tazobactam and avibactam were calculated as the inhibitory concentrations resulting in a 50% reduction of nitrocefin hydrolysis and were determined after a 10-min preincubation with the enzyme at 490 nm. The wavelengths and absorption coefficients used for each antibiotic were as follows: 260 nm/Λ ɛ of −8,660 M−1 cm−1 for ceftazidime, 254 nm/Λ ɛ of −6,810 M−1 cm−1 for ceftolozane, and 297 nm/Λ ɛ of −10,940 M−1 cm−1 for meropenem.
Accession number(s).
The nucleotide sequences described in this work have been deposited in the GenBank database under the following accession numbers: MH986647.1 (blaOXA-681), SRX5389645 (genome sequence data for strain PA34), and SRX5389644 (genome sequence data for strain PA145).
ACKNOWLEDGMENTS
We have no conflicts of interest to declare.
This work was supported by the Ministerio de Economía y Competitividad of Spain, Instituto de Salud Carlos III, through grants PI1800076 to A.O., PI15/00860 and PI18/00501 to G.B., and P14/00059 and P17/01482 to A.B. and through grants from the Spanish Ministry of Economy and Competitiveness (SAF2016-75638-R) and the Xunta de Galicia (ED431G/09) to C.G.-B. This work was also supported by the European Regional Development Fund A Way To Achieve Europe ERDF, through the Spanish Network for Research in Infectious Diseases (RD16/0016). J.C.V.-U. was financially supported by the pFIS program (ISCIII, PI17/01482), and M.M.-G. was financially supported by a Clara Roy grant (SEIMC).
REFERENCES
- 1.van Duin D, Bonomo RA. 2016. Ceftazidime/avibactam and ceftolozane/tazobactam: second-generation β-lactam/β-lactamase inhibitor combinations. Clin Infect Dis 63:234–241. doi: 10.1093/cid/ciw243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fraile-Ribot PA, Cabot G, Mulet X, Periañez L, Martín-Pena ML, Juan C, Pérez JL, Oliver A. 2018. Mechanisms leading to in vivo ceftolozane/tazobactam resistance development during the treatment of infections caused by MDR Pseudomonas aeruginosa. J Antimicrob Chemother 73:658–663. doi: 10.1093/jac/dkx424. [DOI] [PubMed] [Google Scholar]
- 3.MacVane SH, Pandey R, Steed LL, Kreiswirth BN, Chen L. 2017. Emergence of ceftolozane-tazobactam-resistant Pseudomonas aeruginosa during treatment is mediated by a single AmpC structural mutation. Antimicrob Agents Chemother 61:e01183-17. doi: 10.1128/AAC.01183-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Humphries RM, Hindler JA, Wong-Beringer A, Miller SA. 2017. Activity of ceftolozane-tazobactam and ceftazidime-avibactam against beta-lactam-resistant Pseudomonas aeruginosa isolates. Antimicrob Agents Chemother 61:e01858-17. doi: 10.1128/AAC.01858-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poirel L, Gerome P, De Champs C, Stephanazzi J, Naas T, Nordmann P. 2002. Integron-located oxa-32 gene cassette encoding an extended-spectrum variant of OXA-2 β-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother 46:566–569. doi: 10.1128/AAC.46.2.566-569.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans BA, Amyes SGB. 2014. OXA β-lactamases. Clin Microbiol Rev 27:241–263. doi: 10.1128/CMR.00117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fraile-Ribot PA, Mulet X, Cabot G, del Barrio-Tofiño E, Juan C, Pérez JL, Oliver A. 2017. In vivo emergence of resistance to novel cephalosporin–β-lactamase inhibitor combinations through the duplication of amino acid D149 from OXA-2 β-lactamase (OXA-539) in sequence type 235 Pseudomonas aeruginosa. Antimicrob Agents Chemother 61:e01117-17. doi: 10.1128/AAC.01117-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Potron A, Poirel L, Nordmann P. 2015. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: mechanisms and epidemiology. Int J Antimicrob Agents 45:568–585. doi: 10.1016/j.ijantimicag.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 9.Oliver A, Mulet X, López-Causapé C, Juan C. 2015. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist Updat 21–22:41–59. doi: 10.1016/j.drup.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Cabot G, Sánchez-Diener I, Zamorano L, Florit-Mendoza L, Oliver A. 2018. Deciphering β-lactamase-independent β-lactam resistance evolution trajectories in Pseudomonas aeruginosa. J Antimicrob Chemother 73:3322–3331. doi: 10.1093/jac/dky364. [DOI] [PubMed] [Google Scholar]
- 11.Juan C, Conejo MC, Tormo N, Gimeno C, Pascual Á, Oliver A. 2013. Challenges for accurate susceptibility testing, detection and interpretation of β-lactam resistance phenotypes in Pseudomonas aeruginosa: results from a Spanish multicentre study. J Antimicrob Chemother 68:619–630. doi: 10.1093/jac/dks439. [DOI] [PubMed] [Google Scholar]
- 12.Cabot G, López-Causapé C, Ocampo-Sosa AA, Sommer LM, Domínguez MÁ, Zamorano L, Juan C, Tubau F, Rodríguez C, Moyà B, Peña C, Martínez-Martínez L, Plesiat P, Oliver A. 2016. Deciphering the resistome of the widespread Pseudomonas aeruginosa sequence type 175 international high-risk clone through whole-genome sequencing. Antimicrob Agents Chemother 60:7415–7423. doi: 10.1128/AAC.01720-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poole K, Krebes K, McNally C, Neshat S. 1993. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J Bacteriol 175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okamoto K, Gotoh N, Nishino T. 2002. Alterations of susceptibility of Pseudomonas aeruginosa by overproduction of multidrug efflux systems, MexAB-OprM, MexCD-OprJ, and MexXY/OprM to carbapenems: substrate specificities of the efflux systems. J Infect Chemother 8:371–373. doi: 10.1007/s10156-002-0193-7. [DOI] [PubMed] [Google Scholar]
- 15.Vettoretti L, Plésiat P, Muller C, El Garch F, Phan G, Attrée I, Ducruix A, Llanes C. 2009. Efflux unbalance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob Agents Chemother 53:1987–1997. doi: 10.1128/AAC.01024-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Llanes C, Pourcel C, Richardot C, Plésiat P, Fichant G, Cavallo J-D, Mérens A, GERPA Study Group. 2013. Diversity of β-lactam resistance mechanisms in cystic fibrosis isolates of Pseudomonas aeruginosa: a French multicentre study. J Antimicrob Chemother 68:1763–1771. doi: 10.1093/jac/dkt115. [DOI] [PubMed] [Google Scholar]
- 17.Poirel L, Naas T, Nordmann P. 2010. Diversity, epidemiology, and genetics of class D β-lactamases. Antimicrob Agents Chemother 54:24–38. doi: 10.1128/AAC.01512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Danel F, Hall LM, Gur D, Livermore DM. 1997. OXA-15, an extended-spectrum variant of OXA-2 beta-lactamase, isolated from a Pseudomonas aeruginosa strain. Antimicrob Agents Chemother 41:785–790. doi: 10.1128/AAC.41.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dehecq B, Hocquet D, Plésiat P, Colomb M, Courvalin P, Meziane-Cherif D, Belmonte O. 2011. Ceftazidime-hydrolysing β-lactamase OXA-145 with impaired hydrolysis of penicillins in Pseudomonas aeruginosa. J Antimicrob Chemother 66:1745–1750. doi: 10.1093/jac/dkr187. [DOI] [PubMed] [Google Scholar]
- 20.Yoon E-J, Goussard S, Touchon M, Krizova L, Cerqueira G, Murphy C, Lambert T, Grillot-Courvalin C, Nemec A, Courvalin P. 2014. Origin in Acinetobacter guillouiae and dissemination of the aminoglycoside-modifying enzyme Aph(3′)-VI. mBio 5:e01972-14. doi: 10.1128/mBio.01972-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Antunes NT, Lamoureaux TL, Toth M, Stewart NK, Frase H, Vakulenko SB. 2014. Class D β-lactamases: are they all carbapenemases? Antimicrob Agents Chemother 58:2119–2125. doi: 10.1128/AAC.02522-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haidar G, Philips NJ, Shields RK, Snyder D, Cheng S, Potoski BA, Doi Y, Hao B, Press EG, Cooper VS, Clancy CJ, Nguyen MH. 2017. Ceftolozane-tazobactam for the treatment of multidrug-resistant Pseudomonas aeruginosa infections: clinical effectiveness and evolution of resistance. Clin Infect Dis 65:110–120. doi: 10.1093/cid/cix182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barnes MD, Taracila MA, Rutter JD, Bethel CR, Galdadas I, Hujer AM, Caselli E, Prati F, Dekker JP, Papp-Wallace KM, Haider S, Bonomo RA. 2018. Deciphering the evolution of cephalosporin resistance to ceftolozane-tazobactam in Pseudomonas aeruginosa. mBio 9:e02085-18. doi: 10.1128/mBio.02085-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juan C, Zamorano L, Mena A, Albertí S, Pérez JL, Oliver A. 2010. Metallo-β-lactamase-producing Pseudomonas putida as a reservoir of multidrug resistance elements that can be transferred to successful Pseudomonas aeruginosa clones. J Antimicrob Chemother 65:474–478. doi: 10.1093/jac/dkp491. [DOI] [PubMed] [Google Scholar]
- 25.Curran B, Jonas D, Grundmann H, Pitt T, Dowson CG. 2004. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J Clin Microbiol 42:5644–5649. doi: 10.1128/JCM.42.12.5644-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.del Barrio-Tofiño E, López-Causapé C, Cabot G, Rivera A, Benito N, Segura C, Montero MM, Sorlí L, Tubau F, Gómez-Zorrilla S, Tormo N, Durá-Navarro R, Viedma E, Resino-Foz E, Fernández-Martínez M, González-Rico C, Alejo-Cancho I, Martínez JA, Labayru-Echverria C, Dueñas C, Ayestarán I, Zamorano L, Martinez-Martinez L, Horcajada JP, Oliver A. 2017. Genomics and susceptibility profiles of extensively drug-resistant Pseudomonas aeruginosa isolates from Spain. Antimicrob Agents Chemother 61:e01589-17. doi: 10.1128/AAC.01589-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cabot G, Ocampo-Sosa AA, Domínguez MA, Gago JF, Juan C, Tubau F, Rodríguez C, Moyà B, Peña C, Martínez-Martínez L, Oliver A. 2012. Genetic markers of widespread extensively drug-resistant Pseudomonas aeruginosa high-risk clones. Antimicrob Agents Chemother 56:6349–6357. doi: 10.1128/AAC.01388-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Antipov D, Hartwick N, Shen M, Raiko M, Lapidus A, Pevzner PA. 2016. plasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics 32:3380–3387. doi: 10.1093/bioinformatics/btw493. [DOI] [PubMed] [Google Scholar]