Acinetobacter baumannii is an important Gram-negative pathogen in hospital-related infections. However, treatment options for A. baumannii infections have become limited due to multidrug resistance. Bacterial virulence is often associated with capsule genes found in the K locus, many of which are essential for biosynthesis of the bacterial envelope. However, the roles of other genes in the K locus remain largely unknown.
KEYWORDS: ISAba16, K locus, dehydrogenase, morphology, surface polysaccharides
ABSTRACT
Acinetobacter baumannii is an important Gram-negative pathogen in hospital-related infections. However, treatment options for A. baumannii infections have become limited due to multidrug resistance. Bacterial virulence is often associated with capsule genes found in the K locus, many of which are essential for biosynthesis of the bacterial envelope. However, the roles of other genes in the K locus remain largely unknown. From an in vitro evolution experiment, we obtained an isolate of the virulent and multidrug-resistant A. baumannii strain MDR-ZJ06, called MDR-ZJ06M, which has an insertion by the ISAba16 transposon in gnaA (encoding UDP-N-acetylglucosamine C-6 dehydrogenase), a gene found in the K locus. The isolate showed an increased resistance toward tigecycline, whereas the MIC decreased in the case of carbapenems, cephalosporins, colistin, and minocycline. By using knockout and complementation experiments, we demonstrated that gnaA is important for the synthesis of lipooligosaccharide and capsular polysaccharide and that disruption of the gene affects the morphology, drug susceptibility, and virulence of the pathogen.
INTRODUCTION
Acinetobacter baumannii is considered an important opportunistic Gram-negative pathogen, causing a wide range of nosocomial infections as well as septicemia, meningitis, pneumonia, and urinary tract infections. A critical issue for clinical treatment is the increasing number of A. baumannii strains displaying multidrug resistance to most first-line antibiotics (1–3), with reports of strains for which almost all available antimicrobials are ineffective (4, 5). Therefore, new antibiotics against these resistant strains are urgently needed.
The asymmetric architecture of the outer membrane plays a crucial role in Gram-negative bacteria since the impermeability of the outer membrane provides an intrinsic barrier to various antimicrobials. Intact lipopolysaccharide (LPS) is composed of three distinct moieties—the lipid A anchor, core oligosaccharide, and O antigen (6). However, A. baumannii does not contain an O antigen; thus, the surfaces of A. baumannii cells display lipooligosaccharide (LOS) only. The presence of lipid A is essential for the LPS/LOS molecule because it serves as a membrane anchor. Therefore, inhibition of lipid A biosynthesis results in cell death, making proteins involved in this process potential targets for novel antibacterial compounds. Studies have shown that LpxC inhibitors can disturb the first step of lipid A biosynthesis. As a consequence, TLR4 is not activated in host cells, and opsonophagocytic killing of bacteria is enhanced, but LpxC inhibition also results in increased antibiotic susceptibility of bacteria (7–9).
In addition to LOS, capsular polysaccharide (CPS) plays a crucial role in the pathogenicity of bacteria. CPS has been recognized as one of the most important virulence factors (10, 11). The genes for capsule synthesis are located in the K locus in A. baumannii, with the K locus generally displaying high variability. In addition to the genes associated with the formation of CPS, the K locus also contains genes for nucleotide-sugar biosynthesis (12). Although the importance of the K locus for the pathogenicity of bacteria has been widely reported, the genes involved in the synthesis of simple UDP-linked sugars and sugar precursors in A. baumannii have been neglected in most studies.
In this work, we identified a new gene called gnaA (BGV75_06155) in an in vitro evolution experiment using increasing concentrations of tigecycline. We characterized the effects of the gene on antimicrobial susceptibility, bacterial morphology, membrane potential and permeability, LOS formation, and virulence in A. baumannii. Based on the impact that gnaA disruption had on the bacterial phenotype, we propose that the gene product might be a valuable drug target for the development of novel antibacterial compounds.
RESULTS
Gene mutations detected in the mutant strain.
Using the multidrug-resistant A. baumannii strain MDR-ZJ06, we performed a laboratory evolution experiment in which the concentration of tigecycline was doubled every day, starting from 0.5× MIC. After 7 days, at 32× MIC, a culture termed “T7” was obtained, with no growth observed in a subculture into medium with 64× MIC tigecycline. After streaking out a sample from a frozen aliquot of culture T7 on solid medium without tigecycline, we randomly selected one colony and grew a culture from this clone, designated MDR-ZJ06M. To detect potential mutations, we compared the genomes of MDR-ZJ06M (see the supplemental material) and its parental isolate MDR-ZJ06. We found the mutant to be isogenic with MDR-ZJ06, with the exception of an insertion in the gene BGV75_06155, which was verified by PCR amplification and Sanger sequencing. Comparative genomic analysis showed that gnaA (BGV75_06155) was interrupted by the transposon ISAba16 and that this mutation was the only difference between the two strains. gnaA is located in the K locus in A. baumannii and is associated with the synthesis of simple UDP-linked sugars (12). gnaA encodes UDP-N-acetylglucosamine C-6 dehydrogenase, which catalyzes the conversion of UDP-N-acetyl-d-glucosamine to UDP-N-acetyl-d-glucosaminuronic acid.
An 8-bp target site duplication of the gnaA gene encoding the sequence GAATTACA was found at the 5′ and 3′ ends of ISAba16, causing insertional inactivation of gnaA (Fig. 1). As the insertion site was close to the end of the gene, we posed the question of whether the ISAba16 insertion affected the function of the enzyme. Insertion analysis revealed that ISAba16 had the left inverted repeat 5′-GTAAGCATCCGGCTAA-3′ and the right inverted repeat 5′-TTCAGCGGACGCTTAC-3′. The insertion sequence ISAba16 consisted of three open reading frames (ORFs) encoding transposases A, B, and C. ISAba16 belongs to the IS66 family, which displays at least three ORFs (13).
FIG 1.
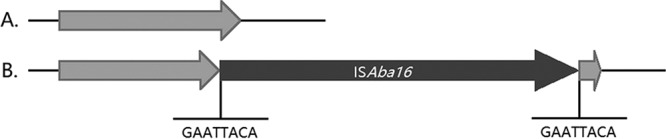
Schematic representation of the gnaA gene. (A) Wild-type gnaA in MDR-ZJ06. (B) A disrupted gnaA gene was generated due to an insertion of the transposon ISAba16. An 8-bp target site duplication of the gnaA gene encoding the sequence GAATTACA was observed at the 5′ and 3′ ends of ISAba16.
A novel gene mutation in the K locus leads to altered antibiotic susceptibility.
Compared with MDR-ZJ06, MDR-ZJ06M (MDR-ZJ06 gnaA::ISAba16) showed increased susceptibility to imipenem, meropenem, ceftazidime, cefepime, colistin, and minocycline. To identify whether gnaA was responsible for susceptibility, both knockout and complementation experiments were performed. MDR-ZJ06M and the MDR-ZJ06 knockout strain (MDR-ZJ06KO) were transformed with a plasmid that encoded wild-type gnaA, which recovered the MICs for imipenem, meropenem, ceftazidime, cefepime, minocycline, and colistin partly or completely (Table 1). Both strains, MDR-ZJ06M and MDR-ZJ06KO, shared similar susceptibility to these drugs. Notably, even though MDR-ZJ06 harbored blaOXA-66, blaOXA-23, and blaADC-25, the inactivation or deletion of gnaA made the strains susceptible to carbapenems and cephalosporins again (compared to the parental strain MDR-ZJ06), as MDR-ZJ06M became sensitive to imipenem and meropenem, and MDR-ZJ06KO became meropenem intermediate. Interestingly, when MDR-ZJ06KO was complemented with the fragment of gnaA found in MDR-ZJ06M, the complementation produced similar findings, indicating that the residual gnaA in MDR-ZJ06M may not produce a functional gene product.
TABLE 1.
Antimicrobial susceptibility of A. baumannii MDR-ZJ06 and gnaA-deficient/deletion strains
| Strain | MIC (mg/liter) for:a
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| IPM | MEM | CAZ | FEP | CST | AMK | GEN | MIN | TGC | |
| MDR-ZJ06 | 32 | 16 | >256 | >256 | 1 | >512 | >512 | >256 | 2 |
| MDR-ZJ06M | 2 | 2 | 32 | 48 | 0.125 | >512 | >512 | 12 | 4 |
| MDR-ZJ06M/pYMAb2-hyg | 2 | 2 | 32 | 32 | 0.0625 | >512 | >512 | 16 | 4 |
| MDR-ZJ06M/pYMAb2-gnaA | 16 | 8 | 64 | 96 | 2 | >512 | >512 | >256 | 4 |
| MDR-ZJ06KO | 8 | 4 | 48 | 64 | 0.0625 | >512 | >512 | 12 | 4 |
| MDR-ZJ06KO/pYMAb2-hyg | 4 | 4 | 48 | 64 | 0.0625 | >512 | >512 | 8 | 4 |
| MDR-ZJ06KO/pYMAb2-part gnaA | 4 | 4 | 32 | 24 | 0.0625 | >512 | >512 | 8 | 4 |
| MDR-ZJ06KO/pYMAb2-gnaA | 16 | 16 | 64 | 32 | 0.5 | >512 | >512 | >256 | 4 |
IPM, imipenem; MEM, meropenem; CAZ, ceftazidime; FEP, cefepime; CST, colistin; AMK, amikacin; GEM, gentamicin; MIN, minocycline; TGC, tigecycline.
By selecting against tigecycline in our laboratory evolution experiment, we would expect a high tolerance toward this antimicrobial compound. After testing the isolate for sensitivity toward tigecycline, albeit under different conditions from those of the in vitro evolution experiment (broth microdilution method), we found that the MICs increased only slightly from 2 to 4 mg/liter; such an increase could be considered a normal variation. We believe that the increased tolerance for the antibiotic was a result of an adaptation process during the evolution experiment (details discussed below).
A. baumannii gnaA plays an important role in membrane integrity and morphology.
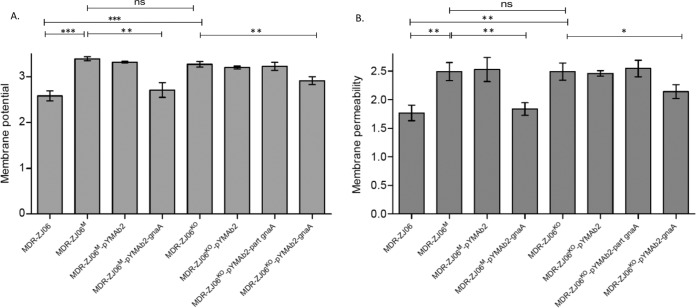
Membrane potential (MP) is an electrochemical gradient across a biological membrane and has been detected in all bacteria (14). MP is vital to the physiology of cells because it is involved in many processes, including the metabolism, autolysis, and survival of a cell. In Gram-negative bacteria, LPS is mainly responsible for the surface charge (15). This study showed that MPs in the gnaA-deficient/deletion strains were significantly higher than those in the wild-type strain (P = 0.0003 and P = 0.0007, respectively). Furthermore, the MP of the gnaA-deficient/deletion strains decreased after complementation with the wild-type gnaA in MDR-ZJ06. After complementation with the residual gnaA in MDR-ZJ06M, the MP of the complemented strain remained at a high level (Fig. 2A).
FIG 2.
Membrane potential and membrane permeability assay. Each strain was tested in triplicate, and the data were calculated based on three independent experiments. A normalized permeability parameter from the ratio of To-Pro-3 fluorescence to green DiOC2(3) fluorescence was derived. The mean ± standard deviation (SD) is shown; the mean differences were analyzed using Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Gram-negative bacteria are resistant to many antimicrobial agents as a result of the effective permeability barrier function of the outer membrane. To evaluate the permeability properties of this barrier, To-Pro-3 was used to assay membrane permeability since it is excluded from cells with intact membranes but stains nucleic acids in cells with damaged membranes. The membrane permeability assay showed that the permeability of the gnaA-deficient/deletion strains was significantly higher than that of MDR-ZJ06 (P = 0.0037 and P = 0.0036, respectively), which correlated with the change in antimicrobial susceptibility. By complementing in trans, the complementation strains (wild-type gnaA) restored the barrier function. However, no significant difference between MDR-ZJ06KO and MDR-ZJ06KO/pYMAb2-part gnaA could be observed (P = 0.6671) (Fig. 2B). Overall, we showed that the membrane of A. baumannii MDR-ZJ06 was significantly influenced by gnaA, as an incomplete gnaA (resulting in the expression of a fragment of the protein) increased the MP and caused damage to the membrane. The effects that the inactivation or fragmentation of gnaA have on the membrane are likely to also lead to the differences that were observed in antimicrobial susceptibility testing.
To determine the effect of gnaA on bacterial morphology, scanning electron microscopy (SEM) was conducted to identify morphological changes caused by gnaA. As shown in Fig. 3, MDR-ZJ06 exhibited a symmetrical, noncurved, and rod-shaped morphology, while the gnaA-deficient/deletion strains looked smaller, bent, and reniform. After complementation with a plasmid-encoded wild-type gnaA, the strains regained the large and straight rod shape, whereas reniform bacteria were rarely observed. In contrast, the MDR-ZJ06KO/pYMAb2-part gnaA strain exhibited a more irregular morphology than that of the gnaA-deficient/deletion strains (see Fig. S2 in the supplemental material).
FIG 3.
Scanning electron microscopy images of MDR-ZJ06 wild-type, mutants, and plasmid-complemented strains (see Materials and Methods). Scale bars are 5 μm on the left and 1 μm on the right. MDR-ZJ06 had a symmetrical and regular rod shape, while the gnaA interruption or deletion strain looked smaller, bent, and reniform; the gnaA plasmid complement strain displayed a larger and more symmetrical morphology.
The gnaA mutation changes the composition of LOS and CPS in A. baumannii.
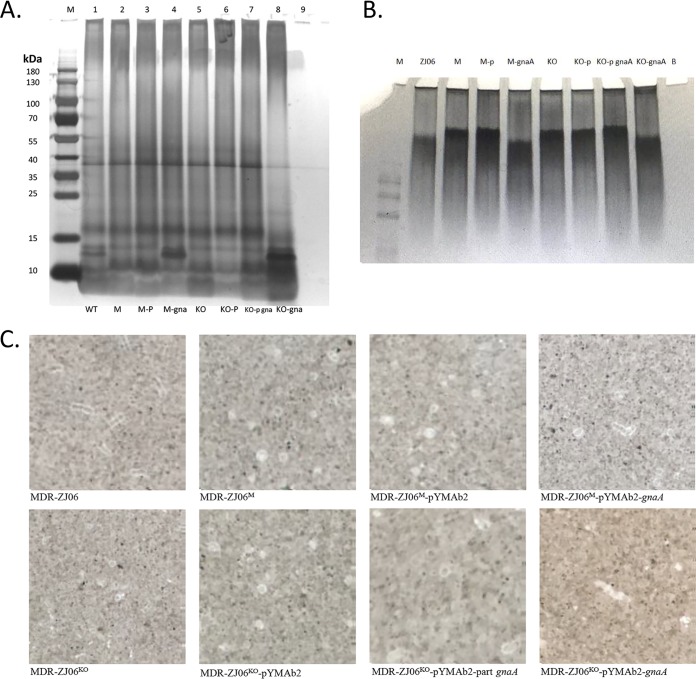
To investigate the change in phenotype described above, we extracted the surface polysaccharides of the strains and separated them on a 10% SDS-polyacrylamide gel and a 4% to 20% Tris-glycine gel. After silver staining, we observed one band that was different between the genotypes. In the gnaA-deficient/deletion strains (lanes 2 and 5), the band was absent but could be observed again when the strain was complemented with wild-type gnaA (lanes 4 and 8), yielding an identical pattern to that of MDR-ZJ06 (lane 1) (Fig. 4A). However, when complemented with a truncated version of gnaA (lane 7), the knockout strain did not result in a full restoration of the LOS pattern. This result suggested that only the full-length gnaA gene is able to allow a wild type-like LOS composition. In addition, when detecting capsular polysaccharides, little difference between wild-type and incomplete gnaA was observed (Fig. 4B). India ink staining also showed that all strains displayed a thin capsule, while no obvious difference was observed by microscopy (Fig. 4C).
FIG 4.
Effects of gnaA on surface polysaccharides, including LOS and CPS. (A) Silver-stained LOS from A. baumannii. Ten microliters of purified LOS was separated on a 10% SDS-polyacrylamide gel and silver stained. The black arrow indicates the band described in the text. (B) Analysis of CPS separated by SDS-PAGE and silver stained. (C) India ink staining of bacteria. Lanes 1 to 9 in panels A and B represent MDR-ZJ06, MDR-ZJ06M, MDR-ZJ06M/pYMAb2, MDR-ZJ06M/pYMAb2-gnaA, MDR-ZJ06KO, MDR-ZJ06KO/pYMAb2, MDR-ZJ06KO/pYMAb2-part gnaA, MDR-ZJ06KO/pYMAb2-gnaA, and SDS buffer, respectively. M, Thermo Scientific PageRuler Prestained Protein Ladder, 10 to 180 kDa.
Virulence assay in Galleria mellonella.
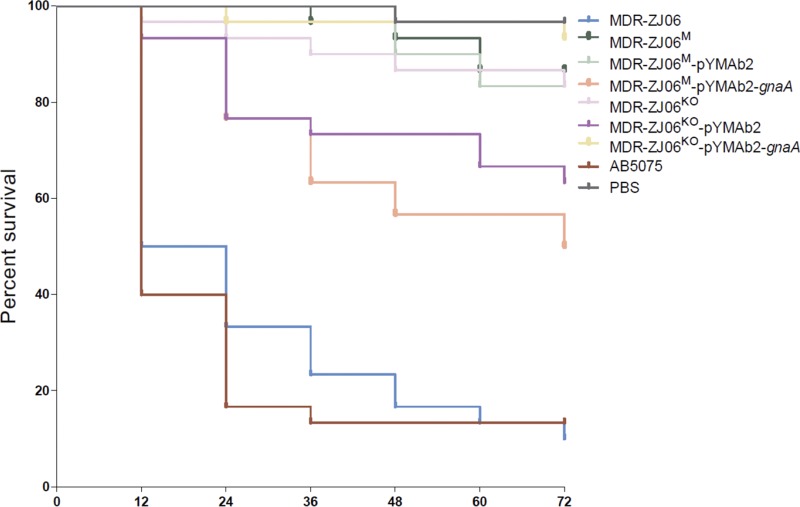
To address the effect of gnaA on LOS and possible implications on the virulence of A. baumannii, we performed experiments using the G. mellonella model. Half of the larvae died by 12 h after injection with MDR-ZJ06, which was similar to the result for the control AB5075 (P = 0.6628) (Fig. 5). However, over 80% of larvae were still alive on day 3 after infection with the mutant or knockout strain of MDR-ZJ06. The gnaA-deficient/deletion strains were both less virulent than MDR-ZJ06 in the G. mellonella model (P < 0.0001). Virulence of the MDR-ZJ06M strain increased after being complemented with a plasmid-encoded wild-type gnaA, with a significant difference between MDR-ZJ06M and MDR-ZJ06M/pYMAb2-gnaA (P = 0.0015). However, the complementation did not restore virulence to the wild-type level. Also, no difference was observed in virulence between MDR-ZJ06KO and MDR-ZJ06KO/pYMAb2-gnaA (P = 0.2244), and their corresponding survival rates were similar.
FIG 5.
Kaplan-Meier survival curve showing the virulence of individual isolates in G. mellonella. G. mellonella larvae (n = 30) were inoculated with 106 CFU of AB5075, MDR-ZJ06, and mutants of MDR-ZJ06. Survival was monitored every 12 h for 3 days.
DISCUSSION
In this study, we obtained the A. baumannii mutant MDR-ZJ06M from an in vivo evolution experiment where we doubled the concentration of tigecycline after each subculturing step. To our surprise, the clone that we isolated from a culture grown at 32× MIC tigecycline did not contain an antibiotic resistance gene or a gene mutation that conferred resistance to the bacterium. In contrast, the isolate contains a transposon insertion in a gene that we found to be involved in the production of the cell wall. MDR-ZJ06M shows only slight tolerance toward tigecycline, which is surprising because it was obtained from a culture grown at a high tigecycline concentration. However, when the selective pressure is removed by subculturing the bacterium in antibiotic-free medium, the bacteriostatic activity of the compound arrests the growth of the nonadapted bacterial culture. We hypothesize that during the in vitro evolution experiment, the bacteria adapted by a yet unidentified mechanism, e.g., by increasing the expression of drug efflux pumps. This explanation remains to be proven, possibly by investigating gene and/or protein expression.
After obtaining this interesting isolate and comparing its sequence to that of the parental strain, we found an insertion in a gene with unknown function, gnaA. In the following work, we delineated the roles of the gnaA gene in the formation of surface polysaccharides and analyzed its role in antimicrobial susceptibility, membrane composition and stability, as well as the morphology and virulence of the bacterium A. baumannii. We also reanalyzed the multidrug resistance of MDR-ZJ06, where we could demonstrate that the wild-type strain is not only carbapenem resistant but also virulent. The gnaA gene has a major impact on both, as its disruption changed the virulence and membrane composition of the bacterium, in turn altering its antibiotic susceptibility.
MDR-ZJ06 is a representative multidrug-resistant isolate that belongs to global clone 2 and is a widely occurring strain in China (16, 17). The whole-genome sequence belonged to one of the earliest submissions to sequence databanks and is frequently used in comparisons in studies involving A. baumannii. However, the sequence contained some sequencing errors due to the technical limitations at that time. Therefore, we recently resequenced the strain using the PacBio platform and assembled and submitted the revised sequence (18), which was used in this study. During our laboratory evolution experiment, a mutation was detected in the gnaA gene, which is located in the K locus of A. baumannii. The gene codes for an enzyme that is responsible for the synthesis of simple UDP-linked sugars (12). While gnaA was found in the K locus in A. baumannii, in other Gram-negative bacteria, homologs of the gene are widely distributed within their genomes. Many studies have focused on the relationship between the K locus and capsule formation in A. baumannii; Russo et al. found that the genes wza and wzc, found in the K locus, are required for a capsule-positive phenotype and that the capsule was an important protectin for bacterial survival (11). The function of gnaA in other Gram-negative bacteria has been found to be associated with the biosynthetic pathway of O antigens. Wang et al. first identified that gnaA was responsible for UDP-d-ManpNAc3NAcA biosynthesis in Escherichia coli, while UDP-d-ManpNAc3NAcA is the activated form of UDP-d-ManpNAc3NAcA first identified in the E. coli O antigen (19). Miller and colleagues found that the homologue of gnaA in Pseudomonas aeruginosa PAO1, wbpA, is involved in the biosynthesis of di-N-acetylated mannosaminuronic acid-derived residues of the B band O antigen, and the substrate UDP-d-GlcNAc is crucial for bacterial polysaccharide biosynthesis (20). The O antigen plays an important role in the colonization of host tissue and resistance or susceptibility in bacteria (21–24). For example, galE, a gene encoding UDP-galactose 4-epimerase that catalyzes the conversion of UDP-galactose to UDP-glucose was confirmed to be involved in biofilm formation and morphology but also the susceptibility to multiple antibiotics and can be correlated with the length of the LPS O antigen in Porphyromonas gingivalis (25).
Due to the lack of an O antigen in A. baumannii, gnaA from this organism has not been conclusively investigated. However, studies on the LOS of other pathogens, including Haemophilus ducreyi, Campylobacter jejuni, and Moraxella bovis, have been instrumental in demonstrating the relevance of LOS in the pathogenesis of the diseases caused by these microbes (26–29). Bauer et al. found that gmhA is essential for the synthesis of LOS in H. ducreyi, and the gene affects virulence in a temperature-dependent way in a rabbit model for experimental chancroid (29). Another study on H. ducreyi found that waaQ and lgtF mutations would produce truncated LOS molecules, which in turn led to pyocin resistance (28). Keo et al. found that LOS of Campylobacter jejuni plays a very important role in protection against cationic antimicrobial peptides and proteins (26). In addition, Singh et al. reported that Moraxella bovis LOS truncation leads to reduced bacterial attachment, increased sensitivity to the bactericidal activity of bovine serum, and a reduction in endotoxin activity (27).
In this study, we described that gnaA affects the formation of surface polysaccharides and that mutations in gnaA cause differences in the degree of virulence of A. baumannii. We demonstrated that gnaA affects the composition of LOS and CPS, an observation that is consistent with the changes observed in the study of Geisinger and Isberg that correlated K-locus gene expression, including gnaA, with the composition of LOS (30), yielding similar patterns of LOS molecules in both studies. In addition, our study showed that membrane potential and permeability are also affected by mutations in the gnaA gene, which in turn is responsible for susceptibility to certain types of antimicrobial agents.
The major component of LOS is lipid A. Modification of the molecule or the deficiency in lipid A synthesis affects bacterial survival and the immune response of the host. Therefore, proteins involved in lipid A synthesis, such as LpxC, have been identified as targets to inhibit lipid A biosynthesis (6). Inhibitors are being developed as antimicrobial agents, and some showed promising antibacterial activity against Gram-negative species, including Pseudomonas aeruginosa, Escherichia coli, and Klebsiella pneumoniae, while exhibiting poor in vitro activity against A. baumannii (31). However, LpxC inhibitors have a positive influence on A. baumannii clearance in vivo, as they increase cell permeability, increasing antibiotic susceptibility (7, 9). Our study showed that gnaA has an important role in the synthesis of LOS in A. baumannii, with drastic consequences on the phenotype of a virulent multidrug-resistant strain. Using inhibitors that inactivate gnaA might be a valuable strategy to combat multidrug-resistant A. baumannii infections. Toward this goal, one of the first steps would be to identify the active sites of the enzyme and to develop a drug-screening assay.
This study had certain limitations, as we did not conduct the complementation experiments in situ. In our work, we could restore phenotypes by complementation experiments, with the exception of the virulence assay in the G. mellonella infection model. The expression of the complementation plasmid is difficult to monitor in G. mellonella, and thus an explanation might be that the plasmid could have been lost under the experimental conditions used. Nonetheless, we still observed a difference in the virulence of the wild-type MDR-ZJ06 strain compared to that of MDR-ZJ06M. To better understand the enzymatic reactions in which gnaA is involved, there is a need to study the entire biosynthesis pathway of LOS in A. baumannii in detail.
In this work, we recharacterized MDR-ZJ06 as a virulent and multidrug-resistant strain. The gene gnaA (BGV75_06155), which we identified in an evolution experiment, had a major impact on the synthesis of LOS and CPS, which also affected the physiology and morphology of the bacterial strain. The gnaA-deficient or deletion strains showed increased sensitivity to multiple drugs and exhibited reduced virulence in a G. mellonella model. Therefore, we think that this gene has the potential to be a valuable drug target for multidrug-resistant A. baumannii infections.
MATERIALS AND METHODS
Bacterial strains, plasmids, and MICs.
The strain MDR-ZJ06 was isolated in a previous study (16) while MDR-ZJ06M was obtained from a laboratory evolution experiment. All isolates used in this study (Table 2) were cultured in Mueller-Hinton agar plates or broth (Oxoid, Hampshire, UK) and Luria-Bertani (LB) broth (Sangon Biotech, Shanghai, China) at 37°C overnight. The MICs of tigecycline and colistin were determined using the broth microdilution method according to the guidelines recommended by the Clinical and Laboratory Standards Institute (CLSI) (32). Other antimicrobial MICs were determined using Etest strips (AB bioMérieux, France) for the following antibiotics: imipenem, meropenem, ceftazidime, cefepime, minocycline, amikacin, and gentamicin.
TABLE 2.
Bacterial strains and plasmids used in this study
| Strain/plasmid | Descriptionc | Source |
|---|---|---|
| Strain | ||
| MDR-ZJ06 | Multidrug-resistant A. baumannii, wild-type strain | 16 |
| MDR-ZJ06M | Selected isolate, gnaA::ISAba16 | This study |
| MDR-ZJ06M/pYMAb2 | MDR-ZJ06 gnaA::ISAba16/pYMAb2-Hygr, as a control | This study |
| MDR-ZJ06M/pYMAb2-gnaA | MDR-ZJ06 gnaA::ISAba16 constitutively expressing wild-type gnaA | This study |
| MDR-ZJ06KO | MDR-ZJ06 ΔgnaA | This study |
| MDR-ZJ06KO/pYMAb2 | MDR-ZJ06 ΔgnaA/pYMAb2-Hygr, as a control | This study |
| MDR-ZJ06KO/pYMAb2-part gnaA | MDR-ZJ06 ΔgnaA expressing residual gnaA from MDR-ZJ06M | This study |
| MDR-ZJ06KO/pYMAb2-gnaA | MDR-ZJ06 ΔgnaA expressing wild-type gnaA from MDR-ZJ06 | This study |
| AB5075 | A virulent clinical isolate | 42 |
| Plasmid | ||
| pACBSR-Hyg | Template for amplification of the hygromycin resistance cassette | 43 |
| pYMAb2 | E. coli-A. baumannii shuttle plasmid, Kmr | 44 |
| pYMAb2-Hygr | Same as pYMAb2 but with an inserted hygromycin resistance cassette | 45 |
| pYMAb2-Hyg-gnaAa | pYMAb2-Hyg derivative expressing gnaA | This study |
| pYMAb2-Hyg-gnaAb | pYMAb2-Hyg derivative expressing residual gnaA | This study |
| pMo130-Telr | Suicide plasmid, Kmr, Telr | 46 |
| pMo130-Hygr | Same as pMo130-Telr, but with the Telr marker replaced by Hygr | This study |
The wild-type gnaA was amplified from MDR-ZJ06 and then cloned into the plasmid pYMAb2-Hygr.
The residual gnaA was amplified from MDR-ZJ06M (MDR-ZJ06 gnaA::ISAba16) and then cloned into the plasmid pYMAb2-Hygr.
Km, kanamycin; Hyg, hygromycin; Tel, tellurite; r, resistance.
Laboratory evolution experiment.
A laboratory evolution experiment was performed as previously described (33). Briefly, a single colony of MDR-ZJ06 was cultured in 2 ml of LB broth overnight at 37°C. The culture was exposed to increasing concentrations of tigecycline, starting at 0.5× MIC and subsequently doubling the amount every 24 h, with concentrations of 1×, 2×, 4×, 8×, 16×, and 32× MIC. All of the overnight cultures were stored at −80°C. After 7 days of serial passage (5 μl of culture in 2 ml of fresh medium), the final concentration reached 32× MIC. This culture was designated “T7” (see Fig. S1 in the supplemental material). No growth was observed after another step, with a concentration of 64× MIC. Bacteria were streaked out from the frozen sample T7 on solid medium without antibiotic, and a single colony (selected at random) was grown in liquid medium, again without antibiotic. This clone, hereafter referred to as MDR-ZJ06M, was stored at −80°C.
Genomic DNA sequencing and analysis.
The genomic DNA of MDR-ZJ06M was extracted using a QIAamp DNA minikit (Qiagen Valencia, CA) following the protocol of the manufacturer. Gel electrophoresis and a NanoDrop 2000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) were used to analyze the quality of genomic DNA. The genome was sequenced on an Illumina HiSeq platform (Illumina, San Diego, CA, USA). In general, more than 300-fold coverage was obtained for the genome sequences. Comparative genome analyses were performed with Breseq and Mauve (34, 35). A putative mutation detected was confirmed by PCR and Sanger sequencing. The sequence type of the strain was analyzed using the PubMLST platform for A. baumannii (https://pubmlst.org/abaumannii/). Resistance genes were detected by ResFinder (https://cge.cbs.dtu.dk/services/ResFinder/).
Gene knockout and complementation experiment.
A knockout experiment was performed according to our previous study (36) with slight modifications. To increase the efficiency of screening for mutants, Telr was replaced by Hygr. Briefly, upstream and downstream sequences of gnaA were amplified from MDR-ZJ06. The fragments containing homologous regions were cloned into the pMo130-Hygr vector using a ClonExpress MultiS one-step cloning kit (Vazyme Biotech Co., Nanjing, China). Then, the resulting plasmid was introduced into MDR-ZJ06 and selected on Mueller-Hinton agar containing 100 mg/liter hygromycin. The deletion mutant was confirmed through PCR and sequenced.
A complementation experiment was performed to evaluate the contribution of the mutation in MDR-ZJ06M. Purified PCR products of the target gene were cloned into the pYMAb2-Hyg plasmid, introducing BamHI and SalІ restriction sites. The recombinant plasmids were then introduced by transformation into the mutant and deletion strains via electroporation and selected on Mueller-Hinton agar containing 100 mg/liter hygromycin. All sequences were confirmed by Sanger sequencing.
Scanning electron microscopy.
SEM was performed according to a procedure described elsewhere (37) with minor modifications. Briefly, strains were grown overnight in Mueller-Hinton broth, diluted 1:100, and incubated in fresh broth to mid-log phase. Cells were harvested by centrifugation at 4,000 rpm for 10 min, washed several times with phosphate-buffered saline (PBS) (pH 7.4), and centrifuged to discard the liquid. Next, cells were fixed with 2.5% glutaraldehyde in the same buffer overnight at 4°C, washed three times with 0.1 M PBS (pH 7.4), and then stained with 1.0% (wt/vol) osmium tetroxide. The samples were again washed three more times and dehydrated in a series of graded ethanol solutions (30% to 100%) before drying in a desiccator under vacuum. Imaging of the samples was performed using Nova NanoSEM 450 (Thermo Fisher, USA).
Membrane potential and membrane permeability assays.
Log-phase bacterial cultures were diluted to approximately 1 × 106 cells/ml in filtered PBS without washing. Membrane potential (MP) was measured as described previously (38). Membrane permeability was measured by 100 nM To-Pro-3 (Invitrogen, Thermo Fisher Scientific, USA), which stains nucleic acids in cells when the membrane is damaged. The following controls were used: an unstained control, a depolarized control, and dead cells, which were prepared by heating at 100°C for 10 min. All samples were incubated for 30 min at room temperature and assayed by flow cytometry (BD). The raw To-Pro-3 fluorescence data were corrected for size variation by calculating a quantity proportional to the logarithm of the ratio of the To-Pro-3 fluorescence to the green DiOC2(3) fluorescence, which is known to be proportional to size (39); the logarithm of green fluorescence was subtracted from the logarithm of To-Pro-3 fluorescence, and a constant was added to keep the values on scale.
Surface polysaccharide extraction and silver staining.
Surface polysaccharides were purified by hot aqueous phenol extraction according to Joanna B. Goldberg’s protocol (40). Ten microliters of extracted samples were separated on a 10% SDS-polyacrylamide gel and a 4% to 20% Tris-glycine gel. The gels were directly stained using a fast silver stain kit (Beyotime Biotechnology, China) according to the manufacturer’s instructions.
Microscopy.
Bacterial capsules were detected by the wet-film India ink method. Images were acquired on a Nikon Eclipse 50i microscope (×100 magnification).
Galleria mellonella infection model.
The G. mellonella survival assay was performed as previously described (41). Log-phase cell cultures were centrifuged and resuspended in PBS to 108 CFU/ml. Ten microliters of bacteria was injected into G. mellonella larvae. Larvae were incubated in dishes at 37°C, and viability was assessed by checking for movement every 12 h. If larvae did not exhibit any movement when prodded with a pipette tip, they were considered dead and typically turned dark brown or black. Survival was monitored for 3 days.
Statistical analysis.
Continuous variables with normally distributed data were compared with Student’s t test, while Wilcoxon rank-sum tests were used for nonnormally distributed data. Kaplan-Meier survival curves were carried out using GraphPad Prism 5, and statistical significance was calculated with a log rank test.
Accession number(s).
The genome of MDR-ZJ06M has been deposited at DDBJ/EMBL/GenBank under the accession number QQQD00000000.
Supplementary Material
ACKNOWLEDGMENTS
We appreciate Howard Shuman (University of Chicago) for providing the strain AB5075, Pep Charusanti (University of California) for offering the plasmid pACBSR-Hyg, and Kim Lee Chua (National University of Singapore) for giving us the plasmid pMo130-Telr. Additionally, we thank Te-Li Chen (National Yang-Ming University) for providing the plasmid pYMAb2. We thank Dandan Song (Center of Cryo Electron Microscopy, Zhejiang University) for helping to perform SEM. Last, we thank Belinda Loh (ZJE) for critically reading the manuscript.
This work was supported by grants from the National Natural Science Foundation of China (grant no. 31670135, 31770142, and 81861138054).
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00694-19.
REFERENCES
- 1.Peleg AY, Seifert H, Paterson DL. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev 21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antunes LCS, Visca P, Towner KJ. 2014. Acinetobacter baumannii: evolution of a global pathogen. Pathogens Dis 71:292–301. doi: 10.1111/2049-632X.12125. [DOI] [PubMed] [Google Scholar]
- 3.Howard A, O’Donoghue M, Feeney A, Sleator RD. 2012. Acinetobacter baumannii: an emerging opportunistic pathogen. Virulence 3:243–250. doi: 10.4161/viru.19700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Göttig S, Gruber TM, Higgins PG, Wachsmuth M, Seifert H, Kempf V. 2014. Detection of pan drug-resistant Acinetobacter baumannii in Germany. J Antimicrob Chemother 69:2578–2579. doi: 10.1093/jac/dku170. [DOI] [PubMed] [Google Scholar]
- 5.Leite GC, Oliveira MS, Perdigão-Neto LV, Rocha CKD, Guimarães T, Rizek C, Levin AS, Costa SF. 2016. Antimicrobial combinations against pan- resistant Acinetobacter baumannii isolates with different resistance mechanisms. PLoS One 11:e0151270. doi: 10.1371/journal.pone.0151270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Powers MJ, Trent MS. 2018. Expanding the paradigm for the outer membrane: Acinetobacter baumannii in the absence of endotoxin. Mol Microbiol 107:47–56. doi: 10.1111/mmi.13872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin L, Tan B, Pantapalangkoor P, Ho T, Baquir B, Tomaras A, Montgomery JI, Reilly U, Barbacci EG, Hujer K, Bonomo RA, Fernandez L, Hancock REW, Adams MD, French SW, Buslon VS, Spellberg B. 2012. Inhibition of LpxC protects mice from resistant Acinetobacter baumannii by modulating inflammation and enhancing phagocytosis. mBio 3:e00312-12. doi: 10.1128/mBio.00312-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomaras AP, McPherson CJ, Kuhn M, Carifa A, Mullins L, George D, Desbonnet C, Eidem TM, Montgomery JI, Brown MF, Reilly U, Miller AA, O'Donnell JP. 2014. LpxC inhibitors as new antibacterial agents and tools for studying regulation of lipid A biosynthesis in Gram-Negative pathogens. mBio 5:e01551-14. doi: 10.1128/mBio.01551-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.García-Quintanilla M, Caro-Vega JM, Pulido MR, Moreno-Martínez P, Pachón J, McConnell MJ. 2016. Inhibition of LpxC increases antibiotic susceptibility in Acinetobacter baumannii. Antimicrob Agents Chemother 60:5076–5079. doi: 10.1128/AAC.00407-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lees-Miller RG, Iwashkiw JA, Scott NE, Seper A, Vinogradov E, Schild S, Feldman MF. 2013. A common pathway for O-linked protein-glycosylation and synthesis of capsule in Acinetobacter baumannii. Mol Microbiol 89:816–830. doi: 10.1111/mmi.12300. [DOI] [PubMed] [Google Scholar]
- 11.Russo TA, Luke NR, Beanan JM, Olson R, Sauberan SL, MacDonald U, Schultz LW, Umland TC, Campagnari AA. 2010. The K1 capsular polysaccharide of Acinetobacter baumannii strain 307-0294 is a major virulence factor. Infect Immun 78:3993–4000. doi: 10.1128/IAI.00366-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kenyon JJ, Hall RM. 2013. Variation in the complex carbohydrate biosynthesis loci of Acinetobacter baumannii genomes. PLoS One 8:e62160. doi: 10.1371/journal.pone.0062160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han CG, Shiga Y, Tobe T, Sasakawa C, Ohtsubo E. 2001. Structural and functional characterization of IS679 and IS66-family elements. J Bacteriol 183:4296–4304. doi: 10.1128/JB.183.14.4296-4304.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shapiro HM. 2008. Flow cytometry of bacterial membrane potential and permeability. Methods Mol Med 142:175–186. doi: 10.1007/978-1-59745-246-5_14. [DOI] [PubMed] [Google Scholar]
- 15.Silhavy T, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb Perspect Biol 2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou H, Zhang T, Yu D, Pi B, Yang Q, Zhou J, Hu S, Yu Y. 2011. Genomic analysis of the multidrug-resistant Acinetobacter baumannii strain MDR-ZJ06 widely spread in China. Antimicrob Agents Chemother 55:4506–4512. doi: 10.1128/AAC.01134-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou H, Yang Q, Yu YS, Wei ZQ, Li LJ. 2007. Clonal spread of imipenem-resistant Acinetobacter baumannii among different cities of China. J Clin Microbiol 45:4054–4057. doi: 10.1128/JCM.00343-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hua X, Xu Q, Zhou Z, Ji S, Yu Y. 2019. Relocation of Tn2009 and characterization of an ABGRI3-2 from re-sequenced genome sequence of Acinetobacter baumannii MDR-ZJ06. J Antimicrob Chemother 74:1153–1155. doi: 10.1093/jac/dky521. [DOI] [PubMed] [Google Scholar]
- 19.Wang Q, Perepelov AV, Beutin L, Senchenkova SN, Xu Y, Shashkov AS, Ding P, Knirel YA, Feng L. 2012. Structural and genetic characterization of the Escherichia coli O180 O antigen and identification of a UDP-GlcNAc 6-dehydrogenase. Glycobiology 22:1321–1331. doi: 10.1093/glycob/cws098. [DOI] [PubMed] [Google Scholar]
- 20.Miller WL, Wenzel CQ, Daniels C, Larocque S, Brisson JR, Lam JS. 2004. Biochemical characterization of WbpA, a UDP-N-acetyl-d-glucosamine 6-dehydrogenase involved in O-antigen biosynthesis in Pseudomonas aeruginosa PAO1. J Biol Chem 279:37551–37558. doi: 10.1074/jbc.M404749200. [DOI] [PubMed] [Google Scholar]
- 21.Camprubí S, Merino S, Guillot JF, Tomás JM. 1993. The role of the O-antigen lipopolysaccharide on the colonization in vivo of the germfree chicken gut by Klebsiella pneumoniae. Microb Pathog 14:433–440. doi: 10.1006/mpat.1993.1042. [DOI] [PubMed] [Google Scholar]
- 22.Harvill ET, Preston A, Cotter PA, Allen AG, Maskell DJ, Miller JF. 2000. Multiple roles for Bordetella lipopolysaccharide molecules during respiratory tract infection. Infect Immun 68:6720–6728. doi: 10.1128/iai.68.12.6720-6728.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nesper J, Lauriano CM, Klose KE, Kapfhammer D, Kraiß A, Reidl J. 2001. Characterization of Vibrio cholerae O1 El Tor galU and galE mutants: influence on lipopolysaccharide structure, colonization, and biofilm formation. Infect Immun 69:435–445. doi: 10.1128/IAI.69.1.435-445.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weber BS, Harding CM, Feldman MF. 2016. Pathogenic Acinetobacter: from the cell surface to infinity and beyond. J Bacteriol 198:880–887. doi: 10.1128/JB.00906-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakao R, Senpuku H, Watanabe H. 2006. Porphyromonas gingivalis galE is involved in lipopolysaccharide O-antigen synthesis and biofilm formation. Infect Immun 74:6145–6153. doi: 10.1128/IAI.00261-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keo T, Collins J, Kunwar P, Blaser MJ, Iovine NM. 2011. Campylobacter capsule and lipooligosaccharide confer resistance to serum and cationic antimicrobials. Virulence 2:30–40. doi: 10.4161/viru.2.1.14752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh S, Grice ID, Peak IR, Frost T, Yue G, Wilson JC. 2018. The role of lipooligosaccharide in the biological activity of Moraxella bovis strains Epp63, Mb25 and L183/2, and isolation of capsular polysaccharide from L183/2. Carbohydr Res 467:1–7. doi: 10.1016/j.carres.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 28.Filiatrault MJ, Munson J, Campagnari AA. 2001. Genetic analysis of a pyocin-resistant lipooligosaccharide (LOS) mutant of Haemophilus ducreyi: restoration of full-length LOS restores pyocin sensitivity. J Bacteriol 183:5756–5761. doi: 10.1128/JB.183.19.5756-5761.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bauer BA, Stevens MK, Hansen EJ. 1998. Involvement of the Haemophilus ducreyi gmhA gene product in lipooligosaccharide expression and virulence. Infect Immun 66:4290–4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geisinger E, Isberg RR. 2015. Antibiotic modulation of capsular exopolysaccharide and virulence in Acinetobacter baumannii. PLoS Pathog 11:e1004691. doi: 10.1371/journal.ppat.1004691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Titecat M, Liang X, Lee CJ, Charlet A, Hocquet D, Lambert T, Pagès JM, Courcol R, Sebbane F, Toone EJ, Zhou P, Lemaitre N. 2016. High susceptibility of MDR and XDR Gram-negative pathogens to biphenyl-diacetylene-based difluoromethyl-allo-threonyl-hydroxamate LpxC inhibitors. J Antimicrob Chemother 71:2874–2882. doi: 10.1093/jac/dkw210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clinical and Laboratory Standards Institute. 2019. Performance standards for antimicrobial susceptibility testing; 29th informational supplement. CLSI M100. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 33.Chen Q, Li X, Zhou H, Jiang Y, Chen Y, Hua X, Yu Y. 2014. Decreased susceptibility to tigecycline in Acinetobacter baumannii mediated by a mutation in trm encoding SAM-dependent methyltransferase. J Antimicrob Chemother 69:72–76. doi: 10.1093/jac/dkt319. [DOI] [PubMed] [Google Scholar]
- 34.Deatherage DE, Barrick JE. 2014. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol 1151:165–188. doi: 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X, Quan J, Yang Y, Ji J, Liu L, Fu Y, Hua X, Chen Y, Pi B, Jiang Y, Yu Y. 2016. Abrp, a new gene, confers reduced susceptibility to tetracycline, glycylcine, chloramphenicol and fosfomycin classes in Acinetobacter baumannii. Eur J Clin Microbiol Infect Dis 35:1371–1375. doi: 10.1007/s10096-016-2674-0. [DOI] [PubMed] [Google Scholar]
- 37.Singh M, Mukhopadhyay K. 2011. C-terminal amino acids of alpha-melanocyte-stimulating hormone are requisite for its antibacterial activity against Staphylococcus aureus. Antimicrob Agents Chemother 55:1920–1929. doi: 10.1128/AAC.00957-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X, Liu L, Ji J, Chen Q, Hua X, Jiang Y, Feng Y, Yu Y. 2015. Tigecycline resistance in Acinetobacter baumannii mediated by frameshift mutation in plsC, encoding 1-acyl-sn-glycerol-3-phosphate acyltransferase. Eur J Clin Microbiol Infect Dis 34:625–631. doi: 10.1007/s10096-014-2272-y. [DOI] [PubMed] [Google Scholar]
- 39.Novo DJ, Perlmutter NG, Hunt RH, Shapiro HM. 2000. Multiparameter flow cytometric analysis of antibiotic effects on membrane potential, membrane permeability, and bacterial counts of Staphylococcus aureus and Micrococcus luteus. Antimicrob Agents Chemother 44:827–834. doi: 10.1128/aac.44.4.827-834.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis MR Jr, Goldberg JB. 2012. Purification and visualization of lipopolysaccharide from Gram-negative bacteria by hot aqueous-phenol extraction. J Vis Exp 63:3916. doi: 10.3791/3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richmond GE, Evans LP, Anderson MJ, Wand ME, Bonney LC, Ivens A, Chua KL, Webber MA, Sutton JM, Peterson ML, Piddock LV. 2016. The Acinetobacter baumannii two-component system AdeRS regulates genes required for multidrug efflux, biofilm formation, and virulence in a strain-specific manner. mBio 7:e00430-16. doi: 10.1128/mBio.00430-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacobs AC, Thompson MG, Black CC, Kessler JL, Clark LP, McQueary CN, Gancz HY, Corey BW, Moon JK, Si Y, Owen MT, Hallock JD, Kwak YI, Summers A, Li CZ, Rasko DA, Penwell WF, Honnold CL, Wise MC, Waterman PE, Lesho EP, Stewart RL, Actis LA, Palys TJ, Craft DW, Zurawski DV. 2014. AB5075, a highly virulent isolate of Acinetobacter baumannii, as a model strain for the evaluation of pathogenesis and antimicrobial treatments. mBio 5:e01076-14. doi: 10.1128/mBio.01076-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang TW, Lam I, Chang HY, Tsai SF, Palsson BO, Charusanti P. 2014. Capsule deletion via a λ-red knockout system perturbs biofilm formation and fimbriae expression in Klebsiella pneumoniae MGH 78578. BMC Res Notes 7:13–18. doi: 10.1186/1756-0500-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuo SC, Yang SP, Lee YT, Chuang HC, Chen CP, Chang CL, Chen TL, Lu PL, Hsueh PR, Fung CP. 2013. Dissemination of imipenem-resistant Acinetobacter baumannii with new plasmid-borne blaOXA-72 in Taiwan. BMC Infect Dis 13:1. doi: 10.1186/1471-2334-13-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Y, Xu Q, Li T, Fu Y, Shi Y, Lan P, Zhao D, Chen Q, Zhou Z, Jiang Y, Peleg AY, Yu Y. 2018. OXA-23 is a prevalent mechanism contributing to sulbactam resistance in diverse Acinetobacter baumannii clinical strains. Antimicrob Agents Chemother 63:e01676-18. doi: 10.1128/AAC.01676-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amin IM, Richmond GE, Sen P, Koh TH, Piddock LV, Chua KL. 2013. A method for generating marker-less gene deletions in multidrug-resistant Acinetobacter baumannii. BMC Microbiol 13:158. doi: 10.1186/1471-2180-13-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.