Carbapenem-resistant Enterobacteriaceae (CRE) can be mechanistically classified into carbapenemase-producing Enterobacteriaceae (CPE) and non-carbapenemase-producing carbapenem nonsusceptible Enterobacteriaceae (NCPCRE). We sought to investigate the effect of antecedent carbapenem exposure as a risk factor for NCPCRE versus CPE. Among all patients with CRE colonization and infection, we conducted a case-control study comparing patients with NCPCRE (cases) and patients with CPE (controls).
KEYWORDS: carbapenem resistance, carbapenem-resistant Enterobacteriaceae, carbapenemase-producing Enterobacteriaceae, risk factors
ABSTRACT
Carbapenem-resistant Enterobacteriaceae (CRE) can be mechanistically classified into carbapenemase-producing Enterobacteriaceae (CPE) and non-carbapenemase-producing carbapenem nonsusceptible Enterobacteriaceae (NCPCRE). We sought to investigate the effect of antecedent carbapenem exposure as a risk factor for NCPCRE versus CPE. Among all patients with CRE colonization and infection, we conducted a case-control study comparing patients with NCPCRE (cases) and patients with CPE (controls). The presence of carbapenemases was investigated with phenotypic tests followed by PCR for predominant carbapenemase genes. We included 843 unique patients with first-episode CRE, including 387 (45.9%) NCPCRE and 456 (54.1%) CPE. The resistance genes detected in CPEs were blaNDM (42.8%), blaKPC (38.4%), and blaOXA-48-like (12.1%). After adjusting for confounders and clustering at the institutional level, the odds of prior 30-day carbapenem exposure was three times higher among NCPCRE than CPE patients (adjusted odds ratio [aOR], 3.48; 95% confidence interval [CI], 2.39 to 5.09; P < 0.001). The odds of prior carbapenem exposure and NCPCRE detection persisted in stratified analyses by Enterobacteriaceae species (Klebsiella pneumoniae and Escherichia coli) and carbapenemase gene (blaNDM and blaKPC). CPE was associated with male gender (aOR, 1.45; 95% CI, 1.07 to 1.97; P = 0.02), intensive care unit stay (aOR, 1.84; 95% CI, 1.24 to 2.74; P = 0.003), and hospitalization in the preceding 1 year (aOR, 1.42; 95% CI, 1.01 to 2.02; P = 0.05). In a large nationwide study, antecedent carbapenem exposure was a significant risk factor for NCPCRE versus CPE, suggesting a differential effect of antibiotic selection pressure.
INTRODUCTION
The emergence and rapid spread of carbapenem-resistant Enterobacteriaceae (CRE) with associated limited antimicrobial options for treatment and poor clinical outcomes (1) is a major threat to safe health care delivery. Mechanisms of carbapenem nonsusceptibility can be divided broadly into carbapenemase production (carbapenemase-producing Enterobacteriaceae [CPE]) and a combination of β-lactamase (extended-spectrum β-lactamase [ESBL] and AmpC) production and dysregulation of porin channels (non-carbapenemase-producing carbapenem nonsusceptible Enterobacteriaceae [NCPCRE]) (2). Carbapenemase genes are diverse and can be located chromosomally or within plasmids, with the latter conferring ease of transmissibility within and between bacterial species.
Orsi and colleagues (3) demonstrated that compared to Klebsiella pneumoniae carbapenemase (KPC)-producing CRE, NCPCRE were associated with prior antibiotic exposure, demonstrating that patient-level risk factors may differ according to mechanisms of resistance. On the other hand, existing evidence suggests that the fitness cost renders NCPCRE less transmissible than CPE (4). The successful Israeli national intervention for CRE control, which instituted more stringent infection prevention and control (IPC) measures (without additional antibiotic stewardship interventions) for CPE than for NCPCRE (5), suggests that CPE may be more likely to emerge through horizontal bacterial or gene transmission than NCPCRE. A more recent study by Simner and colleagues (6) described an association between overnight stay in foreign health care facilities and CPE detection in the United States. However, the authors did not explore the impact of prior carbapenem exposure.
Prior antibiotic exposure, specifically, carbapenem exposure, has long been associated with the emergence of CRE. However, considering the diverse mechanisms of resistance, it is possible that the impact of antibiotic exposure on the occurrence of CRE may vary depending on the resistance mechanism. Identifying potentially different impacts of carbapenem exposure, especially in settings where the bacteria are endemic, is essential for the optimal implementation of infection control and antimicrobial stewardship programs.
To this effect, we aimed to study the role of antecedent carbapenem exposure as a risk factor for NCPCRE and CPE. We hypothesized that antecedent carbapenem exposure was associated more with NCPCRE than with CPE.
RESULTS
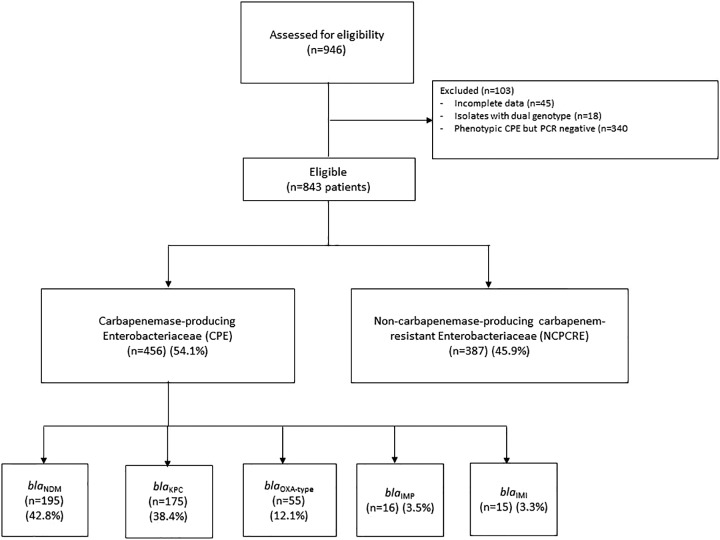
Between September 2010 and April 2015, 843 patients with CRE were recruited. Of these, 456 CPE and 387 NCPCRE patients fulfilled the inclusion criteria (Fig. 1). More than 80% of CPE patients had blaNDM and blaKPC carbapenemase genes. CRE was identified through active surveillance cultures (stool samples, rectal swabs, or perianal swabs) (52.8%), followed by urine (18.4%) and blood (13.8%) (Table 1). K. pneumoniae was the most common Enterobacteriaceae (50.9%), followed by E. coli (25.2%), and Enterobacter sp. (17.6%). Of the CRE isolates, 97.2% were not susceptible to meropenem and/or imipenem and 2.9% were not susceptible to ertapenem only.
FIG 1.
Selection of study subjects.
TABLE 1.
Enterobacteriaceae species, sources, and susceptibility patternsa
| Enterobacteriaceae | No. (%) of patients with: |
||
|---|---|---|---|
| CRE (n = 843) | NCPCRE (n = 387) | CPE (n = 456) | |
| Specimen | |||
| Stool or rectal swabs | 445 (52.8) | 186 (48.1) | 259 (56.8) |
| Blood | 116 (13.8) | 42 (10.9) | 74 (16.2) |
| Urine | 155 (18.4) | 86 (22.2) | 69 (15.1) |
| Wound | 40 (4.7) | 20 (5.2) | 20 (4.4) |
| Intra-abdominal and hepatobiliary specimens | 39 (4.6) | 31 (8.0) | 8 (1.8) |
| Others | 48 (5.7) | 22 (5.7) | 26 (5.7) |
| Bacterial species | |||
| Klebsiella pneumoniae | 429 (50.9) | 241 (62.3) | 188 (41.2) |
| Escherichia coli | 212 (25.2) | 63 (16.3) | 149 (32.7) |
| Enterobacter sp. | 148 (17.6) | 59 (15.3) | 89 (19.5) |
| Citrobacter sp. | 22 (2.6) | 1 (0.3) | 21 (4.6) |
| Klebsiella sp. | 7 (0.8) | 5 (1.3) | 2 (0.4) |
| Proteus sp. | 14 (1.7) | 12 (3.1) | 2 (0.4) |
| Othersb | 11 (1.3) | 6 (1.6) | 5 (1.1) |
| Susceptibility | |||
| Not susceptible to meropenem and/or imipenem | 819 (97.2) | 378 (97.7) | 441 (96.7) |
| Not susceptible to ertapenem only | 24 (2.9) | 9 (2.3) | 15 (3.3) |
Abbreviations: CRE, carbapenem-resistant Enterobacteriaceae; NCPCRE, non-carbapenemase-producing carbapenem-resistant Enterobacteriaceae; CPE, carbapenemase-producing Enterobacteriaceae.
Others include Morganella morganii and Serratia marcescens.
Impact of carbapenem exposure on NCPCRE and CPE.
As summarized in Table 2, on univariate analysis, NCPCRE had higher frequencies of carbapenem exposure than CPE (odds ratio [OR], 2.97; 95% confidence interval [CI], 2.22 to 3.97; P < 0.001). On multivariate analysis, NCPCRE had three times higher odds of carbapenem exposure (adjusted OR [aOR], 3.48; 95% CI, 2.39 to 5.09; P < 0.001) (Table 2). This association strengthened with increasing duration of carbapenem exposure. On multivariate analysis, patients receiving more than 3 days of carbapenems had the highest odds of NCPCRE (aOR, 3.67; 95% CI, 2.44 to 5.54; P < 0.001), followed by patients receiving 1 to 3 days of carbapenems (OR, 2.59; 95% CI, 1.44 to 4.66; P = 0.002) (see Table S1 in the supplemental material).
TABLE 2.
Association between antecedent carbapenem exposure and types of carbapenem-resistant Enterobacteriaceaea
| Variable | Patients with: |
Values by analysis |
||||
|---|---|---|---|---|---|---|
|
NCPCREb (n = 387) |
CPEbc (n = 456) |
Univariate |
Multivariated
|
|||
| OR (95% CI) | P value | aOR (95% CI) | P value | |||
| Exposure of interest | ||||||
| Carbapenem exposed | 197 (50.9) | 118 (25.9) | 2.97 (2.22–3.97) | <0.001 | 3.48 (2.39–5.09) | <0.001 |
| Demographics | ||||||
| Gender, male | 200 (51.7) | 273 (59.9) | 0.72 (0.55–0.94) | 0.02 | 0.69 (0.51–0.94)e | 0.02 |
| Age, median years (IQR) | 68 (57–80) | 67 (58–77) | 0.55 | |||
| Medical history (1 year before culture) | ||||||
| Charlson score, median (IQR) | 5 (3–8) | 6 (3–8) | 0.05 | 0.97 (0.92–1.01) | 0.15 | |
| Hospitalization | 255 (65.9) | 327 (71.7) | 0.76 (0.57–1.02) | 0.07 | 0.70 (0.50–0.99)e | 0.05 |
| Upper gastrointestinal scopes | 80 (20.7) | 108 (23.7) | 0.84 (0.61–1.16) | 0.30 | ||
| Lower gastrointestinal scopes | 47 (12.1) | 53 (11.6) | 1.05 (0.69–1.60) | 0.82 | ||
| Surgery | 241 (62.3) | 305 (66.9) | 0.82 (0.62–1.08) | 0.16 | ||
| Multidrug-resistant organismsf | 152 (39.3) | 167 (36.6) | 1.12 (0.85–1.48) | 0.43 | ||
| Current hospitalization | ||||||
| Time at riskg , median days (IQR) | 12 (2–31) | 8 (1–23.5) | <0.001 | 1.00 (0.99–1.00) | 0.34 | |
| ICU at the time of culture | 85 (22.0) | 124 (27.2) | 0.75 (0.55–1.03) | 0.08 | 0.54 (0.37–0.81)e | 0.003 |
| Feeding tubeh | 144 (37.2) | 157 (34.4) | 1.13 (0.85–1.50) | 0.40 | ||
| Central venous lineh | 164 (42.4) | 160 (35.1) | 1.36 (1.03–1.80) | 0.03 | 0.99 (0.69–1.43) | 0.97 |
| Antibiotic exposure during 30 days before culture (proportions) | ||||||
| Third- and fourth-generation cephalosporins | 122 (31.5) | 125 (27.4) | 1.22 (0.91–1.64) | 0.19 | 1.00 (0.67–1.49) | >0.99 |
| Extended-spectrum penicillins | 228 (58.9) | 258 (56.6) | 1.10 (0.84–1.45) | 0.49 | 1.09 (0.80–1.48) | 0.60 |
| Fluoroquinolones | 100 (25.8) | 87 (19.1) | 1.48 (1.07–2.05) | 0.02 | 1.15 (0.80–1.68) | 0.45 |
| Aminoglycosides | 66 (17.1) | 52 (11.4) | 1.60 (1.08–2.36) | 0.02 | 1.02 (0.64–1.63) | 0.93 |
| Tigecycline | 9 (2.3) | 9 (1.9) | 1.18 (0.46–3.00) | 0.73 | ||
| Trimethoprim-sulfamethoxazole | 37 (9.6) | 17 (3.7) | 2.73 (1.51–4.93) | 0.001 | 2.44 (1.27–4.69) | 0.01 |
| Metronidazole | 94 (24.3) | 89 (19.5) | 1.32 (0.95–1.84) | 0.10 | 1.20 (0.78–1.84) | 0.42 |
| Specimen type | ||||||
| Clinical cultures (versus surveillance cultures) | 224 (57.9) | 201 (44.1) | 1.74 (1.33–2.29) | <0.001 | 1.03 (0.74–1.43) | 0.87 |
Abbreviations: NCPCRE, non-carbapenemase-producing carbapenem-resistant Enterobacteriaceae; CPE, carbapenemase-producing Enterobacteriaceae; ICU, intensive care unit; IQR, interquartile range; OR, odd ratio; aOR, adjusted odds ratio.
Unless otherwise indicated, values are n (%).
CPE is the reference comparison group (control group) unless stated otherwise.
Random effect logistic regression model allowing for clustering within hospitals and adjusted for month of CRE sample collection. Statistically significant values are shown in boldface.
For ease of interpretation of risk factors for CPE, the logistic regression model was repeated with NCPCRE as the reference group and the results were as follows: male gender (aOR,1.45; 95% CI, 1.07 to 1.97; P = 0.02); hospitalization during preceding 1 year (aOR, 1.42; 95% CI, 1.01 to 2.02; P = 0.05), and being in the ICU at the time of culture (aOR, 1.84; 95% CI, 1.24 to 2.74; P = 0.003).
Multidrug-resistant organisms includes methicillin-resistant Staphylococcus aureus (MRSA), carbapenem-resistant Acinetobacter baumannii (CRAB), vancomycin-resistant enterococci (VRE), and carbapenem-resistant Pseudomonas aeruginosa (CRPA).
Days from admission to collection of culture.
During time at risk.
As summarized in Table 3, on stratified analysis by Enterobacteriaceae species, the odds of carbapenem exposure was higher among NCPCR K. pneumoniae (aOR, 5.34; 95% CI, 2.96 to 9.64; P < 0.001) (see Table S2 in the supplemental material) and NCPCRE E. coli (aOR, 5.58; 95% CI, 2.37 to 13.15; P < 0.001) (see Table S3 in the supplemental material) than carbapenemase-producing (CP) K. pneumoniae and CP E. coli, respectively. On stratified analysis by genotype, the odds of carbapenem exposure was higher among KPC-producing Enterobacteriaceae (aOR, 7.04; 95% CI, 3.55 to 13.99; P < 0.001) (see Table S4 in the supplemental material) and NDM-producing Enterobacteriaceae (aOR, 3.74; 95% CI, 2.31 to 6.05; P < 0.001) (see Table S5 in the supplemental material). Carbapenem exposure emerged an independent risk factor for NCPCRE in stratified analysis by specimen type (see Table S6 and S7 in the supplemental material).
TABLE 3.
Association between antecedent carbapenem exposure and the occurrence of carbapenem-resistant Enterobacteriaceae stratified by species and carbapenemase typesa
| Stratified analyses by comparison | Multivariate analysisb aOR (95% CI) |
|---|---|
| NCPCR K. pneumoniae vs CP K. pneumoniaec | 5.34 (2.96–9.64) |
| NCPCR E. coli vs CP E. colid | 5.58 (2.37–13.15) |
| NCPCRE vs KPC-producing Enterobacteriaceaee | 7.04 (3.55–13.99) |
| NCPCRE vs NDM-producing Enterobacteriaceaef | 3.74 (2.31–6.05) |
Abbreviations: NCPCR, non-carbapenemase-producing carbapenem-resistant; CP, carbapenemase-producing; NCPCRE, non-carbapenemase-producing carbapenem-resistant Enterobacteriaceae; KPC, Klebsiella pneumoniae carbapenemase; NDM, New Delhi metallo-beta-lactamase.
Random effect logistic regression model allowing for clustering within hospitals and adjusted for month of CRE sample collection. Note: carbapenemase-producing organisms (CP K. pneumoniae, CP E. coli, KPE-producing Enterobacteriaceae, NDM-producing Enterobacteriaceae) are the base group for odds ratio calculations. All comparisons have P values of <0.001.
Details of univariate and multivariate analyses are available in Table S2.
Details of univariate and multivariate analyses are available in Table S3.
Details of univariate and multivariate analyses are available in Table S4.
Details of univariate and multivariate analyses are available in Table S5.
Other risk factors for NCPCRE and CPE.
In addition to carbapenems, the odds of antecedent trimethoprim-sulfamethoxazole exposure was 2.5 times higher among NCPCRE than CPE (aOR, 2.44; 95% CI, 1.27 to 4.69; P = 0.01) (Table 2). Additionally, independent risk factors for CPE were male gender (aOR, 1.45; 95% CI, 1.07 to 1.97; P = 0.02), hospitalization during preceding 1 year (aOR, 1.42; 95% CI, 1.01 to 2.02; P = 0.05), and being in the intensive care unit (ICU) at the time of culture (aOR, 1.84; 95% CI, 1.24 to 2.74; P = 0.003).
DISCUSSION
In the current analysis involving 843 subjects over 5 years, first-episode NCPCRE detection was associated with carbapenem exposure compared with first-episode CPE detection, an association strengthened by the dose-response relationship with increasing duration of carbapenem exposure. Additionally, carbapenem exposure remained a risk factor for first-episode NCPCRE detection in analyses stratified by bacterial species (E. coli and K. pneumoniae), CP genotype (blaNDM and blaKPC), and specimen type (surveillance cultures and clinical cultures). Conversely, residence in the ICU, hospitalization in the preceding 12 months, and male gender increased the likelihood of first episode CPE detection.
Consistent with our study findings, available epidemiologic studies suggest potentially differing mechanisms of acquisition between NCPCRE with CPE, with NCPCRE possibly arising from de novo mutations or genetic reassortment of carbapenem-sensitive Enterobacteriaceae under antimicrobial selection pressure and CPE being acquired via clonal bacterial spread or horizontal gene transfer (mainly plasmid-mediated), as carbapenemase genes are not endogenously generated in vivo in patients (7–9). Since the occurrence and propagation of resistance mechanisms could be affected by bacterial species and prevailing dominant genotypes, we conducted multiple stratified analyses that showed an association between carbapenem exposure and NCPCRE regardless of the genomic resistance mechanism and Enterobacteriaceae species, as reported before by Cheng et al. (10).
Currently, IPC measures (e.g., hand hygiene, environmental hygiene, and early detection and isolation of CRE carriers) and antibiotic stewardship (ASP) are considered the main pillars of CRE control strategies (11–15), regardless of the prevailing resistance mechanisms. Our findings suggest that antimicrobial stewardship may play a more significant role in preventing NCPCRE than CPE. Although important for the allocation of scarce resources, the impact of CRE resistance mechanisms on the efficacy of different infection prevention and antimicrobial stewardship interventions remains largely undetermined. In the often-cited successful experience of CRE control in Israel, CPE carriers were cohorted with dedicated staff, while NCPCRE carriers were placed under contact precautions without cohorting (5).
In our current analysis, male gender was a risk factor for CPE carriage. Prior studies have demonstrated an association between male gender and methicillin-resistant Staphylococcus aureus (MRSA) (16), as well as health care-associated infections (17). As a first, we found that male gender is also a risk factor for CPE, specifically, NDM-producing Enterobacteriaceae. Community studies of hand hygiene in public restrooms (18) and motorway station restrooms (19) suggest that women show better compliance to hand hygiene and soap use than men. Further studies are needed among hospitalized patients to understand the difference in the occurrence CRE among male and female patients.
Our study has several limitations. First, the lack of a non-CRE control group limits the generalizability of the findings of this study. The association found between NCPCRE and carbapenem exposure is conditional upon the control group being a CPE. Thus, we were unable to investigate the possibility of both NCPCRE and CPE having more carbapenem exposure than a non-CRE or noninfected control groups. Second, we were unable to calculate and include colonization pressure, which is a known risk factor for CRE acquisition (20). Third, molecular typing (e.g., whole-genome sequencing) would have given information regarding the number and sizes of clusters (if any) of CPE and NCPCRE, and this would have strengthened (or weakened) the argument that CPE is acquired exogenously by horizontal transmission, while NCPCRE is acquired endogenously and driven mainly by antibiotic pressure. Fourth, risk factor analyses for antimicrobial resistance at the patient-level and population-level are prone to selection bias and ecological bias, respectively (21); hence, our findings should be interpreted with full consideration of these biases.
To conclude, our study adds credence to the need to identify the heterogeneous resistance mechanisms of CRE to effectively balance antimicrobial stewardship versus infection prevention and control measures in areas with prevalent CPE or NCPCRE. Further studies are needed to explore affordable options for laboratory diagnosis of CRE resistance mechanisms and also the biological and genomic differences between CPE and NCPCRE.
MATERIALS AND METHODS
Study design, setting, and participants.
We conducted a case-control study by using data from the Carbapenemase-Producing Enterobacteriaceae in Singapore (CaPES) network between January 2010 and May 2015. The CaPES study recruited all hospitalized adult patients with colonization and/or infection with CRE. All seven government-funded multidisciplinary hospitals in Singapore, which provide >80% of inpatient medical care in Singapore, participated in this network. Interim results and the methodology of the prospective CaPES study have previously been published and showed that all major genotypes of CPE were endemic in Singapore (22).
Definitions and data collection.
Cases were patients with NCPCRE isolates, which were not susceptible to meropenem, imipenem, and/or ertapenem; were phenotypically noncarbapenemase producers; and tested negative for carbapenemase genes. Controls were defined as patients with CPE isolates, which tested positive for carbapenemases, as described below, and were not susceptible to meropenem, imipenem, and/or ertapenem. For patients with multiple CRE cultures, only the first isolate (clinical or surveillance cultures) was included. All participating hospitals implemented active surveillance for CRE as part of IPC measures. We excluded patients with both CPEs and NCPCREs, mixed CPE genotypes (n = 18), incomplete case record forms, and CRE isolates that were phenotypically carbapenemase producers but were negative for CPE by PCR. Carbapenem exposure (meropenem, imipenem, and ertapenem) within the preceding 30 days was the main exposure of interest.
We documented patient demographics, underlying medical conditions, Charlson comorbidity index score, exposure to third and fourth generation cephalosporins (ceftriaxone, ceftazidime, and cefepime), extended-spectrum penicillins (amoxicillin-clavulanate, ampicillin-sulbactam, and piperacillin-tazobactam), fluoroquinolones (ciprofloxacin and levofloxacin), aminoglycosides (gentamicin and amikacin), tigecycline, metronidazole, trimethoprim-sulfamethoxazole, and polymyxin B during 30 days prior to the CRE culture collection date. Data were collected on history of hospitalization, surgical procedures, upper and/or lower gastrointestinal scopes, and carriage of multidrug-resistant organisms during 12 months before the CRE culture collection date. We also collected data on ICU stay at the time of culture and the presence of a feeding tube and central venous line (CVL) during the time at risk (time from admission to the isolation of CRE). All data were collected from electronic medical records.
Microbiological methods.
Antibiotic susceptibility testing and organism identification of isolates from clinical cultures were conducted at participating institutions. Microbiology laboratories at participating institutions used either Clinical and Laboratory Standards Institute (CLSI) or European Committee on Antimicrobial Susceptibility Testing (EUCAST) methods to detect CREs. Carbapenem MICs were then confirmed by Etest (bioMérieux, Marcy-l’Étoile, France). CREs and isolates suspected of carrying carbapenemase genes were submitted to the National Public Health Laboratory (NPHL) for further phenotypic characterization and PCR-based assays. At NPHL (and at some hospital microbiology laboratories), the presence of carbapenemase production was investigated phenotypically by using diagnostic meropenem tablets of the KPC/metallo-β-lactamase confirmation kit (Rosco Diagnostica A/S, Taastrup, Denmark) and/or Rapidec Carba NP. Characterization of β-lactamase genes was performed by PCR assays targeting class A carbapenemases (blaKPC, blaGES, blaIMI, and blaNMC-A), class B metallo-β-lactamases (blaNDM, blaVIM, and blaIMP) (23), and class D carbapenemases (blaOXA-48-like and blaOXA-23 carbapenemases) (24). Carbapenem nonsusceptibility due to porin loss associated with extended-spectrum β-lactamases and AmpC overproduction was not investigated by molecular techniques.
Statistical method.
For patient-level analysis, the Student’s t test or the nonparametric Wilcoxon signed-rank test was used for continuous variables depending on their distribution. The χ2 or Fisher exact test was used to compare categorical variables.
Covariates identified as apriori confounders were Charlson score, hospitalization during the preceding 1 year, being in ICU at the time of culture, time at risk, presence of central venous line, and exposure to fluoroquinolones, third- or fourth-generation cephalosporins, extended-spectrum penicillins, and aminoglycosides. Additionally, variables with a P value of <0.1 on univariate analysis and clinical plausibility were included in the logistic regression model. For correlated variables (pairwise correlation coefficient, >0.7), only one of the covariates was selected for inclusion into the candidate models by the strength of association. We conducted a random effect logistic regression model allowing for clustering at the institution level and adjusted for month (to account for possibility of unknown outbreaks during certain time periods) of isolation of CRE to identify independent factors associated more with NCPCRE than CPE. A two-sided P value of <0.05 was considered statistically significant. P values were interpreted together with 95% confidence intervals (CIs) for the logistic regression model.
We explored the impact of carbapenem exposure on the occurrence of NCPCRE as a binary variable. We then studied the effect of different durations of carbapenem exposure by grouping patients into three categories (no carbapenem exposure, 1 to 3 days of exposure, and >3 days of exposure). Carbapenem exposure was dichotomized at 3 days, as it is the usual duration for empirical antibiotic therapy and findings would be beneficial for future stewardship application. To explore if the effect of carbapenem exposures on the occurrence of CRE varies by Enterobacteriaceae species and genotypes of CPE, we conducted stratified analysis to study the effect of carbapenem exposure (as a binary variable) on the occurrence of non-carbapenemase-producing carbapenem-resistant (NCPCR) E. coli compared with carbapenemase-producing (CP) E. coli, NCPCR K. pneumoniae compared with CP K. pneumoniae, and NCPCRE compared with NDM-producing Enterobacteriaceae and KPC-producing Enterobacteriaceae. Since more than half of the study populations were colonized rather than infected with CPE, we performed stratified analysis by specimen type, comparing surveillance culture to clinical cultures. Clinical cultures were all cultures sent as part of clinical care of patients.
All analyses were done with STATA 14.0 (StataCorp, Texas, USA).
Ethics approval.
The CaPES study was reviewed and approved by the ethics institutional review board of National Health Group Singapore (DSRB reference no. 2014/00617).
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the infection control units of participating hospitals; Jamie Lo, an epidemiologist at the National Centre for Infectious Diseases for reviewing the manuscript; and Mohammad Abbas from the Infection Control Program, WHO Collaborating Centre for Patient Safety, Geneva University Hospitals, for his input on the statistical analysis.
The members of the CaPES study group include Benjamin Cherng, Douglas Chan Su Gin, Deepak Rama Narayana, De Partha Pratim, Hsu Li Yang, Indumathi Venkatachalam, Jeanette Teo, Kalisvar Marimuthu, Koh Tse Hsien, Nancy Tee, Nares Smitasin, Ng Oon Tek, Ooi Say Tat, Prabha Unny Krishnan, Raymond Fong, Raymond Lin Tzer Pin, Surinder Kaur Pada, Tan Thean Yen, and Thoon Koh Cheng.
This work was supported by National Medical Research Council Clinician-Scientist Individual Research Grant (NMRC/CIRG/1463/2016), Singapore Ministry of Education Academic Research Fund Tier 2 grant (MOE2015-T2-2-096), National Medical Research Council Collaborative grant (NMRC CGAug16C005), National Medical Research Council Clinician Scientist Award (NMRC/CSA-INV/0002/2016), and National Medical Research Clinician Scientist Individual Research Grant (CS-IRG) (CIRG18nov-0034). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00845-19.
Contributor Information
Collaborators: Benjamin Cherng, Douglas Chan Su Gin, Deepak Rama Narayana, Partha Pratim De, Hsu Li Yang, Indumathi Venkatachalam, Jeanette Teo, Kalisvar Marimuthu, Koh Tse Hsien, Nancy Tee, Nares Smitasin, Ng Oon Tek, Ooi Say Tat, Prabha Unny Krishnan, Raymond Fong, Raymond Lin Tzer Pin, Surinder Kaur Pada, Tan Thean Yen, and Thoon Koh Cheng
REFERENCES
- 1.Livorsi DJ, Chorazy ML, Schweizer ML, Balkenende EC, Blevins AE, Nair R, Samore MH, Nelson RE, Khader K, Perencevich EN. 2018. A systematic review of the epidemiology of carbapenem-resistant Enterobacteriaceae in the United States. Antimicrob Resist Infect Control 7:55. doi: 10.1186/s13756-018-0346-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goodman KE, Simner PJ, Tamma PD, Milstone AM. 2016. Infection control implications of heterogeneous resistance mechanisms in carbapenem-resistant Enterobacteriaceae (CRE). Expert Rev Anti Infect Ther 14:95–108. doi: 10.1586/14787210.2016.1106940. [DOI] [PubMed] [Google Scholar]
- 3.Orsi GB, Bencardino A, Vena A, Carattoli A, Venditti C, Falcone M, Giordano A, Venditti M. 2013. Patient risk factors for outer membrane permeability and KPC-producing carbapenem-resistant Klebsiella pneumoniae isolation: results of a double case-control study. Infection 41:61–67. doi: 10.1007/s15010-012-0354-2. [DOI] [PubMed] [Google Scholar]
- 4.Leavitt A, Chmelnitsky I, Colodner R, Ofek I, Carmeli Y, Navon-Venezia S. 2009. Ertapenem resistance among extended-spectrum-beta-lactamase-producing Klebsiella pneumoniae isolates. J Clin Microbiol 47:969–974. doi: 10.1128/JCM.00651-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwaber MJ, Carmeli Y. 2014. An ongoing national intervention to contain the spread of carbapenem-resistant Enterobacteriaceae. Clin Infect Dis 58:697–703. doi: 10.1093/cid/cit795. [DOI] [PubMed] [Google Scholar]
- 6.Simner PJ, Goodman KE, Carroll KC, Harris AD, Han JH, Tamma PD. 2018. Using patient risk factors to identify whether carbapenem-resistant Enterobacteriaceae infections are caused by carbapenemase-producing organisms. Open Forum Infect Dis 5:ofy094. doi: 10.1093/ofid/ofy094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Queenan AM, Bush K. 2007. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev 20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chia JH, Siu LK, Su LH, Lin HS, Kuo AJ, Lee MH, Wu TL. 2009. Emergence of carbapenem-resistant Escherichia coli in Taiwan: resistance due to combined CMY-2 production and porin deficiency. J Chemother 21:621–626. doi: 10.1179/joc.2009.21.6.621. [DOI] [PubMed] [Google Scholar]
- 9.Armand-Lefèvre L, Angebault C, Barbier F, Hamelet E, Defrance G, Ruppé E, Bronchard R, Lepeule R, Lucet J-C, El Mniai A, Wolff M, Montravers P, Plésiat P, Andremont A. 2013. Emergence of imipenem-resistant gram-negative bacilli in intestinal flora of intensive care patients. Antimicrob Agents Chemother 57:1488–1495. doi: 10.1128/AAC.01823-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng VC, Chan JF, Wong SC, Chen JH, Tai JW, Yan MK, Kwan GS, Tse H, To KK, Ho PL, Yuen KY. 2013. Proactive infection control measures to prevent nosocomial transmission of carbapenem-resistant Enterobacteriaceae in a non-endemic area. Chin Med J (Engl) 126:4504–4509. [PubMed] [Google Scholar]
- 11.Magiorakos AP, Burns K, Rodriguez Bano J, Borg M, Daikos G, Dumpis U, Lucet JC, Moro ML, Tacconelli E, Simonsen GS, Szilagyi E, Voss A, Weber JT. 2017. Infection prevention and control measures and tools for the prevention of entry of carbapenem-resistant Enterobacteriaceae into healthcare settings: guidance from the European Centre for Disease Prevention and Control. Antimicrob Resist Infect Control 6:113. doi: 10.1186/s13756-017-0259-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tacconelli E, Cataldo MA, Dancer SJ, De Angelis G, Falcone M, Frank U, Kahlmeter G, Pan A, Petrosillo N, Rodriguez-Bano J, Singh N, Venditti M, Yokoe DS, Cookson B, European S, Of Clinical M. 2014. ESCMID guidelines for the management of the infection control measures to reduce transmission of multidrug-resistant Gram-negative bacteria in hospitalized patients. Clin Microbiol Infect 20:1–55. doi: 10.1111/1469-0691.12427. [DOI] [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention. 2012. CRE toolkit: guidance for control of carbapenem-resistant Enterobacteriaceae (CRE). Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 14.World Health Organization. 2017. Guidelines for the prevention and control of carbapenem-resistant Enterobacteriaceae, Acinetobacter baumannii and Pseudomonas aeruginosa in health care facilities. World Health Organization, Geneva, Switzerland. [PubMed] [Google Scholar]
- 15.Tomczyk S, Zanichelli V, Grayson ML, Twyman A, Abbas M, Pires D, Allegranzi B, Harbarth S. 2018. Control of Carbapenem-resistant Enterobacteriaceae, Acinetobacter baumannii, and Pseudomonas aeruginosa in healthcare facilities: a systematic review and reanalysis of quasi-experimental studies. Clin Infect Dis 68:873–884. doi: 10.1093/cid/ciy752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Humphreys H, Fitzpatick F, Harvey BJ. 2015. Gender differences in rates of carriage and bloodstream infection caused by methicillin-resistant Staphylococcus aureus: are they real, do they matter and why? Clin Infect Dis 61:1708–1714. doi: 10.1093/cid/civ576. [DOI] [PubMed] [Google Scholar]
- 17.Cai Y, Venkatachalam I, Tee NW, Tan TY, Kurup A, Wong SY, Low CY, Wang Y, Lee W, Liew YX, Ang B, Lye DC, Chow A, Ling ML, Oh HM, Cuvin CA, Ooi ST, Pada SK, Lim CH, Tan JWC, Chew KL, Nguyen VH, Fisher DA, Goossens H, Kwa AL, Tambyah PA, Hsu LY, Marimuthu K. 2017. Prevalence of healthcare-associated infections and antimicrobial use among adult inpatients in singapore acute-care hospitals: results from the first national point prevalence survey. Clin Infect Dis 64:S61–S67. doi: 10.1093/cid/cix103. [DOI] [PubMed] [Google Scholar]
- 18.Garbutt C, Simmons G, Patrick D, Miller T. 2007. The public hand hygiene practices of New Zealanders: a national survey. N Z Med J 120:U2810. [PubMed] [Google Scholar]
- 19.Judah G, Aunger R, Schmidt WP, Michie S, Granger S, Curtis V. 2009. Experimental pretesting of hand-washing interventions in a natural setting. Am J Public Health 99:S405–S411. doi: 10.2105/AJPH.2009.164160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papadimitriou-Olivgeris M, Christofidou M, Fligou F, Bartzavali C, Vrettos T, Filos KS, Marangos M, Anastassiou ED. 2014. The role of colonization pressure in the dissemination of colistin or tigecycline resistant KPC-producing Klebsiella pneumoniae in critically ill patients. Infection 42:883–890. doi: 10.1007/s15010-014-0653-x. [DOI] [PubMed] [Google Scholar]
- 21.Harbarth S, Harris AD, Carmeli Y, Samore MH. 2001. Parallel analysis of individual and aggregated data on antibiotic exposure and resistance in gram-negative bacilli. Clin Infect Dis 33:1462–1468. doi: 10.1086/322677. [DOI] [PubMed] [Google Scholar]
- 22.Marimuthu K, Venkatachalam I, Khong WX, Koh TH, Cherng BPZ, Van La M, De PP, Krishnan PU, Tan TY, Choon RFK, Pada SK, Lam CW, Ooi ST, Deepak RN, Smitasin N, Tan EL, Lee JJ, Kurup A, Young B, Sim NTW, Thoon KC, Fisher D, Ling ML, Peng BAS, Teo YY, Hsu LY, Lin RTP, Ong RT, Teo J, Ng OT, Carbapenemase P, for the Carbapenemase-Producing Enterobacteriaceae in Singapore (CaPES) Study. 2017. Clinical and molecular epidemiology of carbapenem-resistant Enterobacteriaceae among adult inpatients in Singapore. Clin Infect Dis 64:S68–S75. doi: 10.1093/cid/cix113. [DOI] [PubMed] [Google Scholar]
- 23.Teo J, Ngan G, Balm M, Jureen R, Krishnan P, Lin R. 2012. Molecular characterization of NDM-1 producing Enterobacteriaceae isolates in Singapore hospitals. WPSAR 3:19–24. doi: 10.5365/wpsar.2011.2.4.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balm MN, Ngan G, Jureen R, Lin RT, Teo JW. 2013. OXA-181-producing Klebsiella pneumoniae establishing in Singapore. BMC Infect Dis 13:58. doi: 10.1186/1471-2334-13-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.