1H-benzo[d]imidazole derivatives exhibit antitubercular activity in vitro at a nanomolar range of concentrations and are not toxic to human cells, but their mode of action remains unknown. Here, we showed that these compounds are active against intracellular Mycobacterium tuberculosis.
KEYWORDS: MmpL3, Mycobacterium tuberculosis inhibitors, benzimidazole, drug resistance, tuberculosis
ABSTRACT
1H-benzo[d]imidazole derivatives exhibit antitubercular activity in vitro at a nanomolar range of concentrations and are not toxic to human cells, but their mode of action remains unknown. Here, we showed that these compounds are active against intracellular Mycobacterium tuberculosis. To identify their target, we selected drug-resistant M. tuberculosis mutants and then used whole-genome sequencing to unravel mutations in the essential mmpL3 gene, which encodes the integral membrane protein that catalyzes the export of trehalose monomycolate, a precursor of the mycobacterial outer membrane component trehalose dimycolate (TDM), as well as mycolic acids bound to arabinogalactan. The drug-resistant phenotype was also observed in the parental strain overexpressing the mmpL3 alleles carrying the mutations identified in the resistors. However, no cross-resistance was observed between 1H-benzo[d]imidazole derivatives and SQ109, another MmpL3 inhibitor, or other first-line antitubercular drugs. Metabolic labeling and quantitative thin-layer chromatography (TLC) analysis of radiolabeled lipids from M. tuberculosis cultures treated with the benzoimidazoles indicated an inhibition of trehalose dimycolate (TDM) synthesis, as well as reduced levels of mycolylated arabinogalactan, in agreement with the inhibition of MmpL3 activity. Overall, this study emphasizes the pronounced activity of 1H-benzo[d]imidazole derivatives in interfering with mycolic acid metabolism and their potential for therapeutic application in the fight against tuberculosis.
INTRODUCTION
Mycobacterium tuberculosis, the causative agent of tuberculosis (TB), is an intracellular pathogen, and its life cycle includes long states of persistence. This pathogen, claiming 1.5 million lives each year, is relatively hard to eradicate and poses a challenge for effective chemotherapy (1). TB treatment lasts six to 24 months depending on the drug susceptibility of the infecting strain and requires a cocktail of at least 4 drugs used simultaneously to prevent the selection of drug-resistant M. tuberculosis mutants. Four first-line anti-TB drugs (isoniazid [INH], rifampin [RMP], ethambutol [EMB], and pyrazinamide [PZA]) are used in the 6-month regimen therapy of TB caused by drug-sensitive strains. Treatment of TB caused by strains resistant to at least isoniazid and rifampin (multidrug-resistant [MDR] strains) requires additional drugs and is often less effective and less tolerated. Additionally, the treatment of MDR TB is much more expensive than standard treatment, the outcomes are several times worse with a high mortality rate (50 to 80%) within 4 months of diagnosis (2), and patients with MDR-TB have twice the risk of relapse after the completion of treatment (3, 4). The therapy of TB caused by an MDR strain is very complex, lasts 2 years, and requires discipline in taking the prescribed drugs, which have long lists of severe side effects, daily for a long period of time (5). Poor treatment management by patients has been postulated as a primary reason for the drastic increase in the number of MDR TB cases observed in recent years. The nearly 600 000 cases of MDR TB estimated to exist worldwide and the phenomenon of HIV/M. tuberculosis coinfection make TB a serious public health challenge worldwide. Taking the above into account, the development of alternative medical strategies based on new generations of the drugs is desperately needed to effectively cure MDR TB, reduce the duration of current therapies, and minimize the toxicity and cost of anti-TB agents (6).
Nearly 50% of antitubercular, clinically relevant drugs available today target the process of biosynthesis of various cell envelope components (7). The mycobacterial cell wall is composed of a complex of peptidoglycan, arabinogalactan (AG), and mycolic acids (MAs) (8, 9). This covalently linked complex is decorated on the surface with trehalose monomycolate (TMM), trehalose dimycolate (TDM), sulfolipids, phenolic glycolipids, phthiocerol dimycocerosates (PDIMs), polysaccharides, and proteins (10–12). Mycolic acids, which are long-chain α-alkyl β-hydroxy fatty acids, are essential components of the M. tuberculosis cell envelope and play a crucial role in the cell wall architecture and impermeability that are responsible for the natural resistance of mycobacteria to most antibiotics (13). Mycolic acids exist as esters of the nonreducing arabinan terminus of AG but are also present as extractable “free” lipids within the cell wall, mainly associated with TDM (9). The fatty acid synthase II complex (FAS2) is the primary target for the first-line anti-TB drug isoniazid as well as for the second-line antitubercular agent ethionamide, resulting in the loss of TMM, TDM, and mycolates attached to cell wall arabinan (14). However, the accumulation of TDM and TMM is observed in the presence of ethambutol (EMB), affecting the arabinogalactan biosynthesis process and thus downregulating the arabinan acceptor sites for the mycolates in the cell wall (15). Methoxy- and keto-mycolic acid synthesis is targeted by delamanid, a dihydro-nitroimidazo-oxazole derivative, which has been conditionally approved by the European Medicines Agency (EMA) for the treatment of MDR TB. Delamanid is used as a prodrug activated within bacilli by the deazaflavin-dependent nitroreductase (Rv3547). A reactive intermediate metabolite, formed between delamanid and the desnitro-imidazo-oxazole derivative, is considered to play a vital role in the inhibition of mycolic acid production (16–19). The mycolic acid modification and elongation process might also be targeted by thiacetazone (TAC), an antitubercular drug that was formerly used in conjunction with isoniazid but was removed from the antitubercular chemotherapeutic arsenal due to toxic side effects. Currently, TAC-derived analogues have shown increased potency against tubercle bacilli and are being considered again as putative antitubercular drugs (20).
Further, peptidoglycan biosynthesis might be affected by cycloserine or β-lactam inhibitors (amoxicillin, meropenem, and imipenem) (21). Additionally, some drugs under development (www.newtbdrugs.org) affect the synthesis of the mycobacterial cell wall. The other highly accessible target in mycobacteria that is involved in the biosynthesis of the cell envelope is MmpL3, the integral membrane flippase responsible for the export of trehalose monomycolate, which is a precursor of the mycobacterial outer membrane (OM) component trehalose dimycolate (TDM) as well as mycolic acids bound to arabinogalactan (22, 23). MmpL3 is one of the targets of the ethambutol analogue SQ109 (24–26) as well as a number of other compounds that are specific M. tuberculosis inhibitors (22, 27) or are active against a broad spectrum of bacterial and fungal pathogens (28–30).
Whole-cell phenotyping (MIC) screens allow the identification of a number of bactericidal compounds that are sometimes potent against tubercle bacilli, yet the mode of action is not always identified even for compounds with strong anti-M. tuberculosis activity. We have recently identified the potent antitubercular activity of 1H-benzo[d]imidazole derivatives even at nanomolar concentrations (31, 32). Here, by selection of drug-resistant mutants, genome sequencing, mmpL3 gene transfer, and quantitative detection of TDM and mycolylated arabinogalactan, we identified the molecular target for these compounds as the TMM transporter, MmpL3.
RESULTS
1H-Benzo[d]imidazole derivatives are effective against M. tuberculosis in human macrophages.
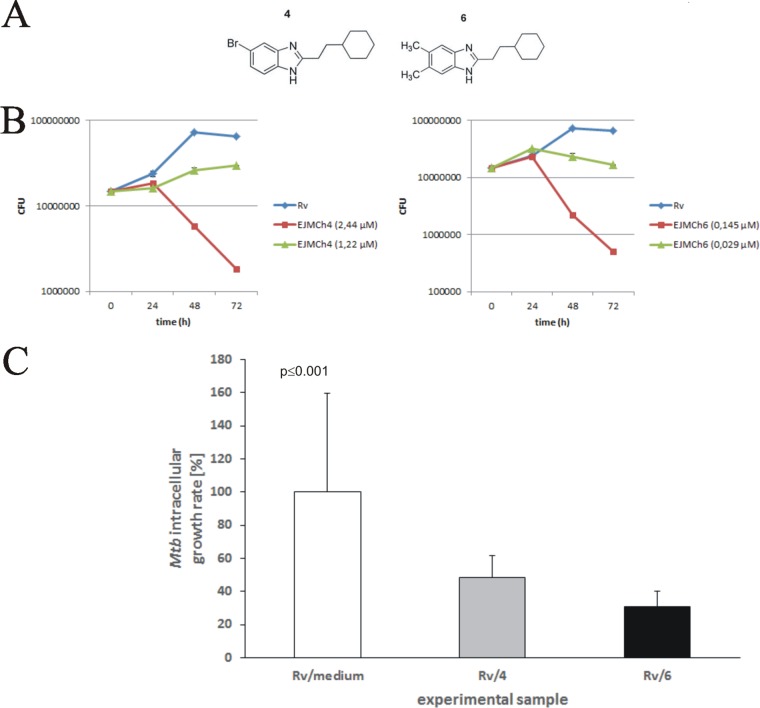
We have previously reported the antitubercular activity of 1H-benzo[d]imidazole derivatives and analogues (32). We found that compounds bearing halogen atoms or methyl groups at the benzimidazole and cyclohexylethyl substituents at the C-2 position show excellent bactericidal activity against tubercle bacilli and low cytotoxicity in the LLC-PK1 pig kidney epithelial cell line (32). Here, we used EJMCh4 {5-bromo-2-(2-cyclohexylethyl)-1H-benzo[d]imidazole} and EJMCh6 {2-(2-cyclohexylethyl)-5,6-dimethyl-1H-benzo[d]imidazole}, the two most promising of these compounds (Fig. 1A). We verified their activity against tubercle bacilli infecting macrophages and identified their mode of action. Both compounds were synthesized from 3-cyclohexylpropanoic acid (2.35 ml, 15 mmol) and the appropriate diamine (10 mmol) in polyphosphoric acid (5 ml) by stirring at 180 to 200°C for 5 h as reported previously (32). The purity of the compounds was confirmed by the thin-layer chromatography (TLC) method and elemental analysis (percent C, H, and N). 1H nuclear magnetic resonance (NMR) spectra in CDCl3 or dimethyl sulfoxide-d6) (DMSO-d6) were recorded on a Varian Gemini (200 MHz) instrument (spectra are shown in Fig. S1 in the supplemental material). They were used to determine their inhibitory effect on the growth of M. tuberculosis in vitro (Fig. 1B; Table 1) (32). Liquid culture growth analyses by optical density at 600 (OD600) and CFU measurements confirmed the strong antitubercular effect of both compounds even at nanomolar concentrations, with the MIC99 as low as 2.44 μM (MIC50, 1.22 μM) for EJMCh4 and 0.145 μM (MIC50, 0.029 μM) for EJMCh6 (Fig. 1B). Determination of the bactericidal activities of the compounds against intracellular bacilli was preceded by estimation of their cytotoxic activities against monocyte-derived macrophages (MDMs) differentiated from peripheral blood mononuclear cells that were isolated from the buffy coats of healthy human blood donors. The 50% inhibitory concentrations (IC50) of both compounds exceeded 128 μM (see Table S1 in the supplemental material). The compounds then were used at the minimal concentrations that inhibited the growth of M. tuberculosis (1× MIC) in broth culture (2.44 μM and 0.145 μM for compounds EJMCh4 and EJMCh6, respectively) to test their activity against intracellular bacilli. The monocyte-derived macrophages of a single donor were infected with M. tuberculosis at a multiplicity of infection (MOI) of 1:10. The bacteria were left undisturbed for 2 h for phagocytosis to occur, after which the extracellular bacilli were removed by intensive washing and any remaining bacilli were killed with gentamicin. Furthermore, the M. tuberculosis-infected cells were exposed to the compounds at 1× MIC and incubated for 48 h. The number of intracellularly located live bacteria was determined by CFU enumeration. The data analysis showed a significant (P ≤ 0.001) reduction in viable bacilli isolated from cells incubated in the presence of either compound (Fig. 1C), with the most substantial effect provided by compound EJMCh6 at 0.145 μM. The number of intracellularly growing mycobacteria decreased by 52% and 69% for compounds EJMCh4 and EJMCh6, respectively.
FIG 1.
(A) Chemical structures of 1H-benzo[d]imidazole derivatives EJMCh4 {5-bromo-2-(2-cyclohexylethyl)-1H-benzo[d]imidazole} (left) and EJMCh6 {2-(2-cyclohexylethyl)-5,6-dimethyl-1H-benzo[d]imidazole} (right). (B) Time-dependent viability of M. tuberculosis at selected concentrations of the 1H-benzo[d]imidazole derivatives EJMCh4 and EJMCh6 based on CFU enumerations from three independent experiments and plotted as the average ± standard deviation. (C) Inhibitory effect of EJMCh4 and EJMCh6 on the intracellular growth of M. tuberculosis in human monocyte-derived macrophages. The intracellular growth of the pathogen in the experimental (1H-benzo[d]imidazole-treated) samples versus the control (non-1H-benzo[d]imidazole-treated) sample is presented as a growth percentage, which represents the ratio of the CFU number determined for the 1H-benzo[d]imidazole-treated sample to the CFU number estimated in the control sample multiplied by 100.
TABLE 1.
MICs of 1H-benzo[d]imidazole derivatives EJMCh4 and EJMCh6 against wild-type M. tuberculosis H37Rv and resistant mutants
| Compound and strain (mutation)a | MIC50 (μM) | MIC90 (μM) |
|---|---|---|
| EJMCh4 | ||
| H37Rv | 1.22 | 2.44 |
| MmpL3 mutants | ||
| 4/1 (V285A) | 13.00 | 19.50 |
| 4/3 (V195A) | 6.50 | 13.00 |
| EJMCh6 | ||
| H37Rv | 0.029 | 0.145 |
| MmpL3 mutants | ||
| 6A (V684A) | 0.77 | 1.93 |
| 6B (V240A) | 0.77 | 1.93 |
| 6C (G253E) | 0.77 | 1.93 |
The 4/1 and 4/3 mutants were selected in the presence of compound EJMCh4 at 2.44 μM. The 6A, 6B, and 6C mutants were selected in the presence of compound EJMCh6 at 0.145 μM.
1H-Benzo[d]imidazole derivative-resistant mutants harbor mutations in mmpL3.
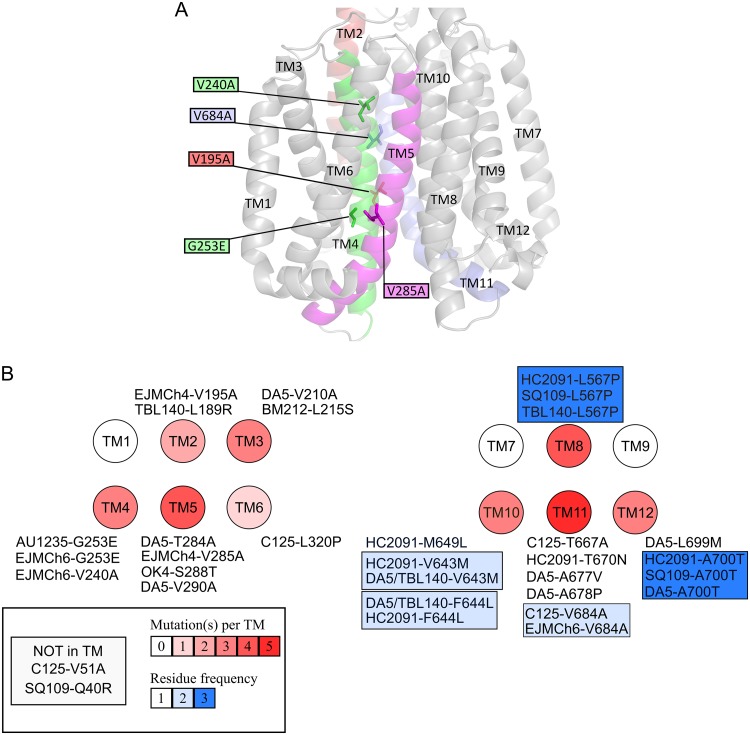
The acquired resistance of M. tuberculosis to anti-TB drugs is typically due to the acquisition of mutations in a gene encoding a drug-target protein or an enzyme activating a given prodrug within the bacilli. The mutation frequency was estimated for acquired resistance to 1H-benzo[d]imidazole derivatives in the presence of 2.6× MIC of EJMCh4 and EJMCh6 as well as rifampin (0.6 μM and 2.4 μM) and streptomycin (STR) (2.8 μM) as controls. Fluctuation analysis was performed as previously described by Ford et al. (33), and then the drug resistance rate was determined using the Ma-Sarkar-Sandri (MSS) method (34). The calculated mutation rate was similar for STR and 1H-benzo[d]imidazole derivatives but was lower than that for both concentrations of RMP (see Table S2 in the supplemental material). To identify a mode of action of the 1H-benzo[d]imidazole derivatives, we decided to select and analyze mutants that were resistant to the investigated compounds. The wild-type M. tuberculosis strain was cultured in the presence of a subinhibitory concentration of EJMCh4 (0.244 μM) or EJMCh6 (0.0145 μM), and the resistant mutants were selected on solid medium supplemented with 2.44 μM compound EJMCh4 and 0.145 or 0.29 μM compound EJMCh6. The resulting mutants were grown in the presence of various concentrations of compound EJMCh4 or EJMCh6 to determine their MIC values (Table 1). Three mutants selected on compound EJMCh4 and three mutants selected on compound EJMCh6, as well as the wild-type strain, were subjected to genomic DNA isolation and sequencing using the NGS Illumina system. Bioinformatics analyses allowed the identification of a number of point mutations in the investigated genomes (see Table S3 in the supplemental material). The data analysis revealed that the gene affected in all investigated mutants was rv0206c, encoding an essential transmembrane transporter, MmpL3. Five of six selected mutants carried diverse mutations in mmpL3 and were processed in further investigations. All mutations in rv0206c resulted in amino acid substitutions localized in the transmembrane helices of MmpL3, as mapped on an MmpL3 three-dimensional (3D) homology model (Fig. 2A). To verify the relationship between the presence of the identified mutations in MmpL3 and resistance to compounds EJMCh4 and EJMCh6, the genes carrying the mutations, V285A, V195A, V684A, V240A, or G253E, were cloned with their putative promoter regions into the pMV306Km integration vector and introduced into the single attB locus of the genome of the wild-type strain, which was sensitive to both compounds. The resultant merodiploid strains carrying both the wild-type and mutated mmpL3 genes were analyzed for their sensitivity to the compounds EJMCh4 and EJMCh6. The transcomplementation of the wild-type strain with mutated mmpL3 conferred resistance to the 1H-benzo[d]imidazole derivatives (Table 2), confirming the role of the investigated mutations in the resistance phenotype.
FIG 2.
(A) Mapping of the mutations found in spontaneous EJMCh4- and EJMCh6-resistant mutants on an M. tuberculosis MmpL3 three-dimensional homology model. Only the transmembrane helices (TM) are depicted. The residues mutated in the spontaneous resistant mutants are shown as sticks, and the mutations are indicated in the colored boxes. All TM domains are in gray, with the exception of those carrying a mutation in resistant mutants, notably TM2 (red), TM4 (green), TM5 (magenta), and TM11 (blue). (B) All mutations found in MmpL3 from resistant mutants selected on several MmpL3 inhibitors. Transmembrane helices are represented by circles. Each mutation as well as which drug it confers resistance to is indicated. The number of mutations affecting TM is shown from white (no mutation) to dark red (five mutations), and the blue color indicates residues that confer cross-resistance to several drugs.
TABLE 2.
MICs of 1H-benzo[d]imidazole derivatives EJMCh4 and EJMCh6 against wild-type M. tuberculosis H37Rv and complemented mutants
| Compound and strain (mutation)a | MIC50 (μM) | MIC90 (μM) |
|---|---|---|
| EJMCh4 | ||
| H37Rv | 1.22 | 2.44 |
| H37Rv + vector | 1.22 | 2.44 |
| H37Rv + MmpL3 mutant 4/1 (V285A) | 6.50 | 32.50 |
| H37Rv + MmpL3 mutant 4/3 (V195A) | 6.50 | 32.50 |
| EJMCh6 | ||
| H37Rv | 0.145 | 0.29 |
| H37Rv + vector | 0.145 | 0.29 |
| H37Rv + MmpL3 mutant 6A (V684A) | 0.77 | 3.90 |
| H37Rv + MmpL3 mutant 6B (V240A) | 0.77 | 3.90 |
| H37Rv + MmpL3 mutant 6C (G253E) | 0.77 | 3.90 |
The wild-type M. tuberculosis was complemented with an empty vector or with mmpL3 genes carrying mutations identified in the resistant mutants.
Mutants resistant to compounds EJMCh4 and EJMCh6 are not cross-resistant to TB drugs or SQ109.
Cross-resistance with current anti-TB drugs would exclude the new compounds as potential drugs. Hence, the spontaneous mutants resistant to the investigated 1H-benzo[d]imidazole derivatives, as well as the wild-type, drug-sensitive M. tuberculosis strain, were investigated for their sensitivity to the selected anti-TB drugs INH, STR, RIF, and EMB. The MIC values determined for all the drugs tested were the same for the wild-type strain and investigated mutants, as presented in Table S4 in the supplemental material. Because SQ109 and our investigated compounds affect the same target, MmpL3, we decided to test their potential cross-resistance. The MIC values of SQ109 were determined for wild-type M. tuberculosis and mutants resistant to the compounds EJMCh4 and EJMCh6, and the same MIC values were obtained. Since the distinct mutations were selected under pressure of compounds EJMCh4 and EJMCh6, we also investigated the cross-resistance between these two compounds (see Table S5 in the supplemental material). As expected, the EJMCh4-resistant M. tuberculosis mutants appeared also to be cross-resistant to EJMCh6 and vice versa, suggesting a common mechanism of action for these two chemicals.
1H-Benzo[d]imidazole derivatives inhibit TDM synthesis and AG mycolylation.
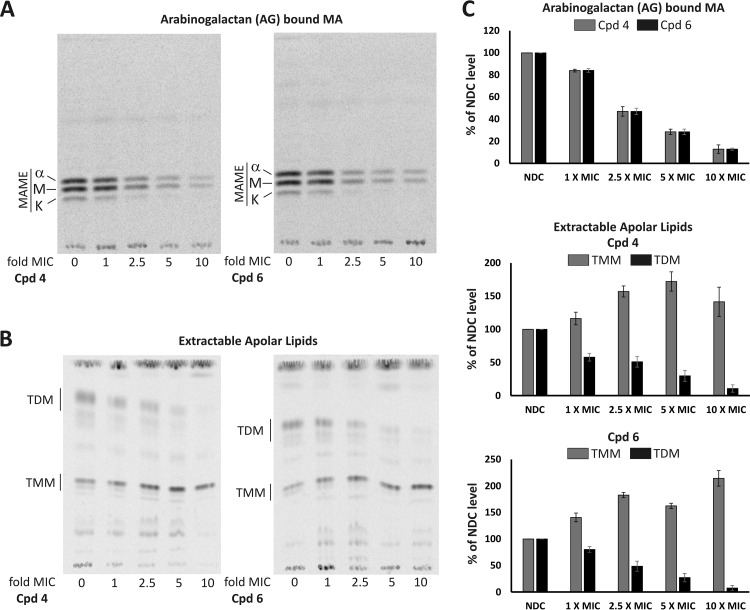
Following synthesis, mycolic acids (MAs) are transported across the cell envelope in the form of TMM and then transferred by the mycolyltransferase Ag85 complex at the outer membrane (OM) either to the arabinogalactan (AG) complex or to another TMM to form TDM (22, 23). Since MmpL3 has been implicated in flipping TMM across the inner membrane (IM) and its depletion reduced formation of TDM and AG-linked mycolates (23, 35), we analyzed whether the effect of 1H-benzo[d]imidazole derivatives on mycobacterial cell wall mycolates reflects the one observed upon MmpL3 depletion. Late-log-phase M. tuberculosis was incubated with increasing concentrations (1×, 2.5×, 5×, and 10× MIC) of compound EJMCh4 or EJMCh6. Cultures were then subjected to radiolabeling of lipids and divided into two volumes. From the first culture volume, total mycolic acid methyl esters (MAMES) and fatty acid methyl esters (FAMES) were extracted. From the second volume, after polar lipid removal, the apolar lipid fraction containing TMM and TDM was obtained prior to derivatization and extraction of AG-bound MAMEs. Quantitative TLC analyses showed that despite the undisturbed pool of total MAs (see Fig. S2 in the supplemental material), treatment of the M. tuberculosis cells with compound EJMCh4 or EJMCh6 resulted in a decrease in AG mycolylation as well as TDM production (Fig. 3). The results clearly show the decrease in transport and concomitant accumulation of TMM in 1H-benzo[d]imidazole derivative-treated cells, confirming inhibition of the MmpL3 flippase. Different levels of AG-bound mycolates and TDM in tubercle bacilli treated with 1H-benzo[d]imidazole derivatives and the untreated control were additionally confirmed by densitometry (Fig. 3).
FIG 3.
(A) Effect of EJMCh4 (Cpd 4) and EJMCh6 (Cpd 6) treatment on the arabinogalactan-bound mycolic acids of M. tuberculosis mc26230. Equal volumes of AG-bound MAME fraction were loaded on a TLC plate and resolved twice using the hexane-ethyl acetate (95:5, vol/vol) solvent system. Unchanged TLC profiles of M. tuberculosis mc26230 total mycolic acid methyl esters (MAMEs) and fatty acid methyl esters (FAMEs) after treatment with Cpd 4 and Cpd 6 are presented in Fig. S2 in the supplemental material. A, alpha-mycolic acid; M, methoxy-mycolic acid; K, keto-mycolic acid. (B) Effect of EJMCh4 (Cpd 4) and EJMCh6 (Cpd 6) treatment on the apolar lipid fraction. Equal counts of apolar lipids were applied on a 1D TLC plate, and TMM and TDM were visualized using the chloroform-methanol-water (40:8:1, vol/vol/vol) solvent system. (C) Densitometric analysis of the results shown in panels A and B, expressed as percentages of results for a no-drug control (NDC). For each analysis densitometry was done on three independent TLC plates, and results represents mean ± standard deviation.
DISCUSSION
The selection of resistant mutants followed by whole-genome sequencing and the observed inhibition of arabinogalactan mycolylation and synthesis of TDM indicate MmpL3 as a molecular target for the antitubercular activity of 1H-benzo[d]imidazole derivatives. MmpL3 belongs to the resistance, nodulation, and division (RND) superfamily of transporters, which require the transmembrane electrochemical proton gradient for activity (36, 37). This protein is essential for transport of TMM from the cytosol to the pseudoperiplasmic space, where TMM is further modified to TDM and incorporated into the membrane (22, 38). More recently, it was demonstrated that MmpL3 acts as a mycolic acid flippase at the plasma membrane of mycobacteria, which might be directly targeted and inhibited by BM212 (1,5-diarylpyrrole) (35). High-density transposon mutagenesis and homologous recombination experiments showed that mmpL3 is required for survival of mycobacteria in vitro as well as in a mouse TB infection model (39–44). The depletion of MmpL3 arrests bacterial cell division, resulting in rapid death. This phenotype makes MmpL3 an attractive target for new putative anti-TB drugs. MmpL3 is inhibited by a large number of compounds with different chemical scaffolds, including the adamantyl urea AU1235 (22), the 1,5-diarylpyrrole derivative BM212 (45), the benzimidazole C215 (46), the diamine SQ109 (38), indoleamides (47), the acetamide E11 (48), the benzothiazole amides (49), and HC2091 {N-[2-(4-chlorophenyl)ethyl]-4-thiophen-2-yloxane-4-carboxamide} (50). The PIPD1-piperidinol-based molecule was identified as an MmpL3 inhibitor that is effective against the nontuberculous mycobacterium M. abscessus (51). Some MmpL3 inhibitors affect bacterial growth by disrupting membrane potential, decreasing intracellular ATP concentration, and inhibiting proton motive force (24, 52, 53). Some of the above-mentioned inhibitors (e.g., SQ109 and BM212) display activity against a broad spectrum of bacterial and fungal pathogens (28–30), while others (e.g., AU1235 and indole carboxamides) are specifically effective against mycobacteria (22, 27). The MmpL3 inhibitors also differ in their activities against nonreplicating tubercle bacilli. Latent bacilli are killed by SQ109 and BM212 (24–26) but not by AU1235, HC2091, and indole carboxamides (22, 27). It was also reported that MmpL3 inhibitors act synergistically with rifampin, bedaquiline, clofazimine, and β-lactams (54).
5-Bromo-2-(2-cyclohexylethyl)-1H-benzo[d]imidazole (EJMCh4) and 2-(2-cyclohexylethyl)-5,6-dimethyl-1H-benzo[d]imidazole (EJMCh6) were selected from a library of 25 derivatives and analogues of 1H-benzo[d]imidazole because they showed the most potent antitubercular activities (32). Both compounds appeared to also be active against intracellular M. tuberculosis residing in human phagocytes. Moreover, cytotoxicity of the investigated compounds toward human macrophages at high concentrations (60× and 512× MIC for compounds EJMCh4 and EJMCh6, respectively) was not observed. EJMCh4 appeared to be inactive against Escherichia coli at concentrations up to 12.5 μM but showed a bactericidal effect against Staphylococcus aureus at a concentration of 3.1 μM. Moreover, EJMCh6 was able to inhibit the growth of E. coli and S. aureus at a concentration of 14.5 μM (see Fig. S3 in the supplemental material). Mycolic acids and the MmpL3 protein are not present in S. aureus and E. coli, so it is likely that an alternative cellular process in these organisms is targeted by the investigated compounds. Similarly, it was reported that SQ109, an inhibitor of MmpL3 in M. tuberculosis, displayed bactericidal activity against Helicobacter pylori and Trypanosoma brucei, organisms lacking the mmpL3 gene (55, 56). Another MmpL3 inhibitor, the pyrrole derivative BM212 (45), was also identified as a compound with good activity against Candida albicans (57). Nonetheless, it was recently shown that BM212 binds MmpL3 directly and inhibits its flippase activity (35). However, SQ109 did not actually inhibit TMM flipping across the IM (35). We also cannot exclude that the 1H-benzo[d]imidazole derivatives investigated here act indirectly by targeting the proton motive force, which drives MmpL3 lipid translocation (26), or that the substitutions identified in MmpL3 in spontaneous mutants resistant to EJMCh4/EJMCh6 are compensatory, masking the inhibition of other cellular targets.
The mode of action of 1H-benzo[d]imidazole derivatives was identified by the selection of resistant mutants, sequencing of their genomes, the transfer of mutated mmpL3 genes identified in the resistant mutants, and the treatment of the wild-type strain with both compounds followed by the quantitative analysis of TDM, TMM, and MAs. The accumulation of mutations in mmpL3 that are responsible for resistance to compound EJMCh4 also affected the resistance to compound EJMCh6 and vice versa, suggesting that the compounds inhibit MmpL3 in the same way. However, the mutations affecting resistance to 1H-benzo[d]imidazole derivatives did not confer resistance in M. tuberculosis to the other MmpL3 inhibitor, SQ109. This finding might suggest that SQ109 and 1H-benzo[d]imidazole derivatives target different regions of the protein. A similar observation was recently made by Zheng et al. (50) while investigating targeting of MmpL3 in tubercle bacilli by HC2091. The mmpL3 mutations responsible for resistance of M. tuberculosis to HC2091 did not significantly affect the sensitivity of M. tuberculosis to SQ109. We were not able to test the potential cross-resistance between the 1H-benzo[d]imidazole derivatives and other inhibitors of mycobacterial MmpL3 that are not commercially available. Among the published inhibitors targeting mycobacterial MmpL3, the one most similar to our investigated compounds is C215 {N-(2,4-dichlorobenzyl)-1-propyl-1H-benzo[d]imidazol-5-amine}, which was identified in whole-cell-based high-throughput screening (46). The common part between C215 and the compounds evaluated in this study is 1H-benzo[d]imidazole. When comparing the two compounds (EJMCh6 and C215), it is worth asking how the presence of the benzimidazole system affects their properties and activity. Taking molecular weights into account, the benzimidazole system in C215 is only 35%, while in compound EJMCh6, it is 45%. From a chemical point of view, C215 is a secondary amine (NH group at C-5 of the benzimidazole system). It should be the most reactive fragment of the molecule and should be sensitive to metabolic changes (e.g., N-dealkylation). In case it is a prodrug, the sensitivity of that region will promote conversion to the active structure. Otherwise, rapid deactivation of the active substance may occur. C215 has two basic nitrogen atoms: the nitrogen atom of the amino group at the C-5 position of the benzimidazole system and nitrogen N-3. Compound EJMCh6 has only one: the nitrogen atom N-3. Each of the compounds still has an N-1 nitrogen atom with a weak acidic character. In C215, the N-1 atom is linked to the propyl substituent, and in compound EJMCh6, it has a proton. In our experience, compounds substituted at this position are associated with a decrease in tuberculostatic activity (32, 58). Both compounds have similar partial coefficient values. The log P for C215 is 4.22, and that for compound EJMCh6 is 4.95. Similarly, the dipole moment values are also close, i.e., 3.01 D for C215 and 3.28 D for EJMCh6. Thus, the bioavailabilities of the two compounds should be similar. However, when comparing molecular electrostatic potential (MEP) maps on the isodensity surface calculated at the B3LYP/6-31G(d) for both compounds, large differences can be noticed in both the spatial structure of the molecules and the distribution of charge (see Fig. S4 in the supplemental material).
In this study, we compared the locations of mutations in MmpL3 that affect resistance to a number of inhibitors (Fig. 2B; see Table S6 in the supplemental material). Five individual amino acid substitutions were identified in MmpL3 of strains resistant to compound EJMCh4 or EJMCh6, with only one (V684A) commonly identified in mutants resistant to C215 (46). Interestingly, the G253E mutation identified in the mutant resistant to compound EJMCh6 was reported in the strain resistant to AU1235 (22) and indole carboxamides (27). This residue is extremely close to Asp251, which was shown to be an essential residue for MmpL3 activity and presumably participates in the proton motive force that energizes the transporter (59, 60). V195 and V288 are in the spatial vicinity of G253, potentially suggesting that they may also affect the proton relay. As presented in the 3D model, V684 and V240 are also within near spatial proximity, suggesting that they may have the same effect on MmpL3 activity.
The mycobacterial cell envelope represents a rich target space for drug discovery. In recent years, a large number of putative MmpL3 inhibitors with diverse chemistry showing a potent bactericidal effect against M. tuberculosis have been discovered, including compounds which have reached phase IIb clinical trials (Sequella). The 1H-benzo[d]imidazole derivatives investigated here are promising candidates for new anti-TB drugs because of their bactericidal activity at nanomolar concentrations and very low cytotoxicity, with IC50 values over 60× and 512× MIC. The data collected so far with both compounds encourage further preclinical investigations to determine their potential to eradicate tubercle bacilli from the infected organism.
MATERIALS AND METHODS
Bacterial growth conditions and susceptibility tests.
The M. tuberculosis H37Rv and M. tuberculosis mc26230 strains (61) were grown at 37°C on Middlebrook 7H10 medium supplemented with 10% oleic acid-albumin-dextrose-catalase (OADC). For liquid culture, Middlebrook 7H9 broth (Difco) supplemented with OADC and 0.05% Tween 80 (pH 7.0) was used. To determine the MIC, the M. tuberculosis liquid cultures (OD600 = 0.1) were supplemented with various concentrations of the 1H-benzo[d]imidazole derivative compounds EJMCh4 and EJMCh6. Both compounds were dissolved in dimethyl sulfoxide (DMSO) and added directly to the growth medium. The final concentration of DMSO in the medium never exceeded 0.1% (vol/vol), and DMSO had no effect on the growth of M. tuberculosis. The growth-inhibitory effect was determined based on cell density (OD600) and CFU at 0, 24, 48, and 72 h after supplementation of M. tuberculosis cultures with the tested compounds by comparison to the control culture (without inhibitors). Colonies were counted after 4 weeks of incubation at 37°C.
The E. coli and S. aureus strains were cultured in Mueller-Hinton medium. The susceptibility of E. coli and S. aureus to the tested compounds was determined by the microdilution method, according to standard CLSI recommendations (62). The analysis was performed in 96-well microplates containing Mueller-Hinton broth. The concentrations of the agents in Mueller-Hinton broth ranged from 12.5 to 0.09 μM for compound EJMCh4 and from 14.5 to 0.11 μM for compound EJMCh6. A DMSO control was included in each experiment. The inoculum density was adjusted to a 0.5 McFarland standard. The microplates were incubated at 37°C for 18 h, and the OD600 was determined for bacterial cultures in the presence and absence of the tested compounds. MIC determinations were repeated at least three times.
Gene cloning strategies.
Standard molecular biology protocols were used for all cloning strategies (63). All PCR products were obtained using thermostable AccuPrime Pfx DNA polymerase (Invitrogen), cloned initially into a blunt vector (pJET1.2; Thermo Fisher), sequenced, and then released by digestion with appropriate restriction enzymes before ligation into the final vectors. All mmpL3 genes (with and without mutations) and their putative promoters (694-bp upstream region) were PCR amplified and cloned into the XbaI/HindIII restriction sites of the pMV306Km integration vector. All plasmids and oligonucleotides used in this work are listed in Tables S7 and S8 in the supplemental material, respectively.
Next-generation sequencing.
The sequencing libraries were prepared using the Nextera XT DNA sample preparation protocol (Illumina, USA). A total of 1 ng of genomic DNA isolated from the wild type and 6 individual M. tuberculosis mutants was used for preparation of paired-end libraries, in accordance with the manufacturer’s instructions. Whole-genome shotgun sequencing was performed on a NextSeq 500 platform at a read length of 2 × 150 bp (300 cycles). In silico/bioinformatical analyses were performed in CLC Biology Workbench 8.0 and 8.5.1 (Qiagen, USA). Raw sequencing reads were subjected to a quality and adapter trimming step and further aligned to the M. tuberculosis H37Rv reference sequence (NC_000962.3) (length fraction = 0.5, similarity fraction = 0.8). Detection of single-nucleotide polymorphism (SNP) variants in sequencing data derived from in vitro-reared resistant strains (carrying mutations) was performed using the Basic Variant Detection algorithm. The results of variant calling for each sample were filtered on the M. tuberculosis H37Rv wild starting strain (count 10<). Low-quality (quality, <10) variants were excluded from further analysis.
Preparation of human MDMs and in vitro cytotoxicity assay.
Human monocytes were isolated from commercially available (Regional Blood Donation Station, Lodz, Poland) and freshly prepared buffy coats from healthy human blood donors using a double density gradient technique employing Histopaque 1077 (Sigma) and 46% isosmotic Percoll (Sigma) as described previously (64). The differentiated human macrophages were extensively washed to remove any nonadherent cells, left resting overnight, and incubated with the culture medium supplemented with various concentrations of the tested compounds, namely, from 1 μM to 128 μM compound EJMCh4 and from 0.125 μM to 128 μM compound EJMCh6. After 48 h of incubation, the viability of macrophages was determined using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (Sigma), as described previously (64). All experimental and control samples were run in quadruplicate.
Evaluation of the bactericidal effect of 1H-benzo[d]imidazole derivatives on intracellularly growing tubercle bacilli.
The differentiated human macrophages were infected with M. tuberculosis at an MOI of 1:10 (see details in reference 64). Two hours after infection, the extracellularly located bacteria were extensively washed out with culture medium without antibiotics, incubated for an additional 1 hour in medium supplemented with 1 mg/ml gentamicin (Sigma), and finally washed three times with Iscove’s medium supplemented with 2% human AB serum (Sigma). The culture medium supplemented or not (control) with the 1H-benzo[d]imidazole derivatives was added to independent samples of the infected macrophages. The tested compounds EJMCh4 and EJMCh6 were applied in quadruplicate at the MICs, namely, 2.44 and 0.145 μM, respectively. The experimental (1H-benzo[d]imidazole derivative-treated) and control samples of the infected phagocytes were incubated at 37°C for 48 h under a humidified atmosphere of 10% CO2–90% air. Subsequently, the macrophages were lysed with 1 ml of 0.1% SDS (Sigma), and appropriate dilutions of the cell lysates were plated onto Middlebrook 7H10 agar supplemented with 10% Middlebrook oleic acid-albumin-dextrose-catalase (OADC) enrichment. After 21 days of incubation at 37°C, the CFU were counted. A one-way analysis of variance (ANOVA) (Holm-Sidak method) was employed for multiple comparisons versus the control samples to determine any significant differences between the mean values from the untreated and EJMCh4/EJMCh6-treated samples of the M. tuberculosis-infected human macrophages. Results were considered to be statistically significant at a P value of <0.05.
Whole-cell radiolabeling and lipid analysis.
To visualize compound-induced changes in the lipid profile, a late-log-phase M. tuberculosis mc26230 culture was incubated with increasing concentrations (1×, 2.5×, 5×, and 10× MIC) of compound EJMCh4 or EJMCh6 for 1 h. Subsequently, bacteria were radiolabeled for 6 h at 37°C by adding 1 μCi/ml of sodium [2-14C]acetate (56 mCi/mmol). The culture was divided into two volumes. One volume was used directly to derivatize and extract methyl esters of total mycolic acids (MAMEs) and fatty acids (FAMEs). From the second volume, polar and apolar lipid fractions were obtained prior to derivatization and extraction of MAMEs covalently bound to arabinogalactan (AG). The extraction procedures are described elsewhere (51, 65, 66). Equal counts (total MA, TMM, and TDM) or equal volumes (AG-bound MA) of each analyzed fraction were applied on a thin-layer chromatography (TLC) plate. Total MAMEs and FAMEs as well as arabinogalactan-bound MAME fractions were resolved twice using hexane-ethyl acetate (95:5, vol/vol). TMM and TDM were resolved once using a chloroform-methanol-water (40:8:1, vol/vol/vol) solvent system.
Mapping of the mutations in MmpL3.
A three-dimensional homology model of M. tuberculosis MmpL3 (Rv0206) was constructed using the Swiss model server (67) and the crystal structure of M. smegmatis MmpL3 as the template (PDB code 6AJF) (60).
Quantum chemical calculations.
Quantum chemical calculations were carried out to study the molecular geometry and electronic structure of benzimidazoles EJMCh6 and C215 using the Gaussian 03W software (Gaussian 03, revision A.1.; Gaussian, Inc., Wallingford, CT, USA). The full optimized geometries of both compounds in vacuum were calculated by the density functional theory-B3LYP method using the diffuse function 6-31G(d) basis set.
Fluctuation analysis.
Fluctuation analysis was performed as previously described (33). M. tuberculosis cultures were inoculated from freezer stocks. From the culture at an OD of 0.7 to 1.1, approximately 300,000 cells were used to inoculate 100 ml of Middlebrook 7H9 supplemented with 10% Middlebrook OADC, 0.0005% Tween 80, and 0.005% glycerol, giving a total cell count of 10,000 cells per 4-ml culture. This volume was immediately divided to start 20 cultures of 4 ml each. Cultures were grown at 37°C until they reached an OD of 1.0. The cultures were then spun at 4,000 rpm for 10 min at 4°C. Cultures were then resuspended in 250 μl of 7H9-OADC-Tween-glycerol and spotted onto 7H10-OADC-Tween-glycerol plates supplemented with 0.6 or 2.4 μM rifampin, 2.8 μM streptomycin, and 6.5 μM EJMCh4 or 0.38 μM EJMCh6. The drug resistance rate was determined by calculating m (the estimated number of mutations per culture) based on the number of mutants (r) observed on each plate using the Ma-Sarkar-Sandri (MSS) method as previously described (34). Dividing m by Nt, the number of cells plated for each culture, gives an estimated drug resistance rate.
Data availability.
Sequences for the three mutants selected on compound EJMCh4 and the three mutants selected on compound EJMCh6 have been deposited in GenBank under accession no. SRX5372947, SRX5372948, SRX5372946, SRX5372945, SRX5372944, and SRX5372943.
Supplementary Material
ACKNOWLEDGMENTS
This study was partially supported by grants from the National Science Centre, Poland (UMO-2014/15/B/NZ7/01002 to J.D. and UMO-2017/25/B/NZ7/00124 to K.G.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00441-19.
REFERENCES
- 1.WHO. 2017. WHO global tuberculosis report. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.WHO. 2015. WHO global tuberculosis report 2014. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 3.Paritala H, Carroll KS. 2013. New targets and inhibitors of mycobacterial sulfur metabolism. Infect Disord Drug Targets 13:85–115. doi: 10.2174/18715265113139990022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang M, Yue J, Yang YP, Zhang HM, Lei JQ, Jin RL, Zhang XL, Wang HH. 2005. Detection of mutations associated with isoniazid resistance in Mycobacterium tuberculosis isolates from China. J Clin Microbiol 43:5477–5482. doi: 10.1128/JCM.43.11.5477-5482.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheepsattayakorn A, Cheepsattayakorn R. 2012. Novel compounds and drugs and recent patents in treating multidrug-resistant and extensively drug-resistant tuberculosis. Recent Pat Antiinfect Drug Discov 7:141–156. doi: 10.2174/157489112801619683. [DOI] [PubMed] [Google Scholar]
- 6.Płocinska R, Korycka-Machala M, Plocinski P, Dziadek J. 2017. Mycobacterial DNA replication as a target for antituberculosis drug discovery. Curr Top Med Chem 17:2129–2142. doi: 10.2174/1568026617666170130114342. [DOI] [PubMed] [Google Scholar]
- 7.Hoagland DT, Liu J, Lee RB, Lee RE. 2016. New agents for the treatment of drug-resistant Mycobacterium tuberculosis. Adv Drug Deliv Rev 102:55–72. doi: 10.1016/j.addr.2016.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brennan PJ. 2003. Structure, function, and biogenesis of the cell wall of Mycobacterium tuberculosis. Tuberculosis (Edinb) 83:91–97. doi: 10.1016/S1472-9792(02)00089-6. [DOI] [PubMed] [Google Scholar]
- 9.Brennan PJ, Nikaido H. 1995. The envelope of mycobacteria. Annu Rev Biochem 64:29–63. doi: 10.1146/annurev.bi.64.070195.000333. [DOI] [PubMed] [Google Scholar]
- 10.Daffe M, Draper P. 1998. The envelope layers of mycobacteria with reference to their pathogenicity. Adv Microb Physiol 39:131–203. [DOI] [PubMed] [Google Scholar]
- 11.Draper P. 1998. The outer parts of the mycobacterial envelope as permeability barriers. Front Biosci 3:D1253–1261. doi: 10.2741/A360. [DOI] [PubMed] [Google Scholar]
- 12.Hancock IC, Carman S, Besra GS, Brennan PJ, Waite E. 2002. Ligation of arabinogalactan to peptidoglycan in the cell wall of Mycobacterium smegmatis requires concomitant synthesis of the two wall polymers. Microbiology 148:3059–3067. doi: 10.1099/00221287-148-10-3059. [DOI] [PubMed] [Google Scholar]
- 13.Pawelczyk J, Kremer L. 2014. The molecular genetics of mycolic acid biosynthesis. Microbiol Spectr 2:MGM2-0003-2013. doi: 10.1128/microbiolspec.MGM2-0003-2013. [DOI] [PubMed] [Google Scholar]
- 14.Vilcheze C, Jacobs WR. 2007. The mechanism of isoniazid killing: clarity through the scope of genetics. Annu Rev Microbiol 61:35–50. doi: 10.1146/annurev.micro.61.111606.122346. [DOI] [PubMed] [Google Scholar]
- 15.Mikusova K, Slayden RA, Besra GS, Brennan PJ. 1995. Biogenesis of the mycobacterial cell wall and the site of action of ethambutol. Antimicrob Agents Chemother 39:2484–2489. doi: 10.1128/aac.39.11.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xavier AS, Lakshmanan M. 2014. Delamanid: a new armor in combating drug-resistant tuberculosis. J Pharmacol Pharmacother 5:222–224. doi: 10.4103/0976-500X.136121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsumoto M, Hashizume H, Tomishige T, Kawasaki M, Tsubouchi H, Sasaki H, Shimokawa Y, Komatsu M. 2006. OPC-67683, a nitro-dihydro-imidazooxazole derivative with promising action against tuberculosis in vitro and in mice. PLoS Med 3:e466. doi: 10.1371/journal.pmed.0030466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gler MT, Skripconoka V, Sanchez-Garavito E, Xiao H, Cabrera-Rivero JL, Vargas-Vasquez DE, Gao M, Awad M, Park SK, Shim TS, Suh GY, Danilovits M, Ogata H, Kurve A, Chang J, Suzuki K, Tupasi T, Koh WJ, Seaworth B, Geiter LJ, Wells CD. 2012. Delamanid for multidrug-resistant pulmonary tuberculosis. N Engl J Med 366:2151–2160. doi: 10.1056/NEJMoa1112433. [DOI] [PubMed] [Google Scholar]
- 19.Singh R, Manjunatha U, Boshoff HI, Ha YH, Niyomrattanakit P, Ledwidge R, Dowd CS, Lee IY, Kim P, Zhang L, Kang S, Keller TH, Jiricek J, Barry CE. 3rd, 2008. PA-824 kills nonreplicating Mycobacterium tuberculosis by intracellular NO release. Science 322:1392–1395. doi: 10.1126/science.1164571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coxon GD, Craig D, Corrales RM, Vialla E, Gannoun-Zaki L, Kremer L. 2013. Synthesis, antitubercular activity and mechanism of resistance of highly effective thiacetazone analogues. PLoS One 8:e53162. doi: 10.1371/journal.pone.0053162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoagland D, Zhao Y, Lee RE. 2016. Advances in drug discovery and development for pediatric tuberculosis. Mini Rev Med Chem 16:481–497. doi: 10.2174/1389557515666150722101723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grzegorzewicz AE, Pham H, Gundi VA, Scherman MS, North EJ, Hess T, Jones V, Gruppo V, Born SE, Kordulakova J, Chavadi SS, Morisseau C, Lenaerts AJ, Lee RE, McNeil MR, Jackson M. 2012. Inhibition of mycolic acid transport across the Mycobacterium tuberculosis plasma membrane. Nat Chem Biol 8:334–341. doi: 10.1038/nchembio.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varela C, Rittmann D, Singh A, Krumbach K, Bhatt K, Eggeling L, Besra GS, Bhatt A. 2012. MmpL genes are associated with mycolic acid metabolism in mycobacteria and corynebacteria. Chem Biol 19:498–506. doi: 10.1016/j.chembiol.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li K, Schurig-Briccio LA, Feng X, Upadhyay A, Pujari V, Lechartier B, Fontes FL, Yang H, Rao G, Zhu W, Gulati A, No JH, Cintra G, Bogue S, Liu YL, Molohon K, Orlean P, Mitchell DA, Freitas-Junior L, Ren F, Sun H, Jiang T, Li Y, Guo RT, Cole ST, Gennis RB, Crick DC, Oldfield E. 2014. Multitarget drug discovery for tuberculosis and other infectious diseases. J Med Chem 57:3126–3139. doi: 10.1021/jm500131s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poce G, Bates RH, Alfonso S, Cocozza M, Porretta GC, Ballell L, Rullas J, Ortega F, De Logu A, Agus E, La Rosa V, Pasca MR, De Rossi E, Wae B, Franzblau SG, Manetti F, Botta M, Biava M. 2013. Improved BM212 MmpL3 inhibitor analogue shows efficacy in acute murine model of tuberculosis infection. PLoS One 8:e56980. doi: 10.1371/journal.pone.0056980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li W, Upadhyay A, Fontes FL, North EJ, Wang YH, Crans DC, Grzegorzewicz AE, Jones V, Franzblau SG, Lee RE, Crick DC, Jackson M. 2014. Novel insights into the mechanism of inhibition of MmpL3, a target of multiple pharmacophores in Mycobacterium tuberculosis. Antimicrob Agents Chemother 58:6413–6423. doi: 10.1128/AAC.03229-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao SP, Lakshminarayana SB, Kondreddi RR, Herve M, Camacho LR, Bifani P, Kalapala SK, Jiricek J, Ma NL, Tan BH, Ng SH, Nanjundappa M, Ravindran S, Seah PG, Thayalan P, Lim SH, Lee BH, Goh A, Barnes WS, Chen Z, Gagaring K, Chatterjee AK, Pethe K, Kuhen K, Walker J, Feng G, Babu S, Zhang L, Blasco F, Beer D, Weaver M, Dartois V, Glynne R, Dick T, Smith PW, Diagana TT, Manjunatha UH. 2013. Indolcarboxamide is a preclinical candidate for treating multidrug-resistant tuberculosis. Sci Transl Med 5:214ra168. doi: 10.1126/scitranslmed.3007355. [DOI] [PubMed] [Google Scholar]
- 28.Sacksteder KA, Protopopova M, Barry CE 3rd, Andries K, Nacy CA. 2012. Discovery and development of SQ109: a new antitubercular drug with a novel mechanism of action. Future Microbiol 7:823–837. doi: 10.2217/fmb.12.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biava M, Porretta GC, Manetti F. 2007. New derivatives of BM212: a class of antimycobacterial compounds based on the pyrrole ring as a scaffold. Mini Rev Med Chem 7:65–78. doi: 10.2174/138955707779317786. [DOI] [PubMed] [Google Scholar]
- 30.Ballell L, Bates RH, Young RJ, Alvarez-Gomez D, Alvarez-Ruiz E, Barroso V, Blanco D, Crespo B, Escribano J, González R, Lozano S, Huss S, Santos-Villarejo A, Martín-Plaza JJ, Mendoza A, Rebollo-Lopez MJ, Remuiñan-Blanco M, Lavandera JL, Pérez-Herran E, Gamo-Benito FJ, García-Bustos JF, Barros D, Castro JP, Cammack N. 2013. Fueling open-source drug discovery: 177 small-molecule leads against tuberculosis. ChemMedChem 8:313–321. doi: 10.1002/cmdc.201200428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gobis K, Foks H, Bojanowski K, Augustynowicz-Kopeć E, Napiórkowska A. 2012. Synthesis of novel 3-cyclohexylpropanoic acid-derived nitrogen heterocyclic compounds and their evaluation for tuberculostatic activity. Bioorg Med Chem 20:137–144. doi: 10.1016/j.bmc.2011.11.020. [DOI] [PubMed] [Google Scholar]
- 32.Gobis K, Foks H, Serocki M, Augustynowicz-Kopeć E, Napiórkowska A. 2015. Synthesis and evaluation of in vitro antimycobacterial activity of novel 1H-benzo[d]imidazole derivatives and analogues. Eur J Med Chem 89:13–20. doi: 10.1016/j.ejmech.2014.10.031. [DOI] [PubMed] [Google Scholar]
- 33.Ford CB, Shah RR, Maeda MK, Gagneux S, Murray MB, Cohen T, Johnston JC, Gardy J, Lipsitch M, Fortune SM. 2013. Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat Genet 45:784–790. doi: 10.1038/ng.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosche WA, Foster PL. 2000. Determining mutation rates in bacterial populations. Methods 20:4–17. doi: 10.1006/meth.1999.0901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu ZJ, Meshcheryakov VA, Poce G, Chng SS. 2017. MmpL3 is the flippase for mycolic acids in mycobacteria. Proc Natl Acad Sci U S A 114:7993–7998. doi: 10.1073/pnas.1700062114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chalut C. 2016. MmpL transporter-mediated export of cell-wall associated lipids and siderophores in mycobacteria. Tuberculosis (Edinb) 100:32–45. doi: 10.1016/j.tube.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 37.Viljoen A, Dubois V, Girard-Misguich F, Blaise M, Herrmann JL, Kremer L. 2017. The diverse family of MmpL transporters in mycobacteria: from regulation to antimicrobial developments. Mol Microbiol 104:889–904. doi: 10.1111/mmi.13675. [DOI] [PubMed] [Google Scholar]
- 38.Tahlan K, Wilson R, Kastrinsky DB, Arora K, Nair V, Fischer E, Barnes SW, Walker JR, Alland D, Barry CE, Boshoff HI. 2012. SQ109 targets MmpL3, a membrane transporter of trehalose monomycolate involved in mycolic acid donation to the cell wall core of Mycobacterium tuberculosis. Antimicrob Agents Chemother 56:1797–1809. doi: 10.1128/AAC.05708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Belardinelli JM, Yazidi A, Yang L, Fabre L, Li W, Jacques B, Angala SK, Rouiller I, Zgurskaya HI, Sygusch J, Jackson M. 2016. Structure-function profile of MmpL3, the essential mycolic acid transporter from Mycobacterium tuberculosis. ACS Infect Dis 2:702–713. doi: 10.1021/acsinfecdis.6b00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lamichhane G, Tyagi S, Bishai WR. 2005. Designer arrays for defined mutant analysis to detect genes essential for survival of Mycobacterium tuberculosis in mouse lungs. Infect Immun 73:2533–2540. doi: 10.1128/IAI.73.4.2533-2540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Griffin JE, Gawronski JD, Dejesus MA, Ioerger TR, Akerley BJ, Sassetti CM. 2011. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog 7:e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li W, Obregon-Henao A, Wallach JB, North EJ, Lee RE, Gonzalez-Juarrero M, Schnappinger D, Jackson M. 2016. Therapeutic potential of the Mycobacterium tuberculosis mycolic acid transporter, MmpL3. Antimicrob Agents Chemother 60:5198–5207. doi: 10.1128/AAC.00826-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Degiacomi G, Benjak A, Madacki J, Boldrin F, Provvedi R, Palu G, Kordulakova J, Cole ST, Manganelli R. 2017. Essentiality of mmpL3 and impact of its silencing on Mycobacterium tuberculosis gene expression. Sci Rep 7:43495. doi: 10.1038/srep43495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rock JM, Hopkins FF, Chavez A, Diallo M, Chase MR, Gerrick ER, Pritchard JR, Church GM, Rubin EJ, Sassetti CM, Schnappinger D, Fortune SM. 2017. Programmable transcriptional repression in mycobacteria using an orthogonal CRISPR interference platform. Nat Microbiol 2:16274. doi: 10.1038/nmicrobiol.2016.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.La Rosa V, Poce G, Canseco JO, Buroni S, Pasca MR, Biava M, Raju RM, Porretta GC, Alfonso S, Battilocchio C, Javid B, Sorrentino F, Ioerger TR, Sacchettini JC, Manetti F, Botta M, De Logu A, Rubin EJ, De Rossi E. 2012. MmpL3 is the cellular target of the antitubercular pyrrole derivative BM212. Antimicrob Agents Chemother 56:324–331. doi: 10.1128/AAC.05270-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stanley SA, Grant SS, Kawate T, Iwase N, Shimizu M, Wivagg C, Silvis M, Kazyanskaya E, Aquadro J, Golas A, Fitzgerald M, Dai HQ, Zhang LX, Hung DT. 2012. Identification of novel inhibitors of M. tuberculosis growth using whole cell based high-throughput screening. ACS Chem Biol 7:1377–1384. doi: 10.1021/cb300151m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lun S, Guo H, Onajole OK, Pieroni M, Gunosewoyo H, Chen G, Tipparaju SK, Ammerman NC, Kozikowski AP, Bishai WR. 2013. Indoleamides are active against drug-resistant Mycobacterium tuberculosis. Nat Commun 4:2907. doi: 10.1038/ncomms3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shetty A, Xu Z, Lakshmanan U, Hill J, Choong ML, Chng SS, Yamada Y, Poulsen A, Dick T, Gengenbacher M. 2018. Novel acetamide indirectly targets mycobacterial transporter MmpL3 by proton motive force disruption. Front Microbiol 9:2960. doi: 10.3389/fmicb.2018.02960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Graham J, Wong CE, Day JS, McFaddin E, Ochsner U, Hoang T, Young CL, Ribble W, DeGroote MA, Jarvis T, Sun XC. 2018. Discovery of benzothiazole amides as potent antimycobacterial agents. Bioorg Med Chem Lett 28:3177–3181. doi: 10.1016/j.bmcl.2018.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng H, Williams JT, Coulson GB, Haiderer ER, Abramovitch RB. 2018. HC2091 kills Mycobacterium tuberculosis by targeting the MmpL3 mycolic acid transporter. Antimicrob Agents Chemother 62:e02459-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dupont C, Viljoen A, Dubar F, Blaise M, Bernut A, Pawlik A, Bouchier C, Brosch R, Guerardel Y, Lelievre J, Ballell L, Herrmann JL, Biot C, Kremer L. 2016. A new piperidinol derivative targeting mycolic acid transport in Mycobacterium abscessus. Mol Microbiol 101:515–529. doi: 10.1111/mmi.13406. [DOI] [PubMed] [Google Scholar]
- 52.Foss MH, Pou S, Davidson PM, Dunaj JL, Winter RW, Pou S, Licon MH, Doh JK, Li Y, Kelly JX, Dodean RA, Koop DR, Riscoe MK, Purdy GE. 2016. Diphenylether-modified 1,2-diamines with improved drug properties for development against Mycobacterium tuberculosis. ACS Infect Dis 2:500–508. doi: 10.1021/acsinfecdis.6b00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feng X, Zhu W, Schurig-Briccio LA, Lindert S, Shoen C, Hitchings R, Li J, Wang Y, Baig N, Zhou T, Kim BK, Crick DC, Cynamon M, McCammon JA, Gennis RB, Oldfield E. 2015. Antiinfectives targeting enzymes and the proton motive force. Proc Natl Acad Sci U S A 112:E7073–E7082. doi: 10.1073/pnas.1521988112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li W, Sanchez-Hidalgo A, Jones V, de Moura VC, North EJ, Jackson M. 2017. Synergistic Interactions of MmpL3 inhibitors with antitubercular compounds in vitro. Antimicrob Agents Chemother 61:e02399-16. doi: 10.1128/AAC.02399-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li K, Wang Y, Yang G, Byun S, Rao G, Shoen C, Yang H, Gulati A, Crick DC, Cynamon M, Huang G, Docampo R, No JH, Oldfield E. 2015. Oxa, thia, heterocycle, and carborane analogues of SQ109: bacterial and protozoal cell growth inhibitors. ACS Infect Dis 1:215–221. doi: 10.1021/acsinfecdis.5b00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Makobongo MO, Einck L, Peek RM Jr, Merrell DS. 2013. In vitro characterization of the anti-bacterial activity of SQ109 against Helicobacter pylori. PLoS One 8:e68917. doi: 10.1371/journal.pone.0068917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Biava M, Cesare Porretta G, Deidda D, Pompei R, Tafi A, Manetti F. 2003. Importance of the thiomorpholine introduction in new pyrrole derivatives as antimycobacterial agents analogues of BM 212. Bioorg Med Chem 11:515–520. doi: 10.1016/S0968-0896(02)00455-8. [DOI] [PubMed] [Google Scholar]
- 58.Gobis K, Foks H, Suchan K, Augustynowicz-Kopeć E, Napiórkowska A, Bojanowski K. 2015. Novel 2-(2-phenalkyl)-1H-benzo[d]imidazoles as antitubercular agents. Synthesis, biological evaluation and structure-activity relationship. Bioorg Med Chem 23:2112–2120. doi: 10.1016/j.bmc.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 59.Bernut A, Viljoen A, Dupont C, Sapriel G, Blaise M, Bouchier C, Brosch R, de Chastellier C, Herrmann JL, Kremer L. 2016. Insights into the smooth-to-rough transitioning in Mycobacterium bolletii unravels a functional Tyr residue conserved in all mycobacterial MmpL family members. Mol Microbiol 99:866–883. doi: 10.1111/mmi.13283. [DOI] [PubMed] [Google Scholar]
- 60.Zhang B, Li J, Yang X, Wu L, Zhang J, Yang Y, Zhao Y, Zhang L, Yang X, Yang X, Cheng X, Liu Z, Jiang B, Jiang H, Guddat LW, Yang H, Rao Z. 2019. Crystal structures of membrane transporter MmpL3, an anti-TB drug target. Cell 176:636–648. doi: 10.1016/j.cell.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 61.Sambandamurthy VK, Derrick SC, Hsu T, Chen B, Larsen MH, Jalapathy KV, Chen M, Kim J, Porcelli SA, Chan J, Morris SL, Jacobs WR. Jr, 2006. Mycobacterium tuberculosis DeltaRDI DeltapanCD: a safe and limited replicating mutant strain that protects immunocompetent and immunocompromised mice against experimental tuberculosis. Vaccine 24:6309–6320. doi: 10.1016/j.vaccine.2006.05.097. [DOI] [PubMed] [Google Scholar]
- 62.Clinical Laboratory Standards Institute. 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 8th ed Clinical Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 63.Sambrook JF, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 64.Korycka-Machala M, Brzostek A, Dziadek B, Kawka M, Poplawski T, Witczak ZJ, Dziadek J. 2017. Evaluation of the mycobactericidal effect of thio-functionalized carbohydrate derivatives. Molecules 22:812–826. doi: 10.3390/molecules22050812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Halloum I, Carrere-Kremer S, Blaise M, Viljoen A, Bernut A, Le Moigne V, Vilcheze C, Guerardel Y, Lutfalla G, Herrmann JL, Jacobs WR Jr, Kremer L. 2016. Deletion of a dehydratase important for intracellular growth and cording renders rough Mycobacterium abscessus avirulent. Proc Natl Acad Sci U S A 113:E4228–E4237. doi: 10.1073/pnas.1605477113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kremer L, Guerardel Y, Gurcha SS, Locht C, Besra GS. 2002. Temperature-induced changes in the cell-wall components of Mycobacterium thermoresistibile. Microbiology 148:3145–3154. doi: 10.1099/00221287-148-10-3145. [DOI] [PubMed] [Google Scholar]
- 67.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T. 2018. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequences for the three mutants selected on compound EJMCh4 and the three mutants selected on compound EJMCh6 have been deposited in GenBank under accession no. SRX5372947, SRX5372948, SRX5372946, SRX5372945, SRX5372944, and SRX5372943.