During chronic biofilm infections, Pseudomonas aeruginosa bacteria are exposed to increased oxidative stress as a result of the inflammatory response. As reactive oxygen species (ROS) are mutagenic, the evolution of resistance to ciprofloxacin (CIP) in biofilms under oxidative stress conditions was investigated.
KEYWORDS: Pseudomonas aeruginosa, antibiotic resistance, biofilm, catalase mutant, oxidative stress
ABSTRACT
During chronic biofilm infections, Pseudomonas aeruginosa bacteria are exposed to increased oxidative stress as a result of the inflammatory response. As reactive oxygen species (ROS) are mutagenic, the evolution of resistance to ciprofloxacin (CIP) in biofilms under oxidative stress conditions was investigated. We experimentally evolved six replicate populations of P. aeruginosa lacking the major catalase KatA in colony biofilms and stationary-phase cultures for seven passages in the presence of subinhibitory levels (0.1 mg/liter) of CIP or without CIP (eight replicate lineages for controls) under aerobic conditions. In CIP-evolved biofilms, a larger CIP-resistant subpopulation was isolated in the ΔkatA strain than in the wild-type (WT) PAO1 population, suggesting oxidative stress as a promoter of the development of antibiotic resistance. A higher number of mutations identified by population sequencing were observed in evolved ΔkatA biofilm populations (CIP and control) than in WT PAO1 populations evolved under the same conditions. Genes involved in iron assimilation were found to be exclusively mutated in CIP-evolved ΔkatA biofilm populations, probably as a defense mechanism against ROS formation resulting from Fenton reactions. Furthermore, a hypermutable lineage due to mutL inactivation developed in one CIP-evolved ΔkatA biofilm lineage. In CIP-evolved biofilms of both the ΔkatA strain and WT PAO1, mutations in nfxB, the negative regulator of the MexCD-OprJ efflux pump, were observed while in CIP-evolved planktonic cultures of both the ΔkatA strain and WT PAO1, mutations in mexR and nalD, regulators of the MexAB-OprM efflux pump, were repeatedly found. In conclusion, these results emphasize the role of oxidative stress as an environmental factor that might increase the development of antibiotic resistance in in vivo biofilms.
INTRODUCTION
Antibiotic resistance is considered a major threat to modern medicine, challenging the treatment of common bacterial infections. The evolution of antibiotic resistance has primarily been studied in liquid cultures; however, in most natural environments and in some chronic infections bacteria grow in biofilms.
Biofilm-grown Pseudomonas aeruginosa experiences heterogeneous physiological and metabolic conditions in a compartmentalized environment which are not encountered in planktonic liquid cultures. These biofilm-specific conditions cause differences in the mutagenesis mechanisms and in the selective pressures acting on biofilm-forming cells compared to those in cells growing in planktonic cultures that might influence the evolution of antimicrobial resistance.
Distinct bacterial subpopulations have been described in biofilms, with a metabolically active population at the biofilm surface and a metabolically inactive population in the deeper layers of the biofilm (1). In addition, a steep oxygen gradient, with decreasing levels from the surface to the deeper layers, is observed in the colony biofilm model (2). The described endogenous oxidative stress in biofilms (3, 4) most probably occurs in the superficial layers of the biofilm, where enough oxygen is present and cells are metabolically active. Under microaerophilic or anaerobic conditions, such as those inside microcolonies, P. aeruginosa is able to respire on nitrogen oxides in the presence of nitrate, and bacterial cells may be exposed to nitrosative stress.
Among antibiotics that promote the evolution of resistance, fluoroquinolones such as ciprofloxacin (CIP) are of particular concern because they directly interfere with DNA replication by binding gyrases and topoisomerase IV and, hence, may encourage replication errors that are the major source of mutations. Gyrases bound with fluoroquinolone molecules result in cross-linked protein-DNA complexes containing broken DNA that induces an SOS response, which can contribute through activation of error-prone DNA polymerases to elevated mutation rates (5). In addition, CIP treatment causes increased intracellular reactive oxygen species (ROS) levels in planktonic cultures (6, 7). In biofilms, antibiotic-induced ROS production has been described in P. aeruginosa (8) and Proteus mirabilis (9) during treatment with quinolones and in Burkholderia cepacia complex during treatment with aminoglycosides (10).
It is widely accepted that patients with cystic fibrosis (CF) and chronic P. aeruginosa lung infection are exposed to increased oxidative stress mainly driven by chronic inflammation with polymorphonuclear leukocytes, a source of ROS, and by a CF-related deficit in antioxidants, such as glutathione, which further increases the inflammatory response, as we have previously shown in animal models (11). Thus, the P. aeruginosa biofilms in the CF lung are exposed to an oxidative stress environment, and our study aims at reproducing in vitro the oxidative stress by investigating the evolution of antibiotic resistance in P. aeruginosa lacking the major catalase KatA, an important antioxidant defense system.
Given that ROS can damage DNA, membranes, lipids, and proteins, P. aeruginosa possesses different antioxidant defenses to survive during aerobic growth and under antibiotic attack, with one of these being the katA-encoded major catalase. P. aeruginosa has three differentially evolved catalase genes, katA, katB, and katE (katC). KatA is the major, constitutively expressed catalase, which is highly produced in all phases of growth but is increased in the stationary-growth phase (12). KatA is important for the resistance of P. aeruginosa growing in planktonic and biofilm states when the bacteria are exposed to H2O2 at high concentrations (12). KatB is induced in both planktonic and biofilm cells in response to H2O2 exposure but plays a relatively small role in biofilm resistance (12, 13).
It has been suggested that the bactericidal effect of beta-lactams, fluoroquinolones, and aminoglycosides has a ROS-dependent component (14) although the significance of this has been challenged (15–17). Biofilms formed by mutants lacking antioxidant systems such as catalases (katA for P. aeruginosa and katB for B. cepacia) showed increased sensitivity to antibiotics (8, 10), and this is regarded as a solid indication of a contribution by ROS to the bactericidal effect of antibiotics.
In addition, it has been shown in biofilms that antioxidant systems, such as catalases and superoxide dismutases, are upregulated by the activation of the stringent response (18, 19). This suggests that inhibiting oxidative stress is an important strategy used by biofilm-forming bacteria to increase antibiotic tolerance. Moreover, the analysis of gene expression in P. aeruginosa biofilms revealed that antioxidant enzymes, including katA, are downregulated during biofilm growth in comparison to the level in planktonic culture (2).
It has been shown that the KatA catalase, in addition to its role in protection against ROS-dependent mutagenesis, also plays a critical role in nitric oxide buffering produced under anaerobic respiration in the presence of nitrate (20). Recently, two different promoters of katA have been identified in P. aeruginosa, one coping with ROS produced under aerobic respiration and the other coping with reactive nitrogen species (RNS) produced under anaerobic respiration (21).
As the antibiotic-tolerant biofilm-grown P. aeruginosa bacteria use both aerobic and microaerophilic respiration, the hypothesized production of mutagenic ROS and RNS in the different biofilm layers might lead to increased mutagenesis and faster development of antibiotic resistance in a ΔkatA mutant.
To investigate the role of KatA in biofilm mutagenesis and the development of resistance to antibiotics, we experimentally evolved a catalase mutant (ΔkatA) of P. aeruginosa (22) in colony biofilm under aerobic and anaerobic conditions in the presence and absence of subinhibitory concentrations of CIP. In addition, evolution in the presence and absence of antibiotic was conducted in stationary-phase aerobic planktonic cultures.
In the present study, we show that in aerobically grown biofilms the CIP-resistant population was larger in the ΔkatA strain than in the WT PAO1 strain. Population sequencing showed a higher number of mutations in ΔkatA populations than in WT PAO1 bacteria, suggesting that lack of catalase KatA might promote development of antibiotic resistance in P. aeruginosa aerobically grown biofilms. However, CIP-resistant mutants occurred also during evolution of WT PAO1 biofilms grown anaerobically, suggesting that development of resistance to CIP can occur even in the absence of oxygen. In addition, we show that, similar to findings of an experimental evolution study of wild-type P. aeruginosa PAO1 (23), a larger CIP-resistant population developed in ΔkatA biofilm than in planktonic cultures, confirming the biofilm mode of growth as a promoter of the development of low-level resistance.
RESULTS
Development of a larger CIP-resistant ΔkatA P. aeruginosa subpopulation in evolved aerobic biofilms than in planktonic cultures.
The development of CIP-resistant subpopulations during experimental evolution in the presence or absence of 0.1 mg/liter CIP (CIP or CTRL culture, respectively) was investigated by plating the evolved biofilm and planktonic ΔkatA populations (six and three replicates, respectively) at different passages on Luria-Bertani (LB) plates containing 0.5, 1, and 2 mg/liter CIP.
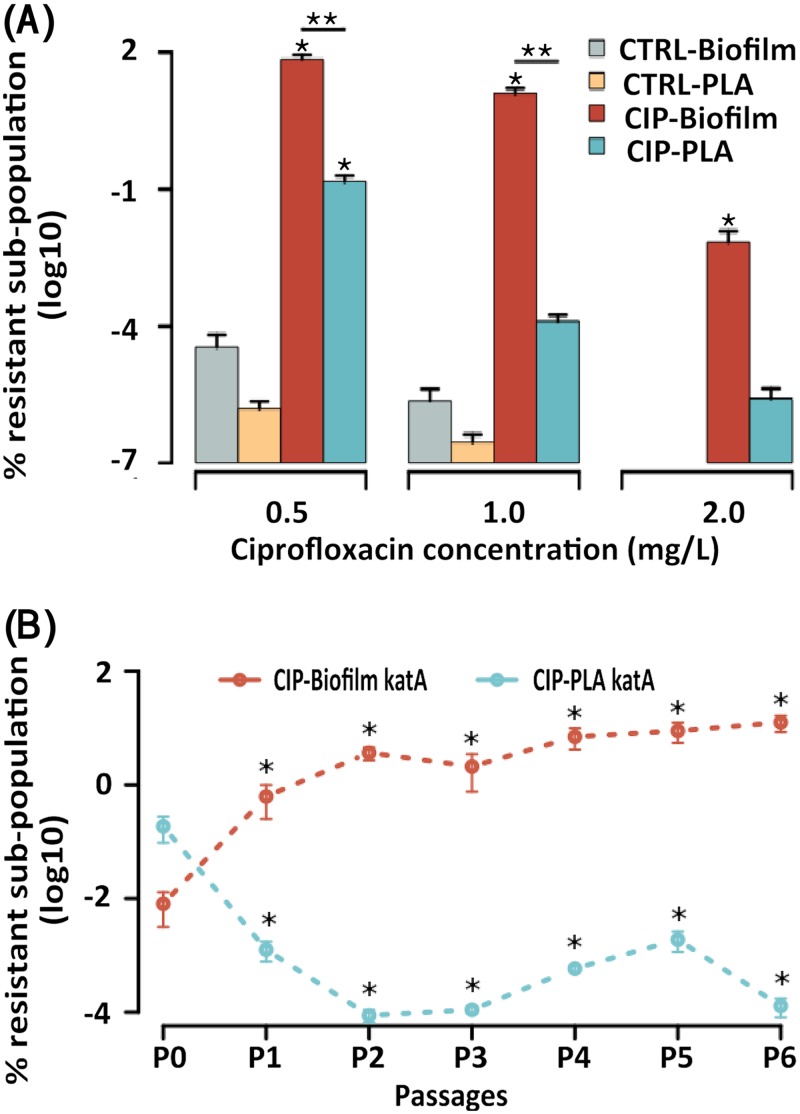
ΔkatA biofilm CIP-evolved populations showed a significantly larger resistant subpopulation on 0.5 and 1 mg/liter CIP (P = 0.002 and 0.003, respectively; t test) than ΔkatA planktonic CIP-evolved populations (Fig. 1A). The dynamics of resistance development during passages showed that in comparison to planktonic ΔkatA CIP populations, biofilm ΔkatA CIP populations developed a significantly larger resistant subpopulation (on 1 mg/liter CIP) at passage 1 (P1; P = 0.04), P2, P3 (P = 0.001), P4 (P = 0.003), and P5 and P6 (P = 0.002 and 0.003, respectively, t test) (Fig. 1B). In ΔkatA CIP biofilm, there was a significant increase in the size of the resistant subpopulation that developed from P0 to P6 on 0.5 and 1 mg/liter CIP (P = 0.0001 and 0.001, t test, respectively).
FIG 1.
(A) The size of P. aeruginosa Δkat biofilm and planktonic (PLA) populations recovered from 0.5, 1, and 2 mg/liter CIP after evolution in the presence of CIP (0.1 mg/l) or in the absence of antibiotic (CTRL). The values represent the means (SEM) of the replicates for each growth condition. *, significantly larger population than in the control population; **, significantly larger population in the biofilm than in the planktonic culture. (B) The development of CIP resistance in ΔkatA biofilm and planktonic (PLA) populations recovered from 1 mg/liter CIP during passages P0 to P6. Under exposure to ciprofloxacin, ΔkatA CIP biofilm populations developed a significantly larger resistant subpopulation than planktonic populations at P1, P2, P3, P4, P5, and P6 (P = 0.04, 0.001. 0.001, 0.003, 0.002, and 0.003, respectively). The values represent the means (SEM) for replicates for each growth condition. *, significantly larger resistant subpopulation in biofilms than in the planktonic population.
Compared to the evolved CTRL populations (eight replicates), CIP-evolved ΔkatA biofilm populations developed a significantly large resistant subpopulation on 0.5, 1 (P < 0.0001), and 2 mg/liter CIP (P = 0.01) (but only on 0.5 mg/liter for the planktonic ΔkatA population) (Fig. 1A).
The ancestor ΔkatA strain, which was used for initiating the evolution experiments, was tested for the preexistence of resistance variants, but no growth on 0.5, 1, and 2 mg/liter CIP was observed, indicating that the CIP-resistant colonies were not present before the initial CIP treatment but developed during the experimental evolution study.
A larger CIP-resistant subpopulation was observed in the ΔkatA strain at passage 3 (P = 0.01), passage 4 (P = 0.02), passage 5 (P = 0.02), and passage 6 (P = 0.03) than in WT PAO1 (23) (see Fig. S1 in the supplemental material).
Evolution of CIP resistance can occur in anaerobically evolved biofilms.
In order to investigate the role of oxygen in development of CIP resistance, we evolved PAO1 and ΔkatA colony biofilms in anaerobic chambers in the presence and absence of 0.1 mg/liter CIP on LB plates supplemented with 1 mM KNO3, a concentration resembling the concentrations in CF sputum (24). The size of the biofilm population was lower under anaerobic than under aerobic conditions, probably due to the consumption of KNO3 during growth (Table S1). We observed occurrence of CIP-resistant colonies (MIC = 1 mg/liter) during anaerobic evolution with CIP in PAO1 (one of the two lineages) but not in the ΔkatA biofilm populations (data not shown). The population analysis of the anaerobically evolved biofilms showed that the percentages of the CIP-resistant subpopulation on 1 mg/liter CIP in one of the WT PAO1 lineages were 0, 0, 0.05, 0.26, 0.65, and 0.33% from passage 1 to passage 6, respectively.
This result suggests that mutagenic mechanisms independent of ROS and RNS formation, such as the SOS response, might play a role in mutagenesis toward ciprofloxacin resistance under anaerobic conditions, similar to the conditions described in CF mucus (25, 26).
Hypermutators evolve in aerobic ΔkatA biofilm populations and attain high MICs.
The mutation frequencies were measured in CIP biofilm and planktonic lineages throughout the passages.
In one out of six independent replicate lineages of ΔkatA biofilm, we observed increased mutation frequencies corresponding to hypermutable phenotypes at the end of the experiment (P6) (>3 × 10−7). Analysis of the mutation frequencies at the different passages in this lineage showed that already after the first 48 h of exposure to ciprofloxacin (passage 0), the bacterial population had a hypermutable phenotype, and this was maintained during passages until the end of the evolution experiment. This was not observed in the other biofilm or planktonic lineages of the ΔkatA strain or WT PAO1.
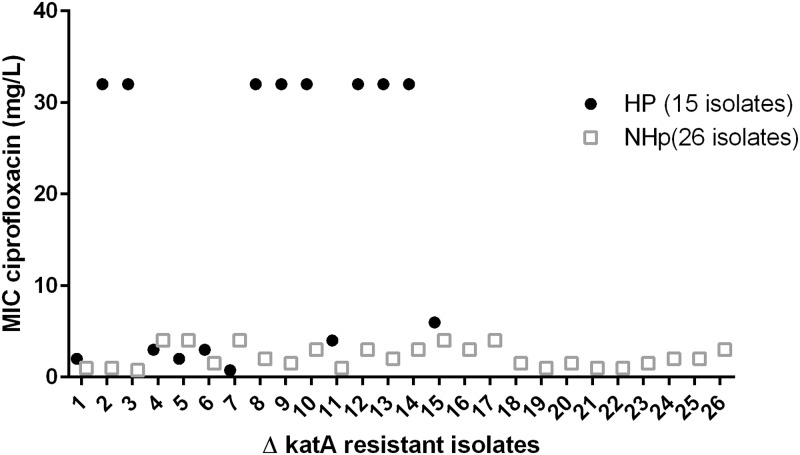
The MICs for ciprofloxacin of the resistant isolates collected at the first and last passages of the experimental evolution of the ΔkatA strain showed that the isolates with the highest MICs were selected from the lineage with the hypermutable phenotype (Fig. 2).
FIG 2.
The MIC of ciprofloxacin for resistant colonies isolated from CIP plates of the ΔkatA P. aeruginosa population from the hypermutable (HP) lineage and the nonhypermutable (NHp) lineage.
Genetic evolution of P. aeruginosa ΔkatA biofilm and planktonic cultures under ciprofloxacin exposure and aerobic conditions.
To get insight into the underlying genetic changes contributing to the accelerated development of antibiotic resistance in biofilms in the absence of catalase, we sequenced the ΔkatA populations at the endpoint of the experimental evolution. A complete list of mutations under the different conditions is presented in Table S3. The identified genetic changes, defined as a minimum variant frequency of 10%, were also compared to those observed in WT PAO1 (23).
The highest number of mutations was observed in CIP-evolved biofilm populations of the ΔkatA strain (Table 1).
TABLE 1.
The types of mutations in evolved populations of WT PAO1 and P. aeruginosa ΔkatA
| Strain and condition | No. (%) of mutations by typea
|
Total no. of mutations | dN/dS | ||||
|---|---|---|---|---|---|---|---|
| Indel | Transition | Transversion | Large indel | Inversion duplication | |||
| PAO1 | |||||||
| CTRL biofilm | 24 (28) | 13 (15) | 48 (56) | 85 | 0.35 | ||
| CTRL planktonic | 24 (26) | 22 (23) | 48 (51) | 94 | 0.25 | ||
| CIP biofilm | 60 (37) | 27 (16) | 77 (47) | 164 | 0.44 | ||
| CIP planktonic | 29 (22) | 44 (34) | 57 (44) | 130 | 0.45 | ||
| ΔkatA strain | |||||||
| CTRL biofilm | 147 (22) | 58 (9) | 452 (69) | 1 | 658 | 1.36 | |
| CTRL planktonic | 141 (24) | 70 (12) | 362 (63) | 5 | 578 | 1.24 | |
| CIP biofilm | 301 (28) | 206 (19) | 557 (52) | 2 | 9 | 1075 | 0.90 |
| CIP planktonic | 150 (22) | 65 (10) | 469 (69) | 6 | 7 | 697 | 2.59 |
The numbers and types of mutations in evolved PAO1 populations were reanalyzed including nonsynonymous mutations in hypothetical proteins (not included in reference 23).
According to gene functional categories, CIP-treated ΔkatA biofilm and planktonic populations have a significantly higher number of mutations than control (CTRL) populations in the category secreted factors (toxins, enzymes) (P = 0.044 and 0.021, respectively, t test) (Fig. S2).
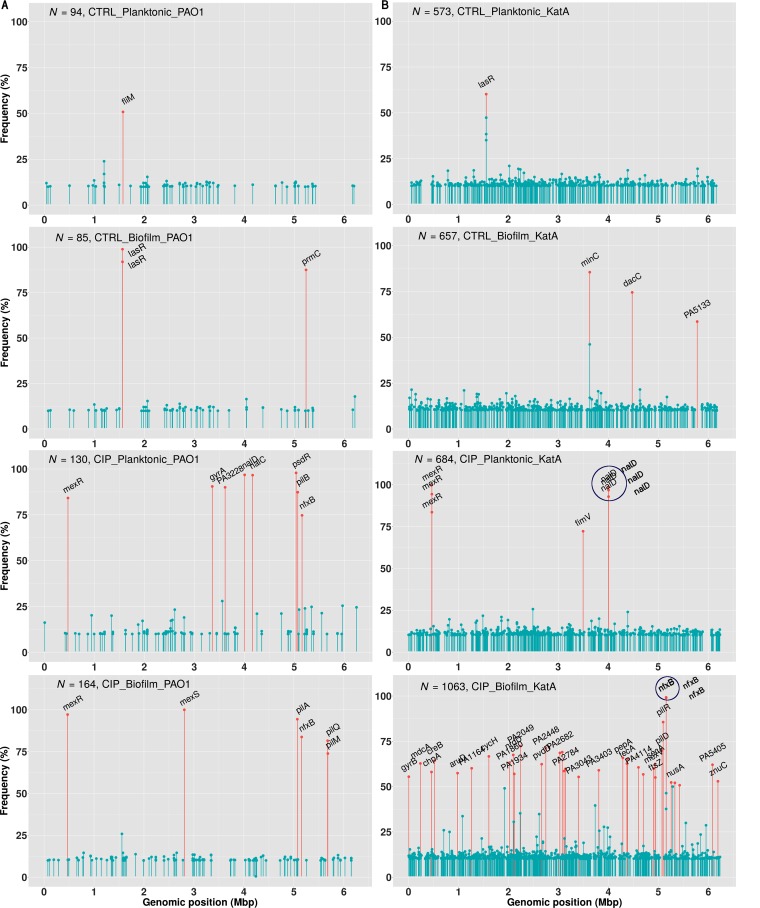
CIP-evolved ΔkatA populations contain a large number of mutations encountered with high frequency compared to levels in the CTRL ΔkatA populations and in the WT PAO1 population evolved under the same conditions (Fig. 3).
FIG 3.
The frequency of each mutation (y axis) and its genomic location (x axis) in the WT PAO1 (A) and ΔkatA strain (B) biofilm and planktonic populations under different conditions. Red bars represent mutations with frequencies of >50%, and blue bars represent mutations with frequencies of <50%. The large dark blue circles mark genes with mutations at the same position occurring in several lineages with similar frequencies.
Large indels, inversions, and duplications were present only in the CIP-evolved ΔkatA populations and were not present in the populations of WT PAO1 evolved in the absence or presence of CIP. The ratio of nonsynonymous to synonymous mutations, dN/dS, was >1 in the evolved ΔkatA population, suggesting positive selection for the described mutations which was not observed in the evolved WT PAO1 populations (dN/dS < 1). Analysis of the mutational spectrum (Table S2) revealed that the most frequent mutation in ΔkatA populations was the transversion A·T→C·G. Mutations which are repeatedly observed after independent exposures to a condition provide strong evidence of adaptive evolution (Fig. 3 and Table S3). This was the case for nfxB, a negative regulator of MexCD-OprJ (27, 28), which was frequently and repeatedly mutated in ΔkatA CIP-evolved biofilm populations, as well as pil genes encoding type IV pili, in contrast to observations in CTRL biofilms. Mutations in other mex genes (mexR, mexD, mexF, and mexT) or RND efflux pumps were also observed in several lineages with lower frequencies. A large replacement of 39 nucleotides with 31 nucleotides was observed in mexT in one of the CIP biofilm lineages (Fig. S3).
The genetic background of the observed hypermutable phenotype in one of the ΔkatA CIP biofilm lineages was shown to be an insertion of 27 nucleotides in the mutL gene (which codes for a DNA mismatch repair protein) (Fig. S3), and, as expected, the number of transitions in this lineage was higher than the levels in the nonhypermutable lineages.
The mutation C1397T in gyrB coding for DNA gyrase subunit B, leading to the amino acid change S466F, was observed in the hypermutable lineages of the CIP-evolved ΔkatA biofilms, correlating with the high CIP MIC of the resistant isolates from this lineage.
Mutations in either mexR or nalD, which are regulators of the MexAB-OprM efflux pump, were frequent in CIP-exposed planktonic ΔkatA lineages. Two different large indels were detected in mexR in two lineages (Fig. S3).
In CIP-evolved biofilm ΔkatA populations, genes related to iron acquisition and transport were mutated in several lineages: genes encoding the siderophores pyoverdin (pvd) and pyochelin (pch), iron transporters (iron transport system permease HitB), and pyrroloquinoline quinone (PQQ) biosynthesis genes (pqq) encoding a redox-sensing protein (Table S2). In evolved biofilm and planktonic ΔkatA populations, mutations in TonB-dependent receptors were found. TonB proteins are essential components in iron-siderophore uptake in bacteria (29), and mutations in these genes were not observed in the evolved populations of WT PAO1.
In the ΔkatA CTRL biofilm compared to planktonic CTRL populations (evolved without CIP exposure), minC, which acts an inhibitor for cell division by inhibiting Z-ring assembly, was observed to be mutated in three different lineages.
Genetic basis of resistance in CIP-resistant colonies isolated from anaerobically evolved biofilms.
The genetic basis of the six CIP-resistant colonies (MIC = 1 mg/liter) isolated from the anaerobically evolved PAO1 biofilm was investigated by sequencing the nfxB gene, and a CTTCT deletion at position 162 leading to frameshift was found in all isolates.
DISCUSSION
Evolution of P. aeruginosa biofilms under exposure to subinhibitory concentrations of CIP resulted in a larger CIP-resistant subpopulation in the ΔkatA than in the WT PAO1 biofilm populations, indicating that the lack of KatA catalase accelerates the evolution of antibiotic resistance under aerobic conditions. Given the role of KatA in the defense against oxidative and nitrosative stress (20), we propose that these stresses play a role in the biofilm-related increased mutagenesis. Respiration by denitrification of a P. aeruginosa biofilm population exposed to low-oxygen tension could be supported by the nitrate present in LB medium (approximately 20 μM NO3−) (24). In addition, it has been shown by transcriptomic (30) and proteomic (31) studies that under CIP treatment P. aeruginosa switches to anaerobic respiration.
In support of the oxidative stress mechanism acting in aerobically evolved populations of the ΔkatA strain but not of the WT PAO1 strain are the repeatedly observed mutations in iron assimilation genes, such as mutations in genes encoding TonB-dependent receptors which are essential for iron-siderophore uptake in P. aeruginosa, mutations in genes encoding the siderophores pyoverdine and pyochelin, and mutations in genes encoding various iron binding and redox proteins (PQQ). Mutations in these genes might represent a protective mechanism against iron uptake, probably as a defense mechanism against increased intracellular production of ROS by the Fenton reaction (32) as increased ROS production was measured in ΔkatA biofilms compared to levels in WT PAO1 biofilms (8). Interestingly, it has been reported that genes encoding TonB-dependent receptors were preferentially deleted in P. aeruginosa isolates from CF patients during adaptation in the CF lung (29).
Although important for occurrence of mutagenic resistance in biofilms, oxidative stress is not the only mechanism, as we have observed CIP-resistant mutants also when PAO1 biofilms were evolved under anaerobic conditions. It is unclear at this time which mutagenesis mechanisms occur under anaerobic conditions, but one might consider the SOS stress response as a possible mechanism.
However, the accelerated development of antibiotic resistance in biofilms under exposure to subinhibitory levels of ciprofloxacin in aerobic biofilms of the ΔkatA strain compared to that in WT PAO1 aerobic biofilms was not observed during the evolution in planktonic cultures (see Fig. S1 in the supplemental material). Although we do not have an explanation for this, we can speculate that this might be due to accumulation of deleterious mutations during evolution in the planktonic ΔkatA population and not in the WT PAO1.
Compared to the planktonic populations, larger CIP-resistant subpopulations were observed in biofilms of both the ΔkatA and PAO1 strains (23), confirming that the biofilm mode of growth promotes development of mutational resistance in experimental evolution studies (Fig. S1).
The genomic analysis of the aerobically evolved populations showed that the highest number of mutations was observed in the CIP-evolved ΔkatA biofilm populations, and this is in agreement with the hypothesis that ciprofloxacin exposure induces mutations either via the SOS response or through increased ROS levels in the catalase-deficient mutants compared to levels in the WT PAO1 (3, 33). Analysis of the mutational spectra revealed that the A·T→C·G transversion was the most frequent mutation type in ΔkatA populations, and this has been previously shown to be related to unrepaired oxidized guanine in the nucleotide pool (34). The genes belonging to the functional category of secreted factors (toxins and enzymes) were also mutated in a significantly higher number in CIP-evolved ΔkatA biofilm and planktonic populations than in CTRL biofilms and planktonic populations, and this is in accordance with our previous observations of loss of virulence factors during evolution in the presence of CIP (30).
During the experimental evolution of ΔkatA biofilms, mutL mutants with a mutator phenotype and high MICs of ciprofloxacin evolved, which corresponds with our previous study on the association in chronically infected CF patients between bacterial hypermutability and chronic inflammation, which is a source of chronic oxidative stress (35). The high MIC levels of ciprofloxacin in the mutator lineage were found to be related to mutations in the CIP target gene, gyrB. Interestingly, a mutation at the same site, C1397T causing the substitution S466F, has been previously found in planktonic experimental evolution of a hypermutator strain due to impaired repair of oxidative lesions (PAOMY-Mgm) (36, 37), and this amino acid change has been described in levofloxacin-nonsusceptible clinical P. aeruginosa isolates (38).
The genes found to be mutated in evolved biofilms or planktonic cultures of both the PAO1 and ΔkatA strains strongly suggest parallel and distinct evolutionary trajectories in the different modes of growth of P. aeruginosa. Confirming previous results, the experimental evolution of biofilm and planktonic ΔkatA P. aeruginosa populations revealed that low-level CIP resistance develops readily in biofilms (23). For example, in CIP-evolved ΔkatA and WT PAO1 biofilms, mutations in the negative regulator, nfxB, of the MexCD-OprJ efflux pump were frequently found. nfxB mutations have previously been identified in P. aeruginosa isolates from CF patients (28). Mutations in pil genes encoding type IV pili were found frequently in CIP-evolved biofilm populations of both the ΔkatA and WT PAO1 strains, while in CIP-evolved planktonic populations of both the ΔkatA and WT PAO1 strains, mutations in the negative regulator mexR and nalD of MexAB-OprM were found. These subpopulations with biofilm-related, low-level resistance may accumulate further mutations which can further increase the MIC of the population. This dynamic of resistance development under exposure to subinhibitory levels of ciprofloxacin emphasizes the importance of treatment of the infections caused by biofilm-growing P. aeruginosa with doses ensuring a high antibiotic concentration at the site of infection that can eliminate the first-step mutants or with combination therapy.
Mutations in the target genes of ciprofloxacin, gyrA and gyrB, were found only in the CIP-evolved planktonic populations of WT PAO1 (23) but not in those of the nonhypermutable ΔkatA lineages, which phenotypically correlated to high MIC levels in planktonic WT PAO1 but not in the ΔkatA populations.
In conclusion, this study of the experimental evolution of the ΔkatA strain in biofilm and planktonic growth complements our earlier findings of the study of evolution in PAO1, emphasizing the role of environmental stresses such as oxidative and nitrosative stresses and SOS responses for the mutational landscape and the development of antibiotic resistance, which might play an important role in vivo during chronic infections (39). A pitfall of all of the correlative analysis between whole-genome sequencing (WGS) and phenotypic susceptibility is that it fails to capture the contribution of gene expression, an important contributor to tolerance and resistance to antibiotics. Complementary analysis such as transcriptome sequencing (RNA-seq) is the next step for correlating the susceptibility phenotype with genetic content.
MATERIALS AND METHODS
Bacterial strains, media and antibiotics.
P. aeruginosa ΔkatA strain (22) (planktonic culture ciprofloxacin MIC of 0.094 mg/liter) was used to test the development of antibiotic resistance during experimental evolution in the colony biofilm model (23, 40) and in planktonic batch cultures. Both biofilms and planktonic cultures were grown in Luria-Bertani (LB) medium and exposed to 0.1 mg/liter CIP (ciprofloxacin hydrochloride; Bayer; Germany). The MIC for a 48-h culture of ΔkatA in stationary-growth phase was 0.2 mg/liter, and the minimal biofilm inhibitory concentration was 0.5 mg/liter. The spontaneous mutation rate to 0.5 mg/liter CIP was 1E−8 for PAO1 and 1.4E−8 for the ΔkatA strain, as measured by a fluctuation test.
Experimental evolution of colony biofilm and planktonic cultures.
The experimental evolution of colony biofilms and planktonic cultures was conducted as previously described (23). In short, a single colony of P. aeruginosa ΔkatA was used to inoculate LB medium for an overnight culture. Five microliters of the diluted overnight culture containing approximately 106 cells (optical density at 600 nm [OD600] adjusted to 0.05 and diluted 1:10) was spot inoculated on the top of polycarbonate membrane filters (Whatman Nuclepore track-etched membranes, 25-mm diameter, 0.2-μm pore size) and incubated at 37°C to form 48-h colony biofilms on LB plates. Membranes bearing 48-h colony biofilms were transferred to fresh LB plates with either 0.1 mg/liter CIP (1/5 minimal biofilm inhibitory concentrations) (CIP culture) or without CIP for 48 h (CTRL culture) (passage 0 consisted of a 4-day-old biofilm, produced by 2 days on LB medium followed by 2 days on LB medium with CIP). Every 48 h the CIP and CTRL membranes were transferred in 10-ml tubes with saline, and biofilms were dispersed by vortexing and sonication. After the OD600 was adjusted to 0.005, the bacterial suspension of CTRL biofilms was used to start new biofilms on LB plates without antibiotics (CTRL). The bacterial suspensions of CIP biofilms were used to start new biofilms on plates with 0.1 mg/liter CIP (CIP).
A total of seven exposures to CIP (from passage 0 [P0] to P6) with six independent lineages were performed.
Two parallel lineages of WT PAO1 and P. aeruginosa ΔkatA were also evolved in an anaerobic chamber (Whitley A85 anaerobic workstation) for 7 passages (as described above). To allow bacterial growth by denitrification, the LB plates were supplemented with 1 mM KNO3.
After each 48 h, the CFU counts for each biofilm population were measured, and the disrupted biofilm populations were kept in 20% glycerol at −80°C until further analysis.
Planktonic experiments were conducted under aerobic conditions with the same experimental design as implemented with biofilm cultures. Briefly, 5 μl of an overnight culture of a single colony of P. aeruginosa ΔkatA was used to inoculate 10 ml of LB medium and incubated for 48 h with shaking on an orbital shaker (180 rpm) at 37°C. From the 48-h culture two flasks were inoculated: one with 0.1 mg/liter CIP (CIP) and one in LB medium (CTRL). From CIP stationary cultures, every 48 h new planktonic cultures were established in flasks with CIP. From CTRL stationary cultures, every 48 h new planktonic cultures were established in flasks with LB medium. This was repeated for six passages (P1 to P6).
Population analysis.
Bacterial populations (100 μl of different dilutions) obtained after sonication and vortexing of membranes containing colony biofilms or of planktonic cultures from each passage and treatment group were plated on LB plates to estimate the size of the bacterial population and on LB plates containing 0.5, 1, and 2 mg/liter CIP to estimate the size of the resistant subpopulations (growing on CIP concentrations higher than the MIC of the strain P. aeruginosa ΔkatA). The size of the resistant population was expressed as a percentage of the total bacterial population and calculated by dividing the number of CFU/milliliter on CIP plates by the number of CFU/milliliter on LB plates and multiplying by 100.
Three colonies were selected from the plates with the highest CIP concentrations allowing growth and passaged twice in antibiotic-free medium, and the MICs of CIP were determined by performing an Etest (bioMérieux) according to the manufacturer’s instructions.
Mutation frequencies and rates determination.
The mutation frequencies of the evolved populations after each passage were investigated on LB plates containing rifampin (300 mg/liter), as previously described (35). A population was considered hypermutable when the mutation frequency was 20-fold higher than the mutation frequency of the reference strain PAO1 (≥3 × 10−7). The mutation rates were determined by a fluctuation test as previously described (30), and calculations were performed using the bz-rates web tool (http://www.lcqb.upmc.fr/bzrates).
Genome sequencing of aerobically evolved bacterial populations.
The genome sequencing was performed as previously described (23). In short, the CIP-evolved populations (after 7 passages in the presence of 0.1 mg/liter CIP) from four P. aeruginosa ΔkatA biofilm lineages and three planktonic lineages were grown on LB plates containing 1 mg/liter CIP for 48 h to enrich for the resistant subpopulation. The CTRL-evolved biofilm and planktonic populations (four lineages/condition) were grown on LB plates. All colonies for each population were collected in 3 ml of saline (0.9% NaCl) for genomic DNA extraction using a Gentra Puregene Yeast/Bact DNA purification kit. The DNA was prepared for sequencing using an Illumina TruSeq DNA Nano kit and sequenced on an Illumina MiSeq system, yielding a coverage of approximately 5 million reads per sample. Sequencing reads were mapped to the reference genome of P. aeruginosa PAO1 (GenBank accession no. NC_002516), and single and multiple nucleotide variants (SNVs and MNVs) were called using CLC Genomics Workbench (Qiagen). Mutations present in the evolved ΔkatA CTRL populations were filtered out from the genome of the CIP-evolved populations (biofilm and planktonic). R (version 3.2.5) was used for further statistical analysis of the mutations detected in each population, and all mutations occurring in >10% of the reads were included in the analysis.
The Pseudomonas Genome Database was used for gene function analysis. The ratio of the rate of nonsynonymous substitutions to the rate of synonymous substitutions, dN/dS, was calculated as a measure of the selection pressure acting on the protein-coding genome, as previously described, assuming that 25% of all single-nucleotide polymorphisms (SNPs) result in synonymous changes (41). The dN/dS is expected to be >1 if natural selection promotes changes in protein sequences and <1 if natural selection suppresses changes.
Sequencing of nfxB was performed on CIP-resistant colonies selected from anaerobic experiments, as described previously (42).
Statistical analysis.
Graphs and statistical analysis were done using GraphPad Prism, version 7, software and R (version 3.2.5). We conducted a D’Agostino-Pearson test to check for normal distribution, and Student's t test was used for comparisons among populations (comparing CIP populations to CTRL populations and CIP biofilm populations to CIP planktonic populations) and to compare the level of resistance at different exposure time points (passages). The differences were considered significant when the P value was ≤0.05. Error bars in all graphs represent the standard errors of the means (SEM).
Supplementary Material
ACKNOWLEDGMENTS
A.P. and M.S. acknowledge support from the Novo Nordisk Foundation under NFF grant number NNF10CC1016517, the European Union H2020 (ERC-2014-STG) under grant agreement 638902, LimitMDR, and the Danish Council for Independent Research Sapere Aude Program DFF 4004-00213. M.N.A. acknowledges financial support from the Egyptian Ministry of Higher Education.
The technical assistance of Tina Wassermann and Janna Becker is highly appreciated.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00766-19.
REFERENCES
- 1.Stewart PS. 2015. Antimicrobial tolerance in biofilms. Microbiol Spectr 3(3):MB-0010-2014. doi: 10.1128/microbiolspec.MB-0010-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borriello G, Werner E, Roe F, Kim AM, Ehrlich GD, Stewart PS. 2004. Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob Agents Chemother 48:2659–2664. doi: 10.1128/AAC.48.7.2659-2664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boles BR, Singh PK. 2008. Endogenous oxidative stress produces diversity and adaptability in biofilm communities. Proc Natl Acad Sci U S A 105:12503–12508. doi: 10.1073/pnas.0801499105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Driffield K, Miller K, Bostock JM, O'Neill AJ, Chopra I. 2008. Increased mutability of Pseudomonas aeruginosa in biofilms. J Antimicrob Chemother 61:1053–1056. doi: 10.1093/jac/dkn044. [DOI] [PubMed] [Google Scholar]
- 5.Blázquez J, Couce A, Rodríguez-Beltrán J, Rodríguez-Rojas A. 2012. Antimicrobials as promoters of genetic variation. Curr Opin Microbiol 15:561–569. doi: 10.1016/j.mib.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 6.Dwyer DJ, Kohanski MA, Collins JJ. 2009. Role of reactive oxygen species in antibiotic action and resistance. Curr Opin Microbiol 12:482–489. doi: 10.1016/j.mib.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gutierrez A, Stokes JM, Matic I. 2018. Our evolving understanding of the mechanism of quinolones. Antibiotics (Basel) 7:32. doi: 10.3390/antibiotics7020032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen PO, Briales A, Brochmann RP, Wang H, Kragh KN, Kolpen M, Hempel C, Bjarnsholt T, Hoiby N, Ciofu O. 2014. Formation of hydroxyl radicals contributes to the bactericidal activity of ciprofloxacin against Pseudomonas aeruginosa biofilms. Pathog Dis 70:440–443. doi: 10.1111/2049-632X.12120. [DOI] [PubMed] [Google Scholar]
- 9.Aiassa V, Barnes AI, Albesa I. 2010. Resistance to ciprofloxacin by enhancement of antioxidant defenses in biofilm and planktonic Proteus mirabilis. Biochem Biophys Res Commun 393:84–88. doi: 10.1016/j.bbrc.2010.01.083. [DOI] [PubMed] [Google Scholar]
- 10.Van AH, Sass A, Bazzini S, De RK, Udine C, Messiaen T, Riccardi G, Boon N, Nelis HJ, Mahenthiralingam E, Coenye T. 2013. Biofilm-grown Burkholderia cepacia complex cells survive antibiotic treatment by avoiding production of reactive oxygen species. PLoS One 8:e58943. doi: 10.1371/journal.pone.0058943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jensen PO, Lykkesfeldt J, Bjarnsholt T, Hougen HP, Hoiby N, Ciofu O. 2012. Poor antioxidant status exacerbates oxidative stress and inflammatory response to Pseudomonas aeruginosa lung infection in guinea pigs. Basic Clin Pharmacol Toxicol 110:353–358. doi: 10.1111/j.1742-7843.2011.00822.x. [DOI] [PubMed] [Google Scholar]
- 12.Elkins JG, Hassett DJ, Stewart PS, Schweizer HP, McDermott TR. 1999. Protective role of catalase in Pseudomonas aeruginosa biofilm resistance to hydrogen peroxide. Appl Environ Microbiol 65:4594–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stewart PS, Roe F, Rayner J, Elkins JG, Lewandowski Z, Ochsner UA, Hassett DJ. 2000. Effect of catalase on hydrogen peroxide penetration into Pseudomonas aeruginosa biofilms. Appl Environ Microbiol 66:836–838. doi: 10.1128/aem.66.2.836-838.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kohanski MA, Dwyer DJ, Collins JJ. 2010. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol 8:423–435. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Imlay JA. 2013. Cell death from antibiotics without the involvement of reactive oxygen species. Science 339:1210–1213. doi: 10.1126/science.1232751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hassett DJ, Imlay JA. 2007. Bactericidal antibiotics and oxidative stress: a radical proposal. ACS Chem Biol 2:708–710. doi: 10.1021/cb700232k. [DOI] [PubMed] [Google Scholar]
- 17.Keren I, Wu Y, Inocencio J, Mulcahy LR, Lewis K. 2013. Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science 339:1213–1216. doi: 10.1126/science.1232688. [DOI] [PubMed] [Google Scholar]
- 18.Khakimova M, Ahlgren HG, Harrison JJ, English AM, Nguyen D. 2013. The stringent response controls catalases in Pseudomonas aeruginosa and is required for hydrogen peroxide and antibiotic tolerance. J Bacteriol 195:2011–2020. doi: 10.1128/JB.02061-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martins D, McKay G, Sampathkumar G, Khakimova M, English AM, Nguyen D. 2018. Superoxide dismutase activity confers (p)ppGpp-mediated antibiotic tolerance to stationary-phase Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 115:9797–9802. doi: 10.1073/pnas.1804525115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su S, Panmanee W, Wilson JJ, Mahtani HK, Li Q, Vanderwielen BD, Makris TM, Rogers M, McDaniel C, Lipscomb JD, Irvin RT, Schurr MJ, Lancaster JR Jr, Kovall RA, Hassett DJ. 2014. Catalase (KatA) plays a role in protection against anaerobic nitric oxide in Pseudomonas aeruginosa. PLoSOne 9:e91813. doi: 10.1371/journal.pone.0091813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chung IY, Kim BO, Jang HJ, Cho YH. 2016. Dual promoters of the major catalase (KatA) govern distinct survival strategies of Pseudomonas aeruginosa. Sci Rep 6:31185. doi: 10.1038/srep31185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hassett DJ, Ma JF, Elkins JG, McDermott TR, Ochsner UA, West SE, Huang CT, Fredericks J, Burnett S, Stewart PS, McFeters G, Passador L, Iglewski BH. 1999. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol Microbiol 34:1082–1093. doi: 10.1046/j.1365-2958.1999.01672.x. [DOI] [PubMed] [Google Scholar]
- 23.Ahmed MN, Porse A, Sommer MOA, Hoiby N, Ciofu O. 2018. Evolution of antibiotic resistance in biofilm and planktonic Pseudomonas aeruginosa populations exposed to subinhibitory levels of ciprofloxacin. Antimicrob Agents Chemother 62:e00320-18. doi: 10.1128/AAC.00320-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Line L, Alhede M, Kolpen M, Kuhl M, Ciofu O, Bjarnsholt T, Moser C, Toyofuku M, Nomura N, Hoiby N, Jensen PO. 2014. Physiological levels of nitrate support anoxic growth by denitrification of Pseudomonas aeruginosa at growth rates reported in cystic fibrosis lungs and sputum. Front Microbiol 5:554. doi: 10.3389/fmicb.2014.00554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Worlitzsch D, Tarran R, Ulrich M, Schwab U, Cekici A, Meyer KC, Birrer P, Bellon G, Berger J, Weiss T, Botzenhart K, Yankaskas JR, Randell S, Boucher RC, Doring G. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J Clin Invest 109:317–325. doi: 10.1172/JCI0213870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolpen M, Hansen CR, Bjarnsholt T, Moser C, Christensen LD, van GM, Ciofu O, Mandsberg L, Kharazmi A, Doring G, Givskov M, Hoiby N, Jensen PO. 2010. Polymorphonuclear leucocytes consume oxygen in sputum from chronic Pseudomonas aeruginosa pneumonia in cystic fibrosis. Thorax 65:57–62. doi: 10.1136/thx.2009.114512. [DOI] [PubMed] [Google Scholar]
- 27.Poole K. 2007. Efflux pumps as antimicrobial resistance mechanisms. Ann Med 39:162–176. doi: 10.1080/07853890701195262. [DOI] [PubMed] [Google Scholar]
- 28.Imamovic L, Ellabaan MMH, Dantas Machado AM, Citterio L, Wulff T, Molin S, Krogh JH, Sommer M. 2018. Drug-driven phenotypic convergence supports rational treatment strategies of chronic infections. Cell 172:121–134. doi: 10.1016/j.cell.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dingemans J, Ye L, Hildebrand F, Tontodonati F, Craggs M, Bilocq F, De VD, Crabbe A, Van HR, Malfroot A, Cornelis P. 2014. The deletion of TonB-dependent receptor genes is part of the genome reduction process that occurs during adaptation of Pseudomonas aeruginosa to the cystic fibrosis lung. Pathogens Dis 71:26–38. doi: 10.1111/2049-632X.12170. [DOI] [PubMed] [Google Scholar]
- 30.Wassermann T, Meinike JK, Ivanyshyn K, Bjarnsholt T, Khademi SM, Jelsbak L, Hoiby N, Ciofu O. 2016. The phenotypic evolution of Pseudomonas aeruginosa populations changes in the presence of subinhibitory concentrations of ciprofloxacin. Microbiology 162:865–875. doi: 10.1099/mic.0.000273. [DOI] [PubMed] [Google Scholar]
- 31.Peng J, Cao J, Ng FM, Hill J. 2017. Pseudomonas aeruginosa develops ciprofloxacin resistance from low to high level with distinctive proteome changes. J Proteomics 152:75–87. doi: 10.1016/j.jprot.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 32.Touati D. 2000. Iron and oxidative stress in bacteria. Arch Biochem Biophys 373:1–6. doi: 10.1006/abbi.1999.1518. [DOI] [PubMed] [Google Scholar]
- 33.Blázquez J, Rodríguez-Beltrán J, Matic I. 2018. Antibiotic-induced genetic variation: how it arises and how it can be prevented. Annu Rev Microbiol 72:209–230. doi: 10.1146/annurev-micro-090817-062139. [DOI] [PubMed] [Google Scholar]
- 34.Sanders LH, Sudhakaran J, Sutton MD. 2009. The GO system prevents ROS-induced mutagenesis and killing in Pseudomonas aeruginosa. FEMS Microbiol Lett 294:89–96. doi: 10.1111/j.1574-6968.2009.01550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ciofu O, Riis B, Pressler T, Poulsen HE, Hoiby N. 2005. Occurrence of hypermutable Pseudomonas aeruginosa in cystic fibrosis patients is associated with the oxidative stress caused by chronic lung inflammation. Antimicrob Agents Chemother 49:2276–2282. doi: 10.1128/AAC.49.6.2276-2282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mandsberg LF, Macia MD, Bergmann KR, Christiansen LE, Alhede M, Kirkby N, Hoiby N, Oliver A, Ciofu O. 2011. Development of antibiotic resistance and up-regulation of the antimutator gene pfpI in mutator Pseudomonas aeruginosa due to inactivation of two DNA oxidative repair genes (mutY, mutM). FEMS Microbiol Lett 324:28–37. doi: 10.1111/j.1574-6968.2011.02383.x. [DOI] [PubMed] [Google Scholar]
- 37.Jørgensen KM, Wassermann T, Jensen PO, Hengzuang W, Molin S, Hoiby N, Ciofu O. 2013. Sublethal ciprofloxacin treatment leads to rapid development of high-level ciprofloxacin resistance during long-term experimental evolution of Pseudomonas aeruginosa. Antimicrob Agents Chemother 57:4215–4221. doi: 10.1128/AAC.00493-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kos VN, Deraspe M, McLaughlin RE, Whiteaker JD, Roy PH, Alm RA, Corbeil J, Gardner H. 2015. The resistome of Pseudomonas aeruginosa in relationship to phenotypic susceptibility. Antimicrob Agents Chemother 59:427–436. doi: 10.1128/AAC.03954-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crabbe A, Jensen PO, Bjarnsholt T, Coenye T. 6 June 2019. Antimicrobial tolerance and metabolic adaptations in microbial biofilms. Trends Microbiol doi: 10.1016/j.tim.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 40.Walters MC III, Roe F, Bugnicourt A, Franklin MJ, Stewart PS. 2003. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob Agents Chemother 47:317–323. doi: 10.1128/AAC.47.1.317-323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang L, Jelsbak L, Marvig RL, Damkiær S, Workman CT, Rau MH, Hansen SK, Folkesson A, Johansen HK, Ciofu O, Høiby N, Sommer MOA, Molin S. 2011. Evolutionary dynamics of bacteria in a human host environment. Proc Natl Acad Sci U S A 108:7481–7486. doi: 10.1073/pnas.1018249108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zaborskyte G, Andersen JB, Kragh KN, Ciofu O. 2016. Real-time monitoring of nfxB mutant occurrence and dynamics in P. aeruginosa biofilm exposed to sub-inhibitory concentrations of ciprofloxacin. Antimicrob Agents Chemother 61:e02292-16. doi: 10.1128/AAC.02292-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.