Although the global deleterious impact of antibiotics on the intestinal microbiota is well known, temporal changes in microbial diversity during and after an antibiotic treatment are still poorly characterized. We used plasma and fecal samples collected frequently during treatment and up to one month after from 22 healthy volunteers assigned to a 5-day treatment by moxifloxacin (n = 14) or no intervention (n = 8).
KEYWORDS: diversity, dysbiosis, intestinal microbiome, metagenomics, moxifloxacin, nonlinear mixed-effect modelling
ABSTRACT
Although the global deleterious impact of antibiotics on the intestinal microbiota is well known, temporal changes in microbial diversity during and after an antibiotic treatment are still poorly characterized. We used plasma and fecal samples collected frequently during treatment and up to one month after from 22 healthy volunteers assigned to a 5-day treatment by moxifloxacin (n = 14) or no intervention (n = 8). Moxifloxacin concentrations were measured in both plasma and feces, and bacterial diversity was determined in feces by 16S rRNA gene profiling and quantified using the Shannon index and number of operational taxonomic units (OTUs). Nonlinear mixed effect models were used to relate drug pharmacokinetics and bacterial diversity over time. Moxifloxacin reduced bacterial diversity in a concentration-dependent manner, with a median maximal loss of 27.5% of the Shannon index (minimum [min], 17.5; maximum [max], 27.7) and 47.4% of the number of OTUs (min, 30.4; max, 48.3). As a consequence of both the long fecal half-life of moxifloxacin and the susceptibility of the gut microbiota to moxifloxacin, bacterial diversity indices did not return to their pretreatment levels until days 16 and 21, respectively. Finally, the model characterized the effect of moxifloxacin on bacterial diversity biomarkers and provides a novel framework for analyzing antibiotic effects on the intestinal microbiome.
BACKGROUND
A large body of data describes the role of the intestinal microbiome in various host processes, including metabolic, nutritional, and immunological processes (1, 2), and in inhibiting the growth of potentially pathogenic microorganisms within the intestines (3, 4). These insights have been unveiled by the growing use of next-generation sequencing techniques, making it possible to investigate the impact of uncultivable bacteria on health and disease. Among such methods, profiling of the 16S rRNA gene consists of sequencing a subset of the 9 hypervariable regions of the bacterial genes encoding the 16S rRNA. Based on their homology, the huge number of sequences obtained can be clustered into operational taxonomic units (OTUs), used for characterizing the composition of the bacterial community (5). This composition can then be summarized for each sample using synthetic indices of diversity, such as the total number of observed OTUs, which characterizes the richness from an ecological perspective, or the Shannon diversity index (6), a composite index of the richness and the distribution of OTUs in the community. These indices of within-sample diversity, also called “alpha diversity,” are now widely used for evaluating the relationships between the microbiome and human health and were recently shown to be highly predictive of mortality in a hamster model of Clostridioides difficile infection (7). This suggests that such markers could be useful to measure the impact of treatment on the microbiota and could possibly be predictive of clinical outcomes (8).
A major cause of microbiome disruption, called dysbiosis, is the administration of antibiotics (9–13). In that respect, fluoroquinolone antibiotics have a particularly severe impact on the gut microbiome due to their high concentrations achieved in feces (14). Through its disruptive effect on the resident bacterial flora, antibiotic administration creates a large space that can be colonized by potentially resistant bacteria. These bacteria may then be disseminated in the environment, making antibiotic-modified gut microbiota the epicenter of bacterial resistance spread (10, 14, 15). In spite of the well-studied impact of repeated administrations of antibiotics on intestinal diversity (13), we still lack a precise characterization of the association between drug concentrations, both in plasma and feces, and temporal changes in the intestinal microbiome (16). This can be done using mathematical models, but parameter estimation of these models is hampered by the high level of both interindividual and intraindividual variability of bacterial diversity, making it difficult to obtain reliable parameter estimates. This difficulty can be in part circumvented by the use of mixed effect models that specifically tease out the two sources of variability and allow precise estimation of parameters, as shown in the context of antibiotic pharmacokinetic/pharmacodynamic assessment (17–19).
Here, we aimed to employ the tools of mathematical and statistical modeling to analyze the relationship between antibiotic gut exposure and temporal changes in the gut microbiome, using the Shannon index and the number of OTUs as synthetic markers of diversity. We focus our analysis on healthy volunteers treated by moxifloxacin, a fluoroquinolone with high oral bioavailability and a high volume of distribution at steady state, which exhibits an excellent penetration in body tissues (20, 21) and whose elimination occurs primarily through the intestinal route (22).
RESULTS
The median age of volunteers was 33.2 years (min, 23.3; max, 59.5), and 8 were males (36.4%). The median weight was 65.2 kg (min, 56.7; max, 86.0). Full characteristics of included subjects can be found in de Gunzburg et al. (23).
Pharmacokinetic analysis of moxifloxacin in plasma and feces.
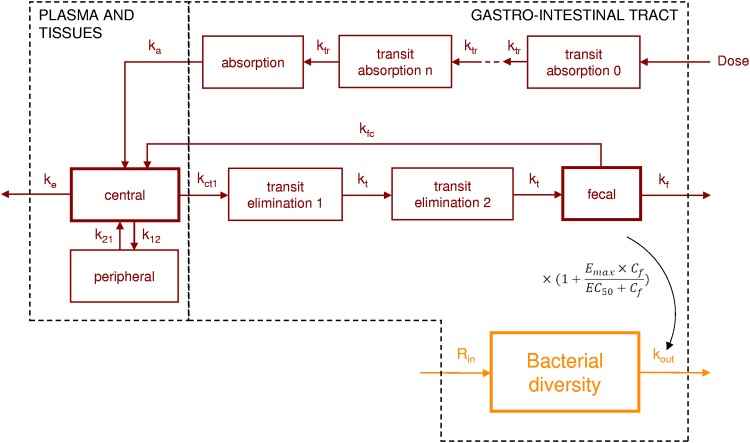
A total of 322 values of total moxifloxacin plasma concentration and 138 values of fecal concentration of free moxifloxacin were collected in the 14 treated patients (see Fig. S1). Plasma concentrations were best described by a two-compartment model with first-order elimination and first-order absorption, using transit compartments to model the absorption delay (Fig. 1). The best fit for the fecal data was a compartmental model that included two transit compartments between plasma and the lower gastrointestinal tract, with enteric recirculation of moxifloxacin from the lower gastrointestinal tract to the central compartment (Fig. 1). Mathematical expressions of the final pharmacokinetic model are presented in Text S2.
FIG 1.
Final compartmental model for plasma and fecal moxifloxacin pharmacokinetics (red) and for bacterial diversity indices (orange). ktr is the transfer rate between each compartment for the absorption delay; ka is the absorption rate to the central compartment; ke is the extraintestinal elimination rate from the central compartment; k12 and k21 are the transfer rates between the central compartment and the peripheral compartment; kct1 is the elimination rate from the central compartment to the intestinal tract; kfc is the transfer rate between the lower gastrointestinal tract and the central compartment; kt is the transfer rate between the intestinal transit compartments; kf is the elimination rate from the lower gastrointestinal tract; Rin is the zero-order constant for production of the diversity index; kout is the first-order elimination rate of the diversity index from the lower gastrointestinal tract. Cf is the concentration in the feces; Emax is the maximal effect of moxifloxacin on the elimination rate of the diversity index; and EC50 is the concentration of moxifloxacin leading to 50% of the maximal effect. Data were available for the 3 compartments with bold boxes. GIT, gastrointestinal tract.
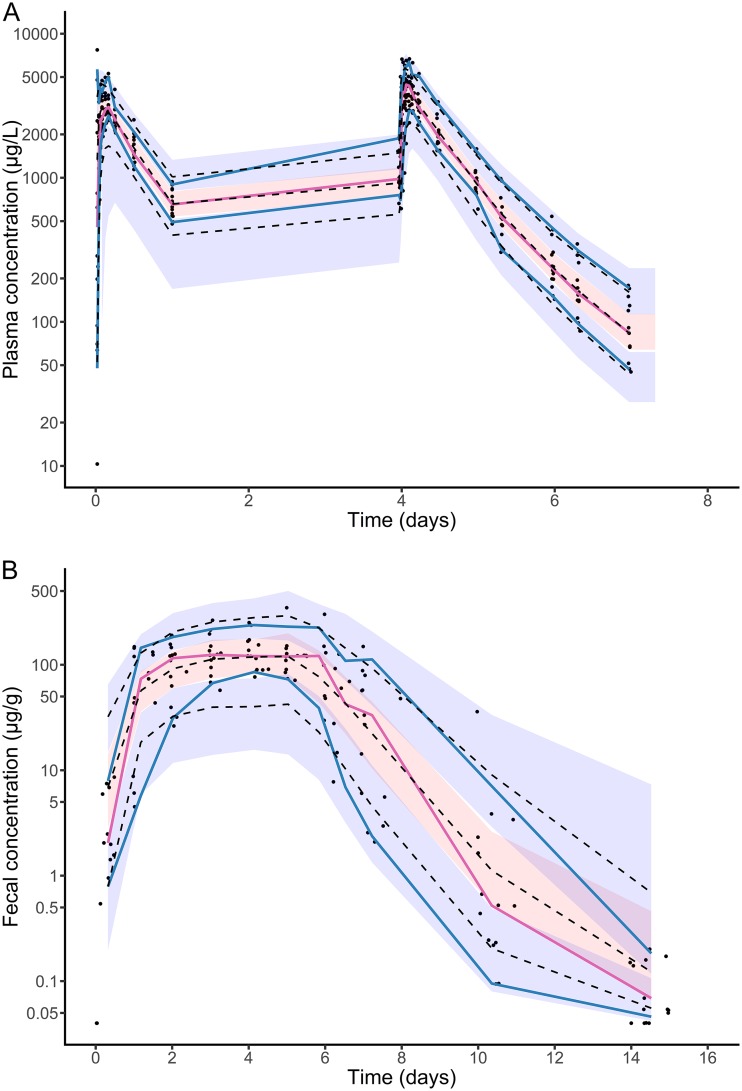
The evolution of moxifloxacin concentration in plasma and feces predicted by the model is presented in Fig. S2A and B. Nearly all parameters of the pharmacokinetic model could be estimated with a good precision (with relative standard error [RSE] below 30%), except for absorption-related parameters ktr (the transfer rate between each compartment for the absorption delay) and ka (the absorption rate to the central compartment) (Table S1). The two rate-constant parameters for the absorption and the elimination transit compartment models were estimated and were not supposed to be identical (which would have led to an increase in the Bayesian information criterion [BIC]). The model well characterized the evolution of both plasma and fecal concentrations of moxifloxacin over time in the 14 treated healthy volunteers (Fig. S3). Other goodness-of-fit plots for this pharmacokinetic model were satisfactory (Fig. 2 and Fig. S4).
FIG 2.
Visual predictive checks for pharmacokinetic model. (A) Plasma concentrations; (B) fecal concentrations. Blue and red lines indicate the observed percentiles (10th, 50th, and 90th percentiles); blue and red ribbons indicate the corresponding 95% confidence intervals. The dashed black lines indicate predicted percentiles. Black points indicate the individual observations.
Of note, the fecal weight (Pf) was assumed to be 200 g/day in all volunteers at all times. As what is observed and fitted are the concentration in the feces (Cf), defined as Af/Pf, a change in Pf results in a similar change in the amount of moxifloxacin in feces (Af) to keep Cf unchanged. As plasma pharmacokinetics (the amount of moxifloxacin in the central compartment [Ac] and the amount of moxifloxacin in the peripheral compartment [Ap]) are not affected by modification of the fecal weight, then kct1 (intestinal elimination from plasma) should increase by the same proportion. As elimination of moxifloxacin from plasma is not affected by the fecal weight, the sum of kct1 and ke (extraintestinal elimination from plasma) is constant. So, when kct1 increases, ke decreases; the first consequence of our assumption on the fecal weight is that it impacts the proportion of the intestinal elimination of moxifloxacin. In addition, for Equation 2 to be similar, kfc (reabsorption from the lower intestinal tract to the plasma) should be divided by that proportion. As the rate of elimination of moxifloxacin from the fecal compartment is not affected by the fecal weight, the sum of kfc and kf (elimination of moxifloxacin from the lower intestinal tract) should be kept constant. Then, when kfc decreases, kf increases; the second consequence of our assumption on the fecal weight is that it impacts the proportion of moxifloxacin reabsorbed from the lower intestinal tract to the plasma.
We performed simulations with fecal weights of 100 g/day, 200 g/day, 300 g/day, and 400 g/day, making the described changes in the population parameters. The simulations resulted in identical concentration-time profiles in both plasma and in feces. The proportions of moxifloxacin eliminated from the plasma by the intestinal route were 94.3%, 88.8%, 83.2%, and 77.7% for fecal weights of 100 g/day, 200 g/day, 300 g/day, and 400 g/day, respectively. The proportions of moxifloxacin reabsorbed from the lower intestinal tract to the plasma were 49.7%, 24.9%, 16.6%, and 12.4% for fecal weights of 100 g/day, 200 g/day, 300 g/day, and 400 g/day, respectively.
The median (min, max) concentration of free moxifloxacin in feces observed 24 h after the first administration of moxifloxacin was 56.9 μg/g (21.7, 95.4), and increased up to 130.4 μg/g (79.2, 250.6) on the fifth day of treatment. Median elimination half-lives from the central compartment and the lower gastrointestinal tract were 0.53 (0.43, 0.67) and 1.0 days (0.76, 1.49), respectively. The median transit time of moxifloxacin between plasma and the lower gastrointestinal tract was 0.59 day (0.35, 1.23). The median time for fecal concentrations of free moxifloxacin to be below the lower limit of quantification was 14.1 days (11.7, 19.3).
Impact of drug concentrations on microbial diversity.
A total of 142 samples were analyzed by 16S rRNA gene profiling (90 in subjects treated by moxifloxacin and 52 in controls). Individual profiles of the Shannon index and number of OTUs are presented in Fig. S5.
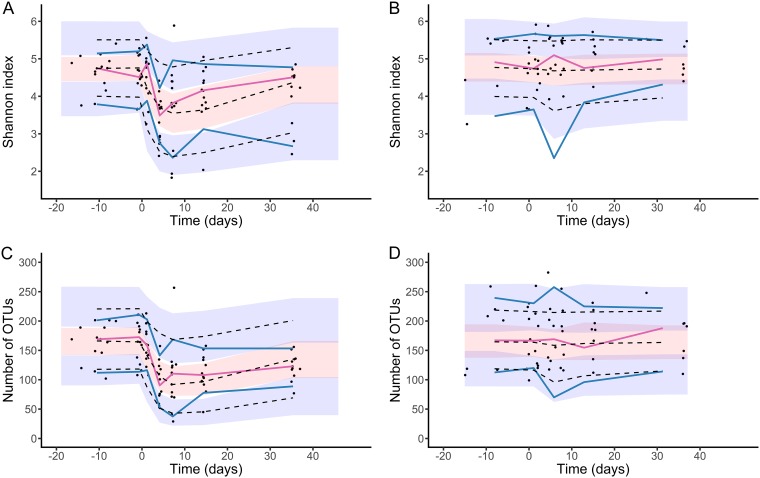
Both indices of bacterial diversity remained constant in the absence of treatment, leading to an estimated mean baseline value of 4.75 Shannon units (RSE, 2%) and 163 OTUs (RSE, 5%) for the Shannon index and the number of OTUs, respectively (Table S2). Initiation of moxifloxacin led to a rapid drop in both diversity indices. This was attributed in our model to a drug concentration-dependent effect in the elimination rate of bacterial diversity from feces (Fig. 1 and Text S2), with a rate increasing up to 38% (RSE, 14%) and 94% (RSE, 4%) for the Shannon and the number of OTUs, respectively. Given the extended presence of moxifloxacin in feces and the susceptibility of bacterial diversity to the drug (as measured by the concentration leading to 50% of the maximal effect [EC50] parameter, which was shown to be similar for both indices; see Materials and Methods), our model predicted that the diversity only slowly returned to pretreatment values after treatment cessation. Of note, EC50 was estimated to 0.13 μg/g of feces and was associated with a large interindividual variability (470%, see Table S2). The model could well fit the evolution of both the Shannon index and the number of OTUs in all individuals (Fig. 3 and Fig. S6 and S7). The model prediction intervals are drawn in Fig. S2 (panels C and D). All parameters were well estimated (Table S2) except the drug EC50, for which likelihood profiling (see Text S1) was used to calculate the 95% confidence interval (0.01 μg/g to 2 μg/g; Fig. S8).
FIG 3.
Visual predictive checks for pharmacodynamic model. The Shannon index is depicted for (A) moxifloxacin-treated subjects and (B) untreated subjects, and the number of OTUs is depicted for (C) moxifloxacin-treated subjects and (D) untreated subjects. Blue and red lines indicate the observed percentiles (10th, 50th, and 90th percentiles); blue and red ribbons indicate the corresponding 95% confidence intervals. The dashed black lines indicate predicted percentiles. Black points indicate the individual observations.
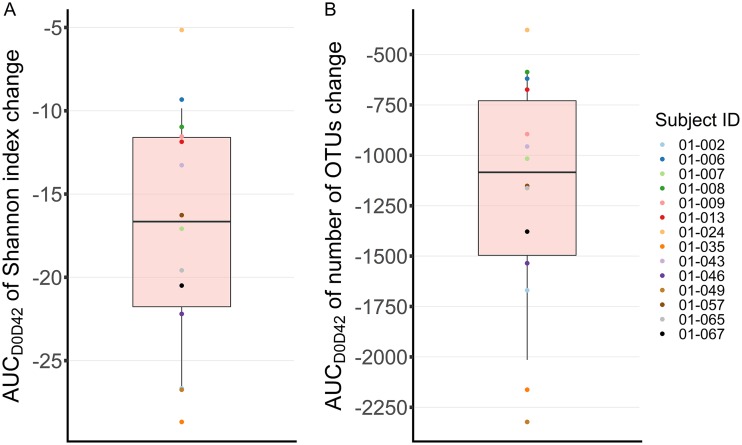
Pharmacodynamic indices derived from the model for the 14 subjects treated by moxifloxacin are presented in Table 1 and Fig. 4. A median loss of 0.6 units of Shannon index (min and max, 0.3 and 0.7) and 26 OTUs (min and max, 11 and 41) was achieved at 24 h after the beginning of treatment. The maximal loss induced by moxifloxacin was 1.2 Shannon units (0.8 and 1.3) and 77 OTUs (47 and 100), achieved 7.0 (5.1 and 11.4) and 8.5 (5.5 and 13.8) days after treatment initiation, respectively. Results obtained for other simulated treatment regimens are presented in Table S3.
TABLE 1.
Measures of the impact of moxifloxacin on gut microbiome diversitya
| Pharmacodynamic index | Shannon index (median [min, max]) | No. of OTUs (median [min, max]) |
|---|---|---|
| Time to maximal loss (days) | 7.0 (5.1, 11.4) | 8.5 (5.5, 13.8) |
| Maximal loss | 1.2 (0.8, 1.3)b | 77 (47, 100)c |
| Maximal loss (% of baseline value) | 27.5 (17.5, 27.7) | 47.4 (30.4, 48.3) |
| Time to return to 95% of baseline value (days) | 16.3 (8.7, 25.3) | 21.3 (13.3, 30.4) |
| AUC between day 0 and day 42 of the change of diversity indices from day 0 | −16.7 (−28.7, −5.2)d | −1,084 (−2,323, −378)e |
Derived from the estimated individual pharmacodynamic parameters in the 14 treated individuals.
Shannon units.
No. of OTUs.
Units/day.
No. of OTUs/day.
FIG 4.
Estimated impact of moxifloxacin on intestinal microbiome in the 14 subjects treated with moxifloxacin. The impact was measured as the area under the curve (AUC) of the change of the Shannon index (A) or number of OTUs (B) from baseline over time, between days 0 and 42 (AUCD0D24). The AUC is a metric which allows a global view of antibiotic impact on the microbiota, as it takes into account both the extent and the duration of dysbiosis. ID, identifier.
DISCUSSION
Here, we characterized for the first time the drug concentration-dependent impact of antibiotic treatment on temporal changes in bacterial diversity in the intestinal microbiome during and after treatment initiation.
Pharmacokinetic analysis revealed that moxifloxacin concentrations in the lower gastrointestinal tract rapidly reached a high plateau, with a median value of 130.4 μg/g at day 5, and only slowly decreased after treatment cessation, with a half-life of 1 day. This slow decrease in the lower gastrointestinal tract was captured in our model, which assumes a reabsorption of moxifloxacin from the lower gastrointestinal tract to the central compartment. Enteric recycling had been previously reported for moxifloxacin (20). Interestingly, reabsorption from the gut to the central compartment did not seem to occur from the upper parts of the gut but rather from the lower gastrointestinal tract. This is consistent with the fact that moxifloxacin concentrations within the colonic mucosa are greater than those measured in the small bowel mucosa (24). This reabsorption process might explain the long-lasting fecal excretion observed after treatment cessation, with concentrations in the lower gastrointestinal tract greater than 2 μg/g up to 5 days (range, 2.9 to 8.4) after the last administration of moxifloxacin. As both indices of diversity were largely susceptible to moxifloxacin (with a similar EC50 value, equal to 0.13 μg/g), diversity was profoundly impacted for up to 10 days after treatment cessation. These slowly decreasing concentrations of moxifloxacin create a replication space for resistant bacteria, consistent with the observation that the posttreatment period is critical for the emergence of fluoroquinolone-resistant strains in the human fecal flora (25). Despite moxifloxacin’s mechanism of action on bacterial growth, in our model, moxifloxacin appeared to increase the rate of elimination of bacterial diversity, with a reduction of the Bayesian information criterion by 60 units.
Despite the complexity of the measures performed, relying on metagenomic analyses, semimechanistic mathematical models provided a good description of the temporal changes of the diversity indices within the intestinal microbiome. Interestingly, the maximal effect of moxifloxacin on the two diversity indices was different, leading to differences in the time needed to return to baseline values. This confirms that the two indices do not exactly measure the same phenomena, and may be complementary for drawing a complete picture of the microbiome dysbiosis induced by an intervention.
Our approach shows that the complexity of the microbiome composition, along with the temporal impact of treatment on it, may be advantageously characterized using semimechanistic mathematical models and synthetic markers of microbiome diversity, such as the Shannon index and the number of OTUs. They can then be used to calculate simple metrics, such as the cumulated loss of bacterial diversity, and may be used to predict the impact of changes in experimental settings. This might prove to be very useful, as microbiome data on multiple days are usually difficult to obtain reliably in the clinical context. The model can also be used to simulate the effect of different dosing regimens, such as that of increasing the daily dose to 800 mg or the treatment duration to 10 days. Given the high susceptibility of bacterial diversity to moxifloxacin, we predicted that increasing the daily dose would result in only a minor additional effect on bacterial diversity compared to that of the dosing regimen studied here. Doubling the treatment duration from 5 to 10 days would result in a 35% increase of the global impact of moxifloxacin on the bacterial diversity, as measured by the area under the curve (AUC) between day 0 and day 42 of the change of diversity indices from day 0. Furthermore, the use of a nonlinear mixed effect model to estimate parameters, a statistical approach that advantageously optimizes the information available by borrowing strength from the between-subject variability, allowed us to precisely estimate parameters despite a limited number of samples per patient.
Our work has some limitations. The main one is that only healthy subjects were included. Model parameters, particularly the interindividual variability of the pharmacodynamic parameters but also possibly that of the gut microbiome, could differ in a clinical context. For instance, it has been reported that parameters such as body weight or the severity of the infection impact the pharmacokinetics of antibiotics, including that of moxifloxacin (26–28). Next, the EC50 parameter could not be precisely estimated and was associated with a large confidence interval, which prevented extrapolation of our findings for lower dosing regimens. Next, we fixed the fecal daily weight to 200 g for all volunteers, whereas there is a high intra- and interindividual variability in feces generation. This assumption affects the values of four elimination rate-constants, kct1, ke, kfc, and kf, whose values should therefore be considered with caution. Furthermore, assuming the same values of daily fecal weight in all individuals at each sampling time, whereas it is known to vary, has an impact on the estimated variabilities of those parameters and on the residual error. It does not, however, have any impact on the pharmacodynamic model, as it was written using fecal concentrations. Finally, we restricted our analysis to a global measure of diversity, and we did not model the specific evolution of OTUs; this will require larger study population in order to account for the variability across patients and the high dependency within OTU dynamics. Such models will also gain from using technologies capable of measuring not only relative counts but absolute counts of OTUs, such as coupling data from next-generation sequencing together with flow cytometric counts of bacterial cells (29).
In summary, we provided here a modeling framework to measure the impact of antibiotics on the intestinal microbiome. Diversity indices obtained through next-generation sequencing offer a simplified view of the dynamics of the bacterial community within the microbiome, and mathematical modeling allowed us to precisely estimate several biomarkers to assess the global dysbiosis induced by moxifloxacin. This encourages extension of these results to the clinical setting for estimating the unwanted effects of drugs on the microbiome. Furthermore, mathematical models make it possible to investigate the effect of various experimental conditions on an outcome and could be helpful to develop strategies aiming to reduce antibiotic impact on the gut microbiota, via, for instance, locally released adsorbents, antibiotic-hydrolyzing enzymes, or optimized dosing regimens of antibiotics.
MATERIALS AND METHODS
Healthy volunteers and sample collection.
We used data from the CL-1002 trial (ClinicalTrials.gov identifier NCT02176005), a prospective, open-label, randomized clinical trial conducted in 2014 at the Clinical Investigation Center of the Bichat Hospital, Paris (France). The trial was approved by French Health Authorities and an independent ethics committee. Full details of the trial have been reported elsewhere (23). We focused here on 22 subjects of this trial who received moxifloxacin alone (n = 14) or who were not treated (negative-control group, n = 8).
Briefly, healthy volunteers over 18 years old and without exposure to antibiotics in the preceding 3 months were prospectively included after obtention of their informed consent. Subjects in the moxifloxacin group received 400 mg of moxifloxacin orally once a day for 5 days (from day 1 [D1] to D5), and all subjects were followed until D37. A total of 24 blood samples were collected in each moxifloxacin-treated volunteer on D1 (just before treatment administration and at 0.5 h, 1 h, 1.5 h, 2 h, 3 h, 4 h, 6 h, 12 h, and 24 h post administration) and at the last day of treatment, i.e., on D5 (just before treatment administration and at 0.5 h, 1 h, 1.5 h, 2 h, 3 h, 4 h, 6 h, 12 h, 24 h, 32 h, 48 h, 56 h, and 72 h post administration). Thirteen fecal samples were collected in all volunteers at screening (between D21 and D3), just before the first administration of moxifloxacin (baseline), once a day from D2 to D9, and then at D12, D16, and D37.
Moxifloxacin assays were performed using specifically developed and validated bioanalytical methods (see Supplemental Methods). Total plasma moxifloxacin concentrations were determined by reverse-phase high-performance liquid chromatography coupled with fluorescence detection (lower limit of quantification, 0.01 μg/ml). Fecal concentrations of free moxifloxacin were determined on the 11 samples collected at baseline, D2, D3, D4, D5, D6, D7, D8, D9, D12, and D16 by tandem mass spectrometry detection (lower limit of quantification, 0.04 μg/g).
Targeted metagenomic analysis of the intestinal microbiota.
The 7 fecal samples collected at screening, baseline, D3, D6, D9, D16, and D37 were analyzed by 16S rRNA gene profiling. Microbial DNA was extracted using an extraction protocol optimized at GenoScreen, partially based on commercially available extraction kits (QIAamp DNA stool lit, Qiagen, Germany), with the addition of chemical and mechanical lysis steps.
The V3 and V4 region of the 16S rRNA gene was then amplified using an optimized and standardized amplicon library preparation protocol (Metabiote; GenoScreen, Lille, France). Positive (artificial bacteria community comprising 17 different bacteria, ABCv2) and negative (sterile water) controls were also included. Briefly, PCRs were performed using 5 ng of genomic DNA and in-house fusion barcoded primers (final concentration, 0.2 μM) with an annealing temperature of 50°C for 30 cycles. PCR products were purified using Agencourt AMPure XP magnetic beads (Beckman Coulter, Brea, CA, USA), quantified according to GenoScreen’s protocol, and mixed in an equimolar amount. Sequencing was performed using 250-bp paired-end sequencing chemistry on the Illumina MiSeq platform (Illumina, San Diego, CA, USA) at GenoScreen.
Raw paired-end reads were then demultiplexed per sample and subjected to the following process: (i) search and removal of both forward and reverse primer using CutAdapt, with no mismatches allowed in the primer sequences; (ii) quality filtering using the PRINSEQ-lite PERL script (30), by truncating bases with a Phred quality score of <30 at the 3′ end; and (iii) paired-end read assembly using FLASH (31), with a minimum overlap of 30 bases and >97% overlap identity.
Diversity analysis was performed using the Metabiote Online v2.0 pipeline (GenoScreen, Lille, France) which is partially based on QIIME software v1.9.1 (32). Following the steps of preprocessing, chimera sequences were detected and eliminated (in-house method based on Usearch v6.1). Then, clustering of similar sequences (97% identity threshold for an affiliation at the genus level on the V3 and V4 regions of the 16S rRNA gene) was performed with Uclust v1.2.22q (33) through an open-reference OTU picking process and complete linkage method, finally creating groups of sequences or OTUs. An OTU cleaning step corresponding to the elimination of singletons was performed. Diversity in each fecal sample was estimated using 2 indices of alpha-diversity, the Shannon diversity index and the number of observed OTUs.
Modeling strategy.
First, we developed a pharmacokinetic model of plasma moxifloxacin using total drug concentrations. Both one- and two-compartment models with first-order absorption and first-order elimination were tested. The absorption delay of moxifloxacin in the central compartment was modelled using either a lag time or absorption transit compartments (34). As moxifloxacin was administered through the oral route, we estimated an apparent volume of distribution after oral administration (V/F, where F is the bioavailability).
Second, the model was extended to account for fecal pharmacokinetics, assuming a fixed fecal weight of 200 g/day in all subjects, allowing us to reconstruct pharmacokinetic profiles in both plasma and feces. Moxifloxacin elimination from the central compartment was divided into an intestinal elimination and an extraintestinal elimination, both being assumed to have first-order rates. Several structural models with various numbers of elimination transit compartments between plasma and the lower gastrointestinal tract were tested. As enteric recirculation had been reported for fluoroquinolones (20), we also tested several models for moxifloxacin reabsorption, occurring either from one of the transit compartments between plasma and the lower gastrointestinal tract or from the lower gastrointestinal tract to the plasma compartment.
Third, a semimechanistic model was developed, which assumes that moxifloxacin impacts microbiome diversity as follows (35). The evolution of the diversity index (D) in the absence of the drug was written as:
| (1) |
where Rin is a zero-order constant for the production of the diversity, and kout is a first-order rate constant for elimination of diversity. At steady state, , and thus in the absence of treatment. The initiation of antibiotic treatment perturbs this equilibrium and can alter either lead to a reduction of Rin or to an increase of kout (k′out, see Equation 2):
| (2) |
with Cf being the fecal concentration of free moxifloxacin, Emax the maximal increase of kout, and EC50 the concentration leading to 50% of the maximal effect of moxifloxacin.
Statistical methods.
We used nonlinear mixed effects models to analyze the evolution of pharmacokinetic and pharmacodynamic data. Data from treated individuals were used for the pharmacokinetic analysis, whereas data from both groups were used for the pharmacodynamic analysis (with fecal concentrations assumed to be 0 in the control group; see Equation 1). Intraindividual correlation of repeated data was taken into account by random effects. Population parameters were estimated by likelihood maximization using the stochastic approximation expectation maximization algorithm (SAEM) (36), implemented in MONOLIX v4.3.2 (Lixoft, Orsay, France). Data below the lower limit of quantification were treated as left-censored data. Their contribution to the likelihood was computed as the probability that these data are indeed below the lower limit of quantification (37). Estimates of the individual parameters were computed as the mode of the a posteriori distribution and used to predict individual pharmacokinetic and pharmacodynamic profiles for the following treatment regimens: moxifloxacin daily doses of 400 mg and 800 mg for 5- and 10-day treatment durations. Model selection at each step of the model building was performed using the Bayesian information criterion (BIC), and model evaluation was conducted by investigating several goodness-of-fit plots. Full details on the statistical model and parameters estimation are available in Text S1.
Measures of antibiotic impact on the microbiome.
The following parameters were computed for moxifloxacin-treated subjects using estimated individual parameters (Text S1): mean transit time between the central compartment and the lower gastrointestinal tract, maximal loss of each bacterial diversity index in the intestinal microbiome after the beginning of treatment (nadir), the time for which this maximal loss was achieved (time to nadir), and the time at which each diversity index returned to 95% of its baseline value. The cumulated impact of moxifloxacin on diversity was obtained by computing the area under the curve for each diversity index up to 42 days after the beginning of treatment.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Da Volterra Company for providing the data. We thank Jean de Gunzburg for fruitful discussions.
Members of the DAV132-CL-1002 study group include B. Ait-Ilalne, L. Alavoine, X. Duval, J. L. Ecobichon, E. Ilic-Habensus, A. Laparra, M. E. Nisus, P. Ralaimazava, S. Raine, S. Tubiana, and V. Vignali, all from the Center of Clinical Investigation 1425 at Bichat Hospital (Paris, France), and E. Chachaty from the Institut Gustave Roussy (Villejuif, France).
Antoine Andremont, Charles Burdet, and France Mentré are consultants for the Da Volterra Company. Thu Thuy Nguyen performed statistical work for the Da Volterra Company through a contract with INSERM UMR 1137.
A.A. and F.M. designed the clinical trial. X.D. included participants. S.F. performed the metagenomic analysis. C.B., T.T.N., J.G., and F.M. performed the statistical analysis of the data. C.B., T.T.N., J.G., and F.M. wrote the paper. All authors agreed on the final version of the manuscript.
The randomized clinical trial was sponsored by Da Volterra (Paris) and funded in part by the European Union Seventh Framework Programme (FP7-HEALTH-2011-single-stage) under grant agreement number 282004 (EvoTAR).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00820-19.
Contributor Information
Collaborators: B. Ait-Ilalne, L. Alavoine, X. Duval, J. L. Ecobichon, E. Ilic-Habensus, A. Laparra, M. E. Nisus, P. Ralaimazava, S. Raine, S. Tubiana, V. Vignali, and E. Chachaty
REFERENCES
- 1.Hollister EB, Gao C, Versalovic J. 2014. Compositional and functional features of the gastrointestinal microbiome and their effects on human health. Gastroenterology 146:1449–1458. doi: 10.1053/j.gastro.2014.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosen CE, Palm NW. 2017. Functional classification of the gut microbiota: the key to cracking the microbiota composition code: functional classifications of the gut microbiota reveal previously hidden contributions of indigenous gut bacteria to human health and disease. Bioessays 39:1700032. doi: 10.1002/bies.201700032. [DOI] [PubMed] [Google Scholar]
- 3.Kim S, Covington A, Pamer EG. 2017. The intestinal microbiota: antibiotics, colonization resistance, and enteric pathogens. Immunol Rev 279:90–105. doi: 10.1111/imr.12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JF, Tijssen JG, Speelman P, Dijkgraaf MG, Keller JJ. 2013. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med 368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 5.Yarza P, Yilmaz P, Pruesse E, Glöckner FO, Ludwig W, Schleifer K-H, Whitman WB, Euzéby J, Amann R, Rosselló-Móra R. 2014. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol 12:635–645. doi: 10.1038/nrmicro3330. [DOI] [PubMed] [Google Scholar]
- 6.Shannon C. 1948. A mathematical theory of communication. Bell Syst Tech J 27:623–656. doi: 10.1002/j.1538-7305.1948.tb00917.x. [DOI] [Google Scholar]
- 7.Burdet C, Sayah-Jeanne S, Nguyen TT, Hugon P, Sablier-Gallis F, Saint-Lu N, Corbel T, Ferreira S, Pulse M, Weiss W, Andremont A, Mentré F, de Gunzburg J. 2018. Antibiotic-induced dysbiosis predicts mortality in an animal model of Clostridium difficile infection. Antimicrob Agents Chemother 62:e00925-18. doi: 10.1128/AAC.00925-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lesko LJ, Atkinson AJ Jr. 2001. Use of biomarkers and surrogate endpoints in drug development and regulatory decision making: criteria, validation, strategies. Annu Rev Pharmacol Toxicol 41:347–366. doi: 10.1146/annurev.pharmtox.41.1.347. [DOI] [PubMed] [Google Scholar]
- 9.Andremont A, Brun-Buisson C, Struelens M. 2001. Evaluating and predicting the ecologic impact of antibiotics. Clin Microbiol Infect 7(Suppl 5):1–6. doi: 10.1046/j.1469-0691.2001.00065.x. [DOI] [PubMed] [Google Scholar]
- 10.Dethlefsen L, Huse S, Sogin ML, Relman DA. 2008. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujisaka S, Ussar S, Clish C, Devkota S, Dreyfuss JM, Sakaguchi M, Soto M, Konishi M, Softic S, Altindis E, Li N, Gerber G, Bry L, Kahn CR. 2016. Antibiotic effects on gut microbiota and metabolism are host dependent. J Clin Invest 126:4430–4443. doi: 10.1172/JCI86674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sullivan A, Edlund C, Nord CE. 2001. Effect of antimicrobial agents on the ecological balance of human microflora. Lancet Infect Dis 1:101–114. doi: 10.1016/S1473-3099(01)00066-4. [DOI] [PubMed] [Google Scholar]
- 13.Dethlefsen L, Relman DA. 2011. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A 108(Suppl 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Lastours V, Fantin B. 2015. Impact of fluoroquinolones on human microbiota. Focus on the emergence of antibiotic resistance. Future Microbiol 10:1241–1255. doi: 10.2217/fmb.15.40. [DOI] [PubMed] [Google Scholar]
- 15.Khanna S, Pardi DS. 2016. Clinical implications of antibiotic impact on gastrointestinal microbiota and Clostridium difficile infection. Expert Rev Gastroenterol Hepatol 10:1145–1152. doi: 10.1586/17474124.2016.1158097. [DOI] [PubMed] [Google Scholar]
- 16.Ruppé E, Burdet C, Grall N, de Lastours V, Lescure FX, Andremont A, Armand-Lefèvre L. 2018. Impact of antibiotics on the intestinal microbiota needs to be re-defined to optimize antibiotic usage. Clin Microbiol Infect 24:3–5. doi: 10.1016/j.cmi.2017.09.017. [DOI] [PubMed] [Google Scholar]
- 17.Harigaya Y, Bulitta JB, Forrest A, Sakoulas G, Lesse AJ, Mylotte JM, Tsuji BT. 2009. Pharmacodynamics of vancomycin at simulated epithelial lining fluid concentrations against methicillin-resistant Staphylococcus aureus (MRSA): implications for dosing in MRSA pneumonia. Antimicrob Agents Chemother 53:3894–3901. doi: 10.1128/AAC.01585-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mouton JW, Vinks AA, Punt NC. 1997. Pharmacokinetic-pharmacodynamic modeling of activity of ceftazidime during continuous and intermittent infusion. Antimicrob Agents Chemother 41:733–738. doi: 10.1128/AAC.41.4.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen TT, Guedj J, Chachaty E, de Gunzburg J, Andremont A, Mentré F. 2014. Mathematical modeling of bacterial kinetics to predict the impact of antibiotic colonic exposure and treatment duration on the amount of resistant enterobacteria excreted. PLoS Comput Biol 10:e1003840. doi: 10.1371/journal.pcbi.1003840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stass H, Kubitza D, Moller JG, Delesen H. 2005. Influence of activated charcoal on the pharmacokinetics of moxifloxacin following intravenous and oral administration of a 400 mg single dose to healthy males. Br J Clin Pharmacol 59:536–541. doi: 10.1111/j.1365-2125.2005.02357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muller M, Stass H, Brunner M, Moller JG, Lackner E, Eichler HG. 1999. Penetration of moxifloxacin into peripheral compartments in humans. Antimicrob Agents Chemother 43:2345–2349. doi: 10.1128/AAC.43.10.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stass H, Kubitza D. 1999. Pharmacokinetics and elimination of moxifloxacin after oral and intravenous administration in man. J Antimicrob Chemother 43(Suppl B):83–90. doi: 10.1093/jac/43.suppl_2.83. [DOI] [PubMed] [Google Scholar]
- 23.de Gunzburg J, Ghozlane A, Ducher A, Le Chatelier E, Duval X, Ruppé E, Armand-Lefèvre L, Sablier-Gallis F, Burdet C, Alavoine L, Chachaty E, Augustin V, Varastet M, Levenez F, Kennedy S, Pons N, Mentré F, Andremont A. 2018. Protection of the human gut microbiome from antibiotics. J Infect Dis 217:628–636. doi: 10.1093/infdis/jix604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wirtz M, Kleeff J, Swoboda S, Halaceli I, Geiss HK, Hoppe-Tichy T, Büchler MW, Friess H. 2004. Moxifloxacin penetration into human gastrointestinal tissues. J Antimicrob Chemother 53:875–877. doi: 10.1093/jac/dkh173. [DOI] [PubMed] [Google Scholar]
- 25.Fantin B, Duval X, Massias L, Alavoine L, Chau F, Retout S, Andremont A, Mentré F. 2009. Ciprofloxacin dosage and emergence of resistance in human commensal bacteria. J Infect Dis 200:390–398. doi: 10.1086/600122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kees MG, Weber S, Kees F, Horbach T. 2011. Pharmacokinetics of moxifloxacin in plasma and tissue of morbidly obese patients. J Antimicrob Chemother 66:2330–2335. doi: 10.1093/jac/dkr282. [DOI] [PubMed] [Google Scholar]
- 27.Zvada SP, Denti P, Sirgel FA, Chigutsa E, Hatherill M, Charalambous S, Mungofa S, Wiesner L, Simonsson US, Jindani A, Harrison T, McIlleron HM. 2014. Moxifloxacin population pharmacokinetics and model-based comparison of efficacy between moxifloxacin and ofloxacin in African patients. Antimicrob Agents Chemother 58:503–510. doi: 10.1128/AAC.01478-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naidoo A, Chirehwa M, McIlleron H, Naidoo K, Essack S, Yende-Zuma N, Kimba-Phongi E, Adamson J, Govender K, Padayatchi N, Denti P. 2017. Effect of rifampicin and efavirenz on moxifloxacin concentrations when co-administered in patients with drug-susceptible TB. J Antimicrob Chemother 72:1441–1449. doi: 10.1093/jac/dkx004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vandeputte D, Kathagen G, D’hoe K, Vieira-Silva S, Valles-Colomer M, Sabino J, Wang J, Tito RY, De Commer L, Darzi Y, Vermeire S, Falony G, Raes J. 2017. Quantitative microbiome profiling links gut community variation to microbial load. Nature 551:507–511. doi: 10.1038/nature24460. [DOI] [PubMed] [Google Scholar]
- 30.Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Magoc T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 34.Savic RM, Jonker DM, Kerbusch T, Karlsson MO. 2007. Implementation of a transit compartment model for describing drug absorption in pharmacokinetic studies. J Pharmacokinet Pharmacodyn 34:711–726. doi: 10.1007/s10928-007-9066-0. [DOI] [PubMed] [Google Scholar]
- 35.Dayneka NL, Garg V, Jusko WJ. 1993. Comparison of four basic models of indirect pharmacodynamic responses. J Pharmacokinet Biopharm 21:457–478. doi: 10.1007/BF01061691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuhn E, Lavielle M. 2005. Maximum likelihood estimation in nonlinear mixed effects models. Comput Stat Data Anal 49:1020–1038. doi: 10.1016/j.csda.2004.07.002. [DOI] [Google Scholar]
- 37.Samson A, Lavielle M, Mentré F. 2006. Extension of the SAEM algorithm to left-censored data in nonlinear mixed-effects model: Application to HIV dynamics model. Comput Stat Data Anal 51:1562–1574. doi: 10.1016/j.csda.2006.05.007. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.