Here, we describe two plasmids carrying mcr-4.3 in two Acinetobacter baumannii strains isolated from imported food and a clinical sample. The comparative analysis of these plasmids, with two other plasmids reported in the NCBI database, highlighted the common origin of the plasmidic structure carrying mcr-4.3. This is the first case of the mcr-4.3 gene in a A. baumannii strain isolated from a clinical case in Europe. We hypothesize that food import is initiating the spread in Czech Republic.
KEYWORDS: Acinetobacter baumannii, WGS, colistin resistance, mcr-4.3
ABSTRACT
Here, we describe two plasmids carrying mcr-4.3 in two Acinetobacter baumannii strains isolated from imported food and a clinical sample. The comparative analysis of these plasmids, with two other plasmids reported in the NCBI database, highlighted the common origin of the plasmidic structure carrying mcr-4.3. This is the first case of the mcr-4.3 gene in a A. baumannii strain isolated from a clinical case in Europe. We hypothesize that food import is initiating the spread in Czech Republic.
INTRODUCTION
The first report of the mcr-4 gene was characterized on a ColE10 plasmid extracted from Salmonella enterica of pig origin in Italy (1). The mcr-4.3 gene, which has two mutations leading to 2 codon changes of V179G and V236F compared with mcr-4 (2), was first reported in 2014 on a ColE10-type plasmid carried by an Enterobacter cloacae isolate of human origin in Singapore (3). Here, we describe two plasmids carrying mcr-4.3 in two Acinetobacter baumannii strains isolated from imported raw food for commercial use and from a clinical sample. Both isolates were selected from an ongoing screening survey for mcr genes in the Czech Republic. The first strain (LEV1449/17Ec) was obtained in 2017 from a frozen turkey liver imported from Brazil, while the second isolate (39741) was recovered in 2017 from a tracheal aspirate of a 94-year-old female patient in Karvina, Czech Republic. (These data have been presented as an oral communication at the ECCMID from 13 April to 16 April 2019.)
Antibiotic susceptibility profiling was performed using the Vitek2 system (bioMérieux, Marcy l’Etoile, France), while colistin susceptibility was determined by broth microdilution assay using EUCAST 2019 breakpoints (http://www.eucast.org/clinical_breakpoints/). A conjugation experiment was conducted using the strain Escherichia coli J62 as a recipient. E. coli DH10B and A. baumannii CIP7010 were transformed by plasmid DNA from the wild-type strains using electroporation (4). Genomic DNA extracted from the two strains was subjected to whole-genome sequencing using the MiSeq (Illumina Inc., San Diego, USA) and Sequel I (Pacific Biosciences, CA, USA) platforms. Reads from both Illumina MiSeq and Sequel sequencing were used to perform hybrid assembly. Sequence types and antibiotic resistance genes were detected upon uploading the assemblies to PubMLST (https://pubmlst.org/abaumannii/) and to ResFinder 3.2 (https://cge.cbs.dtu.dk/services/ResFinder/) (5), respectively. High-quality reads of LEV1449/17Ec were mapped to the 39741 chromosome using Bowtie v2.3.4 (6). Single nucleotide polymorphisms (SNPs) were called from mapped data using VarScan v2.4.3 (7). Detected SNPs were inspected manually by visually checking mapped data via Tablet v1.17 (8). The chromosome of 39741 was annotated using Prokka v1.13 (9) in order to examine the genetic context of the SNPs. Chromosomes of both samples were also compared and similarity statistics were obtained using QUAST tool v5.0.0 (10).
Both strains, namely, 39741 and LEV1449/17Ec, belonged to the sequence types 345 and 747 (ST345/ST747) according to the Pasteur and Oxford scheme, respectively. They exhibited colistin resistance (MIC, >16 mg/liter) while remaining susceptible to almost all other antibiotics tested (see Table S1 in the supplemental material). Moreover, both strains had 3 resistance genes in the chromosome, namely, intrinsic beta-lactamase-encoding genes blaOXA-67 and blaADC-6 and mcr-4.3 gene on a nonconjugative and nontransformable plasmid (see Table S2 in the supplemental material). Comparative analysis of chromosomal DNA showed that these isolates differ in almost 4% of their genetic content. Moreover, 18 regions of clustered SNPs and a total of 130 of nonclustered SNPs were found between their core genome sequences (see Table S3 in the supplemental material). Chromosomal mutations in the pmrCAB operon and in lpxA/lpxC/lpxD genes were screened by comparing the sequences with their respective ones corresponding to the reference strain ATCC 17978 (GenBank accession number CP000521) (11). Most of the amino acid substitutions in PmrCAB have not been previously reported (see Table S4 in the supplemental material). While certain mutations can be lineage specific and have no effect on colistin susceptibility/resistance, the impact of these amino acid substitutions in PmrCAB remains unknown, due to the absence of other genomes belonging to the same sequence type in the GenBank database.
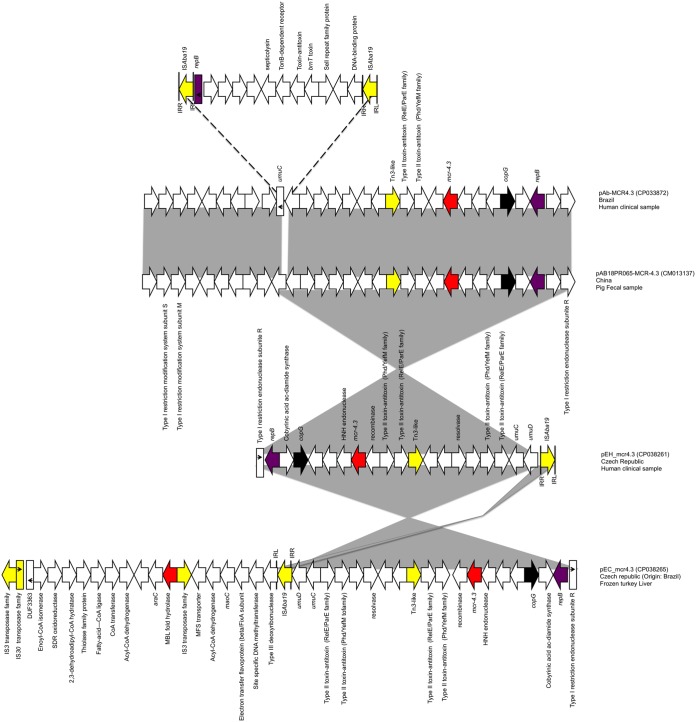
Using long-read sequencing, four complete and circular contigs were obtained for the 39741 strain, namely, a 3,752,576-bp chromosome, a 135,229-bp group 13 plasmid (pEH_gr13), a 25,856-bp group 3 plasmid (pEH_gr3), and another 18,786-bp plasmid harboring the mcr-4.3 gene that did not resemble any known plasmid Inc family (pEH_mcr4.3). The pEH_mcr4.3 plasmid had one copy of ISAba19, two copies of two different type II toxin-antitoxin (TA) systems (PhD/YefM and RelE/ParE), and a parA-like gene (copG).
The LEV1449/17Ec strain had four complete and circular contigs, namely, a 3,708,164-bp chromosome, a 128,013-bp group 13 plasmid (pEC_gr13), a 76,956-bp group 6 plasmid (pEC_gr6), and a 43,093-bp plasmid harboring the mcr-4.3 gene (pEC_mcr4.3). When comparing both mcr-4.3 plasmids, pEH_mcr4.3 was fully represented in pEC_mcr4.3, with the ISAba19 being in the opposite direction. Moreover, pEC_mcr4.3 harbored an extra 24,307-bp segment that mostly contained a set of acyl-coenzyme A (CoA)-related genes which is expressed in low levels under normal conditions but strongly induced under metabolic stress and oxidizing agents (12).
Two isolates carrying plasmids with mcr-4.3 have been recently reported in A. baumannii in the NCBI database; the first one carried a 35,502-bp plasmid (pAb-MCR4.3, GenBank accession number CP033872), which was isolated in 2008 in Brazil from a cerebrospinal fluid sample of a hospitalized patient (A. baumannii ST70/ST233). The second one carried a 25,602-bp plasmid isolated from a pig fecal sample (A. baumannii ST1303/ST1929) in 2018 in China (pAB18PR065, GenBank accession number CM013137) (13). The comparison of the four plasmids showed that pEH_mcr4.3 was the only common segment present within all the 4 plasmids (except for ISAba19) (Fig. 1). The fact that ISAba19 seems to be involved in the addition or removal of the other plasmid segments infers that it plays an important role in the remodeling of the plasmid scaffold, as seen in the 4 discussed plasmids (13). Moreover, the presence of the pEH_mcr4.3 in all of the 4 plasmids suggests that this plasmidic structure is stable. Of note, even though pEH_mcr4.3 carries two copies of the TA systems and was stable within the wild-type strains, it failed to be maintained upon transformation.
FIG 1.
Linear map of the four mcr-4.3-carrying plasmids from Acinetobacter baumannii; arrows show the direction of transcription of open reading frames (ORFs), while rectangles represent truncated ORFs. Replication, partitioning genes, mobile elements, mcr-4.3, and other remaining genes are designated by violet, black, yellow, red, and white, respectively. Gray shaded areas represent similar sequences among the respective regions, while dashed lines represent the insertion point of the complex transposon.
The results of this study highlight the possible role of global food trade in spreading of mcr-carrying plasmids into the human community. We hypothesize that the imported meat from Brazil could be an index case and a route of entry to the Czech Republic. The plasmid was probably not stable enough to keep its initial structure, probably due to the absence of selective pressure, fitness cost, and/or the ease of site-specific recombination of ISAba19, and lost a segment of 20 kb while retaining the rest as it reached the clinical settings. The comparative analysis of the plasmids highlighted the common origin of the plasmidic structure carrying mcr-4.3 among all the reported plasmids. However, the possible risks of transfer of A. baumannii with plasmid-mediated colistin resistance from animals and food to human populations have to be elucidated further. To our knowledge, this is the first case of the mcr-4.3 gene in A. baumannii isolated from a clinical case in Europe.
Data availability.
The nucleotide sequences of the 39741 chromosome (EH), pEH_gr13, pEH_gr3, pEH_mcr4.3, LEV1449/17Ec chromosome (EC), pEC_gr13, pEC_gr6, and pEC_mcr4.3 have been deposited in GenBank under the accession numbers CP038258, CP038259, CP038260, CP038261, CP038262, CP038263, CP038264, and CP038265, respectively, and under BioProject accession number PRJNA529047.
Supplementary Material
ACKNOWLEDGMENTS
We thank the collaborating laboratories involved in the PSMR (Resistance Monitoring Working Group) for their long-term cooperation and sending strains for the survey of mcr genes. Special thanks go to Blanka Ochvatová, M.D., from the Laboratory of Clinical Microbiology, SPADIA Lab, a.s., Ostrava-Poruba, for isolating and sending the Acinetobacter strain. We also thank Ivana Jamborova for performing MiSeq sequencing.
This work was supported by funding from Czech Health Research Council (grant number NV18-09-00605). It was also financed, in part, by the National Sustainability Program I (NPU I; grant number LO1503) and NPU II (grant number LQ1601) provided by the Ministry of Education Youth and Sports of the Czech Republic and Ministry of Agriculture of the Czech Republic, institutional support MZE-RO0518.
We have no conflicts to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01166-19.
REFERENCES
- 1.Carattoli A, Villa L, Feudi C, Curcio L, Orsini S, Luppi A, Pezzotti G, Magistrali CF. 2017. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill 22:30589. doi: 10.2807/1560-7917.ES.2017.22.31.30589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Partridge SR, Di Pilato V, Doi Y, Feldgarden M, Haft DH, Klimke W, Kumar-Singh S, Liu J-H, Malhotra-Kumar S, Prasad A, Rossolini GM, Schwarz S, Shen J, Walsh T, Wang Y, Xavier BB. 2018. Proposal for assignment of allele numbers for mobile colistin resistance (mcr) genes. J Antimicrob Chemother 73:2625–2630. doi: 10.1093/jac/dky262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teo JWP, Kalisvar M, Venkatachalam I, Ng OT, Lin RTP, Octavia S. 2017. mcr-3 and mcr-4 variants in carbapenemase-producing clinical Enterobacteriaceae do not confer phenotypic polymyxin resistance. J Clin Microbiol 56:e01562-17. doi: 10.1128/JCM.01562-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pagano M, Poirel L, Martins AF, Rozales FP, Zavascki AP, Barth AL, Nordmann P. 2015. Emergence of NDM-1-producing Acinetobacter pittii in Brazil. Int J Antimicrob Agents 45:444–445. doi: 10.1016/j.ijantimicag.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 5.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koboldt DC, Zhang Q, Larson DE, Shen D, Mclellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. 2012. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milne I, Stephen G, Bayer M, Cock PJA, Pritchard L, Cardle L, Shaw PD, Marshall D. 2013. Using Tablet for visual exploration of second-generation sequencing data. Brief Bioinform 14:193–202. doi: 10.1093/bib/bbs012. [DOI] [PubMed] [Google Scholar]
- 9.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 10.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oikonomou O, Sarrou S, Papagiannitsis CC, Georgiadou S, Mantzarlis K, Zakynthinos E, Dalekos GN, Petinaki E. 2015. Rapid dissemination of colistin and carbapenem resistant Acinetobacter baumannii in Central Greece: mechanisms of resistance, molecular identification and epidemiological data. BMC Infect Dis 15:559. doi: 10.1186/s12879-015-1297-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsuchiya Y, Zhyvoloup A, Baković J, Thomas N, Yu BYK, Das S, Orengo C, Newell C, Ward J, Saladino G, Comitani F, Gervasio FL, Malanchuk OM, Khoruzhenko AI, Filonenko V, Peak-Chew SY, Skehel M, Gout I. 2018. Protein CoAlation and antioxidant function of coenzyme A in prokaryotic cells. Biochem J 475:1909–1937. doi: 10.1042/BCJ20180043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma F, Shen C, Zheng X, Liu Y, Chen H, Zhong L, Liang Y, Liao K, Xia Y, Tian G-B, Yang Y. 2019. Identification of a novel plasmid carrying mcr-4.3 in an Acinetobacter baumannii strain in China. Antimicrob Agents Chemother 63:e00133-19. doi: 10.1128/AAC.00133-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The nucleotide sequences of the 39741 chromosome (EH), pEH_gr13, pEH_gr3, pEH_mcr4.3, LEV1449/17Ec chromosome (EC), pEC_gr13, pEC_gr6, and pEC_mcr4.3 have been deposited in GenBank under the accession numbers CP038258, CP038259, CP038260, CP038261, CP038262, CP038263, CP038264, and CP038265, respectively, and under BioProject accession number PRJNA529047.