Abstract
Rapid risk-stratification of patients with acute traumatic brain injury (TBI) would inform management decisions and prognostication. The objective of this serum biomarker study (Biomarkers of Injury and Outcome [BIO]-Progesterone for Traumatic Brain Injury, Experimental Clinical Treatment [ProTECT]) was to test the hypothesis that serum biomarkers of structural brain injury, measured at a single, very early time-point, add value beyond relevant clinical covariates when predicting unfavorable outcome 6 months after moderate-to-severe acute TBI. BIO-ProTECT utilized prospectively collected samples obtained from subjects with moderate-to-severe TBI enrolled in the ProTECT III clinical trial of progesterone. Serum samples were obtained within 4 h after injury. Glial fibrillary acidic protein (GFAP), S100B, αII-spectrin breakdown product of molecular weight 150 (SBDP150), and ubiquitin C-terminal hydrolase-L1 (UCH-L1) were measured. The association between log-transformed biomarker levels and poor outcome, defined by a Glasgow Outcome Scale-Extended (GOS-E) score of 1–4 at 6 months post-injury, were estimated via logistic regression. Prognostic models and a biomarker risk score were developed using bootstrapping techniques.
Of 882 ProTECT III subjects, samples were available for 566. Each biomarker was associated with 6-month GOS-E (p < 0.001). Compared with a model containing baseline patient variables/characteristics, inclusion of S100B and GFAP significantly improved prognostic capacity (p ≤ 0.05 both comparisons); conversely, UCH-L1 and SBDP did not. A final predictive model incorporating baseline patient variables/characteristics and biomarker data (S100B and GFAP) had the best prognostic capability (area under the curve [AUC] = 0.85, 95% confidence interval [CI]: CI 0.81-0.89). Very early measurements of brain-specific biomarkers are independently associated with 6-month outcome after moderate-to-severe TBI and enhance outcome prediction.
Keywords: biomarker, GFAP, S100B, traumatic brain injury, UCH-L1
Introduction
Traumatic brain injury (TBI) is the leading cause of death and disability among young adults in the United States and worldwide.1 Rapid risk-stratification of TBI patients is of great importance to provide prognostic information and to make treatment decisions soon after TBI.2 However, existing methods to predict TBI outcome have not had a widespread impact on clinical practice. Existing methods also depend on clinical and imaging findings that are not consistently available in the acute setting. Elements of the neurological exam are often confounded by intoxication, pharmacological sedation or paralysis, or other injuries, and initial computed tomography (CT) interpretation is often limited by the experience and specialization of the reader.
Tissue damage after TBI results in the release of structural proteins into the bloodstream, including astrocytic (S100B protein [S100B], glial fibrillary acidic protein [GFAP]), and neuronal (ubiquitin C-hydrolase-L1 [UCH-L1]) and axonal (αII-spectrin breakdown products [SBDP]) proteins. S100B is a glial-specific protein expressed predominantly in mature astrocytes.3,4 GFAP is an intermediate filament protein also expressed in astrocytes.5–7 Leakage of both S100B and GFAP into blood is thought to reflect blood–brain barrier disruption.8,9 UCH-L1 is a deubiquinating enzyme highly expressed in neurons whose function regulates synaptic structure.10–12 Spectrin is a cytoskeletal protein whose function helps maintain plasma membrane integrity and cytoskeletal structure.13,14 Calpain-induced proteolysis of brain αII spectrin may facilitate cell damage and death in the context of central nervous system (CNS) insult.15 Previous reports suggest that serum blood levels of these proteins may correlate with the extent/type of brain injury and influence prognosis.8,16–18
The goal of this prospective study was to determine whether acute elevations in serum levels of these biomarkers predict unfavorable clinical outcome assessed 6 months after moderate-to-severe acute TBI.
Methods
Design and population
The Progesterone for Traumatic Brain Injury, Experimental Clinical Treatment (ProTECT) III trial was a Phase III, randomized, multi-center trial designed to determine the efficacy of intravenous progesterone administration started within 4 h of TBI, compared with placebo, and was conducted at 49 trauma centers in the United States.19 Eligible subjects had a moderate-to-severe TBI, defined by a Glasgow Coma Scale (GCS) score ranging from 4 to 12 (on a scale of 3–15, with lower scores indicating a lower level of consciousness). Patients with hypoxia (oxygen saturation <90%), hypotension (systolic blood pressure <90 mm Hg), spinal cord injury, status epilepticus, or bilaterally unreactive pupils were not eligible. The ProTECT III protocol was reviewed and approved by each site's institutional review board (IRB), including an amendment allowing blood sample collection, analysis, and storage. The ProTECT III trial began in July 2010 and was stopped early for futility in November 2013, after 882 subjects had been randomized.19 Patients included in this trial received protocol-driven care consistent with the Guidelines for the Management of Severe TBI.20 Biomarkers of Injury and Outcome in ProTECT III (BIO-ProTECT) was designed as an ancillary study to evaluate the association between prospectively collected serum biomarkers of structural injury and recovery in patients enrolled in the ProTECT III trial and was fully embedded into the aforementioned trial design. The first BIO-ProTECT sample was collected in August 2011.
Biomarker sample handling and measurement
Standard operating procedures were used to collect different blood samples, and serum was used for the biomarker analysis. Blood samples were collected within 4 h of injury. Immediately after collection, blood samples were allowed to clot for 15 min on slush ice and then were separated by centrifugation at 2°C within 60 min of phlebotomy. The samples were stored at −80°C until shipped on dry ice to Banyan Biomarkers, Inc. for storage and sample analysis.
All biomarkers' assay performance followed published methods21–23 and samples were batched and run in duplicate at Banyan Laboratories at Banyan Biomarkers, Inc. The lower limit of quantification (LLOQ) and the upper limit of quantification (ULOQ) for GFAP and UCH-L1 assays (defined as limits of 20% precision) are as follows: GFAP: 0.03–50 (ng/mL) and UCH-L1: 0.1–9 (ng/mL). Assay limits for S100B and SBDP-150 represent lower and upper calibrator values as follows: S100B: 0.015–2 (ng/mL) and SBDP-150: 0.02–6.4 (ng/mL). Results of the biomarker analysis were delivered to the Statistical Coordinating Center at the Medical University of South Carolina, where the study database was managed and analyzed by the ProTECT III statistical team (SDY, LF, QP). Clinical investigators and personnel at Banyan Biomarkers, Inc. remained blinded to clinical data and subject allocation.
Outcome measurements
The primary outcome for the ProTECT III study was global neurological recovery at 6 months post-injury, as determined by Glasgow Outcome Scale-Extended (GOS-E).24 Scores on the GOS-E range from 1 (death) and 2 (vegetative state) to 7 and 8 (lower and upper good recovery, respectively), such that higher scores are associated with better outcomes. Unfavorable outcome was defined as a GOS-E score of 1–4 and a favorable outcome, a GOS-E score of 5–8.
Demographic and clinical injury variables
Baseline patient variables/characteristics used to characterize the population and for covariate adjustment included: age, sex, index Glasgow Coma Scale (iGCS) score (highest GCS score recorded prior to enrollment in the ProTECT clinical trial). CNS injury type, based on the Rotterdam CT scoring of the admission head CT,25 was also considered. Polytrauma was identified using the Abbreviated Injury Score (AIS) derived from the Injury Severity Score (ISS).26 Subjects with an AIS value ≥3 on any body region other than head were considered to have polytrauma; subjects who did not meet the criteria for polytrauma were designated as having isolated head injury.
Statistical analysis
Although the ProTECT III trial failed to demonstrate an effect of progesterone,19 the biomarker analyses were first carried out for the progesterone and placebo groups separately. Because the estimated biomarker effects were similar in both groups with no statistically significant interactions between treatment groups and individual biomarkers (Supplementary Fig. S1), data from both treatment groups were pooled together to provide greater power for analyzing the relationship between biomarkers and the primary outcome. Results presented are based on subjects for whom the biomarker levels were within the specified limits of quantification to obtain consistent estimates of their association with outcome.27,28 As a sensitivity analysis, prior to the predictive model building, a single imputation using the ULOQ or the LLOQ was applied to subjects with biomarker levels above or below the specified limits, respectively. Subjects with missing 6-month GOS-E scores were excluded from the analysis, given the small percentage (6%) of missing scores.
Associations between each biomarker and the primary outcome were evaluated with logistic regression models. Biomarker levels were log-transformed in the analytic models to meet the linearity in the logit assumption. Effect estimates for a percentage change in the raw biomarker value (say, e.g., X%) can be obtained by raising the reported odds ratio (OR) to the power ln(1 + [X/100]); that is, the OR for a 10% change in the raw biomarker value can be computed by raising the reported OR to the power ln(1.1). A multi-variable model was then constructed adjusting for covariates age, sex, CT findings according to the Rotterdam CT score and injury severity strata defined according to the index GCS known at randomization into ProTECT III. These variables were included regardless of statistical significance. The multi-variable model was then extended to include all biomarkers, and a backward selection approach was adopted to eliminate biomarkers that failed to significantly improve model fit. Receiver operating characteristic (ROC) curves were constructed, and the area under the curve (AUC) was estimated to assess the ability of each model to discriminate between favorable and unfavorable outcome. These analyses were conducted under a level of significance of 0.05 and performed using statistical analysis software SAS version 9.4.
For the predictive model to be generalizable to future patients with biomarkers outside of quantification limits, model generation was based on all subjects with all four biomarkers available, with the imputation specified above applied. Predictive models were first developed using a split-sample approach based on bootstrapping samples. A training sample with the same size of the original sample was first drawn with replacement from the original data set. Subjects who were not included in the training sample were treated as a test set. For each training sample, predictive models were built using logistic regression and classification-tree based methods, and the AUC, sensitivity, specificity, and predictive error were estimated from the test set. The classification-tree based approaches include Classification and Regression Trees (CARTs)29 and ensemble trees (Boosting, Bagging, and Random Forest).30–32 The CART model recursively partitions a response variable according to the characteristics of predictors until a certain level of homogeneity is achieved within a subgroup, resulting in a single tree. Each of the ensemble methods builds multiple trees, with each tree voting for the final prediction. This process was repeated for 100 iterations.33,34 The approach with the largest average AUC over the 100 repetitions was selected and adopted to construct final predictive models based on all subjects in the data set. An online format that includes key variables for determining outcome is available at https://liqiong.shinyapps.io/BioProTECT.
The internal validity of the final predictive models was evaluated via bootstrapping as described above. This analysis was carried out using software R version 3.0.2 with packages rpart, adabag, randomForest, and shiny.
Using simple logistic regression, a range of cutoff points for biomarker values were established for each individual biomarker among 100 bootstrapping samples of the original data. The cutoffs are calculated by setting the predicted probability of the outcome equal to 0.5, which is the default setup to define a predicted outcome based on logistic regression models.
For each biomarker, a positive exposure was defined as a value greater than or equal to the mean of the cutoff points derived from the above 100 bootstrapping samples. The biomarker score was then calculated as the total number of positive exposures for each subject. The score ranged from 0 (no positive biomarker exposures) to 4 (all four biomarker exposures were positive). This score was used to estimate the likelihood of a poor outcome based solely on biomarker values.
Results
Population description
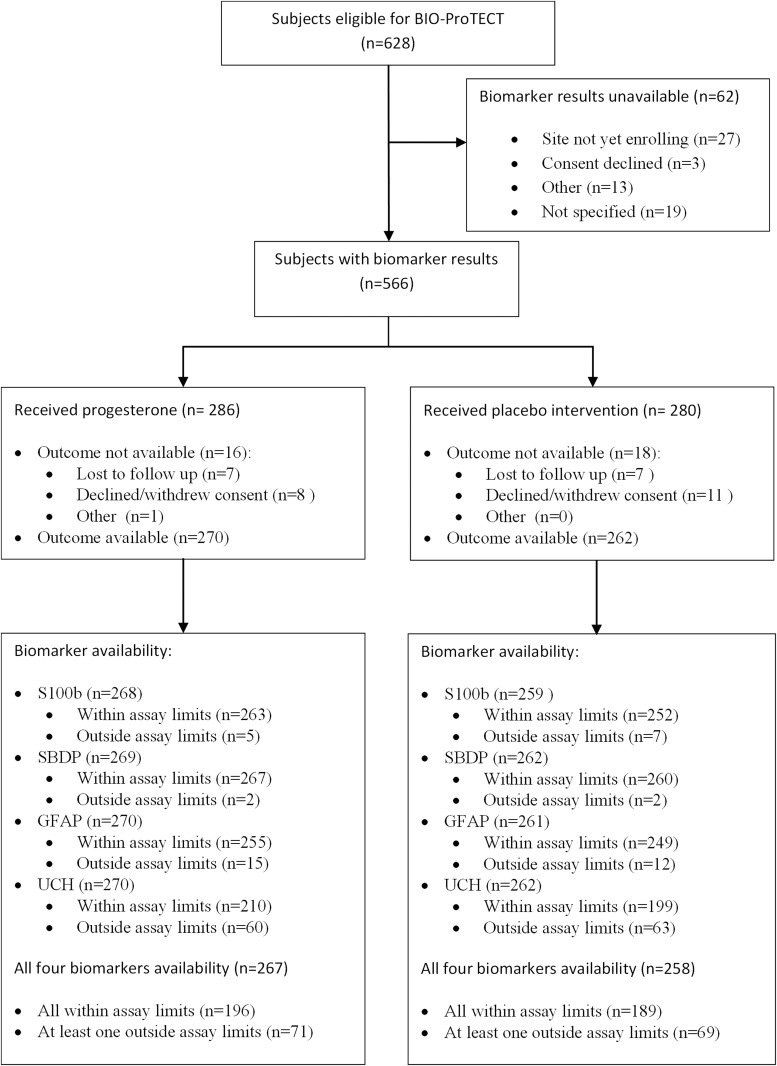
A total of 566 subjects had acute biomarker data available. The percentage of subjects with biomarker values returned within assay limits varied according to biomarker, as shown in Figure 1; 410 subjects had all four biomarker values within reportable ranges and were considered as complete cases. Subject characteristics are described in Table 1. The inclusion of subjects without reportable biomarker values (Imputed Cases) did not change the distribution of baseline characteristics. Among the 566 subjects, 39.6% (n = 224) had poor outcome (GOS-E 1–4), 54.4% (n = 308) had GOS-E 5–8, and 6% (n = 34) were missing the primary outcome at 6 months post-injury.
FIG. 1.
Biomarker availability. Flow chart depicting the number of subjects with acute biomarker data available, within range, and with primary outcome.
Table 1.
Cohort Characteristics
| Complete case | Imputed casea | ||
|---|---|---|---|
| N = 410 | N = 566 | ||
| Age (median [IQR]) | 33 (23 - 50) | 34 (23 - 52) | |
| Sex (men, n [%]) | 317 (77.32%) | 426 (75.27%) | |
| Race | White | 311 (75.85%) | 425 (75.09%) |
| Black/African American | 65 (15.85%) | 84 (14.84%) | |
| Others | 34 (8.29%) | 57 (10.07%) | |
| Initial injury severity | Moderate (iGCS 9-12) | 137 (33.41%) | 179 (31.63%) |
| Moderate severe | 203 (49.51%) | 289 (51.06%) | |
| (iGCS 6-8 / iMOTOR 4-5) | |||
| Most severe | 54 (13.17%) | 81 (14.31%) | |
| (iGCS 4-5 / iMOTOR 2-3) | |||
| Missing | 16 (3.9%) | 17 (3.00%) | |
| Rotterdam CT scoreb | 1&2 | 158 (38.54%) | 203 (35.87%) |
| 3 | 188 (45.85%) | 249 (43.99%) | |
| 4 | 36 (8.78%) | 54 (9.54%) | |
| 5&6 | 28 (6.83%) | 60 (10.60%) | |
| Mechanism of injury | Motor vehicle crash | 153 (37.32%) | 201 (35.51%) |
| Pedestrian struck by moving vehicle | 44 (10.73%) | 69 (12.19%) | |
| Motorcycle/Scooter/ATV crash | 77 (18.78%) | 113 (19.96) | |
| Other | 136 (33.17%) | 183 (32.33%) | |
| AIS head score | Missing | 1 (0.24%) | 1 (0.18%) |
| No injury | 13 (3.17%) | 20 (3.53%) | |
| Minor injury | 3 (0.73%) | 5 (0.88%) | |
| Moderate injury | 41(10.00%) | 55 (9.72%) | |
| Serious injury | 124 (30.24%) | 153 (27.03%) | |
| Severe injury | 108 (26.34%) | 153 (27.03%) | |
| Critical injury | 120 (29.27%) | 178 (31.45%) | |
| Non-survivable injury | 0 (0%) | 1 (0.18%) | |
| Injury Severity Score - mean (SD) | 23.57 (11.08) | 24.62 (11.39) | |
| Hours from injury to sample collected | 3.37 (0.66) | 3.32 (0.67) | |
| S100B | Raw (median [IQR]) | 0.22 (0.11-0.38) | 0.28 (0.13-0.55) |
| Log(S100B) (mean ± SD) | -1.59 ± 0.86 | -1.34 ± 1.04 | |
| GFAP | Raw (median[IQR]) | 2.29 (0.85-5.46) | 2.86 (0.91-8.56) |
| Log(GFAP) (mean ± SD) | 0.74 ± 1.40 | 0.98 ± 1.69 | |
| UCH-L1 | Raw (median[IQR]) | 2.67 (1.66-4.70) | 3.52 (1.89-8.16) |
| Log(UCH-L1) (mean ± SD) | 0.95 ± 0.77 | 1.30 ± 1.07 | |
| SBDP SBDP | Raw (median [IQR]) | 0.13 (0.07-0.21) | 0.16 (0.08-0.29) |
| Log(SBDP) (mean ± SD) | -2.02 ± 0.85 | -1.83 ± 0.93 | |
| GOS-E | Missing | 25 (6.01%) | 34 (6.01%) |
| Dead | 45 (10.98%) | 98 (17.31%) | |
| Vegetative state (vs) | 6 (1.46%) | 7 (1.24%) | |
| Lower severe disability (lsd) | 52 (12.68%) | 76 (13.43%) | |
| Upper severe disability (usd) | 34 (8.29%) | 43 (7.60%) | |
| Lower moderate disability (lmd) | 42 (10.24%) | 51 (9.01%) | |
| Upper moderate disability (umd) | 87 (21.22%) | 105 (18.55%) | |
| Lower good recovery (lgr) | 77 (18.78%) | 95 (16.78%) | |
| Upper good recover (ugr) | 42 (10.24%) | 57 (10.07%) | |
| Favorable outcome (good recovery or moderate disability) | 248 (60.49%) | 308 (54.42%) | |
Biomarker values out of reportable range are imputed with ULOQ or LLOQ as appropriate.
Due to data sparsity, Rotterdam CT scores are regrouped as 1&2, 3, 4, 5&6.
AIS, abbreviated injury score; ATV, all terrain vehicle; CT, computed tomography; GFAP, glial fibrillary acidic protein; GOS-E, Glasgow Outcome Scale-Extended; iGCS, index Glasgow Coma Scale score; iMotor, index Motor Score, IQR, interquartile range; LLOQ, lower limit of quantification; SBDP, spectrin breakdown product; SD, standard deviation; UCH-L1, ubiquitin C-terminal hydrolase-L1; ULOQ, upper limit of quantification.
Associations between individual serum biomarker levels and GOS-E
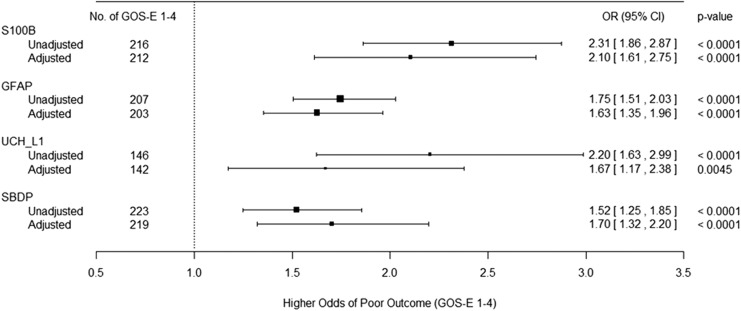
Figure 2 presents the results for the association between individual biomarkers and the outcome with/without adjusting for baseline patient variables/characteristics ; the ORs are estimated based on a 1-unit increase in the logarithm of the biomarker levels. All individual biomarkers are positively associated with the probability of having an unfavorable outcome.
FIG. 2.
Forest plot depicting the unadjusted and adjusted association between individual biomarkers and unfavorable outcome (Glasgow Outcome Scale-Extended [GOS-E] 1–4). The adjusted association includes age, gender, Rotterdam computed tomography (CT) score, and Glasgow Coma Scale (GCS) score.
With a 1-unit increase in log(S100B), log(GFAP), log(UCH-L1), and log(SBDP), the odds of unfavorable outcome at 6 months post-injury are increased by 131%, 75%, 120%, and 52%, respectively; alternatively, with a 10% increase in S100B, GFAP, UCH-L1, and SBDP, the odds of unfavorable outcome are increased by 8.3%, 5.5%, 7.8%, and 4.1%, respectively. Sensitivity analysis using the imputed values for biomarker levels outside of quantification limits does not substantively alter the conclusion. The results do not change substantively after adjusting for specified covariates. When multiple biomarkers are considered, after the backward selection approach to choose among the four biomarkers, the final model includes log(S100B) and log(GFAP) in addition to baseline patient variables/characteristics. A 1-unit increase in the log(S100B) or log(GFAP) increased the odds of unfavorable outcome by 72.4% (OR 1.724, 95% confidence interval [CI]: 1.18-2.52) and 48.8% (OR 1.488, 95%CI: 1.16-1.90), respectively, after controlling for all other covariates in the model; alternatively, a 10% increase in S100B or GFAP increased the odds of unfavorable outcome by 5.3% and 3.9%, respectively, after controlling for all other covariates in the model. These results were not substantively altered by restricting the sample to subjects with isolated head injury (Supplementary Fig. S2).
Comparison of the AUC between the adjusted models for individual biomarkers and the covariate-only model without biomarkers revealed that log(S100B) and log(GFAP) provide additional discriminative capability to a model containing only baseline patient variables/characteristics (Table 2). However, there is insufficient evidence to demonstrate that discrimination is improved with the addition of either log(UCH-L1) or log(SBDP) to the model containing only baseline patient variables/characteristics. The final model (biomarkers plus covariates) outperforms the covariate-only model but is not significantly better than the model that contains one of either log(S100B) or log(GFAP). Thus, excluding either S100B or GFAP from the final model does not significantly decrease discrimination, as long as the other biomarker remains. Sensitivity analysis using the imputed values for biomarker levels outside of quantification limits does not substantively change the reported conclusions; the final model using the imputed data has the same components and similar estimates of association.
Table 2.
Comparison of AUC for Individual Biomarkers before and after Adjusting for Prognostic Covariates
| Models | AUC | SE | 95% Wald | P-value (comparing with covariate only) | ||
|---|---|---|---|---|---|---|
| confidence limits | ||||||
| Covariate only | 0.8087 | 0.0237 | 0.7622 | 0.8551 | ||
| Log(S100B) | Unadjusted | 0.6743 | 0.0292 | 0.6171 | 0.7316 | |
| Adjusted | 0.8310 | 0.0222 | 0.7874 | 0.8745 | 0.0500 | |
| Log(GFAP) | Unadjusted | 0.7030 | 0.0275 | 0.6492 | 0.7569 | |
| Adjusted | 0.8384 | 0.0212 | 0.7968 | 0.8800 | 0.0156 | |
| Log(UCH-L1) | Unadjusted | 0.6605 | 0.0297 | 0.6023 | 0.7187 | |
| Adjusted | 0.8155 | 0.0234 | 0.7696 | 0.8614 | 0.3733 | |
| Log(SBDP) | Unadjusted | 0.5402 | 0.0308 | 0.4798 | 0.6006 | |
| Adjusted | 0.8127 | 0.0236 | 0.7665 | 0.8589 | 0.4276 | |
| Final modela | 0.8437 | 0.0208 | 0.8030 | 0.8844 | 0.0075 | |
The final model includes log(S100B) and log(GFAP) in addition to prognostic covariates among complete cases.
AUC, area under the curve; GFAP, glial fibrillary acidic protein; SBDP, spectrin breakdown product; SE, standard error; UCH-L1, ubiquitin C-terminal hydrolase-L1.
Predictive regression and classification-tree models
The average AUC, predictive error, sensitivity, and specificity of five candidate predictive model approaches over 100 bootstrapping samples revealed that the logistic regression model outperformed all the other tree-based models. The average AUC of the logistic regression model is 0.85 (95% CI: 0.81-0.89) with sensitivity 0.68 (95% CI: 0.58-0.76) and specificity 0.84 (95% CI: 0.78-0.89). It also has the smallest predictive error: 0.23 (95% CI: 0.18-0.27).
After excluding patients with missing data, a total of 509 subjects were included in the predictive model, developed via logistic regression. Three final predictive models were then defined as: 1) a full model, which includes age, sex, Rotterdam CT score, GCS score, S100B, and GFAP; 2) a reduced model, which excludes the Rotterdam CT score and GCS score; and 3) a reduced model, which excludes the biomarker values. Results for each predictive model are presented in Table 3. Predictive ability is increased as the complexity of the model increases. The full model has the best average AUC (0.84). Reduced models, which exclude the GCS and Rotterdam CT score (AUC = 0.79) or the biomarker values (AUC = 0.80), have similar predictive capability.
Table 3.
Final Predictive Models
| Predictors | Coding | Odds ratio (95% CI)a | ||
|---|---|---|---|---|
| Full model | W/o CT and GCS | W/o biomarkers | ||
| Age | 1.05 (1.04-1.07) | 1.05 (1.03-1.06) | 1.05 (1.03-1.06) | |
| Sex | Male | 1.10 (0.66-1.85) | 1.22 (0.75-1.97) | 0.79 (0.49-1.28) |
| Female | Reference (1.0) | Reference (1.0) | Reference (1.0) | |
| Rotterdam | 1&2 | 0.12 (0.04-0.31) | – | 0.05 (0.02-0.11) |
| 3 | 0.24 (0.10-0.60) | – | 0.15 (0.06-0.34) | |
| CT score | 4 | 0.41 (0.14-1.20) | – | 0.33 (0.12-0.92) |
| 5&6 | Reference (1.0) | – | Reference (1.0) | |
| Severityb | Moderate | 0.13 (0.06-0.27) | – | 0.14 (0.07-0.29) |
| Moderate-to-severe | 0.29 (0.15-0.55) | – | 0.29 (0.16-0.54) | |
| Most severe | Reference (1.0) | – | Reference (1.0) | |
| Log(S100B) | 1.59 (1.20-2.10) | 1.67 (1.28-2.17) | – | |
| Log(GFAP) | 1.50 (1.23-1.82) | 1.70 (1.43-2.03) | – | |
| AUC | 0.84 (0.79-0.88) | 0.79 (0.75-0.83) | 0.80 (0.75-0.84) | |
| Sensitivity | 0.67 (0.56-0.78) | 0.60 (0.50-0.72) | 0.64 (0.52-0.75) | |
| Specificity | 0.83 (0.74-0.91) | 0.80 (0.70-0.88) | 0.82 (0.72-0.90) | |
Odds ratios for continuous variable are calculated based on 1-unit increase of the variable.
Severity is defined by GCS (Table 1).
AUC, area under the curve; CI, confidence interval; CT, computed tomography; GCS, Glasgow Coma Scale; GFAP, glial fibrillary acidic protein.
Cutoff points for individual biomarkers
Cutoff points for individual biomarkers were derived based on 100 bootstrapping samples. The range of the cutoff is (−0.95 to −0.76), (1.47-1.69), (1.54-1.73), and (−1.30 to −0.45) for log(S100B), log(GFAP), log(UCH-L1), and log(SBDP), respectively (or [0.39-0.47], [4.35-5.42], [4.66-5.64], and [0.27-0.64], respectively, on the original scale).
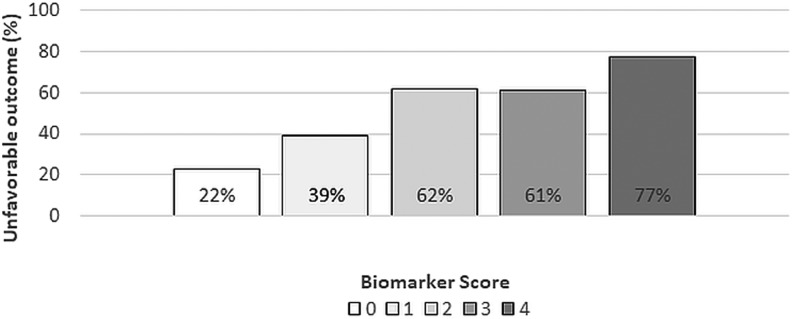
Biomarker score and risk of unfavorable outcome
Figure 3 illustrates the risk of unfavorable outcome (GOS-E 1–4) based on the total number of positive biomarker exposures per subject (0–4). Mean cutoff points upon which exposures are defined are: log(S100B) = −0.8697, log(GFAP) = 1.575, log(UCH_L1) = 1.633, and log(SBDP) = −1.005 (S100B = 0.42, GFAP = 4.83, UCH-L1 = 5.12, and SBDP = 0.37 on the original scale). The percentage of subjects with unfavorable outcome at each biomarker score is shown. For subjects with all four positive biomarker values, 77% experienced unfavorable outcome (95% CI: 0.63-0.92; p < 0.001). Among subjects with 0 biomarker exposures, only 22% (95% CI: 0.17-0.28; p < 0.001) had unfavorable outcome.
FIG. 3.
Rate of unfavorable outcome (Glasgow Outcome Scale-Extended [GOS-E] 1–4) as a function of the biomarker risk score. Rate of unfavorable outcome is the percentage of patients with a GOS-E score of 1–4 at 6 months after acute traumatic brain injury (TBI). The risk score is derived for each patient based on the number of high biomarker values.
Discussion
In this prospective study of serum proteins, we found that very early elevations of S100B, GFAP, UCH-L1, and SBDP150 independently predicted outcome in patients with moderate-to-severe TBI. Among these proteins, S100B and GFAP improved outcome prediction above baseline patient variables/characteristics including age, sex, GCS, and CT findings. A final predictive model that includes both biomarkers and baseline patient variables/characteristics provides an AUC of 84% with sensitivity of 67% and specificity of 83% in predicting poor outcome. In consideration of biomarker levels alone, poor outcome occurred in 77% versus 22% of subjects when all four biomarkers were above or below a cutoff threshold, respectively.
It is of particular interest that the combination of S100B and GFAP biomarker levels remain highly predictive even without baseline patient variables/characteristics in the predictive model. Whereas the combination of baseline patient variables/characteristics and biomarker levels improves outcome prediction, it is more clinically important that very early biomarkers may remain predictive in patients in which reliable clinical exam findings or expert CT interpretation may be limited or variable. Most TBI prognostic models include factors such as age, clinical severity (GCS), pupil reactivity, and CT findings.35 All CT scans in this study were interpreted centrally by the same diagnostic neuroradiologist. Because variability in imaging interpretation will diminish the predictive ability of the CT scan in TBI, biomarker levels may provide clinical utility through greater measurement reliability, particularly in settings where centralized radiological expertise is not readily available 24 h a day.
Similarly, pre-hospital resuscitation interventions (e.g., sedation, pharmacological paralysis and intubation) interfere with the determination of GCS total and subscores and confound clinical prediction tools. In addition, the frequent coexistence of TBI with drug or alcohol intoxication further confounds accurate diagnosis and assessment of TBI severity. Biomarkers appear to provide a less ambiguous assessment of injury and prognosis.
Previous studies have shown an association of these serum proteins with outcome after TBI. Wagner and Zitelli36 have demonstrated a relationship between serum S100B and acute mortality among 80 individuals with severe TBI, whereas Vos and colleagues16 have shown in 79 patients with TBI that, similar to the current study, GFAP and S100B are predictive of global outcome when used together in the same multi-variate model. In addition to outcomes, others have demonstrated some potential utility of serum GFAP and S100B with characterizing injury type and secondary TBI pathophysiology. For example, Herrmann and associates37 found elevated S100B levels correlated with CT imaging findings, whereas Pelinka and co-workers38 found elevated GFAP levels were related to intracranial injury and the development of inflammation, edema, and gliosis. However, none of these previous assessments were limited to consistent, very early sampling. Although the current study focused on the prognostic value of these markers for long-term outcome, future studies may consider large population assessments of how serum GFAP and S100B perform regarding predicting neurological deterioration and other adverse events.
There are several potential study limitations. The choice of biomarkers to measure in this study was not systematic. Instead, we selected four biomarkers with promising published data supporting their predictive ability in TBI. Variability in the kinetics of the expression of the biomarkers may be attributed to both injury phenotype and individual biomarker metabolism.39 Specifically, S100B is known to be released after extracranial injury. Future study may further characterize injury phenotype and expression of individual biomarkers. Additional study is needed to determine if other serum biomarkers not measured in this study, including non-brain specific biomarkers, may further improve outcome prediction. Although this study included subjects with severe TBI, those with a GCS score of 3, hypoxia (oxygen saturation <90%), hypotension (systolic blood pressure <90 mm Hg), spinal cord injury, status epilepticus, or bilaterally unreactive pupils were excluded from the parent study. Likewise, subjects with mild TBI were also excluded. Thus, the findings from this study may not be applicable to patients with TBI who have these clinical characteristics. Although rapid measurements of these biomarker levels are not yet readily available, point-of-care assay platforms may provide access to timely results.
In conclusion, serum biomarkers, particularly S100B and GFAP, may provide clinicians with incremental information, above that obtained by baseline patient variables/characteristics alone, for a more reliable estimate of injury prognosis that will assist in clinical decision making and treatment approach for individuals with moderate-to-severe TBI. Future work should focus on how high biomarker levels may identify patients who are at risk for early neurological deterioration. Such patients might benefit from increased surveillance or novel therapeutics. Lastly, the design of future acute TBI clinical trials may benefit from improved classification of individuals based on the additional information gained from assessing biomarker levels.
Supplementary Material
Acknowledgments
This study received funding from the National Institutes of Neurological Disorders and Stroke 1R01NS071867 (PI: Michael Frankel, Emory University). It was partially supported by grants from the National Institute of Neurological Disorders and Stroke of the National Institutes of Health (NS062778, 5U10NS059032, and U01NS056975, R01 NS071867) and the National Center for Advancing Translational Sciences of the National Institutes of Health (UL1TR000454), and by the Emory Emergency Neurosciences Laboratory in the Department of Emergency Medicine, Emory School of Medicine, and Grady Memorial Hospital.
Author Disclosure Statement/Funding Information
MF reports research support for a clinical trial of intracerebral hemorrhage as co-investigator at Emory: Nico Corporation, Inc. AJ reports being employed by Banyan Biomarkers, Inc, during the conduction of this study. He is also Scientific and Medical Advisor to Quanterix Corp. PV reports consulting fees from Ever Neuropharma during the conduction of this study. DWW reports being listed on a patent related to progesterone for the treatment of traumatic brain injury (U.S. patents 7,473,687, 7,915,244, and 8,455,468), which is owned by Emory University and grants from BHR Pharma during the conduction of this study. The other authors have no competing financial interests.
Supplementary Material
References
- 1. National Center for Health Statistics. (2002). National hospital ambulatory medical care survey. Emergency Department File. Atlanta, GA: Centers for Disease Control and Prevention [Google Scholar]
- 2. Hergenroeder G.W., Redell J.B., Moore A.N., and Dash P.K. (2008). Biomarkers in the clinical diagnosis and management of traumatic brain injury. Mol. Diagn. Ther. 12, 345–358 [DOI] [PubMed] [Google Scholar]
- 3. Isobe T., Takahashi K., and Okuyama T. (1984). S100a0 (alpha alpha) protein is present in neurons of the central and peripheral nervous system. J. Neurochem. 43, 1494–1496 [DOI] [PubMed] [Google Scholar]
- 4. Zomzely-Neurath C.E., and Walker W.A. (1980). Nervous system-specific proteins: 14-3-2 protein, neuron-specific eno-lase, and SlOO protein, in: Proteins of the Nervous System, 2nd ed. Bradshaw R.A., and Schneider D.M. (eds). Raven Press: New York, pps. 1–5 [Google Scholar]
- 5. Pelinka L.E., Kroepfl A., Schmidhammer R., Krenn M., Buchinger W., Redl H., and Raabe A. (2004). Glial fibrillary acidic protein in serum after traumatic brain injury and multiple trauma. J. Trauma 57, 1006–1012 [DOI] [PubMed] [Google Scholar]
- 6. Lumpkins K.M., Bochicchio G.V., Keledjian K., Simard J.M., McCunn M., and Scalea T. (2008). Glial fibrillary acidic protein is highly correlated with brain injury. J. Trauma 65, 778–782; discussion 782–774 [DOI] [PubMed] [Google Scholar]
- 7. Vos P.E., Lamers K.J., Hendriks J.C., van Haaren M., Beems T., Zimmerman C., van Geel W., de Reus H., Biert J., and Verbeek M.M. (2004). Glial and neuronal proteins in serum predict outcome after severe traumatic brain injury. Neurology 62, 1303–1310 [DOI] [PubMed] [Google Scholar]
- 8. Goyal A., Failla M.D., Niyonkuru C., Amin K., Fabio A., Berger R.P., and Wagner A.K. (2013). S100b as a prognostic biomarker in outcome prediction for patients with severe traumatic brain injury. J. Neurotrauma 30, 946–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liedtke W., Edelmann W., Bieri P.L., Chiu F.C., Cowan N.J., Kucherlapati R., and Raine C.S. (1996). GFAP is necessary for the integrity of CNS white matter architecture and long-term maintenance of myelination. Neuron 17, 607–615 [DOI] [PubMed] [Google Scholar]
- 10. Cartier A.E., Djakovic S.N., Salehi A., Wilson S.M., Masliah E., and Patrick G.N. (2009). Regulation of synaptic structure by ubiquitin C-terminal hydrolase L1. J. Neurosci. 29, 7857–7868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu M.C., Akinyi L., Scharf D., Mo J., Larner S.F., Muller U., Oli M.W., Zheng W., Kobeissy F., Papa L., Lu X.C., Dave J.R., Tortella F.C., Hayes R.L., and Wang K.K. (2010). Ubiquitin C-terminal hydrolase-L1 as a biomarker for ischemic and traumatic brain injury in rats. Eur. J. Neurosci. 31, 722–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Papa L., Akinyi L., Liu M.C., Pineda J.A., Tepas J.J., , 3rd, Oli M.W., Zheng W., Robinson G., Robicsek S.A., Gabrielli A., Heaton S.C., Hannay H.J., Demery J.A., Brophy G.M., Layon J., Robertson C.S., Hayes R.L., and Wang K.K. (2010). Ubiquitin C-terminal hydrolase is a novel biomarker in humans for severe traumatic brain injury. Crit. Care Med. 38, 138–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mondello S., Robicsek S., Gabrielli A., Brophy G.M., Papa L., Tepas J., , 3rd, Robertson C., Buki A., Scharf D., Jixiang M., Akinyi L., Muller U., Wang K.K., and Hayes R.L. (2010). αII-spectrin breakdown products (SBDPs): diagnosis and outcome in severe traumatic brain injury patients. J. Neurotrauma 27, 1203–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Riederer B.M., Zagon I.S., and Goodman S.R. (1986). Brain spectrin(240/235) and brain spectrin(240/235E): two distinct spectrin subtypes with different locations within mammalian neural cells. J. Cell Biol. 102, 2088–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huh G.Y., Glantz S.B., Je S., Morrow J.S., and Kim J.H. (2001). Calpain proteolysis of alpha II-spectrin in the normal adult human brain. Neurosci. Lett. 316, 41–44 [DOI] [PubMed] [Google Scholar]
- 16. Vos P.E., Jacobs B., Andriessen T.M., Lamers K.J., Borm G.F., Beems T., Edwards M., Rosmalen C.F., and Vissers J.L. (2010). GFAP and S100B are biomarkers of traumatic brain injury: an observational cohort study. Neurology 75, 1786–1793 [DOI] [PubMed] [Google Scholar]
- 17. Brophy G.M., Mondello S., Papa L., Robicsek S.A., Gabrielli A., Tepas J., 3rd Buki, A., Robertson C., Tortella F.C., Hayes R.L., and Wang K.K. (2011). Biokinetic analysis of ubiquitin C-terminal hydrolase-L1 (UCH-L1) in severe traumatic brain injury patient biofluids. J. Neurotrauma 28, 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mondello S., Papa L., Buki A., Bullock M.R., Czeiter E., Tortella F.C., Wang K.K., and Hayes R.L. (2011). Neuronal and glial markers are differently associated with computed tomography findings and outcome in patients with severe traumatic brain injury: a case control study. Crit. Care 15, R156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wright D.W., Yeatts S.D., Silbergleit R., Palesch Y.Y., Hertzberg V.S., Frankel M., Goldstein F.C., Caveney A.F., Howlett-Smith H., Bengelink E.M., Manley G.T., Merk L.H., Janis L.S., and Barsan W.G. (2014). Very early administration of progesterone for acute traumatic brain injury. New Engl. J. Med. 371, 2457–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bullock M.R., and Povlishock J.T. (2007). Guidelines for the management of severe traumatic brain injury [Editor's Commentary]. J. Neurotrauma 24 Suppl. 1:2, p. preceding S1 [DOI] [PubMed] [Google Scholar]
- 21. Papa L., Akinyi L., Liu MC., Pineda JA., Tepas J.J., , 3rd, Oli M.W., Zheng W., Robinson G., Robicsek S.A., Gabrielli A., Heaton S.C., Hannay H.J., Demery J.A., Brophy G.M., Layon J., Robertson C.S., Hayes R.L., and Wang K.K. (2010). Ubiquitin C-terminal hydrolase is a novel biomarker in humans for severe traumatic brain injury. Crit. Care Med. 38, 138–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Meier T.B., Nelson L.D., Huber D.L., Bazarian J.J., Hayes R.L., and McCrea M.A. (2017). Prospective assessment of acute blood markers of brain injury in sport-related concussion. J. Neurotrauma 34, 3134–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Papa L., Robertson C.S., Wang K.K., Brophy G.M., Hannay H.J., Heaton S., Schmalfuss I., Gabrielli A., Hayes R.L., and Robicsek S.A. (2015). Biomarkers improve clinical outcome predictors of mortality following non-penetrating severe traumatic brain injury. Neurocrit. Care 22, 52–64 [DOI] [PubMed] [Google Scholar]
- 24. Teasdale G.M., Pettigrew L.E., Wilson J.T., Murray G., and Jennett B. (1998). Analyzing outcome of treatment of severe head injury: a review and update on advancing the use of the Glasgow Outcome Scale. J. Neurotrauma 15, 587–597 [DOI] [PubMed] [Google Scholar]
- 25. Maas A.I., Hukkelhoven C.W., Marshall L.F., and Steyerberg E.W. (2005). Prediction of outcome in traumatic brain injury with computed tomographic characteristics: a comparison between the computed tomographic classification and combinations of computed tomographic predictors. Neurosurgery 57, 1173–1182; discussion 1173–1182 [DOI] [PubMed] [Google Scholar]
- 26. Baker S.P., O'Neill B., Haddon W., Jr., and Long W.B. (1974). The injury severity score: a method for describing patients with multiple injuries and evaluating emergency care. J. Trauma 14, 187–196 [PubMed] [Google Scholar]
- 27. Tsimikas J.V., Bantis L.E., and Georgiou S.D. (2012). Inference in generalized linear regression models with a censored covariate. Comput. Stat. Data Analysis 56, 1854–1868 [Google Scholar]
- 28. Sattar A., Sinha S.K., and Morris N.J. (2012). A parametric survival model when a covariate is subject to left-censoring. J. Biom. Biostat., Suppl. 3. doi: 10.4172/2155-6180.S3-002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Breiman L., Friedman J., Stone C.J., and Olshen R.A. (1984). Classification and Regression Trees. Taylor & Francis: New York [Google Scholar]
- 30. Breiman L. (2001). Random forests. Machine Learning 45, 5–32 [Google Scholar]
- 31. Breiman L. (1996). Bagging predictors. Machine Learning 24, 123–140 [Google Scholar]
- 32. Freund Y., Schapire R., and Abe N. (1999). A short introduction to boosting. J. Jpn. Soc. Artificial Intelligence 14, 1612 [Google Scholar]
- 33. Efron B. (1983). Estimating the error rate of a prediction rule: improvement on cross-validation. J. Am. Stat. Assoc. 78, 316–331 [Google Scholar]
- 34. Steyerberg E.W., Harrell F.E., Jr., Borsboom G.J., Eijkemans M.J., Vergouwe Y., and Habbema J.D. (2001). Internal validation of predictive models: efficiency of some procedures for logistic regression analysis. J. Clin. Epidemiol. 54, 774–781 [DOI] [PubMed] [Google Scholar]
- 35. Murray G.D., Butcher I., McHugh G.S., Lu J., Mushkudiani N.A., Maas A.I., Marmarou A., and Steyerberg E.W. (2007). Multivariable prognostic analysis in traumatic brain injury: results from the IMPACT study. J. Neurotrauma 24, 329–337 [DOI] [PubMed] [Google Scholar]
- 36. Wagner A.K., and Zitelli K.T. (2013). A rehabilomics focused perspective on molecular mechanisms underlying neurological injury, complications, and recovery after severe TBI. Pathophysiology 20, 39–48 [DOI] [PubMed] [Google Scholar]
- 37. Herrmann M., Jost S., Kutz S., Ebert A.D., Kratz T., Wunderlich M.T., and Synowitz H. (2000). Temporal profile of release of neurobiochemical markers of brain damage after traumatic brain injury is associated with intracranial pathology as demonstrated in cranial computerized tomography. J. Neurotrauma 17, 113–122 [DOI] [PubMed] [Google Scholar]
- 38. Pelinka L.E., Kroepfl A., Leixnering M., Buchinger W., Raabe A., and Redl H. (2004). GFAP versus S100B in serum after traumatic brain injury: relationship to brain damage and outcome. J. Neurotrauma 21, 1553–1561 [DOI] [PubMed] [Google Scholar]
- 39. Mondello S., Muller U., Jeromin A., Streeter J., Hayes R.L., Wang K.KW. (2011). Blood-based diagnostics of traumatic brain injuries. Expert Rev. Mol. Diagn. 11, 65–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.