Abstract
Objective
We sought to define the prevalence of pre-treatment INSTI resistance and assess the transmission networks of those with pre-treatment INSTI resistance.
Design
A retrospective cohort study of HIV-positive patients with genotypic resistance testing sent to a single referral laboratory in North Carolina between 2010 and 2016.
Methods
We linked genotype and public health data for in-care HIV-positive individuals to determine the prevalence of INSTI resistance among treatment-naïve (defined as those with a first genotype ≤3 months after diagnosis) and treatment-experienced (defined as those with a first genotype >3 months after diagnosis) patients. We performed molecular and phylogenetic analyses to assess whether pre-treatment INSTI resistance mutations represented clustered HIV transmission.
Results
Of 8825 individuals who contributed sequences for protease (PR), reverse transcriptase (RT), or INSTI genotypic resistance testing during the study period, 2784 (31%) contributed ≥1 sequence for INSTI resistance testing. Of these, 840 were treatment-naïve individuals and 20 (2.4%, 95% confidence interval [CI]: 1.5, 3.6%) had INSTI mutations; only two (0.2%, 95%CI: 0.02, 0.9%) had major mutations. Of 1944 treatment-experienced individuals, 9.6% (95%CI: 8.3, 11.0%) had any INSTI mutation and 7.0% (95%CI: 5.9, 8.3%) had major mutations; the prevalence of INSTI mutations among treatment-experienced patients decreased over time (P<0.001). Twelve of 20 individuals with pre-treatment INSTI mutations were part of 10 molecular transmission clusters; only one cluster shared identical minor mutations.
Conclusion
The prevalence of major pre-treatment INSTI resistance is very low. Pre-treatment INSTI mutations do not appear to represent clustered HIV transmission.
Keywords: HIV, integrase strand transfer inhibitors, transmitted drug resistance, cluster analysis, phylogenetic analysis
Introduction
Integrase strand transfer inhibitors (INSTIs) are part of recommended first-line regimens for the treatment of HIV infection.1 As observed with other classes of antiretroviral medications, increasing use of INSTIs and treatment failures on INSTIs may subsequently lead to an increase in pre-treatment INSTI resistance.2,3 While there are two case reports of antiretroviral-naïve individuals with major INSTI mutations in the context of multiclass antiretroviral drug resistance4,5, to date, major INSTI mutations are rare among cohorts of treatment-naïve individuals in Europe6–10, the Middle East11, and the United States.12
There are currently no data on pre-treatment INSTI resistance among HIV-positive individuals in the South, the epicenter of the HIV epidemic in the United States. Presently, the United States Department of Health and Human Services (DHHS) recommends routine pre-treatment reverse transcriptase (RT) and protease (PR) resistance testing but recommends INSTI resistance testing only if transmitted INSTI resistance is a concern (e.g., in the setting of multiclass drug resistance).13 In an analysis of INSTI resistance in the United States, 16.5% of HIV-positive patients in the South with INSTI resistance testing between 2009 and 2012 had a major mutation.14 This analysis, however, did not distinguish between treatment-naïve and treatment-experienced patients. HIV-positive patients initiating anti-retroviral therapy at the University of North Carolina from 1996 to 2014 whose initial regimen contained an INSTI were less likely to discontinue therapy and less likely to experience virologic failure compared to those whose initial regimen did not contain an INSTI.15 The durability of INSTI-containing regimens likely captures their safety, tolerability, and efficacy, factors that might reduce the risk of development and subsequent transmission of resistance compared to other regimens.16
The present study has three main objectives. First, we define the prevalence of INSTI resistance mutations among treatment-naïve and treatment-experienced individuals in North Carolina from 2010 to 2016. Second, we assess the socio-demographic and clinical characteristics of patients with INSTI resistance mutations. Finally, we use HIV sequences to construct transmission clusters and phylogenetic trees to investigate transmission networks of individuals with INSTI resistance mutations.
Methods
Study population
We analyzed HIV-1 sequences derived from samples sent to the largest referral laboratory in North Carolina (Laboratory Corporation of America, Research Triangle Park, North Carolina) for genotypic resistance testing from 16 November 2010 through 22 September 2016. We linked sequence data to the North Carolina State Division of Public Health’s Enhanced HIV/AIDS Reporting System (eHARS) that included age, sex, race/ethnicity, transmission risk category, CD4 count and viral load at the time of genotyping, and dates of diagnosis and genotypic resistance testing. We included individuals who were ≥18 years of age at resistance testing. The final dataset included 8825 individuals with 12,159 sequences for protease (PR), reverse transcriptase (RT), or INSTI resistance testing during the study period; 2784 (31%) of these individuals had 3162 sequences for INSTI resistance testing. With the exception of the cluster analysis described below, the 2784 individuals with INSTI resistance testing represent the population of interest for all analyses.
We examined diagnosis and sequence dates in eHARS to define individuals as treatment-naïve and treatment-experienced. The Centers for Disease Control and Prevention (CDC) estimate that 80% of HIV-positive people linked to care within 3 months of diagnosis in 201117 and that 72% of Blacks and 79% of whites with HIV infection linked to care within 1 month of diagnosis in 2014.18 Thus, we defined an individual as treatment-naïve if their first genotypic resistance test was sent within 3 months of diagnosis to capture the majority of newly diagnosed individuals linking to care for the first time. We classified individuals as treatment-experienced if their first genotypic resistance test was sent more than 3 months after diagnosis.
The University of North Carolina Institutional Review Board (IRB #16–2345) approved this study.
Definition of resistance mutations
Based on the 2015 International Antiviral Society (IAS) Update of the Drug Resistance Mutations in HIV-1, we defined major INSTI mutations as: T66I, E92Q, F121Y, Y143RHC, S147G, Q148HKR, and N155H.19 Minor or accessory mutations were defined as: T66AK, L74M, E92G, T97A, E138AK, G140AS, R263K.
Genotyping and analysis of nucleotide sequence data
Genotypic resistance testing was performed using GenoSure MG (Monogram Biosciences, San Francisco), GenoSure Integrase (Laboratory Corporation of America, Research Triangle Park, NC), and GenoSure PRIme (Monogram Biosciences, San Francisco, SF). We identified INSTI, PR and RT mutations using the Stanford University HIV Drug Resistance Database genotypic resistance interpretation algorithm with Sierra v1.1.20 We confirmed HIV subtypes using the Context-based Modeling for Expeditious Typing (COMET) tool.21
Cluster analysis
We performed a molecular cluster analysis using HIV-TRACE, available at www.hivtrace.org22 to describe the transmission networks of treatment-naïve individuals with INSTI resistance mutations. We included all 8825 individuals with genotypic resistance testing in the cluster analysis. We based the analysis on the partial pol gene (2042 with PR/RT and INSTI genotypes, 6656 with only PR/RT sequences and 127 with only INSTI sequences) using the first available sequence per patient in the 2010–2016 study period. We aligned sequences to HBX2 using MUSCLE and edited sequences manually for gapped positions.23 We identified pairs of sequences whose pairwise genetic distance was ≤0.015 expected substitutions per site divergent based on the Tamura-Nei 93 (TN93) substitution model implemented in HIV-TRACE as putative linkage between individuals.24 These linkages were constructed into clusters composed of ≥2 linked individuals. We counted any matching resolutions in nucleotide ambiguities as a perfect match.
Phylogenetic analysis
We then performed a phylogenetic analysis to identify clades defined by INSTI resistance mutations. For this analysis, we used the first INSTI sequence available during the 2010–2016 study period from the 2784 individuals with INSTI resistance testing. Sequences were aligned as above using MUSCLE and a maximum-likelihood tree was constructed in FastTree v.2.1.4 with the general time reversible model of nucleotide substitution.25,26 The purpose of this analysis was to evaluate for any INSTI resistance mutations circulating in clades at larger genetic thresholds than would be identified in the HIV-TRACE analysis. Statistical support of clades was assessed with local support values (Shimodaira-Hasegawa-like [SH-like] test) in FastTree.
Statistical analysis
We used the chi-squared test to compare the distributions of categorical variables and the Kruskal-Wallis test for continuous variables. We calculated the prevalence of INSTI resistance mutations over the study period and by year for those whose first genotype was sent ≤3 months after diagnosis and for those whose first genotype was sent >3 months after diagnosis. We calculated binomial exact 95% confidence intervals for prevalence estimates. We performed a sensitivity analysis and calculated the prevalence of INSTI resistance mutations using alternative cut-offs of 1 month and 6 months to define the treatment-naïve population. We used logistic regression to determine trends in the prevalence of INSTI resistance mutations over the study period. Statistical significance was defined at the P<0.05 level. We used STATA 14.2 for all analyses (College Station, TX).
Results
Between 2010 and 2016, 2784 individuals contributed 3162 INSTI sequences (2289 [72%] by GenoSure PRIme including PR/RT and INSTI sequencing and 873 [28%] by GenoSure Integrase). Of the 2784 individuals with INSTI testing, 2470 (89%) contributed one sequence and 314 (11%) contributed more than one sequence (range 2–6).
Prevalence of INSTI resistance mutations among those with INSTI resistance testing within 3 months of diagnosis
Both patients who had first resistance testing ≤3 months and >3 months after diagnosis experienced an increase in INSTI resistance testing over time (P for trend < 0.001 for both groups; Table 1). Compared to patients who had first resistance testing >3 months after diagnosis, patients with testing ≤3 months after diagnosis were less likely to have an INSTI sequence from 2010 to 2013; were younger; and, more likely to be male, white or Hispanic, and identify as MSM. Those with first resistance testing ≤3 months after diagnosis had a greater viral load and CD4 count, and a shorter duration to first genotype.
Table 1.
Socio-demographic and clinical characteristics of HIV-positive individuals by timing of first INSTI resistance testing after HIV diagnosis, North Carolina, 2010–2016.
| Individuals with first genotype ≤ 3 months after diagnosis (n = 840) | Individuals with first genotype > 3 months after diagnosis (n = 1944) | P value | |
|---|---|---|---|
| Socio-demographic characteristics | |||
| Year | <0.001 | ||
| 2010 | 0 | 6 (<1) | |
| 2011 | 3 (<1) | 50 (3) | |
| 2012 | 7 (<1) | 76 (4) | |
| 2013 | 13 (1) | 80 (4) | |
| 2014 | 135 (16) | 340 (17) | |
| 2015 | 355 (42) | 672 (34) | |
| 2016 | 327 (39) | 720 (37) | |
| Age, median (IQR) | 30 (24–44) | 43 (32–50) | <0.001 |
| Sex | <0.001 | ||
| Male | 685 (82) | 1378 (71) | |
| Female | 155 (18) | 566 (29) | |
| Race/ethnicity | <0.001 | ||
| Black | 537 (64) | 1452 (74) | |
| White | 190 (23) | 337 (17) | |
| Hispanic/Latino | 85 (10) | 87 (4) | |
| Native American | 8 (<1) | 2 (<1) | |
| Asian, Pacific Islander | 6 (<1) | 8 (<1) | |
| Multiracial | 14 (2) | 57 (3) | |
| Transmission Risk | |||
| MSM (includes MSM/IDU) | 522 (62) | 910 (47) | <0.001 |
| IDU | 16 (2) | 129 (6) | |
| Heterosexual | 104 (12) | 305 (16) | |
| Other | 198 (23) | 600 (31) | |
| Clinical characteristics | |||
| Subtype B | 820 (98) | 1908 (98) | 0.361 |
| HIV viral load, copies/mL, median (IQR) | 43081 (14970–115538) [n=574] | 21981 (3000–79700) [n=1278] | <0.001 |
| CD4 count, cells/mL, median (IQR) | 392 (216–575) [n=574] | 283 (113–488) [n=1084] | <0.001 |
| Time to first sequence, days, median (IQR) | 24 (12–41) | 3639 (1885–5819) | <0.001 |
Data are presented as n (%) unless otherwise specified.
Numbers may not add to total due to missing data.
HET, heterosexual; IDU, intravenous drug use; INSTI, integrase strand transfer inhibitor; IQR, interquartile range; MSM, men who have sex with men
Eight hundred and forty of 2784 (30%) individuals provided their first sample for INSTI resistance testing ≤3 months after diagnosis. Twenty (2.4%, 95% confidence interval [CI]: 1.5, 3.6%) of these individuals had INSTI mutations, 18 (2.1%, 95%CI: 1.3, 3.4%) with only a minor mutation and two (0.2%, 95%CI: 0.02, 0.9%) with a major mutation (Table 2). Both individuals with major INSTI mutations had concomitant RT resistance mutations.
Table 2.
Socio-demographic, clinical, and transmission cluster characteristics of individuals with INSTI mutations captured within 3 months of HIV diagnosis, North Carolina, 2010–2016.
| Subject | Year | Age | Sex | Race | Risk | HIV VL, copies/mL | CD4 cells/mL | Days to genotype | INSTI mutations | RT/PR mutations | Cluster characteristics | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ID, size | Degree | |||||||||||
| 1 | 2012 | 24 | M | Black | MSM | -- | -- | 34 | L74M | None | ||
| 2 | 2014 | 22 | M | Black | MSM | 600 | 387 | 24 | T97A | None | 270, 4 | 1 |
| 3 | 2015 | 45 | M | Black | HET | 141910 | 666 | 42 | T66A, S147G | G190A | 1319, 2 | 1 |
| 4 | 2015 | 40 | M | Black | HET | 6450 | 486 | 30 | T97AT | K103N | 366,14 | 4 |
| 5 | 2015 | 22 | M | Black | MSM | 54635 | 702 | 12 | T97AT | None | 1358, 2 | 1 |
| 6 | 2015 | 42 | M | White | NIR | 6856570 | 5 | 35 | E138K | None | ||
| 7 | 2015 | 20 | M | Black | NIR | 5410 | 805 | 23 | L74M | None | 1299,2 | 1 |
| 8 | 2015 | 32 | M | Black | MSM | 288872 | 383 | 34 | T97A | None | 151,8 | 1 |
| 9 | 2015 | 37 | M | Hispanic | MSM | 11676 | 775 | 54 | T97A | None | ||
| 10 | 2015 | 26 | M | Black | MSM | 46326 | 155 | 23 | T97AT | None | 1452, 2 | 1 |
| 11 | 2015 | 24 | M | Hispanic | MSM | 847 | 760 | 74 | T97AT | None | ||
| 12 | 2015 | 24 | M | Black | MSM | 324720 | 656 | 53 | T97A | K103N | 211, 8 | 3 |
| 13 | 2015 | 19 | M | Black | MSM | 15200 | 836 | 13 | T97A | None | 22, 48 | 11 |
| 14 | 2015 | 52 | F | Black | HET | 23400 | 964 | 39 | L74M | K103N | 222, 3 | 2 |
| 15 | 2016 | 32 | F | Black | NRR | 46630 | 572 | 15 | L74M | K103N | 222, 3 | 1 |
| 16 | 2016 | 29 | M | Black | MSM | 29800 | 538 | 68 | L74LM | None | ||
| 17 | 2016 | 24 | F | White | HET | 1450 | 1148 | 14 | T97A | None | ||
| 18 | 2016 | 65 | M | Black | NRR | 224380 | 13 | 59 | T97A | None | ||
| 19 | 2016 | 42 | F | Black | NRR | 8828 | 732 | 4 | L74M | K103N | 222, 3 | 1 |
| 20 | 2016 | 25 | M | Black | MSM | 30554 | 760 | 20 | N155H | D67N, M184V | ||
-- denotes missing data. HET, heterosexual; INSTI, integrase strand transfer inhibitor; MSM, men who have sex with men; NIR, adult with no identifiable risk; NRR, adult with no reported risk; RT, reverse transcriptase; PR, protease inhibitor; VL, viral load
The median age of the 20 individuals with INSTI mutations was 27.5 (IQR 24–41). Eighty percent were male, 80% identified as Black, and the most common transmission risk was sex with another male. All had subtype B virus. Median HIV viral load was 29800 copies/mL (range: 600 to 6,856,570 copies/mL) and median CD4 count was 666 (range: 5 to 1148 cells/mL). Median time from HIV diagnosis to first genotype was 32 days (range: 4 to 74 days). Eleven (55%) had a T97A/T mutation, 6 (30%) had a L74M mutation, and 1 (5%) had an E138K mutation. Two patients had major mutations: one with an S147G major mutation and a T66A minor mutation (Patient 3) and one with an N155H major mutation without minor mutations (Patient 20). The most common RT mutations were K103N (25%) followed by G190A (5%), M184V (5%), and D67N (5%). There were no major PR mutations. Patient 20 was identified through the acute infection program of the North Carolina Division of Public Health.
Sensitivity analysis of the definition of treatment-naïve individuals
Using a definition of treatment-naïve individuals as those with a genotype ≤1 month after HIV diagnosis, the prevalence of any INSTI mutation was 10/520, or 1.9% (95%CI: 0.9, 3.5%). One patient, Patient 20 who was identified with acute infection, had a major mutation (0.2%, 95%CI: 0.005, 1.1%). Using a cut-off of ≤6 months, the prevalence of any INSTI mutation was 22/908, or 2.4% (95%CI: 1.5, 3.6%). Three had a major mutation (0.3%, 95%CI: 0.07, 1.0%). The additional patient captured by increasing the cut-off to 6 months had an N155H mutation with seven RT mutations: M41L, D67N, V75M, M184V, L210W, T215Y, K103N, and E138Q.
Prevalence of INSTI mutations among those with INSTI resistance testing 3 or more months after diagnosis
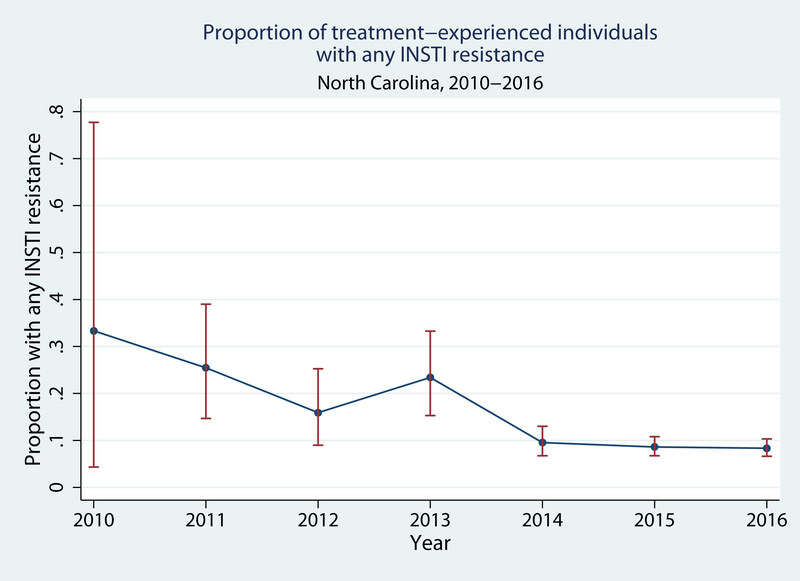
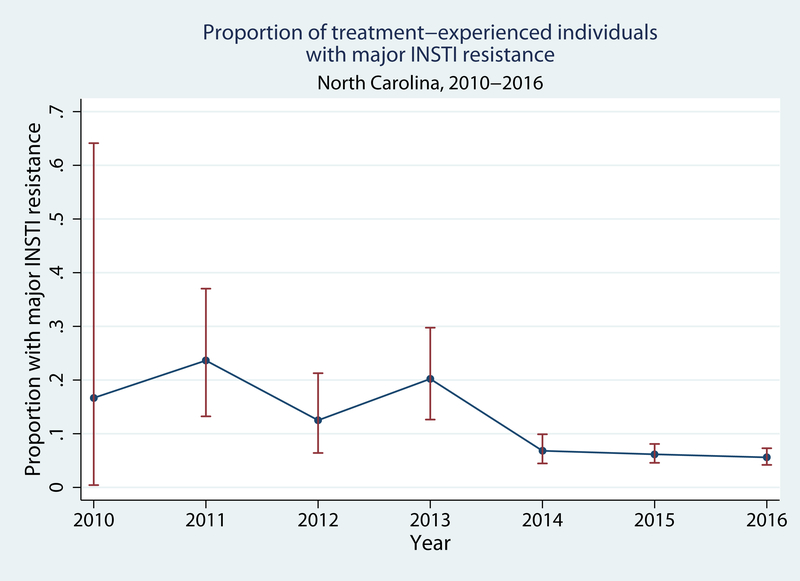
Of the 1944 individuals who provided their first sample for INSTI genotypic resistance testing >3 months after diagnosis, 187 (9.6%, 95%CI: 8.3, 11.0%) had INSTI mutations. Of these 187 individuals, 50 (27%) had only minor mutations, 128 (68%) had one major mutation (with or without minor mutations), and 9 (5%) had two major mutations (with or without minor mutations). Overall, the prevalence of any major mutation was 137/1944, or 7.0% (95%CI: 5.9, 8.3%). The prevalence of any and major INSTI resistance decreased over time. In 2010, 2/6 (33%, 95%CI: 4.3, 78%) had any resistance mutation while in 2016 54/720 had any resistance mutation (7.5%, 95%CI: 5.7, 9.7%; P for trend <0.001; Figure 1A). One of 6 (17%, 95%CI: 0.4, 64%) patients in 2010 and 36/720 (5.0%, 95%CI: 3.5, 6.8%) in 2016 had major mutations (P for trend <0.001; Figure 1B).
Figure 1A.
Prevalence of any INSTI resistance among treatment-experienced patients in North Carolina, 2010–2016.
Figure 1B.
Prevalence of major INSTI resistance among treatment-experienced patients in North Carolina, 2010–2016.
Error bars indicate 95% confidence intervals for each prevalence estimate.
Table 3 describes the socio-demographic and clinical characteristics of those with no, minor, and major mutations detected >3 months after diagnosis. Few patients had two major mutations. Of the 10 patients with two major INSTI mutations, three had S147G, Q148R, and E138K mutations; two had S147 and N155H mutations; and one each had E138EK, S147GS, Q148QR; G140GS, Y143CHRY, Q148HQ; G140S, Q148H, N115H; L74M, T97A, Y143C, S147G; and, T97AT, G149GS, Q148QE, N115HN mutations.
Table 3.
Socio-demographic, clinical, and transmission cluster characteristics of HIV-positive individuals with and without INSTI mutations on genotype testing > 3 months after HIV diagnosis, North Carolina, 2010–2016.
| No INSTI mutations (n = 1757) | Only minor INSTI mutations (n = 50) | Any major INSTI mutation (n = 137) | P value | |
|---|---|---|---|---|
| Socio-demographic characteristics | ||||
| Year | <0.001 | |||
| 2010 | 4 (<1) | 1 (2) | 1 (<1) | |
| 2011 | 38 (2) | 1 (2) | 11 (8) | |
| 2012 | 66 (4) | 2 (4) | 8 (6) | |
| 2013 | 61 (3) | 2 (4) | 17 (12) | |
| 2014 | 307 (17) | 10 (20) | 23 (17) | |
| 2015 | 615 (35) | 16 (32) | 41 (30) | |
| 2016 | 666 (38) | 18 (36) | 36 (26) | |
| Age, median (IQR) | 42 (32–51) | 38 (29–48) | 46 (37–50) | 0.015 |
| Sex | 0.431 | |||
| Female | 512 (29) | 18 (36) | 36 (26) | |
| Male | 1245 (71) | 32 (64) | 101 (74) | |
| Race/ethnicity | 0.904 | |||
| Black | 1311 (75) | 38 (76) | 104 (76) | |
| White | 306 (17) | 11 (22) | 20 (15) | |
| Hispanic/Latino | 80 (4) | 0 | 7 (5) | |
| Native American | 2 (<1) | 0 | 0 | |
| Asian, Pacific Islander | 7 (<1) | 0 | 1 (<1) | |
| Multiracial | 51 (3) | 1 (2) | 5 (4) | |
| Transmission Risk | 0.722 | |||
| MSM (includes MSM/IDU) | 820 (47) | 26 (52) | 64 (47) | |
| IDU | 118 (7) | 2 (4) | 9 (7) | |
| Heterosexual | 271 (15) | 11 (22) | 23 (17) | |
| Other | 548 (31) | 11 (22) | 41 (30) | |
| Clinical characteristics | ||||
| Subtype B | 1727 (98) | 47 (94) | 134 (98) | 0.081 |
| HIV viral load, copies/mL, median (IQR) | 24060 (3055–80520) [n=1176] | 31420 (2930–92350) [n=37] | 6000 (2266–34589) [n=65] | 0.051 |
| CD4, cells/mL, median (IQR) | 285 (113–490) [n=1011] | 276 (95–430) [n=25] | 273 (102–470) [n=48] | 0.590 |
| Years to first sequence, median (IQR) | 9 (5–15) | 9.5 (3–15) | 12 (7–18) | <0.001 |
| NRTI resistance mutations | 262 (15) | 15 (30) | 66 (48) | < 0.001 |
| NNRTI resistance mutations | 381 (22) | 15 (30) | 38 (28) | 0.109 |
| PI resistance mutations | 66 (4) | 2 (4) | 11 (8) | 0.051 |
| Extent of resistance (excludes INSTI resistance) | < 0.001 | |||
| Single class | 355 (20) | 17 (34) | 30 (22) | |
| Dual class | 141 (8) | 6 (12) | 35 (25) | |
| Triple class | 24 (1) | 1 (2) | 5 (4) | |
| INSTI minor mutations | NA | |||
| T66AK | 2 (4) | 0 | ||
| T97A | 32 (64) | 19 (14) | ||
| L74M | 8 (16) | 10 (7) | ||
| E92G | 4 (8) | 4 (3) | ||
| E138AK | 2 (4) | 13 (9) | ||
| G140AS | 1 (2) | 32 (23) | ||
| R263K | 6 (12) | 0 | ||
| INSTI major mutations | ||||
| T66I | 0 | NA | ||
| E92Q | 34 (25) | |||
| F121Y | 1 (<1) | |||
| Y143RHC | 20 (15) | |||
| S147G | 10 (7) | |||
| Q148HKR | 51 (37) | |||
| N155H | 67 (49) | |||
Data are presented as n (%) unless otherwise specified.
Numbers may not add to total due to missing data.
HET, heterosexual; IDU, intravenous drug use; INSTI, integrase strand transfer inhibitor; IQR, interquartile range; MSM, men who have sex with men; NRTI, nucleoside reverse transcriptase inhibitor; NNRTI, non-nucleoside reverse transcriptase inhibitor; PI, protease inhibitor
Cluster analysis
Of the 8825 individuals who contributed sequences between 2010 and 2016, 2899 (33%) comprised 774 clusters. Twelve of the 20 individuals with INSTI mutations in sequences collected ≤3 months of diagnosis were members of 10 distinct clusters with median size 3 (IQR 2–8) and median node degree of 1 (IQR 1–2.5; Figure 2). Forty-one percent (55/93) of cluster members had INSTI resistance testing. We observed two clusters involving ≥2 INSTI mutations (clusters 270 and 222). In cluster 270, the pre-treatment patient had a T97A mutation and the treatment-experienced cluster member had an R263KR mutation. In cluster 222, all three members had L74M and K103N mutations. We identified only one patient with pre-treatment major INSTI resistance in a cluster. Patient 3 with an S147G major mutation was part of a cluster 1319; he and the treatment-experienced cluster member shared a G190A mutation, but the cluster member did not have INSTI genotypic resistance testing. Patient 20, the patient with acute infection and an N155H major mutation, was not a member of a molecular cluster.
Figure 2.
Molecular transmission clusters including individuals with pre-treatment INSTI resistance mutations in North Carolina, 2010–2016.
Phylogenetic analysis
We identified nine clades with ≥2 sequences with identical INSTI resistance mutations. Three clades included pre-treatment patients with INSTI mutations in clusters 211, 222, and 270. Cluster 270 had a patient with a pre-treatment T97A mutation and a treatment-experienced patient with an R263K mutation; phylogenetic analysis included an additional treatment-experienced patient with T97A and F121Y mutations (SH-like test = 0.96). In cluster 211, a cluster member whose first sequence in the study period did not contain an INSTI region, had a subsequent, but initial INSTI sequence with a T97A mutation on phylogenetic analysis (SH-like test = 1.0); this cluster also included three sequences without INSTI mutations. Cluster 222 included no additional sequences or INSTI mutations (SH-like test = 0.96). The six additional clades included treatment-experienced individuals interspersed with individuals without INSTI mutations. The full phylogenetic tree is available as Supplementary Figure 1.
Discussion
We found a low prevalence (0.2%) of major INSTI resistance among patients with genotypes collected within 3 months of an HIV diagnosis. To our knowledge, this is the largest North American sample of individuals with INSTI resistance testing to date. This low prevalence of transmitted major INSTI mutations is consistent with prior reports from Europe6–10 and small sample of patients with primary infection in Seattle, WA.12 A sensitivity analysis of alternate cut-offs to define the treatment-naïve population yielded similarly low prevalence estimates.
Two patients had pre-treatment major INSTI mutations. One individual had an S147G major mutation, a nonpolymorphic mutation associated with elvitegravir resistance, as well as a T66A minor mutation that has also been associated with elvitegravir resistance in combination with other INSTI mutations.27 Neither of these mutations appear to affect susceptibility to dolutegravir or raltegravir. A second patient had an N155H mutation and was diagnosed during acute infection. Viruses with an N155H mutation show high-level resistance to raltegravir and elvitegravir and low-level resistance to dolutegravir.28,29 This patient also had an M184V mutation as well as D67N, a thymidine analogue mutation, or TAM. This particular TAM has been described in treatment-naïve patients as viruses with D67N retain replicative efficiency and can, thus, be transmitted.30,31 Transmitted M184V mutations appear to occur in settings where the viral load of the population of patients who are failing treatment is high due to the lower fitness of viruses with this mutation.3 Only the patient with the S147G major mutation was part of a transmission cluster; this patient and the cluster member shared a G190A mutation. The cluster member did not have INSTI genotypic resistance testing and we cannot know if they also shared INSTI mutations.
Minor INSTI mutations in the treatment-naïve population concentrated among young, Black MSM. The most common minor mutations among treatment-naïve patients were T97A and L74M, natural polymorphisms that have been found in individuals without prior INSTI exposure and prior to the widespread use of INSTIs.32–34 We also observed clustering of three women with both pre-treatment L74M and K103N mutations (cluster 222). While prior studies have documented clustering of individuals with K103N mutations,3,35,36 likely due to preserved or increased replicative fitness of viruses with this mutation, none have documented similar clustering of individuals with L74M mutations. As sexual transmission of HIV between women is rare,37 it is likely that additional (male) members of the transmission cluster are missing in our data (i.e., genotyping was performed before 2010 or performed elsewhere; the individual is not linked to care; or the individual remains undiagnosed).
Phylogenetic analysis revealed three clades that included five patients with pre-treatment INSTI resistance who were also part of clusters identified by molecular cluster analysis. Only cluster 222 represented potentially clustered transmission of minor INSTI mutations (L74M). Other clades and clusters with sequences with identical INSTI mutations were interspersed with sequences with no or other INSTI mutations or with sequences that frequently contained the same RT mutations. For example, in cluster 211, most individuals (75%) had K103N mutations, suggesting clustered transmission of K103N. Phylogenetic analysis showed that this clade included only two individuals with T97A mutations; a pre-treatment patient and a cluster member who only had an initial PR/RT sequence in the molecular cluster analysis and a subsequent, initial INSTI sequence >3 months after diagnosis.
The prevalence of any and major INSTI mutations among those with resistance testing >3 months after diagnosis was 9.6% and 7.0%, respectively. The latter estimate is lower than the 15.6% prevalence of major INSTI mutations in the United States between 2009 and 2012,14 but we also observed a decrease in the prevalence of any and major mutations over time. There are a few explanations for this decrease. First, there has been an increase in the collection of INSTI genotypic resistance testing over time. If the actual number of patients with INSTI mutations remained the same over time (the numerator) but the number of individuals on whom tests were sent increased over time (the denominator), the prevalence would be lower. Second, if the denominator became enriched with treatment-naïve individuals or treatment-experienced patients without INSTI exposure over time, the prevalence of INSTI resistance would also appear lower. The use of GenoSure PRIme resistance testing for PR/RT and INSTI mutations, particularly in pre-treatment patients, may contribute to this explanation. Finally, the decrease may reflect the effectiveness of HIV treatment in patients treated with a regimen with an INSTI backbone, particularly regimens containing dolutegravir,38,39 which have increased over the same time period.40
This study has several limitations. First, without information on treatment history, we have likely misclassified some treatment-naïve individuals as treatment-experienced, particularly newly-diagnosed individuals who present to care or start treatment >3 months after diagnosis. Our sensitivity analysis, however, yielded similar prevalences to the primary analysis. Second, we restricted our cluster and phylogenetic analyses to individuals who provided ≥1 sequence for genotypic resistance testing between 2010 and 2016 from a single laboratory group. Thus, we are missing clusters containing individuals with sequences sent elsewhere for genotypic resistance testing, individuals with genotypes collected outside the study period, and individuals without genotype testing due to lack of care engagement. Finally, there was significant missingness in viral load and CD4 data which may limit the validity of comparisons based on these data.
In a large sample of HIV-positive patients in North Carolina, we found the prevalence of transmitted major INSTI resistance to be very low. Additionally, pre-treatment INSTI resistance is largely due to minor mutations that are natural polymorphisms that are unlikely to impact treatment outcome. These polymorphisms do not appear to indicate clustered HIV transmission in this population. Nonetheless, INSTI mutation surveillance remains important in the setting of increasing use of INSTIs in the United States and worldwide.
Supplementary Material
Supplemental Digital Content Figure 1. Phylogenetic tree of 2784 individuals with INSTI genotype sequences in North Carolina, 2010–2016. Grey boxes indicate clades with two or more members with the same INSTI sequence. Branch labels indicate Shimodaira-Hasegawa-like test branch support. Clades without INSTI mutations are collapsed to improve readability.
Acknowledgments
This work was supported by the National Institute of Allergy and Infectious Diseases [1K08AI112432-01 to A.D.].
References
- 1.HIV/AIDS Treatment Guidelines [Internet]. AIDSinfo. [cited 2016 Aug 30];Available from: https://aidsinfo.nih.gov/ [Google Scholar]
- 2.Hurt CB. Transmitted resistance to HIV integrase strand-transfer inhibitors: right on schedule. Antivir Ther 2011;16(2):137–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang W-L, Kouyos R, Scherrer AU, et al. Assessing the Paradox Between Transmitted and Acquired HIV Type 1 Drug Resistance Mutations in the Swiss HIV Cohort Study From 1998 to 2012. J Infect Dis 2015;212(1):28–38. [DOI] [PubMed] [Google Scholar]
- 4.Boyd SD, Maldarelli F, Sereti I, et al. Transmitted raltegravir resistance in an HIV-1 CRF_AG-infected patient. Antivir Ther 2011;16(2):257–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Volpe JM, Ward DJ, Napolitano L, et al. Five Antiretroviral Drug Class–Resistant HIV-1 in a Treatment-Naïve Patient Successfully Suppressed with Optimized Antiretroviral Drug Selection. J Int Assoc Provid AIDS Care JIAPAC 2015;14(5):398–401. [DOI] [PubMed] [Google Scholar]
- 6.Cossarini F, Boeri E, Canducci F, et al. Integrase and Fusion Inhibitors Transmitted Drug Resistance in Naive Patients With Recent Diagnosis of HIV-1 Infection: JAIDS J Acquir Immune Defic Syndr 2011;56(2):e51–3. [DOI] [PubMed] [Google Scholar]
- 7.Frange P, Assoumou L, Descamps D, et al. HIV-1 subtype B-infected MSM may have driven the spread of transmitted resistant strains in France in 2007–12: impact on susceptibility to first-line strategies. J Antimicrob Chemother 2015;70(7):2084–9. [DOI] [PubMed] [Google Scholar]
- 8.Scherrer AU, Yang W-L, Kouyos RD, et al. Successful Prevention of Transmission of Integrase Resistance in the Swiss HIV Cohort Study. J Infect Dis 2016;214(3):399–402. [DOI] [PubMed] [Google Scholar]
- 9.Tostevin A, White E, Dunn D, et al. Recent trends and patterns in HIV-1 transmitted drug resistance in the United Kingdom. HIV Med 2016; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zoufaly A, Kraft C, Schmidbauer C, Puchhammer-Stoeckl E. Prevalence of integrase inhibitor resistance mutations in Austrian patients recently diagnosed with HIV from 2008 to 2013. Infection 2016;1–6. [DOI] [PubMed] [Google Scholar]
- 11.Jahanbakhsh F, Hattori J, Matsuda M, et al. Prevalence of transmitted HIV drug resistance in Iran between 2010 and 2011. PloS One 2013;8(4):e61864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stekler JD, McKernan J, Milne R, et al. Lack of resistance to integrase inhibitors among antiretroviral-naive subjects with primary HIV-1 infection, 2007–2013. Antivir Ther 2015;20(1):77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drug-Resistance Testing Adult and Adolescent ARV Guidelines [Internet]. AIDSinfo. [cited 2017 Jul 14];Available from: https://aidsinfo.nih.gov/ [Google Scholar]
- 14.Hurt CB, Sebastian J, Hicks CB, Eron JJ. Resistance to HIV integrase strand transfer inhibitors among clinical specimens in the United States, 2009–2012. Clin Infect Dis Off Publ Infect Dis Soc Am 2014;58(3):423–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davy T, Napravnik S, Zakharova O, Eron JJ. Increased persistence of initial ART with INSTI-containing regimens. In: Conference on Retroviruses and Opportunistic Infections Seattle, WA: 2017. [Google Scholar]
- 16.Lepik KJ, Harrigan PR, Yip B, et al. Emergent drug resistance with integrase strand transfer inhibitor-based regimens: AIDS 2017;31(10):1425–34. [DOI] [PubMed] [Google Scholar]
- 17.Bradley H, Hall HI, Wolitski RJ, et al. Vital Signs: HIV Diagnosis, Care, and Treatment Among Persons Living with HIV — United States, 2011. MMWR Morb Mortal Wkly Rep 2014;63(47):1113–7. [PMC free article] [PubMed] [Google Scholar]
- 18.Dailey AF. HIV Care Outcomes Among Blacks with Diagnosed HIV — United States, 2014. MMWR Morb Mortal Wkly Rep [Internet] 2017. [cited 2017 Feb 17];66 Available from: http://www.cdc.gov/mmwr/volumes/66/wr/mm6604a2.htm [DOI] [PMC free article] [PubMed]
- 19.Wensing AM, Calvez V, Günthard HF, et al. 2015 Update of the Drug Resistance Mutations in HIV-1. Top Antivir Med 2015;23(4):132–41. [PMC free article] [PubMed] [Google Scholar]
- 20.Stanford University HIV Drug Resistance Database [Internet]. Available from: https://hivdb.stanford.edu/hivdb/by-sequences/
- 21.Struck D, Lawyer G, Ternes A-M, Schmit J-C, Bercoff DP. COMET: adaptive context-based modeling for ultrafast HIV-1 subtype identification. Nucleic Acids Res 2014;42(18):e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wertheim JO, Leigh Brown AJ, Hepler NL, et al. The global transmission network of HIV-1. J Infect Dis 2014;209(2):304–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004;32(5):1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 1993;10(3):512–26. [DOI] [PubMed] [Google Scholar]
- 25.Price MN, Dehal PS, Arkin AP. FastTree 2 – Approximately Maximum-Likelihood Trees for Large Alignments. PLOS ONE 2010;5(3):e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price MN, Dehal PS, Arkin AP. FastTree: Computing Large Minimum Evolution Trees with Profiles instead of a Distance Matrix. Mol Biol Evol 2009;26(7):1641–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abram ME, Hluhanich RM, Goodman DD, et al. Impact of primary elvitegravir resistance-associated mutations in HIV-1 integrase on drug susceptibility and viral replication fitness. Antimicrob Agents Chemother 2013;57(6):2654–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hardy I, Brenner B, Quashie P, et al. Evolution of a novel pathway leading to dolutegravir resistance in a patient harbouring N155H and multiclass drug resistance. J Antimicrob Chemother 2015;70(2):405–11. [DOI] [PubMed] [Google Scholar]
- 29.Carganico A, Dupke S, Ehret R, et al. New dolutegravir resistance pattern identified in a patient failing antiretroviral therapy. J Int AIDS Soc 2014;17(4 Suppl 3):19749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.García-Lerma JG, MacInnes H, Bennett D, Weinstock H, Heneine W. Transmitted human immunodeficiency virus type 1 carrying the D67N or K219Q/E mutation evolves rapidly to zidovudine resistance in vitro and shows a high replicative fitness in the presence of zidovudine. J Virol 2004;78(14):7545–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.García-Lerma JG. Diversity of thymidine analogue resistance genotypes among newly diagnosed HIV-1-infected persons. J Antimicrob Chemother 2005;56(2):265–9. [DOI] [PubMed] [Google Scholar]
- 32.Llácer Delicado T, Torrecilla E, Holguín Á. Deep analysis of HIV-1 natural variability across HIV-1 variants at residues associated with integrase inhibitor (INI) resistance in INI-naive individuals. J Antimicrob Chemother 2016;71(2):362–6. [DOI] [PubMed] [Google Scholar]
- 33.Abram ME, Ram RR, Margot NA, et al. Lack of impact of pre-existing T97A HIV-1 integrase mutation on integrase strand transfer inhibitor resistance and treatment outcome. PloS One 2017;12(2):e0172206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ceccherini-Silberstein F, Malet I, D’Arrigo R, Antinori A, Marcelin A-G, Perno C-F. Characterization and structural analysis of HIV-1 integrase conservation. AIDS Rev 2009;11(1):17–29. [PubMed] [Google Scholar]
- 35.Mourad R, Chevennet F, Dunn DT, et al. A phylotype-based analysis highlights the role of drug-naive HIV-positive individuals in the transmission of antiretroviral resistance in the UK. AIDS Lond Engl 2015;29(15):1917–25. [DOI] [PubMed] [Google Scholar]
- 36.Drescher SM, von Wyl V, Yang W-L, et al. Treatment-Naive Individuals Are the Major Source of Transmitted HIV-1 Drug Resistance in Men Who Have Sex With Men in the Swiss HIV Cohort Study. Clin Infect Dis 2014;58(2):285–94. [DOI] [PubMed] [Google Scholar]
- 37.Kwakwa HA, Ghobrial MW. Female-to-Female Transmission of Human Immunodeficiency Virus. Clin Infect Dis 2003;36(3):e40–1. [DOI] [PubMed] [Google Scholar]
- 38.Mesplède T, Wainberg MA. Is resistance to dolutegravir possible when this drug is used in first-line therapy? Viruses 2014;6(9):3377–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oliveira M, Mesplède T, Quashie PK, Moïsi D, Wainberg MA. Resistance mutations against dolutegravir in HIV integrase impair the emergence of resistance against reverse transcriptase inhibitors. AIDS Lond Engl 2014;28(6):813–9. [DOI] [PubMed] [Google Scholar]
- 40.Medicare Part D Claims for Tivicay (Dolutegravir) [Internet]. [cited 2017 Feb 22]. Available from: https://projects.propublica.org/checkup/drugs/8149
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Digital Content Figure 1. Phylogenetic tree of 2784 individuals with INSTI genotype sequences in North Carolina, 2010–2016. Grey boxes indicate clades with two or more members with the same INSTI sequence. Branch labels indicate Shimodaira-Hasegawa-like test branch support. Clades without INSTI mutations are collapsed to improve readability.