Abstract
Significant effort has been applied to identify the genome-wide gene expression profiles associated with salivary gland development and pathophysiology. However, relatively little is known about the regulators that control salivary gland gene expression. We integrated data from DNase1 digital genomic footprinting, RNA-seq, and gene expression microarrays to comprehensively characterize the cis- and trans-regulatory components controlling gene expression of the healthy submandibular salivary gland. Analysis of 32 human tissues and 87 mouse tissues was performed to identify the highly expressed and tissue-enriched transcription factors driving salivary gland gene expression. Following RNA analysis, protein expression levels and subcellular localization of 39 salivary transcription factors were confirmed by immunohistochemistry. These expression analyses revealed that the salivary gland highly expresses transcription factors associated with endoplasmic reticulum stress, human T-cell lymphotrophic virus 1 expression, and Epstein-Barr virus reactivation. DNase1 digital genomic footprinting to a depth of 333,426,353 reads was performed and utilized to generate a salivary gland gene regulatory network describing the genome-wide chromatin accessibility and transcription factor binding of the salivary gland at a single-nucleotide resolution. Analysis of the DNase1 gene regulatory network identified dense interconnectivity among PLAG1, MYB, and 13 other transcription factors associated with balanced chromosomal translocations and salivary gland tumors. Collectively, these analyses provide a comprehensive atlas of the cis- and trans-regulators of the salivary gland and highlight known aberrantly regulated pathways of diseases affecting the salivary glands.
Keywords: salivary glands, gene expression regulation, systems biology, gene regulatory networks, messenger RNA, exocrine glands
Introduction
In the past decade, significant scientific effort has been applied to identify the genome-wide gene expression profiles associated with salivary gland development and pathology. Investigators have effectively characterized the temporal differentiation of salivary gland embryonic development and the differential expression patterns associated with oncogenic progression, sexually dimorphic gene expression, normal development, radiation exposure, and Sjögren’s syndrome, to name a few (Chen et al. 2008; Michael et al. 2011; Musselmann et al. 2011; Spiegelberg et al. 2014; Tandon et al. 2017). The extensive use of gene expression microarrays and RNA sequencing has enabled the construction of comprehensive catalogs of differential expression, but little is known about the network connectivity and regulators that drive these expression states. Modern transcriptome engineering techniques offer the potential to reengineer salivary gland pathogenic states via the identification and correction of each state’s regulatory drivers, but these techniques require extensive information about the network of the cell (Cahan et al. 2014; Michael et al. 2016). To address this challenge, we set out to comprehensively identify the cis- and trans-regulators of the salivary gland to enable therapeutic epigenetic reprogramming.
Models for eukaryotic gene regulation divide regulatory control into 2 broad categories: 1) trans-acting regulators, which are typically DNA-binding transcription factors (TFs) that cooperatively interact to form a quaternary structure capable of RNA polymerase recruitment, and 2) cis-acting DNA sequences, which encode the regulatory program to control the temporal and spatial expression of RNA via complex combinatoric logic (Gertz et al. 2009).
Progress in DNA sequencing and chromatin accessibility profiling has enabled the genome-wide characterization of the cis-acting sequences driving gene expression (Tsompana and Buck 2014). Techniques such as DNase1-seq and the assay for transposase-accessible chromatin using sequencing (ATAC-seq) reveal the open chromatin regions of the genome (Sabo et al. 2006; Buenrostro et al. 2013). When deeply sequenced, these techniques can resolve the “footprint” of TFs on the DNA by identifying regions of protection from enzymatic cleavage. When coupled with computational models of TF binding specificity (e.g., position weight matrices [PWMs]), DNase1-seq and ATAC-seq data can be used to generate a model of TF binding across the entire genome. These data provide a comprehensive window into the cis-acting sequences controlling state-dependent gene expression.
RNA sequencing and gene expression microarray data can be integrated with hidden Markov model protein domain predictors to exhaustively identify the trans-regulators expressed within a cell (Zhang et al. 2015). When coupled with gene expression atlases describing the expression of all genes across many tissues, a statistical analysis can identify highly expressed, tissue-enriched trans-factors. Although gene atlases do not typically include tissues of interest to the oral health community, the Human Protein Atlas and the BioGPS Mouse Gene Atlas contain gene expression profiles of salivary gland tissues (Lattin et al. 2008; Uhlen et al. 2015; Wu et al. 2016). These resources offer an opportunity to dissect the gene regulatory pathways driving salivary gland function.
Utilizing RNA sequencing, microarrays, and DNase1-seq data, we thoroughly identify the cis- and trans-components of salivary gland gene regulation (Fig. 1). We identified the highly expressed and tissue-specific TFs for the human and mouse salivary gland and evaluated the expression rank similarity of orthologues across the 2 species. To map cis-sequence utilization, we performed genome-wide DNase1 footprinting sequencing to construct the first genome-wide model of mouse salivary gland gene regulation. Finally, we used immunohistochemistry to confirm protein expression and the salivary gland tissue–type localization of 39 TFs. Collectively, our analyses identify the cis- and trans-regulators driving gene expression in the healthy mouse salivary gland and the trans-regulators of the healthy human submandibular salivary gland.
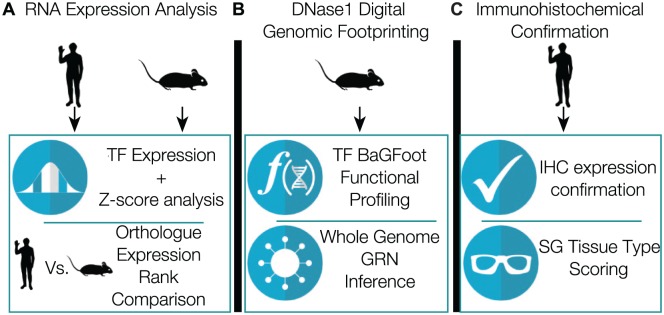
Figure 1.
Integrated genomic analysis approach. (A) Identification of highly expressed and organ-specific transcription factors via analysis of adult mouse and human RNA expression atlases. (B) Utilization of DNase1 digital genomic footprinting for whole genome network inference and transcription factor activity profiling. (C) Immunohistochemical protein expression confirmation and tissue-type scoring. GRN, gene regulatory network; IHC, immunohistochemistry; SG, salivary gland; TF, transcription factor.
Materials and Methods
For details on materials and methods, see the Appendix and Pranzatelli et al. (2018).
Results
Identification of Highly Expressed and Tissue-Specific Salivary Gland TFs in Mus musculus and Homo sapiens
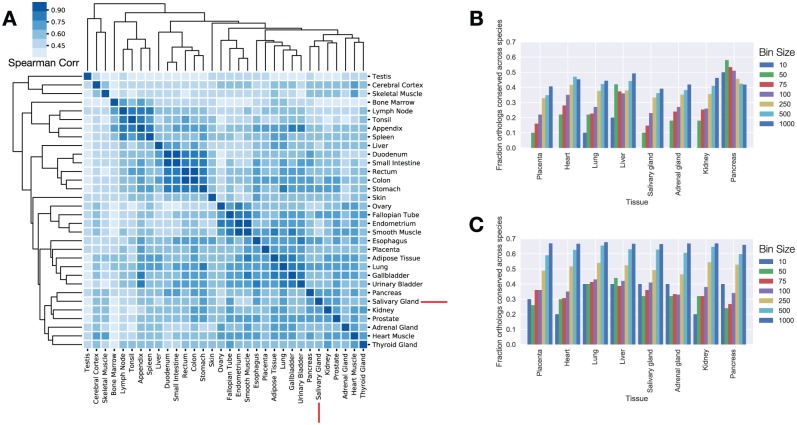
To identify highly expressed and tissue-specific TFs present in the salivary glands, we analyzed gene expression data from the Human Protein Atlas and Mouse Gene Atlas. Z score enrichment analysis was used to identify tissue-enriched TFs, and FPKM rank ordering (fragments per kilobase of a transcript per million mapped reads) was used for the identification of highly expressed factors. Hierarchical clustering with the top 50% highest expressed TFs across the Human Protein Atlas indicated that the salivary gland TF expression was most closely related to the prostate and pancreas (prostate, r = 0.75; pancreas, r = 0.74; Fig. 2A). This pattern was subtly altered in the mouse where RNA expression from additional tissues is available. Mouse salivary TF utilization mapped into an exocrine gland clade containing the salivary glands, pancreas, prostate, and lacrimal glands (Appendix Fig.). Spearman rank correlation coefficients in the mouse between the salivary gland and pancreas indicated similar degrees of TF rank expression (r = 0.72). In mice, the lacrimal gland exhibited the greatest degree of rank conservation with the salivary gland (lacrimal r = 0.89, prostate r = 0.73, pancreas r = 0.72).
Figure 2.
Analysis of gene expression atlases reveals the architecture of transcription factor (TF) expression across multiple organ types and species. (A) Heat map of TF Spearman rank correlations across 37 human tissues. The salivary gland TF expression profile closely matches that of the pancreas. (B) Ortholog expression rank conservation among all expressed human and mouse genes. TF expression ranks for both species were compared in bins of increasing size between species. When all genes are analyzed, highly expressed human genes are unlikely to be equally highly expressed in the mouse. (C) Ortholog rank conservation between mouse and human for TFs only. Forty percent of the top 10 human TFs are also in the top 10 expression ranks within the mouse. The pancreas is a notable exception and exhibits a high degree of rank conservation for all genes.
We evaluated the conservation of the salivary gland gene expression program by examining the TF expression rank of all mouse and human orthologues. In this analysis, the expression rank of each gene in the human was compared with the rank of the nearest ortholog in the mouse across incrementally larger bin sizes (Fig. 2B, C). Within the salivary gland, 40% of the top 10 most expressed TFs were also in the top 10 for mice (i.e., XBP1, ATF4, EHF, and CSDE1). When all expressed salivary gland genes were evaluated, none of the top 10 human orthologs were expressed within the top 10 positions of the mouse. Thus, TF expression ranks are better maintained across the species than the TF target genes. We observed a similar result in 7 of the 8 tissues that had clear human-to-mouse mappings, with the exception of the pancreas, where 50% of the top 10 human orthologs were expressed at similar expression ranks when all genes were evaluated (Fig. 2B).
Our analysis identified the highly expressed, tissue-enriched TFs within all tissues analyzed across both species (Table; Supplemental Data Sets D1–D4, available at https://github.com/ChioriniLab/SG_GRN). Within the human salivary gland, XBP1 was the highest expressed TF. XBP1, also known as X-box binding protein 1, coordinates the cellular response to endoplasmic reticulum (ER) stress by regulating the unfolded protein response in conjunction with ATF4 (Lee et al. 2005). XBP1 was 3.26 standard deviation (SD) and 3.33 SD enriched in the human and mouse salivary glands, respectively. ATF4 was detected at high levels within the salivary gland (rank 3 in humans and rank 10 in mice), suggesting that the salivary glands exhibit high levels of basal ER stress. Both XBP1 and ATF4 were highly expressed in other human exocrine tissues, such as the pancreas and prostate (XBP1: rank 1 in pancreas, rank 8 in prostate; ATF4: rank 3 in the pancreas, rank 9 in prostate).
Table.
Highly Expressed and Tissue-Specific TFs of the Human and Mouse.
| Human SG Highly Expressed
TFs |
Mouse SG Highly Expressed
TFs |
||||||
|---|---|---|---|---|---|---|---|
| Ensembl ID | Gene Symbol | Gene FPKM | Enrichment z Score | Ensembl ID | Gene Symbol | Gene Expression | Enrichment z Score |
| ENSG00000100219 | XBP1 | 398.4 | 3.26 | ENSMUSG00000020484 | Xbp1 | 21,331.93 | 3.33 |
| ENSG00000170345 | FOS | 266.3 | −0.52 | ENSMUSG00000033863 | Klf9 | 19,537.59 | 1.34 |
| ENSG00000128272 | ATF4 | 204 | 0.53 | ENSMUSG00000068823 | Csde1 | 13,942.81 | 1.20 |
| ENSG00000135373 | EHF | 114.9 | 2.06 | ENSMUSG00000059824 | Dbp | 13,481.33 | 4.17 |
| ENSG00000009307 | CSDE1 | 105.8 | −1.13 | ENSMUSG00000020644 | Id2 | 12,490.81 | 1.12 |
| ENSG00000177606 | JUN | 86.5 | 0.42 | ENSMUSG00000012350 | Ehf | 6,578.39 | 3.48 |
| ENSG00000054598 | FOXC1 | 72.6 | 5.41 | ENSMUSG00000052271 | Bhlha15 | 6,121.81 | 3.26 |
| ENSG00000008441 | NFIX | 71 | 1.21 | ENSMUSG00000027230 | Creb3l1 | 4,739.24 | 8.03 |
| ENSG00000043039 | BARX2 | 69.7 | 3.44 | ENSMUSG00000045302 | Preb | 4,588.78 | 3.09 |
| ENSG00000157514 | TSC22D3 | 68.4 | −0.79 | ENSMUSG00000042406 | Atf4 | 3,725.47 | 0.90 |
| ENSG00000120738 | EGR1 | 68 | −0.62 | ENSMUSG00000071076 | Jund | 3,173.16 | 1.74 |
| ENSG00000135374 | ELF5 | 66.4 | 5.41 | ENSMUSG00000004151 | Etv1 | 3,003.68 | 5.40 |
| ENSG00000115415 | STAT1 | 61.4 | 0.48 | ENSMUSG00000008575 | Nfib | 2,938.41 | 2.28 |
| ENSG00000189067 | LITAF | 55.3 | −0.37 | ENSMUSG00000030189 | Ybx3 | 2,793.36 | 0.086 |
| ENSG00000123358 | NR4A1 | 54 | −0.29 | ENSMUSG00000033006 | Sox10 | 2,509.21 | 5.77 |
| ENSG00000072310 | SREBF1 | 52.2 | 2.99 | ENSMUSG00000030199 | Etv6 | 2,314.32 | 2.40 |
| ENSG00000115738 | ID2 | 50.4 | −0.51 | ENSMUSG00000018377 | Vezf1 | 2,178.93 | 0.66 |
| ENSG00000100146 | SOX10 | 49.4 | 4.90 | ENSMUSG00000024515 | Smad4 | 2,091.61 | −0.52 |
| ENSG00000115112 | TFCP2L1 | 47.8 | 3.89 | ENSMUSG00000046532 | Ar | 2,080.42 | 2.06 |
| ENSG00000065978 | YBX1 | 46.1 | −1.19 | ENSMUSG00000046982 | Tshz1 | 1,998.57 | 0.88 |
| Human SG-Enriched TFs | Mouse SG-Enriched TFs | ||||||
| ENSG00000176009 | ASCL3 | 2.8 | 5.55 | ENSMUSG00000035951 | Ascl3 | 1,290.60 | 9.32 |
| ENSG00000136944 | LMX1B | 14.4 | 5.49 | ENSMUSG00000025215 | Tlx1 | 168.18 | 8.87 |
| ENSG00000054598 | FOXC1 | 72.6 | 5.41 | ENSMUSG00000009471 | Myod1 | 5.28 | 8.76 |
| ENSG00000135374 | ELF5 | 66.4 | 5.41 | ENSMUSG00000076431 | Sox4 | 4.68 | 8.22 |
| ENSG00000100146 | SOX10 | 49.4 | 4.90 | ENSMUSG00000027230 | Creb3l1 | 4,739.24 | 8.03 |
| ENSG00000170577 | SIX2 | 10 | 4.61 | ENSMUSG00000032033 | Barx2 | 21.14 | 7.90 |
| ENSG00000115112 | TFCP2L1 | 47.8 | 3.89 | ENSMUSG00000005503 | Evx1 | 6.92 | 7.74 |
| ENSG00000159556 | ISL2 | 0.8 | 3.77 | ENSMUSG00000001497 | Pax9 | 1,354.42 | 7.29 |
| ENSG00000179111 | HES7 | 0.3 | 3.62 | ENSMUSG00000028487 | Bnc2 | 6.86 | 7.16 |
| ENSG00000212993 | POU5F1B | 0.4 | 3.61 | ENSMUSG00000038560 | Sp6 | 7.17 | 7.07 |
| ENSG00000043039 | BARX2 | 69.7 | 3.44 | ENSMUSG00000021506 | Pitx1 | 763.89 | 6.47 |
| ENSG00000170549 | IRX1 | 5.4 | 3.40 | ENSMUSG00000028890 | Mtf1 | 7.47 | 6.10 |
| ENSG00000126778 | SIX1 | 22.9 | 3.33 | ENSMUSG00000013419 | Zfp651 | 8.21 | 5.89 |
| ENSG00000100219 | XBP1 | 398.4 | 3.26 | ENSMUSG00000033006 | Sox10 | 2,509.21 | 5.77 |
| ENSG00000072310 | SREBF1 | 52.2 | 2.99 | ENSMUSG00000004151 | Etv1 | 3,003.68 | 5.40 |
| ENSG00000143867 | OSR1 | 14.7 | 2.99 | ENSMUSG00000059552 | Trp53 | 5.90 | 5.20 |
| ENSG00000180535 | BHLHA15 | 37.1 | 2.96 | ENSMUSG00000048528 | Nkx1-2 | 5.33 | 5.02 |
| ENSG00000100625 | SIX4 | 3.5 | 2.88 | ENSMUSG00000024215 | Spdef | 872.43 | 4.94 |
| ENSG00000006468 | ETV1 | 42.4 | 2.85 | ENSMUSG00000041540 | Sox5 | 6.73 | 4.83 |
| ENSG00000125398 | SOX9 | 37.8 | 2.68 | ENSMUSG00000059824 | Dbp | 13,481.33 | 4.17 |
For humans and mice, the top 20 highly expressed and SG enriched (by z score) in a rank-ordered format. Human gene expression values are represented in units of FPKM. Mouse data are expressed in terms of arbitrary expression units. Expression profiles for all 119 analyzed human and mouse tissues are available in Supplemental Data Sets D1 to D4.
FPKM, fragments per kilobase of a transcript per million mapped reads; SG, salivary gland; TF, transcription factor.
The search for highly expressed and enriched salivary TFs recovered several factors with demonstrated roles in salivary gland development and pathophysiology. In humans and mice, ASCL3 was the most enriched salivary TF and was 5.55 and 9.32 SD enriched for salivary gland expression. ASCL3 was previously utilized as a marker for salivary progenitor cells in the mouse (Bullard et al. 2008). BHLHA15, also known as MIST1, was detected as 2.96 and 3.26 SD enriched for salivary gland expression in humans and mice. BHLHA15 was identified as a TF that promotes a secretory tissue architecture, expression of unfolded protein response genes via coregulation with XBP1, and is a marker for salivary gland acinar cell differentiation (Aure et al. 2015; Hess et al. 2016; Lo et al. 2017). Our results confirmed the analyses by Gluck et al. (2016), finding the enrichment of mouse Ehf (z score = 3.4) and Foxi2 (z score = 2.5) within the salivary gland . Numerous markers for cancer were also detected as highly expressed or enriched by our analysis including SOX10 and ETV1. In addition, our analyses confirmed the results for TFs noted by Gao et al. (2018), including but not limited to Sox10, Etv1, Barx2, Foxi1, Pax9 and Six1.
Bivariate Genomic Footprinting Defines TFs with Increased Salivary Activity
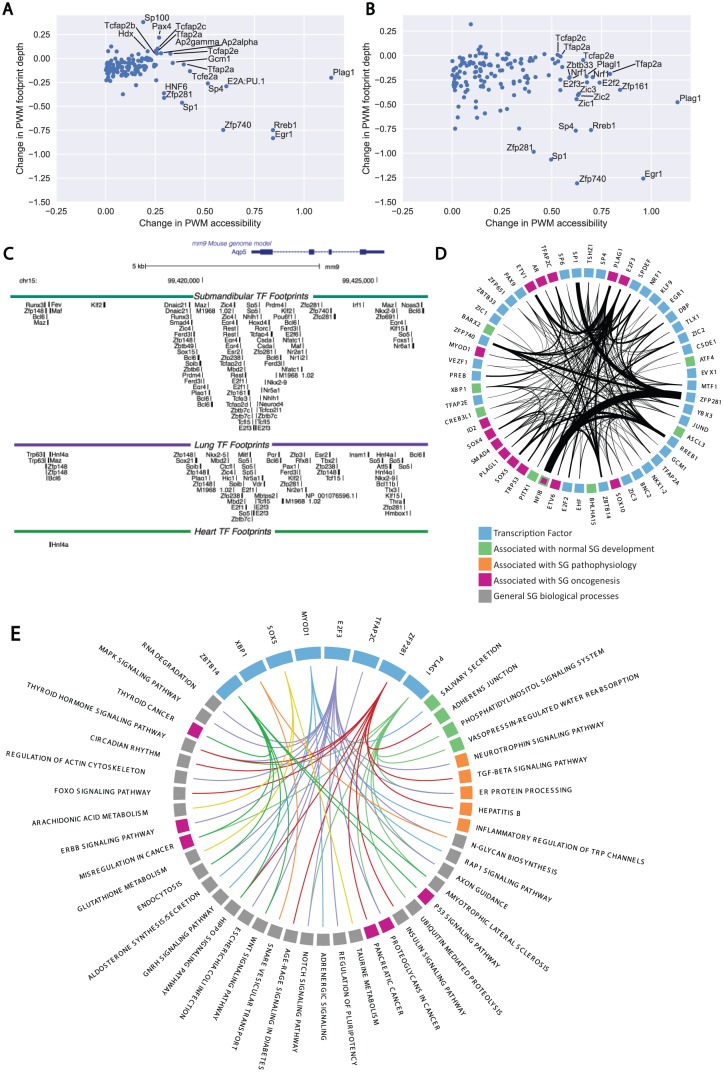
TF activity levels within the mouse salivary gland were assessed with DNase1-seq and bivariate genomic footprinting (BaGFoot) to measure chromatin accessibility and footprint depth of TF binding sites across 2 states (Baek et al. 2017). BaGfoot analysis identified salivary-specific changes in genomic activity by comparing salivary gland DNase1-seq signals against the mouse lung (Fig. 3A), a secretory organ that undergoes branching morphogenesis, and mouse heart (Fig. 3B) as a negative control. Normalized BaGfoot activity levels are presented in the Appendix Table. Interestingly, the pleomorphic adenoma gene 1 (Plag1) exhibited increased levels of PWM accessibility in the salivary gland. Plag1 is commonly rearranged and overexpressed within pleomorphic adenomas, the most common tumor of the salivary glands (Voz et al. 2000). Plag1 is expressed at low levels within the salivary gland (FPKM = 1.5) and is not enriched with the salivary glands of humans or mice (z score: 0.43 in humans, −0.22 in mice). Comparison of expression levels of Plag1 in the lung and heart indicates that the protein is expressed at similar levels to the salivary gland (human salivary gland FPKM = 1.5, human lung FPKM = 1.8, human heart FPKM 1.7). Given this similarity, the increased BaGfoot activity level of PLAG1 within the salivary gland points toward the possibility of extensive posttranscriptional regulation of PLAG1. Increased activity levels of RREB1, EGR1, SP1, ZFP740, ZFP281, and SP100 were also noted in both comparisons (Fig. 3A, B), with only EGR1 showing enriched expression in mouse salivary glands.
Figure 3.
Whole genome network construction and transcription factor (TF) activity inference via DNase1 digital genomic footprinting. (A) Bivariate genomic footprinting (BaGfoot) TF activity profiling of the salivary gland (SG) versus the mouse heart DNase1 profile. PLAG1 and ZFP281 activity levels are clearly increased as compared with the heart. (B) BaGfoot TF activity profile of the SG relative to the mouse lung. Increased activity levels of PLAG1, EGR1, and ZFP281 are observed in both profiles. (C) The complexity of the cis-regulatory architecture for the Aqp5 gene. Ninety-one unique TF footprints are observed within 5 kb of the Aqp5 transcription start site. (D) The core SG gene regulatory network constructed from the DNase1 footprinting. Blue nodes: General TFs. Green nodes: TFs associated with SG development. Orange nodes: TFs associated with salivary pathology. Purple nodes: TFs associated with SG oncogenesis. The network connectivity of the highly expressed, specific, and active TFs was extracted from the DNase1 gene regulatory network and visualized. Increased edge density represents increasing numbers of TF footprints within a gene’s promoter. Sixty-six ZFP281 footprints were detected within 5 kb of the Nfib promoter. (E) Pathway enrichment testing identifies overrepresented connections between TFs and gene pathways. A total of 105 TFs exhibited statistically significant enrichments in connections to specific gene pathways. The connectivity of 8 highly active, expressed, or salivary-specific factors is presented here. ZFP281 regulates 16 gene pathways. PLAG1 was detected as enriched to regulate 10 pathways, including the endoplasmic reticulum protein–processing pathway and the Itpr2/Itpr3 genes. PWM, position weight matrix.
Reconstruction of Salivary Gland Cis-regulation at a Single-Nucleotide Resolution
We utilized the mouse submandibular DNase1-seq data to construct the first genome-wide cis-regulatory model of salivary gland gene regulation with a single-nucleotide resolution. In this model, the TF footprints within a 5-kb window of all 55,419 mm9 mouse transcription start sites were scanned and attributed to a single TF whose PWM best explains that footprint. The analysis results in a genome-wide model that describes a predicted regulator for each detected footprint. In addition to the complete catalog of all annotated transcription start sites, the gene regulatory network incorporated all 44,459 distal enhancers detected by the FANTOM5 consortium. Overall, our algorithms identified 882,991 salivary gland TF footprints across the genome. The cis-regulatory architecture of Aqp5, the primary aquaporin channel for permitting transcellular water flux during the first step of saliva secretion, is presented in Figure 3C. Overall, 54 unique TFs were predicted to regulate the Aqp5 promoter in the salivary gland. Forty-four TFs were predicted to be bound to the Aqp5 promoter in the lung, and only 1 TF was detected in the heart. Of these TFs, 33 were uniquely detected within the salivary gland, and 22 were uniquely identified in the lung. Twenty-one TFs overlapped between the lung and the salivary gland. In addition, our analyses identified 296,271 unique salivary gland TF footprints that were not present in the mouse lung or heart DNase1 data sets (Supplemental Data Set D7). The genomic position and predicted TF for all detected salivary gland footprints across the mouse genome are provided as a BED file for use in future salivary noncoding DNA studies (Supplemental Data Set D5).
Mapping of the Cross-regulation within Highly Expressed and Tissue-Specific Salivary Gland TFs
Following construction of the salivary gland network, we examined the system for cross-regulation within the TFs identified by the RNA and DNase1-seq analyses. Across the 61 TFs, we identified 901 internal connections (Fig. 3D). There was a striking 66 footprints attributed to ZFP281 on the regulatory regions of Nfib, a TF involved in regulating embryonic submandibular salivary gland development and a known translocation partner of MYB, the primary driver translocation for adenoid cystic carcinomas (Mellas et al. 2015). The network indicated that PLAG1, a common translocation partner found in pleomorphic adenomas of the salivary glands, was predicted to regulate 24 core salivary gland network genes, including, but not limited to, Ar, Sox5, Sox9, Sp1, Sp4, and Pitx1. The network also indicated that PLAG1 is a predicted regulator of Etv1 (22 promoter footprints) and Etv6 (4 promoter footprints), 2 genes with known translocation partners involved in prostate, thyroid, breast, and salivary malignancies (Li et al. 2007; Attard et al. 2008; Skalova 2013; Leeman-Neill et al. 2014). We did not observe any internal connectivity between ASCL3 and BHLHA15 across the core network, indicating that these TFs could be activating downstream specialized networks outside the core network or require additional PWM optimization for network representation. Overall, the analysis of the DNase1-seq gene regulatory network implicates ZFP281 and PLAG1 as significant drivers of the core salivary gland gene regulatory program and highlights significant overlap among TFs already identified in salivary oncogenesis. The complete architecture of the salivary gland core network is provided in Supplemental Data Set D5.
Identification of Enriched TF Pathway Targets
To identify the general biological role of each TF, we analyzed the salivary gland gene regulatory network for enriched connectivity with the ENRICHR API and KEGG 2016. In this analysis, all network connections between a TF and all targets were assessed for connectivity enrichment between a TF and all gene pathways. A TF that exhibits higher-than-expected levels of connectivity to a gene pathway is hypothesized to act as a regulator of that pathway. We detected 510 TFs with at least 1 predicted footprint across the genome. Of these, 105 TFs exhibited significant connectivity enrichments to at least 1 biological pathway (a subset of these connections are presented in Fig. 3E). MYOD1-predicted footprints were identified as significantly enriched across the salivary secretion and aldosterone synthesis/secretion pathways. Within the salivary secretion pathway, MYOD1 was predicted to bind the regulatory regions of Gucy1a2, Atp2b3, Calml3, Atp1a1, Atp1b1, Adcy6, Adcy9, Plcb4, Gnaq, Gnas, Calm3, Nos1, Calm1, Kcnn4, and Prkaca. The network also detected PLAG1-predicted binding as enriched across 10 biological pathways, including ER protein processing, vasopressin-mediated water regulation, phosphatidylinositol signaling, and ubiquitin-mediated proteolysis. Examination of the network indicated that predicted Plag1 footprints were detected on the regulatory regions of many genes associated with salivary gland physiology, including Itpr2/Itpr3 and 33 other genes associated with the phosphatidylinositol signaling system (Supplemental Data Set D6). XBP1 was detected as a predicted regulator of N-glycan biosynthesis, general metabolic pathways, and SNARE vesicular interactions. XBP1 was also detected as a predicted regulator of protein processing in the ER, but this interaction did not reach statistical significance (P > 0.05). ZFP281 was detected as a predicted regulator of 16 cellular pathways, including TGF-beta signaling, adherens junctions, and multiple cancer-related pathways. The annotated pathway list for all 510 TFs detected within the network is provided in Supplemental Data Set D6.
Assessment of Salivary Gland TF Tissue Localization and Expression Level with Immunohistochemistry and Immunofluorescence
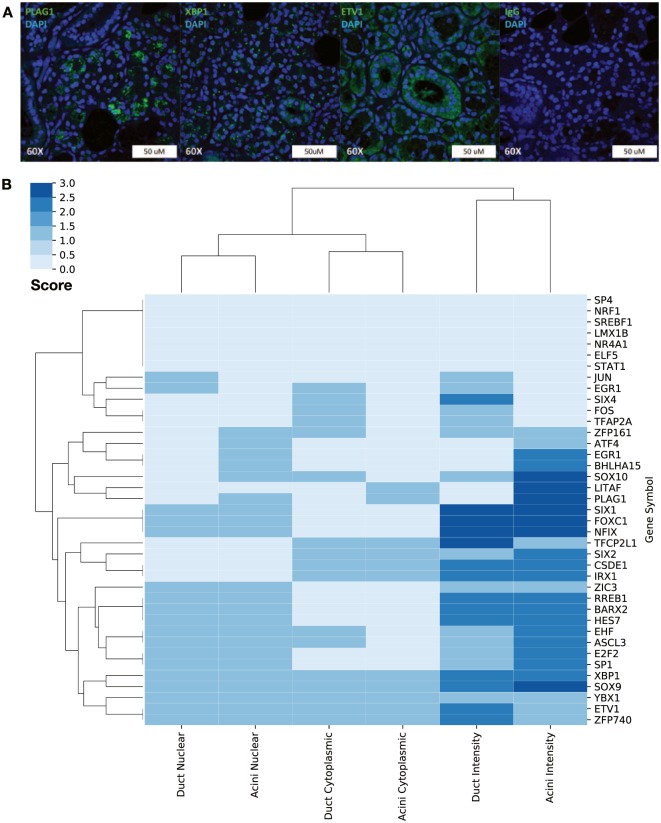
To confirm our RNA studies, we analyzed the expression and tissue localization of 39 TFs with immunohistochemistry and immunofluorescence (staining for selected TFs is presented in Fig. 4A). Each TF’s staining pattern was assigned a score for tissue and subcellular localization, then clustered for visualization (Fig. 4B). This analysis revealed 5 clusters present in the data:
Figure 4.
Immunohistochemical staining of selected transcription factors (TFs) reveals 5 distinct TF expression clusters within the salivary gland. (A) Representative images for PLAG1, XBP1, ETV1, and IgG control. (B) Hierarchical clustering of TF subcellular localization. Five clusters of TF expression were noted within the 39 assessed TFs corresponding to acinar-specific, ductal-specific TFs expressed within both cell types and TFs with a primarily cytoplasmic localization.
Acinar-specific TFs: ZFP161, ATF4, EGR1, BHLHA15, SOX10, LITAF, PLAG1
Ductal-specific TFs: JUN, EGR1, SIX4, FOX, TFAP2A
Acinar/ductal TFs: SIX1, FOXC1, NFIX, ZIC3, RREB1, BARX2, HES7, EHF, ASCL3, E2F2, Sp1, XBP1, SOX9, YBX1, ETV1, ZFP740
Cytoplasmic acinar/ductal TFs: TFCP2L1, SIX2, CSDE1, IRX1
Not detected: SP4, NRF4, SREBF1, LMX1B, NR4A1, ELF5, STAT1
Discussion
By integrating multiple genome-scale data sets and analysis types, we have attempted to comprehensively identify the cis- and trans-components of salivary gland gene regulation. Our analyses identify the highly expressed and tissue-specific trans-factors of the salivary gland. The high degree of conservation between expression rank of salivary gland TF expression and pancreatic TF expression was striking. Mouse and human salivary glands are structurally similar exocrine organs, but our findings demonstrate that the highly expressed genes in the top expression ranks are substantially different. Our analysis indicates that TF RNA expression ranks are more conserved than the expression ranks of the most highly expressed genes selected without regard to gene class. This observation is in line with previous data demonstrating the conservation of gene regulatory network connectivity between humans and mice and supports the mouse as a useful model for the study of salivary gene regulation (Stergachis et al. 2014).
Analysis of the trans-factors revealed that the salivary gland highly expresses XBP1 and ATF4. These factors coordinate the cellular unfolded protein response and were observed at high expression levels in the salivary gland. XBP1 and ATF4 were observed to upregulate the activity of the human T-cell lymphotrophic virus and the Epstein-Barr virus (Tsujimoto et al. 1991; Bhende et al. 2007; Ku et al. 2008; Igoe and Scofield 2013). Elevated transcriptional activity of XBP1 and ATF4 within the exocrine tissues may contribute to the viral-mediated development of exocrinopathies such as Sjögren’s syndrome by creating a transcriptional environment that is permissive toward viral activation and persistence.
Using DNase1-seq, we have developed the first genome-wide model of salivary gland cis-regulation. This computational model describes the genomic position and predicted regulator for all TF footprints detected within the salivary gland. The use of PWMs to analyze footprints for causative TFs is one important limitation of this approach, as each footprint is annotated with a predicted TF that best matches the underlying DNA sequence but may in fact be occupied by an alternative TF with similar sequence specificities. Further studies will be required to verify these predictions via ChIP-seq and discriminate among TFs with similar PWMs (i.e., the Sox TFs implicated in development; Emmerson et al. 2017). As whole salivary glands were used for this analysis, the resulting data represent a weighted average of the constitutive cells of the gland. Reconstruction of the M. musculus gene regulatory network revealed that known drivers of salivary tumors are densely connected to genes known to be involved in development (e.g., PLAG1, ETV6, MYB) and demonstrate increased levels of chromatin accessibility relative to other tissues. This observation highlights the hypothesis that salivary tumorigenesis is the result of dysregulation within the core gene regulatory network.
Collectively, our analyses integrate genome-scale data sets to comprehensively identify the cis- and trans-components of salivary gland gene regulation. The construction of a healthy whole genome salivary gland gene regulatory network is the first step toward enabling the use of transcriptome engineering to therapeutically reprogram salivary gland disease states. From a molecular therapeutics perspective, these results are a useful first step toward the construction of salivary-specific gene therapy promoters, which could utilize the unique cis-regulatory logic and salivary-enriched trans-factors identified here to maximize transgene expression while minimizing off-target effects. The network generated here also represents the first “baseline” state of the salivary epigenome and represents a useful starting point for future studies that examine pathologic states encountered during oncogenesis and autoimmunity. In the future, RNA-seq, DNase1-seq, and other chromatin-profiling strategies, such as ATAC-seq, represent an exciting opportunity to identify the molecular mechanisms underlying the state changes associated with salivary gland development and pathology.
Author Contributions
D.G. Michael, T.J.F. Pranzatelli, B.M. Warner, J.A. Chiorini, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; H. Yin, contributed to conception, design, and data acquisition, drafted and critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplemental Material
Supplemental material, DS_10.1177_0022034518806518 for Integrated Epigenetic Mapping of Human and Mouse Salivary Gene Regulation by D.G. Michael, T.J.F. Pranzatelli, B.M. Warner, H. Yin and J.A. Chiorini in Journal of Dental Research
Acknowledgments
The authors wish to acknowledge Drs. Jim Melvin and Xin Gao for the critical reading of this manuscript.
Footnotes
This study was supported by the Intramural Research Program of the National Institute of Dental and Craniofacial Research, National Institutes of Health (JAC Z01 DE00695). This work utilized the computational resources of the National Institutes of Health High Performance Computation Biowulf supercomputing cluster.
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
A supplemental appendix to this article is available online. Supplemental Data Sets D1 to D7 are publicly available on the Chiorini Lab data repository at https://github.com/ChioriniLab/SG_GRN.
ORCID iD: T.J.F. Pranzatelli  https://orcid.org/0000-0002-5763-1036
https://orcid.org/0000-0002-5763-1036
References
- Attard G, Clark J, Ambroisine L, Mills IG, Fisher G, Flohr P, Reid A, Edwards S, Kovacs G, Berney D, et al. 2008. Heterogeneity and clinical significance of ETV1 translocations in human prostate cancer. Br J Cancer. 99(2):314–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aure MH, Konieczny SF, Ovitt CE. 2015. Salivary gland homeostasis is maintained through acinar cell self-duplication. Dev Cell. 33(2):231–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek S, Goldstein I, Hager GL. 2017. Bivariate genomic footprinting detects changes in transcription factor activity. Cell Rep. 19(8):1710–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhende PM, Dickerson SJ, Sun XP, Feng WH, Kenney SC. 2007. X-box-binding protein 1 activates lytic Epstein-Barr virus gene expression in combination with protein kinase D. J Virol. 81(14):7363–7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 10(12):1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard T, Koek L, Roztocil E, Kingsley PD, Mirels L, Ovitt CE. 2008. Ascl3 expression marks a progenitor population of both acinar and ductal cells in mouse salivary glands. Dev Biol. 320(1):72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahan P, Li H, Morris SA, da Rocha EL, Daley GQ, Collins JJ. 2014. Cellnet: network biology applied to stem cell engineering. Cell. 158(4):903–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Mendez E, Houck J, Fan WH, Lohavanichbutr P, Doody D, Yueh B, Futran ND, Upton M, Farwell DG, et al. 2008. Gene expression profiling identifies genes predictive of oral squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 17(8):2152–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerson E, May AJ, Nathan S, Cruz-Pacheco N, Lizama CO, Maliskova L, Zovein AC, Shen Y, Muench MO, Knox SM. 2017. Sox2 regulates acinar cell development in the salivary gland. Elife. 6:e26620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Oei MS, Ovitt CE, Sincan M, Melvin JE. 2018. Transcriptional profiling reveals gland-specific differential expression in the three major salivary glands of the adult mouse. Physiol Genomics. 50(4):263–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz J, Siggia ED, Cohen BA. 2009. Analysis of combinatorial cis-regulation in synthetic and genomic promoters. Nature. 457(7226):215–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluck C, Min S, Oyelakin A, Smalley K, Sinha S, Romano RA. 2016. RNA-seq based transcriptomic map reveals new insights into mouse salivary gland development and maturation. BMC Genomics. 17(1):923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess DA, Strelau KM, Karki A, Jiang M, Azevedo-Pouly AC, Lee AH, Deering TG, Hoang CQ, MacDonald RJ, Konieczny SF. 2016. Mist1 links secretion and stress as both target and regulator of the unfolded protein response. Mol Cell Biol. 36(23):2931–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igoe A, Scofield RH. 2013. Autoimmunity and infection in Sjogren’s syndrome. Curr Opin Rheumatol. 25(4):480–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku SC, Lee J, Lau J, Gurumurthy M, Ng R, Lwa SH, Lee J, Klase Z, Kashanchi F, Chao SH. 2008. XBP-1, a novel human T-lymphotropic virus type 1 (HTLV-1) tax binding protein, activates HTLV-1 basal and tax-activated transcription. J Virol. 82(9):4343–4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattin JE, Schroder K, Su AI, Walker JR, Zhang J, Wiltshire T, Saijo K, Glass CK, Hume DA, Kellie S, et al. 2008. Expression analysis of G protein–coupled receptors in mouse macrophages. Immunome Res. 4:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Chu GC, Iwakoshi NN, Glimcher LH. 2005. XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. Embo J. 24(24):4368–4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeman-Neill RJ, Kelly LM, Liu PY, Brenner AV, Little MP, Bogdanova TI, Evdokimova VN, Hatch M, Zurnadzy LY, Nikiforova MN, et al. 2014. ETV6-NTRK3 is a common chromosomal rearrangement in radiation-associated thyroid cancer. Cancer. 120(6):799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Tognon CE, Godinho FJ, Yasaitis L, Hock H, Herschkowitz JI, Lannon CL, Cho E, Kim SJ, Bronson RT, et al. 2007. ETV6-NTRK3 fusion oncogene initiates breast cancer from committed mammary progenitors via activation of AP1 complex. Cancer Cell. 12(6):542–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo HYG, Jin RU, Sibbel G, Liu DQ, Karki A, Joens MS, Madison BB, Zhang B, Blanc V, Fitzpatrick JAJ, et al. 2017. A single transcription factor is sufficient to induce and maintain secretory cell architecture. Gene Dev. 31(2):154–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellas RE, Kim H, Osinski J, Sadibasic S, Gronostajski RM, Cho M, Baker OJ. 2015. NFIB regulates embryonic development of submandibular glands. J Dent Res. 94(2):312–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael D, Soi S, Cabera-Perez J, Weller M, Alexander S, Alevizos I, Illei GG, Chiorini JA. 2011. Microarray analysis of sexually dimorphic gene expression in human minor salivary glands. Oral Dis. 17(7):653–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael DG, Maier EJ, Brown H, Gish SR, Fiore C, Brown RH, Brent MR. 2016. Model-based transcriptome engineering promotes a fermentative transcriptional state in yeast. Proc Natl Acad Sci U S A. 113(47):E7428–E7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselmann K, Green JA, Sone K, Hsu JC, Bothwell IR, Johnson SA, Harunaga JS, Wei Z, Yamada KM. 2011. Salivary gland gene expression atlas identifies a new regulator of branching morphogenesis. J Dent Res. 90(9):1078–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pranzatelli TJF, Michael DG, Chiorini JA. 2018. ATAC2GRN: optimized ATAC-seq and DNase1-seq pipelines for rapid and accurate genome regulatory network inference. BMC Genomics. 19(1):563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo PJ, Kuehn MS, Thurman R, Johnson BE, Johnson EM, Hua C, Man Y, Rosenzweig E, Goldy J, Haydock A, et al. 2006. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat Methods. 3(7):511–518. [DOI] [PubMed] [Google Scholar]
- Skalova A. 2013. Mammary analogue secretory carcinoma of salivary gland origin: an update and expanded morphologic and immunohistochemical spectrum of recently described entity. Head Neck Pathol. 7 Suppl 1:S30–S36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelberg L, Swagemakers SM, Van Ijcken WF, Oole E, Wolvius EB, Essers J, Braks JA. 2014. Gene expression analysis reveals inhibition of radiation-induced TGFβ-signaling by hyperbaric oxygen therapy in mouse salivary glands. Mol Med. 20:257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergachis AB, Neph S, Sandstrom R, Haugen E, Reynolds AP, Zhang M, Byron R, Canfield T, Stelhing-Sun S, Lee K, et al. 2014. Conservation of trans-acting circuitry during mammalian regulatory evolution. Nature. 515(7527):365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandon M, Perez P, Burbelo PD, Calkins C, Alevizos I. 2017. Laser microdissection coupled with RNA-seq reveal cell-type and disease-specific markers in the salivary gland of Sjogren’s syndrome patients. Clin Exp Rheumatol. 35(5):777–785. [PubMed] [Google Scholar]
- Tsompana M, Buck MJ. 2014. Chromatin accessibility: a window into the genome. Epigenetics Chromatin. 7(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimoto A, Nyunoya H, Morita T, Sato T, Shimotohno K. 1991. Isolation of cDNAs for DNA-binding proteins which specifically bind to a tax-responsive enhancer element in the long terminal repeat of human T-cell leukemia-virus type-I. J Virol. 65(3):1420–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. 2015. Proteomics. Tissue-based map of the human proteome. Science. 347(6220):1260419. [DOI] [PubMed] [Google Scholar]
- Voz ML, Agten NS, Van de Ven WJM, Kas K. 2000. PLAG1, the main translocation target in pleomorphic adenoma of the salivary glands, is a positive regulator of IGF-II. Cancer Res. 60(1):106–113. [PubMed] [Google Scholar]
- Wu CL, Jin XF, Tsueng G, Afrasiabi C, Su AI. 2016. Biogps: building your own mash-up of gene annotations and expression profiles. Nucleic Acids Res. 44(D1):D313–D316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HM, Liu T, Liu CJ, Song SY, Zhang XT, Liu W, Jia HB, Xue Y, Guo AY. 2015. AnimalTFDB 2.0: a resource for expression, prediction and functional study of animal transcription factors. Nucleic Acids Res. 43(D1):D76–D81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, DS_10.1177_0022034518806518 for Integrated Epigenetic Mapping of Human and Mouse Salivary Gene Regulation by D.G. Michael, T.J.F. Pranzatelli, B.M. Warner, H. Yin and J.A. Chiorini in Journal of Dental Research