Abstract
Basic-helix-loop-helix (bHLH) transcription factors play an important role in various organs’ development; however, a tooth-specific bHLH factor has not been reported. In this study, we identified a novel tooth-specific bHLH transcription factor, which we named AmeloD, by screening a tooth germ complementary DNA (cDNA) library using a yeast 2-hybrid system. AmeloD was mapped onto the mouse chromosome 1q32. Phylogenetic analysis showed that AmeloD belongs to the achaete-scute complex-like (ASCL) gene family and is a homologue of ASCL5. AmeloD was uniquely expressed in the inner enamel epithelium (IEE), but its expression was suppressed after IEE cell differentiation into ameloblasts. Furthermore, AmeloD expression showed an inverse expression pattern with the epithelial cell-specific cell–cell adhesion molecule E-cadherin in the dental epithelium. Overexpression of AmeloD in dental epithelial cell line CLDE cells resulted in E-cadherin suppression. We found that AmeloD bound to E-box cis-regulatory elements in the proximal promoter region of the E-cadherin gene. These results reveal that AmeloD functions as a suppressor of E-cadherin transcription in IEE cells. Our study demonstrated that AmeloD is a novel tooth-specific bHLH transcription factor that may regulate tooth development through the suppression of E-cadherin in IEE cells.
Keywords: yeast two-hybrid assay, ameloblast, E-cadherin, epithelium, cell proliferation, cell migration
Introduction
Mammal tooth development is a classic organogenesis model characterized by reciprocal interactions between the dental epithelium and mesenchyme (Thesleff 1996). In mice, tooth development is initiated at embryonic day 11.5 (E11.5) and followed by a series of morphologically distinctive stages. At E14.5, dental epithelium cells start to differentiate into different dental epithelium cell types, such as the inner enamel epithelium (IEE), outer enamel epithelium, stratum intermedium, and stellate reticulum. IEE cells are progenitors of ameloblasts, and they are able to differentiate into enamel-secreting ameloblasts, a critical process for enamel formation (Miletich and Sharpe 2003; Kuang-Hsien Hu et al. 2014).
Ameloblast development is a stepwise cellular differentiation process. Dental epithelial stem cells, which are marked by the pluripotent factor Sox2, have the potential to differentiate into all the other dental epithelial cell lineages (Juuri et al. 2012; Juuri, Isaksson, et al. 2013; Juuri, Jussila, et al. 2013). In mice, the incisors continuously grow throughout adult life because of the existence of dental epithelial stem cells at the incisor cervical loop region (Kuang-Hsien Hu et al. 2014). IEE cells are highly proliferative and migrate from the cervical loop toward the distal end of the mouse incisor (Wang et al. 2007; Li et al. 2012). Differentiated ameloblasts secrete enamel matrix proteins and form enamel (Fukumoto et al. 2004; Bei 2009). Dental enamel is the hardest tissue in the body and plays significant roles in daily chewing and protecting dentin and the inner pulp tissue (He et al. 2010; He et al. 2011).
Studies with mutant mice have identified a number of factors that function at the different stages of ameloblast development. Sox2, a marker of dental epithelial stem cells, maintains competence for successional tooth formation in mammals (Juuri et al. 2012; Juuri, Jussila, et al. 2013). E-cadherin is dynamically expressed at different stages of ameloblast development and regulates IEE cell migration (Li et al. 2012). The transcription factor epiprofin is one of the key protein factors to promote IEE cell proliferation and differentiation (Nakamura et al. 2004; Nakamura et al. 2008). However, there is limited knowledge available on regulatory mechanisms of stage-specific ameloblast development. Therefore, identification of new factors regulating stage-specific ameloblast development is necessary and important.
In mammals, bHLH proteins are important regulators for neurogenesis, myogenesis, heart development, and hematopoiesis (Ross et al. 2003; Jones 2004). bHLH proteins can be divided into class I and class II: class I proteins are ubiquitously expressed, while class II proteins, such as MyoD and NeuroD, are expressed in a tissue-specific manner. Class I E proteins, such as E12, form a heterodimer with class II bHLH proteins through their HLH domains and activate or suppress gene transcription (Massari and Murre 2000).
In this study, we identified a tooth-specific bHLH protein, AmeloD, which is uniquely expressed in IEE and Hertwig epithelial root sheath (HERS) cells. Overexpression of AmeloD in epithelial cell lines resulted in E-cadherin suppression. Functionally, AmeloD directly binds to the E-cadherin proximal promoter and recruits chromatin repressive complex. Our results suggest that AmeloD is an important regulator for tooth development.
Materials and Methods
Cell Culture and Transfection
CLDE cells were cultured in keratinocyte-SFM medium (Thermo Fisher Scientific) (Yoshizaki et al. 2014). MDCK cells, COS-7 cells, and 293T cells were purchased from ATCC and cultured in Dulbecco’s modified eagle medium (DMEM) with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (Thermo Fisher Scientific). DNA was transfected into cells with Lipofectamine LTX. Small interfering RNAs (siRNAs) were purchased from Dharmacon and transfected into cells with an RNA interference (RNAi) transfection kit (ThermoFisher). For adenovirus-associated expression vectors transfection, CLDE cells were infected at a multiplicity of infection of 100, and MDCK cells were transfected at a multiplicity of infection of 200.
Cell Proliferation Assay and ChIP Analysis
Cell proliferation assay was performed with a CCK-8 kit (Dojindo). For chromatin immunoprecipitation (ChIP), CLDE cells were crosslinked by 1% formaldehyde solution for 10 min at room temperature. Chromatin was sonicated to 200 to 300 bp with a Bioruptor for 22 cycles. Chromatin prepared from 1 × 107 cells was used for each ChIP. The ChIP reactions were done with a ChIP-IT High Sensitivity Kit (Active Motif). DNA purified from ChIP assays was applied for quantitative polymerase chain reaction (qPCR) reactions, and data were normalized by percentage of input.
Detailed methods of yeast two-hybrid (Y2H) screening, complementary DNA (cDNA) library construction and screening, AmeloD full-length sequence and analysis, in situ hybridization, and AmeloD antibody specificity tests are included in the Appendix Materials and Methods.
Results
Isolation of the Novel bHLH Factor AmeloD from a Tooth Germ cDNA Library
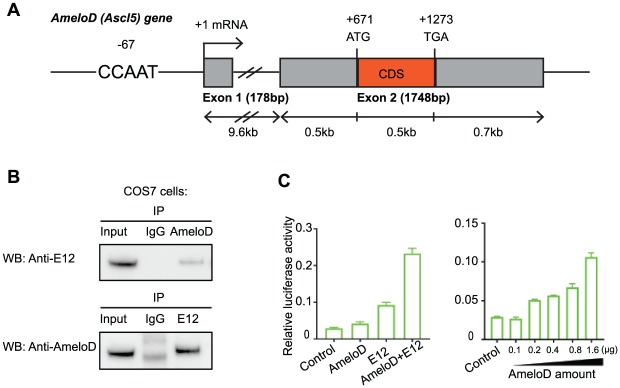
We sought to identify novel tooth-specific bHLH proteins regulating tooth development. For this purpose, we screened an E13.5 rat tooth germ cDNA library using the Y2H system, with the E12 bHLH domain as bait. After 2 rounds of screening, 19 positive clones were identified, and most were previously identified genes, such as MyoD, dHand, and Twist1. One of the positive clones was found to encode a novel bHLH protein, and we named this gene AmeloD (NCBI gene accession MG575629), as the AmeloD transcript was specifically expressed during ameloblast development. To obtain a full-length mouse AmeloD cDNA clone, we screened a mouse E19 molar cDNA library with a rat AmeloD probe. We obtained a mouse AmeloD cDNA clone containing 1,539 bp (Appendix Fig. 1A), including 5’ untranslated sequences, and a coding sequence encoding 188 amino acids, including 3’ untranslated sequences. Phylogenetic analysis showed that AmeloD belongs to the ASCL gene family and is homologous to ASCL5 (Appendix Fig. 1B). ASCL5 has been identified at only the genomic level: its expression patterns and functions are unknown. The ASCL5 (AmeloD) gene structure consists of 2 exons (Fig. 1A): untranslated exon1 (178 bp) and coding sequence containing exon2 (1,748 bp) containing 188 amino acid coding sequences. The AmeloD gene is mapped to mouse chromosome 1 q32 (Appendix Fig. 1C, D). The AmeloD cDNA starts at the residues 210 in exon 2. The 5’ RACE analysis from the 5’ part of the AmeloD cDNA showed the 5’ mRNA extension to the residue 22 in exon1 (data not shown). The amino acid sequences of the AmeloD bHLH domain are highly conserved among those of other ASCL proteins, while 5’ and 3’ sequences are more specific (Appendix Fig. 1B).
Figure 1.
Identification of a novel basic-helix-loop-helix protein AmeloD. (A) Gene structure of AmeloD. The solid boxes represent exons. The location of the CCAAT box is 67 bp upstream of the transcriptional start site. Exon 1 encodes the 5′-untranslated sequence, and exon 2 encodes the coding sequence of AmeloD. (B) Co-immunoprecipitation of AmeloD and E12. Co-transfection of AmeloD and E12 expression plasmid DNA into COS7 cells, followed by co-immunoprecipitation and Western blot. AmeloD and E12 proteins physically interact with each other. (C) MCK-luc reporter assay with AmeloD. AmeloD and E12 synergistically activate the E-box luciferase reporter (left panel), and gradually increasing the AmeloD DNA amount increased the luciferase activity (right panel). Values are presented as mean ± SD (n = 3).
To characterize the function of the AmeloD protein, we developed an AmeloD polyclonal antibody. Western blot analysis showed that the molecular weight of AmeloD protein is about 25 kDa. AmeloD antibody can recognize the exogenous AmeloD protein (Cos7 cells with AmeloD overexpression) and the endogenous AmeloD protein in mouse tooth lysate (Appendix Fig. 2A, B). To test the antibody specificity, we used scrambled and AmeloD-specific peptides to neutralize AmeloD antibody and test the antibody function in Western blot and immunofluorescence staining of postnatal day 1 (P1) mouse incisor. Our results showed that AmeloD-specific peptides totally blocked the AmeloD antibody function, and we did not observe any blocking effect when AmeloD antibody was neutralized by the scrambled peptide (Appendix Fig. 2C, D). Co-immunoprecipitation assay showed that AmeloD and E12 form a heterodimer (Fig. 1B). Co-transfection of HEK293 cells with AmeloD and E12 expression vectors and with a muscle creatine kinase gene reporter (MCK-luc) vector—which is an E-box-containing myogenic enhancer luciferase reporter and can be activated by MyoD-E12 heterodimer (Markus et al. 2002)—showed that AmeloD and E12 synergistically activated the MCK-luc reporter (Fig. 1C, left panel). Gradually increasing the AmeloD expression levels also increased luciferase activity (Fig. 1C, right panel). Together, these results suggest that AmeloD is a novel bHLH protein that physically interacts with E12 protein and binds to the E-box element.
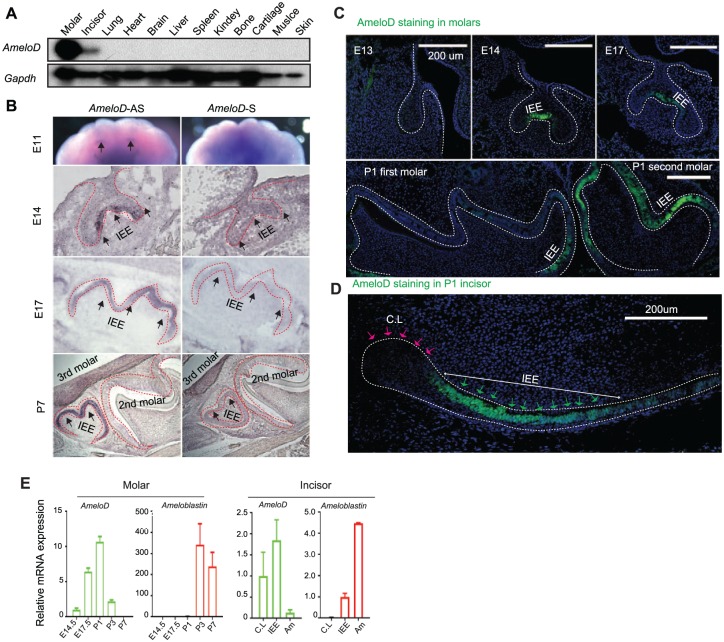
AmeloD Is a Tooth-Specific bHLH Factor and Expressed Uniquely in IEE Cells during Ameloblast Development
Northern blot analysis with RNA from different tissues of P1 mice showed that AmeloD was specifically expressed in molars and incisors (Fig. 2A). In situ hybridization with anti-sense (AmeloD-AS) probes showed that AmeloD expression started in the E11 incisor region (Fig. 2B). Later, AmeloD was strongly expressed in IEE cells of E14 and E17 mouse molar sections. Interestingly, at the P7 stage, AmeloD was detected in only the third molar, not in the second or first molar (Fig. 2B, lower panel). At the P7 stage, IEE cells are already differentiated into ameloblasts in the first and second molars but not yet in the third molars. AmeloD sense (AmeloD-S) probe was used as a control and showed no AmeloD signals in the mouse molar sections at different developmental stages (Fig. 2B, right panel). Immunofluorescence staining of mouse molar sections and the P1 incisor sagittal section also showed that AmeloD was specifically expressed in IEE cells but not in differentiated ameloblasts (Fig. 2C, D). AmeloD is also expressed in the HERS cells during mouse molar development (Figs. 2C, 3A). HERS cells are rapidly proliferating cells located in the cervical loop region of an enamel organ during mouse molar development (Li et al. 2017; Wang and Feng 2017). Immunofluorescence staining with Sox2, a dental epithelium stem cell marker, Ki67, a cell proliferation marker, and amelogenin, an ameloblast differentiation marker, in P2 mouse incisor serial sections showed that a large population of the AmeloD-positive cells were also Ki67 positive; however, AmeloD staining did not co-localize with Sox2 or amelogenin staining (Appendix Fig. 3A). Next, qPCR analysis showed that AmeloD was highly expressed from E14.5 to P1 molars and in the IEE cells of mouse incisor. The AmeloD expression level was dramatically downregulated once cells went into the differentiation stage (Fig. 2E). In contrast, ameloblastin was highly expressed in the P3 and P7 molars. The gene expression analysis also showed that AmeloD has an inverse expression pattern to ameloblastin in incisors and is highly expressed at IEE cells (Fig. 2E). These results suggest that AmeloD may regulate ameloblast development at the progenitor stage.
Figure 2.
Expression pattern of AmeloD. (A) Northern blot analysis of AmeloD expression in different tissues from the postnatal day 1 (P1) mouse, and the results show that AmeloD is uniquely expressed in the mouse molars and incisors. (B) In situ hybridization of AmeloD expression in the embryonic day 11 (E11) maxilla and E14, E17, and P7 mouse molars (secondary and third molars). AS, anti-sense probe; S, sense probe. The arrow indicates the expression of AmeloD transcript. (C) Immunofluorescence staining of AmeloD expression with AmeloD antibody (green) in E13.5, E14.5, E17.5, and P1 molar sections (first and second molars). (D) Immunofluorescence staining of AmeloD with anti-AmeloD antibody (green) in the sagittal section of the P1 mouse incisor. The red arrows indicate the cervical loop (CL) region of mouse incisor, and the green arrows indicate IEE cells. Scale bar: 200 µm. (E) Quantitative polymerase chain reaction gene expression analysis of AmeloD and ameloblastin expression during mouse molar and incisor development. Values are presented as mean ± SD (n = 3). IEE, inner enamel epithelium. Am, ameloblast.
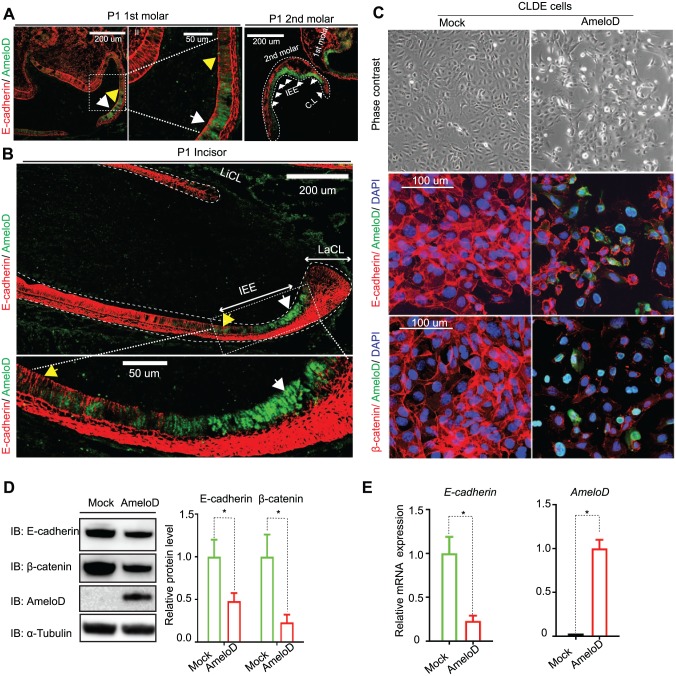
Figure 3.
AmeloD inhibits E-cadherin expression in progenitor ameloblasts. Immunofluorescence staining of AmeloD (green) and E-cadherin (red) in postnatal day 1 (P1) mouse (A) molars and (B) incisors. The white arrows indicate cells that have a high level of AmeloD expression (green color) and a low level of E-cadherin expression (red color), and the yellow arrows indicate cells that have a low level of AmeloD expression and a high level of E-cadherin expression. (C) Overexpression of AmeloD in CLDE cells suppressed E-cadherin expression. Phase contrast images showed that AmeloD overexpression resulted in morphologic changes in CLDE cells (top panel); immunofluorescence staining of AmeloD (green) with E-cadherin (red) or AmeloD (green) with beta-catenin (red) showed that overexpression of AmeloD in CLDE cells resulted in E-cadherin and beta-catenin degradation. (D) CLDE cells were transfected by control and AmeloD constructs, with Western blot analysis of E-cadherin and beta-catenin protein levels in CLDE cells after 72 h of transfection. Quantitative analysis was done by Image J. (E) Quantitative polymerase chain reaction gene expression analysis of E-cadherin and AmeloD expression in CLDE cells 72 h after transfection of control and AmeloD constructs. Data are shown as mean ± SD (n = 3). Statistical significance is shown by a t test. *P < 0.05.
AmeloD Inhibits E-cadherin Expression in IEE Cells
Based on the expression pattern of AmeloD during mouse incisor development, we hypothesize that AmeloD might function in progenitor ameloblast. We used CLDE cells as a model to study the function of AmeloD; CLDE cells are dental epithelium cells derived from the cervical loop region of E17.5 mouse incisor (Yoshizaki et al. 2014). IEE cells are rapidly proliferating cells with high cell motility (Seidel et al. 2010). However, neither overexpression nor siRNA knockdown of AmeloD had an effect on cell proliferation rate, indicating that AmeloD does not regulate cell proliferation (Appendix Fig. 3B). Although CLDE cells have a very low level of AmeloD expression and we could not detect AmeloD protein expression in CLDE cells (data not shown), we did detect AmeloD expression knockdown by siRNAs with qPCR (Appendix Fig. 3C).
E-cadherin expression levels are highly dynamic during ameloblast development and related to the motility function of IEE cells (Li et al. 2012). Immunofluorescence staining of AmeloD and E-cadherin in P1 mouse molar and incisor sections showed that AmeloD and E-cadherin had an inverse expression pattern in IEE cells. In IEE cells, AmeloD was highly expressed, but there was no E-cadherin expression (Fig. 3A, B, white arrowheads). However, when IEE cells were differentiated into ameloblasts, AmeloD expression was downregulated, but the E-cadherin expression level was upregulated again (Fig. 3A, B, yellow arrowheads). The inverse expression pattern of AmeloD and E-cadherin in IEE cells suggests that AmeloD may function as a suppressor of E-cadherin expression during ameloblast development. We tested this hypothesis by infecting CLDE cells with adeno-GFP and adeno-AmeloD expression vectors. Indeed, after AmeloD infection, the morphology of CLDE cells changed from a cuboidal cell shape to spindle-shaped fibroblast-like cells (Fig. 3C, upper panel). Immunofluorescence staining results showed that the overexpression of AmeloD did suppress the expression of the cell–cell adhesion proteins E-cadherin and β-catenin (Fig. 3C, middle and lower panel); this result was further confirmed by Western blot analysis (Fig. 3D) and gene expression analysis (Fig. 3E). Overexpression of adeno-AmeloD in another epithelial cell line, MDCK cells, also resulted in the suppression of E-cadherin and β-catenin expression (Appendix Fig. 4A, B). Thus, AmeloD suppressed E-cadherin expression in CLDE and MDCK cells.
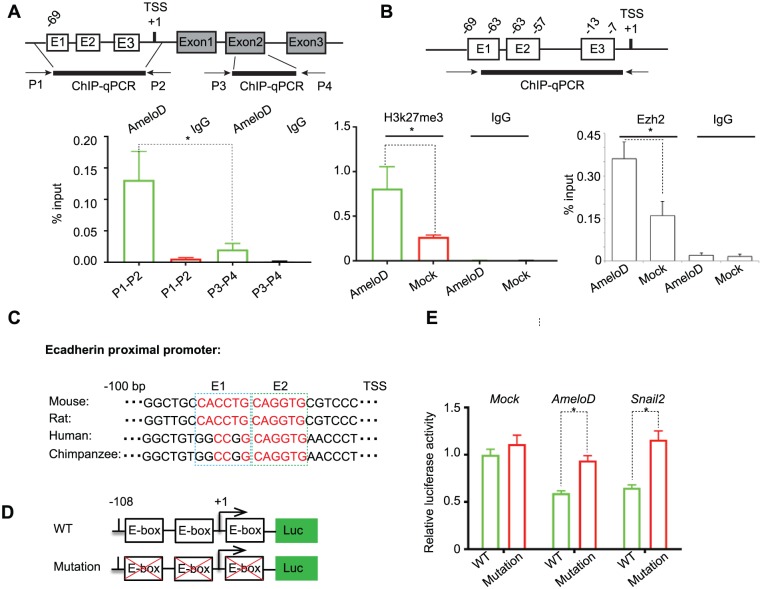
AmeloD Binds to the E-cadherin Proximal Promoter through E-boxes
To investigate the mechanism by which AmeloD suppresses E-cadherin transcription, we performed ChIP analysis using CLDE cells infected with the adeno-AmeloD vector. Two sets of primers were designed for the ChIP analysis: primers P1-P2 were designed to amplify the E-cadherin proximal promoter (−100 bp), which has 2 adjacent E-box elements that are important for binding of bHLH proteins (Yang et al. 2004; Sideridou et al. 2011); primers P3–P4 were designed to amplify part of E-cadherin exon2 and served as a no-binding control. Antibodies against AmeloD and IgG were used for ChIP reactions. We found that AmeloD binds to the E-cadherin proximal promoter at a significantly higher level than that of the no-binding and IgG controls (Fig. 4A). Next, we compared H3K27me3 or PRC2 complex Ezh2 enrichment levels at the E-cadherin proximal promoter in CLDE cells transfected with adeno-GFP and adeno-AmeloD expression vectors. Our results revealed that AmeloD overexpression significantly increased the H3K27me3 and Ezh2 enrichment level at the E-cadherin proximal promoter (Fig. 4B). Since H3K27me3 generally serves as a repressive marker for gene transcription, our results indicate that AmeloD inhibits E-cadherin transcription by recruiting the PRC2 repressive complex to E-cadherin proximal promoter. Co-immunoprecipitation analysis of AmeloD with Ezh2 or Suz12, which are the 2 core components of the PRC2 complex, did not show direct physical interactions between AmeloD and these 2 proteins (data not shown), suggesting that recruiting of the PCR2 complex to E-cadherin promoter was mediated by other factors. In fact, there are conserved E-box elements in the proximal promoter of E-cadherin among different species (Fig. 4C). Furthermore, AmeloD suppressed the activity of the E-cadherin promoter luciferase reporter containing 3 E-boxes but did not suppress the activity of the E-cadherin promoter reporter containing E-box mutations (Fig. 4D). Similar results were obtained when co-transfecting a Snail2 expression vector, a well-known E-cadherin suppressor, with WT and E-box mutant E-cadherin luciferase reporters. These results further demonstrated that the proximal promoter E-box elements are necessary for E-cadherin suppression by AmeloD. In summary, AmeloD directly binds to the E-cadherin proximal promoter through E-box elements and inhibits E-cadherin expression.
Figure 4.
AmeloD binds to E-cadherin proximal promoter and inhibits E-cadherin expression. (A) ChIP-qPCR results showed that AmeloD binds to the E-cadherin proximal promoter. Nuclear extracts were prepared from CLDE cells transfected by adeno-AmeloD for 72 h. Antibodies for AmeloD and IgG were used for ChIP assay. Primer set P1-P2 was designed to amplify the proximal promoter region (−69 bp to TSS) of the mouse E-cadherin gene, and the primer set P3-P4 was designed to amplify 100 bp of the E-cadherin gene exon 2. (B) Left panel: ChIP-qPCR results showed that binding of AmeloD to the E-cadherin promoter increased the repressive marker H3K27me3 enrichment. The H3k27me3 enrichment level was relative to the percentage of input. Nuclear extracts prepared from CLDE cells were transfected by adeno-GFP and adeno-AmeloD expression vectors for 72 h. Antibodies for H3K27me3 and IgG were used for immunoprecipitation. Right panel: ChIP-qPCR results showed that binding of AmeloD to the E-cadherin promoter increased the PCR2 complex component Ezh2 enrichment. The Ezh2 enrichment level was relative to the percentage of input. Nuclear extracts prepared from CLDE cells were transfected by adeno-GFP and adeno-AmeloD expression vectors for 72 h. Antibodies for Ezh2 and IgG were used for immunoprecipitation. (C) The upper panel shows that the consensus E-box element (E1 and E2) appears at the E-cadherin proximal promoter among different species; the lower panel shows the diagram of the wild-type (WT) and mutated E-cadherin luciferase reporter. (D) Luciferase assay showed that AmeloD inhibits the WT E-cadherin luciferase reporter activity but not the mutated E-cadherin luciferase reporter. Transcription factor snail2 served as a positive control, and an empty construct served as a negative control. Data are shown as mean ± SD (n = 3). Statistical significance is shown by a t test. *P < 0.05. ChIP, chromatin immunoprecipitation; qPCR, quantitative polymerase chain reaction.
Discussion
In this study, we identified a novel bHLH protein, AmeloD, which is uniquely expressed in teeth but not in other tissues (Figs. 1, 2A). By in situ hybridization, immunofluorescence staining, and gene expression analysis, we demonstrated that AmeloD is uniquely expressed in proliferating IEE cells and has an inverse expression pattern with E-cadherin (Figs. 2, 3A, B; Appendix Fig. 3A). By overexpression of AmeloD or knockdown AmeloD in CLDE cells, we did not observe that AmeloD regulates cell proliferation (Appendix Fig. 3B, C), suggesting that AmeloD might regulate a different function of IEE cells.
During mouse incisor development, the dental epithelial stem cell-derived progenitor cells migrate from the cervical loop toward the distal region (Dassule et al. 2000; Harada et al. 2002; Klein et al. 2008; Juuri et al. 2012; Juuri, Jussila, et al. 2013). To facilitate this migration process, the epithelium cell-specific cell–cell adhesion protein E-cadherin is partially downregulated in the IEE cells, and its expression level is upregulated again in the differentiated ameloblasts (Li et al. 2012). The inverse expression pattern of AmeloD and E-cadherin prompted us to explore a potential regulatory relationship between them. Functional analysis of AmeloD revealed that AmeloD suppressed E-cadherin expression in CLDE cells; overexpression of AmeloD in CLDE cells resulted in E-cadherin downregulation at the mRNA and protein levels (Fig. 3C–E). ChIP-qPCR analysis showed that AmeloD preferentially binds to the proximal promoter of E-cadherin, which has conserved E-box elements among different species (Fig. 4A, C). In fact, several other E-cadherin repressive factors, such as bHLH proteins Twist1 and E47, also bind to E-box elements at the E-cadherin promoter and repress E-cadherin expression during epithelium-mesenchyme transition (Yang et al. 2004; Yang et al. 2010). Furthermore, by ChIP-qPCR analysis, we found that AmeloD overexpression resulted in a higher enrichment level of histone repressive marker H3K27me3 and PRC2 core complex Ezh2 at the E-cadherin proximal promoter (Fig. 4A, B), suggesting that AmeloD represses E-cadherin expression by recruiting the histone repressive complex to the E-cadherin promoter. By in vitro co-immunoprecipitation analysis of AmeloD with Ezh2 or Suz12 (data not shown), we did not find direct binding of AmeloD to Ezh2 or Suz12, suggesting that recruiting of Ezh2 to the E-cadherin promoter was not directly through AmeloD; instead, some other protein factors mediated an indirect association between AmeloD and the PRC2 complex, and future studies should focus on identifying the protein complex.
In the developing mouse molar, AmeloD is also expressed in HERS cells (Figs. 2C, 3A), which are derived from the elongation of the enamel organ and play an important role in cementum formation and proper root development (Li et al. 2017). Contrary to AmeloD’s inverse expression pattern with E-cadherin in IEE cells, AmeloD and E-cadherin are coexpressed in the progenitor HERS cells, suggesting that AmeloD might have distinct functions in IEE and HERS cells. Crown development and root formation are tightly regulated by multiple signaling pathways, such as Wnt and Tgfβ (Wang and Feng 2017). Transcription factors such as Bcl11b, epiprofin, and Pitx2 have been found primarily to regulate ameloblast differentiation and enamel formation (Nakamura et al. 2008; Golonzhka et al. 2009; Li et al. 2014), while Dkk1, Nfic, and c-Fos were found to regulate root developmental process (Huang and Chai 2012; Jussila and Thesleff 2012; He et al. 2016; Li et al. 2017; Wang and Feng 2017). In our current study, we found that AmeloD suppresses E-cadherin expression and promotes IEE cell motility. Based on its expression pattern and in vitro functional studies, we propose that AmeloD regulates enamel and root development in mice. In a separate study by our group, we showed that AmeloD knockout mice developed mild to moderate tooth developmental defects, such as enamel hypoplasia and short tooth roots (Chiba et al. 2018, unpublished data). Knockout AmeloD protein in mice did not totally block tooth development, indicating that there might be functional compensation between AmeloD and other bHLH proteins. In fact, some other bHLH proteins, such as Mitf and Tfe3, were functionally redundant in osteoclast development (Steingrimsson et al. 2002). One limitation of our current study is that most of the functional studies of AmeloD are through overexpression systems. To fully understand the molecular function of AmeloD, it would be ideal to combine knockdown/knockout experiments in cell lines with in vivo knockout mice studies and study the cell lineage-specific functions (IEE vs. HERS) of AmeloD in mouse tooth development.
In summary, we have identified a novel tooth-specific bHLH factor, AmeloD. AmeloD is uniquely expressed in IEE cells and suppresses E-cadherin expression. Our studies reveal that AmeloD is an important regulator of tooth development.
Author Contributions
B. He, contributed to conception, design, and data acquisition, drafted and critically revised the manuscript; Y. Chiba, S. de Vega, K. Tanaka, C. Rhodes, contributed to design and acquisition, critically revised the manuscript; H. Li, contributed to conception, data acquisition and analysis, critically revised the manuscript; K. Yoshizaki, M. Ishijima, contributed to data acquisition and analysis, critically revised the manuscript; K. Yuasa, M. Ishikawa, K. Sakai, contributed to data acquisition, critically revised the manuscript; P. Zhang, contributed to data analysis and interpretation, critically revised the manuscript; S. Fukumoto, contributed to conception and design, critically revised the manuscript; X. Zhou, contributed to data interpretation, critically revised the manuscript; Y. Yamada, contributed to conception, design, data acquisition and interpretation, drafted and critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplemental Material
Supplemental material, DS_10.1177_0022034518808254 for Identification of the Novel Tooth-Specific Transcription Factor AmeloD by B. He, Y. Chiba, H. Li, S. de Vega, K. Tanaka, K. Yoshizaki, M. Ishijima, K. Yuasa, M. Ishikawa, C. Rhodes, K. Sakai, P. Zhang, S. Fukumoto, X. Zhou and Y. Yamada in Journal of Dental Research
Footnotes
A supplemental appendix to this article is available online.
This work was supported in part by the Intramural Research Program of the National Institute of Dental and Craniofacial Research, National Institutes of Health (Y. Yamada). This work was also supported by the National Natural Science Foundation of China (NSFC81500811 to B. He) and by the Japan Society for the Promotion of Science KAKENHI Grant-in-Aid (15J04116 to Y. Chiba).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Bei M. 2009. Molecular genetics of ameloblast cell lineage. J Exp Zool B Mol Dev Evol. 312B(5):437–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassule HR, Lewis P, Bei M, Maas R, McMahon AP. 2000. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development. 127(22):4775–4785. [DOI] [PubMed] [Google Scholar]
- Fukumoto S, Kiba T, Hall B, Iehara N, Nakamura T, Longenecker G, Krebsbach PH, Nanci A, Kulkarni AB, Yamada Y. 2004. Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J Cell Biol. 167(5):973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golonzhka O, Metzger D, Bornert JM, Bay BK, Gross MK, Kioussi C, Leid M. 2009. Ctip2/Bcl11b controls ameloblast formation during mammalian odontogenesis. Proc Natl Acad Sci U S A. 106(11):4278–4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H, Toyono T, Toyoshima K, Yamasaki M, Itoh N, Kato S, Sekine K, Ohuchi H. 2002. Fgf10 maintains stem cell compartment in developing mouse incisors. Development. 129(6):1533–1541. [DOI] [PubMed] [Google Scholar]
- He B, Huang S, Jing J, Hao Y. 2010. Measurement of hydroxyapatite density and knoop hardness in sound human enamel and a correlational analysis between them. Arch Oral Biol. 55(2):134–141. [DOI] [PubMed] [Google Scholar]
- He B, Huang S, Zhang C, Jing J, Hao Y, Xiao L, Zhou X. 2011. Mineral densities and elemental content in different layers of healthy human enamel with varying teeth age. Arch Oral Biol. 56(10):997–1004. [DOI] [PubMed] [Google Scholar]
- He YD, Sui BD, Li M, Huang J, Chen S, Wu LA. 2016. Site-specific function and regulation of osterix in tooth root formation. Int Endod J. 49(12):1124–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XF, Chai Y. 2012. Molecular regulatory mechanism of tooth root development. Int J Oral Sci. 4(4):177–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S. 2004. An overview of the basic helix-loop-helix proteins. Genome Biol. 5(6):226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jussila M, Thesleff I. 2012. Signaling networks regulating tooth organogenesis and regeneration, and the specification of dental mesenchymal and epithelial cell lineages. Cold Spring Harb Perspect Biol. 4(4):a008425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juuri E, Isaksson S, Jussila M, Heikinheimo K, Thesleff I. 2013. Expression of the stem cell marker, SOX2, in ameloblastoma and dental epithelium. Eur J Oral Sci. 121(6):509–516. [DOI] [PubMed] [Google Scholar]
- Juuri E, Jussila M, Seidel K, Holmes S, Wu P, Richman J, Heikinheimo K, Chuong CM, Arnold K, Hochedlinger K, et al. 2013. Sox2 marks epithelial competence to generate teeth in mammals and reptiles. Development. 140(7):1424–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juuri E, Saito K, Ahtiainen L, Seidel K, Tummers M, Hochedlinger K, Klein OD, Thesleff I, Michon F. 2012. Sox2+ stem cells contribute to all epithelial lineages of the tooth via Sfrp5+ progenitors. Dev Cell. 23(2):317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein OD, Lyons DB, Balooch G, Marshall GW, Basson MA, Peterka M, Boran T, Peterkova R, Martin GR. 2008. An FGF signaling loop sustains the generation of differentiated progeny from stem cells in mouse incisors. Development. 135(2):377–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang-Hsien Hu J, Mushegyan V, Klein OD. 2014. On the cutting edge of organ renewal: identification, regulation, and evolution of incisor stem cells. Genesis. 52(2):79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CY, Cha WH, Luder HU, Charles RP, McMahon M, Mitsiadis TA, Klein OD. 2012. E-cadherin regulates the behavior and fate of epithelial stem cells and their progeny in the mouse incisor. Dev Biol. 366(2):357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JY, Parada C, Chai Y. 2017. Cellular and molecular mechanisms of tooth root development. Development. 144(3):374–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Venugopalan SR, Cao H, Pinho FO, Paine ML, Snead ML, Semina EV, Amendt BA. 2014. A model for the molecular underpinnings of tooth defects in Axenfeld-Rieger syndrome. Hum Mol Genet. 23(1):194–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus M, Du Z, Benezra R. 2002. Enhancer-specific modulation of E protein activity. J Biol Chem. 277(8):6469–6477. [DOI] [PubMed] [Google Scholar]
- Massari ME, Murre C. 2000. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cellular Biol. 20(2):429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miletich I, Sharpe PT. 2003. Normal and abnormal dental development. Hum Mol Genet. 12:R69–R73. [DOI] [PubMed] [Google Scholar]
- Nakamura T, de Vega S, Fukumoto S, Jimenez L, Unda F, Yamada Y. 2008. Transcription factor epiprofin is essential for tooth morphogenesis by regulating epithelial cell fate and tooth number. J Biol Chem. 283(8):4825–4833. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Unda F, de-Vega S, Vilaxa A, Fukumoto S, Yamada KM, Yamada Y. 2004. The Kruppel-like factor epiprofin is expressed by epithelium of developing teeth, hair follicles, and limb buds and promotes cell proliferation. J Biol Chem. 279(1):626–634. [DOI] [PubMed] [Google Scholar]
- Ross SE, Greenberg ME, Stiles CD. 2003. Basic helix-loop-helix factors in cortical development. Neuron. 39(1):13–25. [DOI] [PubMed] [Google Scholar]
- Seidel K, Ahn CP, Lyons D, Nee A, Ting K, Brownell I, Cao T, Carano RA, Curran T, Schober M, et al. 2010. Hedgehog signaling regulates the generation of ameloblast progenitors in the continuously growing mouse incisor. Development. 137(22):3753–3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sideridou M, Zakopoulou R, Evangelou K, Liontos M, Kotsinas A, Rampakakis E, Gagos S, Kahata K, Grabusic K, Gkouskou K, et al. 2011. Cdc6 expression represses E-cadherin transcription and activates adjacent replication origins. J Cell Biol. 195(7):1123–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingrimsson E, Tessarollo L, Pathak B, Hou L, Arnheiter H, Copeland NG, Jenkins NA. 2002. Mitf and Tfe3, two members of the Mitf-Tfe family of bHLH-Zip transcription factors, have important but functionally redundant roles in osteoclast development. Proc Natl Acad Sci U S A. 99(7):4477–4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thesleff I. 1996. Epithelial-mesenchymal signaling regulating tooth morphogenesis. J Cell Sci. 116:1647–1648. [DOI] [PubMed] [Google Scholar]
- Wang J, Feng JQ. 2017. Signaling pathways critical for tooth root formation. J Dent Res. 96(11):1221–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XP, Suomalainen M, Felszeghy S, Zelarayan LC, Alonso MT, Plikus MV, Maas RL, Chuong CM, Schimmang T, Thesleff I. 2007. An integrated gene regulatory network controls stem cell proliferation in teeth. PLoS Biol. 5(6):e159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. 2004. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 117(7):927–939. [DOI] [PubMed] [Google Scholar]
- Yang MH, Hsu DSS, Wang HW, Wang HJ, Lan HY, Yang WH, Huang CH, Kao SY, Tzeng CH, Tai SK, et al. 2010. Bmi1 is essential in Twist1-induced epithelial-mesenchymal transition. Nat Cell Biol. 12(10):982–992. [DOI] [PubMed] [Google Scholar]
- Yoshizaki K, Hu L, Nguyen T, Sakai K, He B, Fong C, Yamada Y, Bikle DD, Oda Y. 2014. Ablation of coactivator Med1 switches the cell fate of dental epithelia to that generating hair. PloS One. 9(6):e99991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, DS_10.1177_0022034518808254 for Identification of the Novel Tooth-Specific Transcription Factor AmeloD by B. He, Y. Chiba, H. Li, S. de Vega, K. Tanaka, K. Yoshizaki, M. Ishijima, K. Yuasa, M. Ishikawa, C. Rhodes, K. Sakai, P. Zhang, S. Fukumoto, X. Zhou and Y. Yamada in Journal of Dental Research