Abstract
Ciliary motility depends on both the precise spatial organization of multiple dynein motors within the 96 nm axonemal repeat and the highly coordinated interactions between different dyneins and regulatory complexes located at the base of the radial spokes. Mutations in genes encoding cytoplasmic assembly factors, intraflagellar transport factors, docking proteins, dynein subunits, and associated regulatory proteins can all lead to defects in dynein assembly and ciliary motility. Significant progress has been made in the identification of dynein subunits and extrinsic factors required for preassembly of dynein complexes in the cytoplasm, but less is known about the docking factors that specify the unique binding sites for the different dynein isoforms on the surface of the doublet microtubules. We have used insertional mutagenesis to identify a new locus, IDA8/BOP2, required for targeting the assembly of a subset of inner dynein arms (IDAs) to a specific location in the 96 nm repeat. IDA8 encodes flagellar-associated polypeptide (FAP)57/WDR65, a highly conserved WD repeat, coiled coil domain protein. Using high resolution proteomic and structural approaches, we find that FAP57 forms a discrete complex. Cryo-electron tomography coupled with epitope tagging and gold labeling reveal that FAP57 forms an extended structure that interconnects multiple IDAs and regulatory complexes.
INTRODUCTION
Cilia and flagella are microtubule-based organelles that play critical roles in cell motility and cell signaling, and defects in ciliary assembly, motility, or signaling can lead to a broad spectrum of diseases known as ciliopathies (reviewed in Reiter and Leroux, 2017). In vertebrates, ciliary motility is essential for the determination of the left–right body axis, development of the heart, movement of fluid in brain ventricles and spinal cord, clearance of mucus and debris in the respiratory tract, and sperm motility. Defects in motility can lead to situs inversus or heterotaxy, hydrocephalus and scoliosis, respiratory disease, and male infertility, symptoms often associated with primary ciliary dyskinesia (PCD) (Mitchison and Valente, 2017). Given the complexity of the microtubule-based 9+2 axonemal structure, motile ciliopathies are often underdiagnosed because of their genetic heterogeneity and multisystem variability (Werner et al., 2015). Yet many proteins of the ciliary axoneme are highly conserved (Li et al., 2004; Pazour et al., 2005; Albee et al., 2013), and so study of motile cilia in model organisms has provided insight into numerous genes and gene products potentially associated with PCD and other ciliopathies (Mitchison and Valente, 2017; Sigg et al., 2017).
Genomic and proteomic strategies have identified more than 600 proteins as structural components of the axoneme, and many other proteins contribute to the preassembly of axonemal complexes in the cytoplasm, their delivery to the basal body region, and their transport through the transition zone and into the ciliary compartment (reviewed in van Dam et al., 2019). Advances in high resolution imaging in combination with the ordered and repetitive nature of the axoneme structure have provided insight into the location of several axonemal complexes (Mizuno et al., 2012), but still only a third or so of the ciliary proteins have been clearly correlated with a specific structure.
Most motile cilia and flagella contain nine doublet microtubules (DMTs) that surround two central pair (CP) singlet MTs. The outer and inner dynein arms (ODAs and IDAs) are multisubunit motors composed of heavy, intermediate, and light chains (DHC, IC, LC) that form two distinct rows on the A-tubule of each DMT and generate the force for microtubule sliding (reviewed in King, 2018). The dynein motors are organized into a 96 nm functional unit that repeats along the length of the axoneme, with four ODAs and seven IDAs (I1/f, a, b, c, e, g, d) found at specific locations within each repeat. Dynein activity is coordinated by mechanical signals from the CP and its associated projections to a series of radial spokes (RSs) that contact the DMTs near the base of the IDAs (Smith and Yang, 2004). The proximal to distal arrangement of the RS (RS1, RS2, RS3, or radial spoke 3 short [RS3S]) and the multiple dyneins in each repeat is specified in part by two proteins (flagellar-associated polypeptide [FAP]59/FAP172 or CCDC39/CCDC40) that form a 96 nm ruler (Oda et al., 2014). The IC/LC complex of the I1/f dynein forms a regulatory node at the base of RS1, and the nexin-dynein regulatory complex (N-DRC) forms a second node at the base of RS2 (Gardner et al., 1994; Nicastro et al., 2006; Bower et al., 2009; Heuser et al., 2009, 2012) that is connected to the base of RS3/RS3S via the calmodulin and spoke-associated complex (CSC). The I1 dynein and N-DRC also connect to other structures in the 96 nm repeat and to the ODAs to coordinate dynein activity, but with the exception of the MIA complex next to the I1 dynein (Yamamoto et al., 2013), the identity of the connectors is largely unknown.
Here we identify a new group of mutations that alter ciliary motility and the assembly of a subset of IDAs in Chlamydomonas. Using plasmid rescue and a chromosome walk, we cloned and mapped the IDA8 gene and found that it is linked to another motility mutation, bop2-1. We then characterized the molecular, biochemical, and structural phenotypes of the ida8/bop2 mutations. We found that IDA8 encodes FAP57, a highly conserved WD repeat and coiled coil protein also found in other species that have motile cilia with IDAs. Biochemical and proteomic analyses indicate that FAP57 is part of a subcomplex required for targeting or stabilizing the binding of a subset of IDAs. Thin-section transmission electron microscopy (TEM) and cryo-electron tomography (cryo-ET) reveal the complexity of structural defects in ida8/bop2 axonemes. Rescue with SNAP-tagged FAP57 constructs followed by streptavidin-gold labeling, subtomogram averaging, and 3D classification suggest that FAP57 forms an extended structure that interconnects multiple regulatory components and IDAs within the 96 nm axoneme repeat.
RESULTS
Characterization of new ida mutations and identification of the IDA8/BOP2 locus
To identify novel genes required for assembly of the IDAs, we screened several collections of motility mutants in Chlamydomonas for strains that exhibited the slow swimming phenotype typical of ida mutants (Brokaw and Kamiya, 1987). Three strains characterized here, ida8-1, ida8-2, and ida8-3, were chosen for further study based on the similarities in their motility phenotypes and inner arm defects. All three strains swam forward with an asymmetric waveform, but their swimming velocities were reduced compared with wild-type (WT) strains (Supplemental Figure S1A), and their motility phenotypes cosegregated with their ability to grow on selective media. Thin-section TEM and 2D image averaging of isolated axonemes showed that structures in the IDA region were reduced (Supplemental Figure S1B). Genomic Southern blots indicated the presence of a single plasmid sequence in ida8-1 and ida8-3 (Supplemental Figure S1C).
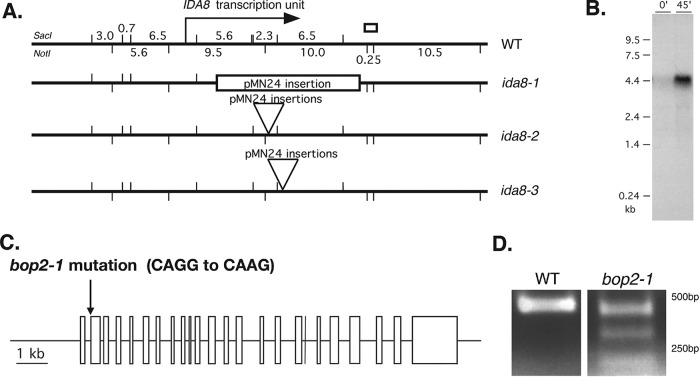
To identify the gene that was disrupted by plasmid insertion, genomic DNA flanking the vector sequences was recovered by plasmid rescue (Materials and Methods). A unique 700-base-pair fragment designated flanking clone 1 (FC1) was recovered from ida8-1 (Figure 1A). Southern blots of genomic DNA probed with FC1 confirmed the presence of a restriction fragment length polymorphism (RFLP) in ida8-1. FC1 was used to screen a phage library and isolate a series of overlapping clones spanning ∼40 kb of genomic DNA. Subclones were tested on Southern blots to determine the extent of DNA rearrangement or deletion caused by the insertion events (Supplemental Figure S1D). The blots showed that the same genomic region was disrupted to varying degrees in all three ida8 strains (Figure 1A). Transformation with a BAC clone (6h9) spanning this region rescued the motility defect (Supplemental Table S1).
FIGURE 1:
Molecular characterization of ida8 and bop2 mutations. (A) Diagram of the ∼40 kb region of genomic DNA around the IDA8 locus in WT, with SacI restriction sites indicated on top and Not1 restriction sites indicated below. The location of the IDA8 transcription unit is shown by the arrow. The site of the genomic fragment recovered by plasmid rescue from ida8-1 is shown by the white box. The next three lines show the sites of pMN24 insertion in each ida8 allele as determined by Southern blotting (Supplemental Figure S1E). (B) Northern blot of total RNA isolated from WT cells before (0) and 45 min after deflagellation and probed with a 6.5 kb SacI restriction fragment that was missing in ida8-1. Other blots probed with the 5.6 and 2.3 kb SacI fragments and several RT-PCR products recognized the same transcript. (C) Diagram of the intron-exon structure of the IDA8 gene showing the bop2-1 mutation in the acceptor splice site of the second exon. (D) RT-PCR products obtained from WT and bop2-1 RNA using primers surrounding the site of the bop2-1 mutation were analyzed on an agarose gel. Sequence analysis identified premature stop codons in all of the RT-PCR products from bop2-1.
To determine the precise location of the IDA8 transcription unit, subclones were used to probe Northern blots of WT RNA isolated before and after deflagellation. These blots defined an ∼12 kb region of genomic DNA that encodes an ∼5 kb transcript whose expression was increased by deflagellation (Figure 1, A and B). Transformation with a subclone containing the complete gene rescued the motility defects (Supplemental Table S1). DNA sequencing and reverse transcriptase-PCR (RT-PCR) revealed that the IDA8 gene contains 24 exons (Figure 1C) that are predicted to encode a polypeptide of 1316 amino acid residues with an estimated molecular weight of ∼146 kDa (Supplemental Figure S2, A and B).
Genetic and molecular mapping further revealed that the IDA8 locus was located on the left arm of Chromosome 4 (Materials and Methods), close to the motility mutation bop2-1 (Dutcher et al., 1988). Sequencing of bop2-1 DNA identified a single-base-pair mutation at nucleotide #605 (CAGG to CAAG) in the acceptor splice site of exon 2 (Figure 1C). Three alternatively spliced transcripts were detected by RT-PCR (Figure 1D), and sequence analysis revealed that all three transcripts contained frameshifts that resulted in premature stop codons. Diploid strains containing bop2-1 and ida8-1 displayed the same motility phenotype as the parental strains, and transformation of bop2-1 with the p59c2 subclone rescued the motility defects (Supplemental Table S1).
The IDA8/BOP2 locus encodes the conserved WD repeat and coiled coil containing polypeptide FAP57
The IDA8/BOP2 gene encodes a polypeptide identified in the flagellar proteome as FAP57 (Pazour et al., 2005). FAP57 is predicted to contain an N-terminal region with several WD repeat domains (residues 1–622) and C-terminal region with several coiled coil domains (residues 640–1188) and more variable, low complexity domains (residues 1206–1314) that are potentially disordered (Figure 2A). Both the polypeptide sequence and structural domains are highly conserved in other species with motile cilia and flagella (Nevers et al., 2017), including a polypeptide identified as WDR65 in vertebrates, with whom it shares almost 42% sequence identity and 61% sequence similarity. Interestingly, FAP57 orthologues are found in organisms that only assemble IDAs, such as Physcomitrella (Table 1), but not in species that only assemble ODAs, such as Thalissiosira.
FIGURE 2:
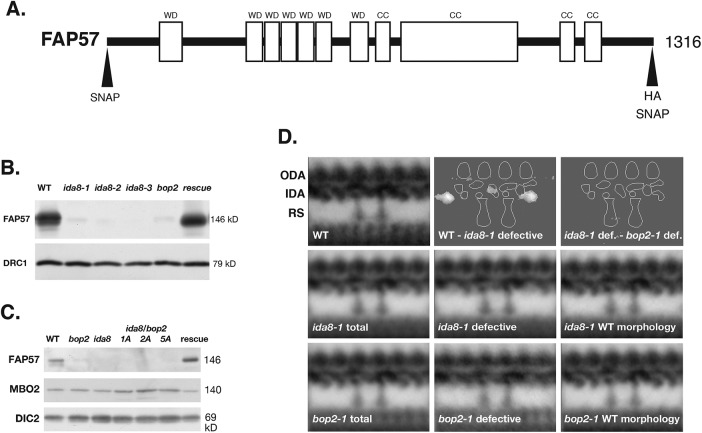
The ida8 and bop2 mutants have similar biochemical and structural defects. (A) Diagram of the IDA8 gene product, FAP57, showing the location of the predicted WD repeat domains (WD) in the first half of the polypeptide and the coiled coil domains (CC) in the second half. Also shown are the positions of the HA and SNAP tags at the N-terminal and C-terminal ends. (B) Western blot of axonemes from WT, three ida8 mutants, bop2-1, and an ida8-1; FAP57 strain (CC-4486, rescued with the 6h9 Bac clone) was probed with antibodies against FAP57 and the N-DRC subunit DRC1 as a loading control. (C) Western blot of axonemes from WT, bop2-1, ida8-1, three ida8-1/bop2-1 diploids (1A, 2A, 5A), and an ida8-1; FAP57 strain (CC-4486) was probed with antibodies against FAP57, MBO2, and the outer arm dynein IC2 (DIC2) subunit, also known as IC69, as a loading control. (D) Averages and difference plots of the 96 nm repeat obtained by TEM of thin sections and image averaging as described in O’Toole et al. (1995). The WT grand average (top left) was obtained from six axonemes with 65 repeats. The approximate locations of the ODA, IDA, and two RS are indicated. The ida8-1 grand average in the second row (ida8-1 total) was obtained from 41 axonemes with 379 repeats. The ida8-1 averages were divided into two classes, those with defective morphology (25 axonemes with 237 repeats) and those with WT morphology (16 axonemes with 142 repeats). The bop2-1 grand average in the third row (bop2-1 total) was obtained from 22 axonemes with 196 repeats. These were also separated into two classes, bop2-1 defective (8 axonemes with 66 repeats) and bop2-1 with WT morphology (14 axonemes with 130 repeats). The difference plots in the top row show identified two densities in the 96 nm repeat that were significantly different (P < 0.05) between the WT and ida8-1 defective grand averages. No significant difference was detected between the ida8-1 defective average and bop2-1 defective average.
TABLE 1:
Orthologues of polypeptides altered in ida8.
| Chlamydomonas | Physcomitrella | Drosophila | Zebrafish | Homo sapiens |
|---|---|---|---|---|
| Name (Cre ID) | Name | Name | Name | Name |
| Length (amino acids) (MW) | Accession number BLAST score (amino acid alignment) | Accession number BLAST score (amino acid alignment) | Accession number BLAST score (amino acid alignment) | Accession number BLAST score (amino acid alignment) |
| FAP57 (Cre04.g217914) 1316 amino acids (146 kDa) | CFAP57 XP_024357376 0.0 (amino acids 1–1201) | CG4329 AAN71097 6e-125 (amino acids 2–1122) | CFAP57 XP_021324203 0.0 (amino acids 1–972) | WDR65/CFAP57 XP_005270577 0.0 (amino acids 11–1177) |
| FAP337 (Cre13.g562800) 999 amino acids (108 kDa) | WDR49-like XP_024383320 2e-75 (amino acids 5–575) | WD40 Y NP_001303588 2e-23 (amino acids 338–826) | WDR95 XP_685972 6e-45 (amino acids 1–855) | WDR49 NP_001335880 9e-48 (amino acids 1–838) EFCAB8 NP_001137439 3e-39 (amino acids 9–830) |
| FBB7 (Cre03.g143827) 1706 amino acids (180 kDa) | CFAP57 XP_024357376 5e-35 (amino acids 467–1269) | CG4329 AAN71097 6e-21 (amino acids 481–1223) | CFAP57 XP_021324203 2e-28 (amino acids 417–1124) 7e-22 (amino acids 16–422) | WDR65/CFAP57 XP_005270577 8e-40 (amino acids 417–1187) |
| FAP331 (Cre06.g308000) 1989 amino acids (205 kDa) | CFAP57-like XP_024388298 4e-07 (amino acids 1005–1339) 2e-05 (amino acids 532–623) |
To assess the location and distribution of the FAP57 polypeptide in WT and mutant Chlamydomonas strains, we generated a specific antibody against a conserved peptide sequence at amino acid residues 460–480. Western blots showed that the affinity purified antibody detected a band of ∼146 kDa present in WT and rescued strains but missing in axonemes from ida8, bop2-1, and ida8-1/bop2-1 diploids (Figure 2, B and C). FAP57 therefore corresponds to the ∼152 kDa band previously described as missing on gels of bop2-1 axonemes (King et al., 1994).
Because previous study of bop2-1 had indicated radial asymmetry in the assembly of uncharacterized inner arm structures (King et al., 1994), we directly compared the defects in ida8-1 and bop2-1 axonemes by thin-section TEM of longitudinal sections and 2D averaging of the 96 nm repeat. As shown in Figure 2D, both mutants showed heterogeneity in the assembly of structures in the inner arm region. When sorted into two classes, a significant number of ida8-1 (37%) and bop2-1 (66%) repeats were similar to WT, but many ida8-1 (63%) and bop2-1 (34%) repeats were also missing two structures, one close to the base of the I1/f dynein between RS1 and RS2 and a second at the distal end of the 96 nm repeat.
FAP57 is located in the basal body region and along the length of the axoneme
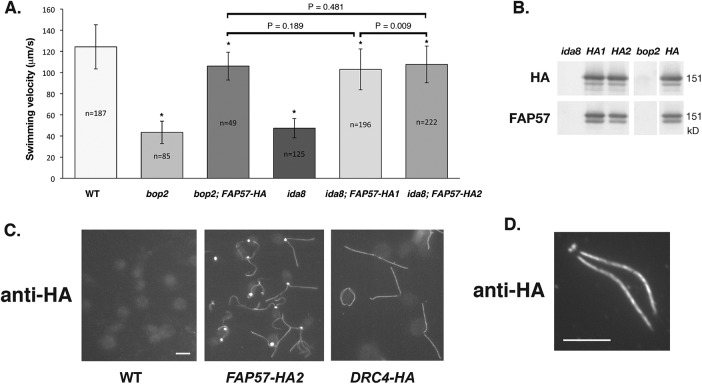
To better understand the role of FAP57 in the assembly of IDA structures, we generated epitope-tagged constructs of FAP57 (Supplemental Figure S2, C and D) and used these constructs and the affinity-purified FAP57 antibody to analyze the distribution of FAP57 in WT, mutant, and rescued cells. Transformation of ida8-1 and bop2-1 with a FAP57-HA construct (Figure 2A) restored near WT motility in both strains (Figure 3A; Supplemental Videos S1–S5). The hemagglutinin (HA)-tagged protein assembled into axonemes and migrated at ∼151 kDa on Western blots, slightly higher than the untagged FAP57 polypeptide (Figure 3B; see also Figure 5, A and B, later in this article). Localization by immunofluorescence microscopy of fixed cells using an HA antibody revealed that FAP57-HA was concentrated near the basal bodies but also found along the entire length of the axoneme (Figure 3C). The intense staining of FAP57-HA at the basal body region was qualitatively different from that observed with another HA-tagged axonemal polypeptide, such as DRC4-HA, one of the subunits of the N-DRC (Figure 3C). Whether FAP57 interacts with additional proteins in the basal body region is unknown. Staining of the isolated nuclear-flagellar apparatus (NFAP) indicated that FAP57 was stably associated with isolated basal bodies and axonemes, but not present in the transition zone (Figure 3D).
FIGURE 3:
Rescue of bop2-1 and ida8-1 with FAP57-HA reveals its subcellular location. (A) Measurements of forward swimming velocity demonstrated that the speed of bop2-1 and ida8-1 transformants increased to ∼85% of WT levels following rescue with the HA-tagged FAP57 gene. The mean velocities of the HA-rescued strains were significantly faster than the mutants but still slower than WT (P < 0.005), as indicated by the asterisks, but not significantly different from one another when analyzed using the Student’s t test. (B) Western blot of axonemes from ida8-1, two ida8-1; FAP57-HA rescued strains (HA1 and HA2), bop2-1, and a bop2-1; FAP57-HA rescued strain (HA) was probed with antibodies against the HA tag and FAP57. (C) Immunofluorescence images of fixed cells stained with an HA antibody revealed background staining of cell bodies in WT and bright staining of the basal body region and two flagella in ida8-1; FAP57-HA rescued cells. Images of pf2-4; DRC4-HA rescued cells showed antibody staining of the two flagella but much weaker staining of the basal body region. (Scale bar = 5 μm.) (D) Immunofluorescence image of a NFAP obtained by autolysin treatment and detergent extraction of an ida8-1; FAP57-HA rescued strain after staining with an HA antibody. (Scale bar = 5 μm.)
FIGURE 5:
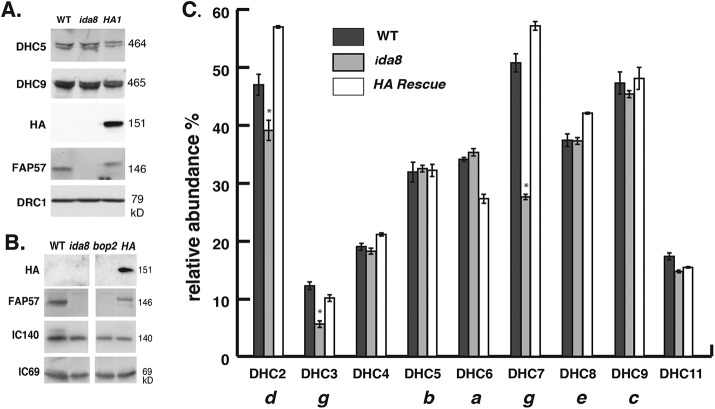
Mass spectrometry reveals defects in the assembly of a subset of inner arm DHCs in ida8. (A) Western blot of axonemes from WT, ida8-1, and an ida8-1; FAP57-HA rescued strain (HA1) was probed with antibodies against two inner arm DHCs (DHC5, DHC9), HA, FAP57, and DRC1. (B) Western blot of axonemes from WT, ida8-1, bop2, and bop2; FAP57-HA was probed with antibodies against HA, FAP57, IC140, and IC69. (C) The axoneme samples shown in A were fractionated by SDS–PAGE, and the DHC region was excised and analyzed in triplicate by tandem MS/MS and spectral counting. The total counts for each DHC were expressed as a percentage of the total counts for the two I1 dynein DHCs. The bars represent the range of the three replicates, and the asterisks represent those DHCs that were reduced more than 15% in ida8-1 relative to both the WT and the HA1 rescued strains.
Video S1.
Video of a wild‐type cell swimming forward with an asymmetric waveform.
Video S2.
Video of an ida8‐1 cell swimming forward with an asymmetric waveform.
Video S3.
Video of a bop2‐1 cell swimming forward with an asymmetric waveform.
Video S4.
Video of an ida8‐1; Fap57‐HA rescued cell swimming forward with an asymmetric waveform.
Video S5.
Video of a bop2‐1, FAP57‐HA rescued cell swimming forward with an asymmetric waveform.
Biochemical fractionation of FAP57 suggests a specific association with a subset of IDA isoforms
To determine whether FAP57 might be associated with a specific subcomplex of axonemal polypeptides, we probed blots of axonemes isolated from several classes of motility mutants. These included outer arm mutants (paralyzed flagella [pf]22, pf28, sup-pf2), inner arm mutants (pf23, pf9, ida4, mia1, mia2), CP mutants (pf19, pf6), N-DRC mutants (pf2, pf3, sup-pf3, sup-pf4), and other motility mutants with more symmetric waveforms (mbo1, pf12). As shown in Supplemental Figure S3, A–C, FAP57 was detected at near WT levels in all of these strains. Because bop2-1 was originally isolated as an extragenic suppressor of pf10 (Dutcher et al., 1988), we also analyzed the phenotypes of the double mutants bop2-1; pf10 and ida8-1; pf10. Western blots confirmed that FAP57 was present in pf10 axonemes but missing in the double mutants (Supplemental Figure S4A). High-speed videos showed that pf10 cells swam in small circles with a more symmetric waveform than the asymmetric breast stroke typically executed by WT, ida8, and bop2 cells. Double mutant cells bop2-1; pf10 and ida8-1; pf10 swam with slightly more asymmetric waveforms than pf10, but still did not make significant forward progress (Supplemental Figure S4B; Supplemental Videos 6–8). As an alternative test for dynein activity, we measured DMT sliding velocities using protease-treated axonemes and an in vitro sliding disintegration assay. As shown in Supplemental Figure S4C, the microtubule sliding velocities of bop2 and ida8-1 axonemes were significantly slower than WT axonemes. Likewise, the sliding velocities of the ida8-1; pf10 and bop2; pf10 double mutants were slower than pf10. These results, taken together with previous epistasis tests indicating that the bop2-1 mutation enhanced the motility defects observed with other dynein or n-drc mutants (King et al., 1994), suggested that FAP57 was part of a previously uncharacterized axonemal subcomplex involved in the assembly, transport, targeting, and/or regulation of IDAs.
Video S6.
Video of a pf10 cell swimming with an abnormal waveform.
Video S7.
Video of an ida8‐1; pf10 cell swimming with a variable waveform.
Video S8.
Video of a bop2‐1; pf10 cell swimming with a variable waveform.
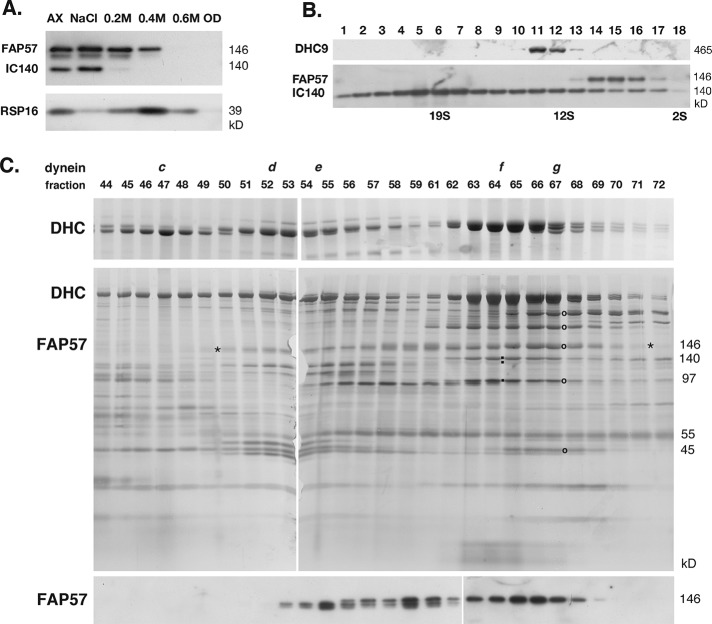
To characterize the biochemical properties of FAP57, we subjected WT flagella to a series of extraction protocols and analyzed the resulting extracts on silver-stained gels and/or Western blots. As shown in the Western blot in Supplemental Figure S3D, FAP57 was readily detected in isolated flagella, but not significantly extracted with either nonionic detergents, which solubilizes membrane plus matrix proteins, or with 10 mM MgATP, which typically extracts intraflagellar transport motor proteins (Cole et al., 1998; Pazour et al., 1999; Perrone et al., 2003). However, extraction of axonemes with 0.6 M NaCl followed by 0.5 M NaI solubilized nearly all of the FAP57 (Supplemental Figure S3D). Sequential treatment of axonemes with 0.2, 0.4, and 0.6 M NaI showed that FAP57 was more resistant to extraction than the I1 dynein subunit IC140, but more readily solubilized than the RS subunit radial spoke protein (RSP)16 (Figure 4A). Fractionation of dynein extracts by sucrose density gradient centrifugation indicated that FAP57 sedimented at ∼8–10 S, which is slower than either I1 dynein (IC140) at ∼20 S or dynein c (DHC9) at ∼12–13 S (Figure 4B). Fractionation of pf28 extracts by FPLC chromatography revealed that FAP57 eluted with a broad profile, cofractionating with IDAs in peaks d, e, f, and g (Figure 4C). The FAP57 polypeptide appeared to split into two distinct bands that fractionated asymmetrically across this region. The two bands may represent alternatively spliced isoforms (Supplemental Figure S2) or possibly proteolytic fragments. The highest concentration of FAP57 coincided with the peak of dynein g. The elution profile of the FAP57 bands was unchanged in dynein extracts obtained from pf10, mbo1, and ida4 axonemes (unpublished data). Because FAP57 was not reduced in mutants that lack dynein d, e, or f (i.e., ida4, pf3, pf9; see Supplemental Figure S3, A and B), it seemed likely that FAP57 is not a bona fide dynein subunit but may be peripherally associated with the IDAs as a docking factor that coelutes during FPLC chromatography.
FIGURE 4:
Biochemical fractionation of FAP57 demonstrates coextraction and coelution with a subset of IDAs. (A) Western blot of WT axonemes (AX) and extracts obtained by sequential treatment with 0.6 M NaCl, 0.2 M, 0.4 M, and 0.6 M NaI, and the final pellet of extracted outer doublets (OD) was probed with antibodies against FAP57, the I1 dynein subunit IC140, and the RS subunit RSP16. (B) A dynein extract was fractionated by centrifugation on a 5–20% sucrose density gradient. Fractions 1–18 were analyzed on a Western blot probed with antibodies against DHC9, FAP57, and IC140. (C) A dynein extract from the outer arm mutant pf28 was fractionated by FPLC chromatography. Fractions 44–72 were analyzed by SDS–PAGE on 3–5% (top) and 5–15% (middle) gels stained with silver or on a Western blot (bottom) stained with the FAP57 antibody. FAP57 eluted in a broad region (see asterisks) that overlapped with the FPLC peaks of dyneins d, e, f, and g. The small black squares indicate the dynein ICs associated with I1/f dynein. The black circles indicate the bands in peak g that were analyzed by MS/MS.
Identification of polypeptides associated with FAP57 by mass spectrometry
To identify other polypeptides that might interact with FAP57, we analyzed FPLC fractions and WT and mutant axonemes by mass spectrometry (MS/MS). For the FPLC samples, fractions spanning the peaks of dynein f and g were separated by SDS–PAGE and silver stained (Figure 4C). Prominent bands were excised from the gel, digested with trypsin, and analyzed by MS/MS. As expected, numerous peptides from FAP57 and copurifying dynein subunits were readily detected. Five other polypeptides coeluted with FAP57; these included FAP44, FAP43, FAP244, FAP159, and FAP75, in order of peptide abundance (Supplemental Table S4). Little is known about FAP159 and FAP75, but FAP43, FAP44, and FAP244 have been identified as subunits of a tether-tether head (T/TH) complex linking the I1 dynein motor domains to the DMT and also interacting with the base of dynein d (Fu et al., 2018; Kubo et al., 2018; Urbanska et al., 2018). FAP43, FAP44, and FAP244 share structural similarity with FAP57 with respect to the arrangement of their WD repeat and coiled coil domains (Fu et al., 2018; Kubo et al., 2018), and FAP43 and FAP44 have been proposed to interact with FAP57 in Tetrahymena cilia based on proximity labeling (Urbanska et al., 2018).
To see whether any of the polypeptides that coeluted with FAP57 might be altered in ida8, axonemes from ida8-1 and an HA-rescued strain were labeled in duplicate using four different isobaric tag for relative and absolute quantitation (iTRAQ) tags, digested, fractionated by liquid chromatography, and analyzed by MS/MS to identify the total complement of polypeptides present in each sample (see Materials and Methods). The protein ratios were then analyzed to identify those polypeptides whose ida8/HA ratios were significantly different (P < 0.05) from the control ratio (HA/HA). Several proteins were reduced to variable degrees, but only two polypeptides, FAP57 and Cre13.g562800, were reproducibly and significantly reduced below 30% in ida8-1 axonemes (Table 2). Cre13.g562800, is an EF hand, WD repeat containing polypeptide identified in the flagellar proteome as FAP337 (Pazour et al., 2005). It is closely related to another protein in Chlamydomonas, Cre07.g313850 (Basic Local Alignment Search Tool [BLAST] score 1e-50), and also shares significant sequence homology with two vertebrate proteins WDR49 and EFCAB8 (Table 1). The iTRAQ ratios of two other proteins were increased more than 50% in ida8-1, FBB7 (Cre03.g143827), and FAP331 (Cre06.g308000). Both contain N-terminal regions with multiple WD repeats and C-terminal regions with coiled coil domains, similar to FAP57. To verify the changes in protein composition predicted by iTRAQ analysis, we also fractionated axonemes from WT, ida8-1, and the ida8-1; FAP57-HA1 rescued strain by SDS–PAGE, cut bands from the appropriate regions, digested the samples with trypsin, and analyzed both the number of unique peptides and total spectra using label free quantification. Spectral counting confirmed that FAP57 and FAP337 were reduced in ida8 (<10% of WT), that FAP331 and FBB7 were increased in ida8 (>30% of WT), and that all were restored to WT levels in FAP57-HA rescued axonemes. The organization of polypeptide domains in each of these proteins is shown diagrammatically in Supplemental Figure S5.
TABLE 2:
iTRAQ protein ratios in ida8 and FAP57-HA axonemes
| Experiment 1 | Experiment 2 | |||||
|---|---|---|---|---|---|---|
| Protein | Peptides | HA/HA | ida8/HA | Peptides | HA/HA | ida8/HA |
| Reduced (<0.30) | ||||||
| FAP57 | 76 (94) | 0.93 | 0.11 | 100 (172) | 0.89 | 0.16 |
| Cre13.g562800 (FAP337) | 11 (13) | 1.00 | 0.26 | 17 (21) | 0.98 | 0.26 |
| Increased (>1.50) | ||||||
| Cre06.g308000 (FAP331) | 55 (58) | 1.12 | 2.01 | 55 (67) | 1.01 | 1.59 |
| Cre03.g143827 (FBB7) | 59 (71) | 1.16 | 2.45 | 56 (69) | 1.03 | 1.83 |
| Dynein heavy chains | ||||||
| 1-α DHC | 318 (368) | 1.01 | 1.02 | 357 (660) | 0.98 | 0.89 |
| 1-β DHC | 328 (419) | 1.02 | 1.00 | 368 (735) | 0.97 | 0.87 |
| DHC2 | 201 (238) | 0.99 | 0.86 | 286 (508) | 0.96 | 0.66 |
| DHC3 | 76 (69) | 0.98 | 0.54 | 99 (113) | 0.95 | 0.45 |
| DHC4 | 144 (122) | 1.02 | 1.16 | 160 (167) | 0.97 | 0.84 |
| DHC5 | 212 (216) | 1.03 | 1.15 | 240 (355) | 0.96 | 0.91 |
| DHC6 | 182 (202) | 1.02 | 1.07 | 237 (395) | 0.97 | 0.77 |
| DHC7 | 209 (235) | 0.99 | 0.63 | 259 (426) | 0.95 | 0.40 |
| DHC8 | 216 (229) | 1.02 | 1.08 | 251 (390) | 0.97 | 0.86 |
| DHC9 | 268 (299) | 1.01 | 1.08 | 290 (355) | 0.98 | 0.91 |
| DHC11 | 146 (135) | 1.03 | 1.05 | 173 (213) | 0.97 | 0.89 |
| DHC12 | 78 (96) | 1.02 | 1.03 | 100 (129) | 0.99 | 0.83 |
| α DHC | 505 (516) | 1.02 | 1.10 | 531 (1084) | 0.98 | 0.88 |
| β DHC | 558 (625) | 1.02 | 1.12 | 605 (1321) | 0.97 | 0.89 |
| γ DHC | 375 (428) | 1.02 | 1.13 | 446 (907) | 0.98 | 0.89 |
| I1 dynein associated | ||||||
| IC140/DIC3 | 68 (119) | 1.01 | 1.04 | 55 (79) | 1.01 | 1.10 |
| IC138/DIC4 | 76 (127) | 0.96 | 1.01 | 64 (77) | 1.04 | 1.29 |
| IC97 | 56 (118) | 0.99 | 1.04 | 41 (62) | 1.00 | 1.15 |
| MIA1/FAP100 | 35 (35) | 1.01 | 1.06 | 42 (53) | 0.99 | 0.93 |
| MIA2/FAP73 | 23 (26) | 1.01 | 0.98 | 23 (38) | 1.01 | 0.99 |
| FAP43 | 87 (107) | 1.03 | 1.06 | 95 (184) | 0.98 | 0.94 |
| FAP44 | 104 (117) | 1.05 | 1.10 | 117 (212) | 0.97 | 0.91 |
| FAP244 | 23 (24) | 0.97 | 0.99 | 80 (122) | 0.97 | 0.86 |
| N-DRC | ||||||
| DRC2 | 36 (54) | 1.00 | 1.03 | 36 (68) | 1.02 | 0.88 |
| DRC3 | 47 (59) | 1.03 | 1.03 | 62 (118) | 0.99 | 0.90 |
| DRC4 | 65 (79) | 1.00 | 0.99 | 73 (138) | 0.99 | 1.03 |
| DRC5 | 21 (18) | 0.96 | 0.96 | 31 (27) | 0.94 | 0.96 |
| DRC6 | 23 (34) | 0.97 | 0.99 | 21 (42) | 0.99 | 0.94 |
| DRC7 | 120 (172) | 1.02 | 1.05 | 133 (304) | 0.98 | 0.92 |
| DRC8 | 17 (25) | 1.05 | 1.00 | 13 (30) | 0.97 | 1.00 |
| DRC9 | 33 (39) | 1.03 | 1.07 | 35 (69) | 1.02 | 0.99 |
| DRC10 | 24 (37) | 0.98 | 0.98 | 29 (56) | 1.00 | 0.98 |
| DRC11 | 72 (93) | 1.00 | 0.95 | 71 (136) | 0.98 | 0.96 |
| MT doublet associated | ||||||
| Rib43 | 52 (52) | 0.95 | 0.86 | 42 (88) | 1.03 | 1.08 |
| Rib72 | 159 (181) | 1.02 | 0.96 | 174 (333) | 0.97 | 0.95 |
| FAP59 | 62 (68) | 1.04 | 0.99 | 61 (117) | 0.96 | 0.90 |
| FAP172 | 67 (71) | 1.01 | 0.95 | 61 (113) | 0.97 | 0.84 |
| MBO2 | 74 (91) | 1.03 | 1.02 | 81 (139) | 0.98 | 0.90 |
| TUA | 449 (440) | 0.98 | 0.98 | 569 (1202) | 1.00 | 1.00 |
| TUB | 584 (632) | 1.02 | 1.03 | 723 (1160) | 0.99 | 1.01 |
| FAP20 | 49 (89) | 1.05 | 1.12 | 37 (36) | 1.01 | 1.12 |
| PACRG | 76 (268) | 0.98 | 1.12 | 68 (95) | 1.01 | 1.13 |
Peptides identified at the 95% confidence interval (peptides used for quantification). The HA/HA ratio represents variation between technical replicates of the same biological sample (typically less than 10%). The ida8/HA ratio is an average of two technical replicates of ida8 relative to the FAP57-HA rescued strain. The two experiments are different biological replicates. The ida8/HA ratios that were significantly different (P < 0.05) in both experiments are highlighted in bold.
Because ida8-1 and bop2-1 displayed defects in the assembly of IDA structures located in the 96 nm repeat, we also analyzed the iTRAQ ratios of several proteins previously localized near the IDAs (Table 2). All polypeptides associated with the two-headed I1/f dynein, which is located at the proximal end of the 96 nm repeat (Piperno et al., 1990; Mastronarde et al., 1992), were present at WT levels. These include the 1α and 1β DHCs, several I1 intermediate chains, the two MIA proteins (FAP73 and FAP100) implicated in the regulation of I1 dynein (Yamamoto et al., 2013), and the three T/TH proteins, FAP43, FAP44, and FAP244, mentioned above. All subunits of the N-DRC, which is located at the distal end of the repeat (Gardner et al., 1994; Heuser et al., 2009), were also present at WT levels. Moreover, the ratios of several coiled coil proteins were also unchanged in ida8-1. These include Rib43a and Rib72, two proteins found inside the lumen of the A-tubule (Stoddard et al., 2018) and the 96 nm ruler complex FAP59/FAP172, which has been identified as elongated structure that is tightly associated with the DMTs, establishes the dimensions of the 96 nm repeat and helps to specify the binding sites of the RSs and IDAs (Oda et al., 2014). No significant changes were observed in any of the outer arm DHCs, DHC4–DHC6, or DHC8–DHC12. However, the iTRAQ ratios of three inner arm DHCs, DHC2, DHC3, and DHC7 were significantly reduced in ida8 (Table 2). To confirm the DHC defects by label free quantification, we excised gel bands containing the DHCs (400–500 kDa) from several axoneme samples (Figure 5, A and B), digested them with trypsin, and analyzed them by MS/MS and spectral counting (Zhu et al., 2010; Bower et al., 2013, 2018; Wirschell et al., 2013). As shown in Figure 5C, DHC2, DHC3, and DHC7 were reduced in ida8-1 and restored to WT levels in the FAP57-HA1 rescued axonemes, consistent with the ratios observed by iTRAQ labeling. (The apparent decrease of DHC6 in the HA1 rescued strain shown in Figure 5C was not observed in a second spectral counting experiment or in the iTRAQ analysis shown in Table 2.) Previous studies have shown that DHC2 elutes in peak d and DHC7 in peak g, that DHC3 is a minor dynein located in the proximal portion of the axoneme and closely related to DHC7, and that both dynein d and g are located at the distal end of the 96 nm repeat (Yagi et al., 2009; Bui et al., 2012). Collectively, these observations strongly suggested that FAP57 targets or stabilizes the attachment of these dyneins to their unique binding sites in the 96 nm repeat.
Cryo-ET of ida8 reveals the complexity of defects in IDA structures
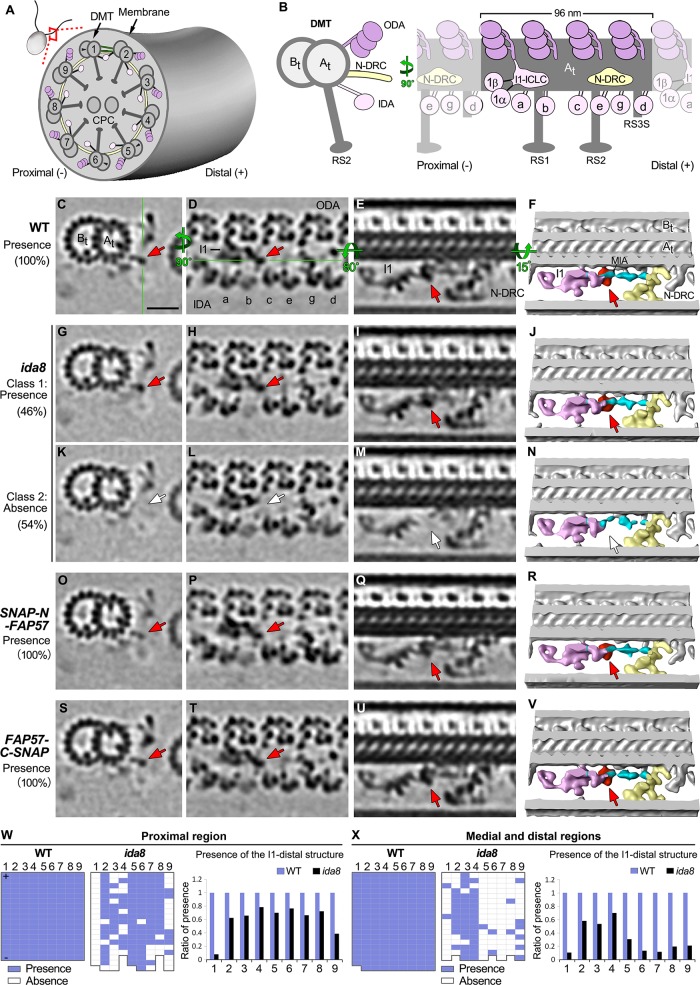
Given the complexity of the biochemical and structural defects observed in ida8, we analyzed WT, ida8, and FAP57-rescued axonemes by cryo-ET and subtomogram averaging to better resolve the defects in the structure of the IDAs. We also rescued ida8 by transformation with N- and C-terminally SNAP-tagged versions of the FAP57 gene to localize FAP57 more precisely and gain insight into the role of FAP57 in the targeting of IDAs. As shown in Supplemental Figure S6, A and B, FAP57 polypeptides with either an N-terminal or a C-terminal SNAP tag were assembled into axonemes and restored the forward swimming velocity of ida8 to near WT levels. Consistent with images obtained by TEM and 2D averaging (Figure 2C and King et al., 1994) but with higher resolution, the average of all ida8 tomograms showed decreases in densities located in two distinct regions of the 96 nm repeat, one located at the distal end of the I1 dynein IC/LC complex (termed the I1-distal structure) and a second corresponding to the locations of the two distal dyneins, IDAs g and d (compare Supplemental Figure S6, C and D with E and F). In addition, the density of IDA b appeared to be weaker in ida8 relative to WT when all ida8 tomograms were compared with all WT tomograms (Supplemental Figure S6, C–F). The reduced densities were restored to WT levels in tomograms from the FAP57 rescued strains (Supplemental Figure S6, G–J).
To better characterize the defects in ida8 axonemes, we performed a classification analysis on each structure of interest. The proximal and medial/distal regions of the axoneme and the identities of DMTs 1–9 were determined by the presence of DMT specific features (Bui et al., 2012; Lin et al., 2012). These analyses not only precisely identified the structural defects in ida8, but it also correlated the defects with a specific region or DMT (Figures 6 and 7). As shown in Figure 6, the I1-distal structure was present in 100% of the WT repeats (Figure 6, C–F), missing in 54% (class 2) of the ida8 repeats (Figure 6, G–N), and recovered to 100% in the repeats from both rescued strains (Figure 6, O–V). The missing structure is located on the distal side of the I1-dynein IC/LC complex, close to the location of the MIA complex (Yamamoto et al., 2013). Based on the iTRAQ results that detected WT levels of MIA proteins in ida8 (Table 2) and the fact that some densities remained in this region in the ida8 tomograms (Figure 6, G–N), the I1-distal structure appears to be distinct from the densities associated with the MIA complex, although the position of the MIA complex may be slightly shifted in the absence of the I1 distal density (Figure 6N). Although defects in the I1-distal structure were observed on all DMTs of ida8, the classification analysis revealed that these defects were asymmetrically distributed both along the length of the axoneme and among the DMTs. More specifically, the I1-distal structure was most significantly reduced on DMTs 1 and 9 in the proximal region and on DMT1 and DMTs 5–9 in the medial/distal region (Figure 6, W and X).
FIGURE 6:
Cryo-ET and class averaging of ida8 reveal defects in a density located between I1 dynein and the MIA complex. (A) Diagram of a Chlamydomonas cell and one of its two flagella shown in cross-section with nine outer DMT1–9 surrounding the CPC. (B) Diagram of a single DMT shown in cross-section view (left) and longitudinal view from the perspective of the neighboring DMT (right), with the A- and B-tubules (At and Bt). The DMT is built up by a series of 96 nm axonemal repeats, each of which contains four three-headed ODAs (dark pink) on top, three RSs (RS1, RS2, and the shorter RS3S) at the bottom, and seven distinct IDAs (light pink) and the N-DRC (yellow) in the middle region. The two-headed I1 dynein is located at the proximal end, with its 1α and 1β dynein heads connected to the I1 IC/LC domain. The six single-headed IDAs (a, b, c, e, g, d) are attached to specific sites along the repeat. (C–E) Tomographic slices of the class average of the 96 nm repeat of WT axonemes showing the I1-distal structure in three different views: a cross-section of 96 nm repeat through the I1-distal structure (C), a longitudinal section of the 96 nm repeat through the dyneins (D), and another longitudinal section that rotates 80 degrees (E). The green lines indicate the locations of the slices shown in next panel. Classification analysis showed that all WT repeats have the I1-distal structure. (F) Iso-surface renderings corresponding to image in E, but with a small rotation for a better 3D view of the I1-distal structure (red) and its neighboring complexes: I1 dynein complex (pink), N-DRC (yellow), MIA complex (cyan). (G–N) Images of the two class averages of the 96 nm repeats in ida8. In Class 1 repeats (∼46%), the I1-distal structure was present (G–J, red arrows); in Class 2 repeats (∼54%), the I1-distal structure was missing (white arrows), and the MIA complex (cyan) appeared slightly shifted in N. (O–V) Images of SNAP-N-FAP57 (O–R) and FAP57-C-SNAP (S–V) rescued axonemes, showing reassembly of the I1-distal structure (red arrows). (W, X) The presence (blue grids) or absence (white grids) of the I1-distal structure in each 96 nm repeat was scored for individual DMTs in the tomograms taken from the proximal (W) or medial/distal regions (X) of the axoneme. Flagellar polarity is indicated by "+" and "–" ends. The WT data set contained five proximal and 20 medial/distal tomograms, whereas the ida8 data set contained 16 proximal and 17 medial/distal tomograms. The averaged histograms on the right depict the ratio of repeats with the I1-distal structure relative to all repeats on the individual DMTs. Scale bar in C is 20 nm.
FIGURE 7:
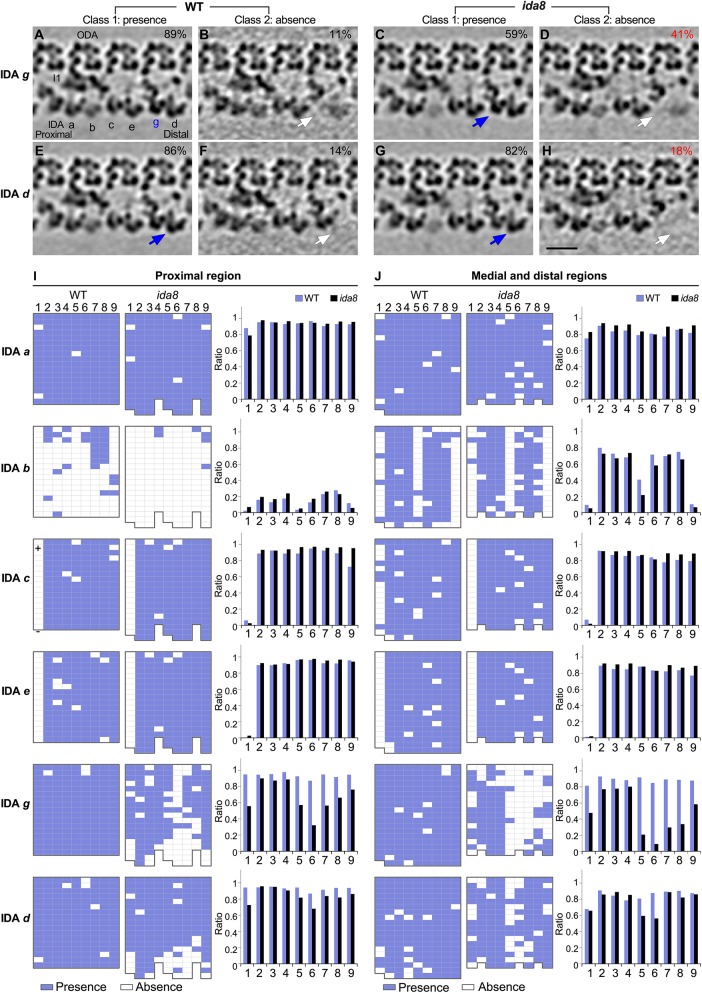
Cryo-ET and class averaging reveal defects in the assembly of IDAs d and g on specific DMTs in ida8. The WT and ida8 repeats were analyzed for the presence (Class 1) or absence (Class 2) of each single-headed IDA (a, b, c, e, g, d). (A–H) Tomographic slices of the class averages of the 96 nm repeat, with four ODAs on top and the single-headed IDAs at the bottom, showing the presence (Class 1, blue arrows) or absence (Class 2, white arrows) of the IDAs g and d, which are reduced in many ida8 repeats. The percentage of subtomograms included in each class average is indicated. (See Supplemental Figure S7 for the class averages of all the single-headed IDAs.) (I, J) The presence (blue grids) or absence (white grids) of the indicated IDA in a 96 nm repeat was scored for each DMT (1–9) in tomograms taken from the proximal (I) or medial and distal (J) regions of the axoneme. The WT data set contained five proximal and 20 medial/distal tomograms, whereas the ida8 data set contained 16 proximal and 17 medial/distal tomograms. The averaged histograms on the right depict the ratio of repeats with the indicated IDA relative to all of the repeats for each DMT (1–9). Classification analysis showed that the assembly of dyneins a, b, c, and e in ida8 was not significantly different from WT. However, more ida8 repeats lacked IDAs g and d than WT (D, H), and the defect in assembly was biased toward DMTs 1 and 5–9. Scale bar in H is 20 nm.
Classification analyses of the individual IDAs (a, b, c, e, g, and d) in the subtomograms revealed significant differences in the assembly of IDAs g and d in ida8 axonemes, but no significant changes in the assembly of IDAs a, b, c, and e (Figure 7; Supplemental Figure S7). In addition, the defects in assembly of IDAs g and d were distributed asymmetrically, similar to defects in the I1-distal structure (Figures 6 and 7). The defect in IDA g was more remarkable: 41% of ida8 repeats lacked IDA g compared with 11% of WT repeats (Figure 7, A–D). This difference was largely due to decreased assembly of dynein g on DMTs 5–8 and to a lesser extent on DMT1 and DMT9 (Figure 7, I and J). IDA d was missing in 18% of the ida8 repeats compared with 14% of WT repeats (Figure 7, E–H). This small change was due to decreased assembly of dynein d on DMTs 1, 5, and 6 (Figure 7, I and J). The missing IDAs g and d were restored to WT levels in the SNAP-tagged, FAP57 rescued axonemes (Supplemental Figure S8).
The classification analyses initially suggested a possible decrease in the assembly of IDA b, because IDA b was missing in 67% of the ida8 repeats versus 52% of the WT repeats (Supplemental Figure S7, E–H). However, further analysis showed that the apparent decrease in IDA b was actually due to the fact that the ida8 data set contained a much higher proportion of tomograms from the proximal region of the axoneme (48%) than the WT data set (20%). As reported previously, IDA b is only rarely seen in the proximal region of WT axonemes, and it is mostly absent from DMTs 1, 5, and 9 in the medial/distal region (Figure 7, I and J; see also Bui et al., 2012; Lin et al., 2012). Therefore, after correlating the IDA b defective repeats with specific regions of the axoneme, no significant difference in the assembly of IDA b was found between ida8 and WT (Figure 7, I and J).
To verify the asymmetric distribution of the structural defects across the nine DMTs identified by classification analysis, we also performed DMT specific averaging on WT, ida8, and FAP57 rescued axonemes. As shown in Supplemental Figure S8, the images of the DMT specific averages are consistent with the classification analyses. The densities of the I1-distal structure, and IDAs g and d on DMTs 1, 5–9 were clearly weaker in ida8 than in WT, but they were not obviously different from WT on DMTs 2–4. In addition, the densities corresponding to the missing structures were all restored to WT levels in the SNAP-tagged, FAP57 rescued axonemes (Supplemental Figure S8).
Localization of FAP57 in the 96 nm repeat by SNA P-tagging and biotin-streptavidin-nanogold labeling
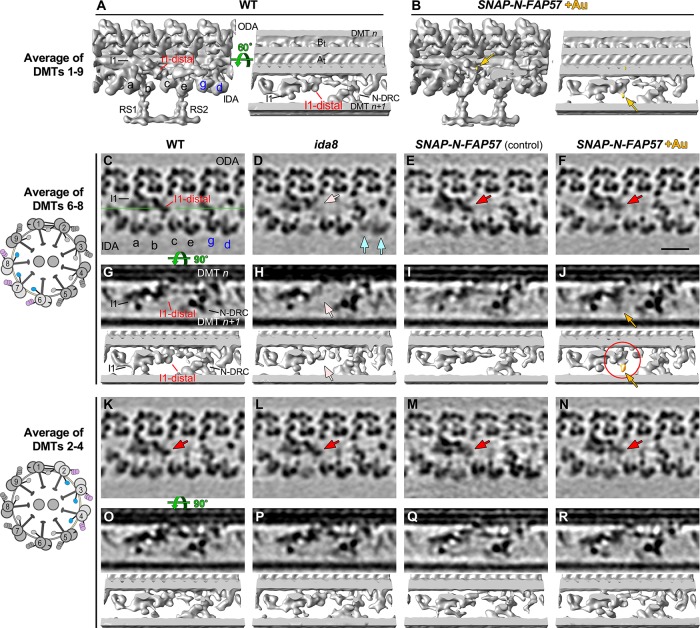
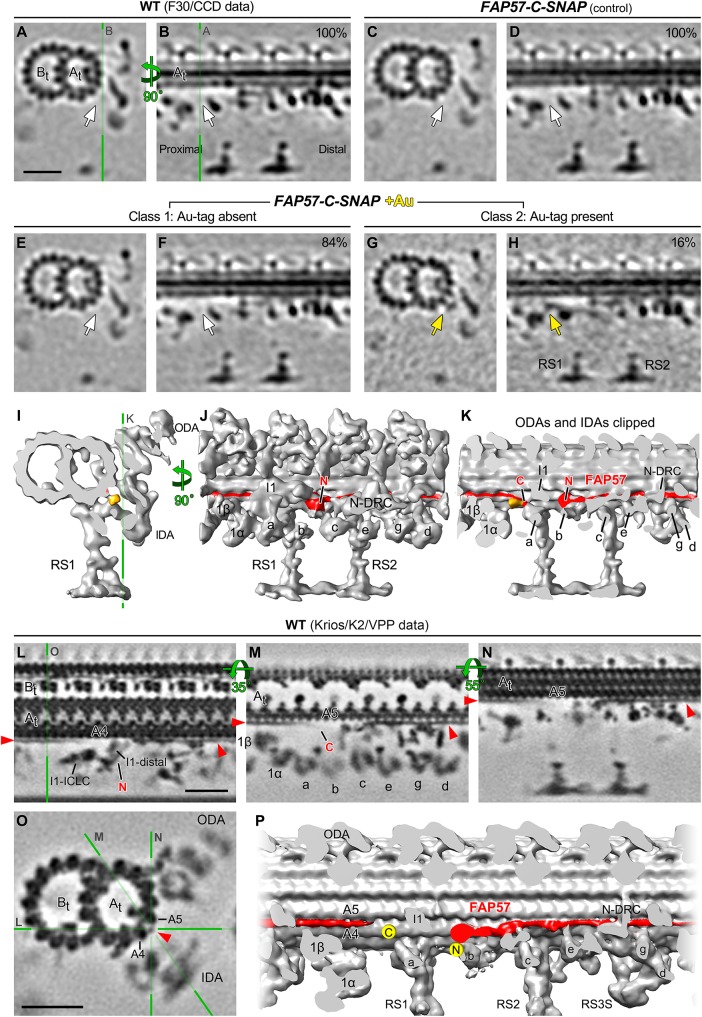
Because FAP57 has two distinct polypeptide domains, an N-terminal region with seven WD repeats and a C-terminal region with several coiled coil domains, we reasoned that these two domains might be arranged along the length of the DMT and facilitate the targeting or stabilization of the different structures missing in ida8. To test this hypothesis, we treated axonemes from the SNAP-tagged rescued strains with biotin and streptavidin-nanogold (+Au) and analyzed the tomograms for presence of additional densities that might reveal the locations of the N- and C-termini of the FAP57 polypeptide (Figures 8 and 9). As shown in Figure 8, A and B, averages of all repeats revealed an additional density in the SNAP-N-FAP57 +Au sample that was located close to the site of the I1-distal structure previously identified as missing in ida8 (Figure 8B, yellow arrows). To enhance the signal-to-noise ratio for detecting the streptavidin-nanogold label, we generated DMT specific averages using subtomograms from DMTs 6–8 (Figure 8, C–J) and DMTs 2–4 (Figure 8, K–R). The DMTs 6–8 averages clearly showed the missing I1-distal structure in ida8 (Figure 8, D and H, pink arrows), its recovery in the SNAP-N-FAP57 strain (Figure 8, E and F, red arrows), and the presence of an additional density in the streptavidin-gold-treated sample (Figure 8, F and J, yellow arrows). Consistent with the classification results (Figure 7; Supplemental Figure S7), the DMT 2–4 averages showed no obvious structural defect in ida8 and no additional density in the streptavidin-gold-treated sample (Figure 8, K–R). These results confirm the asymmetric distribution of the structural defects across the nine DMTs of ida8 and suggest that the N-terminal portion of FAP57, which contains multiple WD repeats, contributes to the assembly of the I1-distal structure.
FIGURE 8:
Streptavidin-gold labeling, cryo-ET, and DMT specific averaging reveal the location of the N-terminus of FAP57. (A, B) Iso-surface renderings of the averaged 96 nm repeats from WT axonemes (A) or from streptavidin–gold–labeled axonemes from SNAP-N-FAP57 rescued strain (B). A new density was detected on the I1-distal structure in the gold-labeled rescued axonemes, as highlighted by yellow arrows. (C–F) Tomographic slices of the averaged 96 nm repeats of DMTs 6–8 from WT (C), ida8 (D), SNAP-N-FAP57 control (E), or gold-labeled SNAP-N-FAP57 axonemes (F; +Au). The SNAP-N-FAP57 control sample is different from the gold-labeled SNAP-N-FAP57 sample because the BG-(PEG)12-Biotin was omitted in the control during the labeling procedure. A diagram of an axoneme cross-section is shown on the left, with DMTs 6–8 highlighted in color. Defects in the I1-distal structure (pink arrows) and IDAs g and d (light blue arrows) were clearly visible as weaker densities in ida8 (D). These densities were restored in both of the rescued samples (red arrows; E and F). (G–J) Tomographic slices (top) and isosurface renderings (bottom) of averaged 96 nm repeat of DMTs 6–8 from WT (G), ida8 (H), SNAP-N-FAP57 control (I), or gold-labeled SNAP-N-FAP57 axonemes (J). The location of the tomographic slice is indicated by a green line in C. The red circle in J highlights the new density on I1-distal structure observed in the gold-labeled sample. This density is noticeably larger than that seen in the average of all nine DMTs in (B). (K–R) Images of averaged 96 nm repeats of DMTs 2–4 from WT (K, O), ida8 (L, P), SNAP-N-FAP57 control (M, Q), or gold-labeled SNAP-N-FAP57 axonemes (N, R). The I1-distal defect was hardly visible in the average of DMTs 2–4 of ida8 (L, P), and a new density was not clearly visible in the gold-labeled axonemes (N, R). Scale bar in F is 20 nm.
FIGURE 9:
Streptavidin-gold labeling, cryo-ET, and class averaging reveal the location of the C-terminus of FAP57, and higher-resolution average directly visualizes the candidate density of FAP57. (A–H) Tomographic slices of class averages of the 96 nm repeat from WT axonemes (A, B), FAP57-C-SNAP control (C, D), or gold-labeled FAP57-C-SNAP axonemes (E–H; +Au). BG-(PEG)12-Biotin was omitted during the labeling protocol in the FAP57-C-SNAP control sample in C and D. The averaged repeats are shown in cross-section (left) and in longitudinal section at a plane close to the surface of the A-tubule (right). The locations of tomographic slices are indicated by green lines. The percentage of subtomograms included in each class average is indicated. A new class (Class 2) was identified in the gold-labeled FAP57-C-SNAP axonemes in G and H. Class 2 contained a novel density (gold arrows) located close to the surface of the A-tubule and proximal to RS1 and the IC/LC complex of the I1 dynein. Such novel density was not observed in the control (C, D) or Class 1 averages (E, F, white arrows). (I–K) Isosurface rendering of the 96 nm repeat from the Class 2 axonemes viewed in cross-sectional (I) and longitudinal (J, K) orientations. The ODAs and IDAs were clipped in K for better visualization of the novel density. The clipping plane is indicated by the green line in K. The predicted arrangement of the FAP57 polypeptide on the surface of A-tubule is shown in red. The proposed locations of the N- and C-termini of FAP57 are denoted based on the streptavidin-gold labeling of SNAP-N-FAP57 (Figure 8) and FAP57-C-SNAP axonemes (this figure), respectively. (L–O) Longitudinal (L–N) and cross-sectional (O) tomographic slices of the 96 nm repeat from WT axonemes that have significantly improved resolution (1.8 nm, 0.5 criteria of FSC). The green lines with letters indicate the locations of the slices shown in corresponding panels. At this significantly improved resolution, a filamentous structure that extends from the I1-distal structure (L), attaches on protofilaments A4 and/or A5 of the A-tubule, and runs along the A-tubule toward the distal side was clearly visible (L–O, red arrowheads). The location of this structure coincides with the location of FAP57 predicted in (J, K), suggesting this structure as a compelling candidate of FAP57. (P) Iso-surface rendering of the higher-resolution average in longitudinal orientation. The candidate density of FAP57 is highlighted in red and the proposed locations of its C- and N-termini were denoted by two yellow dots. Scale bars in A, L, and O are 20 nm; (A) valid for A–H; (L) valid for L–N.
The tomograms obtained from FAP57-C-SNAP axonemes showed recovery of structures missing in ida8, similar to the SNAP-N-FAP57 axonemes (Figure 6, O–V; Supplemental Figures S6, I–J, and S8), but an additional density associated with the streptavidin-nanogold labeling of the C-terminal SNAP tag was much harder to detect than for the N-SNAP tag. These results may indicate that the C-terminus of FAP57 is buried within a complex of other proteins and less accessible to bind the streptavidin-gold in intact axonemes. To increase the signal-to-noise ratio for detection of the C-terminally labeled region of FAP57, we turned again to classification of the subtomogram averages (Figure 9, A–K). This approach identified an additional density in a small subset (∼16%) of subtomograms (Figure 9, G, H, and I–K). This additional density was located close to the surface of the DMT, proximal to the base of RS1, near the base of the I1/f dynein and dynein a.
Direct visualization of the candidate density of FAP57 with recent hardware and software advances for cryo-ET
Aiming at directly resolving the FAP57 structure in 3D, we applied recent hardware and software advances for cryo-ET and reanalyzed the 96 nm repeat in WT axonemes (Materials and Methods). We further improved the resolution of the averaged 96 nm repeat from 3–4 to 1.8 nm (0.5 criteria of FSC; Supplemental Table S5). The significantly higher resolution allowed clear visualization of an extended filamentous structure as a candidate for the FAP57 protein (Figure 9, L–P). This filamentous structure extends from the I1-distal density, near the site of the N-terminus of FAP57, to the outer cleft of protofilaments A4-5 (Figure 9L), from where it runs parallel to the A-tubule toward the distal end of the 96 nm repeat, making attachments to multiple structures such as the N-DRC and tail domains of IDA g and d (Figure 9, L–P). The filament then extends into the next 96 nm repeat and becomes slightly curved, with a weaker density at the end near the base of IDA a, close to the site where the C-terminus of FAP57 was localized (Figure 9M). This weaker density may be a reflection of the greater positional flexibility of the C-terminal region of FAP57, which is predicted to be a low complexity region that is potentially disordered. The flexibility of the FAP57 C-terminus in combination with the crowded molecular environment around the base of both the I1/f dynein and IDA a might explain the lower labeling efficiency with streptavidin-gold (Figure 9, G and H).
DISCUSSION
The BOP2/IDA8 locus encodes a conserved polypeptide required for stabilizing the binding of a subset of IDAs
Our study of the bop2/ida8 mutations has identified a new subcomplex that contributes to the organization of IDAs within the 96 nm repeat. The three ida8 alleles and bop2-1 are null mutations that fail to assemble FAP57 into the axoneme (Figures 1 and 2). Transformation with WT or epitope-tagged versions of FAP57 rescues the motility defects and restores the missing proteins (Figures 2, 3, and 5). FAP57 is a highly conserved polypeptide containing an N-terminal region with multiple WD repeats and a C-terminal region with multiple coiled coil domains (Figure 2). Interestingly, closely related orthologues known as WDR65/CFAP57 have been identified in other organisms with motile cilia, but only in those species that assemble IDAs (Table 1; see also Nevers et al., 2017). The loss of FAP57 in Chlamydomonas is consistently correlated with the absence of a second highly conserved WD repeat protein, FAP337, and reduced assembly of inner arm DHC2, DHC3, and DHC7, as observed by both iTRAQ labeling and spectral counting of multiple axoneme samples (Tables 1 and 2; Figure 5; Supplemental Figure S5). The polypeptide defects observed in ida8 are distinct from those described in other motility mutants (Supplemental Figure S3). Taken together, the results suggest that FAP57 and FAP337 form a distinct subcomplex that is required to stabilize the binding of specific IDAs at the distal end of the 96 nm repeat.
Quantitative MS/MS using both iTRAQ labeling and label-free spectral counting has shown that two other proteins, FAP331 and FBB7, are elevated in ida8 but restored to WT levels in a rescued strain (Tables 1 and 2). The two proteins share a similar overall structural organization of N-terminal, WD repeat domains and C-terminal, coiled coil domains with FAP57 (Supplemental Figure S5). In particular, FBB7 shares significant sequence homology with FAP57 (Table 1). These observations are reminiscent of earlier studies on the ida5 mutants, in which mutations in the conventional Chlamydomonas actin gene (IDA5) were offset by increased expression of a novel actin-related protein (NAP1) (Kato-Minoura et al., 1997, 1998). Actin is an IC subunit of all single-headed IDAs, and ida5 mutations result in the failure to assemble IDAs a, c, d, and e. However, NAP1 substitutes for the missing actin subunit in IDAs b and g and permits their assembly in ida5 mutants. Similar changes in expression were also observed with the redundant I1 tether head subunits FAP43 and FAP244 in fap43 mutants (Fu et al., 2018). One possibility is that FBB7 may partially compensate for the absence of FAP57 and stabilize the binding of the remaining IDAs in the ida8 mutant. Identification of a fbb7 mutant will be required to test this hypothesis.
FAP57 forms an extended structure within the 96 nm repeat that interacts with multiple regulators of IDA activity
Several lines of genetic, biochemical, and structural evidence suggest that FAP57 extends nearly the full length of the 96 nm repeat and interacts with multiple regulators of IDA activity. The first fap57 mutation, bop2-1, was originally isolated as extragenic suppressor of the pf mutant pf10 (Dutcher et al., 1988). Little is known about the PF10 gene product or the identity and location of polypeptides that might be altered in pf10 axonemes. However, the gene product of another pf10 suppressor, BOP5, has been identified as the I1 subunit IC138 (Hendrickson et al., 2004; VanderWaal et al., 2011). IC138 is a WD repeat protein within the IC/LC complex at the base of I1 dynein (Bower et al., 2009; Heuser et al., 2012). Given that two pf10 suppressors have been linked to WD repeat proteins associated with IDAs, pf10 may be directly or indirectly associated with defects in the coordination or regulation of IDAs.
Biochemical studies have also identified potential interactions between FAP57- and I1 dynein-associated proteins. Yamamoto et al. (2013) described FAP57 as one of a few polypeptides that coimmunoprecipitated with the FAP100 subunit of the MIA complex, the IC138 and two DHCs of I1 dynein, and the FAP44 subunit of the I1 dynein tether head after chemical cross-linking. FAP57 has also been linked to the FAP44 subunit of the I1 tether head in Tetrahymena, based on proximity labeling (Urbanska et al., 2018). We found that FAP57 coelutes with the I1 tether head subunits during FPLC fractionation (Figure 4; Supplemental Table S4). Although none of these proteins require FAP57 for assembly into the axoneme (Table 2) nor vice versa (Supplemental Figure S3; see also Urbanska et al., 2018), collectively the data suggest that FAP57 is located in close proximity to both I1 dynein and the MIA complex.
Comparison of WT and mutant axonemes by TEM and cryo-ET has provided even more compelling evidence for a direct physical interaction among FAP57, I1 dynein, and the MIA complex. Loss of FAP57 in ida8 and bop2 results in a defect in the assembly of a globular structure located just distal of the I1 dynein, at the site where the I1 IC/LC complex contacts the MIA complex (Figures 2D and 6). Rescue of ida8 with a SNAP-tagged FAP57 followed by streptavidin-gold labeling confirms that the N-terminus of FAP57 is located within the I1-distal structure (Figures 8 and 10). We propose that the WD repeat domains located within the first half of the FAP57 polypeptide interact with WD repeat domains in FAP337 to form at least part of the I1-distal structure.
FIGURE 10:
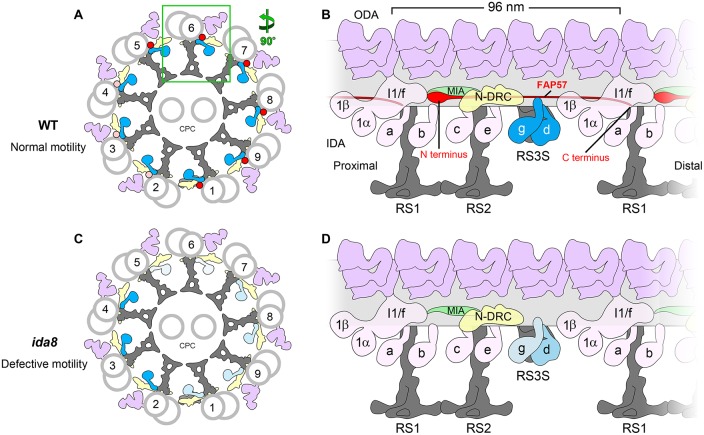
Model for the arrangement of FAP57 in the 96 nm repeat and its role in the assembly of certain IDAs. (A) Diagram of the cross-section of a WT axoneme showing the arrangement of the DMTs and the proposed asymmetric distribution of FAP57 across the nine DMTs: most FAP57 are located on DMTs 1, 5–9 (red dots), while a small number of FAP57 are also located on DMTs 2–4 (pink dots). (B) Diagram of the longitudinal view of a WT DMT showing the proposed location of FAP57. The N-terminal portion of FAP57 containing the WD repeat domains is proposed to form the more globular structure that is located distal to the IC/LC complex of the I1 dynein. The second half of FAP57 containing the coiled coil domains is proposed to extend along the surface of the DMT, passing through or adjacent to the bases of IDAs g and d, and then extend further, with its C terminus located close to the base of RS1. The IDAs g and d are highlighted in blue. Note that FAP57 is proposed to contact multiple structures implicated in the regulation of IDAs, including the IC/LC complex of the I1 dynein, the MIA complex, the N-DRC, and IDAs g and d. (C) Diagram of the cross-section of an ida8 axoneme showing the defects in the assembly of IDAs due to the loss of FAP57. In the absence of FAP57, fewer IDAs are assembled on DMTs 1, 5–9 (light blue) than on DMTs 2–4 (blue). (D) Diagram of the longitudinal view of an ida8 DMT showing the proposed role of FAP57 in stabilizing the assembly of specific IDAs. In the absence of FAP57, the assembly of IDA d is only slightly reduced but IDA g is significantly reduced. The observed increase in FBB7 may compensate in part for the absence of FAP57. The levels of IDA d and g are shown by the intensity of the blue labels, with the lighter blue hues indicating less dynein present. Other labels: ODA, outer dynein arm; IDA, inner dynein arm; N-DRC, nexin-dynein regulatory complex; CPC, central pair complex; RS3S, radial spoke 3 stand-in.
Identifying the location of the C-terminal half of FAP57 (residues 623–1316) has been much more challenging. This region contains an extended coil-coil domain (residues 644–1188) followed by a more variable low complexity domain at the C-terminus (residues 1206–1314) (Figure 2). However, the defects in assembly of IDAs at the distal end of the 96 nm repeat observed in ida8 (Figure 7; Supplemental Figures S6, S7, and S8) suggest that the C-terminal half of FAP57 extends from the I1-distal structure and runs close to the surface of the DMT, above the base of the second RS (RS2) and under the N-DRC, to the sites of attachment for IDAs g and d (Figure 10). Moreover, rescue of ida8 with a FAP57-C-SNAP construct followed by streptavidin-gold labeling and class averaging reveals the presence of an additional density compared with the WT structure that is located near the base of the I1 dynein and IDA a (Figure 9). We propose that the coiled coil domain extends beyond the N-DRC into the next 96 nm repeat and that the additional density identifies the location of the C-terminus of FAP57 (Figure 10). This arrangement would be consistent with the predicted length of the coiled coil region (Surkont et al., 2015; Truebestein and Leonard, 2016) and the observation that FAP57 stabilizes the attachment of IDA g and d. However, FAP57 is not required for docking of the N-DRC.
The challenges in directly visualizing defects in the assembly of a thin filamentous structure on the surface of the DMT are not without precedent. Indeed, localization of the axonemal ruler proteins FAP59/FAP172 relied on the presence of large tags to mark the positions of specific sites, but the proteins themselves could not be directly visualized by conventional cryo-ET (Oda et al., 2014). However, our recent efforts to improve resolution by combining cryo-ET with the methodologies used in single particle cryo-EM have yielded new images of axoneme structures with significantly greater structural detail (Song et al., 2018). This approach made it possible to resolve the FAP59/FAP172 axonemal ruler as a filamentous structure located between protofilaments 2 and 3 of the A-tubule, at the base of the RSs in Tetrahymena axonemes (Song et al., 2018). This study also identified another filamentous structure between protofilaments 4 and 5, at the base of several single-headed IDAs. This structure, named the inner arm ruler, is identical to the extended structure identified in our high-resolution average of the Chlamydomonas axonemal repeat (Figure 9, L–P), with a location that coincides with the location of FAP57 predicted by our labeling results (Figures 9 and 10).
Asymmetry of IDA defects in both bop2 and ida8
A consistent feature of the bop2/ida8 phenotypes is the asymmetry of IDA defects around the circumference of the axoneme, that is, among the DMTs (King et al., 1994; Figures 7 and 10; Supplemental Figure S8). The earlier study of bop2-1 axonemes by TEM and 2D averaging indicated that defects in IDA assembly were limited to DMTs 5, 6, 8, and 9 (King et al., 1994). Here we used cryo-ET, doublet-specific, and class-averaging focused on individual IDAs to show that loss of FAP57 in ida8-1 impacts the assembly of IDA g on DMTs 5–8 and to a lesser extent on DMTs 1 and 9 (Figure 7). Given the nature of the mutations in both strains (Figures 1–5; Table 1), it is clear that although FAP57 is important for stabilizing the binding of IDA g (and to a lesser extent the binding of IDA d), it is not the only factor that specifies these dynein attachment sites. Indeed, we have previously noted defects in the assembly of IDA g and d in n-drc mutants (Heuser et al., 2009; Bower et al., 2013, 2018; Wirschell et al., 2013). As mentioned above, the FAP57-related protein, FBB7, is increased in ida8 axonemes. FBB7 may also contribute to the targeting or stabilization of IDAs on specific DMTs.
Asymmetric defects in the assembly of dynein arms are not unique to bop2/ida8 mutants. For example, a previous study of the sup-pf2 mutations in the γ DHC revealed reduced assembly of ODAs on DMTs 3, 6–9 relative to DMTs 2, 4–5 (Rupp et al., 1996). Reduced assembly of ODAs on DMTs 6–9 was also seen in oda4-s7 and oda2-t, two DHC truncation mutants (Liu et al., 2008). More recently, we observed reduced assembly of IDA b on DMTs 2-4 in the pacrg mutant; PACRG is one component of the inner junction between the A- and B-tubules of the DMT (Dymek et al., 2019). The factors that lead to the destabilization of dyneins on specific DMTs in these mutants are not well understood, but they may be related to the forces experienced by different DMTs during axonemal bending (Liu et al., 2008).
The asymmetry of dynein activity is believed to be essential for the generation of ciliary and flagellar motility. Indeed, a recent study of actively beating sea urchin sperm flagella revealed functionally distinct dynein conformations that alternated in a bend-direction specific manner between DMTs on opposite sides of the flagellum, that is, between DMTs 2–4 and DMTs 7–9 (Lin and Nicastro, 2018). However, the mechanism(s) that switch the dynein activities during flagellar beating are unclear. The asymmetric distribution of FAP57 and its impact on the assembly of IDAs on specific DMTs suggest that FAP57 may be one component of the molecular mechanisms that regulate the pattern of dynein activity in beating cilia.
Function of FAP57/WDR65 in ciliary motility in other species
Although several studies have identified FAP57/WDR65 as a conserved component of motile cilia and flagella (Broadhead et al., 2006; McClintock et al., 2008; Arnaiz et al., 2010; Blackburn et al., 2017; Sigg et al., 2017), functional studies on fap57/wdr65 mutations are very limited. One report identified a missense mutation in WDR65 in a patient with Van der Woude syndrome, a cleft palate disorder, but this work did not establish a clear connection between the mutation and the cleft palate defects, and it is unknown whether this patient was tested for potential defects in ciliary motility or assembly (Rorick et al., 2011). The Drosophila orthologue of FAP57/WDR65, CG4329, is expressed in the chordotonal neurons, which contain 9+0 cilia critical for the auditory response, and mutations in CG4329 lead to moderate hearing impairment (Senthilan et al., 2012). The chordotonal neurons express ODAs, IDAs, and several dynein regulators, and dynein mutations also lead to hearing defects (Senthilan et al., 2012; Karak et al., 2015; zur Lage et al., 2019). Little is known about the motility of the 9+0 cilia in the chordotonal neurons, but the dyneins are thought to act as adaptation motors that amplify mechanical input (Senthilan et al., 2012; Karak et al., 2015). Whether CG4329 regulates either the assembly or the motility of dyneins in these 9+0 cilia remains to be determined. CG4329 is also found in the Drosophila sperm proteome (Wasbrough et al., 2010), but a role in sperm motility has not yet been identified. However, the Drosophila orthologue of FAP337, WDY or CG45799, is expressed in the male reproductive tract and maps to region of the Y chromosome that contains the male fertility factor kl-1 (Vibranovski et al., 2008). Collectively these data hint at a potential role for WDR65 in axonemal motility in Drosophila. FAP57/WDR65 is also highly conserved throughout vertebrate species, and in humans, FAP57 is especially abundant in testes, respiratory tissue, and fallopian tubes (Fagerberg et al., 2014; Blackburn et al., 2017). We suggest that FAP57/WDR65 should be screened as a candidate gene for those patients with PCD whose mutations have not been identified in the more commonly known PCD loci. Indeed, recent work has indicated that a FAP57/WDR65 mutation has been linked to disease in one patient with PCD (Bustamante-Marin et al., 2019).
MATERIALS AND METHODS
Culture conditions, genetic analyses, and strain construction
Strains used in this study (Supplemental Table S1) were maintained on Tris-acetate phosphate (TAP) medium, but occasionally resuspended in liquid minimal medium or 10 mM HEPES, pH 7.6, to facilitate flagellar assembly and mating. The three strains described here, ida8-1 (59c2), ida8-2 (45g11), and ida8-3 (47d7), were isolated by transformation of a nit1-305 strain with the plasmid pMN54 encoding the NIT1 gene (Tam and Lefebvre, 1993; Mitchell and Sale, 1999). To verify that the motility defects were caused by NIT1 insertion, strains were backcrossed to nit1-305 strains with WT motility (either L5 or L8), and ∼120 tetrad progeny were scored for cosegregation of their motility phenotypes and their ability to grow on selective medium (R-NO3) in the absence of added nitrate. Some strains were crossed into an arginine-requiring background (either arg7-2 or arg7-8) for transformation or complementation tests in stable diploids. Haploid arg strains were maintained by addition of 50 μg/ml arginine-HCL to TAP medium, and stable diploids (arg 7-2/arg 7-8) were selected after mating by plating on arginine-free media. Transformants were selected by cotransformation with pARG7.8 (Debuchy et al., 1989) and plating on arginine-free media or cotransformation with pSI103 (encoding the aphVIII gene) (Sizova et al., 2001) and plating on media containing 10 μg/ml paromomycin. Double mutant strains were recovered from progeny of nonparental ditype tetrads and confirmed by their motility phenotypes and Western blot analyses. The strains used for generating the higher-resolution average of the 96 nm repeat in a pseudo WT strain (Figure 9) include cw15 (CC-4533) and the CP-mutant strains fap76-1, fap81, fap92, fap216, and fap76-1; fap81, in which the DMT structure is undistinguishable from WT (Fu et al., 2019). The latter are insertional mutants that were obtained from the Chlamydomonas Library Project (CLiP; www.chlamylibrary.org; Li et al., 2016, 2019).
Characterization of plasmid insertion sites and recovery of the IDA8 gene
Purification of genomic, phage, and BAC DNA; restriction enzyme digests; agarose gels; isolation of RNA; preparation of cDNA, PCR, and RT-PCR; and Southern and Northern blots were performed as previously described (Perrone et al., 2000, 2003; Rupp et al., 2001; Rupp and Porter, 2003; Bower et al., 2013). Genomic DNA from each strain and several tetrad progeny was digested with a series of restriction enzymes and probed on Southern blots with 32P-labeled pUC119 DNA to estimate the number of plasmid insertions (Supplemental Figure S1C). The plasmid DNA cosegregated with a slow swimming phenotype in 15 random progeny from a cross between ida8-1 and nit1-305. To identify the sites of plasmid insertion, genomic DNA was isolated from each strain, digested with restriction enzymes to release vector and flanking genomic DNA, treated with T4 ligase to recircularize the plasmid, and transformed by electroporation into Escherichia coli DH5α. The resulting plasmids were purified, digested, blotted, and probed with both pUC119 and NIT1 DNA to identify restriction fragments that contained only flanking genomic DNA. An ∼700-base-pair Sau3AI band was recovered and subcloned as FC1. Southern blots of genomic DNA probed with FC1 confirmed the presence of a RFLP in ida8-1.
FC1 was used to screen a genomic phage library (Schnell and Lefebvre, 1993) by colony hybridization and plaque purification. DNA isolated from positive clones was restriction mapped, subcloned, and used to rescreen the library to extend the chromosome walk in both directions. Subclones were tested on Southern blots to determine the extent of DNA rearrangement or deletion caused by each insertion event (Supplemental Figure S1D). Subclones were also tested on Northern blots of total WT RNA isolated before and after deflagellation to identify the number and locations of the transcription units in the region. Six phage clones spanning ∼40 kb were tested for their ability to rescue the motility defects by cotransformation of ida8-1; arg7-2 with the plasmid pArg7.8 and selection on medium lacking arginine. Over 500 Arg+ transformants were screened per clone, but none of the clones rescued the motility defect. Because the complete transcription unit might not be contained within a single phage insert, a BAC library was also screened (www.chlamycollection.org/product/bac/). Five positive BAC clones (16p22, 34a14, 35d14, 6h9, and 7h7) were recovered and restriction mapped, and together they formed a contig of ∼117 kb. Four strains with WT motility were recovered out of 400 Arg+ positive colonies following cotransformation with BAC clone 6h9 (∼1% rescued). An ∼12.7 kb genomic clone containing the full-length IDA8 transcription unit was subcloned using EcoRV and Nde1, ligated into SmaI digested pUC119 after partial fill-in, and transformed into E. coli DH10b cells by electroporation to generate the plasmid p59c2. Cotransformation of ida8-1; arg7-2 with p59c2 rescued the motility defects (9/131 transformants or 6.9% rescue). Because the IDA8 gene was cloned prior to completion of the Chlamydomonas genome project, the genomic DNA and predicted cDNA sequences of the IDA8 transcription unit were determined by PCR and RT-PCR (Supplemental Table S2). Sequence files were analyzed using the Sequencher (Gene Codes, Ann Arbor, MI) and MacVector (Apex, NC) software packages.
Mapping of the IDA8 gene and identification of bop2-1 as an ida8 allele
To place the IDA8 locus on the genetic map, a genomic fragment was used to identify an EcoRI/XhoI RFLP between two strains, 137c and S1-D2 (Gross et al., 1988). The fragment was then hybridized to a series of mapping filters containing DNA isolated from the tetrad progeny of crosses between multiply marked Chlamydomonas reinhardtii strains and S1-D2. The segregation of the RFLP was analyzed relative to the segregation of more than 42 genetic and molecular markers (Porter et al., 1996; Kathir et al., 2003). The IDA8 sequence was linked to the genetic marker pyr1 (PD:NPD:TT = 7:0:7, ∼49 map units) and a molecular marker for α2 tubulin (PD:NPD:TT = 26:0:2, ∼3.6 map units) on the left arm of Linkage group IV, close to the predicted location of the bop2-1 mutation (Dutcher et al., 1988). This distance was consistent with the later sequence assembly of Chromosome 4, with α2 tubulin (Cre04.g216850) at nucleotides 144030–147635 and FAP57 (Cre04.g217914) at nucleotides 370547–382328. To determine whether bop2-1 might be an IDA8 mutation, genomic DNA and RNA were isolated from bop2-1 and analyzed by PCR, RT-PCR, and DNA sequencing (Bower et al., 2013, 2018). Cotransformation of bop2-1; arg7-8 with p59c2 rescued the motility defects (3/60 transformants or ∼5% rescued).
Characterization of the IDA8 gene product and generation of a specific antibody
The predicted amino acid sequence was compared with predicted sequences found in several versions of the Chlamydomonas genome project (https://phytozome.jgi.doe.gov/pz/portal.html). Predicted domains were identified using programs available at www.expasy.org/proteomics and https://iupred2a.elte.hu/. To identify a peptide that could be used to generate a specific antibody, regions of the amino acid sequence were analyzed for antigenicity using MacVector. Potentially immunogenic peptides were searched against all sequences available in the genome project to increase the likelihood that the chosen peptide would be unique. Peptides were also searched against predicted amino acid sequences in other species to identify regions of high sequence conservation. The peptide NLRGHNGKVRSVAWSPDDSKL (corresponding to amino acid residues 460–480) was synthesized, conjugated to KLH, and used to immunize two rabbits (Research Genetics, Huntsville, AL). Immune sera were tested by enzyme-linked immunosorbent assay and affinity purified against the peptide.
Epitope tagging of FAP57
For epitope-tagging of the C-terminus of FAP57, a 1.2 kb SpeI-EcoRI fragment spanning the predicted stop codon was isolated from p59c2 and subcloned into pBluescript. A NdeI site was created in the stop codon (TAA to TAT) using the Quick Change II XL Mutagenesis kit (Stratagene) and primers listed in Supplemental Table S2. A triple-HA epitope tag was amplified from the plasmid p3HA (gift of Carolyn Silflow, University of Minnesota, Saint Paul, MN), and a SNAP tag was amplified from a codon optimized SNAP plasmid (Song et al., 2015) using primers with NdeI sites (Supplemental Table S2). Both tags were inserted into the NdeI site of the 1.2 kb SpeI/EcoRI fragment and sequenced to verify orientation and reading frame. The tagged fragments were reinserted back into the original p59c2 plasmid to make pFAP57-HA and pFAP57-C-SNAP. To tag the N-terminus of FAP57, a 453-base-pair genomic fragment spanning the start codon was removed from the original 59c2 plasmid using two unique restriction sites, AfiII and AvrII. A 1032-base-pair version of this sequence was synthesized with the SNAP tag sequence inserted prior to the start codon (GeneWiz, South Plainfield, NJ) and then ligated back into the original p59c2 plasmid to make pSNAP-N-FAP57. All constructs were verified by sequencing in both directions and then linearized with SspI prior to cotransformation into ida8-1 or bop2-1. Rescue of motility defects by cotransformation with the epitope-tagged FAP57 constructs was similar to that seen with the WT gene (∼3–6% rescued). The predicted amino acid sequences of the epitope-tagged FAP57 polypeptides are shown in Supplemental Figure S2.
Phase contrast and fluorescence microscopy and measurements of swimming velocity and microtubule sliding
Motility phenotypes were assessed by phase contrast microscopy using a 20× or 40× objective on a Zeiss Axioskop microscope. Initial measurements of swimming velocities were made from video recordings using a C2400 Newvicon camera and Argus 10 video processor (Hammamatsu Photonic Systems, Bridgewater, NJ) calibrated with a stage micrometer (Myster et al., 1997, 1999; Perrone et al., 1998, 2000). More recent assays used a Rolera-MGi EM-CCD camera (Q-imaging, Surrey, BC, Canada) and Metamorph software (Molecular Devices, San Jose, CA) (VanderWaal et al., 2011; Bower et al., 2013, 2018; Reck et al., 2016). To compare flagellar waveforms, a pco.1200HS camera with Camware software (Cooke Corporation, Londonderry, NH) was used to capture high-speed images (500 fps) of motile cells and create videos at 30 fps (Dymek et al., 2019). Videos and montages were created in ImageJ (Schneider et al., 2012).
Measurements of microtubule sliding velocities in protease-treated axonemes were performed as previously described (Okagaki and Kamiya, 1986; Dymek et al., 2019). A Zeiss AxioSkop 2 microscope equipped for dark-field optics with a Plan-Apochromat 403 oil immersion objective lens with iris and an ultra-dark field oil immersion condenser was used for imaging. Images were captured and analyzed using an ORCA-Flash 4.0 V2 (Hamamatsu) camera and Nikon NIS Elements Advanced Research Software (Tokyo, Japan). Data are presented as the mean ± SEM. At least three independent experiments were performed for each strain. The Student’s t test was used for comparisons between different strains.
Cells were fixed for immunofluorescence microscopy using ice-cold methanol (Sanders and Salisbury, 1995), stained with a rat monoclonal antibody to HA (Roche clone 3F10) and an Alexafluor-488–conjugated secondary antibody (Molecular Probes, Eugene, OR), and processed as previously described (Bower et al., 2013, 2018; Reck et al., 2016). Images were collected on a Zeiss Axioscop using a 100×/1.3 NA Plan Neuofluor objective, a CoolSnap ES CCD camera (Photometrics, Tuscon, AZ), and Metamorph software. Selected images were cropped, rotated, and labeled in ImageJ and Adobe Photoshop (San Jose, CA).
Isolation and fractionation of axonemes, SDS–PAGE, and Western blot analyses
Chlamydomonas whole cell lysates, isolated flagella, and demembranated axonemes were prepared as previously described (Witman, 1986; Bower et al., 2013, 2018; Reck et al., 2016) using 0.1–1.0% Nonidet-P-40 to remove membrane and matrix proteins. Purified axonemes were resuspended in HMEEN (10 mM HEPES, pH 7.4, 5 mM MgSO4, 1 mM ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid [EGTA], 0.1 mM EDTA, 30 mM NaCl) plus 1 mM dithiothreitol (DTT) and 0.1 μg/ml protease inhibitors (leupeptin, aprotinin, pepstatin) and extracted with HMEEN containing 10 mM MgATP, 0.6 M NaCl, 0.2 M NaI, 0.4 M NaI, or 0.6 M NaI. The 0.6 M NaCl extracts (containing most of the axonemal dyneins and FAP57) were dialyzed against HMEEN, clarified by centrifugation, and fractionated on 5–20% sucrose density gradients (Bower et al., 2013, 2018) or diluted 10-fold and fractionated by Mono-Q ion-exchange FPLC chromatography (Gardner et al., 1994). Samples were separated on 5–15% polyacrylamide gradient gels and silver stained or transferred to Immobilon P and probed with different antibodies (Bower et al., 2013, 2018). Antibody sources and dilutions are listed in Supplemental Table S3.
Preparation of samples for iTRAQ labeling and tandem MS/MS
Isolated axonemes were washed in 10 mM HEPES, pH 7.4, to remove salt, DTT, and protease inhibitors, then resuspended in 0.5 M triethylammonium bicarbonate pH 8.5 and processed for trypsin digestion and iTRAQ labeling as described in detail (Bower et al., 2013, 2018; Reck et al., 2016). Duplicate aliquots of axonemes (50–60 μg each) from each strain were reacted with 4-plex iTRAQ reagents (114–117; AB Sciex, Foster City, CA) to obtain two technical replicates per biological sample. The four labeled aliquots were mixed together and processed to remove excess trypsin, unreacted iTRAQ reagents, and buffer. The combined sample (containing two control aliquots with different iTRAQ labels and two mutant aliquots with different iTRAQ labels) was fractionated offline using high pH, C18 reversed phase chromatography (Reck et al., 2016). The column fractions were then further processed and loaded in 1–1.5 µg aliquots for capillary LC using a C18 column at low pH. The C18 column was mounted in a nanospray source directly in line with a Velos Orbitrap mass spectrometer (Thermo Fisher Scientific, Waltham, MA). Online capillary LC, MS/MS, database searching, and protein identification were performed as previously described (Lin-Moshier et al., 2013; Reck et al., 2016) using ProteinPilot software version 5.0 (AB Sciex, Foster City, CA) and the most recent version of the Chlamydomonas database (https://phytozome.jgi.doe.gov/pz/portal.html). The bias factors for all samples were normalized to α and β tubulin (Reck et al. 2016). The relative amount of protein in each aliquot was compared with that present in the control aliquot to obtain a protein ratio. The WT/WT or HA/HA ratios indicated the variability in labeling and protein loading between technical replicates of the same sample (typically less than 10% for all proteins). All iTRAQ experiments were repeated with a second set of samples for independent biological replicates. A total of 1060 proteins were identified at a 1% false discovery rate in the first experiment, and 1098 proteins were identified in the second experiment. The protein lists were filtered using a minimum of six peptides per protein, yielding 603 proteins from the first set of samples and 554 proteins from the second.
Because inner arm DHCs vary widely in abundance, purified axonemes from WT, ida8-1, and the FAP57-HA rescued strain were also fractionated by SDS–PAGE, stained briefly with Coomassie blue, and the DHC region was excised from the gel to improve the signal to noise for the DHCs (Bower et al., 2013). Following extraction and trypsin digestion, three to five replicates per sample were analyzed by MS/MS, and both the total number of peptides and total number of assigned spectra per DHC isoform were determined. The relative abundance of each DHC was estimated by spectral counting (Zhu et al., 2010) and expressed as a percentage of the total spectra identified for the 1-α and 1-β DHCs of the I1 dynein (Bower et al., 2013, 2018; Wirschell et al., 2013). Bands containing other proteins identified as significantly altered in the iTRAQ experiments were also analyzed by spectral counting to confirm the results with additional biological replicates. In addition, a subset of FPLC fractions was analyzed by MS/MS to identify polypeptides that cofractionated with the FAP57 polypeptide. Samples were analyzed by SDS–PAGE, stained briefly with silver (Bower et al., 2013), and selected bands were excised from the gel and analyzed as described above (Supplemental Table S4).
Thin-section electron microscopy and image averaging
Isolated axonemes were fixed, resin-embedded, and sectioned for imaging by TEM as previously described (O’Toole et al., 1995, 2012). Cross-sectional and longitudinal views of axonemes were 2D averaged using the image processing software developed by the Boulder Laboratory for 3D microscopy and available at http://bio3d.colorado.edu/. To control for possible variations in staining, three to five biological replicates were processed for each sample. For averages of DMTs in cross-section, 183–659 DMTs were used to obtain a grand average for each strain. For averages of the 96 nm repeat in longitudinal section, individual averages containing multiple repeats were obtained for each axoneme, and 6–40 axonemes were analyzed for each strain to obtain a grand average (see Figure 2D).
Cryo-ET and image processing
The purified axoneme pellet was resuspended in HMEEK buffer (30 mM HEPES, pH 7.4, 5 mM MgSO4, 1 mM EGTA, 0.1 mM EDTA, 25 mM KCl), and the suspension was directly used for cryo-sample preparation. For precise localization of the amino and carboxyl terminal ends of FAP57, streptavidin-nanogold labeling was performed on axonemes from strains rescued with SNAP-tagged versions of the FAP57 as previously described (Song et al., 2015). Briefly, 1 µl of 1 mM BG-(PEG)12-biotin (New England Biolabs; PEG linker available on request) was added to 200 µl of axonemes. A control sample was also prepared without added BG-(PEG)12-biotin. Both suspensions were incubated overnight at 4°C, followed by three cycles of resuspension with HMEEK buffer and centrifugation at 10,000 × g for 10 min at 4°C. The axoneme pellets were resuspended in 200 µl of buffer, and then 5 µl of 1.4-nm-sized streptavidin-nanogold particles (strep-Au 1.4 nm, Nanoprobes) was added, and the two suspensions were incubated at 4°C in the dark for 3 h with rotation. The samples were then diluted with 1 ml of HMEEK buffer, pelleted by centrifugation at 10,000 × g for 10 min at 4°C, carefully resuspended in 200 µl of HMEEK buffer, and used for cryo-sample preparation.
Cryo-sample preparation, cryo-ET, and image processing were done as previously described (Nicastro et al., 2006, 2009; Heuser et al., 2009; Lin et al., 2014; Fu et al., 2018). Briefly, Quantifoil copper grids (Quantifoil Micro Tools, Jena, Germany) with a holey carbon film (R2/2, 200 mesh) were glow discharged for 30 s at –40 mA and loaded with 3 µl of axoneme sample and 1 µl of fivefold-concentrated and BSA-coated 10 nm colloidal gold (Sigma-Aldrich, St. Louis, MO) (Iancu et al., 2006). After brief mixing, grids were blotted from the back with Whatman No.1 filter paper for 1.5–3 s and plunge frozen in liquid ethane using a homemade plunger. Vitrified samples were cryo-transferred to a Tecnai F30 transmission electron microscope (Thermo Fisher Scientific, Waltham, MA) for imaging. Single-axis tilt series of noncompressed, intact axonemes were acquired using the software package SerialEM (Mastronarde, 2005). Typically, 50–70 images were recorded at 13,500-fold magnification (∼1 nm pixel size) with –6 to –8 µm defocus while the specimen was tilted from about –65 to +65° in 1.5–2.5° increments. The microscope was operated in low dose mode at 300 keV and the cumulative electron dose for each tilt series was restricted to ∼100 e-/Å2 to minimize radiation damage. Electron micrographs were recorded digitally with a 2k × 2k CCD camera (Gatan) after passing a postcolumn energy filter (Gatan) in zero-loss mode with a slit width of 20 eV.
For higher-resolution 3D structure of a pseudo WT axonemal repeat, vitrified axoneme samples from WT and CLiP mutants fap76-1, fap81, fap92, fap216, and a fap76-1; fap81 double mutant were imaged by cryo-ET using a Titan Krios transmission electron microscope (Thermo Fisher Scientific, Waltham, MA) equipped with a K2 direct electron detection camera (Gatan, Pleasanton, CA) operated in counting mode, and a Volta-Phase-Plate (Danev et al., 2014). Tilt series were recorded at the magnification of 26,000 (∼5.5 Å pixel size) and –0.5 µm defocus using SerialEM with a dose-symmetric tilting scheme (Hagen et al., 2017). At each tilt angle, a video stack of 15 frames was collected with a total exposure time of 6 s and an electron dose rate of 8 electrons/pixel/s. Motion correction of the frames was later performed on the video stacks with a script extracted from IMOD (Kremer et al., 1996), and the resulting images of individual tilts were assembled into the tilt series.
Three-dimensional tomograms were reconstructed from the recorded tilt series using fiducial alignment and weighted back projection in IMOD. Subtomograms containing the highly repetitive 96 nm repeat units were further aligned and averaged with particle estimation for electron tomography (PEET) (Nicastro et al., 2006), resulting in 3D structures with compensated missing wedge, reduced noise, and thus increased resolution. For doublet-specific averaging, the nine DMTs were identified based on DMT-specific features (Bui et al., 2012; Lin et al., 2012), and repeats from individual DMTs were averaged. To further analyze structural defects that appeared heterogeneous or to identify the sites labeled with streptavidin-nanogold, classification analyses were performed on the aligned subtomograms using the PEET program (Heumann et al., 2011). Appropriate masks were applied to focus the classification analyses on specific regions of interest. Subtomograms containing the same structures were grouped into class averages. The structures were mapped onto their respective locations in the raw tomograms to determine the distribution of the different classes within the axonemes. The numbers of tomograms, subtomograms analyzed and the resolutions of the resulting averages are summarized in Supplemental Table S5. The resolution was estimated at the center of the DMT of axonemal repeat using the Fourier shell correlation method with a criterion of 0.5. The structures were visualized as 2D tomographic slices and 3D isosurface renderings using IMOD and UCSF Chimera (Pettersen et al., 2004), respectively.
Supplementary Material
Acknowledgments
We thank LeeAnn Higgins, Todd Markowski, Bruce Witthun, and Alan Zimmerman from the Center for Mass Spectrometry and Proteomics at the University of Minnesota (UMN) for expert assistance with iTRAQ labeling, mass spectrometry, and spectral counting. This center is supported by multiple grants including the National Science Foundation major research instrumentation grants 9871237 and 0215759 as described at https://cbs.umn.edu/cmsp/about. We also thank Matt Laudon and the Chlamydomonas Resource Center ( https://www.chlamycollection.org/) for strains. This center is supported by the National Science Foundation Living Stock Collections for Biological Research program (National Science Foundation grants NSF-0951671 and NSF-00017383). The Porter laboratory also acknowledges the dedicated assistance of multiple UMN undergraduates including Aimee DeCathelineau, Jasjeet Sekhon, Jared Rieck, Shada Ahrar, and Alexandria Schauer and Clare Palmer from Wesleyan University. Expert assistance with TEM and image analysis was also provided by Amanda Bednarz, Thomas Giddings, and David Mastronarde at the Boulder Laboratory for 3D Fine Structure (University of Colorado). We also thank Chen Xu (Brandeis University) and Daniel Stoddard (UT Southwestern Medical Center) for dedicated training and maintenance of EM facilities. The UTSW cryo-electron microscope facility is funded in part by a CPRIT Core Facility Award (RP170644). Richard Linck (UMN), Toshiki Yagi (Prefectural University of Hiroshima), Win Sale (Emory University), Ritsu Kamiya (Tokyo University), Paul Lefebvre (UMN), and Pinfen Yang (Marquette University) generously supplied antibodies as listed in Supplemental Table S3. Preliminary reports of this work were presented at the American Society for Cell Biology meetings and the International Conference on the Cell and Molecular Biology of Chlamydomonas. This work was supported by National Institutes of Health grants to M.E.P. (GM-055667), D.N. (GM-088122), and E.F.S. (GM-112050).
Abbreviations used:
- CLiP
Chlamydomonas Library Project
- CP
central pair
- cryo-ET
cryo-electron tomography
- CSC
calmodulin- and spoke-associated complex
- DHC
dynein heavy chain
- DIC
differential interference contrast
- DMT
doublet microtubule
- FAP
flagellar-associated polypeptide
- FC1
flanking clone 1
- HA
hemagglutinin
- IC
intermediate chain
- IDA
inner dynein arm
- iTRAQ
isobaric tag for relative and absolute quantitation
- LC
light chain
- MS/MS
tandem mass spectrometry
- N-DRC
nexin-dynein regulatory complex
- ODA
outer dynein arm
- PCD
primary ciliary dyskinesia
- PEET
particle estimation for electron tomography
- pf
paralyzed flagella
- RFLP
restriction fragment length polymorphism
- RS
radial spoke
- RSP
radial spoke protein
- RS3S
radial spoke 3 short
- TAP
Tris-acetate-phosphate
- TEM
transmission electron microscopy
- T/TH
tether-tether head
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-07-0367) on September 4, 2019.
REFERENCES
- Albee AJ, Kwan AL, Lin H, Granas D, Stormo GD, Dutcher SK. (2013). Identification of cilia genes that affect cell-cycle progression using whole-genome transcriptome analysis in Chlamydomonas reinhardtii . G3: Genes, Genomes, Genetics , 979–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaiz O, Gout JF, Betermier M, Bouhouche K, Cohen J, Duret L, Kapusta A, Meyer E, Sperling L. (2010). Gene expression in a paleopolyploid: a transcriptome resource for the ciliate Paramecium tetraurelia . BMC Genomics , 547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn K, Bustamante-Marin X, Yin W, Goshe MB, Ostrowski LE. (2017). Quantitative proteomic analysis of human airway cilia identifies previously uncharacterized proteins of high abundance. J Proteome Res , 1579–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower R, Tritschler D, Mills KV, Heuser T, Nicastro D, Porter ME. (2018). DRC2/CCDC65 is a central hub for assembly of the nexin-dynein regulatory complex and other regulators of ciliary and flagellar motility. Mol Biol Cell , 137–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower R, Tritschler D, Vanderwaal K, Perrone CA, Mueller J, Fox L, Sale WS, Porter ME. (2013). The N-DRC forms a conserved biochemical complex that maintains outer doublet alignment and limits microtubule sliding in motile axonemes. Mol Biol Cell , 1134–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower R, VanderWaal K, O’Toole E, Fox L, Perrone C, Mueller J, Wirschell M, Kamiya R, Sale WS, Porter ME. (2009). IC138 defines a subdomain at the base of the I1 dynein that regulates microtubule sliding and flagellar motility. Mol Biol Cell , 3055–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadhead R, Dawe HR, Farr H, Griffiths S, Hart SR, Portman N, Shaw MK, Ginger ML, Gaskell SJ, McKean PG, Gull K. (2006). Flagellar motility is required for the viability of the bloodstream trypanosome. Nature , 224–227. [DOI] [PubMed] [Google Scholar]
- Brokaw CJ, Kamiya R. (1987). Bending patterns of Chlamydomonas flagella: IV. Mutants with defects in inner and outer dynein arms indicate differences in dynein arm function. Cell Motil Cytoskeleton , 68–75. [DOI] [PubMed] [Google Scholar]
- Bui KH, Yagi T, Yamamoto R, Kamiya R, Ishikawa T. (2012). Polarity and asymmetry in the arrangement of dynein and related structures in the Chlamydomonas axoneme. J Cell Biol , 913–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante-Marin X, Horani A, Stoyanova M, Charng W-L, Sears P, Bottier MM, Daniels LA, Bowen H, Conrad D, Knowles MR, et al (2019). Mutation of CFAP57 causes primary ciliary dyskinesia by disrupting the asymmetric targeting of a subset of ciliary inner dynein arms. bioRxiv, http://biorxiv.org/cgi/content/short/773028v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, Rosenbaum JL. (1998). Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol , 993–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danev R, Buijsse B, Khoshouei M, Plitzko JM, Baumeister W. (2014). Volta potential phase plate for in-focus phase contrast transmission electron microscopy. Proc Natl Acad Sci USA , 15635–15640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debuchy R, Purton S, Rochaix J-D. (1989). The arginosuccinate lyase gene of Chlamydomonas reinhardtii: an important tool for nuclear transformation and for correlating the genetic and molecular maps of the ARG7 locus. EMBO J , 2803–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher SK, Gibbons W, Inwood WB. (1988). A genetic analysis of suppressors of the PF10 mutation in Chlamydomonas reinhardtii . Genetics , 965–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymek EE, Lin J, Fu G, Porter M, Nicastro D, Smith EF. (2019). PACRG and FAP20 form the inner junction of axonemal doublet microtubules and regulate ciliary motility. Mol Biol Cell , 1805–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, et al (2014). Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics , 397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu G, Wang Q, Phan N, Urbanska P, Joachimiak E, Lin J, Wloga D, Nicastro D. (2018). The I1 dynein-associated tether and tether head complex is a conserved regulator of ciliary motility. Mol Biol Cell , 1048–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu G, Zhao L, Dymek E, Hou Y, Song K, Phan N, Shang Z, Smith EF, Witman GB, Nicastro D. (2019). Structural organization of the C1a-e-c supercomplex within the ciliary central apparatus. bioRxiv, https//.org/10.1101/773416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner LC, O’Toole E, Perrone CA, Giddings T, Porter ME. (1994). Components of a “dynein regulatory complex” are located at the junction between the radial spokes and the dynein arms in Chlamydomonas flagella. J Cell Biol , 1311–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross CH, Ranum LP, Lefebvre PA. (1988). Extensive restriction length polymorphisms in a new isolate of Chlamydomonas reinhardtii . Curr Genet , 503–508. [DOI] [PubMed] [Google Scholar]
- Hagen WJH, Wan W, Briggs JAG. (2017). Implementation of a cryo-electron tomography tilt-scheme optimized for high resolution subtomogram averaging. J Struct Biol , 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson TW, Perrone CA, Griffin P, Wuichet K, Mueller J, Yang P, Porter ME, Sale WS. (2004). IC138 is a WD-repeat dynein intermediate chain required for light chain assembly and regulation of flagellar bending. Mol Biol Cell , 5431–5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heumann JM, Hoenger A, Mastronarde DN. (2011). Clustering and variance maps for cryo-electron tomography using wedge-masked differences. J Struct Biol , 288–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser T, Barber CF, Lin J, Krell J, Rebesco M, Porter ME, Nicastro D. (2012). Cryoelectron tomography reveals doublet-specific structures and unique interactions in the I1 dynein. Proc Natl Acad Sci USA , E2067–E2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser T, Raytchev M, Krell J, Porter ME, Nicastro D. (2009). The dynein regulatory complex is the nexin link and a major regulatory node in cilia and flagella. J Cell Biol , 921–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iancu CV, Tivol WF, Schooler JB, Dias DP, Henderson GP, Murphy GE, Wright ER, Li Z, Yu Z, Briegel A, et al (2006). Electron cryotomography sample preparation using the Vitrobot. Nat Protoc , 2813–2819. [DOI] [PubMed] [Google Scholar]
- Karak S, Jacobs JS, Kittelmann M, Spalthoff C, Katana R, Sivan-Loukianova E, Schon MA, Kernan MJ, Eberl DF, Gopfert MC. (2015). Diverse roles of axonemal dyneins in Drosophila auditory neuron function and mechanical amplification in hearing. Sci Rep , 17085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathir P, LaVoie M, Brazelton WJ, Haas NA, Lefebvre PA, Silflow CD. (2003). Molecular map of the Chlamydomonas reinhardtii nuclear genome. Eukaryot Cell , 362–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato-Minoura T, Hirono M, Kamiya R. (1997). Chlamydomonas inner-arm dynein mutant, ida5, has a mutation in an actin-encoding gene. J Cell Biol , 649–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato-Minoura T, Uryu S, Hirono M, Kamiya R. (1998). Highly divergent actin expressed in a Chlamydomonas mutant lacking the conventional actin gene. Biochem Biophys Res Commun , 71–76. [DOI] [PubMed] [Google Scholar]
- King SM. (2018). Composition and assembly of axonemal dyneins. In Dyneins: The Biology of Dynein Motors, ed. King S., Cambridge, UK: Academic Press; 162–201. [Google Scholar]
- King SJ, Inwood WB, O’Toole ET, Power J, Dutcher SK. (1994). The bop2–1 mutation reveals radial asymmetry in the inner dynein arm region of Chlamydomonas reinhardtii . J Cell Biol , 1255–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. (1996). Computer visualization of three-dimensional image data using IMOD. J Struct Biol , 71–76. [DOI] [PubMed] [Google Scholar]
- Kubo T, Hou Y, Cochran DA, Witman GB, Oda T. (2018). A microtubule-dynein tethering complex regulates the axonemal inner dynein f (I1). Mol Biol Cell , 1060–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, Li H, Blacque OE, Li L, Leitch CC, et al (2004). Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell , 541–552. [DOI] [PubMed] [Google Scholar]
- Li X, Patena W, Fauser F, Jinkerson RE, Saroussi S, Meyer MT, Ivanova N, Robertson JM, Yue R, Zhang R, et al (2019). A genome-wide algal mutant library and functional screen identifies genes required for eukaryotic photosynthesis. Nat Genet , 627–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang R, Patena W, Gang SS, Blum SR, Ivanova N, Yue R, Robertson JM, Lefebvre PA, Fitz-Gibbon ST, et al (2016). An indexed, mapped mutant library enables reverse genetics studies of biological processes in Chlamydomonas reinhardtii. Plant Cell , 367–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Heuser T, Song K, Fu X, Nicastro D. (2012). One of the nine doublet microtubules of eukaryotic flagella exhibits unique and partially conserved structures. PLoS One , e46494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Nicastro D. (2018). Asymmetric distribution and spatial switching of dynein activity generates ciliary motility. Science , eaar1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Yin W, Smith MC, Song K, Leigh MW, Zariwala MA, Knowles MR, Ostrowski LE, Nicastro D. (2014). Cryo-electron tomography reveals ciliary defects underlying human RSPH1 primary ciliary dyskinesia. Nat Commun , 5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Moshier Y, Sebastian PJ, Higgins L, Sampson ND, Hewitt JE, Marchant JS. (2013). Re-evaluation of the role of calcium homeostasis endoplasmic reticulum protein (CHERP) in cellular calcium signaling. J Biol Chem , 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Takazaki H, Nakazawa Y, Sakato M, Yagi T, Yasunaga T, King SM, Kamiya R. (2008). Partially functional outer-arm dynein in a novel Chlamydomonas mutant expressing a truncated gamma heavy chain. Eukaryot Cell , 1136–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN. (2005). Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol , 36–51. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN, O’Toole ET, McDonald KL, McIntosh JR, Porter ME. (1992). Arrangement of inner dynein arms in wild-type and mutant flagella of Chlamydomonas . J Cell Biol , 1145–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock TS, Glasser CE, Bose SC, Bergman DA. (2008). Tissue expression patterns identify mouse cilia genes. Physiol Genomics , 198–206. [DOI] [PubMed] [Google Scholar]
- Mitchell DR, Sale WS. (1999). Characterization of a Chlamydomonas insertional mutant that disrupts flagellar central pair microtubule-associated structures. J Cell Biol , 293–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison HM, Valente EM. (2017). Motile and non-motile cilia in human pathology: from function to phenotypes. J Pathol , 294–309. [DOI] [PubMed] [Google Scholar]
- Mizuno N, Taschner M, Engel BD, Lorentzen E. (2012). Structural studies of ciliary components. J Mol Biol , 163–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myster SH, Knott JA, O’Toole E, Porter ME. (1997). The Chlamydomonas Dhc1 gene encodes a dynein heavy chain subunit required for assembly of the I1 inner arm complex. Mol Biol Cell , 607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myster SH, Knott JA, Wysocki KM, O’Toole E, Porter ME. (1999). Domains in the 1alpha dynein heavy chain required for inner arm assembly and flagellar motility in Chlamydomonas . J Cell Biol , 801–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevers Y, Prasad MK, Poidevin L, Chennen K, Allot A, Kress A, Ripp R, Thompson JD, Dollfus H, Poch O, Lecompte O. (2017). Insights into ciliary genes and evolution from multi-level phylogenetic profiling. Mol Biol Evol , 2016–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro D. (2009). Cryo-electron microscope tomography to study axonemal organization. Methods Cell Biol , 1–39. [DOI] [PubMed] [Google Scholar]
- Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, McIntosh JR. (2006). The molecular architecture of axonemes revealed by cryoelectron tomography. Science , 944–948. [DOI] [PubMed] [Google Scholar]
- Oda T, Yanagisawa H, Kamiya R, Kikkawa M. (2014). A molecular ruler determines the repeat length in eukaryotic cilia and flagella. Science , 857–860. [DOI] [PubMed] [Google Scholar]
- Okagaki T, Kamiya R. (1986). Microtubule sliding in mutant Chlamydomonas axonemes devoid of outer or inner dynein arms. J Cell Biol , 1895–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole ET, Giddings TH, Jr, Porter ME, Ostrowski LE. (2012). Computer-assisted image analysis of human cilia and Chlamydomonas flagella reveals both similarities and differences in axoneme structure. Cytoskeleton , 577–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole E, Mastronarde D, McIntosh JR, Porter ME. (1995). Computer-assisted image analysis of flagellar mutants. Methods Cell Biol , 183–191. [DOI] [PubMed] [Google Scholar]
- Pazour GJ, Agrin N, Leszyk J, Witman GB. (2005). Proteomic analysis of a eukaryotic cilium. J Cell Biol , 103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Dickert BL, Witman GB. (1999). The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J Cell Biol , 473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone CA, Myster SH, Bower R, O’Toole ET, Porter ME. (2000). Insights into the structural organization of the I1 inner arm dynein from a domain analysis of the 1beta dynein heavy chain. Mol Biol Cell , 2297–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone CA, Tritschler D, Taulman P, Bower R, Yoder BK, Porter ME. (2003). A novel dynein light intermediate chain colocalizes with the retrograde motor for intraflagellar transport at sites of axoneme assembly in Chlamydomonas and mammalian cells. Mol Biol Cell , 2041–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone CA, Yang P, O’Toole E, Sale WS, Porter ME. (1998). The Chlamydomonas IDA7 locus encodes a 140-kDa dynein intermediate chain required to assemble the I1 inner arm complex. Mol Biol Cell , 3351–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. (2004). UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem , 1605–1612. [DOI] [PubMed] [Google Scholar]
- Piperno G, Ramanis Z, Smith EF, Sale WS. (1990). Three distinct inner dynein arms in Chlamydomonas flagella: molecular composition and location in the axoneme. J Cell Biol , 379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ME, Knott JA, Myster SH, Farlow SJ. (1996). The dynein gene family in Chlamydomonas reinhardtii . Genetics , 569–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reck J, Schauer AM, VanderWaal Mills K, Bower R, Tritschler D, Perrone CA, Porter ME. (2016). The role of the dynein light intermediate chain in retrograde IFT and flagellar function in Chlamydomonas . Mol Biol Cell , 2404–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter JF, Leroux MR. (2017). Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol , 533–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorick NK, Kinoshita A, Weirather JL, Peyrard-Janvid M, de Lima RL, Dunnwald M, Shanske AL, Moretti-Ferreira D, Koillinen H, Kere J, et al (2011). Genomic strategy identifies a missense mutation in WD-repeat domain 65 (WDR65) in an individual with Van der Woude syndrome. Am J Med Genet A , 1314–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp G, O’Toole E, Gardner LC, Mitchell BF, Porter ME. (1996). The sup-pf-2 mutations of Chlamydomonas alter the activity of the outer dynein arms by modification of the gamma-dynein heavy chain. J Cell Biol , 1853–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp G, O’Toole E, Porter ME. (2001). The Chlamydomonas PF6 locus encodes a large alanine/proline-rich polypeptide that is required for assembly of a central pair projection and regulates flagellar motility. Mol Biol Cell , 739–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp G, Porter ME. (2003). A subunit of the dynein regulatory complex in Chlamydomonas is a homologue of a growth arrest-specific gene product. J Cell Biol , 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders MA, Salisbury JL. (1995). Immunofluorescence microscopy of cilia and flagella. Methods Cell Biol , 163–169. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods , 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell RA, Lefebvre PA. (1993). Isolation of the Chlamydomonas regulatory gene NIT2 by transposon tagging. Genetics , 737–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthilan PR, Piepenbrock D, Ovezmyradov G, Nadrowski B, Bechstedt S, Pauls S, Winkler M, Mobius W, Howard J, Gopfert MC. (2012). Drosophila auditory organ genes and genetic hearing defects. Cell , 1042–1054. [DOI] [PubMed] [Google Scholar]
- Sigg MA, Menchen T, Lee C, Johnson J, Jungnickel MK, Choksi SP, Garcia G, 3rd, Busengdal H, Dougherty GW, Pennekamp P, et al (2017). Evolutionary proteomics uncovers ancient associations of cilia with signaling pathways. Dev Cell , 744–762.e711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizova I, Fuhrmann M, Hegemann P. (2001). A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii . Gene , 221–229. [DOI] [PubMed] [Google Scholar]
- Smith EF, Yang P. (2004). The radial spokes and central apparatus: mechano-chemical transducers that regulate flagellar motility. Cell Motil Cytoskeleton , 8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K, Awata J, Tritschler D, Bower R, Witman GB, Porter ME, Nicastro D. (2015). In situ localization of N and C termini of subunits of the flagellar nexin-dynein regulatory complex (N-DRC) using SNAP tag and cryo-electron tomography. J Biol Chem , 5341–5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K, Shang Z, Fu X, Lou X, Grigorieff N, Nicastro D. (2018). Structure of the ciliary axoneme at nanometer resolution reconstructed by TYGRESS. bioRxiv. 10.1101/363317. [DOI] [Google Scholar]
- Stoddard D, Zhao Y, Bayless BA, Gui L, Louka P, Dave D, Suryawanshi S, Tomasi RF, Dupuis-Williams P, Baroud CN, et al (2018). Tetrahymena RIB72A and RIB72B are microtubule inner proteins in the ciliary doublet microtubules. Mol Biol Cell , 2566–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surkont J, Diekmann Y, Ryder PV, Pereira-Leal JB. (2015). Coiled-coil length: size does matter. Proteins , 2162–2169. [DOI] [PubMed] [Google Scholar]
- Tam LW, Lefebvre PA. (1993). Cloning of flagellar genes in Chlamydomonas reinhardtii by DNA insertional mutagenesis. Genetics , 375–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truebestein L, Leonard TA. (2016). Coiled-coils: the long and short of it. Bioessays , 903–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanska P, Joachimiak E, Bazan R, Fu G, Poprzeczko M, Fabczak H, Nicastro D, Wloga D. (2018). Ciliary proteins Fap43 and Fap44 interact with each other and are essential for proper cilia and flagella beating. Cell Mol Life Sci , 4479–4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam TJP, Kennedy J, van der Lee R, de Vrieze E, Wunderlich KA, Rix S, Dougherty GW, Lambacher NJ, Li C, Jensen VL, et al (2019). CiliaCarta: an integrated and validated compendium of ciliary genes. PLoS One , e0216705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanderWaal KE, Yamamoto R, Wakabayashi K, Fox L, Kamiya R, Dutcher SK, Bayly PV, Sale WS, Porter ME. (2011). bop5 Mutations reveal new roles for the IC138 phosphoprotein in the regulation of flagellar motility and asymmetric waveforms. Mol Biol Cell , 2862–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibranovski MD, Koerich LB, Carvalho AB. (2008). Two new Y-linked genes in Drosophila melanogaster . Genetics , 2325–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasbrough ER, Dorus S, Hester S, Howard-Murkin J, Lilley K, Wilkin E, Polpitiya A, Petritis K, Karr TL. (2010). The Drosophila melanogaster sperm proteome-II (DmSP-II). J Proteomics , 2171–2185. [DOI] [PubMed] [Google Scholar]
- Werner C, Onnebrink JG, Omran H. (2015). Diagnosis and management of primary ciliary dyskinesia. Cilia , 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirschell M, Olbrich H, Werner C, Tritschler D, Bower R, Sale WS, Loges NT, Pennekamp P, Lindberg S, Stenram U, et al (2013). The nexin-dynein regulatory complex subunit DRC1 is essential for motile cilia function in algae and humans. Nat Genet , 262–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman GB. (1986). Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol , 280–290. [DOI] [PubMed] [Google Scholar]
- Yagi T, Uematsu K, Liu Z, Kamiya R. (2009). Identification of dyneins that localize exclusively to the proximal portion of Chlamydomonas flagella. J Cell Sci , 1306–1314. [DOI] [PubMed] [Google Scholar]
- Yamamoto R, Song K, Yanagisawa HA, Fox L, Yagi T, Wirschell M, Hirono M, Kamiya R, Nicastro D, Sale WS. (2013). The MIA complex is a conserved and novel dynein regulator essential for normal ciliary motility. J Cell Biol , 263–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Smith JW, Huang CM. (2010). Mass spectrometry-based label-free quantitative proteomics. J Biomed Biotechnol , 840518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zur Lage P, Newton FG, Jarman AP. (2019). Survey of the ciliary motility machinery of Drosophila sperm and ciliated mechanosensory neurons reveals unexpected cell-type specific variations: a model for motile ciliopathies. Front Genet , 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.