Abstract
Mammalian cells express two oligosaccharyltransferase complexes, STT3A and STT3B, that have distinct roles in N-linked glycosylation. The STT3A complex interacts directly with the protein translocation channel to mediate glycosylation of proteins using an N-terminal–to–C-terminal scanning mechanism. N-linked glycosylation of proteins in budding yeast has been assumed to be a cotranslational reaction. We have compared glycosylation of several glycoproteins in yeast and mammalian cells. Prosaposin, a cysteine-rich protein that contains STT3A-dependent glycosylation sites, is poorly glycosylated in yeast cells and STT3A-deficient human cells. In contrast, a protein with extreme C-terminal glycosylation sites was efficiently glycosylated in yeast by a posttranslocational mechanism. Posttranslocational glycosylation was also observed for carboxypeptidase Y–derived reporter proteins that contain closely spaced acceptor sites. A comparison of two recent protein structures indicates that the yeast OST is unable to interact with the yeast heptameric Sec complex via an evolutionarily conserved interface due to occupation of the OST binding site by the Sec63 protein. The efficiency of glycosylation in yeast is not enhanced for proteins that are translocated by the Sec61 or Ssh1 translocation channels instead of the Sec complex. We conclude that N-linked glycosylation and protein translocation are not directly coupled in yeast cells.
INTRODUCTION
Asparagine-linked glycosylation is a prominent protein modification reaction for proteins in eukaryotic cells. The enzyme oligosaccharyltransferase (OST) transfers a preassembled high-mannose oligosaccharide onto acceptor sites (NXT/S/C where X ≠ P) in nascent polypeptides that enter the lumen of the endoplasmic reticulum. The yeast OST is a heterooctamer composed of an active-site subunit (STT3) plus seven accessory subunits (Kelleher and Gilmore, 2006). Full activity of the yeast OST requires an oxidoreductase subunit (either Ost3p or Ost6p), which has been proposed to delay protein folding of segments in the vicinity of glycosylation acceptor sites (Schulz and Aebi, 2009; Schulz et al., 2009; Poljak et al., 2018). Most metazoan organisms express two OST complexes, which are composed of an active-site subunit (STT3A or STT3B), six shared subunits, and complex-specific accessory subunits (Cherepanova et al., 2014; Shrimal et al., 2017). Phylogenetic analysis of metazoan and fungal STT3 proteins indicates that fungal STT3 proteins are more closely related to the STT3B clade than to the STT3A clade of metazoan STT3 proteins (Shrimal et al., 2013b). Either MagT1 or TUSC3, which are orthologues of yeast Ost3p/Ost6p, is incorporated into the mammalian STT3B complex (Cherepanova et al., 2014). Thus, from an evolutionary and composition standpoint, the yeast OST is equivalent to the mammalian STT3B complex.
N-linked glycosylation is typically viewed as a cotranslational reaction in mammalian cells, as there is abundant evidence that N-linked glycans are added in a synchronized manner to the elongating nascent chain (Rothman and Lodish, 1977; Chen et al., 1995) when the acceptor site is roughly 65–70 residues from the peptidyltransferase site on the ribosome (Whitley et al., 1996; Nilsson et al., 2003; Deprez et al., 2005). Insight into the coupling between protein translocation and N-linked glycosylation was provided by the discovery that the STT3A complex includes accessory subunits (DC2 and KCP2) that mediate a direct interaction between the STT3A complex and the protein translocation channel (Shrimal et al., 2017; Braunger et al., 2018). DC2(–/–) HEK293 cells assemble a catalytically active STT3A complex that lacks DC2 and KCP2. The DC2/KCP2 deficient STT3A complex does not interact with the Sec61 complex and is unable to glycosylate STT3A-dependent acceptor sites (Shrimal et al., 2017). The mammalian STT3B complex, which does not interact with the protein translocation channel in the presence or absence of the STT3A complex (Shrimal et al., 2017; Braunger et al., 2018), can mediate cotranslational or posttranslocational glycosylation of a subset of acceptor sites that have been skipped by STT3A (Ruiz-Canada et al., 2009; Shrimal et al., 2013b).
High-resolution structures of the yeast OST obtained by cryo–electron microscopy (Bai et al., 2018; Wild et al., 2018) indicate that Ost3p is located in the same position that is occupied by DC2 in the mammalian STT3A complex (Braunger et al., 2018). This shared location led to the hypothesis that Ost3p or Ost6p positions the yeast OST adjacent to the protein translocation channel (Bai et al., 2018). However, yeast has three protein translocation channels with different subunit compositions and different roles in protein translocation. The heterotrimeric Sec61 and Ssh1 complexes are involved in cotranslational translocation of proteins that are targeted to the ER membrane by the SRP–SRP receptor-targeting pathway (Finke et al., 1996; Ng et al., 1996; Jiang et al., 2008). The heptameric Sec complex, which is composed of the heterotetrameric Sec62/Sec63 complex plus the Sec61 complex, is involved in posttranslational translocation of proteins with less hydrophobic signal sequences (Panzner et al., 1995; Ng et al., 1996; Plath et al., 1998).
The split-ubiquitin system has been used to investigate potential interactions between yeast OST subunits and the Sec61p, Sbh1p, and Sss1p subunits of the Sec61 heterotrimer (Scheper et al., 2003; Chavan et al., 2005). However, the high-resolution structure of the yeast OST (Bai et al., 2018; Wild et al., 2018) is incompatible with the conclusions of these earlier studies (Chavan et al., 2005) based on the location of the tagging sites relative to the membrane surface. For example, the tagging sites on Swp1p and Wbp1p are more than 60 Å away from the surface of the yeast OST (Ost3p) that has been proposed to interact with the Sec61 complex (Bai et al., 2018). One possible explanation for the multiple contacts between the Sec61 subunits and OST subunits that were observed with the split ubiquitin assay is that the interactions occurred during membrane integration of the OST subunits by the Sec61 complex. It was also proposed that OST complexes containing Ost3p interact with the Sec61 complex via Sbh1p, while OST complexes containing Ost6p interact with the Ssh1 complex via Sbh2p (Yan and Lennarz, 2005). However, the ubiquitin tags appended to Ost3p, Ost6p, Sbh1p, and Sbh2p were actually attached to lumenal C-terminal segments; hence, the split ubiquitin interactions that were sensed in the cytosol were generated from unassembled or misfolded protein subunits. Using correctly designed split ubiquitin constructs (Scheper et al., 2003), the Römisch lab detected an interaction between the N-terminal amphipathic helix of Sss1p and the C-terminal cytoplasmic segment of Wbp1p. Examination of the yeast OST structure (Wild et al., 2018) reveals that the C-terminus of Wbp1p is located more than 60 Å from the closest cytoplasmically exposed segment of Ost3p; thus the putative Wbp1p-Sss1p interaction would localize the OST on the opposite side of the Sec61 heterotrimer, with the acceptor peptide–binding pocket in Stt3p facing away from the lateral gate of the Sec61 complex. For these reasons, we believe that the previous studies, which reported interactions between the yeast OST and protein translocation channels, are largely inconsistent with the subsequently determined structures of protein translocation channels and the oligosaccharyltransferase. Isolation of complexes that contain the yeast OST, a translocation channel, and a ribosome-nascent chain complex for cryo–electron microscopy analysis has also proven to be elusive despite attempts to assemble these complexes from purified components (Harada et al., 2009).
The structural homology between the yeast OST and the STT3B complex raises the possibility that N-linked glycosylation in yeast occurs by a mechanism that does not involve direct interactions between the OST and the three yeast protein–translocation channels. To address this possibility, we first compared glycosylation of several proteins in HEK293 cells and yeast to determine whether the yeast OST can glycosylate previously established STT3A-dependent or STT3B-dependent acceptor sites. Second, we used pulse–chase-labeling procedures to determine whether posttranslocational N-glycosylation can be detected in yeast cells. Third, we solubilized yeast cell extracts to determine whether the yeast OST complex is associated with ribosome bound–protein translocation channels. All three lines of evidence led to the conclusion that protein translocation and N-glycosylation of polypeptides are uncoupled reactions in the yeast endoplasmic reticulum.
RESULTS
Sec63p and DC2 bind to the same surface of Sec61p and Sec61α
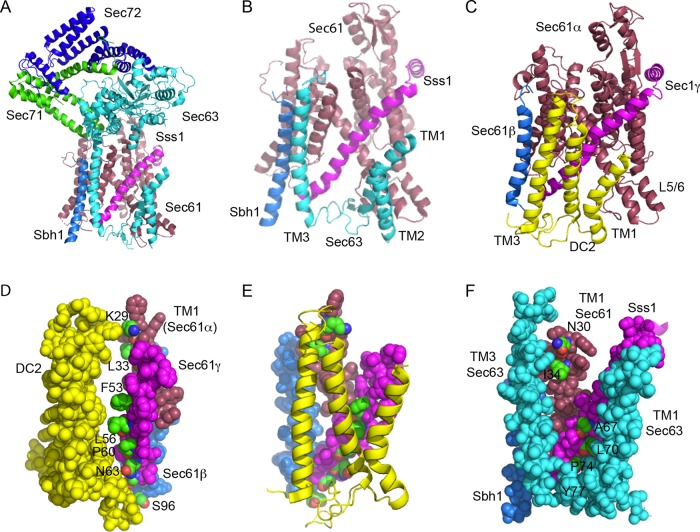
The recently solved structures of the yeast heptameric Sec complex (Itskanov and Park, 2019; Wu et al., 2019) have well-resolved density for the Sec61 heterotrimer, Sec63p, Sec71p, and Sec72p (Figure 1A). The three transmembrane (TM) spans of Sec63 (cyan helices) pack against the TM1 and TM5 of Sec61p and against the single TM spans of the β (Sbh1p) and γ (Sss1p) subunits of the Sec61 heterotrimer (Figure 1B). Importantly, this is precisely the same surface as that of the mammalian Sec61 heterotrimer that interacts with the DC2 protein to promote interaction between the Sec61 heterotrimer and the STT3A complex (Figure 1C). Similarly to Sec63p, TM3 of DC2 contacts TM1 of Sec61α and the C-terminal membrane–embedded regions of Sec61β and Sec61γ, while TM1 of DC2 contacts TM5 of Sec61α. The orientation of Sec63 (3TM Nlum-Ccyt) is the opposite of DC2 (3TM Ncyt-Clum), and TM2 of Sec63p and DC2 occupy nonoverlapping positions.
FIGURE 1:
Comparison of the Sec63p and DC2 binding sites on the yeast and mammalian Sec61 complexes. (A) Structure of the yeast Sec complex. Sec61p is raspberry, Sbh1p is marine, Sss1 is magenta, Sec63p is cyan, Sec71p is green, and Sec72p is blue. (B) Enlarged view of the interactions between the three TMs of Sec63p and the Sec61 heterotrimer, using the same color code as in panel A. The cytoplasmic domains of Sec63p, Sec71p, and Sec72p have been removed for clarity. The cytosolic (L1/2) and lumenal (L2/3) loops of Sec63p were not completely resolved. (C) Interaction between DC2 (yellow) and the mammalian Sec61 heterotrimer (color codes as in A). The other OST subunits are omitted for clarity, as the STT3A protein binds to the opposite face of DC2 from the Sec61 complex. (D) Space-filling model showing the interaction surface of DC2 and the TM spans of Sec61β, Sec61γ, and TM1 of Sec61α. Proteins are color-coded as in C. Residues in the Sec61 complex that compose the interaction surface (K29 and L33 in Sec61α, F53, L56, P60, and N63 in Sec61γ, and S96 in Sec61β) are color-coded by element and are labeled. (E) DC2-Sec61 complex interaction surface color-coded as in D. (F) Space-filling model of the interface between Sec63p (cyan; TM1, TM3, and the resolved section of L2), Sss1p (magenta), Sbh1p (slate), and TM1 of Sec1p (raspberry). The residues in the yeast Sec61 complex subunits that align with residues shown in panels D and E (Supplemental Table S1) are color-coded by element and are labeled. P74 and Y77 in Sss1p are completely buried by Sec63p, while other interaction residues are partially buried. The figure was made with PYMOL v2.1 software and PDB files 6N3Q (A, B, F) and 6FTI (C–E).
For a more detailed analysis of the interaction surfaces, we identified residues in Sec61α, Sec61β, and Sec61γ that define the binding surface for DC2 (Figure 1, D and E; Supplemental Table S1). On the basis of sequence alignments of yeast and mammalian Sec61 complex subunits, we asked whether the corresponding residues in Sec61p, Sbh1p, and Sss1p were surface- exposed in the Sec complex, or were instead providing the binding site for Sec63p. As shown in Figure 1F, five of the six aligned yeast residues are either partially or fully buried by Sec63 in the Sec complex. TM2 of Sec63, which is not shown in Figure 1F for clarity, would sterically block Ost3p from contacting the remaining residue of Sec61γ (A67) that does not directly contact Sec63p. Thus, the potential Ost3p interaction surface in the Sec complex is occluded by Sec63p.
The yeast secretome lacks cysteine-rich glycoproteins
Cotranslational glycosylation of human proteins by the translocation channel–associated STT3A complex is important for glycosylation of sequons that are located in cysteine-rich protein domains (Cherepanova et al., 2019). To determine whether there are cysteine-rich yeast glycoproteins, we compiled a list of yeast N-glycosylation sites by merging four databases from glycoproteomics studies that utilized different glycopeptide enrichment or detection procedures (Zielinska et al., 2012; Chen et al., 2014a,b). The glycoproteomics studies utilized N-glycanase–dependent deamidation of lectin-enriched glycopeptides (Zielinska et al., 2012), chemical (TMSF)- or enzymatic (Endo H)-dependent generation of GlcNAc-modified peptides (Chen et al., 2014b), or N-glycanase–dependent deamidation of boronic acid–enriched glycopeptides (Chen et al., 2014a). Owing to the variety of methods used to enrich and detect N-glycopeptides, a combined data set should have excellent coverage of the yeast N-glycoproteome. From each data set, we excluded peptides that lack a canonical sequon (N-X-T/S/C; X ≠ P), as such sites, if modified in human cells, show very low occupancy (Valliere-Douglass et al., 2009). We also excluded peptides that are derived from proteins that do not contain a signal sequence or a transmembrane span, using a yeast secretome database as a resource (Supplemental Table S5 from Jan et al., 2014). In ambiguous cases where an ER targeting signal may have been overlooked, protein sequences were also analyzed using the SignalP 4.1 server (Petersen et al., 2011) to detect signal sequences and the ΔG prediction server for TM span identification (Hessa et al., 2007). Peptides derived from carboxy-terminal tail-anchored (TA) membrane proteins were excluded, as the putative glycopeptides are located in the cytosol. After merger of the four data sets, we obtained a list of 830 experimentally detected acceptor sites, roughly 60% of which were present in at least two data sets (Figure 2A [overlap]; Supplemental Table S2A). It should be noted that the merged data set is not a complete list of verified yeast glycosylation sites, as we have not attempted to augment Supplemental Table S2A by including acceptor sites that have been identified by nonglycoproteomics methods. For example, only two of the four known glycosylation sites in carboxypeptidase Y (CPY, PRC1 gene) were detected by LC-MS/MS (Supplemental Table S2A). The 830 N-linked glycosylation sites are derived from 251 yeast proteins (Supplemental Table S2B). Roughly 17% of the yeast secretome proteins included in Supplemental Table S2B are based on a single glycopeptide that was present in only one of the four data sets. Although the glycoprotein list may well include false positives, we did not want to overlook any cysteine-rich yeast glycoproteins. The 251 proteins have an average cysteine content of 1.3%, with no protein having a cysteine content that exceeds 3.5% (Figure 2B). Labeled arrows indicate the cysteine content of several yeast glycoproteins that have been used to evaluate N-glycosylation in previous studies (Reddy et al., 1988; Silberstein et al., 1995b; Poljak et al., 2018).
FIGURE 2:
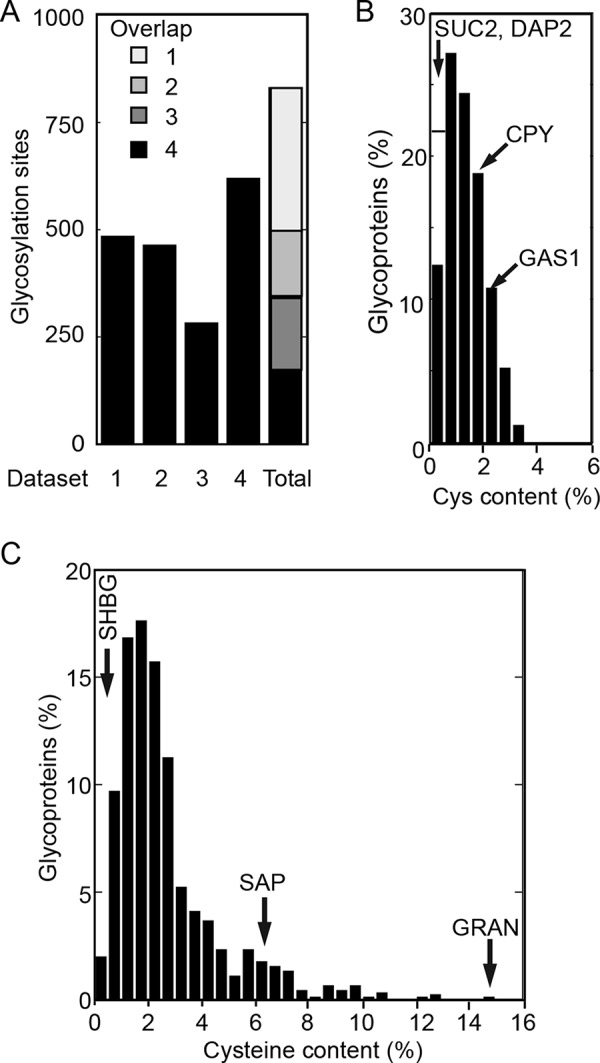
The yeast secretome does not contain cysteine-rich glycoproteins. (A) Yeast glycosylation sites were identified in the following mass spectrometry studies: data set 1, Class 1 sites, Supplemental Table S2 from Zielinska et al. (2012); data set 2, Supplemental Table S1 from Chen et al. (2014b); data set 3, Supplemental Table S2 from Chen et al. (2014b); data set 4, Supplemental Table S3 from Chen et al. (2014a). Data sets were trimmed by excluding sites that did not match a consensus glycosylation site (NXT/S/C) or were derived from nonsecretome proteins. The data sets were merged to obtain 830 glycosylation sites that were present in one to four of the initial data sets, as indicated in the overlap column. The 830 glycosylation sites were derived from 251 yeast secretome proteins. (B) The distribution of cysteine content in 251 yeast glycoproteins. The cysteine content of several glycoproteins is indicated by labeled arrows. (C) The distribution of cysteine content in a collection of 892 human glycoproteins (Cherepanova et al., 2019). The cysteine content of an STT3B-dependent (SHBG) and two STT3A-dependent (SAP and GRAN) substrates is indicated by labeled arrows.
The cysteine content of a collection of 892 HEK293 cell glycoproteins with experimentally verified glycosylation sites (Cherepanova et al., 2019) was also determined (Figure 2C; Supplemental Table S2C). The average cysteine content of the human proteins was twofold higher (2.7%), with 21.5% of the proteins having a cysteine content that was higher than those of all yeast glycoproteins. Prosaposin (SAP) and granulin (GRAN) are cysteine-rich proteins that are STT3A-dependent substrates (Ruiz-Canada et al., 2009; Shrimal et al., 2013a). We conclude that yeast lacks cysteine-rich glycoproteins.
Hypoglycosylation of prosaposin in yeast
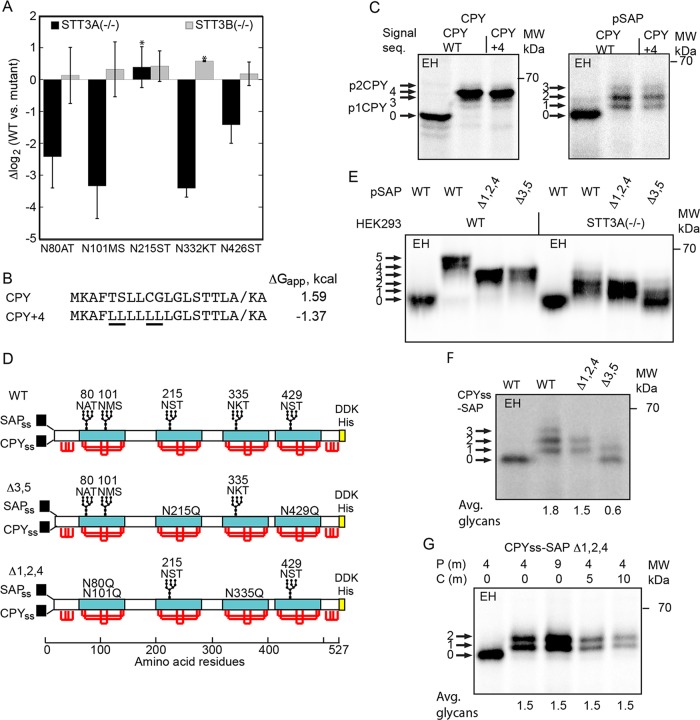
Prosaposin is a cysteine-rich mammalian glycoprotein that contains five acceptor sites that are located in 80-residue cysteine-rich protein domains. Synthesis of prosaposin in STT3A(–/–) cells yields prosaposin glycoforms having on average two glycans (Cherepanova and Gilmore, 2016; Shrimal et al., 2017). A SILAC-based quantitative glycoproteomics analysis of STT3A(–/–) and STT3B(–/–) cells yielded glycosylation-site occupancy data for roughly 1000 N-X-T/S/C, sites including all five prosaposin sites (Cherepanova et al., 2019). Of the five sites in prosaposin, glycosylation of three acceptor sites was reduced four- to eightfold, and one site was reduced roughly 2.5-fold, while glycosylation of the N215ST site was not reduced in the STT3A(–/–) cell line (Figure 3A).
FIGURE 3:
Glycosylation of human prosaposin in yeast and human cells. (A) SILAC-based glycoproteomic analysis of prosaposin glycosylation in HEK293-derived cells that lack either the STT3A or STT3B complex. Site occupancy is expressed as Δlog2, where a negative value indicates reduced glycosylation in the mutant cells relative to the wild-type cells. Error bars designate standard deviations (n = 3–7) or individual data points (*, n = 2). (B) Signal sequences for wild-type CPY and a more hydrophobic derivative (CPY+4). The underlined leucine residues in CPY+4 replace marginally hydrophobic amino acids. The diagonal line designates the signal sequence cleavage site. (C) CPY and pSAP constructs that have the CPY signal sequence or the CPY+4 signal sequence were pulse-labeled for 7 min with Tran-35S label in wild-type yeast cells. (D) Diagrams of the SAP, SAPΔ3,5, and SAPΔ1,2,4 constructs for expression of prosaposin derivatives in yeast and human cells. The signal sequences for human prosaposin (SAPss) and yeast CPY (CPYss) are for expression in HEK293 cells and yeast, respectively. Saposin domains are designated by cyan rectangles. Red lines designate disulfides. The disulfide bonding pattern of the N-terminal and C-terminal flanking domains is not known. A C-terminal DDK-His tag was appended for immunoprecipitation with anti-DDK sera. (E) Pulse labeling (10 min) of pSAP, pSAPΔ1,2,4, and pSAPΔ3,5 in wild-type and STT3A-deficient HEK293 cells. (F) Pulse labeling (7 min) of SAP, SAPΔ3,5, and SAPΔ1,2,4 in yeast. (G) Pulse–chase labeling of CPYss-SAP (Δ1,2,4) in yeast using the indicated pulse (P) and chase (C) intervals. (F, G) Quantified values are of the experiment shown, which is representative of two or more replicates. In C and E–G, labeled arrows indicate the number of glycans. EH indicates digestion with endoglycosidase H.
Yeast proteins that are cotranslationally translocated by the Sec61 or Ssh1 heterotrimers have more hydrophobic signal sequences than carboxypeptidase Y (CPY), which is translocated via the heptameric Sec complex (Ng et al., 1996). Replacement of four weakly hydrophobic residues in the CPY signal sequence with leucine residues yielded the CPY+4 signal sequence (Figure 3B). The calculated ΔGapp values (Hessa et al., 2007) indicate that the CPY+4 signal sequence has a hydrophobicity comparable to that of the TM spans of the proteins (DPAPB and Pho8), which are cotranslationally integrated in yeast cells (Ng et al., 1996). Previously we showed that the CPY+4 protein is cotranslationally translocated by the Sec61 complex or the Ssh1 complex (Trueman et al., 2012).
Translocation of CPY into the ER lumen is accompanied by cleavage of the N-terminal signal sequence and by the addition of four N-linked glycans to yield the ER (p1) form of CPY. Mannosylation of the N-linked glycans in the Golgi yields the p2 form of CPY, which migrates slightly more slowly than p1CPY when resolved by SDS–PAGE. Proteolytic removal of the propeptide in the vacuole yields mature CPY. When yeast cells are pulse-labeled for 7 min, p1-CPY is the major form of CPY that is resolved by SDS–PAGE (Cheng et al., 2005; Jiang et al., 2008; Trueman et al., 2012), though p2CPY was detected as a faint band (Figure 3C). Endoglycosidase H (EH) digestion of p1CPY removes the N-linked glycans, demonstrating that the predominant form of p1CPY has four N-linked glycans, as expected (Figure 3C). The signal sequence and the protein translocation channel had no impact upon N-glycosylation of CPY (Figure 3C).
To investigate glycosylation of prosaposin in yeast, we replaced the prosaposin signal sequence with the CPY or CPY+4 signal sequence and expressed the CPYss-SAP and CPY+4-SAP constructs under control of the glyceraldehyde 3-phosphate (GPD) promoter in a wild-type yeast strain. A pulse-labeling experiment revealed prosaposin glycoforms containing one to three glycans, with the two-glycan form being most abundant (Figure 3C). Glycosylation of prosaposin in yeast was not affected by the targeting pathway or the protein translocation channel. Thus, occupation of the potential Ost3p binding site in the yeast Sec complex by Sec63p does not reduce glycosylation of CPY or prosaposin when these proteins are translocated via the Sec complex.
On the basis of the mass spectrometry results (Figure 3A), we designed two prosaposin reporters, one of which includes the three sites that are more dependent on STT3A (pSAP Δ3,5), while a second reporter (pSAP Δ1,2,4) contained the two sites that are less dependent on STT3A (Figure 3D). Our objective was to determine whether the same prosaposin sites that are hypoglycosylated in STT3A-deficient human cells are the pSAP sites that are poorly glycosylated in yeast cells. The pSAP Δ1,2,4 and pSAP Δ3,5 reporters were characterized by expressing them in wild-type and STT3A(–/–) HEK293 cells (Figure 3E). Glycosylation of pSAP and of pSAP Δ1,2,4 were quite similar in the STT3A(–/–) cell line, consistent with the mass spectrometry results. The pSAP Δ3,5 reporter is glycosylated in wild-type cells, but is very poorly glycosylated in STT3A(–/–) cells. When CPYss-SAP, CPYss-SAP Δ1,2,4, and CPYss-SAP Δ3,5 were expressed in yeast, we detected prosaposin glycoforms that were remarkably similar to what we observed for the mammalian pSAP reporters in the STT3A-deficient cells (compare Figure 3, E and F). The yeast OST is unable to glycosylate the three prosaposin sites that need to be cotranslationally glycosylated in human cells by the translocation channel–associated STT3A complex (Figure 2F, CPYss-SAP Δ3,5).
Owing to the oxidizing environment of the ER lumen, cysteine residues in translocated proteins begin to form native and nonnative disulfide bonds upon entry into the ER lumen. Oxidation of the cysteine residues in prosaposin is particularly rapid in mammalian cells. In less than 5 min, prosaposin becomes resistant to PEG-maleimide modification following pulse labeling (Ruiz-Canada et al., 2009); hence the four saposin domains quickly acquire a disulfide-stabilized conformation. A pulse–chase-labeling experiment was conducted to determine whether the acceptor sites in the CPYss-pSAP Δ1,2,4 reporter could be fully glycosylated upon extended incubation (Figure 3G). We did not detect any change in glycosylation during the chase interval, indicating that the incompletely modified site (presumably N426ST) is intractable to glycosylation by the end of the 4-min pulse-labeling period in yeast cells. Given the conservation of disulfide bond–formation pathways in eukaryotic cells (Sevier and Kaiser, 2006; Bulleid and van Lith, 2014) and the critical role of the protein sequence and secondary structural elements in the kinetics of protein folding, oxidation of prosaposin in the yeast ER should be rapid. Transport of disulfide-bonded proteins such as CPY from the ER to the Golgi is blocked when the ER lumen is reduced by DTT treatment (Jämsä et al., 1994); hence the rapid intracellular transport of CPY in untreated cells is diagnostic of rapid disulfide bond oxidation in the yeast ER. We propose that formation of disulfides in the yeast ER minimizes posttranslocational glycosylation of prosaposin by the yeast OST.
Posttranslocational glycosylation of sex hormone–binding in yeast
Sex hormone–binding globulin (SHBG) has two extreme C-terminal sites (Figure 4A) that are posttranslocationally glycosylated by the STT3B complex in human cells (Shrimal et al., 2013b; Cherepanova and Gilmore, 2016). Here, we tested whether the yeast OST can glycosylate these STT3B-dependent acceptor sites. Incomplete glycosylation of the N380RS site is responsible for the glycoform doublet (Figure 4B) that is detected in wild-type or STT3A(–/–) cells (Shrimal et al., 2013b). A similar glycoform doublet was detected when CPYss-SHBG was expressed in yeast cells (Figure 4B), indicating that the yeast OST can glycosylate a human protein that has STT3B-dependent sites. In human cells, glycosylation of SHBG occurs by a posttranslocational mechanism, as shown by pulse–chase-labeling experiments (Shrimal et al., 2013b). Pulse–chase-labeling of SHBG in yeast cells revealed clear evidence for increased glycosylation of SHBG during the chase incubation (Figure 4C). The yeast OST, unlike the human STT3A complex (Figure 4B), can glycosylate acceptor sites after the completed protein has entered the ER lumen.
FIGURE 4:
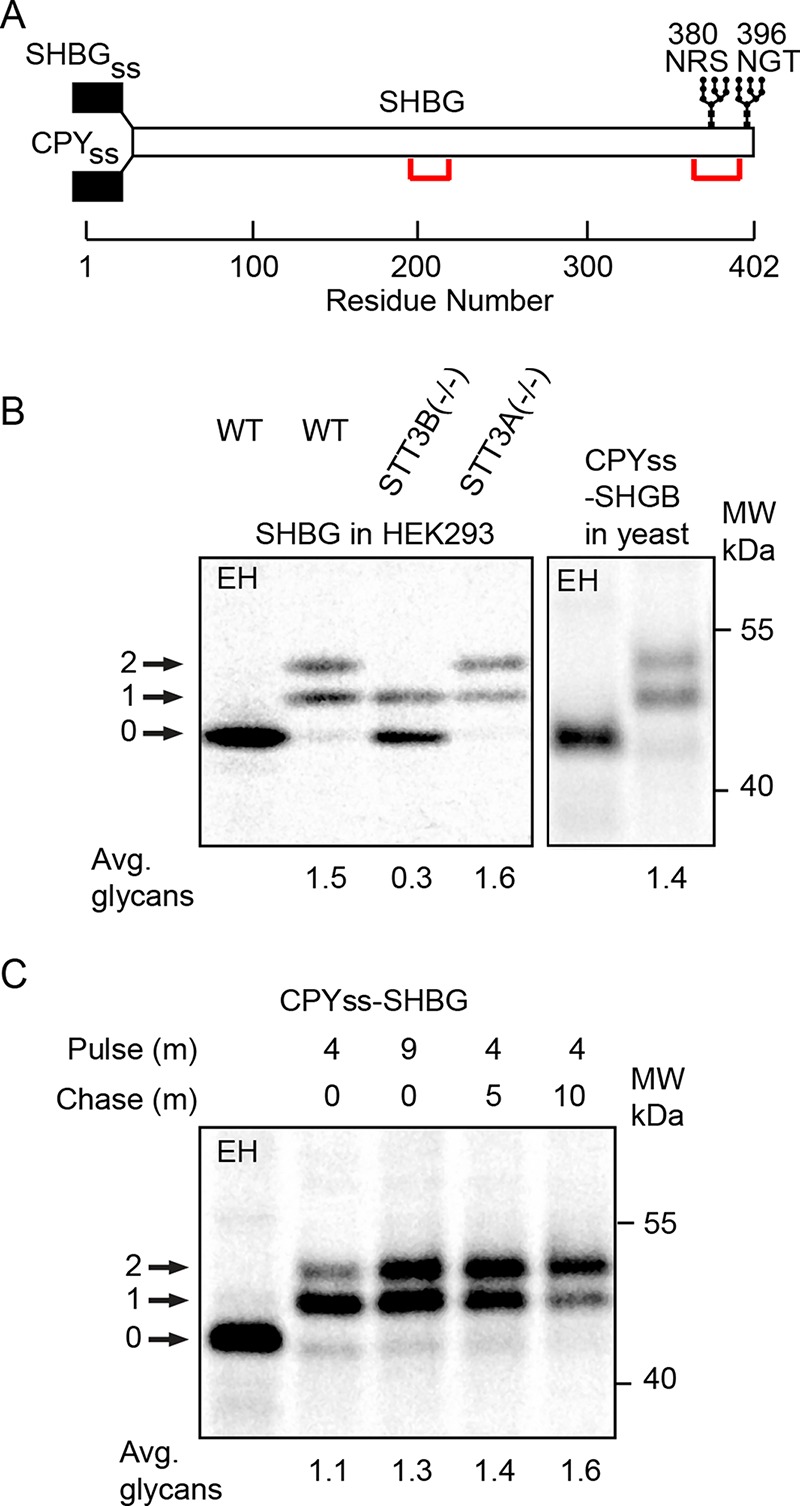
Pulse labeling of SHBG in human and yeast cells. (A) Diagram of SHBG showing signal sequences for expression of SHBG in human and yeast cells, the location of disulfide bonds, and the C-terminal glycosylation sites. (B) Pulse–chase labeling of SHBG in wild-type and mutant HEK293 cells (left panel, 5 min pulse, 20 min chase). Pulse labeling of CPYss-SHBG for 8 min in yeast cells (right panel). (C) Pulse–chase labeling of CPYss-SHBG in yeast using the indicated pulse (P) and chase (C) intervals. In B and C, quantified values are of the experiment shown, which is representative of two replicates. EH indicates digestion with endoglycosidase H.
Glycosylation of closely spaced acceptor sites
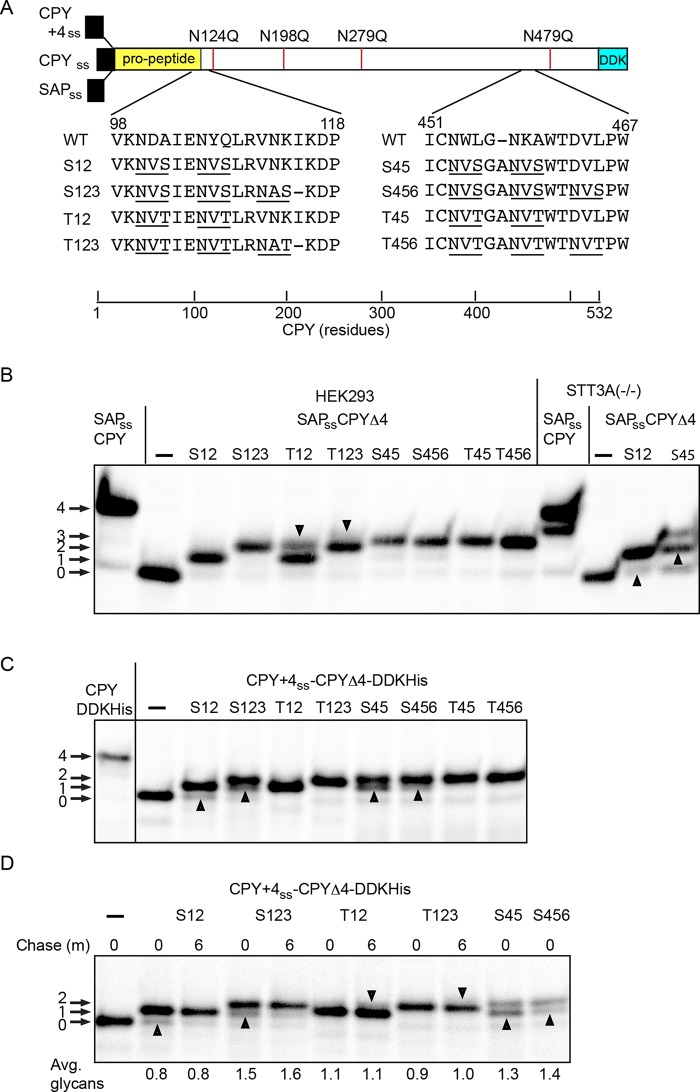
N-terminal to C-terminal scanning of the nascent polypeptide for acceptor sites by the translocation channel–associated STT3A complex appears to be important for N-glycosylation of closely spaced acceptor sites in human cells (Shrimal and Gilmore, 2013). The STT3A complex can glycosylate certain NXT sites that are adjacent (e.g., NHTNAT), while NHS sites were incompletely modified unless separated by two (e.g., NHSAANAS) or more residues (Shrimal and Gilmore, 2013). To compare glycosylation of closely spaced sites in yeast and human cells, we introduced glycosylation sites into a CPY derivative that lacks the four sites present in wild-type CPY (Winther et al., 1991). The introduced glycosylation sites were positioned near the propeptide–mature CPY boundary, or roughly 80 residues from the C-terminus, as sets of either two or three NXT or NXS sites that are separated by two spacer residues (Figure 5A). The CPY+4 signal sequence was used for expression in yeast, while the prosaposin signal sequence was used for expression in HEK293 cells. A wild-type CPY construct was also included to provide a marker for CPY with four glycans. When it was expressed in HEK293 cells, we detected a single glycan on the S12 and two glycans on the S123, S45, T45, S456, and T456 reporters. The T12 and T123 reporters yielded doublets, consistent with partial modification of two and three sites, respectively (Figure 5B, downward-pointing arrowheads). Suboptimal flanking residues are preventing complete modification of one acceptor site in several constructs. In the case of the S456 and T456 acceptor sites, bioinformatic analysis indicates that a proline residue at the +3 position of a sequon is unfavorable but not forbidden (Gavel and Von Heijne, 1990). In vitro assays using proline containing synthetic peptides have shown that a proline residue at the +3 position reduces peptide glycosylation (Bause, 1984).
FIGURE 5:
Glycosylation of CPY derivatives in yeast and human cells. (A) Diagram of CPY derivatives with closely spaced acceptor sites. The wild-type signal sequence of CPY was replaced with either the prosaposin signal sequence (SAPss) for expression in human cells or the CPY+4 sequence to direct cotranslational translocation in yeast. The four glycosylation sites in CPY were eliminated by the indicated N to Q mutations to create the SAPss-CPYΔ4 and CPY+4ss-CPYΔ4 constructs. Two or three closely spaced NXS or NXT sites were created at the indicated sites to yield eight reporters. The introduced glycosylation sites are underlined. (B) Pulse labeling of the SAPss-CPY and SAPss-CPYΔ4 derivatives in wild-type and STT3A(–/–) HEK293 cells. Downward-pointing arrowheads indicate the fully glycosylated form of the T12 and T123 reporters. Upward-pointing arrowheads indicate hypoglycosylated forms of S12 and S45 reporter that were detected in STT3A(–/–) cells. (C) Pulse labeling (4 min) of CPY+4ss-CPY and CPY+4ss-CPYΔ4 derivatives expressed in yeast. Upward-pointing arrowheads designate hypoglycosylated forms of the reporters that are either less abundant or not detected in wild-type HEK293 cells. Note the absence of the fully glycosylated T12 and T123 reporters. (D) Pulse–chase labeling of selected CPY+4ss-CPYΔ4 derivatives in yeast. The pulse period was 2 min for all samples. Upward-pointing arrowheads designate hypoglycosylated forms of NXS containing reporters that are more prominent before the chase period. The downward-pointing arrowheads designate trace amounts of the fully glycosylated T12 and T123 reporters that were visible after the chase. The quantified values below the gel lanes are for the experiment shown, which is representative of two similar experiments.
Expression of the S12 and S45 constructs in STT3A-deficient cells yielded faster-migrating products that lacked an additional glycan (Figure 5B, upward-pointing arrowheads), highlighting the importance of a cotranslational mechanism for glycosylation of closely spaced sites. All of the reporters with NXS sites migrated as doublets when expressed in yeast, due to the incomplete modification of one of the sites (Figure 5C, upward-pointing arrowheads). Notably, the glycoform doublets for the S12 and S45 reporters resembled the products synthesized in STT3A-deficient HEK293 cells. Fully glycosylated forms of the T12 and T123 reporters were absent when yeast cells were pulse-labeled for 4 min. Thus, all reporters except T45 and T456 were hypoglycosylated in yeast cells relative to HEK293 cells. The yeast OST has a reduced ability to glycosylate closely spaced acceptor sites compared with that of the STT3A complex.
A pulse–chase-labeling experiment using a shorter pulse period (2 min) was conducted to determine whether the acceptor sites in the CPY derivatives were being glycosylated by a strictly cotranslational mechanism (Figure 5D). Although glycosylation of the CPY derivatives was rapid, we detected modest increases in glycan occupancy after the chase incubation. The glycoform doublets for the reporters with NXS sites were more pronounced after a 2-min pulse period (Figure 5D, upward-pointing arrowheads) and diminished during the subsequent 6-min chase period. Traces of the fully glycosylated T12 and T123 reporters were also visible after the chase incubation (Figure 4D, downward-pointing arrowheads). Slow glycosylation of these internal acceptor sites (e.g., S123) provides evidence that glycosylation of acceptor sites is not occurring by an N– to C–terminal scanning mechanism as the nascent polypeptide passes through the protein translocation channel.
The yeast OST is not a ribosome-associated membrane protein
In an effort to detect OST complexes that interact with the Sec61 or Ssh1 heterotrimers, we used a ribosome-associated membrane protein (RAMP) isolation procedure (Figure 6A) that had been developed to characterize the interaction between mammalian ER proteins and the Sec61 complex (Görlich et al., 1992; Wang and Dobberstein, 1999; Shibatani et al., 2005). Treatment of mammalian microsomes with the nonionic detergent digitonin in the presence of a physiological–ionic strength buffer dissolves the membrane, releasing complexes between translating ribosomes and the Sec61 protein translocation channel. The STT3A complex, but not the STT3B complex, is a prominent component in a RAMP preparation (Shibatani et al., 2005; Shrimal et al., 2017).
FIGURE 6:
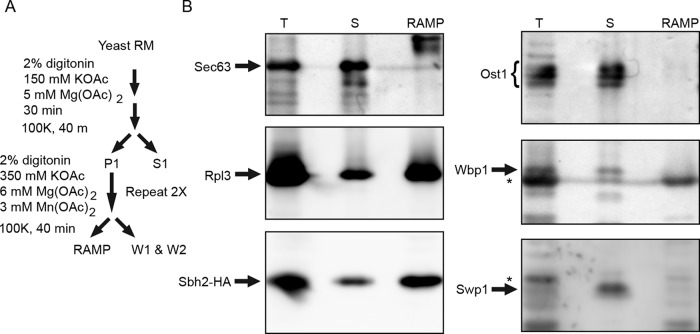
The yeast OST does not cosediment with ribosome translocation channel complexes. (A) Microsomes isolated from RGY1455, which expresses His6-FLAG-Sbh2p, were used to prepare a RAMP fraction as outlined in the diagram. (B) The total membrane extract (T), the combined supernatant fraction (S = S1+W1+W2), and the pellet fraction (RAMP) were analyzed by protein immunoblot analysis using the indicated antisera. Sample loads for the three fractions were derived from equal amounts of yeast microsomes. Asterisks designate nonspecific immunoreactive products on the Wbp1 and Swp1p blots.
Yeast microsomal membranes were subjected to the RAMP isolation procedure (Figure 6A). Immunoblotting was used to detect proteins in the total detergent extract (T), the RAMP fraction (RAMP), and a supernatant fraction (S), which was obtained by combining the S1, W1, and W2 fractions. Ribosomes, as detected using antisera specific for a large ribosomal subunit protein (Rpl3p), and Ssh1 heterotrimers, as indicated by the Sbh2-HA immunoreactivity, were primarily recovered in the RAMP fraction (Figure 6B). We used the Sbh2 subunit as the positive control for the RAMP isolation, as the Ssh1 complex is a cotranslational translocation channel (Jiang et al., 2008) that binds ribosome–nascent chain complexes (Becker et al., 2009). Sec61p and Sbh1p would report on the Sec complex in addition to the RNC-associated Sec61 heterotrimer. Cosedimentation of Sbh2p (or Shb1p) with the ribosome is strictly dependent on evolutionarily conserved basic residues in cytoplasmic loop 8/9 of Ssh1p or Sec61p (Cheng et al., 2005; Becker et al., 2009; Mandon et al., 2018). The yeast Sec complex does not bind ribosomes (Prinz et al., 2000) because cytosolic loop L6/7 in Sec61p is covered by the cytosolic domain of Sec63p (Figure 1A). As expected, Sec63p was recovered in the supernatant fraction. The protein immunoblots were probed with antibodies to three of the yeast OST subunits (Ost1p, Wbp1p, and Swp1p). Ost1p migrates as a triplet containing two to four glycans due to incomplete N-glycosylation of the four acceptor sites in Ost1p (Kelleher and Gilmore, 1994; Silberstein et al., 1995b). Ost1p was recovered exclusively in the supernatant fraction (Figure 6B). Wbp1p and Sw1p were also recovered in the supernatant fractions, indicating that the yeast OST does not interact with Sec61 or Ssh1 heterotrimers with sufficient avidity to cosediment with ribosome nascent chain complexes.
Two previous publications provide additional evidence that the yeast OST does not form stable complexes with protein translocation channels. Blue native PAGE (BN-PAGE) of yeast microsomes has been used to resolve the yeast OST into two complexes that contain either Ost3p or Ost6p as a subunit (Spirig et al., 2005). These yeast OST complexes have a mobility on BN-PAGE gels that is very similar to that of the mammalian STT3B complex (∼500 kDa) (Spirig et al., 2005; Roboti and High, 2012; Shrimal et al., 2017). The mammalian STT3A complex (∼470 kDa on BN-PAGE gels) is assembled into larger complexes that contain the Sec61 heterotrimer (∼700 kDa by BN-PAGE; Shibatani et al., 2005; Conti et al., 2015; Shrimal et al., 2017). The yeast OST is not present in larger complexes with a lower mobility on BN-PAGE gels (Spirig et al., 2005). Native immunoprecipitation of the yeast OST from uniformly radiolabeled yeast microsomes using an epitope-tagged subunit (OST3-HA or STT3-HA) as the precipitating antigen provided evidence for 1:1 stoichiometry of the eight OST subunits, but did not disclose additional unidentified proteins that could be subunits of the protein translocation channel (Karaoglu et al., 1997).
DISCUSSION
Previous investigators have assumed that N-linked glycosylation is directly coupled to protein translocation in budding yeast, based on the view that a cotranslational scanning mechanism for protein glycosylation would be conserved between metazoa and fungi. The yeast proteins that have been used to monitor protein translocation reactions in yeast are N-linked glycoproteins including CPY, Gas1p, dipeptidylaminopeptidase B (DPAPB), and invertase (Suc2p; Ng et al., 1996; Trueman et al., 2011). Typically, a 5- to 7-min pulse-labeling interval is used to obtain sufficient radiolabeled protein for detection of the nonglycosylated cytoplasmic precursors and fully glycosylated translocated products. Hypoglycosylated glycoprotein variants are not typically detected in wild-type yeast, but are prominent products in strains bearing mutations in OST subunits (te Heesen et al., 1993; Silberstein et al., 1995b). Thus, standard labeling procedures utilizing endogenous yeast glycoproteins as substrates had not revealed evidence for posttranslocational glycosylation. Here, we have been able to detect posttranslocational glycosylation in part due to the choice of substrates like SHBG, and in part due to utilizing shorter pulse-labeling intervals.
Hypoglycosylation of STT3A substrates by the yeast OST
In mammalian glycoproteins, the sequons that cannot be efficiently modified by the STT3B complex, such as those in prosaposin, are classified as being STT3A-dependent. Strikingly, glycosylation of prosaposin by the yeast OST was remarkably similar to glycosylation of prosaposin in STT3A-deficient human cells. We speculate that the difference in STT3A sensitivity of the acceptor sites in prosaposin is related to the folding kinetics of the individual saposin domains and the affinity of STT3B for the acceptor sites. Prosaposin contains a single NXS site (N101MS), so the observed differences in STT3A dependence are not explained simply by the preference of the OST for NXT sites relative to NXS sites.
Efficient glycosylation of closely spaced acceptor sites in human glycoproteins is facilitated by cotranslational scanning of the nascent protein by the translocation channel–associated STT3A complex (Shrimal and Gilmore, 2013). The CPY derivatives containing closely spaced acceptor sites were more prone to hypoglycosylation when expressed in yeast or in STT3A(–/–) cells, particularly when the reporters contained tandem NXS sites. The pulse–chase-labeling experiment conducted in yeast provided evidence for posttranslocational glycosylation of the closely spaced acceptor sites even when the sequons were located in the N-terminal half of the protein.
Posttranslocational glycosylation of extreme C-terminal glycosylation sites
Previously we had reported that murine glycoproteins contain a lower density of sequons and experimentally verified acceptor sites in the extreme C-terminal 70 residues of the protein (Shrimal et al., 2013b). In contrast, yeast glycoproteins have a more uniform distribution of sequons and experimentally verified N-glycosylated acceptor sites (Shrimal et al., 2013b). Here we observed that the yeast OST glycosylates SHBG, a previously established STT3B substrate, with an efficiency similar to that displayed by the human STT3B complex. Glycosylation of SHBG was incomplete after a brief pulse labeling period, but increased during the subsequent chase incubation, consistent with posttranslocational glycosylation. The rather slow but effective posttranslocational glycosylation of SHBG by the yeast OST contrasts markedly with the ineffective glycosylation of pSAP Δ3,5 and the rapidly achieved plateau value for glycosylation of pSAP Δ1,2,4. Thus, posttranslocational glycosylation of proteins by the yeast OST appears to be limited by the folding status of the polypeptide in the vicinity of the glycosylation site. The active-site disulfide in the oxidoreductase subunit (Ost3p or Ost6p) of the yeast OST is in the oxidized state in the cell (Schulz et al., 2009), so formation of a mixed disulfide with a free thiol in a substrate is possible. As Ost3p and Ost6p lack a second active-site disulfide, they lack protein disulfide isomerase activity (Schulz et al., 2009) and are not able to reduce preformed disulfides in cysteine-rich protein domains such as those in prosaposin.
N-glycosylation efficiency in yeast is not influenced by translocation channel identity
The identity of the translocation channel (Sec complex vs. Sec61 or Ssh1 heterotrimers) does not influence CPY or prosaposin glycosylation in yeast, indicating that a cotranslational scanning mechanism for N-glycosylation is not restricted to the heterotrimeric-protein translocation channels that retain the potential OST interaction site. We were unable to detect any interaction between the yeast OST and ribosome-bound translocation channels that persists after detergent solubilization of yeast microsomes. The distribution of incompletely modified sites in prosaposin and the closely spaced CPY reporters rules out the possibility that the OST is recruited to a translocation channel after the nascent polypeptide enters the ER lumen.
The yeast oligosaccharyltransferase lacks determinants that promote interactions with translocation channels
DC2 and to a lesser extent KCP2 are the accessory subunits that promote interaction between the STT3A complex and the mammalian Sec61 complex (Shrimal et al., 2017). Eukaryotic organisms (e.g., Saccharomyces cerevisiae) that lack a gene encoding STT3A also lack the genes encoding DC2 and KCP2. OST3 and DC2 occupy identical positions in the yeast OST and the mammalian STT3A complex due to a shared binding surface on STT3 and STT3A, respectively. Moreover, DC2 and TM2-4 of Ost3p have a similar folded structure and are evolutionarily related, based on the presence of short blocks of sequence identity (Braunger et al., 2018; Wild et al., 2018). A second interaction between the STT3A complex and the RNC-Sec61 complex is mediated by the C-terminal four-helix bundle of ribophorin I (RPN1), which contacts eL28, rRNA H25, and rRNA H19/20 on the large ribosomal subunit (Braunger et al., 2018). Ost1p, the yeast orthologue of RPN1, lacks the cytoplasmic extension that forms this four-helix bundle (Silberstein et al., 1995b). Consequently, the yeast OST lacks both of the binding surfaces that promote the interaction between the STT3A complex and the Sec61-RNC complex in mammalian cells.
Several papers relying on split-ubiquitin reporter constructs have reported interactions between subunits of the yeast Sec61 complex and the OST (Scheper et al., 2003; Chavan et al., 2005). While the putative interactions are structurally incompatible with the hypothesis that Ost3p mediates the interaction of the OST with the Sec61 heterotrimer (Bai et al., 2018), an alternative interaction surface remains a formal possibility. Cryo–electron tomographic analysis of yeast microsomes will be needed to resolve this issue, as such analysis has been very useful for detecting Sec61-associated protein complexes in the mammalian ER (Pfeffer et al., 2014, 2017; Braunger et al., 2018).
The oxidoreductase subunits (Ost3p/Ost6p) have an N-terminal signal sequence, a lumenal thioredoxin domain, and four TM spans, so they adopt a 4TM Nlum-Clum topology, while DC2, which lacks a thioredoxin domain, has a 3TM Ncyt-Clum topology. As TM1 of Ost3p and the lumenal thioredoxin domain were not well resolved in the cryo–electron microscopy structures of the yeast OST (Bai et al., 2018; Wild et al., 2018), it is likely that the thioredoxin domain can adopt several conformations with respect to STT3. We have proposed that TM1 and the thioredoxin domain of Ost3p (or Ost6p) act as negative determinants to prevent the interaction between the Sec61 complex and the yeast OST (Shrimal and Gilmore, 2019). The STT3B complex, which also contains an oxidoreductase subunit (MagT1 or TUSC3), does not interact with the Sec61-RNC complex in STT3A-deficient human cells (Shrimal et al., 2017; Braunger et al., 2018).
STT3 duplication and glycoproteome expansion
The 830 glycosylation sites listed in Supplemental Table S2A can be viewed as a lower limit for the total number of yeast glycosylation sites. Nonetheless, it is clear from previous glycoproteomics analysis of model organisms that fungal glycoproteomes (S. cerevisiae and Schizosaccharomyces pombe) are considerably smaller than metazoan glycoproteomes (Zielinska et al., 2010, 2012). Below, we present a rationale for why the duplication of the OST active site subunit (STT3) to give rise to the STT3A and STT3B complexes is one factor that allowed glycoproteome expansion.
Prosaposin, which was the first identified STT3A-dependent substrate (Ruiz-Canada et al., 2009), is an example of a cysteine-rich glycoprotein that contains relatively small cysteine-rich domains. Quantitative glycoproteomic analysis of STT3A-deficient HEK293 cells (Cherepanova et al., 2019) indicated that the STT3A-dependent sites in cysteine-rich proteins are located within cysteine-rich protein domains, while sequons located in cysteine-deficient domains show little STT3A dependence. Rapid formation of both native and nonnative disulfides within a cysteine-rich domain will yield protein conformations that are incompatible with insertion of an acceptor site into the peptide-binding pocket of an OST active site, as shown by the x-ray crystal and cryo–electron microscopy structures of eubacterial, archaebacterial and eukaryotic OSTs (Lizak et al., 2011; Matsumoto et al., 2017; Wild et al., 2018). Cotranslational scanning of the nascent polypeptide by the mammalian STT3A complex provides a mechanism for N-glycosylation of acceptor sites before disulfides form between nearby cysteine residues.
Metazoan organisms, with the exception of Caenorhabditis species, express both STT3A and STT3B. The Caenorhabditis elegans genome does not encode STT3A, DC2, or KCP2. The absence of the STT3A complex in C. elegans gives rise to an interesting question concerning the expansion of the glycoproteome. Glycoproteomic analysis of C. elegans identified a large number of N-glycosylation sites (Zielinska et al., 2012), including those located within cysteine-rich proteins. Would targeted glycoproteomic analysis indicate that the C. elegans cysteine-rich glycoproteins are hypoglycosylated, or are the acceptor sites mainly located in cysteine-deficient domains?
Quantitative glycoproteomic analysis of STT3A(–/–) cells revealed that the STT3A complex is important for glycosylation of sequons with suboptimal flanking sequences (Cherepanova et al., 2019). We considered residues between the -2 position and the +3 position relative to the glycosylated asparagine, as there is evidence that flanking sequence residues impact the glycosylation efficiency of NXS sites. The OST has a higher affinity for NXT peptides than for NXS peptides (Bause and Legler, 1981). Glycosylation of NXS sequons is reduced when the +1 position (X residue) has a polar or bulky side chain (Shakin-Eshleman et al., 1996; Malaby and Kobertz, 2014). To a first approximation, suboptimal sequons can be viewed as NXS sites that have bulky or polar X residues, or NXT/NXS sites that have a proline at the +3 position.
Organisms that express the ER lectins involved in the glycoprotein quality control pathway display strong positive selection for NXT sequons in secretome proteins (Cui et al., 2009). Although weaker positive selection is apparent for NXS sequons than for NXT sequons in mammalian glycoproteins, S. cerevisiae and C. elegans both display negative selection for NXS sites in their glycoproteins. Positive selection for NXS sites in mammalian glycoproteins may reflect an enhanced ability of the translocation channel-associated STT3A complex to glycosylate suboptimal NXS sites. Thus, suboptimal sequons and cysteine-rich glycoproteins represent two examples of how the presence of distinct STT3A and STT3B complexes may have aided expansion of the metazoan glycoproteome by enhancing the modification efficiency of challenging glycosylation sites. In contrast, glycosylation sites in yeast proteins have likely been optimized for efficient glycosylation by an OST complex that does not interact directly with protein translocation channels.
MATERIALS AND METHODS
Yeast and mammalian expression vectors
The yeast expression plasmid pEM497 (pRS316 ppCPY-T7 URA3) encoding ppCPY with an appended T7 epitope tag has been described (Trueman et al., 2011). The plasmid pEM519 (pRS316 ppCPY+4-T7 URA3) encoding the ppCPY+4-T7 derivative that has four mutations in the signal sequence (T5L S6L C9L G10L) has been described (Trueman et al., 2012). Mammalian expression vectors encoding pSAP-DDKHis (Shrimal and Gilmore, 2015) and SHBG (Shrimal et al., 2013b) have been described. The plasmid pJW373 encoding unglycosylated CPY (ug-CPY; N124Q, N198Q, N279Q, N479Q) was a generous gift from Jakob R. Winther (University of Copenhagen) (Winther et al., 1991).
Recombinant PCR using the pSAP-DDK-His vector and either pEM497 or pEM519 was used to replace the proCPY coding sequence (amino acids [a.a.] 19–532) in pEM497 or pEM519 with the prosaposin coding sequence (a.a. 22–527) and the C-terminal DDKHis tag to obtain pSS101 (pRS316 CPYss-pSAP-DDKHis URA3) and pSS102 (pRS316 CPY+4-pSAP-DDKHis URA3). Oligonucleotides encoding asparagine-to-glutamine substitutions were used as primers together with pSS102 in recombinant PCRs to produce pSS103 (pRS316 CPYss-SAPΔ1,2,4 DDKHis URA3) and pSS104 (pRS316 CPYss-SAPΔ3,5 DDKHis URA3). Recombinant PCR using the mammalian SHBG expression vector and the pEM497 was used to replace the coding sequence for pro-CPY (a.a. 19–532) with the coding sequence for the mature region of SHBG (a.a. 30–402) to obtain plasmid pSS105 (PRS316 CPYss-SHBG URA3). Expression of prosaposin or SHBG in yeast using pSS101-pSS105 was not optimal for very short pulse-labeling experiments. To increase expression, the protein coding sequences and 3′UTR regions were subcloned into a plasmid (pRS416 GDPp URA3) containing a glyceraldehyde 3-phosphate dehydrogenase (GPD) promoter (GPDp) to obtain plasmids pSS106-110 (e.g., pSS106 (pRS416 GPDp-CPYss-pSAP-DDKHis URA3).
Recombinant PCR using pJW373 encoding unglycosylated CPY and the pSS107 vector (pRS416 GPDp-CPY+4-pSAP-DDKHis URA3) was used to replace the wild-type CPY coding sequence with the ug-CPY coding sequence to obtain pSS111. Recombinant PCR using the mammalian pSAP-DDKHis expression vector and pJW373 was used to replace the prosaposin coding sequence (a.a. 22–527) with the ug-proCPY coding sequence. Recombinant PCR using oligonucleotides encoding point mutations was used to construct eight additional constructs that contain sets of two or three tandem NXT or NXS sites for expression in yeast or mammalian cells (see diagram in Figure 5A).
Cell culture and transfection
The HEK293-derived STT3A(–/–) cell line was characterized previously (Cherepanova and Gilmore, 2016). HEK293 cells were cultured at 37°C in 60-mm dishes in DMEM (GIBCO), 10% fetal bovine serum with penicillin (100 U/ml), and streptomycin (100 µg/ml). Cells that were seeded at up to 80% confluency were transfected with reporter plasmids (6 µg) using Lipofectamine 2000 following a protocol from the manufacturer (Invitrogen) and were processed after 24 h. Standard yeast synthetic media containing dextrose (SD), supplemented as noted, were used for growth and transformations (Sherman, 1991).
Antibodies and protein immunoblotting
Rabbit polyclonal antibodies to Ost1p (Silberstein et al., 1995b), Wbp1p (te Heesen et al., 1992), Swp1p (te Heesen et al., 1993), CPY (Silberstein et al., 1995a), and Sec63p (Feldheim et al., 1992) were characterized previously. A goat polyclonal antibody to SHBG (R&D Systems, VFJ01), a mouse monoclonal anti-HA antibody (Roche, 11867423001), and the mouse monoclonal anti-DDK antibody (Sigma, F3165 anti-FLAG M2) were from commercial sources. A mouse monoclonal antibody to S. cerevisiae RPL3 (ScRPL3) was obtained from the Developmental Studies Hybridoma Bank at the University of Iowa.
Pulse–chase radiolabeling and immunoprecipitation
Mammalian cells were pulse or pulse–chase labeled using 35S Trans label (Perkin-Elmer) as described previously (Shrimal et al., 2013b) using the following pulse and pulse–chase intervals: pSAP, pSAP derivatives, and CPY derivatives, 10 min pulse; SHBG, 5 min pulse, 20 min chase.
Yeast cells that were grown at 30°C in SD media to mid-log phase (0.4–0.6 OD at 600 nm) were collected by centrifugation and resuspended in fresh SD media at a density of 6 A600/ml. The yeast cells were allowed to recover for 10 min before pulse-labeling with Tran-35S-label (100 µCi/OD). Pulse labeling at 30°C was for 7 min unless noted otherwise in the figure legends or charts. Radiolabeling experiments were terminated by dilution of the culture with an equal volume of ice-cold 20 mM NaN3, followed by freezing in liquid nitrogen. Rapid lysis of cells with glass beads and the immunoprecipitation of the pulse-labeled proteins was done as described (Rothblatt and Schekman, 1989) followed by SDS–PAGE to resolve protein glycoforms. As indicated, immunoprecipitated proteins were digested with endoglycosidase H (New England Biolabs). Dry gels were exposed to a phosphor screen (Fujifilm), scanned in Typhoon FLA 9000, and quantified using ImageQuant.
Ribosome-associated membrane protein isolation from yeast microsomes
A yeast strain (RGY1455; Becker et al., 2009) that expresses His6-FLAG-Sbh2p was grown at 30°C and used to prepare microsomal membranes as described previously (Cheng et al., 2005). The membranes were solubilized by 30 min incubation on ice after adjustment to 2% digitonin, 50 mM triethanolamine acetate, pH 7.5 (TEA-OAc), 150 mM KOAc, 5 mM Mg(OAc)2. Centrifugation at 100,000 × g for 40 min at 4°C yielded a ribosome–membrane protein pellet fraction (P1) and a supernatant fraction (S1). The pellet fraction was resuspended in 50 mM TEA-OAc, 350 mM KOAc, 6 mM Mg(OAc)2, and 3 mM Mn(OAc)2 and centrifuged at 100,000 × g for 40 min at 4°C to obtain a high salt-washed pellet (P2) and wash fraction (W1). This previous step was repeated to obtain a RAMP and a second wash fraction (W2). The first supernatant fraction (S1) and the two wash fractions (W1 and W2) were combined for protein immunoblot analysis.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award GM43768. We thank Randy Schekman for the anti-Sec63 sera and Markus Aebi for anti-Wbp1p and anti-Swp1p sera.
Abbreviations used:
- a.a.
amino acids
- CPY
carboxypeptidase Y
- EH
endoglycosidase H
- ER
endoplasmic reticulum
- OST
oligosaccharyltransferase
- pSAP
prosaposin
- RAMP
ribosome-associated membrane protein
- RNC
ribosome nascent-chain complex
- SHBG
sex hormone-binding globulin
- SILAC
stable isotope labeling by amino acids
- SRP
signal recognition particle
- TEA-OAc
triethanolamine acetate, pH 7.5
- TM
transmembrane
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-06-0330) on August 21, 2019.
REFERENCES
- Bai L, Wang T, Zhao G, Kovach A, Li H. (2018). The atomic structure of a eukaryotic oligosaccharyltransferase complex. Nature , 328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bause E. (1984). Model studies on N-glycosylation of proteins. Biochem Soc Trans , 514–517. [DOI] [PubMed] [Google Scholar]
- Bause E, Legler G. (1981). The role of the hydroxy amino acid in the triplet sequence asn-xaa-thr(ser) for the N-glycosylation step during glycoprotein biosynthesis. Biochem J , 639–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Bhushan S, Jarasch A, Armache JP, Funes S, Jossinet F, Gumbart J, Mielke T, Berninghausen O, Schulten K, et al (2009). Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science , 1369–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunger K, Pfeffer S, Shrimal S, Gilmore R, Berninghausen O, Mandon EC, Becker T, Forster F, Beckmann R. (2018). Structural basis for coupling protein transport and N-glycosylation at the mammalian endoplasmic reticulum. Science , 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulleid NJ, van Lith M. (2014). Redox regulation in the endoplasmic reticulum. Biochem Soc Trans , 905–908. [DOI] [PubMed] [Google Scholar]
- Chavan M, Yan A, Lennarz WJ. (2005). Subunits of the translocon interact with components of the oligosaccharyl transferase complex. J Biol Chem , 22917–22924. [DOI] [PubMed] [Google Scholar]
- Chen W, Helenius J, Braakman I, Helenius A. (1995). Cotranslational folding and calnexin binding during glycoprotein synthesis. Proc Natl Acad Sci USA , 6229–6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Smeekens JM, Wu R. (2014a). A universal chemical enrichment method for mapping the yeast N-glycoproteome by mass spectrometry (MS). Mol Cell Proteomics , 1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Smeekens JM, Wu R. (2014b). Comprehensive analysis of protein N-glycosylation sites by combining chemical deglycosylation with LC-MS. J Proteome Res , 1466–1473. [DOI] [PubMed] [Google Scholar]
- Cheng Z, Jiang Y, Mandon EC, Gilmore R. (2005). Identification of cytoplasmic residues of Sec61p involved in ribosome binding and cotranslational translocation. J Cell Biol , 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanova NA, Gilmore R. (2016). Mammalian cells lacking either the cotranslational or posttranslocational oligosaccharyltransferase complex display substrate-dependent defects in asparagine linked glycosylation. Sci Rep , 20946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanova NA, Shrimal S, Gilmore R. (2014). Oxidoreductase activity is necessary for N-glycosylation of cysteine-proximal acceptor sites in glycoproteins. J Cell Biol , 525–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanova NA, Venev SV, Leszyk JD, Shaffer SA, Gilmore R. (2019). Quantitative glycoproteomics reveals new classes of STT3A- and STT3B-dependent N-glycosylation sites. J Cell Biol , 2782–2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti BJ, Devaraneni PK, Yang Z, David LL, Skach WR. (2015). Cotranslational stabilization of Sec62/63 within the ER Sec61 translocon is controlled by distinct substrate-driven translocation events. Mol Cell , 269–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Smith T, Robbins PW, Samuelson J. (2009). Darwinian selection for sites of Asn-linked glycosylation in phylogenetically disparate eukaryotes and viruses. Proc Natl Acad Sci USA , 13421–13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deprez P, Gautschi M, Helenius A. (2005). More than one glycan is needed for ER glucosidase II to allow entry of glycoproteins into the calnexin/calreticulin cycle. Mol Cell , 183–195. [DOI] [PubMed] [Google Scholar]
- Feldheim D, Rothblatt J, Schekman R. (1992). Topology and functional domains of Sec63p, an endoplasmic reticulum membrane protein required for secretory protein translocation. Mol Cell Biol , 3288–3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finke K, Plath K, Panzer S, Prehn S, Rapoport TA, Hartmann E, Sommer T. (1996). A second trimeric complex containing homologues of the Sec61p complex functions in protein transport across the ER membrane of S. cerevisiae . EMBO J , 1482–1494. [PMC free article] [PubMed] [Google Scholar]
- Gavel Y, Von Heijne G. (1990). Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites: implications for protein engineering. Protein Eng , 433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Prehn S, Hartmann E, Kalies K-U, Rapoport TA. (1992). A mammalian homologue of Sec61p and SecYp is associated with ribosomes and nascent polypeptides during translocation. Cell , 489–503. [DOI] [PubMed] [Google Scholar]
- Harada Y, Li H, Lennarz WJ. (2009). Oligosaccharyltransferase directly binds to ribosome at a location near the translocon-binding site. Proc Natl Acad Sci USA , 6945–6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessa T, Meindl-Beinker NM, Bernsel A, Kim H, Sato Y, Lerch-Bader M, Nilsson I, White SH, von Heijne G. (2007). Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature , 1026–1030. [DOI] [PubMed] [Google Scholar]
- Itskanov S, Park E. (2019). Structure of the posttranslational Sec protein-translocation channel complex from yeast. Science , 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jämsä E, Simonen M, Makarow M. (1994). Selective retention of secretory proteins in the yeast endoplasmic reticulum by treatment with a reducing agent. Yeast , 355–370. [DOI] [PubMed] [Google Scholar]
- Jan CH, Williams CC, Weissman JS. (2014). Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science , 1257521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Cheng Z, Mandon EC, Gilmore R. (2008). An interaction between the SRP receptor and the translocon is critical during cotranslational protein translocation. J Cell Biol , 1149–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaoglu D, Kelleher DJ, Gilmore R. (1997). The highly conserved Stt3 protein is a subunit of the yeast oligosaccharyltransferase and forms a subcomplex with Ost3p and Ost4p. J Biol Chem , 32513–32520. [DOI] [PubMed] [Google Scholar]
- Kelleher DJ, Gilmore R. (1994). The Saccharomyces cerevisiae oligosaccharyltransferase is a protein complex composed of Wbp1p, Swp1p, and four additional polypeptides. J Biol Chem , 12908–12917. [PubMed] [Google Scholar]
- Kelleher DJ, Gilmore R. (2006). An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology , 47–62. [DOI] [PubMed] [Google Scholar]
- Lizak C, Gerber S, Numao S, Aebi M, Locher KP. (2011). X-ray structure of a bacterial oligosaccharyltransferase. Nature , 350–355. [DOI] [PubMed] [Google Scholar]
- Malaby HL, Kobertz WR. (2014). The middle x residue influences cotranslational N-glycosylation consensus site skipping. Biochemistry (Mosc) , 4884–4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandon EC, Butova C, Lachapelle A, Gilmore R. (2018). Conserved motifs on the cytoplasmic face of the protein translocation channel are critical for the transition between resting and active conformations. J Biol Chem , 13662–13672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto S, Taguchi Y, Shimada A, Igura M, Kohda D. (2017). Tethering an N-glycosylation sequon-containing peptide creates a catalytically competent oligosaccharyltransferase complex. Biochemistry , 602–611. [DOI] [PubMed] [Google Scholar]
- Ng DTW, Brown JD, Walter P. (1996). Signal sequences specify the targeting route to the endoplasmic reticulum. J Cell Biol , 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson I, Kelleher DJ, Miao Y, Shao Y, Kreibich G, Gilmore R, Von Heijne G, Johnson AE. (2003). Photocross-linking of nascent chains to the STT3 subunit of the oligosaccharyltransferase complex. J Cell Biol , 715–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzner S, Dreier L, Hartmann E, Kostka S, Rapoport TA. (1995). Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell , 561–570. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods , 785–786. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Dudek J, Gogala M, Schorr S, Linxweiler J, Lang S, Becker T, Beckmann R, Zimmermann R, Forster F. (2014). Structure of the mammalian oligosaccharyl-transferase complex in the native ER protein translocon. Nat Commun , 3072. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Dudek J, Schaffer M, Ng BG, Albert S, Plitzko JM, Baumeister W, Zimmermann R, Freeze HH, Engel BD, Forster F. (2017). Dissecting the molecular organization of the translocon-associated protein complex. Nat Commun , 14516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K, Mothes W, Wilkinson BM, Stirling CJ, Rapoport TA. (1998). Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell , 795–807. [DOI] [PubMed] [Google Scholar]
- Poljak K, Selevsek N, Ngwa E, Grossmann J, Losfeld ME, Aebi M. (2018). Quantitative profiling of N-linked glycosylation machinery in yeast Saccharomyces cerevisiae. Mol Cell Proteomics , 18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz A, Behrens C, Rapoport TA, Hartmann E, Kalies KU. (2000). Evolutionarily conserved binding of ribosomes to the translocation channel via the large ribosomal RNA. EMB0 J , 1900–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy VA, Johnson RS, Biemann K, Williams RS, Ziegler FD, Trimble RB, Maley F. (1988). Characterization of the glycosylation sites in yeast external invertase. I. N-linked oligosaccharide content of the individual sequons. J Biol Chem , 6978–6985. [PubMed] [Google Scholar]
- Roboti P, High S. (2012). Keratinocyte-associated protein 2 is a bona fide subunit of the mammalian oligosaccharyltransferase. J Cell Sci , 220–232. [DOI] [PubMed] [Google Scholar]
- Rothblatt J, Schekman R. (1989). A hitchhiker’s guide to the analysis of the secretory pathway in yeast. Methods Cell Biol , 3–36. [DOI] [PubMed] [Google Scholar]
- Rothman JE, Lodish HF. (1977). Synchronized transmembrane insertion and glycosylation of a nascent membrane protein. Nature , 775–780. [DOI] [PubMed] [Google Scholar]
- Ruiz-Canada C, Kelleher DJ, Gilmore R. (2009). Cotranslational and posttranslational N-glycosylation of polypeptides by distinct mammalian OST isoforms. Cell , 272–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheper W, Thaminy S, Kais S, Stagljar I, Romisch K. (2003). Coordination of N-glycosylation and protein translocation across the endoplasmic reticulum membrane by Sss1 protein. J Biol Chem , 37998–38003. [DOI] [PubMed] [Google Scholar]
- Schulz BL, Aebi M. (2009). Analysis of glycosylation site occupancy reveals a role for Ost3p and Ost6p in site-specific N-glycosylation efficiency. Mol Cell Proteomics , 357–364. [DOI] [PubMed] [Google Scholar]
- Schulz BL, Stirnimann CU, Grimshaw JP, Brozzo MS, Fritsch F, Mohorko E, Capitani G, Glockshuber R, Grutter MG, Aebi M. (2009). Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency. Proc Natl Acad Sci USA , 11061–11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevier CS, Kaiser CA. (2006). Conservation and diversity of the cellular disulfide bond formation pathways. Antioxid Redox Signal , 797–811. [DOI] [PubMed] [Google Scholar]
- Shakin-Eshleman SH, Spitalnik SL, Kasturi L. (1996). The amino acid at the X position of an Asn-X-Ser sequon is an important determinant of N-linked core-glycosylation efficiency. J Biol Chem , 6363–6366. [DOI] [PubMed] [Google Scholar]
- Sherman F. (1991). Getting started with yeast. Methods Enzymol , 3–21. [DOI] [PubMed] [Google Scholar]
- Shibatani T, David LL, McCormack AL, Frueh K, Skach WR. (2005). Proteomic analysis of mammalian oligosaccharyltransferase reveals multiple subcomplexes that contain Sec61, TRAP, and two potential new subunits. Biochemistry , 5982–5992. [DOI] [PubMed] [Google Scholar]
- Shrimal S, Cherepanova NA, Gilmore R. (2017). DC2 and KCP2 mediate the interaction between the oligosaccharyltransferase and the ER translocon. J Cell Biol , 3625–3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrimal S, Gilmore R. (2013). Glycosylation of closely spaced acceptor sites in human glycoproteins. J Cell Sci , 5513–5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrimal S, Gilmore R. (2015). Reduced expression of the oligosaccharyltransferase exacerbates protein hypoglycosylation in cells lacking the fully assembled oligosaccharide donor. Glycobiology , 774–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrimal S, Gilmore R. (2019). Oligosaccharyltransferase structures provide novel insight into the mechanism of asparagine-linked glycosylation in prokaryotic and eukaryotic cells. Glycobiology , 288–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrimal S, Ng BG, Losfeld ME, Gilmore R, Freeze HH. (2013a). Mutations in STT3A and STT3B cause two congenital disorders of glycosylation. Hum Mol Genet , 4638–4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrimal S, Trueman SF, Gilmore R. (2013b). Extreme C-terminal sites are posttranslocationally glycosylated by the STT3B isoform of the OST. J Cell Biol , 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberstein S, Collins PG, Kelleher DJ, Gilmore R. (1995a). The essential OST2 gene encodes the 16-kD subunit of the yeast oligosaccharyltransferase, a highly conserved protein expressed in diverse eukaryotic organisms. J Cell Biol , 371–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberstein S, Collins PG, Kelleher DJ, Rapiejko PJ, Gilmore R. (1995b). The alpha subunit of the Saccharomyces cerevisiae oligosaccharyltransferase complex is essential for vegetative growth of yeast and is homologous to mammalian ribophorin I. J Cell Biol , 525–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirig U, Bodmer D, Wacker M, Burda P, Aebi M. (2005). The 3.4 kDa Ost4 protein is required for the assembly of two distinct oligosaccharyltransferase complexes in yeast. Glycobiology , 1396–1406. [DOI] [PubMed] [Google Scholar]
- te Heesen S, Janetzky B, Lehle L, Aebi M. (1992). The yeast WBP1 is essential for oligosaccharyltransferase activity in vivo and in vitro. EMBO J , 2071–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Heesen S, Knauer R, Lehle L, Aebi M. (1993). Yeast Wbp1p and Swp1p form a protein complex essential for oligosaccharyl transferase activity. EMBO J , 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueman SF, Mandon EC, Gilmore R. (2011). Translocation channel gating kinetics balances protein translocation efficiency with signal sequence recognition fidelity. Mol Biol Cell , 2983–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueman SF, Mandon EC, Gilmore R. (2012). A gating motif in the translocation channel sets the hydrophobicity threshold for signal sequence function. J Cell Biol , 907–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valliere-Douglass JF, Kodama P, Mujacic M, Brady LJ, Wang W, Wallace A, Yan B, Reddy P, Treuheit MJ, Balland A. (2009). Asparagine-linked oligosaccharides present on a non-consensus amino acid sequence in the CH1 domain of human antibodies. J Biol Chem , 32493–32506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Dobberstein B. (1999). Oligomeric complexes involved in translocation of proteins across the membrane of the endoplasmic reticulum. FEBS Lett , 316–322. [DOI] [PubMed] [Google Scholar]
- Whitley P, Nilsson IM, von Heijne G. (1996). A nascent secretory protein may traverse the ribosome/endoplasmic reticulum translocase complex as an extended chain. J Biol Chem , 6241–6244. [DOI] [PubMed] [Google Scholar]
- Wild R, Kowal J, Eyring J, Ngwa EM, Aebi M, Locher KP. (2018). Structure of the yeast oligosaccharyltransferase complex gives insight into eukaryotic N-glycosylation. Science , 545–550. [DOI] [PubMed] [Google Scholar]
- Winther JR, Stevens TH, Kielland-Brandt MC. (1991). Yeast carboxypeptidase Y requires glycosylation for efficient intracellular transport, but not for vacuolar sorting, in vivo stability, or activity. Eur J Biochem , 681–689. [DOI] [PubMed] [Google Scholar]
- Wu X, Cabanos C, Rapoport TA. (2019). Structure of the post-translational protein translocation machinery of the ER membrane. Nature , 136–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan A, Lennarz WJ. (2005). Two oligosaccharyl transferase complexes exist in yeast and associate with two different translocons. Glycobiology , 1407–1415. [DOI] [PubMed] [Google Scholar]
- Zielinska DF, Gnad F, Schropp K, Wisniewski JR, Mann M. (2012). Mapping N-glycosylation sites across seven evolutionarily distant species reveals a divergent substrate proteome despite a common core machinery. Mol Cell , 542–548. [DOI] [PubMed] [Google Scholar]
- Zielinska DF, Gnad F, Wisniewski JR, Mann M. (2010). Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell , 897–907. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.